Abstract
Streptomyces thermoviolaceus OPC-520 secretes two types of xylanases (StxI and StxII), an acetyl xylan esterase (StxIII), and an α-l-arabinofuranosidase (StxIV) in the presence of xylan. Xylan degradation products (mainly xylobiose) produced by the action of these enzymes entered the cell and were then degraded to xylose by an intracellular β-xylosidase (BxlA). A gene cluster involved in xylanolytic system of the strain was cloned and sequenced upstream of and including a BxlA-encoding gene (bxlA). The gene cluster consisted of four different open reading frames organized in the order bxlE, bxlF, bxlG, and bxlA. Reverse transcriptase PCR analysis revealed that the gene cluster is transcribed as polycistronic mRNA. The deduced gene products, comprising BxlE (a sugar-binding lipoprotein), BxlF (an integral membrane protein), and BxlG (an integral membrane protein), showed similarity to components of the bacterial ATP-binding cassette (ABC) transport system; however, the gene for the ATP binding protein was not linked to the bxl operon. The soluble recombinant BxlE protein was analyzed for its binding activity for xylooligosaccharides. The protein showed high-level affinity for xylobiose (Kd = 8.75 × 10−9 M) and for xylotriose (Kd = 8.42 × 10−8 M). Antibodies raised against the recombinant BxlE recognized the detergent-soluble BxlE isolated from S. thermoviolaceus membranes. The deduced BxlF and BxlG proteins are predicted to be integral membrane proteins. These proteins contained the conserved EAA loop (between the fourth and the fifth membrane-spanning segments) which is characteristic of membrane proteins from binding-protein-dependent ABC transporters. In addition, the bxlR gene located upstream of the bxl operon was cloned and expressed in Escherichia coli. The bxlR gene encoded a 343-residue polypeptide that is highly homologous to members of the GalR/LacI family of bacterial transcriptional regulators. The purified BxlR protein specifically bound to a 4-bp inverted sequence overlapping the −10 region of the bxl operon. The binding of BxlR to the site was inhibited specifically by low concentrations of xylobiose. This site was also present in the region located between stxI and stxIV and in the upstream region of stxII. BxlR specifically bound to the regions containing the inverted sequence. These results suggest that BxlR might act as a repressor of the genes involved not only in the uptake system of xylan degradation products but also in xylan degradation of S. thermoviolaceus OPC-520.
Streptomyces bacteria are gram positive, soil inhabiting, and filamentous, with a high G+C content in their DNA. They produce a number of secondary metabolites and extracellular proteins, including enzymes hydrolyzing different types of polysaccharides such as xylan, cellulose, and chitin (12). Unlike cellulose and chitin, xylans have a relatively complex structure consisting of a β-1,4-linked d-xylose polymer replaced with l-arabinofuranosyl, glucuronyl, 4-O-methlglucuronyl, and acetyl groups (37). For complete hydrolysis of xylan, many xylanolytic microorganisms coordinately synthesize the multiple groups of xylanolytic enzymes, such as endo-β-1,4-xylanases (EC 3.2.1.8), β-xylosidase (EC 3.2.1.37), α-l-arabinofuranosidases (EC 3.2.1.55), and acetylxylan esterases (EC 3.1.1.6). The microbial hydrolysis of xylan is central to the recycling of photosynthetically fixed carbon and plays a pivotal role in the turnover of abundant organic molecules in the biosphere (24).
Streptomyces thermoviolaceus OPC-520, a thermophilic actinomycete isolated from decayed wood, grows actively on xylan as a sole carbon source and does not have cellulase activity (29). The biosynthesis of xylanolytic enzymes in S. thermoviolaceus OPC-520 was induced by xylan or xylobiose (which is the smallest molecule to induce the production of xylanolytic enzymes) and repressed by readily metabolized sugars such as glucose (31). The bacterium produces four extracellular enzymes (designated StxI through StxIV) in the presence of xylan (29-32). StxI and StxII are endo-β-1,4-xylanases, StxIII is an acetylxylan esterase, and StxIV is a α-l-arabinofuranosidase. These enzymes effectively convert xylan into xylooligosaccharides (mainly xylobiose, which is a major product of xylan degradation). The generated xylobiose and small amounts of xylooligosaccharides enter the cells and are further hydrolyzed to xylose by an intracellular β-d-xylosidase (BxlA) (31). We have cloned and sequenced the genes involved in the xylan degradation of the strain (30-32). Furthermore, they have been expressed in Streptomyces lividans or Escherichia coli and the biochemical properties of each recombinant protein have been investigated (30-32). A variety of enzymes that can degrade xylans have been identified, and the corresponding genes have been cloned from saprophytic prokaryotes (5, 8, 10, 23) and eukaryotes (4, 11, 33). However, little is known about the uptake system for xylan degradation products and the molecular mechanisms of the gene regulation in Streptomyces species.
Recently, sequence analysis of the upstream region of the bxlA gene revealed a gene cluster consisting of four complete open reading frames (ORFs) organized in the order bxlE, bxlF, bxlG, and bxlA. Upstream of the bxlE gene, furthermore, the bxlR gene (which had an opposite orientation) was found. In the present work, we show that the clustered genes encode an ATP-binding cassette (ABC)-type transporter for xylooligosaccharides and an intracellular β-D-xylosidase required for hydrolysis of xylooligosaccharides. The substrate-binding protein (BxlE) of the ABC transporter was expressed in E. coli and showed the highest level of affinity for xylobiose among xylooligosaccharides from dimer to hexamer. Furthermore, we report that BxlR is a common regulatory protein that specifically binds to the same 4-bp inverted repeat located upstream of bxlE, stxI, stxII, and stxIV.
MATERIALS AND METHODS
Bacterial strains, vectors, and culture conditions.
S. thermoviolaceus OPC-520 was grown at 50°C in medium (1% glucose, 0.5% proteose peptone, 0.1% yeast extract, 0.1% K2HPO4, 0.02% MgSO4· • 7H2O) and was used as the source of chromosomal DNA. To extract total RNA, 500-ml flasks containing 100 ml of minimum medium [NNMP; 0.2% (NH4)2SO4, 0.5% Casamino Acids, 0.06% MgSO4· • 7H2O, 5% polyethylene glycol 6000, 0.001% each of ZnSO4· • 7H2O, MnCl2· • 4H2O, and CaCl2, 0.01 M phosphate buffer (pH 6.8)] supplemented with either oat spelt xylan (Sigma) or glucose were inoculated with 1 ml of mycelium grown for 24 h in YPG medium (29). The flasks were incubated at 50°C with vigorous shaking. The E. coli strains employed in this study were JM109 and BL21(DE3) pLysS. E. coli cells were grown in Luria broth supplemented with appropriate antibiotics at 37°C. The vectors used were pUC18, pUC19 (Takara Biochemicals, Shiga, Japan), pThioHis C (Invitrogen Co.), and pGEX-6P-1 (Amersham Biosciences).
General recombinant DNA techniques.
S. thermoviolaceus chromosomal DNA was isolated by the method of Hopwood et al. (6). Restriction enzymes, T4 DNA ligase, and other modifying enzymes were purchased from Toyobo (Osaka, Japan). Agarose gel electrophoresis, plasmid DNA preparation, transformation of E. coli, and Southern hybridization were performed as described by Sambrook and Russell (15). Nucleotide sequencing was carried out by a dideoxy chain termination method (16) using a DYEnamic ET terminator cycle sequencing premix kit (Amersham Biosciences) on a DNA sequencer (ABI Prism 310 genetic analyzer; Applied Biosystems). Sequence data were analyzed using a GENETYX-WIN program (Software Development Co., Ltd.).
Cloning of the 5′ upstream region of bxlA.
We carried out cloning of the 5′ upstream region of the bxlA gene by the DNA-probing method with a 0.30-kb PstI-BamHI fragment of pBXL3 encoding an intracellular β-d-xylosidase as a probe. The fragment was labeled with alkaline phosphatase (AlkPhos DIRECT; Amersham Biosciences) according to the manufacturer's instructions. Chromosomal DNA of S. thermoviolaceus was digested with various restriction enzymes and electrophoresed on a 0.6% agarose gel. Southern hybridization revealed that the probe hybridized with the 2.7-kb chromosomal fragment digested with BamHI. The DNA fragments corresponding to 2.7 kb were excised from the gel and purified with a GenElute gel extraction kit (Sigma). These were ligated into the dephosphorylated BamHI site of pUC19, and the recombinant plasmids were introduced into competent E. coli JM109. The library was screened by colony hybridization with the labeled probe as previously described (31). The resulting plasmid was designated pBXL3.1. To clone the 5′ upstream region of the 2.7-kb BamHI-BamHI fragment of pBXL3.1, the second colony hybridization was performed using a 0.18-kb BamHI-BglII fragment as a probe. The resulting plasmid was designated pBXL3.2. The 2.7-kb SphI-BamHI fragment of pBXL3.1 and 4.4-kb BglII-SphI fragment of pBXL3.2 were ligated together, and the resulting 7.1-kb fragment was inserted into the corresponding sites of pUC19. The resulting plasmid was named pBXL3.3.
Construction of expression plasmids.
The expression plasmid pThioHis-BxlE, coding for BxlE, was constructed as follows. Two oligonucleotide primers, P1 and P2 (Table 1), were synthesized and were modified to contain XhoI and PstI recognition sites to facilitate cloning in frame into pThioHis C. The bxlE gene was amplified by PCR with the primers, with plasmid pBXL3.3 as the template. PCR was performed for 30 cycles consisting of 97°C for 15 s, 63°C for 30 s, and 68°C for 80 s. The amplified DNA was digested by XhoI and PstI, and the resulting fragment was inserted into the corresponding sites of pThioHis C. On the other hand, a DNA fragment of bxlR, coding for BxlR, was prepared by PCR with plasmid pBXL3.3 as the template and the primers P3 and P4 (Table 1). PCR was performed for 30 cycles consisting of 97°C for 15 s, 55°C for 30 s, and 68°C for 80 s. The amplified DNA was digested by BglII and EcoRI, and the resulting fragment was inserted in frame into the glutathione S-transferase (GST) fusion protein expression vector pGEX-6P-1. The resulting expression plasmid was designated pGEX-BxlR. The nucleotide sequences of the junctions between vectors and inserts and the whole amplified DNA were confirmed with a DYEnamic ET terminator cycle sequencing premix kit with synthesized primers.
TABLE 1.
Sequences of primers used in this study
| Primer | Sequencea | Location |
|---|---|---|
| P1 | 5′-CCGGTCTGAGCTCCTGCGGGTC-3′ | 3957-3978b |
| P2 | 5′-GCCCGTCCCTGCAGTCTCACCG-3′ | 5262-5241b |
| P3 | 5′-GTCGTGCAGATCTACGATCCGGA-3′ | 3749-3727b |
| P4 | 5′-GCGCTGATCGAATTCCTCAT-3′ | 2371-2390b |
| P5 | 5′-CGACCAGCTGTGGGTCGAAG-3′ | 7471-7452b |
| P6 | 5′-GTAGTCGGTGTTCATCTGCT-3′ | 7236-7217b |
| P7 | 5′-AAGCTGAACCTGTACGACAG-3′ | 6581-6600b |
| P8 | 5′-TTGTAGTTCTCCAGGGTGGG-3′ | 6357-6338b |
| P9 | 5′-TCAAGAGCTACGACTTCGGC-3′ | 5969-5988b |
| P10 | 5′-ACGACAGATAGAAGGCCAG-3′ | 5350-5332b |
| P11 | 5′-GACTTCGCCAAGAAGGATCT-3′ | 4784-4803b |
| P12 | 5′-GCGAACACCGACTTGTTGTA-3′ | 4398-4379b |
| P13 | 5′-CTTCATGGTCCGGATCGTATC-3′ | 3719-3739b |
| P14 | 5′-CCCGGAACATTCGACATCAC-3′ | 3782-3801b |
| P15 | 5′-GACACCTCGAGCGTCGACCC-3′ | 3834-3815b |
| P16 | 5′-TCTCGGGCGATCTCGGCAAGTGTGG-3′ | 3666-3690b |
| P17 | 5′-CGACCCGAATGTATCGCATGTGATG-3′ | 3820-3796b |
| P18 | 5′-GCGTCGACCCGACTGTTTCGCATGT-3′ | 3824-3800b |
| P19 | 5′-AGCGTCGACCCGCATGTTTCGCATG-3′ | 3825-3801b |
| P20 | 5′-CCCGACAGGATGATTGAGACTTG-3′ | 1956-1934c |
| P21 | 5′-AGGAGACCTTCCCTGTCCTCGTC-3′ | 1784-1806c |
| P22 | 5′-ACGACGTGAGTGTTGGCCTGAC-3′ | 223-202d |
| P23 | 5′-CTCTTGAGCTGCGACTAGCCAG-3′ | 112-133d |
| P24 | 5′-GGCGTCCACGGTACAGACACCC-3′ | 1396-1375d |
| P25 | 5′-GGCTGAGCGGCGTACGGACAAC-3′ | 1263-1284d |
| P26 | 5′-CAGCGAGCGGTGGAACGACAAC-3′ | 1520-1541c |
| P27 | 5′-CGTCACACACCGGCAAACACAA-3′ | 1618-1597c |
| P28 | 5′-CCTTGTCCTGGACCTTCACC-3′ | 4058-4039b |
Purification of recombinant BxlE and BxlR.
E. coli TOP10 cells harboring pThioHis-BxlE were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at the mid-exponential growth phase and further incubated for 3 h at 37°C. The cells were harvested by centrifugation, washed, and resuspended with 20 mM phosphate buffer (pH 7.4). The cells were disrupted by sonication, and the lysate were centrifuged at 10,000 × g for 30 min. The fusion protein was purified from the supernatant by affinity chromatography with a nickel-charged Sepharose resin (ProBond resin; Invitrogen Co.). The purified fusion protein was treated with enterokinase (Invitrogen Co.) for 16 h at 37°C to obtain BxlE. The recombinant BxlE included the extra six amino acid residues in the N-terminal portion. On the other hand, E. coli BL21(DE3) pLysS cells harboring pGEX-BxlR were induced with 1 mM IPTG at the mid-exponential growth phase and further incubated for 2 h at 37°C. The lysate was prepared in the same manner as the E. coli TOP10 cells harboring pThioHis-BxlE. The GST-fusion protein was purified from the lysate by affinity chromatography with glutathione-Sepharose 4B (Amersham Biosciences). The purified fusion protein was treated with PreScission protease (Amersham Biosciences) for 4 h at 4°C to obtain BxlR. The N-terminal amino acid sequences of BxlE and BxlR were confirmed by protein sequencing (Procise 491 HT protein sequencer; Applied Biosystems).
RT-PCR.
S. thermoviolaceus OPC-520 was grown for 24 h at 50°C in NMMP containing 1% xylan, 1% xylobiose, or 1% glucose. Total RNA was extracted from 1.0-ml suspensions of S. thermoviolaceus OPC-520 cells with an SV total RNA isolation system (Promega) in accordance with the manufacturer's instructions. Total RNA (2 μg) and primer P5 were used to reverse the bxl transcripts. The reaction was carried out at 42°C for 60 min with Moloney murine leukemia virus reverse transcriptase (RT) (RNase H minus; Promega) and terminated by heating at 70°C for 15 min. The RT products were used as a template for PCR, and the following primer pairs were designed: primers P6 and P7, primers P8 and P9, primers P10 and P11, and primers P12 and P13 (Table 1). PCR was performed for 30 cycles consisting of 97°C for 15 s, 55°C for 30 s, and 68°C for 80 s. As negative controls, the reactions were performed in the absence of RT or RNA template.
Preparation of membrane proteins.
S. thermoviolaceus OPC-520 was grown on NNMP (10 ml) supplemented with 1% xylan or 1% glucose at 50°C for 12 h. Mycelia were harvested by centrifugation (5,000 × g for 15 min), washed twice in 50 mM Tris-HCl buffer (pH 7.5) containing 0.2 M NaCl, and suspended in 10 ml of the same buffer. The mycelia were disrupted by sonication, and membranes were isolated by centrifugation (10,000 × g for 1 h at 4°C). Membrane proteins were extracted for 30 min with 1% N-lauroylsarcosine in 50 mM Tris-HCl buffer (pH 7.5) containing 0.2 M NaCl and centrifuged at 10,000 × g for 20 min at 4°C. The supernatant was used for Western blotting.
Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done by the method of Laemmli (9). Proteins on the gel were transferred to a Sequi-Blot polyvinylidene difluoride membrane. The membrane was incubated for 1 h at room temperature with anti-BxlE polyclonal mouse serum diluted to 1:1,000 in phosphate-buffered saline containing 2.0% skim milk (Difco). Bound antibody was detected as described previously (14).
Surface plasmon resonance analyses.
BIAcore X system and carboxymethylated dextran (CM5) sensor chips were purchased from Pharmacia Biosensor. To immobilize BxlE to the CM5 sensor chip surface, carboxyl groups along the carboxymethylate-dextran chains of the sensor chip surface were activated by exposure (35 μl at 5 μl/min) to a mixture of 0.1 M N-hydroxysuccinimide and 0.1 M N′-(3-diethylaminopropyl) carbodiimide hydrochloride (1:1[vol/vol]). BxlE was injected over the surface at 50 μg/ml in coupling buffer (10 mM sodium acetate buffer, pH 3.0). After coupling, unreacted surface ester groups were blocked by exposure to 1 M ethanolamine (pH 8.5). Bovine serum albumin was used as the control protein. The interaction of BxlE with xylooligosaccharides (at concentrations ranging from 3 × 10−9 to 2 × 10−4 M) was analyzed at a flow rate of 10 μl/min in 10 mM HEPES buffer (pH 7.4) containing 150 mM NaCl and 3 mM EDTA. The association time was 3.5 min, and the dissociation time was 5 min. The kinetic parameters were determined with a BIA evaluation program (version 3.0).
Gel retardation assay.
The upstream region of bxlE (corresponding to positions −114 to −61 [taking A of the initiation codon of bxlE as position +1]) was amplified by PCR using P14 and P15. The oligonucleotide-directed mutagenesis of the region was performed by PCR using primer pairs P16 and P17, P16 and P18, and P16 and P19. The upstream regions of stxI (corresponding to positions −418 to −246 [taking the initiation codon of stxI as position +1]), stxII (corresponding to positions −149 to −38 [taking the initiation codon of stxII as position +1]), stxIII (corresponding to positions −40 to +94 [taking the initiation codon of stxIII as position +1]), and stxIV (corresponding to positions −78 to +21 [taking the initiation codon of stxIV as position +1]) were amplified by PCR using primer pairs P20 and P21, P22 and P23, P24 and P25, and P26 and P27 (Table 1), respectively. PCR was performed for 30 cycles consisting of 97°C for 15 s, 53°C for 30 s, and 68°C for 80 s. The amplified fluorescein isothiocyanate (FITC)-labeled DNA fragments were used in the gel retardation assay. Binding reactions contained 10 ng of FITC-labeled DNAs, 10 to 100 ng of purified BxlR, and 10 ng of poly(dI-dC) in a final volume of 20 μl of buffer consisting of 10 mM HEPES-KOH buffer (pH 7.5), 1 mM MgCl2, 0.5 mM dithiothreitol, and 50 mM KCl. Binding reaction mixtures were allowed to equilibrate for 30 min at 37°C and immediately loaded onto 5% polyacrylamide gels. The gels were electrophoresed at 200 V for 20 min by using a Mini-Protean II apparatus (Bio-Rad) in Tris-acetate-EDTA buffer. The gels were visualized by a fluoro image analyzer (FLA-2000; Fujifilm, Tokyo, Japan).
Primer extension.
Total RNA was extracted from 1.5 ml of cell suspensions of S. thermoviolaceus OPC-520 grown in NNMP supplemented with 1% xylan by using an SV total RNA isolation system (Promega). About 10.0 μg of RNA was used to map the 5′ end of the bxlE transcript. Reverse transcription was initiated from P28 (the FITC-labeled primer) complementary to the 5′ end of the bxlE coding region. The reaction was carried out at 50°C for 60 min with Moloney murine leukemia virus RT (RNase H minus; Promega). The primer extension and the sequencing reaction products were analyzed on a 6.0% denaturing polyacrylamide gel by a DNA sequencer (Hitachi SQ3000). The sequence reaction was performed with the same primer.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession numbers AB110643 (stxI and stxIV), AB110644 (stxII and stxIII), and AB110645 (bxlR, bxlE, bxlF, bxlG, and bxlA).
RESULTS
Cloning and sequence analysis of the gene cluster.
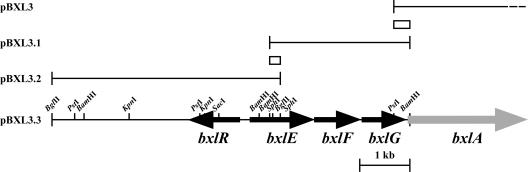
Previous study of an intracellular β-d-xylosidase (BxlA) in S. thermoviolaceus OPC-520 led to the identification and characterization of bxlE, bxlF, bxlG, and bxlR genes (31). A 0.3-kb PstI-BamHI fragment of pBXL3 (Fig. 1) coding for the COOH-terminal end of BxlG (last hydrophobic membrane-spanning region) was used as a probe to screen a S. thermoviolaceus BamHI gene library constructed with pUC19. Among 800 transformants, only one clone (pBXL3.1) (which hybridized with the probe) was isolated by colony hybridization (Fig. 1). Analyses by restriction enzyme digestion and sequencing of the fragment revealed that the insert of pBXL3.1 and that of pBXL3 shared a 0.3-kb PstI-BamHI region. Analysis of the entire nucleotide sequence of pBXL3.1 led to the prediction of two complete ORFs (bxlF and bxlG) and one truncated frame (bxlE) (Fig. 1). Then the 5′ upstream region of the insert of pBXL3.1 (designated pBXL3.2) was further cloned in the second colony hybridization, using the 0.18-kb BamHI-BglII fragment as a probe. The 2.7-kb SphI-BamHI fragment of pBXL3.1 and 4.4-kb BglII-SphI fragment of pBXL3.2 were ligated together, and the resulting 7.1-kb fragment was inserted into the corresponding sites of pUC19. The resulting plasmid was named pBXL3.3 (Fig. 1).
FIG. 1.
Restriction map of bxlR, bxlE, bxlF, bxlG, and bxlA. The hybridization probes are represented by boxes. The arrows indicate the ORFs and directions of transcription.
The nucleotide sequence of the gene cluster of pBXL3.3 was determined. The overall G+C content of the sequenced fragment was 74%. This value is in agreement with the G+C content of Streptomyces (38). Upstream of the bxlA gene, three ORFs (bxlE, bxlF, and bxlG) were found which were carried on the same strand and had the same directions of transcription. On the other hand, the bxlR gene was located in an opposite orientation 171 bp upstream of the bxlE gene. There are only 44 nucleotides between the TGA termination codon of bxlG and the ATG initiation codon of bxlA, 4 nucleotides between bxlF and bxlG, and 17 nucleotides between bxlE and bxlF.
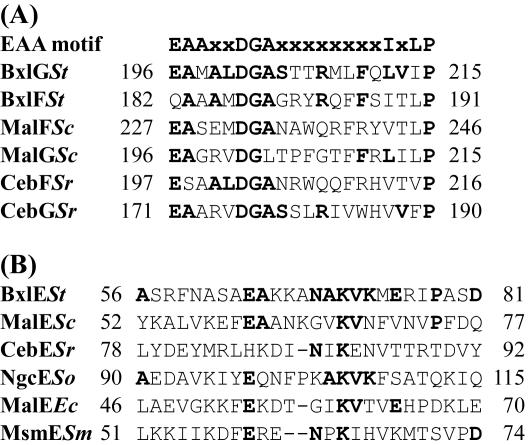
The bxlG gene consists of 906 nucleotides encoding a protein of 301 amino acids with a predicted molecular mass of 32,428 Da. The deduced amino acid sequence of the encoded protein (BxlG) was compared with those of the other proteins. A search of the BLAST database found that BxlG had similarity with a putative binding protein-dependent transport protein from Streptomyces coelicolor A3 (2) (accession no. CAB88163) (54% identity), BxlG from S. lividans (accession no. AAC99627) (52% identity), a hypothetical protein from Thermobifida fusca (accession no. ZP_00056972) (43% identity), and a putative sugar ABC transporter integral membrane protein from S. coelicolor A3 (2) (accession no. CAD55434) (41% identity). The bxlF gene consists of 876 nucleotides encoding a protein of 291 amino acids with a predicted molecular mass of 31,789 Da. The deduced BxlF was closely related to BxlF from S. lividans (accession no. AAC99626) (53% identity), a putative binding protein-dependent transport protein from S. coelicolor A3 (2) (accession no. CAB88162) (49% identity), and a hypothetical protein from Thermobifida fusca (accession no. ZP_00056971) (45% identity). When their hydrophobicity profiles were analyzed, both BxlG and BxlF were predicted to span the membrane six times (data not shown). Furthermore, they contain the consensus sequence EAAX2DGAX8IXLP between the fourth and the fifth membrane-spanning segments (which is characteristic of membrane proteins from binding protein-dependent ABC transporters) (17) (Fig. 2A).
FIG. 2.
Comparison of BxlG, BxlF, and BxlE sequences with those of other proteins. (A) The consensus sequences conserved in membrane proteins of ABC transporters and the corresponding regions of BxlG and BxlF are shown. The number of the first amino acid in each line is shown on the left. Residues that are identical are indicated by boldface letters. St, S. thermoviolaceus OPC-520; Sc, S. coelicolor A3 (2); Sr, S. reticuli. (B) A sequence in BxlE is compared with the signature sequences of sugar-binding proteins of cluster 1. St, S. thermoviolaceus OPC-520; Sc, S. coelicolor A3 (2); Sr, S. reticuli; So, S. olivaceoviridis; Ec, E. coli; Sm, Streptococcus mutans.
The bxlE gene consists of 1,311 nucleotides encoding a protein of 436 amino acids with a predicted molecular mass of 46,661 Da. The deduced amino acid sequence of BxlE showed sequence homology with several sugar-binding proteins, such as a putative sugar-binding lipoprotein from S. coelicolor A3 (2) (accession no. CAB88161) (43% identity), BxlE from S. lividans (accession no. AAC99625) (43% identity), and a hypothetical protein from Thermobifida fusca (accession no. ZP_00056970) (38% identity). The deduced N-terminal portion of BxlE (MQSYSRRWFLGAGATTLISAAGLTACG) includes positively charged residues and the consensus sequence [L(S,A)(A,G)C(S,G)] (which corresponds to the sites cleaved by lipoprotein-specific signal peptidases in gram-positive bacteria) (27). These results suggest that Cys-26 is at the amino terminus of the mature form and is covalently modified by the typical ester-linked and amide-linked acylation of lipoproteins. BxlE has the signature sequence (cluster 1) of binding proteins specific for multiple sugars and glycerol phosphate (28) (Fig. 2B). The highly conserved lysine residue of the signature sequence is also conserved in BxlE. These results suggest that BxlE serves as a substrate-binding protein of the components comprising an ABC transporter system.
The bxlR gene consists of 1,032 nucleotides encoding a protein of 343 amino acids with a predicted molecular mass of 36,706 Da. The deduced amino acid sequence of BxlR showed sequence homology with transcriptional repressors classified into the GalR/LacI family. The N-terminal helix-turn-helix DNA-binding motif (TLAEIAREAGVSAPTVSKVLNG) (located between amino acid 13 and amino acid 34) was found at the amino-terminal portion of BxlR. BxlR showed sequence homology with a putative transcriptional regulator from S. coelicolor A3 (2) (accession no. CAA20410) (86% identity), a probable LacI-family transcriptional regulator from S. avermitilis (accession no. BAC72695) (58% identity), and BxlR from S. lividans (accession no. AAC99624) (55% identity). The amino acid sequence (residues 200 to 204, 251 to 254, and 277 to 281) of BxlR also shares some homology with that of the sugar-binding sites of the GalR/LacI proteins.
The bxlEFG and bxlA genes are polycistronically transcribed.
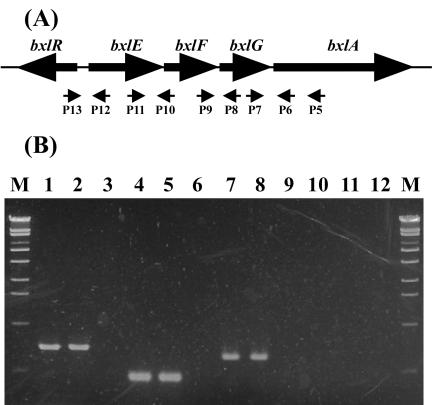
We performed RT-PCR to examine transcription of the gene cluster by using the primers indicated in Fig. 3. When total RNA isolated from S. thermoviolaceus mycelia grown in the presence of xylan or xylobiose was used, the expected sizes of DNA fragments (655 bp [lanes 1 and 2], 388 bp [lanes 4 and 5], and 566 bp [lanes 7 and 8]) were amplified. However, when total RNA from glucose-grown mycelia was used, DNA fragments were not amplified (lanes 3, 6, and 9). These results indicate that the gene cluster comprising bxlEFG and bxlA is induced in the presence of xylan or xylobiose and is polycistronically transcribed. On the other hand, the DNA fragments between P12 and P13 were not amplified (lanes 10, 11, and 12) when total RNAs prepared from the medium containing xylan, xylobiose, or glucose were used.
FIG. 3.
RT-PCR analysis of bxlE, bxlF, bxlG, and bxlA RNA. (A) The schematic locations of oligonucleotide primers (P5 to P13) used in RT-PCR analysis are shown (for more details, see Table 1). The primer P5 was used for RT reactions. (B) Agarose gel electrophoresis of RT-PCR products. RT-PCR analysis was performed with total RNA isolated from S. thermoviolaceus OPC-520 in medium containing 1% xylan (lanes 1, 4, 7, and 10), 1% xylobiose (lanes 2, 5, 8, and 11), or 1% glucose (lanes 3, 6, 9, and 12). Lanes: 1, 2, and 3, cDNA products by P6 and P7; 4, 5, and 6, cDNA products by P8 and P9; 7, 8, and 9, cDNA products by P10 and P11; 10, 11, and 12, cDNA products by P12 and P13; M, molecular size standards.
Expression and purification of BxlE and BxlR.
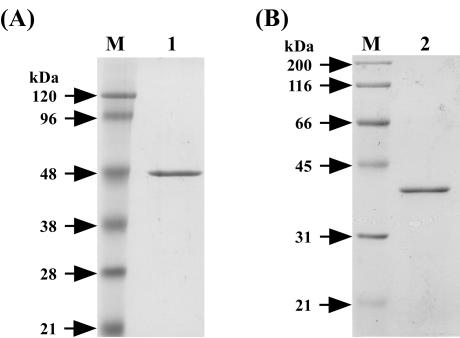
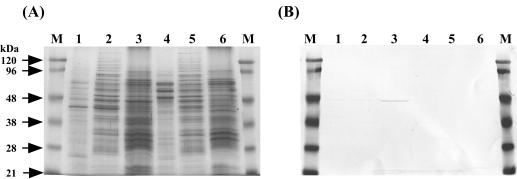
The sequence analysis of the gene cluster suggests that BxlE, BxlF, and BxlG are components of an ABC transporter system and that BxlR is a transcriptional repressor of bxlEFGA. To clarify the roles of these proteins in the xylanolytic system of S. thermoviolaceus OPC-520, BxlE and BxlR were expressed in E. coli with the procedure described in Materials and Methods. The six-His-tagged BxlE was purified by HisTrap column chromatography. On the other hand, the fusion protein (GST-BxlR) was purified by affinity chromatography with glutathione-Sepharose 4B. The purified GST-BxlR was treated with PreScission protease, and then BxlR was purified with glutathione-Sepharose 4B. The molecular masses of BxlE and BxlR calculated from each of the amino acid sequences were in reasonable agreement with those estimated by SDS-PAGE (Fig. 4). On the other hand, gel filtration chromatography (Superdex 200; Amersham Biosciences) showed that BxlR had an apparent molecular mass of 75 kDa (data not shown). This result suggests that BxlR forms a dimer.
FIG. 4.
SDS-PAGE of BxlE and BxlR. (A) Lanes: M, molecular size standards; 1, BxlE. (B) Lanes: M, molecular size standards; 2, BxlR.
BxlE is a sugar-binding protein.
To clarify the role of BxlE in an ABC transporter system, recombinant BxlE was immobilized on a BIAcore sensor chip. Xylose or each of xylooligosaccharides (from dimer to hexamer) was passed over the sensor chip, and the binding to BxlE was monitored directly by surface plasmon resonance detection. Sensorgrams for the interaction of various concentrations of ligands with BxlE were examined (data not shown). The equilibrium dissociation constants (Kd) were calculated according to the ratio [Kd = the dissociation rate constant (koff)]/[the association rate constant (kon)]. Among the sugars tested, xylobiose showed the highest affinity towards BxlE (Kd = 8.75 × 10−9 M) and xylotriose showed the second highest affinity (Kd = 8.42 × 10−8M), as shown in Table 2. The affinity of xylooligosaccharides towards BxlE showed a tendency to decrease with increases in the degree of polymerization. However, xylose and glucose showed weak affinity towards BxlE.
TABLE 2.
Kinetic parameters for binding of various saccharides to BxlE
| Saccharide | Results for kinetic parameter:
|
||
|---|---|---|---|
| kon (M−1 s−1)a | koff (s−1)b | Kd (M) | |
| Xylose | 4.33 × 102 | 7.60 × 10−3 | 1.76 × 10−5 |
| Xylobiose | 9.48 × 105 | 8.30 × 10−3 | 8.75 × 10−9 |
| Xylotriose | 3.31 × 105 | 2.78 × 10−2 | 8.42 × 10−8 |
| Xylotetraose | 4.69 × 105 | 5.03 × 10−2 | 1.07 × 10−7 |
| Xylopentaose | 1.95 × 105 | 1.06 × 10−1 | 5.44 × 10−7 |
| Xylohexaose | 1.03 × 105 | 1.20 × 10−1 | 1.16 × 10−6 |
| Glucose | 1.50 × 103 | 4.20 × 10−3 | 2.80 × 10−6 |
Rate constant for association.
Rate constant for disassociation.
In gram-positive bacteria, solute-binding proteins are located at the surface of the cytoplasmic membrane by a lipid anchor (27). To examine the distribution of native BxlE in S. thermoviolaceus OPC-520, Western blotting analysis was performed. Membrane vesicles from S. thermoviolaceus OPC-520 were prepared from mycelia grown in NNMP supplemented with xylan or glucose. Native BxlE was detected only in membrane fraction from mycelia grown in the presence of xylan and not in the culture supernatant and cytoplasm (Fig. 5). Taken together, these findings show that BxlE located at the cytoplasmic membrane is a sugar-binding protein which serves as one of the components of an ABC transporter for xylobiose and larger oligosaccharides.
FIG. 5.
SDS-PAGE (A) and Western blot analysis (B) of BxlE. S. thermoviolaceus OPC-520 was grown at 50°C for 12 h in the presence of 1.0% xylan (lanes 1, 2, and 3) or 1.0% glucose (lanes 4, 5, and 6). Lanes: M, prestained molecular mass marker; 1 and 4, culture supernatant; 2 and 5, cytosol; 3 and 6, membrane fraction.
BxlR binds to the inverted repeat sequence.
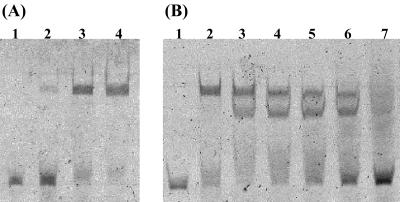
To investigate whether recombinant BxlR binds specifically to the region located between the bxlR and bxlE genes, gel retardation assays were performed. The perfect inverted repeat sequence (5′-CGAA-Nx-TTCG-3′) was found in the intergenic region. To identify the transcriptional start site of the bxl operon, primer extension analysis was carried out for total RNA prepared from the cells of S. thermoviolaceus OPC-520. The primer extension revealed that the inverted repeat sequence and the −10 region overlap each other (Fig. 6). Then, a FITC-labeled 54-bp DNA fragment containing the inverted repeat sequence was amplified by PCR using P14 and P15. As shown in Fig. 7, the purified BxlR was found to bind to the amplified intergenic bxlR-bxlE. As the amount of BxlR increased, the amounts of DNA-protein complex increased. We reported that the smallest molecule to induce the production of both xylanase and β-xylosidase in S. thermoviolaceus OPC-520 was xylobiose (31). Then, we examined the effect of the presence of xylobiose on BxlR protein-DNA interaction. The binding of BxlR to the FITC-labeled DNA fragment was weakened by xylobiose at concentrations of 1 to 250 mM and was lost by the addition of high concentrations (500 mM) of xylobiose (Fig. 7). However, the presence of xylose and glucose had no effect on binding even at a concentration of 500 mM (data not shown). These results indicate that BxlR is a transcriptional repressor of the bxl operon.
FIG. 6.
Determination of the transcription start site of bxl operon. (A) Primer extension and nucleotide sequencing were performed with the same FITC-labeled primer. The nucleotide sequence around the transcription start site is shown in lanes A, C, G, and T. The transcriptional start site is shown by an arrow (lane P). (B) Nucleotide sequence of the 5′ upstream region of bxlE. The deduced amino acid sequences of BxlR and BxlE are shown below the nucleotide sequence. The transcriptional start site is indicated with +1. The putative −35 and −10 regions are shown by boxes.
FIG. 7.
Gel retardation assays of BxlR. (A) The FITC-labeled DNA fragment (10 ng) was incubated with increasing amounts of BxlR. Lanes: 1, no protein; 2, 10 ng of BxlR; 3, 50 ng of BxlR; 4, 100 ng of BxlR. (B) Inhibition of BxlR protein binding to the FITC-labeled DNA fragment by xylobiose. Lanes: 1, no protein; 2, 100 ng of BxlR; 3 to 7, 100 ng of BxlR and xyobiose (1.0, 10, 100, 250, or 500 mM).
To investigate whether BxlR specifically binds a 4-bp inverted sequence, oligonucleotide-directed mutagenesis was performed. An A-to-T transversion (5′-CGAA-Nx-TTCG-3′; the A is indicated by underlining) resulted in loss of the binding of BxlR to the amplified DNA fragment containing the inverted sequence. Two additional base changes (a T-to-G transversion [5′-CGAA-Nx-TTCG-3′] and a T-to-G transversion [5′-CGAA-Nx-TTCG-3′]; the T is indicated by underlining) also resulted in loss of BxlR-DNA interaction (data not shown). These data suggest that BxlR specifically binds to a 4-bp inverted repeat sequence.
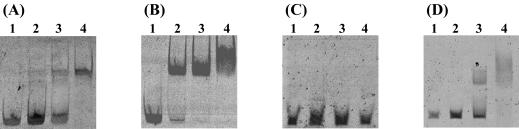
Alignment of the upstream regions of xylanase (stxI and stxII), acetylxylan esterase (stxIII), and α-l-arabinofuranosidase (stxIV) genes in S. thermoviolaceus OPC-520 revealed that (like bxlE) stxI, stxII, and stxIV had the sequence 5′-CGAA-Nx-TTCG-3′ in the putative promoter regions (Fig. 8). To clarify whether BxlR regulates transcription of these genes, DNA fragments containing the inverted repeat were amplified by PCR and interactions of BxlR with each of the amplified DNA fragments were examined by gel retardation assays. As shown in Fig. 9, BxlR specifically binds to DNA fragments containing the perfect inverted repeat sequence but not to DNA fragments from stxIII. The tandem organization of stxII and stxIII genes forms an operon, as in the case of the gene cluster bxlE, bxlF, bxlG, and bxlA (unpublished data). These results suggest that BxlR is a common transcriptional regulator not only of the bxl operon but also of the stxI, stxII, stxIII, and stxIV genes dispersed throughout the genome of S. thermoviolaceus OPC-520.
FIG. 8.
Alignment of the upstream regions of the genes involved in xylanolytic system of S. thermoviolaceus OPC-520. The conserved sequences (5′-CGAA-Nx-TTCG-3′) are shown by white letters on a black background. The inverted repeat sequences are indicated by convergent arrows. The transcriptional start site of the bxl operon is indicated with +1. The putative −35 and −10 regions of the bxl operon are shown by boxes.
FIG. 9.
Binding of BxlR to the upstream regions of the genes involved in xylan degradation. Each of the upstream regions of stxI (A), stxII (B), stxIII (C), and stxIV (D) was incubated with increasing amounts of BxlR. Lanes: 1, no protein; 2, 10 ng of BxlR; 3, 50 ng of BxlR; 4, 100 ng of BxlR.
DISCUSSION
The presented data suggest that xylooligosaccharides are specifically transported to the cytoplasm through an ABC transporter system and that xylooligosaccharides are then degraded to xylose by an intracellular β-xylosidase. In addition to these findings, we clarified that BxlR is a regulator not only of the bxl operon for xylooligosaccharide uptake and degradation and but also of the genes involved in xylan degradation in S. thermoviolaceus OPC-520.
The bxl operon is composed of four genes encoding xylooligosaccharide binding protein (bxlE), two permeases (bxlF and G), and an intracellular β-xylosidase (bxlA). Analysis of the deduced amino acid sequence of BxlE showed that the protein is a membrane-associated lipoprotein probably serving as a solute-binding protein in an ABC transport system. Solute-binding proteins have been classified into eight clusters (28). The sequence from Ala-61 to Asp-81 of BxlE shows similarity to the signature sequence characteristic of cluster 1 binding proteins. These include MalE (essential for import of maltose in S. coelicolor A3) (2, 35), CebE (essential for import of cellobiose and cellotriose in Streptomyces reticuli) (20), and NgcE (essential for import of N-acetylglucosamine and N,N′-diacetylchitobiose in S. olivaceoviridis) (40).
To investigate the substrate specificity of the identified ABC transporter system, BxlE was expressed in E. coli and analyzed (using surface plasmon resonance) for the kinetics of sugar binding. The association (kon) and dissociation (koff) rate constants of various sugars were determined for BxlE, and the equilibrium dissociation constant (Kd) was calculated. Among the sugars tested, xylobiose showed the highest affinity (Kd = 8.75 × 10−9 M) followed by xylotriose (Kd = 8.42 × 10−8 M). The lowest value was measured for xylohexaose (Kd = 1.16 × 10−6 M). Kinetic parameters for bacterial sugar-binding proteins so far reported have been determined by several methods, such as equilibrium dialysis (19), sugar uptake by whole cells (39), and stopped-flow techniques (13). A sugar-binding protein of S. reticuli showed the highest affinity (Kd = 1.5 × 10−6 M) for cellobiose and cellotriose (19). The dissociation constant of recombinant trehalose/maltose-binding protein from Thermococcus litoralis was determined to be 1.6 × 10−7 M (7). Recently, the equilibrium dissociation constants (Kd) of NgcE purified from S. olivaceoviridis were ascertained (using surface plasmon resonance) for N-acetylglucosamine (Kd = 8.28 × 10−9 M) and chitobiose (Kd = 2.87 × 10−8 M) (40). These values were very close to the Kd values of xylobiose and xylotriose, although the recombinant BxlE includes the extra amino acid residues (VPGMLSS) in the N-terminal portion (CGSGSGS). In the case of trehalose/maltose-binding protein of T. litoralis, the N-terminally truncated protein was expressed in E. coli, resulting in a soluble protein exhibiting the same binding characteristics as the wild-type protein whose N-terminal cysteine is covalently modified by lipid (3). Therefore, it seems likely that the lipid anchor of BxlE does not influence the binding affinity (since binding is brought about by the movement of the two soluble lobes forming the binding site between them) (26). These results indicate that BxlE is a sugar-binding protein involved in xylan metabolism of S. thermoviolaceus OPC-520 (which shows a high specificity for xylobiose and larger oligosaccharides).
Both BxlG and BxlF contain the conserved EAA cytoplasmic loop that is found in all other integral membrane components of binding protein-dependent transport systems (27) and that may interact with the membrane-associated ATP-hydrolyzing subunit. However, we could not discover the identity of the gene encoding an ATP-hydrolyzing subunit in the vicinity of the bxl operon. That the gene encoding an ATP-hydrolyzing subunit is absent also holds for the cellobiose operon from S. reticuli (18), the maltose operons from S. lividans (22) and S. coelicolor (35), and the N-acetylglucosamine operon from S. olivaceoviridis (40). Schlösser has shown that the Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport systems as a general ATP-hydrolyzing subunit (18). Thus, MsiK (or an MsiK-like protein which is homologous to the ATP-hydrolyzing subunit Malk in the maltose transport system in E. coli) (1, 2) seems to be encoded elsewhere on the chromosome of S. thermoviolaceus OPC-520.
The bxlR genes occur upstream of the bxl operon organized in the order bxlE, bxlF, bxlG, and bxlA. The gene organization (bxlR-bxlE-bxlF-bxlG-bxlA) is the same as those for the mal operon (malR-malE-malF-malG-aglA) from S. coelicolor A3 (2) (35) and the ceb operon (cebR-cebE-cebF-cebG-bglC) from S. reticuli (20). The mal operon and the ceb operon correspond to the maltose and cellobiose-cellotriose import systems, respectively. These operons are regulated by MalR and CebR belonging to GalR/LacI family (21, 34, 35). The deduced protein BxlR is related to members of the GalR/LacI regulatory family. Thus, the bxlR gene was expressed in E. coli to investigate whether BxlR is the transcriptional regulator of the bxl operon. The purified BxlR was found to bind specifically to the 61-bp region located between the bxlR and bxlE. The GalR/LacI regulators bind to their target DNA sites as homodimers, and their operator sequences are inverted repeats (25). Since the BxlR protein shares a number of common features with other members of GalR/LacI family, we investigated whether there are the inverted repeats within the 61-bp region located between the bxlR and bxlE. Computer analysis revealed that the sequence required for recognition by BxlR appeared to be a 4-bp inverted repeat (5′-CGAA-Nx-TTCG-3′) located in the −10 region of the bxl operon. Several base changes within this sequence resulted in loss of the binding activity of BxlR, indicating that BxlR recognizes a 5′-CGAA-Nx-TTCG-3′ sequence as an operator. Most proteins of the GalR/LacI family bind carbohydrate or nucleoside effectors (36). Our results showed that BxlR-DNA interaction was weakened in the presence of low concentrations of xylobiose (1 to 10 mM) and was not affected by the high concentrations of xylose or glucose (500 mM). The synthesis of β-xylosidase (BxlA) and sugar-binding protein (BxlE) in S. thermoviolaceus OPC-520 was induced by the presence of xylobiose but not that of xylose. Therefore, it is presumed that xylobiose is the true inducer of the bxl operon which leads to release of the BxlR from the operator upstream of the bxlE gene. In the absence of xylobiose, BxlR presumably binds to the inverted repeat and blocks the transcription of the bxl operon.
Xylanase production (StxI and StxII) in S. thermoviolaceus OPC-520 is also induced in the presence of xylobiose. These results suggest that xylanase genes and the bxl operon might be coordinately controlled by the same regulatory system. As expected, the inverted repeat sequences corresponding to the BxlR binding motif were identified in the regions located between stxI and stxIV and in the upstream region of stxII. Indeed, BxlR specifically bound to the regions containing a 4-bp inverted repeat (5′-CGAA-Nx-TTCG-3′). Our results resemble those of investigations of the regulatory system of the cellulase genes in Thermomonospora fusca that is controlled by CebR, a member of GalR/LacI family (25). The 14-bp inverted repeat is present in the regions upstream of all six cellulase genes in T. fusca. CebR specifically binds to the inverted repeat as a repressor and is released from the binding site through a direct interaction of CelR with cellobiose. These results indicate that the expression of genes encoding enzymes involved in xylan degradation and of an ABC transporter required for the uptake of xylan degradation products is controlled by BxlR. This simple regulation may allow quick adaptation of the strain to various environments and avoidance of the unnecessary production of proteins involved in the xylan degradation system. Furthermore, the 5′-CGAA-Nx-TTCG-3′ sequence was found in the upstream regions of the genes involved in xylan degradation systems from different Streptomyces species (such as S. coelicolor A3 [2] and S. lividans) containing ORFs very similar to that of S. thermoviolaceus OPC-520. These findings suggest that the genes involved in the xylan degradation systems of different Streptomyces species might be also regulated in the same manner as those of the system regulating S. thermoviolaceus OPC-520. Our aim is to investigate our major remaining question: whether bxlR gene disruption leads to the loss of catabolite repression by glucose of many genes involved in xylan metabolism of S. thermoviolaceus OPC-520 (although the strain is not amenable to general recombinant DNA techniques).
REFERENCES
- 1.Boos, W., and J. M. Lucht. 1996. Periplasmic binding protein-dependent ABC transporters, p. 1175-1209. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington D.C.
- 2.Boos, W., and H. Schuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diez, J., K. Diederichs, G. Greller, R. Horlacher, W. Boos, and W. Welte. 2001. The crystal structure of a liganded trehalose/maltose-binding protein from the hyperthermophilic archaeon Thermococcus litoralis at 1.85 Å. J. Mol. Biol. 305:905-915. [DOI] [PubMed] [Google Scholar]
- 4.Gielkens, M. M. C., J. Visser, and L. H. de Graaff. 1997. Arabinoxylan degradation by fungi: characterization of the arabinoxylan-arabinofuranohydrolase encoding genes from Aspergillus niger and Aspergillus tubingensis. Curr. Genet. 31:22-29. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert, H. J., D. A. Sullivan, G. Jenkins, L. E. Kellett, N. P. Minton, and J. Hall. 1988. Molecular cloning of multiple xylanase genes from Pseudomonas fluorescens subsp. cellulosa. J. Gen. Microbiol. 134:3239-3247. [DOI] [PubMed] [Google Scholar]
- 6.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 7.Horlacher, R., K. B. Xavier, H. Santos, J. DiRuggiero, M. Kossmann, and W. Boos. 1998. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 180:680-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irwin, D., E. D. Jung, and D. B. Wilson. 1994. Characterization and sequence of a Thermomonospora fusca xylanase. Appl. Environ. Microbiol. 60:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 10.Luthi, E., D. R. Love, J. McAnulty, C. Wallance, A. Caughey, D. Saul, and P. L. Bergquist. 1990. Cloning, sequence analysis, and expression of genes encoding xylan-degrading enzymes from the thermophilic Caldocellum saccharolyticum. Appl. Environ. Microbiol. 56:1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margolles-Clark, E., M. Ilmén, and M. Penttilä. 1997. Expression patterns of ten hemicellulase genes of the filamentous Trichoderma reesei on various carbon sources. J. Biotechnol. 57:167-179. [Google Scholar]
- 12.McCarthy, A. J., and S. T. Williams. 1992. Actinomycetes as agents of biodegradation in the environment—a review. Gene 115:189-192. [DOI] [PubMed] [Google Scholar]
- 13.Miller, D. M., J. S. Olson, J. W. Pflugrath, and F. A. Quiocho. 1983. Rates of ligand binding to periplasmic proteins involved in bacterial transport and chemotaxis. J. Biol. Chem. 258:13665-13672. [PubMed] [Google Scholar]
- 14.Miyamoto, K., E. Nukui, H. Itoh, T. Sato, T. Kobayashi, C. Imada, E. Watanabe, Y. Inamori, and H. Tsujibo. 2002. Molecular analysis of the gene encoding a novel chitin-binding protease from Alteromonas sp. strain O-7 and its role in the chitinolytic system. J. Bacteriol. 184:1865-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Sanger, F., S. Nicklen, and A. R. Coulson. 1997. DNA sequencing with chain-termination inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saurin, W., W. Köster, and E. Dassa. 1994. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol. Microbiol. 12:993-1004. [DOI] [PubMed] [Google Scholar]
- 18.Schlösser, A. 1999. MsiK-dependent trehalose uptake in Streptomyces reticuli. FEMS Microbiol. Lett. 184:187-192. [DOI] [PubMed] [Google Scholar]
- 19.Schlösser, A., and H. Schrempf. 1996. A lipid-anchored binding protein is a component of an ATP-dependent cellobiose/cellotriose-transport system from the cellulose degrader Streptomyces reticuli. Eur. J. Biochem. 242:332-338. [DOI] [PubMed] [Google Scholar]
- 20.Schlösser, A., J. Jantos, K. Hackmann, and H. Schrempf. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl. Environ. Microbiol. 65:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlösser, A., T. Aldekamp, and H. Schrempf. 2000. Binding characteristics of CebR, the regulator of the ceb operon required for cellobiose/cellotriose uptake in Streptomyces reticuli. FEMS Microbiol. Lett. 190:127-132. [DOI] [PubMed] [Google Scholar]
- 22.Schlösser, A., A. Weber, and H. Schrempf. 2001. Synthesis of the Streptomyces lividans maltodextrin ABC transporter depends on the presence of the regulator MalR. FEMS Microbiol. Lett. 196:77-83. [DOI] [PubMed] [Google Scholar]
- 23.Shareck, F., C. Roy, M. Yamaguchi, R. Morosoli, and D. Kluepfel. 1991. Sequence of three genes specifying xylanases in Streptomyces lividans. Gene 107:75-82. [DOI] [PubMed] [Google Scholar]
- 24.Simpson, P. J., D. N. Bolam, A. Cooper, A. Ciruela, G. P. Hazlewood, H. J. Gilbert, and M. P. Williamson. 1999. A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity. Structure 7:853-864. [DOI] [PubMed] [Google Scholar]
- 25.Spiridonov, N. A., and D. B. Wilson. 1999. Characterization and cloning of CelR, a transcriptional regulator of cellulose genes from Thermomonospora fusca. J. Biol. Chem. 274:13127-13132. [DOI] [PubMed] [Google Scholar]
- 26.Spurlino, J. C., L. E. Rodseth, and F. A. Quiocho. 1992. Atomic interactions in protein-carbohydrate complexes-tryptophan residues in the periplasmic maltodextrin receptor for active transport and chemotaxis. J. Mol. Biol. 226:15-22. [DOI] [PubMed] [Google Scholar]
- 27.Sutcliffe, I. C., and R. R. B. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsujibo, H., K. Miyamoto, T. Kuda, K. Minami, T. Sakamoto, T. Hasegawa, and Y. Inamori. 1992. Purification, properties, and partial amino acid sequences of thermostable xylanases from Streptomyces thermoviolaceus OPC-520. Appl. Environ. Microbiol. 58:371-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsujibo, H., T. Ohtsuki, T. Iio, I. Yamazaki, K. Miyamoto, M. Sugiyama, and Y. Inamori. 1997. Cloning and sequence analysis of genes encoding xylanases and acetyl xylan esterase from Streptomyces thermoviolaceus OPC-520. Appl. Environ. Microbiol. 63:661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsujibo, H., C. Takada, A. Tsuji, M. Kosaka, K. Miyamoto, and Y. Inamori. 2001. Cloning, sequencing, and expression of the gene encoding an intracellular β-d-xylosidase from Streptomyces thermoviolaceus OPC-520. Biosci. Biotechnol. Biochem. 65:1824-1831. [DOI] [PubMed] [Google Scholar]
- 32.Tsujibo, H., C. Takada, Y. Wakamatsu, M. Kosaka, A. Tsuji, K. Miyamoto, and Y. Inamori. 2002. Cloning and expression of an alpha-l-arabinofuranosidase gene (stxIV) from Streptomyces thermoviolaceus OPC-520, and characterization of the enzyme. Biosci. Biotechnol. Biochem. 66:434-438. [DOI] [PubMed] [Google Scholar]
- 33.van Peji, N. N. M. E., J. Brinkmann, M. Vrsanská, J. Visser, and L. H. de Graaff. 1997. β-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzymes spectrum. Eur. J. Biochem. 245:164-173. [DOI] [PubMed] [Google Scholar]
- 34.van Wezel, G. P., J. White, P. Young, P. W. Postma, and M. J. Bibb. 1997. Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3 is controlled by malR, a member of the lacI-galR family of regulatory genes. Mol. Microbiol. 23:537-549. [DOI] [PubMed] [Google Scholar]
- 35.van Wezel, G. P., J. White, M. J. Bibb, and P. W. Postma. 1997. The malEFG gene cluster of Streptomyces coelicolor A3(2): characterization, distribution and transcriptional analysis. Mol. Gen. Genet. 254:604-608. [DOI] [PubMed] [Google Scholar]
- 36.Weickert, M. J., and S. Adhya. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267:15869-15874. [PubMed] [Google Scholar]
- 37.Whistler, R. L., and E. L. Richards. 1970. Hemicellulose, pp. 447-469. In W. Pigman and D. Horton (ed.), The carbohydrates—chemistry and biochemistry, 2nd ed, vol. 2A. Academic Press, New York, N.Y.
- 38.Wright, F., and M. J. Bibb. 1992. Codon usage in the G+C-rich Streptomyces genome. Gene 113:55-65. [DOI] [PubMed] [Google Scholar]
- 39.Xavier, K., B., L. O. Martins, R. Peist, M. Kossmann, W. Boos, and H. Santos. 1996. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 178:4773-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao, X., F. Wang, A. Saito, J. Majka, A. Schlösser, and H. Schrempf. 2002. The novel Streptomyces olivaceoviridis ABC transporter Ngc mediates uptake of N-acetylglucosamine and N,N′-diacetylchitobiose. Mol. Genet. Genomics 267:429-439. [DOI] [PubMed] [Google Scholar]