Abstract
The DNA binding proteins ArgR and AhrC are essential for regulation of arginine metabolism in Escherichia coli and Bacillus subtilis, respectively. A unique property of these regulators is that they form hexameric protein complexes, mediating repression of arginine biosynthetic pathways as well as activation of arginine catabolic pathways. The gltS-argE operon of Lactococcus lactis encodes a putative glutamate or arginine transport protein and acetylornithine deacetylase, which catalyzes an important step in the arginine biosynthesis pathway. By random integration knockout screening we found that derepression mutants had ISS1 integrations in, among others, argR and ahrC. Single as well as double regulator deletion mutants were constructed from Lactococcus lactis subsp. cremoris MG1363. The three arginine biosynthetic operons argCJDBF, argGH, and gltS-argE were shown to be repressed by the products of argR and ahrC. Furthermore, the arginine catabolic arcABD1C1C2TD2 operon was activated by the product of ahrC but not by that of argR. Expression from the promoter of the argCJDBF operon reached similar levels in the single mutants and in the double mutant, suggesting that the regulators are interdependent and not able to complement each other. At the same time they also appear to have different functions, as only AhrC is involved in activation of arginine catabolism. This is the first study where two homologous arginine regulators are shown to be involved in arginine regulation in a prokaryote, representing an unusual mechanism of regulation.
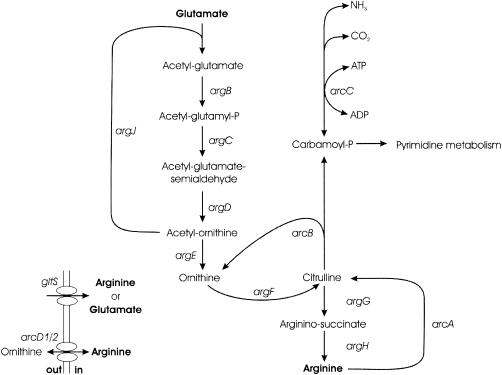
Arginine, a nonessential amino acid in the lactic acid bacterium Lactococcus lactis, is synthesized de novo from glutamate in eight enzymatic steps (Fig. 1). The recent publication of the L. lactis genome sequence (5) has revealed that the putative arginine biosynthesis genes are encoded by the three operons argCJDBF, gltS-argE, and argGH. The products of these genes all show homology to known arginine biosynthetic enzymes, except for that of gltS, which has been annotated as a putative glutamate or arginine ABC transporter (5). While the biosynthetic genes have been shown to be regulated by the presence of arginine in other organisms, this has not been investigated in lactic acid bacteria (LAB). The activities of the biosynthetic enzymes have been shown to be repressed by arginine in Lactobacillus plantarum (6), but regulatory studies on the transcriptional level have not been performed on LAB.
FIG. 1.
Schematic representation of arginine metabolism in L. lactis. Genes encode enzymes as follows: argB, N-acetylglutamate 5-phosphotransferase; argC, N-acetylglutamate 5-semialdehyde dehydrogenase; argD, N2-acetylornithine 5-aminotransferase; argJ, ornithine acetyltransferase; argE, acetylornithine acetyltransferase; argF, ornithine carbamoyltransferase; argG, argininosuccinate synthetase; argH, argininosuccinase; arcA, arginine deiminase; arcB, ornithine carbamoyltransferase; arcC, carbamate kinase; gltS, arginine or glutamate transporter.
Mechanisms for arginine catabolism vary among organisms (1). In L. lactis, complete degradation of arginine into ornithine, ammonium, and carbon dioxide takes place via the arginine deiminase pathway (ADI pathway) in three enzymatic steps catalyzed by arginine deiminase (ArcA), ornithine carbamoyltransferase (ArcB), and carbamate kinase (ArcC) (Fig. 1). The genes arcA, arcB, arcC1, and arcC2 encoding these enzymes are located in the arcABD1C1C2TD2 gene cluster. L. lactis harbors an extra arcC homologue, called arcC3, which is located distant from the remainder of the arginine-related genes in the chromosome. The genes arcD1 and arcD2 encode antiporter proteins, allowing ATP-independent 1:1 arginine-ornithine exchange (37), while arcT specifies an aminotransferase.
It has long been known that carbon metabolism and arginine catabolism are closely connected in L. lactis (9). However, the presence of arginine has a higher regulatory effect than the available carbon source does (37). The ADI pathway enzymes and amino acid transport systems are more stable during starvation than are enzymes of glycolysis (23). Thus, the ADI pathway plays an important role in supplying the cells with energy during recovery from starvation without energy expenditure. Additionally, glycolysis enzymes are more sensitive for low pH than the ADI enzymes are. Consequently, the ADI pathway represents an additional source of ATP production, combats acid stress by production of ammonium, and finally supplies carbamoyl phosphate, which is essential for de novo synthesis of pyrimidines. The identification of two putative cre (catabolite recognition element) sites in the arcA promoter of Lactobacillus sake (52) strongly suggests that carbon source-dependent regulation of the arginine catabolic genes is mediated by the major carbon catabolite repressor CcpA in this organism.
Arginine metabolism has been shown to be regulated by a transcriptional regulator called ArgR or AhrC in several diverse organisms (10, 12, 25, 34, 41). In this respect arginine regulation deviates from the “rule” of attenuation regulation of amino acid metabolism in prokaryotes (8, 39, 51). Regulation of amino acid metabolism in LAB via the direct action of a DNA binding protein has been observed only in the case of CmbR, which activates expression of the sulfur-related metC-cysK operon in response to acetylserine in L. lactis (13).
Several characteristic features of ArgR-AhrC-type regulators have been described: (i) they form hexaoligomeric complexes (12, 25), (ii) they have a winged helix-turn-helix DNA binding domain (44), and (iii) ArgR plays a role as an accessory factor in multimer resolution of ColE1 plasmids in Escherichia coli (17, 43). ArgR and AhrC repress their own expression (25) and activate the transcription of arginine catabolic genes by interacting with other regulation factors, such as ANR and RocR of E. coli and Bacillus subtilis, respectively (14, 27, 50).
ArgR and AhrC monomers consist of two domains, an N-terminal DNA binding domain containing the winged helix-turn-helix structure and a C-terminal domain involved in arginine binding and subunit multimerization (44). Investigation of the hexameric structure by crystallization has shown that six arginine molecules bind in the interphase between the C-terminal domains of two trimers (49) and that arginine thereby functions as a corepressor.
ArgR and AhrC homohexamers bind to operator sites (called ARG boxes) in regions of biosynthetic and catabolic arginine promoters. The ARG box is an 18-bp imperfect palindromic sequence, the consensus of which varies slightly among organisms (11, 24, 30, 33). The number of boxes was shown to correlate with the observed regulation. Thus, repression is stronger when two or three ARG boxes are present, as seen in the E. coli biosynthetic promoters, than when only a single box is present, as in the argR promoter of E. coli (10).
The publication of the entire Lactococcus lactis subsp. lactis IL1403 genome (5) has led to the identification of two ArgR-AhrC orthologues. Multiple putative arginine regulators have also been found in the genomes of other bacteria (3), but the function of these and the reason for the presence of more than one regulator in one organism remain to be established.
In this paper we show that Lactococcus lactis subsp. cremoris MG1363 harbors two functional arginine regulators. They cooperate in the repression of arginine biosynthesis but have different functions in the activation of arginine catabolism.
MATERIALS AND METHODS
Bacterial strains and media.
Strains of L. lactis used in this study are listed in Table 1. L. lactis was grown at 30 or 37°C in M17 medium (45) with 0.5% glucose as carbon source (GM17). A chemically defined medium (CDM) was made as described earlier (31) with Casitone (Difco, West Molesey, United Kingdom) in concentrations of 0.1 or 4%. CDM buffer containing 15 free amino acids (CDM15) was made as described previously (22), omitting arginine unless stated otherwise. Arginine stock solutions were made in distilled H2O; pH was set to 7.0 with HCl. For solid media, agar was added to a concentration of 15 g · liter−1. The following components were added when needed: erythromycin, 4 μg · ml−1 for selection of plasmids or 1 μg · ml−1 for maintaining TnNuc integrations; tetracycline, 2 μg · ml−1; chloramphenicol, 4 μg · ml−1; and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 40 μg · ml−1. Antibiotics were purchased from Sigma Chemical Co. (St. Louis, Mo.), and X-Gal was from Roche Molecular Biochemicals (Mannheim, Germany).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| MG1363 | L. lactis subsp. cremoris, plasmid-free derivative of NCDO 712 | 15 |
| MG1614 | MG1363, Strr Rifr | 15 |
| C17 | MG1614, chromosomal pTnNuc insertion in gltS-argE | This work |
| gdm strains | C17; pGh8::ISSI random integration mutants | This work |
| gdm-ex strains | gdm; pGh8::ISSI excised from chromosome | This work |
| MGΔargR | MG1363; chromosomal deletion of argR | This work |
| MGΔahrC | MG1363; chromosomal deletion of ahrC | This work |
| MGΔargRahrC | MG1363; chromosomal deletions of argR and ahrC | This work |
| Plasmids | ||
| pTnNuc | Camr Eryr Tetr Ampr; contains promoterless Staphylococcus aureus nuc gene, transcriptionally fused to lacZ | 38 |
| pGh8::ISS1 | Tetrori(Ts), random integration vector | 29 |
| pORI13 | Eryrori+RepA−; promoterless lacZ | 40a |
| pORI280 | Eryrori+RepA−; lacZ expressed constitutively via promoter P32 | 23a |
| pIL252 | Eryr; low-copy-number cloning vector | 41a |
| pVE6007 | Camrori(Ts) | 28 |
| p280ΔargR | Eryr; pORI280 containing argR deletion construct | This work |
| p280ΔahrC | Eryr; pORI280 containing ahrC deletion construct | This work |
| pORI13P32 | Eryr; P32 cloned upstream of lacZ in pORI13 | This work |
| pILORI4 | Eryr; pIL252 carrying the multiple cloning site and promoterless lacZ of pORI13 | This work |
| pILORI4::PargC | Eryr; pILORI4 carrying argC-1/argC-2 PCR fragment | This work |
| pILORI4::ParcA-1 | Eryr; pILORI4 carrying arcA-1/arcA-2 PCR fragment | This work |
| pILORI4::ParcA-3 | Eryr; pILORI4 carrying arcA-2/arcA-3 PCR fragment | This work |
| pILORI4::ParcA-4 | Eryr; pILORI4 carrying arcA-2/arcA-4 PCR fragment | This work |
| pILORI4::ParcA-5 | Eryr; pILORI4 carrying arcA-2/arcA-5 PCR fragment | This work |
| pILORI4::ParcA-6 | Eryr; pILORI4 carrying arcA-2/arcA-6 PCR fragment | This work |
| pILORI4::ParcA-7 | Eryr; pILORI4 carrying arcA-2/arcA-7 PCR fragment | This work |
| pILORI4::ParcA-8 | Eryr; pILORI4 carrying arcA-2/arcA-8 PCR fragment | This work |
DNA isolation and manipulations.
Chromosomal and plasmid DNAs were isolated from L. lactis according to the methods of Johansen and Kibenich (20) and Birnboim (4), respectively. DNA was manipulated essentially as described by Sambrook et al. (40), and lactococcal strains were transformed with plasmid DNA by electroporation (18).
Chromosomal deletion mutants were made using pVE6007 (28) as helper plasmid for single-crossover integration of p280ΔargR and p280ΔahrC in L. lactis MG1363 grown at 37°C. Excision of pORI280, leaving the deletion constructs in the chromosome of strain MG1363, was performed at 37°C without antibiotic selection. Excissants grown on solid medium were screened by PCR, and mutants were confirmed with Southern blotting. Probe labeling, hybridization, and detection were performed using the ECL direct nucleic acid labeling system according to the specifications of the manufacturer (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Restriction enzymes were purchased from New England BioLabs (Beverly, Mass.). DNA was amplified using specific primers as listed in Table 2. PCR products were purified with the High Pure PCR product purification kit (Roche Molecular Biochemicals). Taq DNA polymerase (Roche Molecular Biochemicals) was used for colony PCR, and Pwo DNA polymerase (Roche Molecular Biochemicals) was used for DNA constructs.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence | Purpose |
|---|---|---|
| pORI13m2 | GGCAATTGAAGGCAGCTGATCTCAAC | Construction of pILORI4 |
| pORI13s2 | GGACTAGTAGATCTAATCGATGCATGC | As above |
| P32-1 | GCTCTAGACTTGTTTTTCGTGTGC | Construction of pORI13P32 |
| P32-2 | GCTCTAGACATTTCAAAATTCCTCCG | As above |
| ISS1-For | ATTGTAAAACGACGGCCAGTGTTCATTGATATATCCTCGCTGTC | Inverse PCR of MG1614::ISS1 integrants |
| ISS1-T7 | ACCTAATACGACTCACTATAGGGCTACTGAGATTAAGGTCTTAATGGG | As above |
| argR-1 | GAAGATCTAATCTTCTTTAGCTTCCG | Construction of MG1363 argR deletion mutant |
| argR-2 | CGGAATTCTTCTAATCTTTTATCTCT | As above |
| argR-3 | CGGAATTCGCAAATATTTTGACAGC | As above |
| argR-4 | GCTCTAGAGATATGACAGATGTTGC | As above |
| ahrC-1 | GAAGATCTTAGAAAAAGCGCTCAAAG | Construction of MG1363 ahrC deletion mutant |
| ahrC-2 | CGGAATTCCTTTTCATAGTTCTTCGC | As above |
| ahrC-3 | CGGAATTCAGAGTTTTAAATTTACTG | As above |
| ahrC-4 | GCTCTAGATTGACTGTCATGTTGACC | As above |
| argC-1 | CGGAATTCTGGAACATAATAAAGCG | Cloning of argC promoter in pILORI4 |
| argC-2 | GCTCTAGATATAACCTCTAATTCCG | As above |
| arcA-1 | CGGAATTCATTCTTGCTGATGAGAG | Cloning of arcA promoter fragments |
| arcA-2 | GCTCTAGAATTTCCCAATTTCTGAG | As above |
| arcA-3 | CGGAATTCAAATATTTTGTAAAATAAG | As above |
| arcA-4 | CGGAATTCGAATCCCATGATAAGC | As above |
| arcA-5 | CGGAATTCAACGTGAAATTGTCAG | As above |
| arcA-6 | CGGAATTCTATAAATGAATAAACC | As above |
| arcA-7 | CGGAATTCAAAATATGCATAGATG | As above |
| arcA-8 | CGGAATTCGCTTGACAAAAAATATGC | As above |
Nucleotide sequencing reactions were performed on a DNA Labstation 625 (Vistra DNA System) with the Thermo Sequenase primer cycle sequencing kit (Amersham Pharmacia). Fragments were separated and detected with the ALFexpress II gel system (Amersham Pharmacia).
Construction of lacZ expression plasmids.
The constitutive lactococcal promoter P32 was amplified using primers P32-1 and P32-2 and cloned in pORI13, resulting in pORI13P32. With this plasmid as template, a PCR product containing the multiple cloning site and lacZ of pORI13 was obtained using the pORI13m2 and pORI13s2 primers. The PCR fragment was inserted as an MfeI/SpeI restriction fragment in the EcoRI/XbaI sites of pIL252, yielding plasmid pILORI4. Only very low intrinsic β-galactosidase activity could be measured in cells carrying the empty pILORI4 vector.
The promoter fragment to be analyzed for expression was amplified from chromosomal DNA of L. lactis MG1363 by PCR with the primers listed in Table 2 and cloned in the low-copy-number expression vector pILORI4.
Isolation of mutants derepressed in arginine metabolism.
L. lactis C17 (gltS-argE::lacZ) was transformed with pGh8::ISS1 (29) and submitted to random integration screening on CDM containing erythromycin, tetracycline, X-Gal, and 4% Casitone at the nonpermissive temperature (37°C). Integrants showing a clear gltS-argE::lacZ derepression phenotype were isolated for further characterization. pGh8::ISS1 was cured from the strains by repeated 1,000-fold dilution and growth in GM17 plus erythromycin at the permissive temperature (28°C).
Enzyme assays.
β-Galactosidase activity assays were performed on cell suspensions that were permeabilized by chloroform as described previously (19).
Data analysis.
The Clustal W program was used for protein sequence alignments (46). Clone Manager 6.0 was used for free energy calculations of palindromic DNA structures.
Nucleotide sequence accession numbers.
The new sequences generated in this work have been given the accession numbers AY518512 (argR), AY518513 (ahrC), AY518514 (PargC), and AY518515 (ParcA).
RESULTS
gltS-argE derepression mutations in L. lactis target to two ArgR-AhrC-type regulators.
A pTnNuc integration library of L. lactis MG1614, an isogenic L. lactis subsp. cremoris MG1363 derivative (38), was plated on GM17 plates containing erythromycin and X-Gal. Colonies were screened by replica plating onto CDM plates containing erythromycin, X-Gal, and 4 or 0.1% Casitone. An L. lactis MG1614 strain called C17, showing Casitone-dependent β-galactosidase activity, had lacZ of TnNuc integrated in the C-terminal part of argE, the second gene of the arginine biosynthetic gltS-argE operon. Expression of gltS-argE was high in the 0.1% Casitone medium and low in the 4% Casitone medium. Random ISS1 transposon integration screening using pGh8::ISS1 (29) was performed in L. lactis C17, to identify genes involved in Casitone-dependent regulation of gltS-argE. Approximately 14,000 colonies were screened, and 18 integrants (called gdm for gltS-argE derepression mutation) that were clearly derepressed on a rich medium containing X-Gal were isolated. Chromosomal ISS1 integration sites were determined for nine of the integrants by sequencing of inverse PCR products. The resultant target genes of seven of these are presented in Table 3.
TABLE 3.
Characterization of L. lactis gdm-ex mutants
| Strain | ISS1 target gene (insertion site relative to start of gene) |
gltS-argE expression determined as specific β-galactosidase activity (Miller units), at the in- dicated arginine concna
|
Repression ratio (0.1/10 mM)b | ||
|---|---|---|---|---|---|
| 0.1 mM | 1 mM | 10 mM | |||
| C17 | None | 1.1 | 0.5 | 0.3 | 3.7 |
| C17(gdm24ex) | argR (231 bp) | 116.6 | 96.9 | 88.3 | 1.3 |
| C17(gdm25ex) | argR (411 bp) | 85.5 | 14.3 | 3.6 | 23.8 |
| C17(gdm28ex) | argR (59 bp) | 91.1 | 63.9 | 64.9 | 1.4 |
| C17(gdm1ex) | ahrC (105 bp) | 87.3 | 79.1 | 73.2 | 1.2 |
| C17(gdm26ex) | ahrC (28 bp) | 88.2 | 84.8 | 80.3 | 1.1 |
| C17(gdm8ex) | arcD2c | 17.5 | 10.0 | 0.6 | 29.2 |
| C17(gdm29ex) | arcD2 | 14.8 | 8.9 | 0.8 | 18.5 |
Activity was measured in cells from two independent cultures in CDM15 harvested during exponential growth phase.
Specific β-galactosidase activity in CDM15 with 0.1 mM l-arginine divided by that in CDM15 with 10 mM l-arginine.
Exact location in arcD2 not determined.
The chromosomally integrated copy of pGh8::ISS1 was cured from the C17(gdm) strains by growing them in GM17 without tetracycline selection (29). In this way only the ISS1 element was left at the chromosomal integration site (strains designated “gdm-ex”), allowing for a direct comparison between the cured strains and the parental strain L. lactis C17 under the same culturing conditions. Excision of pGh8::ISS1 was confirmed by Southern blotting, PCR on chromosomal DNA, and testing for tetracycline sensitivity.
In strains C17(gdm8) and C17(gdm29) the integration sites could be localized to an open reading frame with high homology to arcD2 of L. lactis IL1403. This gene encodes a putative arginine-ornithine antiporter and is the last gene of the arginine catabolic pathway operon arcABD1C1C2TD2. The chromosomal ISS1 targets of the six remaining C17(gdm) strains are all located in or upstream of either of two open reading frames the products of which show high homology to ArgR-AhrC-type DNA binding proteins in other organisms. Growth of the integrants was found to be strongly reduced in the absence of arginine, and the experiments described below were all performed with cells grown in the presence of (different concentrations of) arginine.
The gltS-argE operon of L. lactis is strongly derepressed in both argR and ahrC mutants.
ISS1 of pGh8::ISS1 had integrated in the very N-terminal part of ahrC in strains C17(gdm1ex) and C17(gdm26ex), resulting in strong derepression of gltS-argE expression. Surprisingly, the expression of gltS-argE in strains C17(gdm1ex) and C17(gdm26ex) was much higher than that observed for strain C17 even at very low arginine concentrations (Table 3). Strain C17(gdm27ex) showed the same derepression phenotype as that of strains C17(gdm1ex) and C17(gdm26ex), but ISS1 insertion had occurred in the yiiB gene located just upstream of ahrC (data not shown). The genes yiiB and ahrC overlap by 4 bp, suggesting that they are transcriptionally coupled. Homology searches predict YiiB to be a 23S rRNA methyltransferase, with some homology to an S4 RNA binding domain and to an FtsJ-like methyltransferase (E values of 5.9e-3 and 1.8e-5, respectively), not known to have any influence on arginine metabolism. The observed derepression in strain C17(gdm27ex) is probably caused by a polar effect on ahrC expression rather than inactivation of the yiiB gene product. The fact that derepression reached the same levels as those measured for the other ahrC integration knockouts is in accordance with this hypothesis (data not shown). The ahrC gene is followed by a terminator structure with a calculated free energy of −13.0 kcal. A recN homologue is present downstream of ahrC, with an intergenic spacing of 180 bp. A weak putative promoter (TTGTGC-18N-TATAAT) and ribosomal binding site (AGAAAGGAAAT) precede recN. Considering the genetic structure of the ahrC region, disruption of ahrC expression alone is expected to cause the derepression of gltS-argE expression in strains C17(gdm1ex) and C17(gdm26ex) and possibly also in strain C17(gdm27ex).
The C17(gdm24ex), C17(gdm25ex), and C17(gdm28ex) strains all carry ISS1 in a 459-bp gene annotated as argR in L. lactis IL-1403. The strains differed with respect to the extent to which gltS-argE was derepressed. Strain C17(gdm28ex), in which argR is disrupted in the start of the gene, showed a complete gltS-argE derepression phenotype similar to that of the ahrC knockout strains C17(gdm1ex) and C17(gdm26ex) (Table 3). In strain C17(gdm24ex) the insertion had taken place in the center of argR (Table 3). Interestingly, disruption of argR in this region resulted in a drastic growth inhibition, with growth rates of 0.39 h−1 in CDM15 with 0.1 mM arginine to 0.31 h−1 in CDM15 with 10 mM arginine, compared to growth rates between 0.5 and 0.63 h−1 for the other strains. Finally, with ISS1 insertion at the very end of argR, strain C17(gdm25ex) showed maximum derepression to a level comparable to that in strain C17(gdm28ex) but differing in that it had maintained the ability to sense and respond to arginine availability (Table 3). Two transcriptional terminator structures with calculated free energies of −12.5 and −14.4 kcal, respectively, are located in the argR-murC intergenic region. The argR gene is located in a divergent orientation with argS (encoding arginyl-tRNA synthetase) and is separated from this gene by a putative promoter region of only 67 bp. A consensus extended −10 box (TGGTATAAT) is located upstream of argR, but no clear ribosome binding site could be identified. As argR is in opposite orientation with respect to the neighboring argS and murC genes, disruption of argR is expected to be the sole cause of gltS-argE derepression in the strains C17(gdm24ex), C17(gdm25ex), and C17(gdm28ex).
Regulation of the arginine biosynthesis argCJDBF operon in L. lactis.
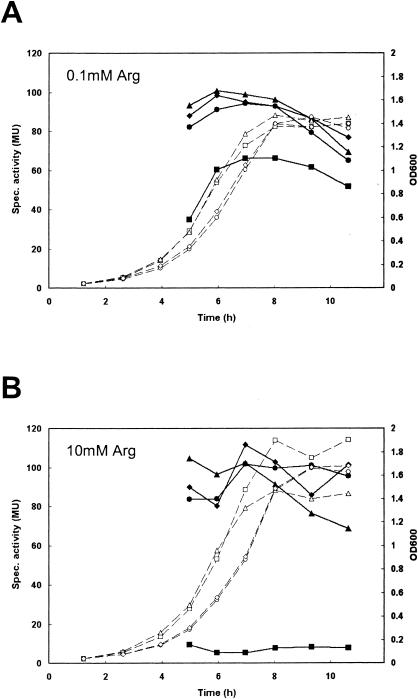
A fragment of 296 bp containing the entire argC promoter (PargC) was cloned upstream of lacZ in the promoter expression vector pILORI4. This expression construct was introduced in the wild-type strain L. lactis MG1363, as well as its single isogenic regulator mutants L. lactis MGΔargR and MGΔahrC and the double regulator mutant L. lactis MGΔargRahrC. Expression of lacZ from this promoter was investigated during growth on CDM (CDM15) containing different concentrations of arginine. Clear arginine-dependent repression was observed in the wild-type strain MG1363 (Fig. 2). In each of the single regulator mutants, arginine repression was no longer seen and β-galactosidase expression reached the same levels as that in the double regulator mutant (Fig. 2). As was observed for the expression of the gltS-argE operon in the argR::ISS1 or ahrC::ISS1 knockout strains, disruption of a single regulator gene resulted in complete derepression of expression from PargC. Thus, it appears that the two regulators, ArgR and AhrC, have a corepressing effect rather than a cumulative effect on repression of the argCJDBF arginine biosynthetic operon in L. lactis.
FIG. 2.
Growth (dashed lines and open symbols) and β-galactosidase activities (solid lines and symbols) of L. lactis MG1363 (squares), MGΔargR (circles), MGΔahrC (triangles), and MGΔargRahrC (diamonds), all harboring p4::PargC, in CDM15 with 0.1 mM (A) or 10 mM (B) l-arginine. MU, Miller units.
ArgR and AhrC have different roles in regulation of the arginine catabolism operon of L. lactis.
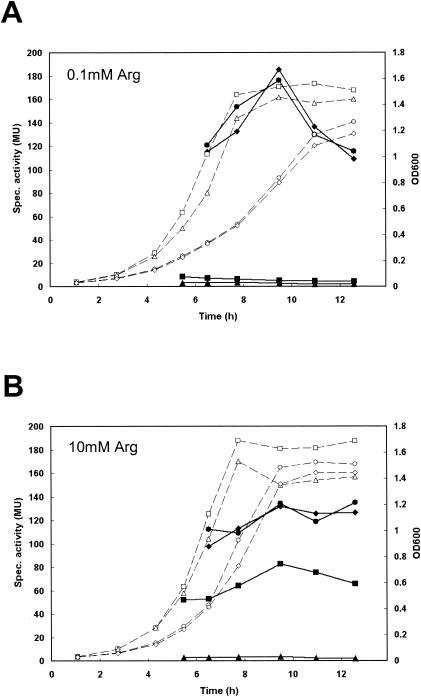
In the light of the regulation of the arginine biosynthetic gltS-argE and argCJDBF operons, we decided to examine the role of the regulators in the expression of the arginine catabolic arc gene cluster. To that end, the arcA promoter region (ParcA) up to 260 bp upstream of the arcA start codon (same construct as ParcA-1 in Fig. 4) was cloned in the expression vector pILORI4, which was then introduced in L. lactis MG1363 and its isogenic regulator deletion mutants. As shown in Fig. 3, clear arginine-dependent regulation was observed in L. lactis MG1363, with expression from ParcA increasing with an increase in the arginine concentration. Deletion of ahrC resulted in no or only low expression from ParcA even at a high arginine concentration in the medium. In contrast, expression was constitutively high in the argR mutant and in the argR ahrC double deletion strain. This would suggest that activation of arginine catabolism in L. lactis MG1363 is mediated by AhrC and that a repressing effect is exerted by ArgR. Alternatively, the high ParcA expression in the argR deletion mutants could be caused by a constant high intracellular level of arginine resulting from the derepression of arginine biosynthesis. However, in the double mutant the function of AhrC seems to be overruled by the removal of ArgR.
FIG. 4.
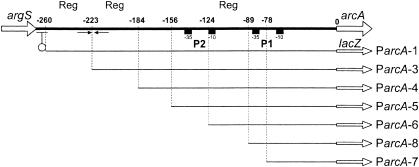
Schematic representation of the argS-arcA intergenic region. Numbers on the top line refer to the positions relative to the AUG start codon of arcA (0). The indicated fragments were cloned in the pILORI4 promoter expression vector in transcriptional fusion with lacZ. Fragment names are shown on the right. The putative argS transcriptional terminator is indicated by a lollipop, −10 and −35 boxes of the putative promoters P1 and P2 are shown by boxes, and a putative regulatory palindromic structure is shown with arrows. “Reg” denotes regions involved in arginine-dependent regulation (see text for details).
FIG. 3.
Growth (dashed lines and open symbols) and β-galactosidase activities (solid lines and symbols) of L. lactis MG1363 (squares), MGΔargR (circles), MGΔahrC (triangles), and MGΔargRahrC (diamonds), all harboring p4::ParcA, in CDM15 with 0.1 mM (A) or 10 mM (B) l-arginine. MU, Miller units.
The argS-arcA intergenic region contains several features (Fig. 4): a putative transcription terminator with a calculated free energy of −9.1 kcal composed of a dyad symmetry followed by a stretch of thymidine residues, starting 19 bp downstream of argS; two core promoter structures, P1 (5′-TTGACA-17N-TATAAT) and P2 (5′-TTGTCA-17N-TATAAA), located at 56 to 84 bp and at 118 to 146 bp, respectively, from the start of arcA; and a characteristic ribosomal binding site (5′-AAAGGA) 9 bp upstream of arcA. In order to identify possible operator sites involved in the observed regulation, deletion derivatives of the arcA promoter region were transcriptionally fused to lacZ in pILORI4 (Fig. 4). β-Galactosidase activities of these promoter fragments were measured in the wild-type strain MG1363 grown in CDM15 with 0.1 or 10 mM arginine (Table 4).
TABLE 4.
Expression of ParcA subclones
| ParcA promoter construct | Specific β-galactosidase activity (Miller units), at the indicated arginine concna
|
Fold regulation (10/0.1 mM)b | |
|---|---|---|---|
| 0.1 mM | 10 mM | ||
| pILORI4::ParcA-1 | 7.0 (±0.7) | 51.0 (±15.8) | 7.3 |
| pILORI4::ParcA-3 | 32.6 (±5.3) | 97.5 (±1.1) | 3.0 |
| pILORI4::ParcA-4 | 84.1 (±16.6) | 67.0 (±8.0) | 0.8 |
| pILORI4::ParcA-5 | 11.9 (±2.2) | 35.1 (±8.7) | 2.9 |
| pILORI4::ParcA-6 | 16.7 (±1.0) | 37.5 (±8.2) | 2.2 |
| pILORI4::ParcA-8 | 11.8 (±2.3) | 9.6 (±3.1) | 0.8 |
| pILORI4::ParcA-7 | 1.0 (±0.6)c | 0.4 (±0.1)c | 0.4 |
Activity was measured in cells from three independent cultures in CDM15 harvested during the transition phase of growth. Standard deviations are shown in parentheses.
Specific β-galactosidase activity in CDM15 with 10 mM arginine divided by that in CDM15 with 0.1 mM arginine.
Corresponds to the low intrinsic β-galactosidase activity from the empty pILORI4 vector (data not shown).
Removing the −35 box of P1 (Fig. 4, compare ParcA-7 to ParcA-8) resulted in a severe decrease of expression of lacZ (Table 4). Fragment ParcA-8 gave arginine-independent expression, defining P1 as the minimal promoter, lacking operators involved in arginine regulation. Partial arginine-dependent activation, compared to ParcA-1 containing the entire promoter region, took place in ParcA-5 and ParcA-6, suggesting the presence of an operator(s) of arginine regulation in this region. Including the entire putative promoter P2 (the region up to −156 bp upstream of arcA) did not result in increased β-galactosidase activity, questioning the functionality of P2. However, the region included in ParcA-4 just upstream of the P2 structure had a dramatic effect on expression, resulting in high arginine-independent expression. Regulation was restored only by including sequences further upstream, 223 to 260 bp from arcA, with maximal arginine-dependent regulation taking place with the largest fragment, ParcA-1.
The lactococcal arginine regulators lack conserved amino acid residues.
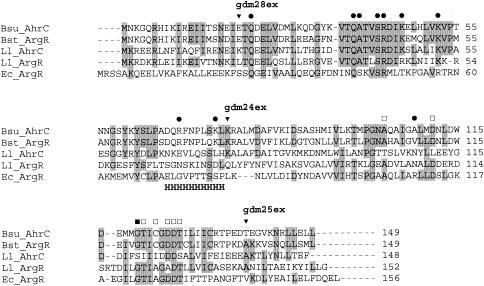
The argR gene of L. lactis subsp. cremoris MG1363 (which is isogenic to strain MG1614 [15]) encodes a putative protein of 152 amino acids, called ArgRLl hereafter, while ahrC specifies a putative protein of 148 amino acids, named AhrCLl. The two regulators show mutual identity for 50 amino acid residues (32%) and are homologous to well-known arginine regulators, like ArgR of E. coli, AhrC of B. subtilis, and ArgR of Bacillus stearothermophilus (Fig. 5). All these proteins contain an N-terminal DNA binding domain, a central hinge region, and a C-terminal arginine-sensing and subunit multimerization domain (Fig. 5). Mutagenesis studies of the arginine regulators of E. coli (ArgREc) (7, 26, 48) and B. stearothermophilus (ArgRBst) (21) have identified amino acid residues that are essential for regulator functionality. Of these residues, Ser47 and Arg48 of ArgREc are conserved in both lactococcal regulators (Fig. 5). However, other residues known to play a role in operator-regulator interaction have changed in ArgRLl and AhrCLl. Ser44 of ArgREc has changed to Ala, Thr51 is replaced by Lys or Arg, and Arg57 has changed to Lys in the regulators of the aligned gram-positive organisms in Fig. 5. A range of residues in the N-terminal part of the arginine regulators of the gram-positive bacteria is highly conserved, but less so in the gram-negative bacterial ArgREc, e.g., amino acid residues 36 to 45 of AhrCLl show a highly conserved VTQATVSRDI motif. In the C-terminal domains of the proteins there appears to be higher similarity between the gram-negative E. coli regulator and the gram-positive bacterial regulators, and in most cases, residues known to be essential for subunit multimerization and arginine binding have been conserved. However, it is noteworthy that, of the conserved GTI-X-GDDT motif (residues 123 to 130 of ArgREc), only the Ile and the double Asp residues are maintained in AhrCLl. Whereas most of these residues are preserved in ArgRLl, it should be noted that Asp128, which is essential for arginine binding in ArgREc, is replaced by an Ala residue. The possible significance of these changes will be discussed below.
FIG. 5.
Clustal W-aligned sequences of arginine regulators from B. subtilis 168 (Bsu_AhrC), B. stearothermophilus (Bst_ArgR), L. lactis MG1363 (Ll_ArgR and Ll_AhrC), and E. coli K-12 (Ec_ArgR). Shaded residues are identical in more than 50% of the sequences. “H” indicates the hinge region residues connecting the C- and N-terminal domains, as determined from the B. stearothermophius ArgR crystal structure (34). Functions of specific residues are specified as follows: involved in operator recognition and binding (•), involved in subunit multimerization (▪), and involved in arginine binding (□). ISS1 integration sites in ArgRLl and integrant strain names are indicated by ▾.
DISCUSSION
In this work we have investigated the regulation of arginine metabolic genes in L. lactis and have shown that two ArgR-AhrC-type regulators are required for repression of the arginine biosynthetic gltS-argE operon. Chromosomal argR and ahrC deletion mutants of L. lactis MG1363 were made to confirm that repression of the central arginine biosynthesis operon argCJDBF is also dependent on the presence of both regulators. Arginine-dependent regulation of the catabolic arcABD1C1C2TD2 gene cluster was also abolished in the regulator mutants. However, in this case the mutations had different effects, as the lack of ArgR resulted in high and arginine-independent expression while lack of AhrC resulted in constitutive low expression. Until now, the function of arginine regulators has been investigated only for organisms carrying a single arginine regulator (e.g., ArgR in E. coli and AhrC in B. subtilis). What has mainly caught our interest is the fact that two functional, homologous regulators are involved in and necessary for arginine-dependent gene regulation in L. lactis.
The presence of two homologous regulators suggests that (i) the regulators are paralogs, able to perform the same function(s) and to complement each other, or that (ii) they have different functions, e.g., one regulating arginine biosynthesis and the other regulating arginine catabolism, as proposed by Guèdon et al. (16). Neither supposition holds true for the arginine regulators of L. lactis. The results for the regulation of the gltS-argE and argCJDBF biosynthetic operons clearly demonstrate that the two regulators are not complementary. Not only did the ISS1 integration knockout screening allow identification of both regulators, which would not be the case could any one of them perform the action of the other, but also arginine-dependent regulation was abolished in both of the single regulator deletion mutants. Both regulators have different functions with respect to regulation of the arginine catabolic pathway, but neither of the single regulators could be shown to be responsible for the arginine-dependent regulation of arginine catabolism observed in the wild-type strain. Another surprising observation was that expression of gltS-argE in the wild-type strain, although regulated in dependence on arginine availability, was much lower than that in either of the regulator knockout strains. A similar observation was made in the study of ArgR in two different E. coli strains, K-12 and B (47). Only a single amino acid substitution differentiates ArgR of E. coli K-12, which showed strong arginine-dependent regulation, from ArgR of E. coli B, which mediated only weakly arginine-dependent regulation, resulting in so-called superrepression of arginine biosynthesis (42, 47). Both ways to regulate arginine metabolism are effective, and a mechanism of superrepression as observed for ArgR of E. coli B might be utilized by L. lactis. This putative superrepression in the wild-type L. lactis MG1363 was not observed in the promoter expression studies, but this may be explained by the possibly low levels of ArgRLl and AhrCLl in the cell: the multicopy vector situation may, to some extent, dilute the regulator proteins relative to the plasmid-located operators, despite pILORI4 being a low-copy-number vector. Alternatively, the difference in the level of regulation between the argC and gltS promoters could be explained by the presence of only one ARG box upstream of the gltS operon as opposed to two in the argC operon (see below), as the number of ARG boxes is known to correlate with the level of regulation in E. coli (10).
The three different ISS1 integration sites in argR yielded entirely different growth characteristics or gltS-argE expression patterns, which allowed us to confirm the functions of the ArgRLl subdomains. Integration in the putative hinge region of ArgR, disrupting the C-terminal part, caused not only arginine-independent derepression but also a considerable growth inhibition (Table 3). As seen for ArgREc and AhrCBs, this suggests that the C terminus of ArgRLl is essential for arginine sensing. Additionally, the N-terminal part may have some intrinsic DNA binding capacity, disturbing other metabolic functions of the cell. The reappearance of arginine sensing when disruption takes place in the very C-terminal region of the regulator confirms the sensory function of this domain. The more pronounced derepression of gltS-argE caused by the latter mutation compared to the wild type is most likely the result of incorrect arginine sensing.
The arcD1 and arcD2 genes most likely encode the arginine-ornithine antiporter described by Poolman et al. (37). The gene arcD2 is the last gene in the catabolic arc operon and, therefore, the only gene the expression of which was affected by the ISS1 insertion in strains C17(gdm8ex) and C17(gdm29ex). The observed effect on gene regulation is probably indirect: derepression of gltS-argE expression as a result of arcD2 disruption is probably caused by low arginine uptake rates, leading to endogenous arginine deficiency with subsequent increased expression of the arginine biosynthetic genes. In these integrants (gdm8ex and gdm29ex) gltS-argE was still regulated as a function of arginine availability, presumably via the ArgR and AhrC proteins that are present in these strains. However, only in the highest extracellular concentration of arginine tested was gltS-argE expression restored to wild-type level.
Regulation mediated by ArgR-AhrC-type regulators suggests the presence of ARG box operators. Indeed, operators similar to ARG boxes of E. coli and B. subtilis, 5′-WNTGAATWWWWATTCANW (26) and 5′-CATGAATAAAAATKCAAK (32), respectively, are present in the promoter regions of the argCJDBF and gltS-argE operons: gltSO, 5′-AATGTATAATTATACTTA (at −43 to −26 bp from the start of gltS); argCO1, 5′-AAAGTATAATAATACATA (at −82 to −65 bp from argC); and argCO2, 5′-AGTGTATAAAAATACATA (at −32 to −15 bp from argC), where positions identical to the E. coli ARG box are underlined. gltSO and argCO2 are both located in the putative core promoters of gltS and argC, respectively. The 32-bp spacing of argCO1 and argCO2 is unusual, as double ARG boxes are generally only 3 bp apart (26). Still, this organization would be in accordance with repression of these promoters taking place via direct interaction between the arginine regulators and the ARG box operators. This possibility is further supported by the fact that the N-terminal DNA binding domains of both lactococcal arginine regulators show high mutual similarity and similarity to those of ArgREc, ArgRBst, and AhrCBsu (Fig. 5).
A catabolite-responsive element (cre site) overlaps the core promoter of arcA, which is in agreement with the previously described carbon source-dependent regulation of arginine degradation in L. lactis (9). Subcloning of the arcA promoter allowed us to locate regions involved in the observed arginine-dependent regulation. However, in none of these regions could consensus ARG boxes be identified. Regions of regulatory importance localized to three different parts of the argS-arcA intergenic region (Fig. 4). The region just upstream of P1 partially restored arginine-dependent regulation of arcA, suggestive of an element activating expression from P1. The high arginine-independent expression observed by including the region upstream of P2 could be the result of activation via an upstream operator lacking regulatory capacity and inducing expression from P1 or P2 or both. That the regulatory capacity was restored by including the entire promoter region points to operators being involved in arginine-dependent control by a repressing mechanism. This pattern of regulation is intriguing and reveals a rather complex regulatory scheme, involving activation as well as repression. An A/T-rich palindromic structure (5′-TCTTTTTTAAAATATTTTGTAAAATA, 206 to 231 bp upstream of the start of arcA; nucleotides of the palindrome are underlined) that lacks features of a typical transcriptional terminator is present in the region upstream of P2 (Fig. 4). Approximately half of the structure is included in ParcA-3, and the complete structure is present in ParcA-1. Whether this structure in reality is involved in regulation of ParcA remains to be verified. The fact that the arginine degradative pathway is involved in a range of diverse cellular functions such as energy production, acid stress resistance, and pyrimidine biosynthesis could explain the presence of such a complex regulatory circuit. Interestingly, O'Connell-Motherway et al. (36) have reported on an essential two-component system that is involved in activation of arginine degradation. Whether and how this system is responsible for some of the effects described above remain to be elucidated.
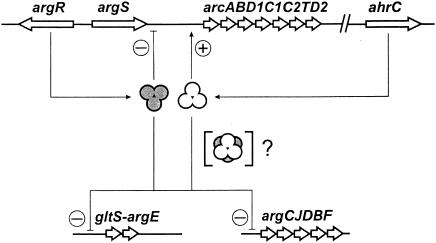
Whereas the N termini of ArgRLl and AhrCLl are highly similar, greater divergence is seen between the C-terminal domains, in particular between those of AhrCLl and the other regulators aligned in Fig. 5. The lack of conservation is especially intriguing for those residues with known functions in the B. stearothermophilus, B. subtilis, and E. coli regulators (7, 21, 35, 48): whereas, e.g., ArgRLl lacks one of the C-terminal Asp residues directly involved in arginine binding (34), AhrCLl harbors an extra Asp residue at the equivalent location (Fig. 5). The fact that both regulators are essential for regulation and that the missing conserved arginine-binding Asp residue of ArgRLl seems to be complemented in AhrCLl has led us to postulate a working hypothesis in which both proteins are thought to interact to form heterohexameric complexes, consisting of one ArgRLl trimer interacting with one AhrCLl trimer (Fig. 6).
FIG. 6.
Working model of the possible regulatory mechanism exerted by ArgR and AhrC of L. lactis. Circled plus and minus signs at promoter regions indicate positive and negative regulation, respectively. For details, see the text.
The presence of two arginine regulator homologues in Enterococcus faecalis has recently been described (2). Only a single Asp residue, as is the case for ArgRLl, is present in the putative arginine-binding region of both E. faecalis homologues, leading to the suggestion that these regulators may bind metabolites other than arginine. However, the functionality of the E. faecalis gene products remains to be investigated.
A gene regulatory mechanism of the type that we have described in this paper is, to our knowledge, unprecedented in prokaryotes and is the focus of ongoing research.
Acknowledgments
We are grateful to Peter Ravn, Biotechnological Institute, Hørsholm, Denmark, for providing the L. lactis MG1614 TnNuc integration library used in this study.
REFERENCES
- 1.Abdelal, A. T. 1979. Arginine catabolism by microorganisms. Annu. Rev. Microbiol. 33:139-168. [DOI] [PubMed] [Google Scholar]
- 2.Barcelona-Andres, B., A. Marina, and V. Rubio. 2002. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 184:6289-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky, B. R. 2002. Biosynthesis of amino acids of the glutamate and aspartate families, alanine and polyamines, p. 203-231. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 4.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 5.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bringel, F., L. Frey, S. Boivin, and J. C. Hubert. 1997. Arginine biosynthesis and regulation in Lactobacillus plantarum: the carA gene and the argCJBDF cluster are divergently transcribed. J. Bacteriol. 179:2697-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke, M., A. F. Merican, and D. J. Sherratt. 1994. Mutant Escherichia coli arginine repressor proteins that fail to bind l-arginine, yet retain the ability to bind their normal DNA-binding sites. Mol. Microbiol. 13:609-618. [DOI] [PubMed] [Google Scholar]
- 8.Chopin, A. 1993. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol. Rev. 12:21-37. [DOI] [PubMed] [Google Scholar]
- 9.Crow, V. L., and T. D. Thomas. 1982. Arginine metabolism in lactic streptococci. J. Bacteriol. 150:1024-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunin, R., T. Eckhardt, J. Piette, A. Boyen, A. Pierard, and N. Glansdorff. 1983. Molecular basis for modulated regulation of gene expression in the arginine regulon of Escherichia coli K-12. Nucleic Acids Res. 11:5007-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czaplewski, L. G., A. K. North, M. C. Smith, S. Baumberg, and P. G. Stockley. 1992. Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. Mol. Microbiol. 6:267-275. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardan, R., G. Rapoport, and M. Debarbouille. 1995. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J. Mol. Biol. 249:843-856. [DOI] [PubMed] [Google Scholar]
- 15.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guèdon, E., E. Jamet, and P. Renault. 2002. Gene regulation in Lactococcus lactis: the gap between predicted and characterized regulators. Antonie Leeuwenhoek 82:93-112. [PubMed] [Google Scholar]
- 17.Hodgman, T. C., H. Griffiths, and D. K. Summers. 1998. Nucleoprotein architecture and ColE1 dimer resolution: a hypothesis. Mol. Microbiol. 29:545-558. [DOI] [PubMed] [Google Scholar]
- 18.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 19.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen, E., and A. Kibenich. 1992. Isolation and characterization of IS1165, an insertion sequence of Leuconostoc mesenteroides subsp. cremoris and other lactic acid bacteria. Plasmid 27:200-206. [DOI] [PubMed] [Google Scholar]
- 21.Karaivanova, I. M., P. Weigel, M. Takahashi, C. Fort, A. Versavaud, G. Van Duyne, D. Charlier, J. N. Hallet, N. Glansdorff, and V. Sakanyan. 1999. Mutational analysis of the thermostable arginine repressor from Bacillus stearothermophilus: dissecting residues involved in DNA binding properties. J. Mol. Biol. 291:843-855. [DOI] [PubMed] [Google Scholar]
- 22.Kunji, E. R., I. Mierau, B. Poolman, W. N. Konings, G. Venema, and J. Kok. 1996. Fate of peptides in peptidase mutants of Lactococcus lactis. Mol. Microbiol. 21:123-131. [DOI] [PubMed] [Google Scholar]
- 23a.Leenhouts, K., G. Buist, A. Bolhuis, A. ten Berge, J. Kiel, I. Mierau, M. Dabrowska, G. Venema, and J. Kok. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217-224. [DOI] [PubMed]
- 23.Kunji, E. R., E. J. Smid, R. Plapp, B. Poolman, and W. N. Konings. 1993. Di-tripeptides and oligopeptides are taken up via distinct transport mechanisms in Lactococcus lactis. J. Bacteriol. 175:2052-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, H., V. Rhodius, C. Gross, and E. D. Siggia. 2002. Identification of the binding sites of regulatory proteins in bacterial genomes. Proc. Natl. Acad. Sci. USA 99:11772-11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim, D. B., J. D. Oppenheim, T. Eckhardt, and W. K. Maas. 1987. Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc. Natl. Acad. Sci. USA 84:6697-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maghnouj, A., A. A. Abu-Bakr, S. Baumberg, V. Stalon, and W. C. Vander. 2000. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol. Lett. 191:227-234. [DOI] [PubMed] [Google Scholar]
- 28.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova, K. S., A. A. Mironov, and M. S. Gelfand. 2001. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2:RESEARCH0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mierau, I., A. J. Haandrikman, O. Velterop, P. S. Tan, K. L. Leenhouts, W. N. Konings, G. Venema, and J. Kok. 1994. Tripeptidase gene (pepT) of Lactococcus lactis: molecular cloning and nucleotide sequencing of pepT and construction of a chromosomal deletion mutant. J. Bacteriol. 176:2854-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, C. M., S. Baumberg, and P. G. Stockley. 1997. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol. Microbiol. 26:37-48. [DOI] [PubMed] [Google Scholar]
- 33.Mironov, A. A., E. V. Koonin, M. A. Roytberg, and M. S. Gelfand. 1999. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 27:2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni, J., V. Sakanyan, D. Charlier, N. Glansdorff, and G. D. Van Duyne. 1999. Structure of the arginine repressor from Bacillus stearothermophilus. Nat. Struct. Biol. 6:427-432. [DOI] [PubMed] [Google Scholar]
- 35.Niersbach, H., R. Lin, G. D. Van Duyne, and W. K. Maas. 1998. A superrepressor mutant of the arginine repressor with a correctly predicted alteration of ligand binding specificity. J. Mol. Biol. 279:753-760. [DOI] [PubMed] [Google Scholar]
- 36.O'Connell-Motherway, M., D. van Sinderen, F. Morel-Deville, G. F. Fitzgerald, S. D. Ehrlich, and P. Morel. 2000. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935-947. [DOI] [PubMed] [Google Scholar]
- 37.Poolman, B., A. J. Driessen, and W. N. Konings. 1987. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J. Bacteriol. 169:5597-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravn, P., J. Arnau, S. M. Madsen, A. Vrang, and H. Israelsen. 2000. The development of TnNuc and its use for the isolation of novel secretion signals in Lactococcus lactis. Gene 242:347-356. [DOI] [PubMed] [Google Scholar]
- 39.Renault, P., J. J. Godon, N. Goupil, C. Delorme, G. Corthier, and S. D. Ehrlich. 1995. Metabolic operons in lactococci. Dev. Biol. Stand. 85:431-441. [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40a.Sanders, J. W., G. Venema, J. Kok, and K. Lennhouts. 1998. Identification of a sodium chloride-regulated promoter in Lactococcus lactis by single-copy chromosomal fusion with a reporter gene. Mol. Gen. Genet. 257:681-685. [DOI] [PubMed]
- 41.Savchenko, A., P. Weigel, D. Dimova, M. Lecocq, and V. Sakanyan. 1998. The Bacillus stearothermophilus argCJBD operon harbours a strong promoter as evaluated in Escherichia coli cells. Gene 212:167-177. [DOI] [PubMed] [Google Scholar]
- 41a.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 42.Soutar, A., and S. Baumberg. 1996. Implication of a repression system, homologous to those of other bacteria, in the control of arginine biosynthesis genes in Streptomyces coelicolor. Mol. Gen. Genet. 251:245-251. [DOI] [PubMed] [Google Scholar]
- 43.Stirling, C. J., G. Szatmari, G. Stewart, M. C. Smith, and D. J. Sherratt. 1988. The arginine repressor is essential for plasmid-stabilizing site-specific recombination at the ColE1 cer locus. EMBO J. 7:4389-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunnerhagen, M., M. Nilges, G. Otting, and J. Carey. 1997. Solution structure of the DNA-binding domain and model for the complex of multifunctional hexameric arginine repressor with DNA. Nat. Struct. Biol. 4:819-826. [DOI] [PubMed] [Google Scholar]
- 45.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gilson. 2003. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4674-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian, G., D. Lim, J. D. Oppenheim, and W. K. Maas. 1994. Explanation for different types of regulation of arginine biosynthesis in Escherichia coli B and Escherichia coli K12 caused by a difference between their arginine repressors. J. Mol. Biol. 235:221-230. [DOI] [PubMed] [Google Scholar]
- 48.Tian, G., and W. K. Maas. 1994. Mutational analysis of the arginine repressor of Escherichia coli. Mol. Microbiol. 13:599-608. [DOI] [PubMed] [Google Scholar]
- 49.Van Duyne, G. D., G. Ghosh, W. K. Maas, and P. B. Sigler. 1996. Structure of the oligomerization and l-arginine binding domain of the arginine repressor of Escherichia coli. J. Mol. Biol. 256:377-391. [DOI] [PubMed] [Google Scholar]
- 50.Winteler, H. V., and D. Haas. 1996. The homologous regulators ANR of Pseudomonas aeruginosa and FNR of Escherichia coli have overlapping but distinct specificities for anaerobically inducible promoters. Microbiology 142:685-693. [DOI] [PubMed] [Google Scholar]
- 51.Yanofsky, C. 1981. Attenuation in the control of expression of bacterial operons. Nature 289:751-758. [DOI] [PubMed] [Google Scholar]
- 52.Zuniga, M., M. Champomier-Verges, M. Zagorec, and G. Perez-Martinez. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]