Abstract
When Bacillus subtilis is subjected to phosphate starvation, the Pho regulon is activated by the PhoP-PhoR two-component signal transduction system to elicit specific responses to this nutrient limitation. The response regulator, PhoP, and its cognate histidine sensor kinase, PhoR, are encoded by the phoPR operon that is transcribed as a 2.7-kb bicistronic mRNA. The phoPR operon is transcribed from two σA-dependent promoters, P1 and P2. Under conditions where the Pho regulon was not induced (i.e., phosphate-replete conditions or phoR-null mutant), a low level of phoPR transcription was detected only from promoter P1. During phosphate starvation-induced transition from exponential to stationary phase, the expression of the phoPR operon was up-regulated in a phosphorylated PhoP (PhoP∼P)-dependent manner; in addition to P1, the P2 promoter becomes active. In vitro gel shift assays and DNase I footprinting experiments showed that both PhoP and PhoP∼P could bind to the control region of the phoPR operon. The data indicate that while low-level constitutive expression of phoPR is required under phosphate-replete conditions for signal perception and transduction, autoinduction is required to provide sufficient PhoP∼P to induce other members of the Pho regulon. The extent to which promoters P1 and P2 are activated appears to be influenced by the presence of other sigma factors, possibly the result of sigma factor competition. For example, phoPR is hyperinduced in a sigB mutant and, later in stationary phase, in sigH, sigF, and sigE mutants. The data point to a complex regulatory network in which other stress responses and post-exponential-phase processes influence the expression of phoPR and, thereby, the magnitude of the Pho regulon response.
Bacillus subtilis responds to phosphate starvation by inducing or repressing genes of the phosphate stimulon, comprising: (i) the phosphate starvation-specific Pho regulon, (ii) the σB-dependent general stress (σB-GS) regulon, and (iii) PhoP-PhoR/σB-independent phosphate starvation-inducible genes (2, 12, 15, 22). The σB-GS regulon has ∼200 members (29, 34), while the Pho regulon presently has 31 members. Of the latter, five operons (phoPR, phoB-ydhF, pstSCA-pstBA-pstBB, phoD-tatAD, and tuaABCDEFGH) and seven monocistronic genes (glpQ, phoA, tatCD, ykoL, yhaX, yhbH, and yttP) are induced in response to phosphate starvation, and two operons (tagAB and tagDEF) are repressed. The alkaline phosphatases (APases) PhoA and PhoB (4, 16), the phosphodiesterase-APase PhoD (6), and the glycerophosphodiesterase GlpQ (2) generate new sources of inorganic phosphate (Pi) from organic sources, such as deacylated phospholipids, nucleic acids, and teichoic acid (3). PhoD is secreted by the twin-arginine transporter (Tat) pathway, some components of which are encoded by members of the Pho regulon (19). Pst, a high-affinity phosphate ABC transporter (35), facilitates the uptake of Pi at low Pi concentrations. Concomitant repression of the teichoic acid operons (tagAB and tagDEF) and induction of teichuronic acid operon (tuaA to tuaH) conserves phosphate by bringing about the controlled replacement of the phosphate-containing cell wall polymer teichoic acid with the non-phosphate-containing teichuronic acid (21, 24, 28). The functions of five putative Pho regulon genes (ydhF, ykoL, yhaX, yhbH, and yttP) are presently unknown (2, 32, 36).
During phosphate starvation, genes of the Pho regulon are regulated by the PhoP-PhoR two-component signal transduction system (39, 40). The PhoP response regulator is activated by its cognate sensor kinase, PhoR. Phosphorylated PhoP (PhoP∼P) induces the expression of the phoPR operon about threefold from a low constitutive level of expression (17, 30, 32) and is required for the induction or repression of other members of the Pho regulon (15).
Phosphate starvation also induces the σB-mediated general stress response, and the Pho and σB-GS regulons interact to modulate the levels to which each is activated. In the absence of the regulator of one of these regulons, the expression of the other regulon is activated to a higher level (2, 32). For maximal induction of the Pho regulon, the respiration signal transduction system, ResD-ResE, is required (15). If, despite these responses, phosphate starvation persists, a third response regulator, Spo0A, initiates sporulation and terminates the phosphate response by repressing phoPR transcription via AbrB and ResD-ResE (15, 17).
The induction or repression of Pho regulon genes is mediated by the binding of PhoP∼P to Pho box sequences: direct repeats of TT(A/T/C)ACA with a 5 ± 2-bp spacer (7). For efficient binding, four TT(A/T/C)ACA-like sequences with an 11-bp periodicity are required. In the case of genes induced by PhoP∼P, the PhoP-binding sites are located on the coding strand of the promoter region and on the noncoding strand of the promoter regions of PhoP∼P-repressed genes (25).
In the work described here we have used a combination of Northern hybridization, primer extension analyses, gel shift assays, and DNase I footprinting to analyze the transcriptional regulation of the phoPR operon. We compared the binding of PhoP and PhoP∼P to the promoter region of phoPR with that of two other putative members of the Pho regulon, namely ykoL and yhaX. In addition, the transcription of phoPR was studied in phoR, sigB, and abrB mutants as well as in a number of mutants deficient in various stages of sporulation. The data confirm the role of PhoP in the regulation of phoPR and identify two sigma A-like promoters (P1 and P2) with associated Pho boxes. Moreover, the extent to which P1 and P2 are activated appears to be influenced by the presence of other sigma factors, possibly due to competition between sigma factors for binding to core RNA polymerase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
B. subtilis strains and plasmids are listed in Table 1. Strains were grown in Luria Bertani (LB) medium, low-phosphate medium (LPM; 0.42 mM Pi), or high-phosphate medium (5.0 mM Pi) (31). E. coli XL1-Blue (Stratagene Europe, Amsterdam, The Netherlands) was used as the host for plasmid constructions, and E. coli BL21(λD3) (Novagen, Madison, Wis.) was used for the production of PhoP-His6 and PhoR231-His6. When required, the concentrations of antibiotics were the following: for E. coli, 100 μg of ampicillin (Ap) per ml and 25 μg of kanamycin (Km) per ml; for B. subtilis, 6 μg of chloramphenicol per ml, 0.3 μg of erythromycin per ml, 25 μg of lincomycin per ml, 10 μg of Km per ml, and 12.5 μg of tetracycline per ml. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at 1 mM.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Sourcea or reference |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | 1 |
| 168-PR | trpC2 phoRΔBalI::Tcr | 30 |
| ML6 | trpC2 sigBΔHindIII-EcoRV::Cmr | 18 |
| 168-PR-SB | trpC2 phoRΔBalI::TcrsigBΔHindIII-EcoRV::Cmr | ML6→168-PR |
| SWV119 | trpC2 pheA1 abrB::Tcr | 44 |
| 168-AB | trpC2 abrB::Tcr | SWV119→168 |
| SWV215 | trpC2 pheA1 spo0A::Kmr | 44 |
| 168-0A | trpC2 spo0A::Kmr | SWV215→168 |
| BHI | trpC2 pheA1 spo0HΔHindIII-EcoRI::Cmr | 11 |
| 168-SH | trpC2 spo0HΔHindIII-EcoRI::Cmr | BHI→168 |
| 650 | trpC2 ilvB2 leuB16 spoIIAABC::Cmr | J. Errington |
| 168-SF | trpC2 spoIIAABC::Cmr | 650→168 |
| 901 | trpC2 spoIIGA::Kmr | 43 |
| 168-SE | trpC2 spoIIGA::Kmr | 901→168 |
| Plasmids | ||
| pBluescript II KS(+) | Phagemid cloning vector Apr (2.961 kb) | Stratagene |
| pNHP | pBluescript II KS(+) containing a 621-bp insert of phoP Apr (3.558 kb) | This study |
| pNHR | pBluescript II KS(+) containing a 1,530-bp insert of phoR Apr (4.467 kb) | This study |
| pPE | pBluescript II KS(+) containing a 451-bp insert of phoP Apr (3.4 kb) | This study |
| pET2816 | 164-bp XbaI-BlpI fragment of pET16b replaced with the 255-bp XbaI-BlpI fragment of pET28a(+) (Novagen) Apr (5.802 kb) | T. Msadek |
| pET-PhoP | pET2816 containing a 722-bp insert of phoP Apr (6.386 kb) | This study |
| pET-PhoR231 | pET2816 containing a 1,049-bp insert of phoR Apr (6.713 kb) | This study |
Arrows indicate transformation from donor to recipient.
DNA manipulations and general methods.
Plasmid and chromosomal DNA extraction, restriction endonuclease digestion, agarose gel electrophoresis, transformation of E. coli cells, PCR, and bioinformatical analyses were carried out as described previously (30, 33). Enzymes, molecular size markers, and deoxynucleotides were purchased from Roche Diagnostics, Ltd. (Lewes, United Kingdom), and from Amersham Pharmacia Biotech, Ltd. (Little Chalfont, United Kingdom).
Construction of plasmids.
Plasmids pNHP and pNHR (Table 1) were constructed to prepare digoxigenin-labeled RNA probes for phoP and phoR, respectively. Primers NHP-FOR and NHP-REV (Table 2) were used for PCR amplification of a 621-bp fragment of phoP, and primers NHR-FOR and NHR-REV were used to amplify a 1,530-bp fragment of phoR. The PCR fragments were cloned into HindIII- and BamHI-digested pBluescript II KS(+) and were transformed into E. coli XL1-Blue. The resulting plasmids, pNHP and pNHR, were confirmed by sequencing the inserted DNA and adjacent vector sequences.
TABLE 2.
Primersa
| Primer | Sequence (5′→3′) | Position (range) |
|---|---|---|
| NHP-FOR | CGCGAAGCTTGCACAGCATGAACAAGA | 2977568 to 2977552 |
| NHP-REV | CGCGGATCCTGCACATCAACAATTCTC | 2976948 to 2976965 |
| NHR-FOR | CGCGAAGCTTGGAAAGCAGAGGAACAC | 2976737 to 2976721 |
| NHR-REV | CGCGGATCCAATGCTTGACAATCGCTA | 2975208 to 2975225 |
| PE-FOR | CGCGGATCCACAGACTATGAAAGAGCG | 2977844 to 2977827 |
| PE-REV | CCGGAATTCAAGCATCACATCAAGCAC | 2977394 to 2977411 |
| PEPH1 | GATTCTTCATCATCCACAACTA | 2977524 to 2977545 |
| PEPH2 | GTAATGACATCATAGCCTGACC | 2977470 to 2977491 |
| PP-FOR | TCATCATGAACAAGAAAATTTTAGTTGTGGATGATGAAG | 2977557 to 2977528 |
| PP-REV | CTCCTCGAGTTCATTCATTTTTGGCTCCTCCAGTTTATACC | 2976842 to 2976873 |
| R231-FOR | GGTGGTCTCCCATGCAGCGGGATCGGCTGCTGAC | 2976156 to 2976134 |
| R231-REV | CTCCTCGAGGGCGGACTTTTCAGCGGCCCGTTTCAG | 2975110 to 2975136 |
| PhoP-FOR | GAGAGAAAGGCTTGCTTAATAC | 2977766 to 2977745 |
| PhoP-REV | AAATTTTCTTGTTCATGCTGTG | 2977546 to 2977567 |
| YhaX-FOR | TAACGATATTAGGGAGAATGGC | 1055846 to 1055867 |
| YhaX-REV | GAAGCAGCGCTCCATCTATATT | 1056056 to 1056035 |
| YkoL-FOR | TGAAATGCTGGAGACGTTTATG | 1397308 to 1397329 |
| YkoL-REV | TTTTCTAAAGCGGATTTCAATA | 1397520 to 1397499 |
Positions of the primers specific for B. subtilis 168 are with respect to the entire genome as noted in the SubtiList database (http://genolist.pasteur.fr/SubtiList) (20). The 5′ ends of primers included a 9- or a 10-bp linker with a BamHI, BsaI, BspHI, EcoRI, XhoI, or HindIII restriction site (underlined).
Plasmid pPE (Table 1) was constructed to generate a sequencing ladder for primer extension analysis. A 451-bp fragment from the 5′ end of phoP was amplified by using primers PE-FOR and PE-REV (Table 2) and was cloned into BamHI- and EcoRI-digested pBluescript II KS(+). The structure of pPE was confirmed by sequencing the inserted DNA and adjacent vector sequences.
For the production and purification of PhoP-His6 and PhoR231-His6 proteins a 734-bp fragment encoding PhoP and a 1,067-bp fragment encoding the cytoplasmic region of PhoR (from amino acid 231 to the C terminus) were amplified by using primer pairs PP-FOR/PP-REV and R231-FOR/R231-REV, respectively (Table 2). The BspHI-XhoI-digested PCR fragment containing phoP and the BsaI-XhoI-digested PCR fragment containing phoR231 were ligated into NcoI-XhoI-digested pET2816 (Table 1), and the mixtures were used to transform E. coli XL1-Blue. The structures of resulting plasmids pET-PhoP and pET-PhoR231 (Table 1), respectively, were confirmed by sequencing the inserted DNA and adjacent vector sequences. The PhoP and PhoR231 proteins encoded by these plasmids contained His6 tags at their C termini (i.e., PhoP-His6 and PhoR231-His6).
RNA extraction, Northern hybridization, and primer extension.
Total RNA of the B. subtilis strains was extracted with phenol (27). Digoxigenin-labeled RNA probes specific for phoP and phoR were synthesized in vitro with T7 RNA polymerase, using HindIII-linearized pNHP and pNHR (Table 1), respectively, and the DIG Northern Starter kit (Roche Diagnostics GmbH, Mannheim, Germany).
For primer extension analysis, primers PEPH1 and PEPH2 (Table 2), complementary to the region encoding the N terminus of phoP, were 5′ end labeled with [γ-32P]ATP (Amersham Pharmacia Biotech, Ltd.) by using T4 polynucleotide kinase (Promega). Total RNA (4 μg), 32P-labeled primer, and SuperScript II RNase H− reverse transcriptase (Invitrogen Ltd., Paisley, United Kingdom) were used for the primer extension reaction. The primers used for reverse transcription were also used to prime dideoxy sequencing reactions from the corresponding phoP region of plasmid pPE (Table 1) as described previously (33).
Production and purification of PhoP-His6 and PhoR231-His6.
Proteins PhoP-His6 and PhoR231-His6 were produced from E. coli BL21(λD3) carrying pET-PhoP or pET-PhoR231, respectively, as described previously (5).
Phosphorylation of PhoP by PhoR231.
Purified PhoR231 (8 μM) was incubated in phosphorylation buffer (0.1 M Tris-HCl [pH 8], 0.2 M KCl, 4 mM MgCl2, 8 mM CaCl2, 0.5 mM dithiothreitol, 0.1 mM EDTA, 3.7% glycerol) in the presence of PhoP (24 μM) for 10 min at room temperature (RT), followed by incubation with 2.5 μM (40 μCi) [γ-32P]ATP for 20 min at RT. The reaction was stopped by the addition of 1/5 volume of sodium dodecyl sulfate (SDS) blue loading buffer. The samples were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The dried gels were exposed to Hyperfilm ECL X-ray film (Amersham Pharmacia Biotech). The radioactive gel images were scanned with a PhosphorImager (Storm 860; Molecular Dynamics) and were quantified by using Quantity One software (version 4.3; Bio-Rad Laboratories, Hercules, Calif.).
Gel shift assay.
For the preparation of DNA probes, fragments of the phoPR, yhaX, and ykoL promoter regions were amplified with primer pairs PhoP-FOR/PhoP-REV, YhaX-FOR/YhaX-REV, and YkoL-FOR/YkoL-REV. In each case the 5′ ends of the forward (FOR) primers were labeled with [γ-32P]ATP by using T4 polynucleotide kinase (Promega). In the gel shift reactions, 4 μM PhoR231 and 0.1 μg of poly(dI-dC) (Sigma) per μl were incubated with 0, 24, 47, 71, and 95 μM PhoP in the presence or absence of 5 mM ATP for 15 min at RT (23). After addition of the DNA probe (1,000 cpm per μl) the mixture was incubated for a further 30 min. The samples were analyzed on a 6% native polyacrylamide gel by using Tris-glycine-EDTA buffer (38).
DNase I footprinting.
The coding DNA strand was labeled as described for the gel shift assay. The noncoding strand was labeled with [32P] by using reverse PCR primers. In the DNA binding reactions, a solution of 4 μM PhoR231, 0.05 μg of bovine serum albumin per μl, and 0.1 μg of poly(dI-dC) per μl was incubated with 0, 19, 38, 57, and 76 μM PhoP in the presence or absence of 5 mM ATP at RT for 15 min in binding buffer (20 mM sodium phosphate buffer [pH 8], 50 mM NaCl, 2 mM MgCl2, 1 mM dithiothreitol, 10% glycerol). After addition of the DNA probe (500 cpm per μl) the mixture was incubated for a further 30 min. DNase I (0.1 U in 10 mM MgCl2-5 mM CaCl2) was added to the reaction mixture, and digestion was conducted for 1 min. The reactions were stopped with DNase I stop solution (0.4 M Na-acetate, 50 μg of calf thymus DNA [Sigma] per ml, and 2.5 mM EDTA). The samples were analyzed on a 6% polyacrylamide gel containing 7 M urea. A Maxam and Gilbert sequencing ladder (cleavage reactions at purine residues [A+G]) (38) was loaded on the same gel.
Protein assay.
Protein concentration was determined by using the Bio-Rad protein assay kit.
SDS-PAGE.
SDS-PAGE was carried out as described previously (38): a 16% separating gel in Tris-glycine buffer was used for the detection of PhoP-His6 and PhoR231-His6 proteins, with low-molecular-size-range prestained protein standards as size markers (Bio-Rad). The gels were stained with GelCode blue stain reagent (Pierce).
Enzyme and Pi assays.
APase production (32) and Pi concentration (10) were determined as described previously.
RESULTS
Transcription of the phoPR operon.
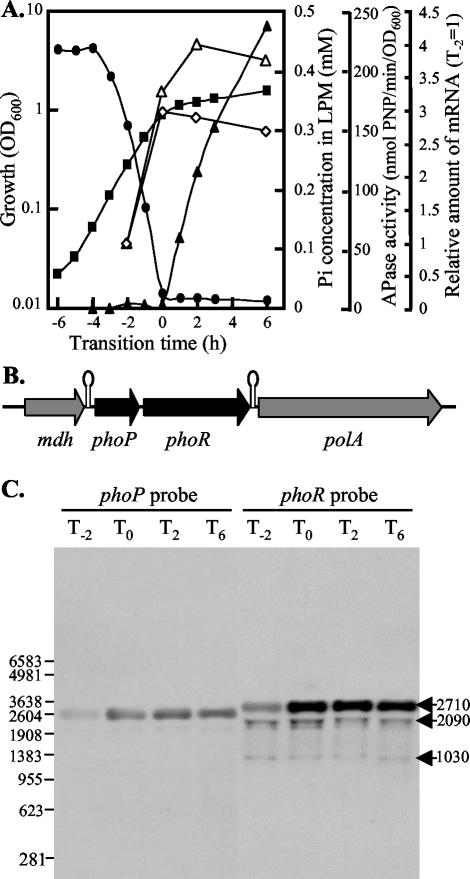
lacZ transcriptional reporter studies indicated that the phoPR operon exhibits constitutive low-level expression during exponential growth (i.e., Pi-replete conditions) and is induced in response to phosphate starvation (32): 4 h after the transition from exponential to stationary phase, the expression of phoPR was threefold higher than that in the exponential phase (17, 30). To confirm these data and to determine the size of phoPR transcript(s), Northern blot analyses were performed on RNA extracted from B. subtilis 168 at various times during growth and phosphate starvation-induced stationary phase (Fig. 1). The phoPR operon is located on a 2,742-bp region of the chromosome (Fig. 1B) between two terminators, Tmdh [bp 2977773 to 2977730 (20); ΔG = −19.8 kcal/mol] and TphoPR [bp 2975014 to 2974989 (20); ΔG = −13.8 kcal/mol]. Using probes specific for phoP and phoR, a single prominent band was detected with an estimated size of 2.7 kb (Fig. 1C). Compared with data for exponential phase, the amounts of this transcript were approximately threefold higher during transition phase and in stationary phase. Minor bands at 2.1 and 1 kb, seen only with the phoR probe, are likely to represent degradation products that are missing regions homologous to the phoP probe. The putative processing sites at the 3′ ends of the 1-kb and the 2.1-kb products coincide with inverted repeat sequences located 132 bp (AAGAAGCAAA and TTCTTCGCTT) and 1,221 bp (TGCTCGATCT and ACGAGCTAAA) downstream of the translation start site of phoR.
FIG. 1.
Transcriptional activity of the phoPR operon. (A) B. subtilis 168 was grown in LPM. (▪), optical density at 600 nm (OD600); (▴), APase activities; (•), concentration of Pi in the medium. Also shown is the relative amount of mRNA as detected with probes specific for phoP (◊) and phoR (Δ). The relative amount of mRNA was calculated by using the intensities of the bands in the Northern hybridization experiments shown in panel C, normalizing the intensities with respect to the samples at T−2. (B) Schematic representation of the phoPR region of B. subtilis. Filled thick arrows indicate structural genes while putative Rho-independent terminators are shown as stem-loop structures. (C) Total RNA, isolated 2 h before (T−2) or at 0 (T0), 2 (T2), and 6 h (T6) after entry into phosphate starvation-induced stationary growth phase, was used for Northern blot experiments with digoxigenin-labeled RNA probes specific for phoP (left-hand side) and phoR (right-hand side). The length (in nucleotide bases) of the molecular size marker is shown on the left-hand side of the images; the sizes (in nucleotide bases) of the three mRNA species of the phoPR operon are on the right-hand side. PNP, p-nitrophenyl.
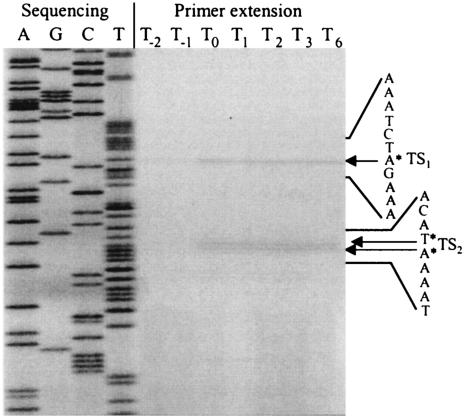
Primer extension analysis identified two major transcriptional start sites for the phoPR operon (Fig. 2). The upstream transcriptional start site (TS1) was mapped to a single nucleotide (A) located 70 bp upstream of the phoP translational start site, while the second transcriptional start site (TS2) mapped to two nucleotides (T and A) 49 and 48 bp upstream of the phoP translational start site. TS1 was preceded by the sequence 5′ TTGTCG-N14-CGCTAAAAT 3′ (P1) (see Fig. 5), which is similar to the consensus sequence for a σA promoter (13). TS2 was preceded by the sequence 5′ TAAAAT-N14-TGTTAAGAT 3′ (P2) (see Fig. 5), which has a −10 region similar to the consensus sequence for a σA promoter. However, the −35 region of P2, which overlaps the −10 region of P1, showed no homology to the σA consensus sequence. Because both promoters were induced in response to phosphate starvation (Fig. 2), the region upstream of the operon was analyzed for the presence of putative PhoP binding sites. Three TT(A/T/C)ACA-like sequences were located from 7 to 33 bp upstream of the −35 region of the P1 promoter, while the P2 promoter was associated with four TT(A/T/C)ACA-like sequences (see Fig. 5). In the latter case the location of the Pho box-like sequences from 28 bp upstream to 3 bp downstream of TS2 is unusual for a PhoP-regulated promoter.
FIG. 2.
Primer extension analysis of phoPR mRNA. Total RNA was isolated from B. subtilis 168 grown in LPM from T−2 to T6 and was used as template for reverse transcriptase. The oligonucleotide primer PEPH1, used for reverse transcription, was also used to prime dideoxy sequencing reactions from the corresponding pPE plasmid (lanes A, G, C, and T). Positions of transcription start sites (TS1 and TS2) are labeled with asterisks and arrows.
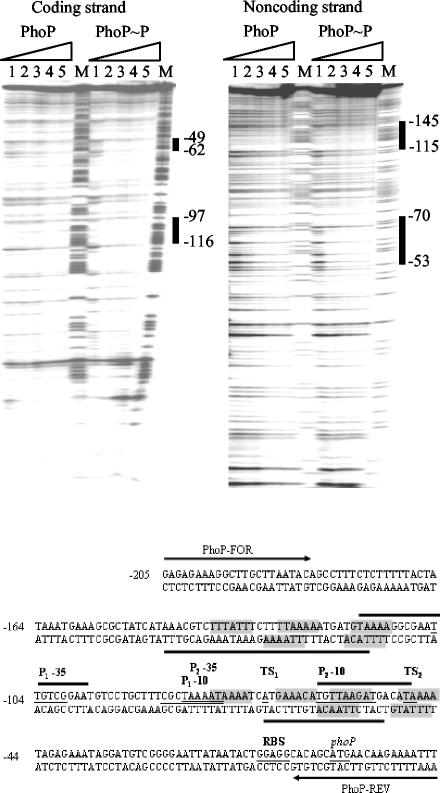
FIG. 5.
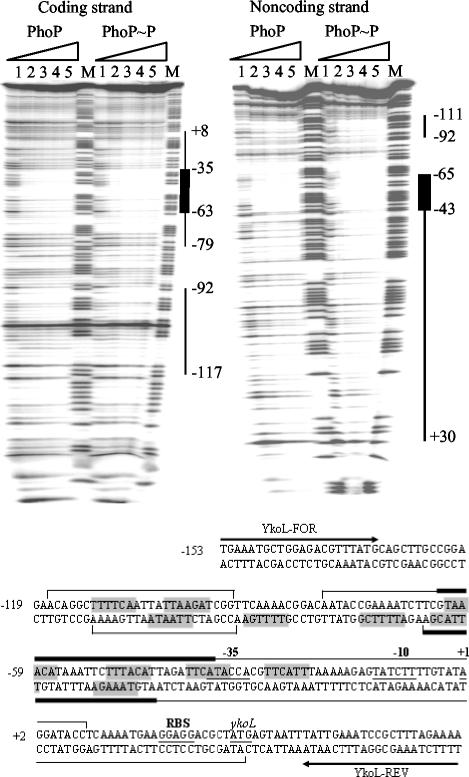
DNase I footprinting assay of the phoPR promoter by using PhoP and PhoP∼P. A PCR fragment corresponding to the phoPR promoter region (−205 to +16 relative to the translational start site of phoP) was used as the DNA probe. Coding strand footprinting 32P-labeled PhoP-FOR primer and the noncoding strand footprinting 32P-labeled PhoP-REV primer were used in the PCR to prepare the DNA probes. Increasing amounts (0, 19, 38, 57, and 76 μM; lanes 1 to 5) of PhoP were incubated with PhoR231 (4 μM) in the presence or absence of ATP and were mixed with the DNA probe. The thick black vertical lines show the regions where PhoP and PhoP∼P bound. The numbers indicate the positions of the PhoP binding sites relative to the translational start site. M is the A+G Maxam and Gilbert sequencing reaction lane used as size markers. In the sequence of the phoPR promoter (lower part of the figure), the translational start site, ribosome binding site (RBS), transcriptional start sites (TS), and corresponding −35 and −10 sequences of the P1 and P2 promoters are underlined and labeled. Grey shading indicates direct repeats of TT(A/T/C)ACA for putative binding of the PhoP dimer (5 ± 2-bp spacer and maximum of two mismatches). The positions of PhoP-FOR and PhoP-REV primers are shown by thick arrows and are labeled.
Binding of PhoP and PhoP∼P to the region of the phoPR promoter.
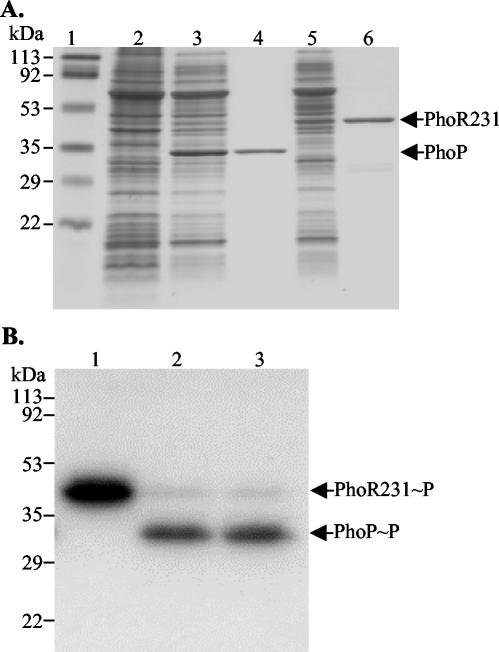
Gel shift assays and DNase I footprinting experiments were used to analyze the binding of PhoP to the promoter region upstream of phoPR. Because preparations of PhoP and PhoR were required for these experiments, PhoP-His6 and PhoR231-His6 variants of these proteins were overexpressed and purified from E. coli BL21(λD3) carrying plasmids pET-PhoP and pET-PhoR231, respectively (Table 1). Both protein preparations exhibited greater than 95% homogeneity, as determined by SDS-PAGE with the GelCode blue stain reagent (Fig. 3A). The functional activities of the purified proteins were determined with an in vitro phosphorylation assay. This confirmed that PhoR231-His6 was autophosphorylated in the presence of [γ-32P]ATP (Fig. 3B, lane 1) and was able to phosphorylate PhoP-His6 (Fig. 3B, lanes 2 and 3).
FIG. 3.
Production and purification of PhoP-His6 and PhoR231-His6 and phosphorylation assay. (A) E. coli BL21(λD3) with pET-PhoP or pET-PhoR231 was grown in LB, and PhoP-His6 and PhoR231-His6 production was induced with IPTG. Purified proteins were analyzed by SDS-PAGE, and the gel was stained with GelCode blue stain reagent. An amount of 1 μg of protein was loaded in lanes 4 and 6, and 10 μg of protein was loaded in lanes 2, 3, and 5. Shown are prestained protein standards (lane 1), supernatant fraction of the whole-cell sonication lysate from the IPTG-induced E. coli BL21(λD3) carrying pET-PhoP (lane 3) or pET-PhoR231 (lane 5), and eluates of PhoP-His6 (lane 4) and PhoR231-His6 (lane 6) by 30 to 300 mM imidazole gradient. Supernatant fraction of the whole-cell sonication lysate from the noninduced E. coli BL21(λD3) carrying pET-PhoP (lane 2) was the negative control. (B) Phosphorylation of PhoP-His6 by PhoR231-His6 in the presence of ATP. PhoR231 (8 μM) was incubated with 40 μCi of [γ-32P]ATP isotope in the absence (lane 1) or the presence (lanes 2 and 3) of 24 μM PhoP for 5 min (lane 1), 10 min (lane 2), and 20 min (lane 3) at RT. The samples were subjected to SDS-PAGE and, after being dried, the gel was exposed to an X-ray film.
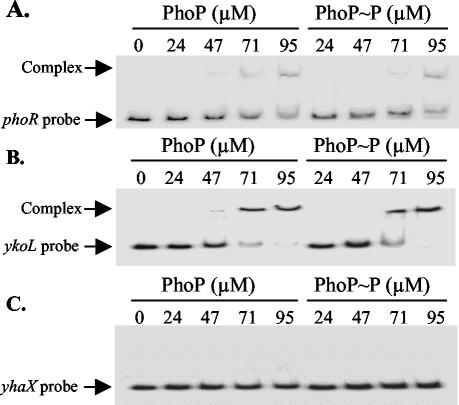
Gel shift assays were performed to determine whether PhoP is able to bind to the promoter region of phoPR (Fig. 4A). PhoP and PhoP∼P decreased the mobility of the 32P-labeled phoPR promoter probe in a concentration-dependent manner. No significant differences were observed in the retardation of the probe when PhoP was phosphorylated, indicating that PhoP and PhoP∼P bind to the phoPR promoter region in vitro with similar efficiencies. Compared to ykoL (Fig. 4B), relatively low amounts of the phoP probe were retarded by PhoP and PhoP∼P, reflecting their relative in vitro levels of expression (∼200 nmol o-nitrophenyl (ONP)/min/optical density unit for ykoL-lacZ and ∼18 nmol ONP/min/optical density unit for phoP-lacZ at T4) and induction ratios (∼250-fold for ykoL and ∼3-fold for phoPR at T4) (32).
FIG. 4.
Gel shift assays of the phoPR, ykoL, and yhaX promoter regions using PhoP and PhoP∼P. PhoR231 (4 μM) was incubated with PhoP (0, 24, 47, 71, and 95 μM) in the absence (left-hand lanes) or presence (right-hand lanes) of ATP at RT for 15 min. 32P-labeled phoPR (A), ykoL (B), and yhaX (C) promoter probes were added and, after binding (30 min), the samples were loaded onto a 6% native polyacrylamide gel to separate free DNA probe from DNA-protein complexes. After being dried the gel was exposed to X-ray film. The amounts of PhoP and PhoP∼P added to the reaction mixtures are indicated above each lane.
DNase I footprinting was performed on both the coding and noncoding strands (Fig. 5) to locate the PhoP binding site on the phoPR promoter region. PhoP and PhoP∼P protected the regions between −49 to −62 bp and −97 to −116 bp on the coding strand and between −53 to −70 bp and −115 to −145 bp on the noncoding strand. These regions correspond to the −10 sequence of promoter P2 and the −35 sequence of promoter P1. In addition, relatively more probe was bound by PhoP∼P than by PhoP.
Binding of PhoP and PhoP∼P to the promoters of ykoL and yhaX.
The binding of PhoP and PhoP∼P to the promoter region of phoPR was compared with that of two other putative members of the Pho regulon, namely ykoL and yhaX (32, 36). Gel shift assays and DNase I footprinting experiments were performed to determine if PhoP binds to the ykoL and yhaX promoter regions. PhoP and PhoP∼P retarded the mobility of the ykoL promoter probe (Fig. 4B) in a concentration-dependent manner and to a greater extent than was observed for the phoPR promoter probe (Fig. 4A). DNase I footprinting of the ykoL promoter region showed that regions protected by PhoP and PhoP∼P were located between +8 to −79 and −92 to −117 on the coding strand and between +30 to −65 and −92 to −111 on the noncoding strand (Fig. 6). Both PhoP and PhoP∼P bound more efficiently to a region located at −35 to −63 on the coding strand and at −43 and −65 on the noncoding strand. In this region of the ykoL promoter, just upstream of the −35 sequence, the level of protection was significantly higher than was observed with the phoPR promoter (Fig. 5). The levels of protection afforded by the phosphorylated and nonphosphorylated forms of PhoP were similar. In contrast, the mobility of the yhaX probe was not influenced by PhoP and PhoP∼P in the gel shift assay (Fig. 4C), and these proteins showed no protective effect in the DNase I footprinting experiments (data not shown), indicating that there are no PhoP binding sites in the region of the yhaX promoter.
FIG. 6.
DNase I footprinting assay of the ykoL promoter using PhoP and PhoP∼P. A PCR fragment corresponding to the ykoL promoter region (−153 to +60 relative to the transcriptional start site of ykoL) was used as a DNA probe. Coding strand footprinting 32P-labeled YkoL-FOR primer and the noncoding strand footprinting 32P-labeled YkoL-REV primer were used in the PCR to prepare the DNA probes. Increasing amounts (0, 19, 38, 57, and 76 μM; lanes 1 to 5) of PhoP were incubated with PhoR231 (4 μM) in the presence or absence of ATP and were mixed with the DNA probe. The black vertical lines show the regions that were bound by PhoP and PhoP∼P. The thick black lines show the regions where PhoP and PhoP∼P bound with a higher affinity. The numbers indicate the positions of the PhoP binding sites relative to the transcriptional start site M is the A+G Maxam and Gilbert sequencing reaction lane used as size markers. In the sequence of the ykoL promoter the translational start site, RBS, transcriptional start site (+1), and corresponding −35 and −10 sequences of the promoter are underlined and labeled. Grey shading indicates direct repeats of TT(A/T/C)ACA for putative binding of PhoP dimer (5 ± 2-bp spacer and a maximum of two mismatches). The positions of YkoL-FOR and YkoL-REV primers are shown by thick arrows and are labeled.
The effect of null mutations in the phoR and sigB genes on the transcription of the phoPR operon.
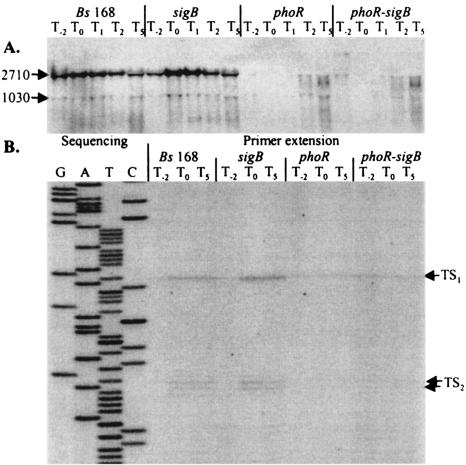
Transcriptional studies with the lacZ reporter gene have shown that the induction of phoPR in response to phosphate starvation is enhanced threefold in a sigB-null mutant (32). In vitro transcription analyses were performed to determine the promoter(s) involved in this phenomenon. B. subtilis strains 168 (wild type), 168-PR (phoR), ML6 (sigB), and 168-PR-SB (phoR/sigB) were grown in LPM, and RNA was extracted throughout the growth cycle for Northern hybridization and primer extension analyses. Using a phoR-specific mRNA probe, Northern hybridization analysis showed that the level of induction of phoPR was twofold higher at T0 and T1 in the absence of SigB (Fig. 7A). Primer extension analysis (Fig. 7B) supported this result and showed that the P1 and P2 promoters were both up-regulated at T0 and T5. The data also confirmed that promoter P1 was primarily responsible for the transcription observed during exponential growth (i.e., Pi-replete conditions) and in the absence of PhoR (i.e., phoR-null and phoR/sigB-null mutants). Evidence from the Northern hybridization experiments that the phoPR promoters were slightly induced at T2 and T5 in the phoR-null and phoR/sigB-null mutants (Fig. 7A) were not confirmed by the primer extension data (Fig. 7B). It is likely that the observed weak transcription was due to the presence in the phoR-null mutant of the tetracycline resistance gene (17).
FIG. 7.
The effect of phoR-null sigB-null mutations on the transcription of the phoPR operon. (A) For Northern hybridization analyses the phoR-specific mRNA probe and total RNA (4 μg), isolated from B. subtilis strains 168, 168-PR, ML6, and 168-PR-SB, were used. Bacteria were grown in LPM, and samples were taken 2 h before (T−2) and at 0 (T0), 1 (T1), 2 (T2), and 5 h (T5) after entry into phosphate starvation-induced stationary growth. The sizes (in nucleotide bases) of the phoPR mRNA species are indicated on the left-hand side. (B) Primer extension analysis of phoPR mRNA. Total RNA, isolated from T−2, T0, and T5, was used as template for reverse transcriptase. The oligonucleotide primer PEPH1 used for reverse transcription was also used to prime dideoxy sequencing reactions from the corresponding pPE plasmid (lanes G, A, T, and C). The positions of transcription start sites (TS1 and TS2) are labeled with arrows.
The effect of mutations in abrB, spo0A, sigH, sigF, and sigE on the transcription of the phoPR operon.
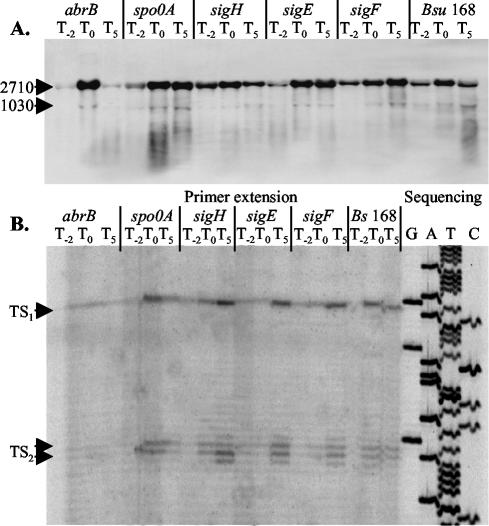
Transcription of phoPR is controlled by a regulatory network that includes, in addition to the PhoP and PhoR proteins, at least two other signal transduction systems, namely ResD-ResE and Spo0A (15). When activation of the Pho regulon fails to overcome the phosphate deficiency, phosphorylated Spo0A (Spo0A∼P) initiates sporulation and indirectly represses phoPR transcription via AbrB and ResD-ResE (17). We therefore studied the effects on phoPR expression of mutations in genes encoding the transition state regulator AbrB and regulators controlling various stages of sporulation (Spo0A, SigH, SigF, and SigE). Northern hybridization analyses revealed that at the phosphate starvation-induced transition stage (T0), phoPR transcription was induced about fourfold in the abrB-null mutant and two- to threefold in the wild type and sporulation-deficient mutants (Fig. 8A). In late stationary phase (T5) the transcription of phoPR decreased slightly in the wild type and decreased markedly in the abrB-null mutant (Fig. 8A). However, the activities of both promoters P1 and P2 were induced in the sporulation-specific mutants (Fig. 8).
FIG. 8.
Influence of mutations in abrB, spo0A, sigH, sigF, and sigE on the transcription of phoPR. (A) For Northern hybridization analyses the phoR-specific mRNA probe and total RNA (4 μg), isolated from B. subtilis strains 168, 168-AB, 168-0A, 168-SH, 168-SE, and 168-SF, were used. Bacteria were grown in LPM, and samples were taken 2 h before (T−2) and at 0 (T0) and 5 h (T5) after entry into phosphate starvation-induced stationary growth. The sizes (in nucleotide bases) of the phoPR mRNA species are indicated on the left-hand side. (B) Primer extension analysis of phoPR mRNA. Total RNA, isolated from T−2, T0, and T5, was used as template for reverse transcriptase. The oligonucleotide primer PEPH1 used for reverse transcription was also used to prime dideoxy sequencing reactions from the corresponding pPE plasmid (lanes G, A, T, and C). The positions of transcription start sites (TS1 and TS2) are labeled with arrows.
DISCUSSION
In B. subtilis the phoPR operon encodes the response regulator and histidine sensor kinase responsible for activating or repressing genes of the Pho regulon in response to phosphate starvation. This bicistronic operon encodes a single major transcript of 2.7 kb (Fig. 1C). Primer extension analysis revealed the presence of two transcriptional start sites, TS1 and TS2 (Fig. 2), which correspond to promoters P1 and P2. The −35 and −10 sequences of P1 promoter and the −10 sequence of P2 promoter were similar to the consensus sequence, TTGACA-N14-TGNTATAAT, for σA promoters of B. subtilis (13). Our data indicate that the P1 and P2 promoters were active in sigB (Fig. 7B), sigH, sigF, and sigE (Fig. 8B) mutants, indicating that they are recognized by σA and not by these alternative sigma factors.
There is evidence that σ factors compete for a limiting pool of core RNA polymerase (9, 14, 26, 37), and data presented here and in previous work (32) are consistent with sigma factor competition affecting the expression of genes in the Pho regulon. For example, the Pho and σB-GS regulons are induced in response to phosphate starvation, and the cognate sigma factors σA and σB appear to compete for the core enzyme, because σA-dependent transcription of phoPR was enhanced twofold in a sigB mutant (Fig. 7A). Later in stationary phase σH, σF, and σE may compete with σA for the core enzyme, because the transcriptional activities of the two σA-dependent promoters of phoPR were significantly higher in the spo0A, sigH, sigF, and sigE mutants (stage 0, I, and II sporulation mutations) at T5 than in the wild type (Fig. 8) and sigB mutants (Fig. 7A). However, we have not been able to provide direct evidence for sigma factor competition, and it is important to recognize that the inactivation of alternative sigma factors is likely to have pleiotropic effects on gene regulation that could also account for the data.
Transcription of the phoPR operon was higher in the spo0A-null mutant than in the wild type (Fig. 8A). This could be due to (i) direct repression by Spo0A∼P at putative 0A-boxes (TGNCGAA) (41) located at the −35 sequences of P1 promoter (98 to 105 bp and 106 to 112 bp upstream of the translational start site of phoP), (ii) competition between Spo0A∼P- and PhoP∼P-activated regulons for EσA holoenzyme, or (iii) indirect repression by Spo0A∼P via both AbrB and ResDE (17, 42).
The P1 and P2 promoters were induced at T0 (Fig. 2), when the phosphate concentration in the medium decreased to below 0.1 mM (Fig. 1A). Gel shift assays and DNase I footprinting showed that both PhoP and PhoP∼P were able to bind in the region of phoPR promoters in vitro (Fig. 4A and 5). However, in vivo only PhoP∼P was able to induce the transcription of P1 and P2 under phosphate starvation, because constitutive low-level transcription from the P1 promoter was observed in the phoR-null mutant (Fig. 7B).
The binding of PhoP and PhoP∼P to the phoPR promoter region was compared to that with the ykoL and yhaX promoter regions. Transcriptional reporter studies (32) showed that the σA-dependent promoter of ykoL (36) was induced 250-fold in response to phosphate starvation, while the σE-dependent promoter of yhaX (8) was induced 21-fold. The binding of PhoP and PhoP∼P to the ykoL promoter was very much more efficient than that to the weakly expressed phoPR promoters (Fig. 4A and B and 5 and 6). It was previously reported that yhaX was induced in response to phosphate starvation in a PhoP- and PhoR-dependent manner (32). However, the present studies failed to demonstrate in vitro binding of PhoP and PhoP∼P in the region of the yhaX promoter (Fig. 4C). This indicates either that yhaX is activated by PhoP indirectly via another regulatory pathway or that binding of PhoP∼P to the yhaX promoter region requires an additional factor(s).
Acknowledgments
We thank J. Errington (University of Oxford) for the gift of strains SWV215, BHI, 650, and 901 and M. A. Strauch for strain SWV119.
This work was funded by the European Commission (QLG2-CT-1999-01455) and the UK Biotechnology and Biological Sciences Research Council (13/PRES/12179).
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure. Synthesis and turn over, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular biology. American Society for Microbiology, Washington, D.C.
- 4.Bookstein, C., C. W. Edwards, N. V. Kapp, and F. M. Hulett. 1990. The Bacillus subtilis 168 alkaline phosphatase III gene: impact of a phoAIII mutation on total alkaline phosphatase synthesis. J. Bacteriol. 172:3730-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat-shock regulatory mechanisms in Staphylococcus aureus and other gram-positive bacteria. Mol. Microbiol. 47:1061-1073. [DOI] [PubMed] [Google Scholar]
- 6.Eder, S., L. Shi, K. Jensen, K. Yamane, and F. M. Hulett. 1996. A Bacillus subtilis secreted phosphodiesterase/alkaline phosphatase is the product of a Pho regulon gene, phoD. Microbiology 142:2041-2047. [DOI] [PubMed] [Google Scholar]
- 7.Eder, S., W. Liu, and F. M. Hulett. 1999. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J. Bacteriol. 181:2017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 9.Farewell, A., K. Kvint, and T. Nyström. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 10.Harwood, C. R., R. D. Coxon, and I. C. Hancock. 1990. The Bacillus cell envelope and secretion, p. 327-390. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 11.Healy, J., J. Weir, I. Smith, and R. Losick. 1991. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor σH in Bacillus subtilis. Mol. Microbiol. 5:477-487. [DOI] [PubMed] [Google Scholar]
- 12.Hecker, M., and U. Völker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the σB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 13.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 14.Hicks, K. A., and A. D. Grossman. 1996. Altering the level and regulation of the major sigma subunit of RNA polymerase affects gene expression and development in Bacillus subtilis. Mol. Microbiol. 20:201-212. [DOI] [PubMed] [Google Scholar]
- 15.Hulett, F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 16.Hulett, F. M., C. Bookstein, and K. Jensen. 1990. Evidence for two structural genes for alkaline phosphatase in Bacillus subtilis. J. Bacteriol. 172:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hulett, F. M., J. Lee, L. Shi, G. Sun, R. Chesnut, E. Sharkova, M. F. Duggan, and N. Kapp. 1994. Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis. J. Bacteriol. 176:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Igo, M., M. Lampe, C. Ray, W. Schafer, C. P. Moran, Jr., and R. Losick. 1987. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J. Bacteriol. 169:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jongbloed, J. D., U. Martin, H. Antelmann, M. Hecker, H. Tjalsma, G. Venema, S. Bron, J. M. van Dijl, and J. Müller. 2000. TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J. Biol. Chem. 275:41350-41357. [DOI] [PubMed] [Google Scholar]
- 20.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 21.Lahooti, M., and C. R. Harwood. 1999. Transcriptional analysis of the Bacillus subtilis teichuronic acid operon. Microbiology 145:3409-3417. [DOI] [PubMed] [Google Scholar]
- 22.Lahooti, M., Z. Prágai, and C. R. Harwood. 2000. Phosphate regulation, p. 237-244. In W. Schumann, S. D. Ehrlich, and N. Ogasawara. (ed.), Functional analysis of bacterial genes: a practical manual. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 23.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, W., and F. M. Hulett. 1998. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology 144:1443-1450. [DOI] [PubMed] [Google Scholar]
- 25.Liu, W., S. Eder, and F. M. Hulett. 1998. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for Pho∼P. J. Bacteriol. 180:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lord, M., D. Barillà, and M. D. Yudkin. 1999. Replacement of vegetative σA by sporulation-specific σF as a component of the RNA polymerase holoenzyme in sporulating Bacillus subtilis. J. Bacteriol. 181:2346-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majumdar, D., Y. J. Avissar, and J. H. Wyche. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA - a new approach for isolating DNA. BioTechniques 11:94-101. [PubMed] [Google Scholar]
- 28.Müller, J. P., Z. An, T. Merad, I. C. Hancock, and C. R. Harwood. 1997. Influence of Bacillus subtilis phoR on cell wall anionic polymers. Microbiology 143:947-956. [DOI] [PubMed] [Google Scholar]
- 29.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prágai, Z., C. Eschevins, S. Bron, and C. R. Harwood. 2001. Bacillus subtilis NhaC, an Na+/H+ antiporter, influences expression of the phoPR operon and production of alkaline phosphatases. J. Bacteriol. 183:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prágai, Z., and C. R. Harwood. 2000. Screening for mutants affected in their response to phosphate, p. 245-249. In W. Schumann, S. D. Ehrlich, and N. Ogasawara. (ed.), Functional analysis of bacterial genes: a practical manual. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 32.Prágai, Z., and C. R. Harwood. 2002. Regulatory interactions between the Pho and σB-dependent general stress regulons of Bacillus subtilis. Microbiology 148:1593-1602. [DOI] [PubMed] [Google Scholar]
- 33.Prágai, Z., H. Tjalsma, A. Bolhuis, J. M. van Dijl, G. Venema, and S. Bron. 1997. The signal peptidase II (lsp) gene of Bacillus subtilis. Microbiology 143:1327-1333. [DOI] [PubMed] [Google Scholar]
- 34.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 35.Qi, Y., Y. Kobayashi, and F. M. Hulett. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in the regulation of the Pho regulon. J. Bacteriol. 179:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robichon, D., M. Arnaud, R. Gardan, Z. Prágai, M. O'Reilly, G. Rapoport, and M. Debarbouille. 2000. Expression of a new operon from Bacillus subtilis, ykzB-ykoL, under the control of the TnrA and PhoP-PhoR global regulators. J. Bacteriol. 182:1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rollenhagen, C., H. Antelmann, J. Kirstein, O. Delumeau, M. Hecker, and M. D. Yudkin. 2003. Binding of σA and σB to core RNA polymerase after environmental stress in Bacillus subtilis. J. Bacteriol. 185:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Seki, T., H. Yoshikawa, H. Takahashi, and H. Saito. 1987. Cloning and nucleotide sequence of phoP, the regulatory gene for alkaline phosphatase and phosphodiesterase in Bacillus subtilis. J. Bacteriol. 169:2913-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seki, T., H. Yoshikawa, H. Takahashi, and H. Saito. 1988. Nucleotide sequence of the Bacillus subtilis phoR gene. J. Bacteriol. 170:5935-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strauch, M., V. Webb, G. Spiegelman, and J. A. Hoch. 1990. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. USA 87:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, G., S. M. Birkey, and F. M. Hulett. 1996. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:941-948. [DOI] [PubMed] [Google Scholar]
- 43.Wu, L. J., and J. Errington. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]
- 44.Xu, K., and M. A. Strauch. 1996. Identification, sequence, and expression of the gene encoding γ-glutamyltranspeptidase in Bacillus subtilis. J. Bacteriol. 178:4319-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]