Abstract
Conjugative transfer and replacement of hundreds of kilobases of a bacterial chromosome can occur in vitro, but replacements in nature are either an order of magnitude smaller or involve the movement of mobile genetic elements. We discovered that two lineages of Staphylococcus aureus, including a pandemic methicillin-resistant lineage, were founded by single chromosomal replacements of at least ∼244 and ∼557 kb representing ∼10 and ∼20% of the chromosome, respectively, without the obvious involvement of mobile genetic elements. The replacements are unprecedented in natural populations of bacteria because of their large size and unique structure and may have a dramatic impact on bacterial evolution.
The parasexual mechanisms of transformation, transduction, and conjugation provide the means by which bacteria exchange genetic material. It is thought that these mechanisms facilitate the replacement of small portions of the chromosome, a process referred to as localized sex (24). Transformation is the uptake of DNA from the environment by competent bacteria and results in small replacements of <10 kb in nature (10). Transduction, the most common form of genetic exchange, involves the packaging of host DNA by phage and can result in replacements of tens of kilobases in vitro (17). Conjugation involves cell-to-cell contact and the movement of host DNA by conjugative plasmids or transposons and can result in the largest replacements of hundreds of kilobases in vitro (15, 17). With both transduction and conjugation, the donor DNA is frequently abridged by endonuclease cutting and exonuclease shortening before incorporation into the recipient chromosome (17).
The size of recombined genetic material in natural populations of bacteria is seldom examined; more is known about the frequency of recombination (9) and the size of mobile and accessory genetic elements (3). For example, studies of Escherichia coli strain MG1655 have revealed that gene content can be accounted for by 67 insertions and deletions ranging in size from <2.5 to >20 kb with a bias toward smaller sizes (18). The largest mobile genetic elements in bacteria include the ∼60-kb staphylococcal cassette chromosome mec (SCCmec) antibiotic resistance island of Staphylococcus aureus (12), the ∼105-kb clc biodegradation island of Pseudomonas sp. strain B13 (21), the ∼190-kb PAI-II pathogenicity island of E. coli (2), and the ∼500-kb symbiosis islands of the rhizobia (25). Mobile genetic elements have a characteristic structure of flanking repeats and a recombinase for excision and integration but ultimately depend on transformation, transduction, and conjugation for movement between bacteria.
It is unknown whether rare large chromosomal replacements are as important in the evolution of a bacterial species as frequent small replacements and the movement of mobile genetic elements. Here, we address this question on the basis of observations of S. aureus, a human pathogen responsible for a significant burden of disease worldwide that has evolved resistance to all antibiotic classes (7). While clones of S. aureus arise more frequently by point mutation than by recombination (8), our findings suggest that the long-term evolution of S. aureus can be influenced by unusually large chromosomal replacements that leave little evidence of their mechanism of exchange.
MATERIALS AND METHODS
Bacterial isolates.
We screened a total of 220 isolates for chromosomal replacements by using partial sequence data from 15 genes, including 7 housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL), 7 surface protein-encoding genes (sasA, sasB, sasD, sasE, sasF, sasH, and sasI), and the gene that encodes immunoglobulin G-binding protein A (spa). One set of 147 isolates representing the major lineages of hospital-acquired methicillin-resistant S. aureus (MRSA) was examined previously (22). Another set of 73 isolates representing lineages of methicillin-sensitive S. aureus and lineages of community-acquired MRSA was examined here. Details of the isolates studied are available on the S. aureus MLST website (saureus.mlst.net). All isolates were stored at −80°C and grown overnight on blood agar plates at 37°C.
PCR and DNA sequencing.
Chromosomal DNA was isolated with the DNeasy kit (Qiagen). PCR and DNA sequencing on both strands were performed as previously described (22), with an ABI3700 automated sequencer (PE Applied Biosystems). The 15 primer sets used to screen the 220 isolates for chromosomal replacements are found in references 6 and 22. The 53 primer sets used to characterize the replacements to a base pair resolution are provided in supplementary Table 1 (http://staff.bath.ac.uk/bsspaw/supptable1.doc).
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in the GenBank database and assigned accession numbers AY442690 to AY442811 (sas alleles) and AY442390 to AY442506 (other alleles).
RESULTS AND DISCUSSION
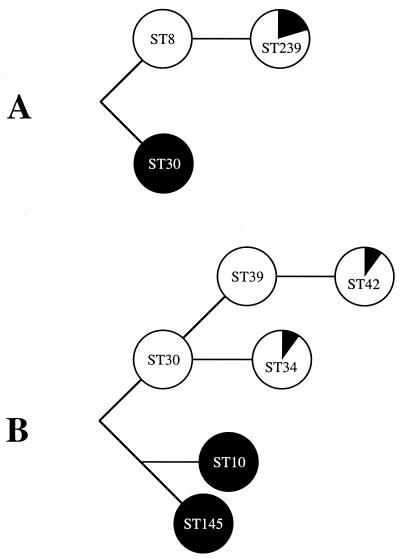
Multilocus sequencing of seven housekeeping genes was used previously to study the evolutionary events resulting in pandemic clones of MRSA (7). To resolve events between closely related clones, we chose to sequence an additional set of eight surface protein-encoding genes that evolve more rapidly than housekeeping genes. Partial sequencing of these 15 genes was applied to 147 isolates representing common hospital-acquired MRSA (22). Multilocus sequence type 8 (ST8) and ST239 were thought to be closely related because they differed at one of seven housekeeping genes (arcC) (7) but were found to differ at four of seven sas genes (sasA, sasD, sasF, and sasH) (22) and had extensively different spa repeat sequences (4, 22). The six genes that distinguished ST8 from ST239 differed at multiple nucleotide sites, and the differing alleles were almost exclusive to ST239 and an unrelated clone named ST30. The hypothesized evolutionary relationships of the involved STs are shown in Fig. 1A. The six genes that distinguished ST239 from ST8 were found to be contiguous and spanned the origin of replication on the strains whose genomes have been sequenced COL (www.tigr.org) and MRSA252 (www.sanger.ac.uk), which are recent descendants of ST8 and ST30, respectively (22) (Fig. 2A). Two hypotheses have been presented to explain these data, including (i) parallel evolution of multiple nucleotide sites within multiple genes located within a specific region of the chromosome and (ii) genetic exchange between unrelated clones (22).
FIG. 1.
Hypothesized evolutionary relationships of STs involved in chromosomal replacements. The approximate contributions of each parent chromosome to the mosaic chromosomes are shown with black and white sectors. (A) The ST239 mosaic has descended from ST8 and ST30 parents. (B) The ST34 mosaic has descended from ST30 and ST10/ST145 parents. The ST42 mosaic has descended from ST39 and ST10/ST145 parents.
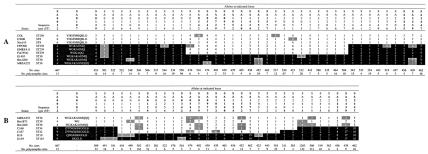
FIG. 2.
Allelic differences showing large chromosomal replacements. Loci are numbered according to the finished genome sequence of strain N315 (13). The 15 loci used to screen 220 isolates for chromosomal replacements are in bold. Parent alleles are shown with black and white backgrounds. A grey background indicates alleles that differ from the parent allele by a single base pair, probably because of a de novo point mutation. One exception involves locus SA0325 allele 3, which differs from the parent allele by 2 bp. Each letter of the spa repeat sequence, SA0107, represents a unique 24-bp sequence named in accordance with Shopsin et al. (23). The symbol ‡ indicates the presence of insertions-deletions. COL and MRSA252 genomic reference sequences are included where appropriate to show their close genetic relationship to the strains that we sequenced. The gene order of the examined loci was conserved in the COL, MRSA252, and N315 genomes. (A) Alleles from 41 loci and nine strains showing the ST239 mosaic. (B) Alleles from 33 loci and seven strains showing the ST34 and ST42 mosaics.
To test the hypothesis of genetic exchange between ST8 and ST30, we sequenced portions of an additional 26 genes from seven strains and determined whether the ST239 sequences were similar to ST8 or ST30. Of the total of 41 genes, 22 genes asymmetrically oriented around the origin of replication were similar for ST239 and ST30 and 19 genes outside of this region were similar for ST239 and ST8 (Fig. 2A). No strains of ST239 were similar to ST8 near the origin of replication, and likewise, no strains of ST239 were similar to ST30 outside of this region. This pattern converged within the sgaT and mmpL genes, which encode putative transport proteins. An ∼1-kb region surrounding the junctions of the pattern was sequenced. The nucleotide polymorphisms clearly showed that the pattern converged within sgaT and mmpL (Fig. 3A). These data were consistent with the genetic exchange hypothesis.
FIG. 3.
Polymorphic nucleotide sites spanning the junctions of the chromosomal replacements. Loci are numbered in accordance with the finished genome sequence of strain N315 (13). Parent nucleotides are shown with black and white backgrounds. A grey background indicates unique nucleotides. (A) Polymorphic sites from 995- and 1,049-bp fragments spanning the left and right junctions, respectively, of the ST239 mosaic. (B) Polymorphic sites from 1,199-, 1,918-, and 973-bp fragments spanning the left and both right junctions, respectively, of the ST34 and ST42 mosaics.
The left junction within sgaT was identical for all of the strains examined (Fig. 3A). However, the right junction within mmpL occurred between sites 85 and 317 for strains EMRSA11 and Fin75541 and between sites 528 and 633 for strain FFP200 (Fig. 3A). These data suggested that a secondary replacement may have occurred at the right junction. Strain FFP200 is a representative of the so-called Brazilian clone, which is a major cause of MRSA disease in Brazil and Portugal (5). As of July 2003, all of the strains of ST239 recorded in the MLST database (www.mlst.net) were methicillin resistant. Resistance arises from carriage of the mobile genetic element called SCCmec. While most strains of ST239 carry SCCmec type III, the Brazilian clone carries variant SCCmec type IIIA (20). It is likely that the Brazilian clone is a derivative of the ST239 lineage and that the slightly different right junction seen with strain FFP200 represents a secondary replacement.
On the basis of the COL and MRSA252 genome sequences, we infer that the ST239 mosaic chromosome has ∼557 kb spanning the origin of replication (oriC) from its ST30 parent and has ∼2,220 kb spanning the terminus of replication (terC) from its ST8 parent. From these data, we cannot determine which parent was the donor and which was the recipient. However, we can infer that the minimal replacement size was ∼557 kb or ∼20% of the chromosome. Neither flanking repeats nor known mobile genetic elements were apparent in the sequences that we obtained, nor were such elements apparent near the junctions in the COL or MRSA252 genome sequences.
On the basis of data from Okuma et al. (19), the average (± standard deviation) doubling times of ST239, ST30, and ST8 are 42.2 ± 6.5, 27.0 ± 0.6, and 28.7 ± 2.3 min, respectively, after removal of an outlier of 61.0 min from ST30. The longer doubling time of ST239 could be the result of mixing large portions of unrelated genomes in which gene regulatory circuits have separately evolved. It is difficult to understand how ST239 could have survived with a doubling time much longer than that of its parents without a counterbalancing selective advantage. However, it is clear that ST239 has thrived to become a pandemic lineage of MRSA represented by numerous clones, including the EMRSA-1, -4, -7, -9, -11, Brazilian, Portuguese, and Vienna clones (5, 16, 26). We note that neither ST8 nor ST30 carries SCCmec type III, which suggests that ST239 acquired its SCCmec element elsewhere. SCCmec type III is larger by ∼10 kb or more than other SCCmec genes and carries resistance to multiple antibiotics, antiseptics, and heavy metals (12). Thus, we speculate that this antibiotic resistance island provides a selective advantage to ST239 in hospitals.
To determine whether such a large chromosomal replacement is common in S. aureus, we partially sequenced the seven housekeeping genes, seven sas genes, and spa from a further 73 isolates representing lineages of methicillin-sensitive S. aureus and community-acquired MRSA. In the total of 220 isolates in which we have sequenced these 15 gene fragments, we observed one additional case in which the putative replacement would span a region of the chromosome similar to that in the case characterized above. This case involved multiple replacements within the same lineage. The hypothesized evolutionary relationships of the STs involved are shown in Fig. 1B. ST30 and ST34 differed at one of seven housekeeping genes (arcC), they differed at three of seven sas genes (sasA, sasF, and sasH), and they differed in spa repeat sequences. The five genes that distinguished ST30 from ST34 differed at multiple nucleotide sites, the differing alleles were shared by a pair of clones named ST10 and ST145, and the genes were contiguous and spanned oriC on the strains whose genomes have been sequenced (Fig. 2B). Likewise, ST39 and ST42 differed at two of seven housekeeping genes (arcC and gmk), they differed at three of seven sas genes (sasA, sasF, and sasH), and they differed in spa repeat sequences. gmk differed by only a single base pair, and this was probably due to a point mutation in ST42. The remaining five genes that distinguished ST39 from ST42 were the same as those that distinguished ST30 from ST34 (Fig. 2B).
To test the hypothesis of genetic exchange between ST30 and ST10/ST145 and between ST39 and ST10/ST145, we sequenced portions of an additional 18 genes from six strains. The results from the total of 33 genes are summarized in Fig. 2B. The left junction for both ST34 and ST42 occurred within a gene that is predicted to encode a cysteine synthase. The right junction for ST34 occurred within cysJ, which encodes a component of a sulfite reductase. The right junction for ST42 occurred ∼12 kb further upstream, between betA and betB, which encode a choline dehydrogenase and a glycine betaine aldehyde dehydrogenase, respectively. The nucleotide polymorphisms from ∼1- to 2-kb sequences clearly defined the junctions (Fig. 3B). These data were consistent with the genetic exchange hypothesis. On the basis of the MRSA252 genome sequence, we infer that the ST34 and ST42 mosaic chromosomes, respectively, have ∼244 and ∼256 kb spanning oriC from their ST10/ST145 parent and have ∼2,659 and ∼2,647 kb spanning terC from their ST30 and ST39 parents. Again, neither flanking repeats nor known mobile genetic elements were apparent near the junctions.
What mechanism of genetic exchange could account for these novel replacements in S. aureus? Conjugation in gram-negative bacteria is a well-characterized process and has been shown to exchange large fragments of the chromosome in vitro (15, 17). Although much is unknown about conjugation in gram-positive bacteria (11), we consider it to be a candidate for the mechanism that leads to the replacements characterized here because of the sizes involved. A role for conjugative plasmids, transposons, and phage is unlikely because they would have been precisely excised from the junctions of the replacements, perhaps on multiple occasions. Moreover, the replacements represent the exchange of essential genetic material, not the insertion or deletion of accessory genetic material that is carried on mobile genetic elements. Although phage-induced transformation (1) and electrotransformation (14) of S. aureus can occur in the laboratory, we know of no reports of natural transformation in this species. Protoplasm fusion driven by cell wall-acting antibiotics is also an unlikely mechanism of genetic exchange because ST34 and ST42 are invariably methicillin sensitive and would be killed by antibiotic treatment.
We note that in both replacements, the left junction was common to all of the strains examined but the right junction varied among strains. This observation suggests that a specificity occurs in the left junction that is not present in the right junction. We also note that since both replacements involved the ST30 lineage, these events may not occur randomly throughout the species and hence it follows that expression of lineage-specific genes may be required for the replacements to occur. As genomic sequencing has been limited to a few strains of any one species, our methodology based on comparative sequencing of well-characterized strains would more efficiently reveal the extent of large replacements in S. aureus and in other bacterial species.
Acknowledgments
We thank Brian Spratt, Ed Feil, Laurence Hurst, and Angus Buckling for comments on the manuscript and Paul Wilkinson for technical assistance.
This work was supported by the Wellcome Trust. M.C.E. is a Royal Society University Research Fellow.
REFERENCES
- 1.Birmingham, V. A., and P. A. Pattee. 1981. Genetic transformation in Staphylococcus aureus: isolation and characterization of a competence-conferring factor from bacteriophage 80 alpha lysates. J. Bacteriol. 148:301-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschape, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 4.de Sousa, M. A., M. I. Crisostomo, I. S. Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2003. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Sousa, M. A., I. S. Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 14.Lee, J. C. 1993. Electrotransformation of staphylococci, p. 209-212. In J. A. Nickloloff (ed.), Methods in molecular biology. Humana Press, Totowa, N.J.
- 15.Lloyd, R. G., and C. Buckman. 1995. Conjugational recombination in Escherichia coli: genetic analysis of recombinant formation in Hfr × F− crosses. Genetics 139:1123-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marples, R. R., and E. M. Cooke. 1985. Workshop on methicillin-resistant Staphylococcus aureus held at the headquarters of the Public Health Laboratory Service on 8 January 1985. J. Hosp. Infect. 6:342-348. [PubMed] [Google Scholar]
- 17.Milkman, R., E. A. Raleigh, M. McKane, D. Cryderman, P. Bilodeau, and K. McWeeny. 1999. Molecular evolution of the Escherichia coli chromosome. V. Recombination patterns among strains of diverse origin. Genetics 153:539-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochman, H., and I. B. Jones. 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 21.Ravatn, R., S. Studer, D. Springael, A. J. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, J. M., C. G. Dowson, and B. G. Spratt. 1991. Localized sex in bacteria. Nature 349:29-31. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witte, W. 1999. Antibiotic resistance in gram-positive bacteria: epidemiological aspects. J. Antimicrob. Chemother. 44(Suppl. A):1-9. [DOI] [PubMed] [Google Scholar]