Abstract
The environmental strain Bacillus amyloliquefaciens FZB42 promotes plant growth and suppresses plant pathogenic organisms present in the rhizosphere. We sampled sequenced the genome of FZB42 and identified 2,947 genes with >50% identity on the amino acid level to the corresponding genes of Bacillus subtilis 168. Six large gene clusters encoding nonribosomal peptide synthetases (NRPS) and polyketide synthases (PKS) occupied 7.5% of the whole genome. Two of the PKS and one of the NRPS encoding gene clusters were unique insertions in the FZB42 genome and are not present in B. subtilis 168. Matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis revealed expression of the antibiotic lipopeptide products surfactin, fengycin, and bacillomycin D. The fengycin (fen) and the surfactin (srf) operons were organized and located as in B. subtilis 168. A large 37.2-kb antibiotic DNA island containing the bmy gene cluster was attributed to the biosynthesis of bacillomycin D. The bmy island was found inserted close to the fen operon. The responsibility of the bmy, fen, and srf gene clusters for the production of the corresponding secondary metabolites was demonstrated by cassette mutagenesis, which led to the loss of the ability to produce these peptides. Although these single mutants still largely retained their ability to control fungal spread, a double mutant lacking both bacillomycin D and fengycin was heavily impaired in its ability to inhibit growth of phytopathogenic fungi, suggesting that both lipopeptides act in a synergistic manner.
The rhizosphere colonizing Bacillus amyloliquefaciens strain FZB42 is distinguished from the related model organism Bacillus subtilis 168 by its ability to stimulate plant growth and to suppress plant pathogenic organisms (12, 14). However, the basis for successful mutualistic colonization of plant rhizosphere by some Bacillus strains is still unknown. We assume that rhizosphere competence and biocontrol function in bacilli are partly caused by nonribosomally produced cyclic lipopeptides acting against phytopathogenic viruses, bacteria, fungi, and nematodes. These lipopeptides are synthesized at modular multienzymatic templates (33) and consist of a β-amino or β-hydroxy fatty acid component that is integrated into a peptide moiety.
Some of these lipopeptides have been studied in greater detail, including surfactin, fengycins, and several iturins. Surfactin is a heptapeptide with an LLDLLDL chiral sequence linked, via a lactone bond, to a β-hydroxy fatty acid with 13 to 15 carbon atoms. Surfactin exerts its antimicrobial and antiviral effect by altering membrane integrity (30). Fengycin and the closely related plipastatin are cyclic lipodecapeptides containing a β-hydroxy fatty acid with a side chain length of 16 to 19 carbon atoms. Four d-amino acids and ornithine (a nonproteinogenic residue) have been identified in the peptide portion of fengycin. It is specifically active against filamentous fungi and inhibits phospholipase A2 (26). Members of the iturin family, such as mycosubtilin, bacillomycin D, and iturin A, contain one β-amino fatty acid and seven α-amino acids. The peptide moiety of the iturin lipopeptides contains a tyrosine in the d-configuration at the second amino acid position and two additional d-amino acids at positions 3 and 6. The members of the iturin family exhibit strong antifungal and hemolytic activities and a limited antibacterial activity (21).
Previous matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) analysis of an environmental B. subtilis strain revealed expression of surfactin-, fengycin-, and iturin-like compounds (36). Parallel production of the lipopeptides iturin and surfactin by B. subtilis RB14 in a sterilized vermiculite-soil system (2) and of viscosinamide, tensin, and amphisin by Pseudomonas fluorescens (25) in bulk soil and the sugar beet rhizosphere were detected and illustrate production of these peptide antibiotics by biocontrol strains in their natural environment. In addition, gene sequences encoding enzymes for mycosubtilin and iturin A biosynthesis have been reported (9, 35). The antifungal iturin lipopeptide bacillomycin D is produced by several Bacillus strains (24, 28), but the corresponding gene sequences were until now still unknown. The model organism B. subtilis 168 contains two genetic loci coding for large multifunctional peptide synthetases (srf and pps [16]) but does not produce any lipopeptide biosurfactant (20) due to a defect in 4′-phosphopantetheine transfer from coenzyme A onto peptidyl carrier proteins caused by a mutation in the sfp gene (23).
Comparison of the whole genome from an environmental strain able to synthesize a wide variety of antibacterial and antifungal metabolites to that of the laboratory model strain should allow identification of additional genetic elements involved in the complex network responsible for rhizosphere competence. We present a preliminary comparison of the B. subtilis 168 genome to sample sequences of the genome of FZB42, primarily focused on biosynthesis of biologically active cyclic lipopeptides. Gene clusters involved in surfactin, bacillomycin D, and fengycin synthesis were identified in the FZB42 genome. We found that bacillomycin D and fengycin act in a synergistic manner, enabling FZB42 to cope with competing organisms within plant rhizosphere.
(The results of this study were presented in part at the Functional Genomics of Gram-Positive Microorganisms Meeting, 12th International Conference on Bacilli, 22 to 27 June, 2003, Baveno, Italy.)
MATERIALS AND METHODS
Strains, plasmids, growth conditions.
The strains and plasmids used in this study are listed in Table 1. Strains were cultivated routinely on Luria broth (LB) medium solidified with 1.5% agar. For biosurfactant production and MALDI-TOF-MS characterization, the bacteria were grown either in Landy medium (18) or sucrose-ammonium citrate medium (ACS) (10). To prepare surface cultures, the strains were grown in petri dishes containing 1.5% Landy agar for 24 h at 37°C and stored at room temperature prior to MALDI-TOF-MS analysis. Fermentation in liquid media was carried out in 500-ml flasks at 30°C and 180 rpm in a New Brunswick shaker (New Brunswick Scientific Co., Edison, N.Y.). The media and buffer used for DNA transformation of Bacillus cells were prepared according to the method of Kunst and Rapoport (15).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Descriptiona | Source and/or reference |
|---|---|---|
| B. amyloliquefaciens | ||
| FZB42 | Producer of surfactin, fengycin, and bacillomycin D | FZB Biotech GmbH Berlin (11) |
| AK1 | ΔbmyA::Emr | This study |
| AK2 | ΔfenA::Cmr | This study |
| AK3 | ΔbmyA::Emr ΔfenA::Cmr | This study |
| CH1 | ΔsrfA::Emr | This study |
| Plasmids | ||
| pMX39 | Apr Emr Tetr | Derivative of pDB101 (3) |
| pDG364 | amyE; Apr Cmr | Integrative vector (6) |
| pGEM-T | Cloning vector; Apr | Laboratory stock |
| pAK1 | pGEM-T carrying a 1.2-kb fragment from bmyA; Apr | This study |
| pAK2 | pGEM-T carrying bmyA::Emr; Apr Emr | This study |
| pAK3 | pGEM-T carrying a 1.3 kb fragment from fenA; Apr | This study |
| pAK4 | pGEM-T carrying fenA::Cmr; Apr Cmr | This study |
| pCH1 | pGEM-T carrying a 2.3-kb fragment from srfA::Emr; Apr | This study |
Apr, ampicillin resistance; Emr, erythromycin resistance; Tetr, tetracycline resistance.
DNA transformation.
Competent cells were prepared according to the method of Kunst and Rapoport (15), with slight modifications, as follows. FZB42 cells were grown in 10 ml of glucose-casein hydrolysate-potassium phosphate buffer (GCHE) medium under vigorous shaking (220 rpm) at 37°C until an optical density at 600 nm of 1.4 was reached. Then, 10 ml of GC medium without casein hydrolysate was added, and the culture was incubated under the same conditions for 1 h. The cells were harvested, and the pellet was resuspended in 1 ml of supernatant containing 0.5% glucose. Subsequently, 100 ng of DNA was added to 0.2 ml of cell suspension and incubated for 20 min. Finally, the cells were cultivated in LB medium with inducing (sublethal) concentrations of the appropriate antibiotics for 90 min before they were plated on selective agar.
Sequencing strategy.
The genome of B. amyloliquefaciens FZB42 was sequenced by using the random shotgun approach. Total genomic DNA was sheared randomly or partially digested with Sau3AI, and DNA fragments 1 to 3 kb in size were cloned into pTZ19R or pCR2.1 TOPO (Invitrogen) to establish a shotgun library. The inserts of the recombinant plasmids were sequenced from both ends by using MegaBACE DNA Sequencing Systems 1000 and 4000 (Amersham Biosciences) and ABI Prism 377 sequencers (Applied Biosystems) with dye terminator chemistry.
Approximately 39,500 sequences were processed with PHRED, assembled into contigs by using the PHRAP assembling tool (11), and edited with GAP4, which is part of the STADEN package software (32). The resulting contigs of B. amyloliquefaciens FZB42 were sorted by using the genome of B. subtilis 168 as a scaffold. PCR-based techniques and primer walking on recombinant plasmids were applied in order to close remaining sequence gaps.
MS analysis.
For the detection of the lipopeptide products from whole cells, B. amyloliquefaciens FZB42 was grown on agar plates with the Landy medium. To record mass spectra, cell material was picked from the agar plate, spotted onto the target, and covered with matrix medium, i.e., a saturated solution of α-cyanocinnamic acid in 40% acetonitrile-0.1% trifluoroacetic acid, air dried, and analyzed by MALDI-TOF-MS as previously described (20). Alternatively, a small sample of the freeze-dried culture filtrate was extracted with 70% acetonitrile-0.1% trifluoroacetic acid. The extract was mixed 1:1 (vol/vol) in a vial with matrix medium. A 1-μl aliquot was spotted onto the target. The samples were air dried prior to MS measurement (36). Postsource decay (PSD) mass spectra were obtained with the same samples. Monoisotopic mass numbers were recorded.
SSH.
Suppression subtractive hybridization (SSH) was performed essentially as described elsewhere (1, 8). B. amyloliquefaciens FZB42 genomic DNA was used as the tester, and B. subtilis 168 was used as the driver. The PCR-Select bacterial genome subtraction kit (Clontech Laboratories, Heidelberg, Germany) was used according to the manufacturer's instructions. Basically, genomic DNAs from the two strains were digested separately by RsaI, yielding fragments of 100 to 1,000 bp. The tester DNA was subdivided into two lots, each of which was ligated with a different adaptor. A large excess of driver DNA was then hybridized to each adaptor-ligated tester lot, resulting mainly in hybridized double-stranded DNA enriched for tester DNA sequences. The two hybridized lots were then mixed together without denaturating, allowing hybridization of tester DNA with different adaptors on each end. The samples were then amplified by PCR primers Ssh1 and Ssh2 in order to enrich for tester-specific sequences. Finally, the subtracted DNAs were cloned into pGEM-T vector and sequenced. The following adaptor-specific oligonucleotide primers were used: Ssh1 (5′-TCGAGCGGCCGCCCGGGCAGGT), Ssh2 (5′-AGCGTGGTCGCGGCCGAGGT), Adaptor 1 (5′- CTAATACGACTCACTATAGGGCTCGAGCGGCGCCCGGGCAGGTGGCCCGTCCA), Adaptor 2 (5′- CACTATAGGGCAGCGTGGTCGCGGCCGAGGTGCCGGCTCCA), Gem 1 (5′-CCCGACGTCGCATGCTCCCG), and Gem 2 (5′-CCCATATGGTCGACCTGCAGGCG).
Construction of mutants deficient in lipopeptide synthesis.
B. amyloliquefaciens FZB42 mutants were generated according to a modified protocol originally developed for B. subtilis 168 (15). The bmyA gene was disrupted by insertion of an erythromycin cassette. In detail, a 1.2-kb fragment was amplified by PCR by using the primers BmyAa (5′-AAAGCGGCTCAAGAAGCGAAACCC) and Bmyab (5′- CGATTCAGCTCATCGACCAGGTAGGC) and cloned into vector pGEM-T, generating pAK1. The latter was digested with AvaI, which cuts in the middle of the PCR fragment. Simultaneously, pMX39 was digested with the same enzyme to obtain the erythromycin cassette (1.5 kb), which was then ligated to pAK1, resulting in pAK2. This was subsequently cut by ApaI, and the linearized plasmid was transformed into the naturally competent B. amyloliquefaciens FZB42, where it was introduced into the genome via double-crossover homologous recombination (7). The disruption of bmyA was demonstrated in the resistant colonies obtained by PCR with appropriate primers BmyAa and BmyaB and by Southern hybridization.
Gene disruption into the fenA gene was achieved by insertion of a chloramphenicol cassette. A PCR product of 1.4 kb, obtained with the primers FenAa (5′-AAGAGATTCAGTAAGTGGCCCATCCAG) and FenAb (5′-CGCCCTTTGGGAAGAGGTGC), was cloned into pGEM-T, resulting in pAK3. The central 100-bp HindIII KpnI fragment was removed. Simultaneously, the chloramphenicol cassette was PCR amplified from plasmid pDG364 by using the primers Cm1KpnI (5′-TGAGGTACCATGTTTGACAGCTTATCATCGGC) and Cm2HindIII (5′-TATGCCAAGCTTTTCTTCAACTAACGGGGCAGG). The cassette was digested by HindIII/KpnI and ligated to pAK3, resulting in pAK4. The latter was linearized by PstI and transformed into FZB42. After selection of chloramphenicol-resistant colonies and confirmation of the desired insertion by PCR and Southern hybridization, the mutant AK2 was selected. To obtain a bacillomycin fengycin double mutant, the linearized plasmid pAK4 was transformed into the ΔbmyA::Emr mutant AK1. The desired mutant genotype was confirmed by PCR with appropriate primers and by Southern hybridization.
The srfAA gene was disrupted by insertion mutagenesis with an erythromycin cassette derived from pMX39 as described above. A 2.3-kb PCR product from the srfAA gene region was amplified with the primers Srfkn-1 (5′-AGCCGTCCTGTCTGACGACG) and Srfkn-2 (5′-TCTGCTGCCATACCGCATAGTC) and inserted into a pGEM-T vector. After digestion with HindIII, the erythromycin cassette was inserted into the 2.3-kb PCR fragment. The ApaI-linearized construct was transformed into competent B. amyloliquefaciens FZB42 cells. Chromosomal DNAs obtained from erythromycin-resistant colonies were proved for correct integration of the gene cassette by PCR with the primers Srfkn-1 and Srfkn-2 and by Southern hybridization.
Nucleotide sequence accession number.
The nucleotide sequences of two contigs containing the surfactin, fengycin, and bacillomycin D operon of FZB42 have been deposited in the EMBL nucleotide sequence database under accession numbers AJ575417 and AJ575642.
RESULTS AND DISCUSSION
Analysis of the sampled genome of B. amyloliquefaciens FZB42.
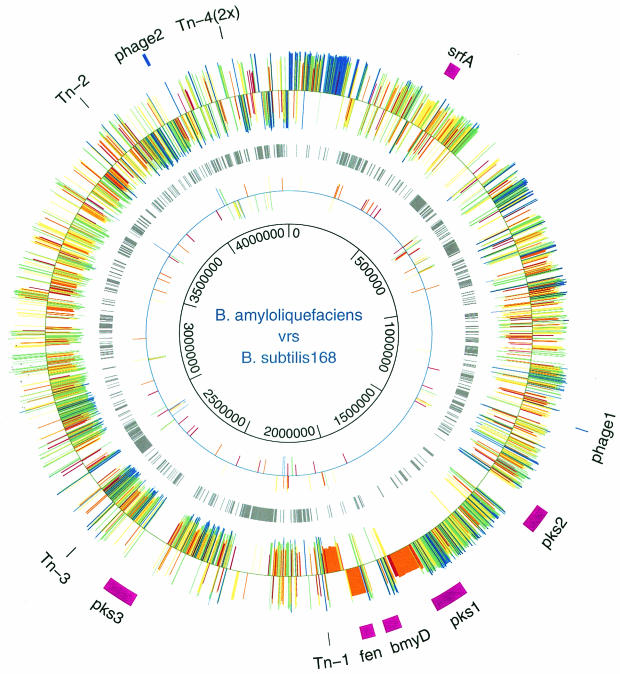
We obtained 411 contigs by assembling the 39,850 sequence reads (5.76 × coverage) from our shot gun approach (see Materials and Methods). The total length of the nonredundant sequence formed by all contigs was 3,818 kb, which is slightly less than the size of the B. subtilis 168 genome of 4,214 kb (16). We identified 2,980 genes (72.7%) on the FZB42 genome encoding proteins with more than 50% amino acid identity to B. subtilis 168. However, 194 of these genes had been rearranged on the chromosome compared to B. subtilis 168 during divergent evolution of both genomes. A weak homology of between 30 and 50% was revealed for additional 174 FZB42 genes. A total of 970 B. subtilis genes, most of them without an assigned function, were not detected in the FZB42 genome sample. Many of the missing genes were found to be substituted for by genes not present in B. subtilis 168. Based on an analysis of FZB42 genomic macrorestriction digest separated by pulsed-field gel electrophoresis (12), we conclude that our genome sample contained at least 90% of the whole FZB42 genome. Given this at least 20% of the B. subtilis genes are missing in B. amyloliquefaciens. The presence of orthologs of all genes involved in the development of competence in B. subtilis 168 in our genome sample indicated a high degree of completeness in our data collection (data not shown). However, in light of the present state of sequencing, it cannot be ruled out that some genome parts that are not easily clonable are still missing. Large parts of the FZB42 genome, ca. 60 to 70%, were colinear with B. subtilis 168. However, regions with high structural similarity are frequently interrupted by regions with a low degree of similarity or FZB42-specific regions (Fig. 1). A preliminary analysis of these variable regions revealed that some of them are due to phage or prophage sequences distributed over the B. amyloliquefaciens genome. A similar result was recently reported for the B. licheniformis genome (19).
FIG. 1.
Colinear scaffold of the B. amyloliquefaciens FZB42 genome over the B. subtilis 168 genome. DNA sequences from FZB42 were compared by TBLASTN with the proteins of B. subtilis 168. The locations of NRPS and PKS gene clusters are indicated by purple bars. Insertions of transposon (red) and phage (blue) sequences into FZB42 are also indicated. The outer circle shows FZB42 genes homologous and colinear with B. subtilis 168 genes. The region adjacent to the bidirectional origin of replication from 0 to 195 kb displayed highest homology to B. subtilis 168. Second circle (gray) shows B. subtilis 168 genes without orthologs in FZB42 (cutoff, <30% amino acid identity). Two hotspots of missing genes are close to the terminator region, which is indicated by a sudden change in the direction of genes and at a region corresponding to 265 to 2.77 Mb of B. subtilis 168. These three areas are mainly occupied by phage-like sequences with unknown function in B. subtilis 168. The third circle shows conserved genes that have been rearranged in FZB42. The color code indicates amino acid identities of >90% (blue), 80% (green), 70% (yellow), 60% (orange), and 50% (red). The inner circle, with coordinates in base pairs, shows the B. subtilis 168 genome, enlarged by the 306-kb sequence containing additional NRPS and PKS operons, identified in the FZB42 genome.
SSH.
A rapid screening for genes without homology to B. subtilis 168 was performed by SSH (8). This method is a powerful tool for the rapid identification of gene differences between closely related bacterial strains (1). Chromosomal DNA from B. amyloliquefaciens FZB42 (tester) and from B. subtilis 168 (driver) were digested to fragments of between 100 and 1,000 bp. The majority of the clones obtained after subtraction did not hybridize with chromosomal DNA of B. subtilis 168 in subsequent Southern blot analyses. A total of 65 clones were selected for sequence analysis. Sequences were validated with the sampled genome of FZB42, and their putative function was deduced by basic local alignment search tool (BLAST) analysis. The results are presented in Table 2. Homology to transposases was found in two clones. Four insertions with IS-like sequences were detected, and the positions of the respective genes on the FZB42 genome corresponded to the B. subtilis 168 genome kilobase positions 2017, 2580, 3910, and 4066. The sequences displayed similarity to an IS231-type transposase from B. thuringiensis (IS sequence between proH and yxoD; P12249), to an IS1627-related transposase from B. anthracis (IS sequences between yqgG and yqgH and between yxeB and yxeA; NC_003980.1 [22]), and to a putative IS3-like transposase recently identified in B. licheniformis (IS sequence inserted in ywcH; AF459921.1 [18]). B. subtilis 168 does not contain transposases, and it is assumed that horizontal gene transfer is mainly achieved by bacteriophages. This is obviously not the case for B. amyloliquefaciens. Three SSH clones harboring phage-like sequences were also detected in our library (Table 2).
TABLE 2.
FZB42 strain-specific SSH clones corresponding to chromosomal genesa
| Clone | Gene size (bp) | Flanking gene(s) (genome position in kb)b | Putative function (homology) |
|---|---|---|---|
| Transposons | |||
| C6 | >5,640 | ywcH, yxeA (3,898, 4,070) | IS3 transposase of B. licheniformis |
| C6 | >1,472 | yxdK, yqgH (2,577) | Two copies of IS1627s1 transposase B. anthracis |
| 50 | >5,303 | yxxD (4,035) | Transposase of Listeria monocytogenes |
| X3 | 5,269 | proH, yoxD (2,017) | IS231 transposase of B. thuringiensis |
| Phages | |||
| C14 | >3,559 | yndL (1,914) | Phage related |
| AK91 | 1,229 | xtmA, xkdX (1,324) | Phage related |
| AM83 | >12,329 | ywdH (3,895) | Phage related |
| Pks/Nrps | |||
| C11 | 37,241 | xynD, yngA (1,943) | bmyD (bacillomycin D biosynthesis) |
| AK25 | 37,241 | xynD, yngA (1,943) | bmyB (bacillomycin D biosynthesis) |
| AK24 | 72,773 | yqjM, proI (2,475) | pks3 (polyketide synthesis) |
| AK250 | 72,773 | yqjM, proI (2,475) | pks3 (polyketide synthesis) |
| AK266 | 72,773 | yqjM, proI (2,475) | pks3 (polyketide synthesis) |
| AK51 | 54,413 | ykyA, pdhA (1,526) | pks2 (polyketide synthesis) |
| AK202 | 54,413 | ykyA, pdhA (1,526) | pks2 (polyketide synthesis) |
| AK46 | 77,808 | ymcC, ymzB (1,781) | pks1 (polyketide synthesis) |
ORF identification was done by BLASTX search within the sampled genome of FZB42.
The sequence was directly identified from the sample genome data.
Sequence analysis revealed that 9 of 65 SSH clones were similar to genes involved in synthesis of cyclic lipopeptides and polyketides. Two of them were assigned to the 37.2-kb gene cluster bmy in the sampled genome, displaying the highest degree of homology to the iturin A operon of B. subtilis RB14 (35). No similar sequences were detected in B. subtilis 168. The other seven clones were assigned to three different gene clusters—pks1, pks2, and pks3—encoding modular type I polyketide synthases (PKS). The B. subtilis 168 genome contains only one pks operon, which displays some similarity to the pks1. In summary, the genome of FZB42 contains three large gene clusters—pks2, pks3, and bmy—involved in polyketide and peptide synthesis, which are not present in the B. subtilis 168 genome.
Overall, the six FZB42 gene clusters involved in nonribosomal peptide and polyketide synthesis—bmy, fen, srf, pks1, pks2, and pks3—span more than 306 kb, representing ca. 7.5% of the total genome (Fig. 1). This accentuates the potential of FZB42 to produce an array of bioactive compounds by processes not based on conventional translation.
MS identification of the lipopeptide products of B. amyloliquefaciens FZB42.
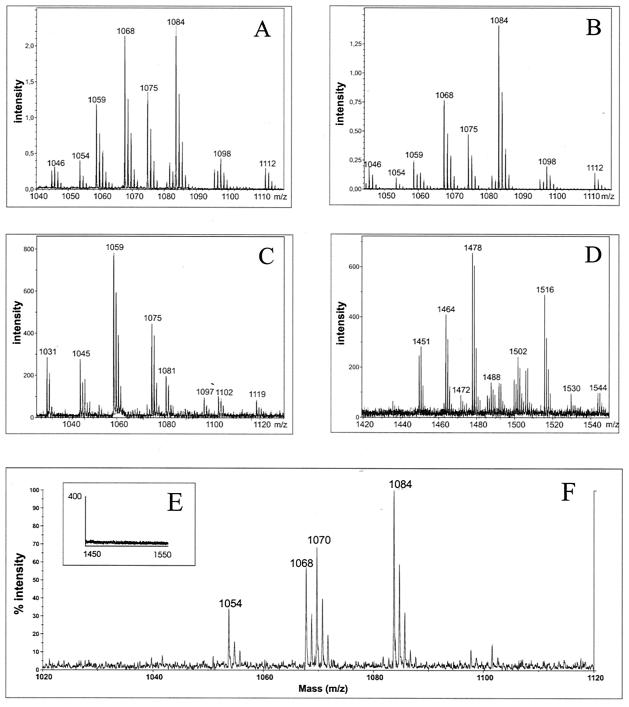
In order to functionally characterize the gene clusters involved in lipopeptide synthesis, the lipopeptide products of B. amyloliquefaciens FZB42 were investigated by MALDI-TOF-MS of culture filtrate extracts and of whole cells of this organism as described previously (20, 36). The spectra obtained by both methods were found identical and three groups of mass peaks were detected (Fig. 2A, B, and D). Their mass numbers are summarized in Table 3. The lipopeptide species of ensembles 1 and 3 have been identified as surfactins and fengycins by comparing their mass data with those previously obtained by MS analysis of the lipopeptide products of numerous B. subtilis strains (36). B. amyloliquefaciens produces C13 to C15 surfactins and fengycins with fatty acid side chains of 15 to 17 carbon atoms. The known Ala/Val dimorphy in position 6 of the fengycin isoforms (36) was confirmed, but we did not observe in FZB42 cultivated in Landy or ACS medium an Ala/Val dimorphy exchange as described for [Ala4]surfactin produced by B. subtilis cells grown in l-Ala-containing medium without supplemented amino acids (29). This pattern of lipopeptides corresponds to the metabolite spectra found for most of the surfactin- and fengycin-producing B. subtilis strains (20, 36).
FIG. 2.
MALDI-TOF-MS analysis of lipopeptides from B. amyloliquefaciens FZB42 and the mutant strains. (A) Detection of surfactin and bacillomycin D mass peaks in extracts prepared from the lyophilysate of the culture filtrate of FZB42 wild-type cells grown in the ACS medium. Panels B to F show mass spectra from intact whole cells grown on agar plates by using Landy medium. (B) Detection of surfactin and bacillomycin D mass peaks in FZB42 wild-type cells; (C) detection of surfactin mass peaks but not of bacillomycin D in mutant AK1 (ΔbmyA::Emr); (D) detection of fengycin mass peaks in FZB42 wild-type cells; (E) AK2 (ΔfenA::Cmr) was deficient in production of fengycin; (F) mutant CH1 (srfA::Emr) was deficient in production of surfactin, but mass peaks indicating bacillomycin D production are still remaining. This sample was analyzed by using a Voyager DE-Pro instrument (Applied Biosystems/Applera Deutschland GmbH, Darmstadt, Germany). For peak identifications, refer to Table 3. The same patterns were detected in samples prepared from culture filtrates from cells growing in Landy and ACS medium (data not shown).
TABLE 3.
Lipopeptide products of B. amyloliquefaciens FZB42 detected by MALDI-TOF-MSa
| Product and observed mass peaks (m/z) | Assignment |
|---|---|
| Surfactin | |
| 1,030.8*, 1,046.8 | C13-surfactin [M + Na, K]+ |
| 1,044.8*, 1,060.8 | C14-surfactin [M + Na, K]+ |
| 1,058.8*, 1,074.8* | C15-surfactin [M + Na, K]+ |
| Bacillomycin D | |
| 1,031.7, 1,053.7*, 1,069.7 | C14-bacillomycin D [M + H, Na, K]+ |
| 1,045.7*, 1,067.7*, 1,083.7 | C15-bacillomycin D [M + H, Na, K]+ |
| 1,059.7, 1,081.7*, 1,097.7*, 1,095.7*, 1,111.7* | C16-bacillomycin D [M + H, Na, K]+ |
| C17-bacillomycin D [M + Na, K]+ | |
| Fengycin | |
| 1,449.9, 1,471.9*, 1,487.9 | Ala-6-C15 fengycin [M + H, Na, K]+ |
| 1,463.9*, 1,485.9*, 1,501.9* | Ala-6-C16 fengycin [M + H, Na, K]+ |
| 1,477.9*, 1,499.9*, 1,515.9* | Ala-6-C17 fengycin [M + H, Na, K]+ |
| 1,491.8, 1,513.9*, 1,529.9* | Val-6-C16 fengycin [M + H, Na, K]+ |
| 1,505.8*, 1,527.8*, 1,543.8* | Val-6-C17 fengycin [M + H, Na, K]+ |
The data are compiled from whole cells grown on LB agar and supernatants from cells grown in Landy medium or ACS medium (see text). Peaks presented in Fig. 2 are indicated by an asterisk.
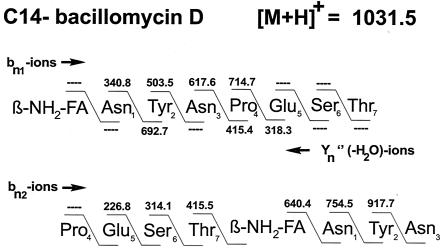
The lipopeptide products of ensemble 2 were identified as bacillomycin D by evaluation of the fragment spectra obtained from PSD-MALDI-TOF-MS (36). In the mass spectra obtained for whole cells and surface extracts the mass peaks of the sodium and potassium adducts dominate, whereas the protonated species always appeared with minor intensities. However, they are preferred for sequence analysis because they decompose into fragments more readily than the alkali adducts. For example, the lipopeptide with a mass number of m/z 1,031.5 produced by B. amyloliquefaciens FZB42 was identified as the protonated form of a bacillomycin D isoform with a fatty chain side chain of 14 carbon atoms. Its sequence (Fig. 3) was determined from series of bn1-, Yn"(−H2O)-, and proline-directed bn2 fragment ions. The peptide ring of this bacillomycin D was cleaved both at the peptide bond between its amino fatty acid residue and threonine at position 7 as well as at the N terminus of proline-5. In the first case series of bn1 and Yn"(−H2O) fragment ions were detected. In addition, bn2 ions of high intensity were observed. Based on all of these data, this lipopeptide was identified as the protonated form of a C14-bacillomycin D. The obtained sequence was corroborated by bn1 ions of dipeptide fragments at m/z values of 171.4, 212.3, 226.8, and 278.4, indicating nearest-neighbor relationships in the peptide ring of this lipopeptide for ES(−H2O), NP, PE, and NY, respectively.
FIG. 3.
In situ structural analysis of the lipopeptide product of B. amyloliquefaciens FZB42 with an m/z of 1,031.5 by PSD-MALDI-TOF-MS of whole cells of B. amyloliquefaciens FZB42. The structure was derived from a series of N- and C-terminal fragments [bn and Yn(−H2O) ions, as well as proline-directed Pn fragments]. FA, fatty acid.
Appearance of lipopeptides during growth of FZB42.
The appearance of lipopeptide species during growth in liquid culture (ACS medium) was followed by MALDI-TOF-MS (Table 4). Since MALDI-TOF-MS is not suitable for determining the exact concentration of the lipopeptide products of B. amyloliquefaciens, mainly because of inhomogeneities in the analytical distribution in the crystalline matrix and different ionization efficiencies of the investigated compounds, the ratio of the different species using intensity values can be estimated. Surfactins and bacillomycins were present at similar intensities but peaked in different stages of growth. Whereas maximum levels of surfactin appeared in samples obtained after 10 to 40 h of growth and dropped after 60 h, bacillomycin D lipopeptide species displayed maximum intensity after 40 and 60 h of culture. The time course of fengycin production resembled that of surfactin, but its intensity level was clearly less than in both other lipopeptides.
TABLE 4.
Lipopeptide production of B. amyloliquefaciens FZB42 grown in ACS mediuma
| Lipopeptide | m/z | Species | Intensity at:
|
|||
|---|---|---|---|---|---|---|
| 10 h | 20 h | 40 h | 60 h | |||
| Surfactin | 1,044.8 | C14 [M + Na]+ | 15,500 | 10,300 | 10,100 | 2,500 |
| 1,058.8 | C15 [M + Na]+ | 16,500 | 11,700 | 10,000 | 2,400 | |
| Bacillomycin C | 1,053.7 | C14 [M + Na]+ | 3,800 | 6,600 | 11,700 | 12,300 |
| 1,067.7 | C15 [M + Na]+ | 2,500 | 2,500 | 7,600 | 7,000 | |
| Fengycin | 1,485.9 | C16 [M + Na]+ | 3,500 | 2,950 | 3,630 | 470 |
| 1,499.9 | C17 [M + Na]+ | 1,800 | 1,810 | 2,510 | 170 | |
MALDI-TOF-MS measurements were made by using samples drawn at various growth times. Only the main mass peaks (m/z) of each lipopeptide were selected for analysis.
Presence and organization of nonribosomal peptide synthetases (NRPS) and PKS gene clusters on the FZB42 chromosome.
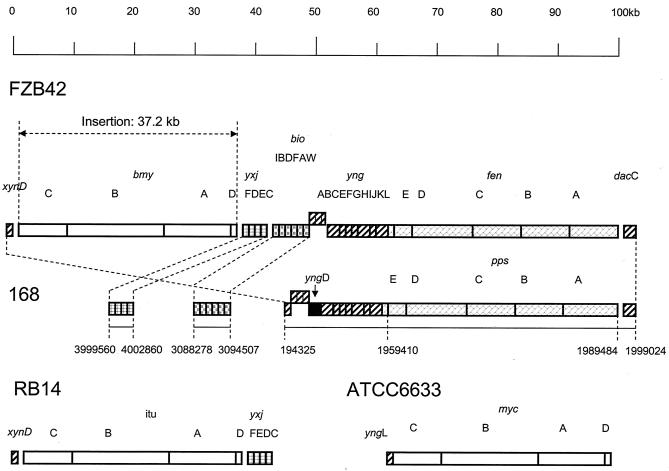
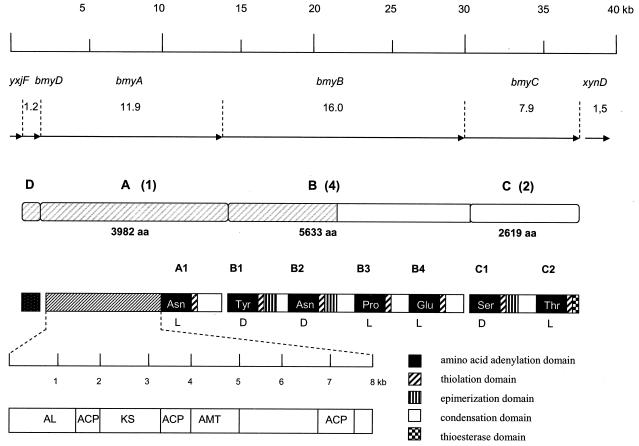
The FZB42 genome contained operons srf, fen, and bmy, which are responsible for the synthesis of the three lipopeptide types surfactin, fengycin, and bacillomycin D, respectively, and three large gene clusters involved in synthesis of hitherto-unidentified polyketides (pks1, pks2, and pks3). The sequences of all six gene clusters are completely available in the FZB42 sample sequence. Three of the six gene clusters (bmy, pks2, and pks3) are FZB42-specific DNA islands. The fengycin and bacillomycin D operons are close to each other on the chromosome (Fig. 4). Regions flanking the large gene clusters are characterized by DNA rearrangements joining the antibiotic DNA islands with sequences originally present in different regions of the Bacillus chromosome. Interestingly, the 37.2-kb bmy gene cluster was inserted, together with two rearranged gene clusters yxjCDEF, located at kilobase position 4000000, and bioIBDFAW, located at kilobase positions 3088278 to 3094507 in B. subtilis 168, at the same position (kilobase position 1943 according to B. subtilis 168) as the iturin A gene cluster in B. subtilis RB14 (Fig. 4). Many DNA rearrangements were detected left from the bmy insertion site in which the DNA regions located in B. subtilis 168 at kilobase positions 1910 to 1943 (yndG, bglC, ynfJ, and xynD) are shuffled with sequences occurring in B. subtilis 168 at kilobase positions 3405 to 3407 (yvrGH), 1303 to 1306 (yjmD, uxuA, and exuT), and 2322 to 2323 (kdgKA), indicating high variability within this area (a 122,883-bp sequence under GenBank accession number AJ575417).
FIG. 4.
ORF organization of the bacillomycin and fengycin operon in B. amyloliquefaciens FZB42 (AJ575417). The intersecting dotted lines indicate events of insertion and rearrangement in FZB42 compared to the respective B. subtilis 168 genome region. The organization and positions of the homologous gene clusters of B. subtilis 168 (fengycin biosyntheses [pps]), B. subtilis RB14 (iturin A biosynthesis [itu]), and B. subtilis ATCC 6633 (mycosubtilin biosynthesis [myc]) were drawn by referring to references 9 and 35.
The fen locus in FZB42 was related to the pps operon in B. subtilis 168 and corresponded to the region from kb 1959 to 1998 kb, which was about 25 kb distant from the bmy gene cluster (Fig. 4). The pps gene cluster encodes a peptide-forming multienzyme system (16). Because of its similarity to the fen gene cluster of the fengycin producer B. subtilis F29-3 (6), the pps operon was assigned to fengycin biosynthesis, although B. subtilis does not produce this lipopeptide. A five-gene cluster (fen1 to fen5) homologous to the fen and pps operons was also detected in the B. subtilis A1/3 genome (34). Interestingly, in the genome of B. subtilis ATCC 6633, the mycosubtilin biosynthesis gene cluster devoted to synthesis of an iturin-like compound, is found in exactly the same location (9) (Fig. 4), suggesting that additional NRPS operons could be integrated in different ways in this area either as an insertion or as a substitution of existing NRPS operons.
The 41,884-bp sequence AJ575642 present in our genome sample contained the srfA operon of FZB42. The 26.5-kb surfactin region is located between kb 376 and 402 of the B. subtilis 168 chromosome and is flanked by sequences partially conserved in both bacilli. The srf genes exhibited between 72% (srfAA) and 83% (srfAC) identity on an amino acid level to the respective B. subtilis 168 genes. As in B. subtilis 168, the comS gene, encoding a competence signal molecule, is embedded within the srfAB sequence. On the right flank of the srfA gene cluster, the B. subtilis 168 ycxAB genes were substituted by two open reading frames (ORFs) with unknown function. The sfp gene, located 4 kb downstream of the srfA operon, is essential for the production of surfactin. Sfp is a 4′-phosphopantetheinyl transferase that functions as a primer of nonribosomal peptide synthesis via phosphopantetheinylation of thiotemplates (23). The amino acid homology of sfp-FZB42 to the B. subtilis 168 sfp gene was 70%.
Disruption of bmyA, fenA, and srfA genes yielded a lipopeptide-deficient phenotype.
To confirm that the bmy, fen, and srf gene cluster is directing bacillomycin D, fengycin, and surfactin biosynthesis, we disrupted the bmyA, fenA, and srfA genes by cassette mutagenesis taking advantage of the natural competence of FZB42 (see Materials and Methods). PCR control by using primers flanking the expected integration sites and Southern hybridization confirmed correct insertion of the antibiotic cassettes within the target gene sequences (data not shown). Analysis of the mutant strains by MALDI-TOF-MS confirmed that strain ΔbmyA::Emr was deficient in bacillomycin D production, that strain ΔfenA::Cmr was deficient in fengycin production, and that the double mutant ΔbmyA::Emr ΔfenA::Cmr failed to produce both lipopeptides. Disruption of the srfA gene in mutant ΔsrfA::Emr lead to inability to produce surfactin (Fig. 2C, E, and F). Based on these results, we concluded that the gene clusters are responsible for biosynthesis of the respective lipopeptides in FZB42.
Analysis of functional domains in the bmy operon.
The assembly of the multifunctional proteins of the peptide synthetases involved in surfactin, fengycin, mycosubtilin, iturin A, or bacillomycin D biosynthesis is reflected in its genetic organization following the colinearity rule (9, 35). The first ORF of the bmy operon, bmyD, encodes a putative malonyl coenzyme A transacylase, similar to FabD, which participates in fatty acid synthesis. BmyD is nearly identical to FenF of B. subtilis ATCC 6633 and B. subtilis RB14. It has been shown that this enzyme is indispensable for iturin production (35). The ORFs encoding BmyA (3,982 amino acids), BmyB (5,633 amino acids), and BmyC (2,619 amino acids) are organized like their respective counterparts in the iturin A and mycosubtilin operons. They showed strong sequence similarity with these components and consist of an ordered arrangement of domains involved in condensation, adenylation, and thiolation (Fig. 5). Seven amino acid-activating modules can be distinguished: A1, located in BmyA; BmyB1, BmyB2, BmyB3, and BmyB4, located in BmyB; and C1 and C2, located in BmyC. The modules B1, B2, and C1 also contain epimerization domains, directing conversion of amino acids 2, 3, and 6 in a d-configuration. The last domain of this multienzyme system is a thioesterase domain, which is presumably required for release and circularization of the synthesized lipopeptide molecule. This structure is identical to the one described for iturin A and mycosubtilin biosynthesis operons (9, 35) in B. subtilis isolates, Therefore, biosynthesis of bacillomycin D also follows the multiple carrier thiotemplate mechanism of nonribosomal synthesis, as first proposed for gramicidin S (17, 33) and meanwhile specified for nonribosomal biosynthesis of many bioactive lipopeptides (4, 13) including mycosubtilin (9). The adenylation domains responsible for activation of the amino acid chosen to be linked with the nascent peptide moiety play an important role in this process. Sequence comparison of bacillomycin D with the other iturins shows that sequence variations begin with amino acid 4, although iturin A and mycosubtilin proteins differ only by a reversion at position 6 and 7 (9). We found that adenylation domains within the first three modules of the bacillomycin D operon showed >97% amino acid identity to the iturin A operon (Table 5). Homology to the respective domains in the mycosubtilin operon was less pronounced but still >70%. However, homologies were less pronounced for the adenylation domains responsible for activation of the amino acids 4 to 7, a finding that corresponds to the variability in the sequence order of the synthesized peptide. The best homology among the last four adenylation domains was obtained between Bmy_C1, the putative Ser_6-activating domain, and the corresponding domain of the mycosubtilin operon, which also activates Ser in position 6. The other adenylation domains possibly involved in activating amino acids Pro-4, Glu-5, and Thr-7, which are unique for bacillomycin D, displayed less homology (Table 5). Prediction of adenylation domain specificity determining residues revealed that Pro-4, Glu-5, Ser-6, and Thr-7 are activated by the adenylation domains in modules B3, B4, C1, and C2, respectively. These domains contained the corresponding selectivity determining amino acids (Table 5).
FIG. 5.
Schematic representation of the bacillomycin D operon of FZB42 comprising the ORFs bmyD (malonyl coenzyme A transacylase), bmyA, bmyB, and bmyC. The deduced domain organizations of the different proteins specified by the operon are indicated. The module numbers are given in parentheses. The region which is highly similar to the iturin operon of RB14 is shaded. AL, acyl coenzyme A ligase domain; ACP, acyl carrier protein domain; KS, β-ketoacyl synthetase domain; AMT, aminotransferase domain.
TABLE 5.
Homologies and selectivity-conferring code of the amino acid-specific adenylation domains (A domains) of the bacillomycin D (bmy) operon compared to the appropriate A domains extracted from the iturin A (itu) and mycosubtilin (myc) gene clusters
| A domain | Amino acidb | % Identityc | Selectivity-conferring amino acida at position:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | |||
| bmy_A1_Asn | Asn (1) | 100 | D | L | T | K | I | G | E | V | G | K |
| itu_A1_Asn | Asn (1) | 98.6 | D | L | T | K | I | G | E | V | G | K |
| myc_A1_Asn | Asn (1) | 80.5 | D | L | T | K | I | G | E | V | G | K |
| BacC, TycCd | Asn | D | L | T | K | I | G | E | V | G | K | |
| bmy_B1_Tyr | Tyr (2) | 100 | D | A | L | S | V | G | E | V | V | K |
| itu_B1_Tyr | Tyr (2) | 99.5 | D | A | L | S | V | G | E | V | V | K |
| myc_B1_Tyr | Tyr (2) | 85.2 | D | A | L | S | V | G | E | V | V | K |
| TycB, TycCa | Tyr | D | A | L | V | T | G | A | V | V | K | |
| bmy_B2_Asn | Asn (3) | 100 | D | L | T | K | I | G | E | V | G | K |
| itu_B2_Asn | Asn (3) | 97.7 | D | L | T | K | I | G | E | V | G | K |
| myc_B2_Asn | Asn (3) | 80.1 | D | L | T | K | I | G | E | V | G | K |
| BacC, TycCd | Asn | D | L | T | K | I | G | E | V | G | K | |
| bmy_B3_Pro | Pro (4) | 100 | D | V | Q | F | I | A | H | V | V | K |
| myc_B4_Pro | Pro (5) | 44.8 | D | V | Q | F | I | A | H | V | V | K |
| itu_B4_Pro | Pro (5) | 42.7 | D | V | Q | F | I | A | H | V | V | K |
| Pps4d | Pro | D | V | Q | F | I | A | H | V | V | K | |
| bmy_B4_Glu | Glu (5) | 100 | D | A | K | D | L | G | V | V | D | K |
| myc_B3_Gln | Gln (4) | 59.8 | D | A | Q | D | L | G | V | V | D | K |
| itu_B3_Gln | Gln (4) | 58.2 | D | A | Q | D | L | G | V | V | D | K |
| SrfAAd | Glu | D | A | K | D | L | G | V | V | D | K | |
| bmy_C1_Ser | Ser (6) | 100 | D | V | W | H | F | S | L | I | D | K |
| myc_C1_Ser | Ser (6) | 81.4 | D | V | W | H | F | S | L | I | D | K |
| itu_A_C2 | Ser (7) | 72.4 | D | V | W | H | F | S | L | I | D | K |
| EntF, Cdald | Ser | D | V | W | H | F | S | L | I | D | K | |
| itu_C1_Asn | Asn (6) | 43.8 | D | L | T | K | I | G | E | V | G | K |
| bmyC2_Thr | Thr (7) | 100 | D | F | W | N | I | G | M | V | H | K |
| FenD, Pps2, PvDd | Thr | D | F | W | N | I | G | M | V | H | K | |
| itu_A_C2 | Ser (7) | 50.2 | D | V | W | H | F | S | L | I | D | K |
| mycC2_Asn | Asn (7) | 47.6 | D | L | T | K | I | G | E | V | G | K |
As determined by Stachelhaus et al. (31). Domains and conserved residues lining the substrate-binding pockets of adenylation domains of assigned functions are indicated in boldface.
The positions of the activated amino acid within the respective lipopeptides are given in parentheses.
That is, the overall homology of the whole adenylation domain, about 440 amino acids, compared to the respective domain of the bmy operon.
Domains and residues lining the substrate-binding pocket are compiled as described in Challis et al. (4).
Biological activity of wild-type and mutant strains.
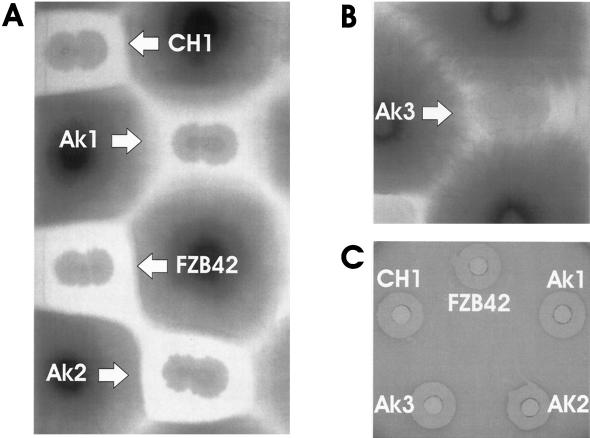
B. amyloliquefaciens FZB42 is able to inhibit growth of phytopathogenic fungi such as Fusarium oxysporum. The mutants deficient in production of bacillomycin D (ΔbmyA) and fengycin (ΔfenA) were affected differently in their biocontrol capacity. Although the bacillomycin D producer strain ΔfenA suppressed growth of F. oxysporum in a manner similar to that of the wild type, strain ΔbmyA was less efficient in fungus growth inhibition, suggesting that bacillomycin D is contributing to the antifungal activity of B. amyloliquefaciens FZB42. In contrast, abolishing surfactin synthesis in the ΔsrfA mutant did not affect the capacity of FZB42 to suppress fungal growth (Fig. 6A). Surprisingly, the ΔbmyA ΔfenA double mutant did not repress F. oxysporum grown on Waksman agar (Fig. 6B), indicating a synergistic action of both antibiotics against the target microorganism, a phenomenon until now only described for secondary metabolites produced by actinomycetes. The effect has been interpreted as an adaptation evolved due to the “sessile” lifestyle of the production organism in order to compete with other microorganisms (5). Due to low concentration of fengycin compared to bacillomycin D (Table 4), the observed synergistic action of both antifungal iturin-like compounds that produced here by a motile soil bacterium is unexpected.
FIG. 6.
Biological activity of supernatants drawn from FZB42 and the mutants impaired in the biosynthesis of surfactin (CH1), bacillomycin D (AK1), fengycin (AK2), or both bacillomycin D and fengycin (AK3). A volume of 2 μl of a 20-h culture of FZB42 or the respective mutant strains grown in Landy medium was dropped onto Waksman agar plates with regularly arranged actively growing F. oxysporum f. sp. cucumerinum DSMZ 62313 cultures. (A and B) The plates were incubated for 3 days at 27°C. (C) Inhibition of S. coelicolor DSMZ 40233 by FZB42 and mutant cultures grown for 20 h in Landy medium. The S. coelicolor indicator strain was mixed with LB soft agar (0.3%) and poured onto LB agar dishes. Supernatants (300 μl) obtained from the respective Bacillus strains were applied and incubated overnight at 37°C.
Here we present evidence for two adjacent gene clusters directing biosynthesis of the synergistically acting but chemically different lipopeptides, fengycin and bacillomycin D in FZB42, suggesting that this phenomenon might also occur in motile bacteria, such as bacilli.
The ability of the ΔbmyA::Emr ΔfenA::Cmr mutant to suppress the growth of F. oxysporum was restored by the addition 10 μg of bacillomycin D/ml, purified from B. amyloliquefaciens DSM 10273 (kindly provided by FZB Biotech, Berlin, Germany [results not shown]). The inhibitory activity of FZB42 against Streptomyces spp. was not impaired in the mutant strains (Fig. 6C), suggesting that antibiotics different from the nonribosomal lipopeptides analyzed in the present study are important for the antibacterial activity of FZB42.
Conclusions.
By combining whole-genome sampling and SSH, we were able to characterize the genetic capacity of rhizobacterium B. amyloliquefaciens FZB42 to deal with competing soil microorganisms. Genetic differences between the model B. subtilis 168, cultivated in the laboratory for decades, and the related environmental B. amyloliquefaciens strain, recently isolated from the rhizosphere (14), were detected by SSH. The results obtained by SSH were subsequently validated by comparison of the B. subtilis genome with the sampled genome of B. amyloliquefaciens FZB42. Both strains share a common genomic scaffold that is interrupted by variable regions due to numerous events of rearrangements, deletions, substitutions, and insertions during the divergent evolution of both genomes. In contrast to B. subtilis 168, the genome of FZB42 contained several transposases belonging to IS structures previously described in other bacilli (19, 22). In addition to the phages, the IS elements present in B. amyloliquefaciens FZB42 may also be involved in events of horizontal gene transfer.
The most striking property of the genome of FZB42 is that a significant part of the genome (∼7.5%, 306 kb, organized in six operons) is devoted to the biosynthesis of polyketides and peptides, enabling this bacterium to cope with competing organisms within the plant rhizosphere. Two gene clusters encoding PKS and one large gene cluster involved in nonribosomal peptide synthesis (bmy) have been identified. The impressive genetic capacity of environmental FZB42 for the production of secondary metabolites, such as lipopeptides and polyketides, exceeds twice that of the laboratory model organisms B. subtilis 168 (37) and Streptomyces coelicolor (4) and has been until now comparable only to Streptomyces avermitilis, which is well known for its production of a wide range of secondary metabolites and in which 6.4% of the entire genome is devoted to the production of secondary metabolites (27).
Strain FZB42 is naturally competent for DNA uptake and homologous recombination ideally suitable for genetic approaches in analyzing its metabolic capacity e.g., by targeted construction of mutants impaired in the synthesis of cyclic lipopeptides. Here we demonstrated that disruption of one of the bmy, fen, and srf genes prevented production of the respective lipopeptides, providing evidence that these gene clusters are involved in their biosynthesis. A double mutant that was unable to produce bacillomycin D and fengycin retained its antibacterial activity directed against Streptomyces spp. but did not develop antifungal activity, suggesting that both lipopeptides might act synergistically in order to intensify suppression of fungal growth. A phenomenon until now only described in actinomycetes. Microcosm experiments with wild-type and mutant strains are necessary to clarify role of the cyclic lipopeptides in biocontrol function within plant rhizosphere.
The present study characterized the production of the three cyclic lipopeptides surfactin, fengycin, and bacillomycin D by B. amyloliquefaciens FZB42. The sequence obtained for the first time for a gene cluster involved in bacillomycin D biosynthesis reflects perfectly colinearity in nonribosomal PKS domain order and its peptide synthesis function. Adenylation domains specifying amino acids different from other iturin-like peptides displayed a high degree of variability, but their functional amino acids lining the substrate binding pocket matched perfectly with the known selectivity code of the respective amino acids compiled for NRPS (4, 31). The description of the putative polyketide products directed by the three gene clusters pks1, pks2, and pks3 awaits further investigation.
Acknowledgments
This study was done within the GenoMik program of the BMBF, the German ministry for education and research.
We thank the Göttingen Genomics Laboratory and especially G. Gottschalk for continuous support of this project. We are very grateful to M. Meixner, B. Krebs, and B. Hoeding from FZB Berlin for advice and support in performing the SSH experiments and biological activity tests. We are indebted to Nicolas Grammel and Ariane Zwintscher of the ActinoDrug GmbH for the intensive cooperation in MALDI-TOF-MS analysis. We also thank Steffen Porwollik, San Diego, Calif., and the unknown referees for improving the manuscript by many corrections and suggestions.
REFERENCES
- 1.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asaka, O., and M. Shoda. 1996. Biocontrol of Rhizoctonia solani damping-off of tomato with Bacillus subtilis RB14. Appl. Environ. Microbiol. 62:4081-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceglowski, P., and J. C. Alonso. 1994. Gene organization of the Streptococcus pyogenes plasmid pDB101: sequence analysis of the ORF eta-copS region. Gene 145:33-39. [DOI] [PubMed]
- 4.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 5.Challis, G. L., and D. A. Hopwood. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14555-14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C.-L., L.-K. Chang, Y.-S. Chang, S.-T. Liu, and J. S.-M. Tschen. 1995. Transposon mutagenesis and cloning of the genes encoding the enzymes of fengycin biosynthesis in Bacillus subtilis. Mol. Gen. Genet. 248:121-125. [DOI] [PubMed] [Google Scholar]
- 7.Cutting, S. M., and P. B. van der Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. Wiley Interscience, Chichester, United Kingdom.
- 8.Diatchenko, L., Y.-F. Ch. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ulrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC 6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebata, M., K. Miyazaki, and Y. Takahashi. 1969. Studies on subsporin. I. Isolation and characterization of subsporins A, B, and C. J. Antibiot. 22:467-472. [PubMed] [Google Scholar]
- 11.Ewing, B., L. Hillier, and P. Green. 1998. Base-calling of automated sequencer traces using phred. 1. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 12.Idriss, E. S. E., O. Makarewicz, A. Farouk, K. Rosner, R. Greiner, H. Bochow, T. Richter, and R. Borriss. 2002. Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology 148:2097-2109. [DOI] [PubMed] [Google Scholar]
- 13.Kallow, W., M. Pavela-Vrancic, R. Dieckmann, and H. von Döhren. 2002. Nonribosomal peptide synthetases: evidence for a second ATP binding site. Biochim. Biophys. Acta 1601:93-99. [DOI] [PubMed] [Google Scholar]
- 14.Krebs, B., B. Höding, S. M. Kübart, A. Workie, H. Junge, G. Schmiedeknecht, P. Grosch, H. Bochow, and M. Heves. 1998. Use of Bacillus subtilis as biocontrol agent. 1. Activities and characterization of Bacillus subtilis strains. J. Plant Dis. Prot. 105:181-197. [Google Scholar]
- 15.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunst, F., N. Ogasawara, I. Moszer, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 17.Laland, S. G., and T. L. Zimmer. 1973. The protein thiotemplate mechanism of synthesis for the peptide antibiotics produced by Bacillus brevis. Essays Biochem. 9:31-57. [PubMed] [Google Scholar]
- 18.Landy, M., G. H. Warren, S. B. Roseman, and L. G. Colio. 1948. Bacillomycin, an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc. Soc. Exp. Biol. Med. 67:539-541. [DOI] [PubMed] [Google Scholar]
- 19.Lapidus, A., N. Galleron, J. T. Andersen, P. L. Jorgensen, S. D. Ehrlich, and A. Sorokin. 2002. Co-linear scaffold of the Bacillus licheniformis and Bacillus subtilis genomes and its use to compare their competence genes. FEMS Microbiol. Lett. 209:23-30. [DOI] [PubMed] [Google Scholar]
- 20.Leenders, F., T. H. Stein, B. Kablitz, P. Franke, and J. Vater. 1999. Rapid typing of Bacillus subtilis strains by their secondary metabolites using matrix-assisted laser desorption/ionisation mass spectrometry of intact cells. Rapid Commun. Mass Spectrom. 13:943-949. [Google Scholar]
- 21.Maget-Dana, R., and F. Peypoux. 1994. Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology 87:151-174. [DOI] [PubMed] [Google Scholar]
- 22.Mahillon, J., R. Rezsohazy, B. Hallet, and J. Delcour. 1994. IS231 and other Bacillus thuringiensis transposable elements: a review. Genetica 93:13-26. [DOI] [PubMed] [Google Scholar]
- 23.Mootz, H. D., R. Finking, and M. Marahiel. 2001. 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 276:37289-37298. [DOI] [PubMed] [Google Scholar]
- 24.Moyne, A.-L., R. Shelby, T. E. Cleveland, and S. Tuzun. 2001. Bacillomycin D: an iturin with antifungal activity against Aspergillus flavus. J. Appl. Microbiol. 90:622-629. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen, T. H., and J. Sorensen. 2003. Production of cyclic lipopeptides by Pseudomonas fluorescens strains in bulk soil and in the sugar beet rhizosphere. Appl. Environ. Microbiol. 69:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishikori, T., H. Naganawa, Y. Muraoka, T. Aoyagi, and H. Umezawa. 1986. Plipastatins; new inhibitors of phospholipase A2, produced by Bacillus cereus BMG302-fF67. III. Structural elucidation of plipastatins. J. Antibiot. 39:755-761. [DOI] [PubMed] [Google Scholar]
- 27.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peypoux, F., M. T. Pommier, B. C. Das, F. Besson, L. Delcambe, and G. Michel. 1984. Structures of bacillomycin D and bacillomycin L peptidolipide antibiotics from Bacillus subtilis. J. Antibiot. 37:1600-1604. [DOI] [PubMed] [Google Scholar]
- 29.Peypoux, F., J. M. Bonmatin, H. Labbe, I. Grangemard, B. C. Das, M. Ptak, J. Wallach, and G. Michel. 1994. [Ala4]surfactin, a novel isoform from Bacillus subtilis studied by mass and NMR spectroscopies. Eur. J. Biochem. 224:89-96. [DOI] [PubMed] [Google Scholar]
- 30.Peypoux, F., J. M. Bonmatin, and J. Wallach. 1999. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51:553-563. [DOI] [PubMed] [Google Scholar]
- 31.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 32.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 33.Stein, T., J. Vater, V. Krufts, A. Otto, B. Wittmann-Liebold, P. Franke, M. Panico, R. McDowell, and H. R. Morriss. 1996. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J. Biol. Chem. 271:15428-15435. [DOI] [PubMed] [Google Scholar]
- 34.Steller, S., D. Vollenbroich, F. Leenders, T. Stein, B. Conrad, J. Hofemeister, P. Jacques, P. Thonaert, and J. Vater. 1999. Structural and functional organization of the fengycin synthetase multienzyme system from Bacillus subtilis b213 and A1/3. Chem. Biol. 6:31-41. [DOI] [PubMed] [Google Scholar]
- 35.Tsuge, T., T. Akiyama, and M. Shoda. 2001. Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 183:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vater, J., B. Kablitz, C. Wilde, P. Franke, N. Mehta, and S. S. Cameotra. 2002. Matrix assisted laser desorption ionization-time of flight mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of Bacillus subtilis C-1 isolated from petroleum sludge. Appl. Environ. Microbiol. 68:6210-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wipat, A., and C. R. Harwood. 1999. The Bacillus subtilis genome sequence: the molecular blueprint of a soil bacterium. FEMS Microbiol. Ecol. 28:1-9. [Google Scholar]