Abstract
Helicobacter pylori causes gastroduodenal disease, which is mediated in part by its outer membrane proteins (OMPs). To identify OMPs of H. pylori strain 26695, we performed a proteomic analysis. A sarcosine-insoluble outer membrane fraction was resolved by two-dimensional electrophoresis with immobilized pH gradient strips. Most of the protein spots, with molecular masses of 10 to 100 kDa, were visible on the gel in the alkaline pI regions (6.0 to 10.0). The proteome of the OMPs was analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Of the 80 protein spots processed, 62 spots were identified; they represented 35 genes, including 16 kinds of OMP. Moreover, we identified 9 immunoreactive proteins by immunoblot analysis. This study contributes to the characterization of the H. pylori strain 26695 proteome and may help to further elucidate the biological function of H. pylori OMPs and the pathogenesis of H. pylori infection.
Helicobacter pylori is a spiral-shaped, microaerophilic gram-negative bacterium that causes acute and chronic gastritis, gastroduodenal ulcers, and gastric cancer (3, 7, 21, 40). More than half of the world's population has suffered from H. pylori infection (4, 5, 46). Surface proteins, including flagella, urease, and adhesin, are known to be involved in the pathogen-host relationship between H. pylori and the human gastric mucosa. A correlation between the motility state of some H. pylori isolates, and their ability to colonize the gastric epithelium has been established in experiments with gnotobiotic piglets (18). Urease enables H. pylori to survive in the acidic environment of the stomach (13) and plays a key role in colonizing the gastric mucosa (17). Adhesins, including BabA (25), AlpA/AlpB (42), HopZ (43), and SabA (26), are known to adhere to gastric epithelial cells.
The genomes of two H. pylori strains have been sequenced (2, 49) and extensively compared (1). Of 64 theoretically predicted outer membrane proteins (OMPs), at least 8, including adhesins and porins, have been confirmed experimentally. However, it is unclear whether all of the predicted OMPs are expressed.
Several methodological approaches have been applied to the identification of H. pylori surface proteins, including OMPs. Sarbarth et al. (48) selectively biotinylated intact H. pylori with the hydrophilic reagent sulfosuccinimidyl-6-(biotinamido)-hexanoate and purified the labeled proteins by membrane isolation, solubilization, and affinity chromatography. Exner et al. (19) purified OMP fractions by sucrose gradient centrifugation and identified heat-mobile OMPs, which may be porins, by using two-dimensional (2-DE) gel electrophoresis. Doig et al. (15) identified six OMPs in a sarcosine-insoluble OMP fraction and by using monoclonal antibodies, demonstrated that these proteins are located within or are associated with the outer membrane. In addition, by comparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of outer membrane fractions isolated by different isolation procedures such as the use of sarcosine, a sucrose gradient ultracentrifuge, Triton X-100, and Triton X-114, eight major protein species with 6 to 10 minor proteins were identified. The outer membrane fraction prepared by sarcosine differential solubilization exhibited a higher level of these proteins than those of the other preparations. Moreover, it was demonstrated previously that the outer membrane fraction was insoluble in sarcosine, whereas the cytoplasmic membrane was totally soluble (20).
2-DE analysis of bacterial OMPs has proven to be impractical because of technical difficulties associated with the solubilization of membrane proteins and with OMP preparation. Recent advances in the solubilization of intractable proteins have prompted the proteomic analysis of bacterial OMPs (37). Specifically, proteomic analysis of Escherichia coli (37), Salmonella enterica serovar Typhimurium (38), Klebsiella pneumoniae (38), Caulobacter crescentus (38), and Leptospira interrogans serovar Lai (12) OMPs has been completed.
We sought here to identify the OMPs of H. pylori strain 26695 by using the sarcosine-insoluble outer membrane fraction. We identified 62 spots, including 16 OMPs, on 2-DE gels and identified 9 immunogenic proteins by immunoblot analysis.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
H. pylori strain 26695 was incubated on brucella agar plates containing 10% bovine serum. The bacterial cells were cultivated overnight at 37°C in an atmosphere of 10% CO2and 100% humidity.
Sarcosine preparation of H. pylori OMPs.
The sarcosine-insoluble outer membrane fraction of H. pylori was prepared as described previously (15) with minor modification. H. pylori cells were harvested by centrifugation (12,000 × g, 20 min, 4°C) and washed three times with 20 mM Tris-HCl (pH 7.5). The cells were suspended in 20 mM Tris-HCl (pH 7.5) and then disrupted with an ultrasonicator (Sonics & Materials, Inc., Danbury, Conn.). DNase and RNase (20 μg/ml each) were added to the cell suspension, and the mixture was incubated at room temperature for 30 min. The unbroken cells were removed by centrifugation (12,000 × g, 20 min, 4°C), and the supernatant was retained. Total membrane proteins were then collected by centrifugation (40,000 × g, 30 min, 4°C), resuspended in 20 mM Tris-HCl (pH 7.5) containing 2.0% (wt/vol) sodium lauryl sarcosine, and incubated at room temperature for 30 min. OMPs were collected by centrifugation (40,000 × g, 30 min, 4°C) and washed three times with distilled water. The pellet was resuspended in distilled water, divided into aliquots, and stored at −20°C until use.
Protein quantification.
Protein concentrations were determined by the Bradford method (8), with bovine serum albumin as a standard.
2-DE electrophoresis.
Isoelectric focusing (IEF) was performed by using IPG strips (Bio-Rad, Hercules, Calif.) (50). Portions (300 μg) of the OMPs were applied to strips of pH ranges of 3.0 to 10.0 and 6.0 to 11.0. The samples were diluted by incubation in a rehydration solution containing 7 M urea, 2 M thiourea, 2 mM tributyl phosphine (Sigma-Aldrich, St. Louis, Mo.), 40 mM Tris base, 1% Triton X-100, and 0.5% ampholyte (pH 3.0 to 10.0 [Bio-Rad] and pH 6.0 to 11.0 [Amersham]) overnight in a reswelling tray (Bio-Rad). The strips were rehydrated under the following passive conditions: 0 V, 20°C, and a 14- to 16-h reaction time in a Protean IEF cell (Bio-Rad). Three preset programs were executed with slight modifications such that focusing conditions comprised the conditioning step, voltage ramping, and final focusing. The purpose of the conditioning step (250 V for 15 min) was to remove salt ions and charged contaminants, and this was followed by linear voltage ramping for 3 h at 10,000 V. In the final focusing step, the maximum voltage of the ramp step was maintained (up to 80,000 V · h), and the current did not exceed 50 μA/strip. After IEF, the strips were equilibrated in 0.375 M Tris buffer (pH 8.8) containing 6 M urea, 2% SDS, 20% glycerol, 2% dithiothreitol, and 0.01% bromophenol blue, followed by the addition of the same buffer supplemented with 2.5% iodoacetamide. SDS-PAGE was performed according to the Laemmli method (32) with a 12.5% resolving polyacrylamide gel (20 by 30 cm) without a stacking gel. Separation in the second dimension was carried out at 30 mA/gel at 4°C until the running dye reached the bottom.
Silver staining and gel drying.
Proteins resolved on gels were visualized by using a silver staining method (25, 27 [see also http://prospector.ucsf.edu]) with slight modifications. Briefly, the gel was fixed in a solution containing 50% methanol, 12% acetic acid, and 0.5 ml of 37% formaldehyde for 1.5 h. All incubations were performed in a shaker with gentle shaking. After a fixing step, the gel was washed with 50% ethanol twice for 20 min and then washed again with double-distilled water (dDW) for 20 min. The gel was pretreated with Na2S2 · 5H2O (0.1 g/liter) for 1 min and washed again with dDW. The gel was impregnated with silver by incubation in AgNO3(2 g/liter) and 0.75 ml of 37% formic acid for 30 min, and it was then rinsed with dDW three times for 20 s each time. A developing solution consisting of Na2CO3(60 g/liter), Na2S2 · 5H2O (4 mg/liter), and 0.5 ml of 37% formic acid was prepared ahead of time and preserved in ice slurry. The visualization was performed by incubating the gel in the developing solution until a clear image was observed. When clear spots appeared, the gel was washed with dDW twice for 20 s each time, after which the reaction was stopped by adding 50% methanol and 12% acetic acid for 10 min. The visualized gel was dried with cellophane paper.
Image analysis.
A gel image was obtained by scanning the silver stained gels with the Fluor-S MultiImager (Bio-Rad). The image was documented through the PDQUEST 2-D gel analysis software (version 6; Bio-Rad).
Destaining and in-gel digestion of proteins.
Silver-stained spots were excised from 2-DE gels and transferred into microcentrifuge tubes. Silver-stained proteins were destained with chemical reducers as described previously (22). The chemical reducer mixture was freshly prepared and comprised a 1:1 ratio of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate. A portion (100 μl) of the mixture was added to the microcentrifuge tube, and this was vortexed occasionally until the brownish color disappeared. Gel pieces were rinsed three times with DW to stop the reaction, and 500 μl of 200 mM ammonium bicarbonate was added to cover the gel for 20 min and then discarded. Gel pieces were dehydrated with 100 μl of acetonitrile and dried in a vacuum centrifuge. An in-gel digestion was carried out by the method of O'Connell and Stalts (41). Gel pieces containing proteins were rehydrated by adding a digestion buffer containing 12.5 ng of trypsin/ml for 45 min on ice. The enzyme solution was removed and replaced with 20 μl of the buffer without the enzyme, such that the gel pieces were kept wet overnight at 37°C. The gel pieces were subjected to vigorous vortexing for 30 min, after which 20 μl of the digested solution was transferred into a clean microcentrifuge tube and dried under vacuum. The resulting samples were dissolved in 2 μl of 0.1% trifluoroacetic acid.
Peptide mass fingerprinting.
A matrix solution composed of α-cyano-4-hydroxy cinnamic acid (40 mg/ml) in 50% acetonitrile and 0.1% trifluoroacetic acid was prepared for peptide mass fingerprinting. The, 2 μl each of the matrix solution and sample solution were mixed, applied to the target well, rapidly dried, and washed with deionized water. The solution mixture was dried for 10 min at room temperature and subjected to a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF-MS) operation by using the Voyager Biospectrometry Workstation (PE Biosystems) with the following parameters: 20-kV accelerating voltage, 75% grid voltage, 0.02% guide wire voltage, 70-ns delay, and a mass gate of 800 to 2,500.
Identification of proteins.
Peptide mass fingerprints were analyzed by using the MS-FIT ProteinProspecter program developed by the UCSF Mass Spectrometry Faculty (http://prospector.ucsf.edu). Helicobacter proteins in the NCBI database were searched to identify proteins. Monoisotopic peptide masses were used to search the database, allowing a molecular mass range for 2-DE analyses of ±15%, a peptide mass accuracy of 50 ppm, and one partial cleavage. If matched proteins were absent, the molecular mass window was extended. Pyroglutamic acid modification of N-terminal glutamine, oxidation of methionine, and acrylamide modification of cysteine were considered. Matches were defined by the number of homologous peptides and the percentage of total translated ORF sequence covered by those peptides, in comparison to other database entries. Identified proteins were deemed identical if they produced the same results from the same site spots of more than five independent 2-DE gels.
Immunoblot analysis.
OMPs were transferred from the 2-DE gels onto a nitrocellulose membrane (PROTRAN; Schleicher & Schuell) with a blotting buffer containing 39 mM glycine, 48 mM Tris base, 20% methanol, and 0.037% SDS and running conditions of 15 V constant voltage for 2 h. The membrane was blocked with 1% bovine serum albumin in Tris-buffered saline containing 0.05% Tween 20 (TBST) for 1 h at room temperature. A pool of 300 sera obtained from seropositive patients in Gyeongsang National University Hospital, Jinju, Korea, was used as an antibody source, and a pool of 13 sera from H. pylori-uninfected persons was used as a negative control; in each case the presence or absence of antibodies had been confirmed previously by Western blot (3, 51). The membrane was incubated for 30 min at room temperature with the pooled sera, which were diluted 10-fold in TBST. After the membrane was washed with TBST, an alkaline phosphatase-conjugated rabbit anti-human immunoglobulin A (IgA), IgG, IgM (diluted by 1/1,000; Dako) was added, and the membrane was incubated for 1 h at room temperature. After a wash with TBST, the bound antibody was detected by using BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazoliuum (ImmunoPure; Pierce).
Duplicate 2-DE gels were simultaneously prepared under identical conditions, one for Ponceau-S staining and immunoblotting and the other for silver staining. The spots profile of immunoblot membrane was compared to that of the silver-stained gel and Ponceau-S-stained membrane after electrotransfer by using PDQUEST software for further identification of immunoreactive spots.
RESULTS AND DISCUSSION
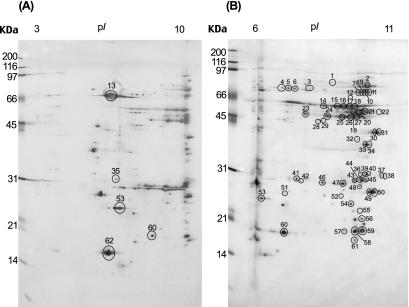
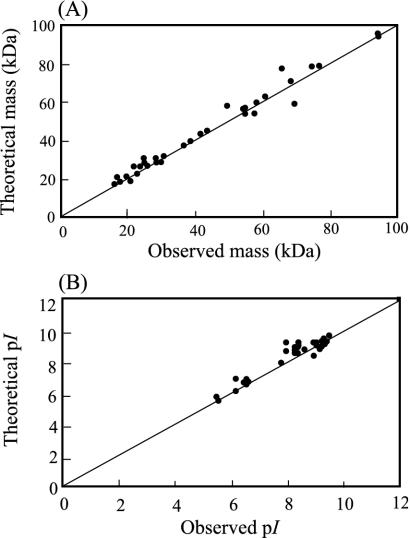
We analyzed the OMP proteome of the H. pylori 26695 strain by using the sarcosine-insoluble outer membrane fraction. This fraction was loaded onto precast IPG strips with a pH gradient of 3.0 to 10.0 for separation in the first dimension. The strips were then loaded onto a 12.5% acrylamide gel of 20 by 30 cm for electrophoretic separation in the second dimension, and separated spots were visualized by silver staining. Sarcosine-treated proteins were enriched in the alkaline pI regions, and their molecular masses were between approximately 10 and 100 kDa (Fig. 1A). Because the spots within alkaline pI regions tended to be located in close proximity, it was difficult to identify them with a pH 3.0 to 10.0 IPG strip since the alkaline region of that strip was too small to separate alkaline proteins with good resolution. Thus, to resolve these protein spots the sarcosine-insoluble fraction was applied to an IPG strip with a narrower pH range of 6.0 to 11.0. Using this approach, more than 80 protein spots were visualized on the corresponding silver-stained 2-DE gel (Fig. 1B). These spots were numbered, excised, destained, and then digested in the gel with trypsin for peptide fingerprinting (Fig. 1). The mass of the resulting peptide mixtures was measured by MALDI-TOF-MS. The theoretical or observed pI values of the majority of the H. pylori OMPs identified in the present study were higher than 8.0 (Fig. 2), which is higher than that of other bacterial OMPs (12, 37, 38). It has been reported that the pI ranges of OMPs of E. coli (37), Salmonella enterica serovar Typhimurium (38), Klebsiella pneumoniae (38), Caulobacter crescentus (38), and Leptospira interrogans serovar Lai (12) are pI 4 to 7, which is consistent with the theoretical pI values predicted from their genome databases. In contrast to these bacterial OMPs, H. pylori OMPs were detected in the alkaline pI region of the 2-DE gel. This may reflect evolutionary pressure for high alkaline proteins because of the acidic environment of the organism.
FIG. 1.
Sarcosine-insoluble fraction of H. pylori 26695 were separated by 2-DE with an IPG strip, followed by SDS-12.5% PAGE. Spots were detected by silver staining. The circled proteins were identified by MALDI-TOF-MS. (A) IPG strip, pH 3.0 to 10.0; (B) IPG strip, pH 6.0 to 11.0. Strain 26695 was grown as described in Materials and Methods, and 300 μg of sarcosine-insoluble fraction was loaded in the first dimension. Identified proteins are indicated by spot numbers in Table 1. Molecular size markers are shown on the left in kilodaltons.
FIG. 2.
Comparison of gel-estimated and calculated molecular weights and pI values of the protein spots of H. pylori 26695. (A) Molecular masses; (B) pI values. The theoretical values were referred from the NCBI database of H. pylori strain 26695.
In the present study, we identified 62 protein spots that represented 35 genes (Table 1). Of these 35 proteins, several have already been predicted to be surface-exposed in H. pylori based on results from various independent methods. Antibody staining indicated that UreA, UreB, catalase, and a homologue of HP0410 are present on the cell surface (16, 33, 44). Neutrophil-activating protein (NapA), phosphoglycerate dehydrogenase (SerA), glutamine synthetase (GlnA), and alkyl hydroperoxide reductase (TsaA) have previously been identified in whole-cell extracts of H. pylori (10). In addition, surface localization of NapA has been demonstrated (39). Glutamine sythetase (GlnA) from Azotobacter vinelandii is attached to the plasma membrane (ca. 30%), while the main fraction is found in the cytosol (31), as is the case for H. pylori urease (44). Even though these proteins may be cytoplasmic proteins, they did appear in the sarcosine-insoluble fraction. These results suggested that the surface properties of H. pylori could promote adsorption of cytoplasmic proteins.
TABLE 1.
List of identified proteins in the sarcosine-insoluble fraction of H. pylori strain 26695
| Spot no. | Annotation | Source or referencea | TIGR locus |
Mr
|
pI
|
No. of matching peptides | Sequence coverage (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| Observed | Theoretical | Observed | Theoretical | ||||||
| 1 | Hypothetical protein | HP0205 | 94,000 | 94,347 | 8.4 | 8.4 | 7 | 11.0 | |
| 2 | OMP (Omp2)b | HP0025 | 77,000 | 77,665 | 9.2 | 9.2 | 17 | 30.0 | |
| 3 | OMP (Omp19) | HP0896 | 75,000 | 77,693 | 8.0 | 8.6 | 5 | 16.0 | |
| 4 | Phosphoglycerate dehydrogenase (SerA) | 10 | HP0397 | 70,000 | 57,906 | 6.5 | 6.7 | 4 | 14.0 |
| 5 | Phosphoglycerate dehydrogenase (SerA) | 10 | HP0397 | 70,000 | 57,906 | 6.6 | 6.7 | 5 | 13.0 |
| 6 | Phosphoglycerate dehydrogenase (SerA) | 10 | HP0397 | 70,000 | 57,906 | 6.7 | 6.7 | 4 | 19.0 |
| 7 | OMP (Omp27) | HP1177 | 69,000 | 69,692 | 9.0 | 9.2 | 11 | 27.0 | |
| 8 | OMP (Omp27) | HP1177 | 69,000 | 69,692 | 9.1 | 9.2 | 4 | 18.0 | |
| 9 | OMP (Omp27) | HP1177 | 69,000 | 69,692 | 9.2 | 9.2 | 7 | 14.0 | |
| 10 | OMP (Omp27) | HP1177 | 69,000 | 69,692 | 9.3 | 9.2 | 5 | 15.0 | |
| 11 | OMP (Omp27) | HP1177 | 69,000 | 69,692 | 9.3 | 9.2 | 6 | 17.0 | |
| 12 | OMP (Omp25) | HP1177 | 66,000 | 76,346 | 9.0 | 9.1 | 5 | 17.0 | |
| 13 | Urease beta subunit (UreB) | 10, 28, 30 | HP0072 | 61,000 | 61,671 | 5.6 | 5.6 | 10 | 23.0 |
| 14 | Catalase | 48 | HP0875 | 59,000 | 58,630 | 8.3 | 8.7 | 14 | 32.0 |
| 15 | Catalase | 48 | HP0875 | 59,000 | 58,630 | 8.4 | 8.7 | 12 | 27.0 |
| 16 | Catalase | 48 | HP0875 | 59,000 | 58,630 | 8.5 | 8.7 | 12 | 30.0 |
| 17 | Catalase | 48 | HP0875 | 59,000 | 58,630 | 8.6 | 8.7 | 14 | 32.0 |
| 18 | Catalase | 48 | HP0875 | 59,000 | 58,630 | 8.7 | 8.7 | 6 | 20.0 |
| 19 | OMP (Omp20) | 29, 35 | HP0912 | 55,000 | 55,932 | 9.0 | 9.1 | 8 | 23.0 |
| 20 | OMP (Omp20) | 29, 35 | HP0912 | 55,000 | 55,932 | 9.1 | 9.1 | 9 | 20.0 |
| 21 | OMP (Omp20) | 29, 35 | HP0912 | 55,000 | 55,932 | 9.2 | 9.1 | 7 | 13.0 |
| 22 | OMP (Omp6) | HP0229 | 55,000 | 55,932 | 9.5 | 9.2 | 4 | 15.0 | |
| 23 | OMP (Omp21) | 29 | HP0913 | 50,000 | 57,063 | 8.0 | 9.2 | 5 | 13.0 |
| 24 | OMP (Omp21) | 29 | HP0913 | 50,000 | 57,063 | 8.4 | 9.2 | 5 | 12.0 |
| 25 | OMP (Omp21) | 29 | HP0913 | 50,000 | 57,063 | 8.5 | 9.2 | 4 | 11.0 |
| 26 | OMP (Omp21) | 29 | HP0913 | 50,000 | 57,063 | 9.0 | 9.2 | 6 | 14.0 |
| 27 | OMP (Omp21) | 29 | HP0913 | 50,000 | 57,063 | 9.2 | 9.2 | 6 | 14.0 |
| 28 | Hypothetical protein | HP1349 | 44,000 | 44,396 | 8.3 | 8.9 | 4 | 14.0 | |
| 29 | Hypothetical protein | HP1349 | 44,000 | 44,396 | 8.3 | 8.9 | 4 | 14.0 | |
| 30 | OMP (Omp32) | HP1501 | 42,000 | 42,976 | 9.5 | 9.5 | 9 | 33.0 | |
| 31 | OMP (Omp32) | HP1501 | 42,000 | 42,976 | 9.5 | 9.5 | 6 | 22.0 | |
| 32 | Hypothetical protein | HP0052 | 39,000 | 39,246 | 9.0 | 8.3 | 6 | 22.0 | |
| 33 | Iron(III) ABC transporter, periplasmic iron-binding protein (CeuE) | 48 | HP1561 | 37,000 | 37,552 | 9.4 | 9.2 | 4 | 19.0 |
| 34 | Iron(III) ABC transporter, periplasmic iron-binding protein (CeuE) | 48 | HP1561 | 37,000 | 37,552 | 9.4 | 9.2 | 4 | 16.0 |
| 35 | Glutamine ABC transporter, periplasmic glutamine-binding protein | HP1172 | 31,000 | 31,222 | 6.2 | 6.9 | 4 | 16.0 | |
| 36 | OMP (Omp24) | HP1113 | 31,000 | 31,896 | 9.2 | 9.2 | 4 | 17.0 | |
| 37 | OMP (Omp4) | HP0127 | 31,000 | 31,890 | 9.5 | 9.6 | 5 | 24.0 | |
| 38 | OMP (Omp4) | HP0127 | 31,000 | 31,890 | 9.6 | 9.6 | 5 | 17.0 | |
| 39 | Amino acid ABC transporter, periplasmic binding protein (Yckk) | HP0940 | 30,000 | 28,737 | 9.3 | 9.4 | 5 | 24.0 | |
| 40 | Amino acid ABC transporter, periplasmic binding protein (Yckk) | HP0940 | 30,000 | 28,737 | 9.4 | 9.4 | 4 | 17.0 | |
| 41 | Putative neuraminyllactose-binding hemagglutinin homolog (HpaA) | 10 | HP0410 | 29,000 | 28,349 | 7.8 | 7.9 | 5 | 23.0 |
| 42 | OMP (Omp10) | HP0324 | 29,000 | 28,767 | 7.8 | 7.8 | 5 | 24.0 | |
| 43 | OMP (Omp15) | HP0706 | 29,000 | 29,929 | 9.1 | 9.0 | 8 | 27.0 | |
| 44 | OMP (Omp15) | HP0706 | 29,000 | 29,929 | 9.2 | 9.0 | 5 | 26.0 | |
| 45 | OMP (Omp15) | HP0706 | 29,000 | 29,929 | 9.3 | 9.0 | 8 | 31.0 | |
| 46 | Urease alpha subunit (UreA) | 29, 48 | HP0073 | 28,500 | 26,540 | 8.3 | 8.5 | 10 | 54.0 |
| 47 | Urease alpha subunit (UreA) | 29, 48 | HP0073 | 28,500 | 26,540 | 8.5 | 8.5 | 6 | 44.0 |
| 48 | OMP (Omp14) | HP0671 | 27,000 | 30,347 | 9.2 | 9.2 | 8 | 36.0 | |
| 49 | OMP (Omp31) | HP1469 | 26,000 | 28,152 | 9.4 | 9.5 | 8 | 37.0 | |
| 50 | OMP (Omp31) | HP1469 | 26,000 | 28,152 | 9.5 | 9.5 | 9 | 44.0 | |
| 51 | Conserved hypothetical secreted protein | HP0139 | 25,500 | 27,492 | 6.6 | 6.9 | 4 | 22.0 | |
| 52 | Flagellar basal-body L-ring protein (FlgH) | HP0325 | 25,000 | 26,469 | 8.5 | 8.7 | 4 | 22.0 | |
| 53 | Alkyl hydroperoxide reductase (TsaA) | 10, 28, 30 | HP1563 | 24,500 | 22,235 | 6.2 | 6.2 | 8 | 50.0 |
| 54 | OMP (Omp23) | HP1107 | 22,000 | 25,698 | 9.1 | 9.1 | 4 | 31.0 | |
| 55 | Membrane fusion protein (MtrC) | HP0606 | 21,000 | 25,957 | 9.2 | 8.8 | 4 | 26.0 | |
| 56 | Glutamine synthetase (GlnA) | 10, 30 | HP0512 | 20,000 | 18,920 | 9.3 | 9.0 | 4 | 30.0 |
| 57 | Hypothetical protein | 9 | HP1173 | 20,000 | 20,586 | 8.5 | 8.9 | 8 | 28.0 |
| 58 | Hypothetical protein | 9 | HP1173 | 20,000 | 20,586 | 9.2 | 8.9 | 4 | 20.0 |
| 59 | Hypothetical protein | 9 | HP1173 | 20,000 | 20,586 | 9.3 | 8.9 | 5 | 42.0 |
| 60 | (3R)-Hydroxymyristoyl-(acyl carrier protein) dehydratase (FabZ) | HP1376 | 18,000 | 18,196 | 6.6 | 6.6 | 4 | 35.0 | |
| 61 | OMP (Omp11) | HP0472 | 17,000 | 20,925 | 9.2 | 9.2 | 6 | 38.0 | |
| 62 | Neutrophil-activating protein (NapA) | 10, 28, 30 | HP0243 | 16,000 | 16,835 | 5.5 | 5.8 | 6 | 46.0 |
That is, the reference numbers of previously identified proteins.
Refers to the 26695 genome annotation (www.tigr.org).
We identified five hypothetical proteins (HP0205, HP1349, HP0052, HP1173, and HP0139). These proteins are not theoretically OMPs, and their localization is unclear. HP1173 is secreted into the extracellular medium (9), but its functions are not yet known. Of the other proteins we found, the flagellar basal-body L-ring protein (HP0325) is encoded by the flgH gene and is located in the outer membrane (1, 11). The iron ABC transporter (CeuE) is likely to be localized in the periplasm based on data from Campylobacter coli (47) and other ABC transporters, such as the amino acid ABC transporter, glutamine ABC transporter, and iron(III) ABC transporter are theoretically predicted to be periplasmic binding proteins (49). (3R)-Hydroxymyristol-(acyl carrier protein) dehydratase (FabZ) is involved in fatty acid synthesis such that it efficiently catalyzed the dehydration of short-chain β-hydroxyacyl-acyl carrier proteins and unsaturated β-hydroxyacyl-acyl carrier proteins in E. coli (36). However, the localization of H. pylori FabZ has not been reported yet. MtrC is an outer membrane decahaem c-type cytochrome that appears to be required for the activity of the terminal Fe(III) reductase from Shewanella putrefaciens (6). Although cellular localization of H. pylori MtrC has not been reported, it may be localized to the outer membrane due to its functional similarity to S. putrefaciens MtrC.
It has been well documented that cytoplasmic and periplasmic components and inner membrane proteins are present as contaminants in the outer membrane preparation. This partially reflects their surface localization (e.g., urease, catalase, and neutrophil-activating protein), as well as the tight association between the inner and outer membranes (15). The present study focused on whether or not theoretically predicted OMPs were expressed and exposed on the H. pylori surface. A total of 16 OMPs were identified in the present study, whereas previously 33 ORFs have been assigned as putative OMPs in the H. pylori 26695 genome (49). Five horizontally aligned spots (69 kDa each) were identified as Omp27. The horizontal separation may be due to posttranslational modifications that result in differentially charged side chains on the amino acids residues of one species of protein. Maguire et al. (34) reported that horizontally aligned spots may represent the same protein isoforms in different phosphorylation states in the phosphotyrosine proteome from thrombin-activated platelets. Omp4, Omp15, Omp20, Omp21, Omp31, and Omp32 showed similar horizontal arrays of spots.
The functions of at least six OMPs have been already predicted. Omp19 is a homologue of BabB, which is a Lewis B binding adhesin (25). Omp6, Omp21, Omp20, Omp2, and Omp15 have previously been designated HopA, HopB, HopC, HopD, HopE, respectively, and function as porins (14, 19). In particular, Omp20 (HopC) has been reported to be expressed in this strain (35). Omp20 and Omp21, which have been reported to be enriched in the supernatant when H. pylori was grown in the absence of nalidixic acid (29), were also identified in the present study. It was reported that Omp4 might be associated with bacterial adherence due to its sequence similarity to established adhesins although this has not been proven. The expression level of Omp4 was reduced in both the virB11 mutant and the fliI mutant, revealing that Omp4 transport was dependent on a flagellum export apparatus and virulence factor export (45). The functions of the other identified outer membrane proteins remain to be elucidated. Therefore, our results may serve as a first step toward further functional characterization of H. pylori OMPs.
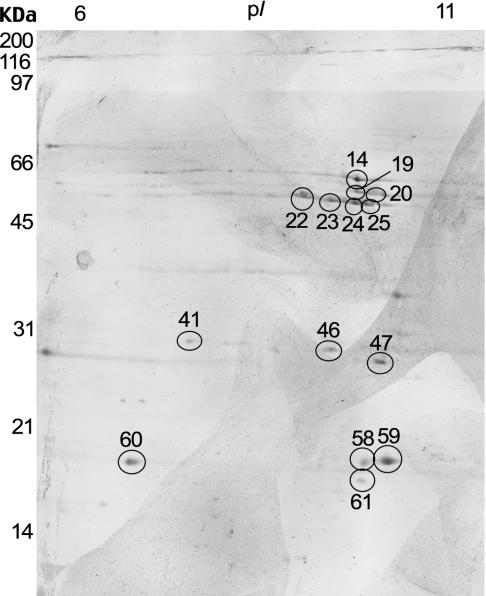
After 2-DE within a pH range of 6.0 to 11.0 IPG, proteins were transferred to nitrocellulose membranes for immunoblotting. We used a pool of 300 sera obtained from H. pylori-infected patients for immunoblotting, and the antibody reactivities with the sarcosine-insoluble fraction are shown in Fig. 3. The pooled sera from the infected patients bound to at least 10 spots. Of these, nine proteins were identified: catalase, Omp11, HP1173, UreA, a putative neuraminyllactose-binding hemagglutinin homolog (HP0410), (3R)-hydroxymyristoyl-(acyl carrier protein) dehydratase (FabZ), Omp14, Omp20, and Omp21 (Table 2). The presence of catalase, UreA, a putative neuraminyllactose-binding hemagglutinin homolog (HP0410), and Omp20 have previously been demonstrated by immunoblot analysis (23, 35). Therefore, in the present study, we identified five new immunoreactive proteins: hypothetical protein (HP1173), (3R)-hydroxymyristoyl-(acyl carrier protein) dehydratase (FabZ), Omp11, Omp14, and Omp21. The pooled sera from uninfected persons that was used as a negative control did not exhibit any immunoreactivity (data not shown).
FIG. 3.
2-DE immunoblot with the sarcosine-insoluble fraction of H. pylori resolved by IEF in the pH 6 to 11 and SDS-12.5% PAGE gel. The membrane was probed with a pool of 300 sera from seropositive patients (dilution 1:10). Numbers correspond to the identified proteins in Table 1. Molecular size markers are shown on the left.
TABLE 2.
Immunoreactive proteins in the sarcosine-insoluble fraction of H. pylori 26695
| Spot no.a | Mr | pI | Annotation | TIGR locus name |
|---|---|---|---|---|
| 14 | 58,630 | 8.7 | Catalase | HP0875 |
| 19 | 55,932 | 9.1 | OMP (Omp20) | HP0912 |
| 20 | 55,932 | 9.1 | OMP (Omp20) | HP0912 |
| 22 | 57,063 | 9.2 | OMP (Omp21) | HP0913 |
| 23 | 57,063 | 9.2 | OMP (Omp21) | HP0913 |
| 24 | 57,063 | 9.2 | OMP (Omp21) | HP0913 |
| 25 | 57,063 | 9.2 | OMP (Omp21) | HP0913 |
| 41 | 28,349 | 7.9 | Putative neuraminyllactose-binding hemagglutinin homolog (HpaA) | HP0410 |
| 46 | 26,540 | 8.5 | Urease alpha subunit (UreA) | HP0073 |
| 47 | 30,347 | 9.2 | OMP (Omp14) | HP0671 |
| 58 | 20,586 | 8.9 | Hypothetical protein | HP1173 |
| 59 | 20,586 | 8.9 | Hypothetical protein | HP1173 |
| 60 | 18,196 | 6.6 | (3R)-Hydroxymyristoyl-(acyl carrier protein) dehydratase (FabZ) | HP1376 |
| 61 | 20,925 | 9.2 | OMP (Omp11) | HP0472 |
The numbers correspond to the identified proteins listed in Table 1.
Our results contribute to the characterization of H. pylori OMPs and may help to identify new target proteins for vaccine development and drug therapy.
Acknowledgments
This study was supported by grant 02-PJ1-PG10-20201-005 from the Ministry of Health and Welfare of Korea. J.-Y.S., K.-M.K., S.-M.S., D.-S.K., J.-S.J., S.-G.L., and J.-U.P. were supported by the Brain Korea 21 program from the Ministry of Education of Korea. J.-U.P. was supported by a Korea Research Foundation Grant (KRF-2002-050-E00002).
REFERENCES
- 1.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Baik, S. C., H. S. Yoon, M. H. Chung, W. K. Lee, M. J. Cho, G. H. Ko, C. K. Park, H. Kasai, and K. H. Rhee. 1996. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 56:1279-1282. [PubMed] [Google Scholar]
- 4.Baik, S. C., J. B. Kim, M. J. Cho, Y. C. Kim, C. K. Park, H. H. Ryou, H. J. Choi, and K. H. Rhee. 1990. Prevalence of Helicobacter pylori infection among normal Korean adults. J. Korean Soc. Microbiol. 455-462. 6:
- 5.Banatvala, N. K., K. Mayo, F. Megraud, R. Jennings, J. J. Deeks, and R. A Feldman. 1993. The cohort effect and Helicobacter pylori. J. Infect. Dis. 168:219-221. [DOI] [PubMed] [Google Scholar]
- 6.Beliaev, A. S., D. A. Saffarini, J. L. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. [DOI] [PubMed] [Google Scholar]
- 7.Blaser, M. J. 1987. Gastric Campylobacter-like organisms, gastritis, and peptic ulcer disease. Gastroenterology 93:371-383. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Bumann, D., S. Aksu, M. Wendland, K. Janek, U. Zimny-Arndt, N. Sabarth, T. F. Meyer, and P. R. Jungblut. 2002. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect. Immun. 70:3396-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, M. J., B. S. Jeon, J. W. Park, T. S. Jung, J. Y. Song, W. K. Lee, Y. J. Choi, S. G. Park, J. U. Park, M. Y. Choe, S. A. Jung, E. Y. Byun, S. C. Baik, H. S. Youn, G. H. Ko, D. B. Lim, and K. H. Rhee. 2002. Identifying the major proteome components of Helicobacter pylori strain 26695. Electrophoresis 231:161-1173. [DOI] [PubMed] [Google Scholar]
- 11.Christopher, J. J., M. Homma, and M. M. Robert. 1989. l-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J. Bacteriol. 171:3890-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen, P. A., S. J. Cordwell, D. M. Bulach, D. A. Haake, Adler, B. 2002. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 70:2311-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cussac, V., R. L. Ferrero, and A. Labigne. 1992. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 174:2466-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doig, P., M. M. Exner, R. E. Hancock, and T. J. Trust. 1995. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J. Bacteriol. 177:5447-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doig, P., and T. J. Trust. 1994. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect. Immun. 62:4526-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn, B. E., and S. H. Phadnis. 1998. Structure, function, and localization of Helicobacter pylori urease. Yale J. Biol. Med. 71:63-73. [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton, K. A., C. L. Brooks, D. R. Morgan, and S. Krakowka. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 59:2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonization of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 19.Exner, M. M., P. Doig, T. J. Trust, and R. E. Hancock. 1995. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 63:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forman, D., D. G. Newell, F. Fullerton, J. W. Yarnell, A. R. Stacey, N. Wald, and F. Sitas. 1991. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. BMJ 302:1302-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gharahdaghi, F., C. R. Weinberg, D. A. Meagher, B. S. Imai, and S. M. Mische. 1999. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20:601-605. [DOI] [PubMed] [Google Scholar]
- 23.Haas, G., G. Karaali, K. Ebermayer, W. G. Metzger, S. Lamer, U. Zimny-Arndt, S. Diescher, U. B. Goebel, K. Vogt, A. B. Roznowski, B. J. Wiedenmann, T. F. Meyer, T. Aebischer, and P. R. Jungblut. 2002. Immunoproteomics of Helicobacter pylori infection and relation to gastric disease. Proteomics 2:313-324. [DOI] [PubMed] [Google Scholar]
- 24.Heukeshoven, J., and R. Dernick. 1988. Improved silver staining procedure for fast staining in Phast system development unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis 9:28-32. [DOI] [PubMed] [Google Scholar]
- 25.Ilver, D., A. Arnqvist, J. Ogren, I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 26.Jafar, M., S. Berit, H. Marina, O. O. Farzad, F. Lina, R. Niamh, A. Jonas, L. Thomas, T. Susann, K. Karl-Anders, A. Siiri, W. Torkel, K. Dangeruta, E. B. Douglas, D. Andre, P. Christoffer, M. Karl-Eric, N. Thomas, L. Frank, B. L. Bertil, A. Anna, H. Nennart, and B. Tomas. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Nature 297:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung, T. S., S. C. Kang, Y. J. Choi, B. S. Jeon, J. W. Park, S. A. Jung, J. Y. Song, S. H. Choi, S. G. Park, M. Y. Choe, B. S. Lee, E. Y. Byun, S. C. Baik, W. K. Lee, M. J. Cho, H. S. Youn, G. H. Ko, and K. H. Rhee. 2000. Two-dimensional gel electrophoresis of Helicobacter pylori for proteomic analysis. J. Korean Soc. Microbiol. 35:97-108. [Google Scholar]
- 28.Jungblut, P. R., D. Bumann, G. Haas, U. Zimny-Arndt, P. Holland, S. Lamer, F. Siejak, A. Aebischer, and T. F. Meyer. 2000. Comparative proteome analysis of Helicobacter pylori. Mol. Microbiol. 36:710-725. [DOI] [PubMed] [Google Scholar]
- 29.Kim, N., D. L. Weeks, J. M. Shin, D. R. Scott, M. K. Young, and S. George. 2002. Proteins released by Helicobacter pylori in vitro. J. Bacteriol. 184:6155-6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimmel, B., A. Bosserhoff, R. Frank, R. Gross, W. Goebel, and D. Beier. 2000. Identification of immunodominant antigens from Helicobacter pylori and evaluation of their reactivities with sera from patients with different gastroduodenal pathologies. Infect. Immun. 68:915-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinschmidt, J. A., and D. Kleiner. 1978. The glutamine synthetase from Azotobacter vinelandii: purification, characterization, regulation and localization. Eur. J. Biochem. 89:51-60. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lundstrom, A. M., K. Blom, V. Sundaeus, and I. Bolin. 2001. HpaA shows variable surface localization but the gene expression is similar in different Helicobacter pylori strains. Microb. Pathog. 31:243-253. [DOI] [PubMed] [Google Scholar]
- 34.Maguire, P. B., K. J. Wynne, D. F. Harney, N. M. O'Donoghue, G. Stephens, and D. J. Fitzgerald. 2002. Identification of the phosphotyrosine proteome from thrombin activated platelets. Proteomics 2:642-648. [DOI] [PubMed] [Google Scholar]
- 35.McAtee, C. P., M. Y. Lim, K. Fung, M. Velligan, K. Fry, T. Chow, and D. E. Berg. 1998. Identification of potential diagnostic and vaccine candidates of Helicobacter pylori by two-dimensional gel electrophoresis, sequence analysis, and serum profiling. Clin. Diagn. Lab. Immunol. 5:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohan, S., T. M. Kelly, S. S. Eveland, C. R. Raetz, and M. S. Anderson. 1994. An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase: relation to fabA and suppression of mutations in lipid A biosynthesis. J. Biol. Chem. 269:32896-32903. [PubMed] [Google Scholar]
- 37.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871-2881. [DOI] [PubMed] [Google Scholar]
- 38.Molloy, M. P., N. D. Phadke, J. R. Maddock, and P. C. Andrews. 2001. Two-dimensional electrophoresis and peptide mass fingerprinting of bacterial outer membrane proteins. Electrophoresis 22:1686-1696. [DOI] [PubMed] [Google Scholar]
- 39.Namavar, F., M. Sparrius, E. C. Veerman, B. J. Appelmelk, and C. M. Vandenbroucke-Grauls. 1998. Neutrophil-activating protein mediates adhesion of Helicobacter pylori to sulfated carbohydrates on high-molecular-weight salivary mucin. Infect. Immun. 66:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomura, A., G. N. Stemmermann, P. H. Chyou, I. Kato, G. I. Perez-Perez, and J. Blaser. 1991. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N. Engl. J. Med. 325:1132-1136. [DOI] [PubMed] [Google Scholar]
- 41.O'Connell, K. L., and J. T. Stalts. 1997. Identification of mouse liver proteins on two-dimensional electrophoresis gel by matrix-associated larser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis 18:349-359. [DOI] [PubMed] [Google Scholar]
- 42.Odenbreit, S., M. Till, D. Hofreuter, G. Faller, and R. Haas. 1999. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 31:1537-1548. [DOI] [PubMed] [Google Scholar]
- 43.Peck, B., M. Ortkamp, K. D. Diehl, E. Hundt, and B. Knapp. 1999. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 27:3325-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phadnis, S. H., M. H. Parlow, M. Levy, D. Ilver, C. M. Caulkins, J. B. Connors, and B. E. Dunn. 1996. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect. Immun. 64:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porwollik, S., B. Noonan, and P. W. O'Toole. 1999. Molecular characterization of a flagellar export locus of Helicobacter pylori. Infect. Immun. 67:2060-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhee, K. H., H. S. Yoon, S. C. Baik, W. K. Lee, M. J. Cho, H. J. Choi, K. Y. Maeng, and K. W. Ko. 1990. Prevalence of Helicobacter pylori infection in Korea. J. Korean Soc. Microbiol. 6:475-490. [Google Scholar]
- 47.Richardson, P. T., and S. F. Park. 1995. Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology 141:3181-3191. [DOI] [PubMed] [Google Scholar]
- 48.Sabarth, N., S. Lamer, U. Zimny-Arndt, P. R. Jungblut, T. F. Meyer, and D. Bumann. 2002. Identification of surface proteins of Helicobacter pylori by selective biotinylation, affinity purification, and two-dimensional gel electrophoresis. J. Biol. Chem. 277:27896-27902. [DOI] [PubMed] [Google Scholar]
- 49.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 50.Weekes, J., C. H. Wheeler, J. X. Yan, J. Weil, T. Eschenhagen, G. Scholtysik, and M. J. Dunn. 1999. Bovine dilated cardiomyopathy: proteomic analysis of an animal model of human dilated cardiomyopathy. Electrophoresis 20:898-906. [DOI] [PubMed] [Google Scholar]
- 51.Youn, H. S., S. C. Baik, Y. K. Cho, H. O. Woo, Y. O. Ahn, K. Kim, M. J. Cho, W. K. Lee, G. H. Ko, K. Okada, K. Ueda, and K. H. Rhee. 1998. Comparison of Helicobacter pylori infection between Fukuoka, Japan and Chinju, Korea. Helicobacter 9-14. 3: [DOI] [PubMed]