Abstract
The Drosha-DGCR8 complex (Microprocessor) is required for microRNA (miRNA) biogenesis. DGCR8 recognizes the RNA substrate, whereas Drosha functions as the endonuclease. High-throughput sequencing and crosslinking immunoprecipitation (HITS-CLIP) was used to identify RNA targets of DGCR8 in human cells. Unexpectedly, miRNAs were not the most abundant targets. DGCR8-bound RNAs also comprised several hundred mRNAs as well as snoRNAs and long non-coding RNAs. We found that the Microprocessor controls the abundance of several mRNAs as well as of MALAT-1. By contrast, DGCR8-mediated cleavage of snoRNAs is independent of Drosha, suggesting the involvement of DGCR8 in cellular complexes with other endonucleases. Interestingly, binding of DGCR8 to cassette exons, acts as a novel mechanism to regulate the relative abundance of alternatively spliced isoforms. Collectively, these data provide new insights in the complex role of DGCR8 in controlling the fate of several classes of RNAs.
MicroRNAs (miRNAs) are small non-coding RNAs that negatively regulate the expression of target mRNAs and affect a great diversity of biological processes (reviewed by1). Their biogenesis involves a co-transcriptional processing event in the cell nucleus that is catalyzed by the RNase III enzyme Drosha resulting in the production of stem loop precursors, termed pre-miRNAs2-5. Subsequently, pre-miRNAs are exported by Exportin 5 to the cytoplasm6-8, where they are further processed by the type III ribonuclease Dicer into mature miRNAs9, 10.
Human Drosha is part of the Microprocessor, a large multiprotein complex, which comprises DGCR8, a double-stranded RNA-binding protein that is deleted in the DiGeorge syndrome11,12, and several RNA-associated proteins including RNA helicases and hnRNP proteins2,3,13,14. In vitro studies have shown that the minimal catalytically active Microprocessor comprises Drosha and DGCR814. The proteins present in the larger Microprocessor complex may act to modulate its activity on specific subsets of miRNAs either directly or via the recruitment of additional regulatory factors. Indeed, miRNA biogenesis is heavily regulated at the post-transcriptional level (for recent reviews see15-17). DGCR8, which contains two double-stranded RNA (dsRNA)-binding domains, interacts with the pri-miRNA and functions as the molecular anchor that measures the distance from the dsRNA-ssRNA junction18 directing the cleavage by the endonuclease Drosha 11bp away from this junction2. The efficiency of Drosha cleavage has been shown to be increased by the presence of heme that promotes the formation of highly ordered DGCR8 structures upon binding to RNA19,20.
Recent evidence has pointed out to a more extended role of Drosha and/or the Microprocessor in the regulation of RNA processing. For this reason, we set out to identify endogenous cellular RNA targets of the Microprocessor. Here, we have used high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP) to identify endogenous RNA targets of the Microprocessor component DGCR8 in HEK 293T cells. Since DGCR8 is the Microprocessor component that recognizes and binds directly RNA, it represents the best candidate to study new targets for this complex and elucidate whether the Microprocessor is involved in the processing of cellular RNAs other than miRNAs. This technique has been successfully used to map in vivo RNA-protein interactions for several RNA binding proteins, such as the neuronal splicing factor NOVA21, the SR protein family SRSF122, as well as to define miRNA-mRNA interactions mediated by Argonaute (Ago) proteins in both mouse brain and C. elegans, respectively23,24. More recently, several variations of this protocol have also been introduced (iCLIP, PAR-CLIP)25,26.
As expected, most of the miRNAs known to be expressed in HEK 293T cells were identified, as well as, the DGCR8 mRNA, which was known to be a target of the Microprocessor complex27-29. However, these did not represent the most abundant targets. We found that DGCR8 binds to a large number of different RNAs, including several hundred mRNAs, small nucleolar RNAs (snoRNAs) and long non-coding RNAs. Whereas DGCR8 together with Drosha controls the abundance of several mRNAs and specific alternatively spliced isoforms, the DGCR8-mediated cleavage of snoRNAs is independent of Drosha. This suggests the involvement of DGCR8 in cellular complexes with other endonucleases. Altogether, these data suggests an expanded role for the Microprocessor and/or DGCR8 in the control of RNA processing.
RESULTS
HITS-CLIP identifies novel targets of the Microprocessor
In order to determine whether the Microprocessor is involved in the processing of cellular RNAs (other than miRNAs) that contain hairpin structures, we focused on the identification of endogenous RNA targets of DGCR8. For this, we used the Cross-Linking Immunoprecipitation protocol coupled to high-throughput sequencing (HITS-CLIP). This technique relies on ultraviolet irradiation to induce protein covalent crosslinks in protein-RNA complexes in situ. This allows the use of highly stringent immunoprecipitation and washing conditions so that only those RNAs directly bound to the protein of interest are selected (reviewed by30,31). We performed two replicate HITS-CLIP experiments on UV-irradiated HEK293T cells. Each replicate comprised two independent experiments: one involved immunoprecipitation (IP) of endogenous DGCR8 protein (Supplementary Fig 1a, left) whereas the second one involved IP of a transiently transfected epitope-tagged DGCR8 protein (pCG T7-DGCR8) (Supplementary Fig 1a, right and 1b). We focused our analyses on the second replicate, since it gave higher depth of coverage and better mapping to the genome. Results from the first replicate are shown on Supplementary Material (Supplementary Fig. 1c-e). In the second HITS-CLIP experiment we obtained 37 million reads for the endogenous DGCR8 protein, and 36 million reads for the epitope-tagged T7-DGCR8 and 93% and 98% of the reads were mapped to the genome, respectively (Supplementary Table 1). After removing read duplicates, uniquely mapped reads from each of the datasets were clustered according to their genomic positions. In order to identify significant clusters we applied a modified false discovery rate (mFDR) to each cluster, as previously described24. We assessed the reproducibility of the DGCR8 targets identified in CLIP experiments for both endogenous and overexpressed DGCR8 proteins. Analysis of the reads of these two samples revealed a strong correlation (Pearson correlation co-efficient R >0.8) (Fig. 1a).
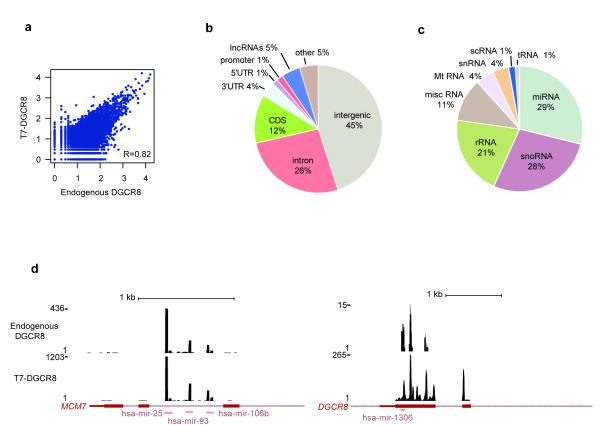
Figure 1.
Cross-linking Immunoprecipitation (CLIP) for endogenous and overexpressed DGCR8 in HEK 293T cells. (a) Reproducibility of all DGCR8 binding sites, when comparing endogenous and overexpressed DGCR8 CLIP experiments. The axes show the amount of reads in each of the multisample clusters in log10 scale. (b) Distribution of reproducible DGCR8 significant clusters (FDR<0.01) at the genomic level. (c) Location of significant clusters (FDR<0.01) in non-coding RNAs. (d) Graphical representation of DGCR8 binding sites for both endogenous (top) and overexpressed protein (bottom) on two well-characterized Microprocessor targets, the miR-106b-25 cluster on the left and, DGCR8 mRNA on the right. A list with all DGCR8 binding sites and the predicted RNA secondary structures can be found at http://regulatorygenomics.upf.edu/Data/DGCR8.
In order to describe DGCR8 bona fide targets, we focused on overlapping significant clusters between these two samples (for a complete list go to http://regulatorygenomics.upf.edu/Data/DGCR8). In this way, we found 8,196 common targets (harboring one or more significant clusters) at the genomic level that were distributed as follows: 45% mapped to intergenic regions, 43% to protein coding genes and 5% to long non-coding RNAs (lncRNAs) (Fig. 1b). We also found that 30% of the genomic clusters mapped to repetitive elements, mainly LINE-1 and LTR retroelements, but these were excluded from the graphical representation. When analyzing protein-coding genes, we observed that the majority of the clusters were located in introns (26%, including miRNAs), followed by coding exons (12%). Interestingly, we also observed some clusters in 3′ and 5′UTRs (Fig. 1b). As expected, due to the canonical function of the Microprocessor, we identified 117 miRNAs (Fig. 1d, left), with the let-7 family being the most abundant family. We also identified the only so far described non-canonical RNA substrate for the Microprocessor, the DGCR8 mRNA (Fig. 1d, right)27.We also observed CLIP tags mapping to other non-coding RNAs, such as rRNA, snRNAs, and snoRNAs (Fig. 1c). In addition for each of the clusters, an RNA secondary structure was predicted in order to select those clusters overlapping with structures resembling that of pri-miRNAs (see http://regulatorygenomics.upf.edu/Data/DGCR8 and Supplementary Methods). Importantly, we found that more than 60% of the DGCR8 binding sites were overlapping with stable RNA secondary structures (Supplementary Table 1). In addition, we observed that between 11-20% of the DGCR8 binding sites overlap with small RNAs harboring a 5′phosphorylated end, which are proposed to be generated by endonucleolytic activity32.
The Microprocessor regulates the abundance of MALAT1
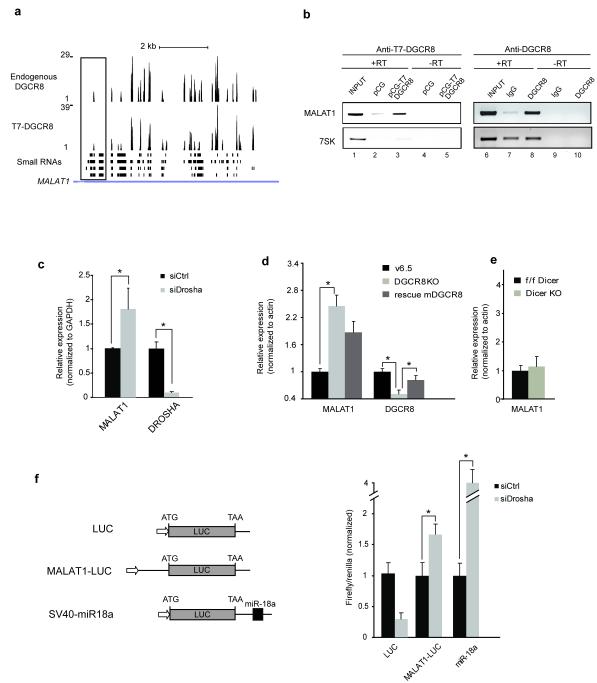
MALAT1 (metastasis-associated lung adenocarcinoma transcript 1, also known as Neat2) is a mammalian abundant, long non-coding RNA that was initially identified due to its elevated expression in several cancers33. We found that DGCR8 binds to MALAT1 and several other long non-coding RNAs (Fig. 1b and Fig. 2a and http://regulatorygenomics.upf.edu/Data/DGCR8). We further confirmed this interaction by immunoprecipitating overexpressed and endogenous DGCR8 protein followed by analysis of the associated RNA by semi-quantitative RT-PCR (Fig. 2b). In order to study the effect of Microprocessor binding to the MALAT1 RNA, we determined its steady-state levels upon siRNA-mediated depletion of Drosha in HeLa cells. We observed a two-fold increase in the levels of MALAT1 RNA when Drosha levels were reduced (Fig. 2c). This was recapitulated in DGCR8-deficient mouse ES cells (Dgcr8 KO) (Fig. 2d), but not in Dicer KO cells (Fig. 2e), suggesting that the Microprocessor controls MALAT1 RNA levels, independently of the activity of miRNAs. In order to test direct cleavage of the MALAT1 mRNA in vivo, we fused the 5′end of MALAT1, which harbors significant DGCR8 clusters (Fig. 2a, indicated with a box) to a Luciferase reporter gene and transfected this construct into HeLa cells. Reduction of Drosha levels resulted in an increased expression of this reporter, suggesting that MALAT1 can be cleaved in vivo by the Microprocessor (Fig 2f). In agreement, we observed small RNA reads mapping to the majority of DGCR8 clusters present on the MALAT1 sequence (small RNA dataset from32), also suggesting that DGCR8 binding could indeed induce RNA cleavage; however, it still remains unknown whether small RNAs generated from these loci behave like miRNAs (Fig. 2a).
Figure 2.
DGCR8 binds and controls the stability of the long intergenic non-coding RNA MALAT1 (a) Schematic representation of DGCR8 clusters on MALAT1 and overlap of the binding sites with small RNA libraries. (b) Overexpressed and endogenous DGCR8 protein (left and right, respectively) bind to endogenous MALAT1 RNA, as shown by IP RT-PCR. (c) Si-RNA mediated depletion of Drosha results in MALAT1 accumulation in HeLa cells. (d) Mouse embryonic stem cells (mES) lacking DGCR8 also display an upregulation of MALAT1 that is partially reversed upon reintroduction of mouse DGCR8 (mDGCR8). (e) mES cells lacking Dicer do not show changes in MALAT1 expression as compared to the parental mES line (where Dicer was floxed but not removed, f/f). All the values shown are averages of at least three independent experiments, and P<0.05 (t test) were deemed significant and marked with asterisks. Errors bars represent standard deviation. (f) A construct containing 500 bp of the 5′end of MALAT1 RNA (depicted with a box, Fig. 2a) was fused to Firefly Luciferase ORF (LUC) and transfected into Drosha (siDrosha) or mock (siCtrl) depleted HeLa cells. Levels of luciferase were analyzed relative to the cotransfected Renilla luciferase reporter. An SV40 construct containing a single miR-18a target site (SV40-miR18a) in its 3′UTR was used to monitor miRNA activity. Error bars indicate standard deviation (n=3, except for LUC siCtrl, n=2), and P<0.05 (t test) were deemed significant and marked with asterisks (right).
DGCR8 regulates snoRNA stability independently of Drosha
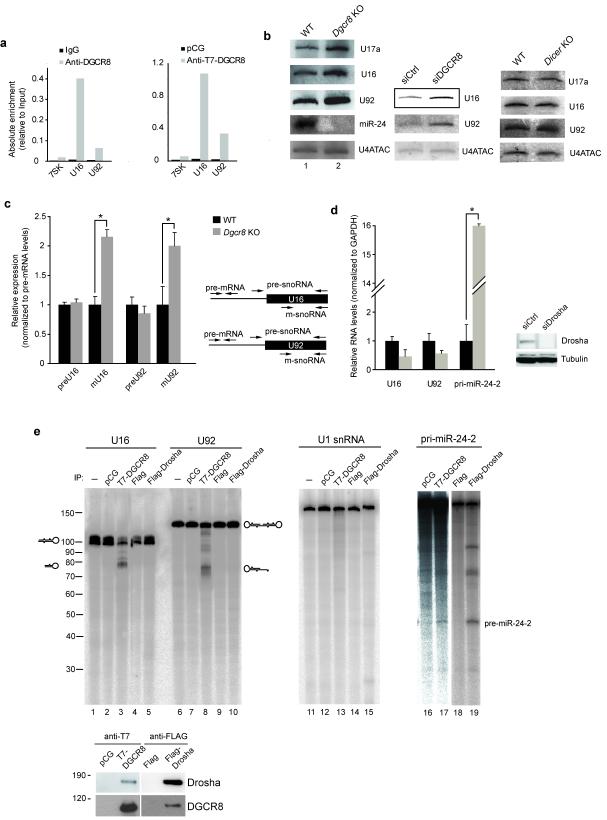
Another class of RNAs bound by DGCR8 corresponds to small nucleolar RNAs (snoRNAs) (Fig. 1c). These small RNAs are responsible for guiding many chemical modifications of other RNAs, such as rRNA, tRNA and snRNAs34. First, we confirmed the binding of endogenous and overexpressed DGCR8 to snoRNAs observed by CLIP using immunoprecipitation followed by qRT-PCR in HEK 293T cells (Fig. 3a). This reported binding to snoRNAs has a physiological consequence as seen by the accumulation of U17a, U16 and U92, which represent different subclasses of snoRNAs, in mouse cells lacking DGCR8 (Fig. 3b, left panel), as well as in HeLa cells knocked-down for DGCR8 (Fig. 3b, middle panel) or when overexpressing a dominant negative form of DGCR8 in HEK293T cells (Supplementary Fig. 2a-c). Importantly, mature snoRNA levels did not change in the absence of Dicer (Fig. 3b, right panel). Thus, DGCR8 can affect the stability of the mature forms of an H/ACA box snoRNA (U17a), a C/D box snoRNA (U16) and a small cajal RNA (scaRNA U92) (Fig. 3b). SnoRNAs are mostly intronic and transcribed as part of the host gene. Their biogenesis involves trimming of the host introns from the 5′ and 3′ end following pre-mRNA splicing and release of the mature snoRNA form that is being protected from degradation by the core snoRNP components (reviewed by35). We determined that DGCR8 affects the levels of mature snoRNAs but not of their precursor forms (Fig. 3c), indicating that DGCR8 acts at a post-synthesis level. To further elucidate the role of the Microprocessor in snoRNA stability in human cells, we studied the effect of either depleting Drosha or overexpressing a dominant negative form of Drosha. To our surprise we could not detect any significant change in mature snoRNA levels in these situations (Fig. 3d and Supplementary Fig. 2d). These data suggested a new DGCR8 function that operates independently of Drosha, strongly indicating that DGCR8 might associate to another nuclease/s to cleave and destabilize snoRNAs. To test this hypothesis we incubated radiolabeled mature snoRNAs with immunoprecipitated T7-DGCR8 and observed the appearance of smaller bands, corresponding to digested products (Fig. 3e, lanes 3 and 8). Importantly, the addition of immunoprecipitated Flag-Drosha did not result in the formation of these subproducts (Fig. 3e, lanes 5 and 10), despite Drosha being active in the processing of a pri-miRNA precursor (pri-miR-24-2) (Fig. 3e, lane 19). This DGCR8-mediated effect was specific for snoRNAs, since U1 snRNA was not cleaved by the DGCR8 complex (Fig. 3e, lane 13). In vitro cleavage analyses revealed the precise location of the snoRNA cleavages, which occur towards their 3′end; potentially generating 3′end fragments of 20nt long small RNAs for C/D box snoRNAs (U16) and 40nt long for H/ACA (U92) (Supplementary Fig. 3a,b). Furthermore, we observed stabilization of the 70nt long fragment when overexpressing dominant negative forms of DGCR8 in human cells (Supplementary Fig. 2e,f). This observation agrees with previous bioinformatic analyses of Dgcr8 KO and Dicer KO small RNA libraries36 whereby snoRNA-encoded small RNAs (sdRNAs) are affected by the loss of DGCR8 and Dicer. Here, we observed that the loss of DGCR8 decreases the amount of small RNAs originated from both the 5′ and the 3′ ends for both C/D box and H/ACA snoRNAs (Supplementary Fig. 3c,d) and these sites do not coincide with the cleavage sites previously described for H/ACA box snoRNAs (at the H box), which are required for the production of miRNA-like molecules in a Drosha-independent and Dicer-dependent pathway37. Collectively, these data demonstrates a role for DGCR8 in controlling the abundance of mature snoRNAs, in a Drosha-independent manner. This strongly suggests that DGCR8 associates to another endonuclease/s to specifically destabilize snoRNAs and/or to produce new types of small RNAs
Figure 3.
Binding of DGCR8 to snoRNAs affects their abundance independently of Drosha (a) Endogenous and epitope-tagged DGCR8 proteins bind to U16 and U92 snoRNA, as shown by IP-qRT-PCR. 7SK was used as a negative control. (b) In the absence of DGCR8 (Dgcr8 KO mES cells) mature forms of U17a, U16 and U92 snoRNAs accumulate, as detected in Northern blot analysis (left). Transient DGCR8 knock-down in HeLa cells also resulted in upregulation of the mature forms of these snoRNAs (middle panel), whereas Dicer depletion had no effect (right panel). (c) DGCR8 absence only affects the levels of mature snoRNAs (m-snoRNA), whereas snoRNA precursors remain unchanged (noted as preU16 and preU92). Specific primers were used to differentiate prefrom m-snoRNA forms, as seen in the right panel. (d) Drosha knock-down in HeLa cells, as shown by Western blot, does not affect mature snoRNA levels, but results in the accumulation of pri-miR-24-2. All the values are average of at least three independent experiments, and P<0.05 (t test) were deemed significant and marked with asterisks. Errors bars represent standard deviation. (e) Mature snoRNAs can be endonucleotically cleaved by immmmunopurified DGCR8 (lanes 3 and 8), but not Drosha (lanes 5 and 10). This effect is specific to snoRNAs, as U1snRNA cannot be cleaved by the DGCR8 complex (lane 13), which was shown to be functional as it can cleave a canonical pri-miRNA (lane 17). Western blot analysis reveals the level of Drosha and DGCR8 proteins in the immunoprecipitated material (lower panel).
The Microprocessor cleaves the mRNA of protein-coding genes
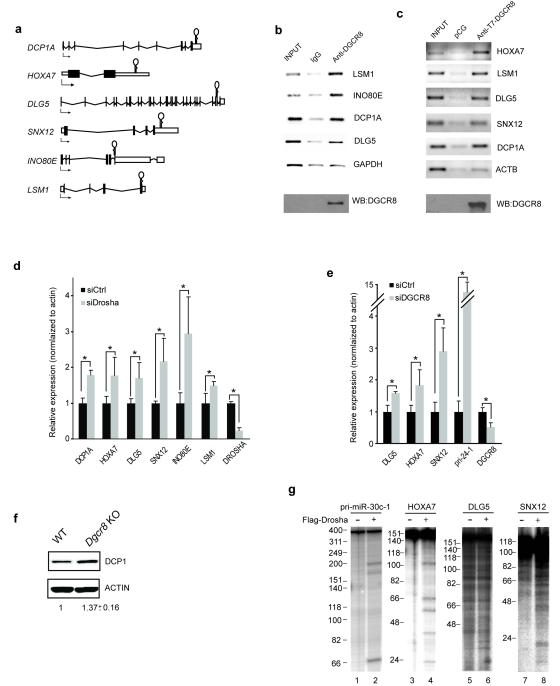
We obtained DGCR8 CLIP-tags mapping to 2,256 different mRNAs, comprising 3,610 significant clusters, indicating that some mRNAs harbor more than one DGCR8 binding site. We also found DGCR8 binding sites at the 5′UTR and first exon of the DGCR8 mRNA and in agreement with previous data, these clusters overlapped hs-miR-130627 (Fig. 1d). We confirmed by IP RT-PCR that both DGCR8 and Drosha bind to the DGCR8 mRNA and that Drosha knock-down increases steady-state levels of DGCR8 mRNA (Supplementary Fig. 4a-b). Next, we chose 6 different mRNAs with exonic DGCR8 clusters in both CLIP samples (endogenous and overexpressed) and harboring a predicted RNA secondary structure that resembles that of pri-miRNAs (Fig. 4a and Supplementary Fig. 4c). First, we confirmed binding of these mRNAs by IP-RT-PCR to both endogenous and overexpressed DGCR8 (Fig. 4b, c, and data not shown. Quantitation of these data is shown in Supplementary Fig. 4d-g). Si-RNA mediated depletion of Drosha and/or DGCR8 in HeLa cells resulted in an increase of their mRNA levels, suggesting that these mRNAs could be direct targets of the Microprocessor complex (Fig. 4d,e) and also affect the corresponding protein levels (Fig. 4f). Accordingly, the upregulation observed in Microprocessor-deficient cells (DGCR8 KO) was not detected in Dicer KO cells, suggesting a direct role of the Microprocessor in processing these mRNAs (Supplementary Fig. 4h). In vitro analysis of radioactively labeled mRNAs (comprising regions of those mRNAs that contain the DGCR8 binding sites) with immunopurified Microprocessor (Flag-Drosha) resulted in the cleavage of HOXA7, DLG5 and SNX12 (Fig. 4g, lanes 4, 6 and 8), as well as of the positive control, pri-miR-30c-1 (Fig 4g, lane 2).
Figure 4.
Microprocessor binding to mRNAs. (a) Schematic representation of DGCR8 binding sites on several mRNAs, symbolized by hairpin structures. (b) mRNAs chosen for further validation bind to endogenous DGCR8 as shown by IP-RT-PCR. GAPDH was used as a control and quantitation by qRT-PCR is shown in Supplementary Fig. 4d,e. (c) The same mRNAs are shown to be specifically bound to overexpressed DGCR8 in HEK 293T cells, as compared to the negative control, ACTB and quantitation by qRT-PCR is shown in Supplementary Fig. 4f,g. (d) SiRNA-mediated depletion of Drosha in HeLa cells results in the upregulation of selected mRNAs. The levels of Drosha depletion are also shown. All the values shown are averages of at least three independent experiments and P<0.05 (t test) were deemed significant and marked with asterisks. Errors bars represent standard deviation (e) The up-regulation of selected mRNAs was also observed when knocking-down DGCR8 in HeLa cells (f) The up-regulation of the mRNA levels correlates with the level of the encoded proteins, as shown for Dcp1p protein in Dgcr8 KO cells, which displayed an average of 1.37-fold increase in protein level (n=2). (g) mRNAs that are upregulated in the absence of Drosha can also be cleaved in vitro when incubated with the Microprocessor (lanes, 4, 6 and 8) as well as the positive control pri-miR-30c-1 (lane 2).
Modulation of alternative spliced isoforms by the Microprocessor
Analysis of the HITS-CLIP data revealed DGCR8 binding to several predicted cassette exons (n=241) (Fig. 5a). This would suggest the interesting possibility that Microprocessor-mediated cleavage of these cassette exons would affect the relative abundance of alternatively spliced (AS) isoforms. A global analysis of alternative splicing by exon junction arrays in mouse cells lacking Dgcr8 revealed 318 changes in cassette exons, as well as 40 alternative 5′/3′splice site (ss) events and 18 changes in mutually exclusive exons (Supplementary Fig. 5a), suggesting a role for this complex in AS regulation.
Figure 5.
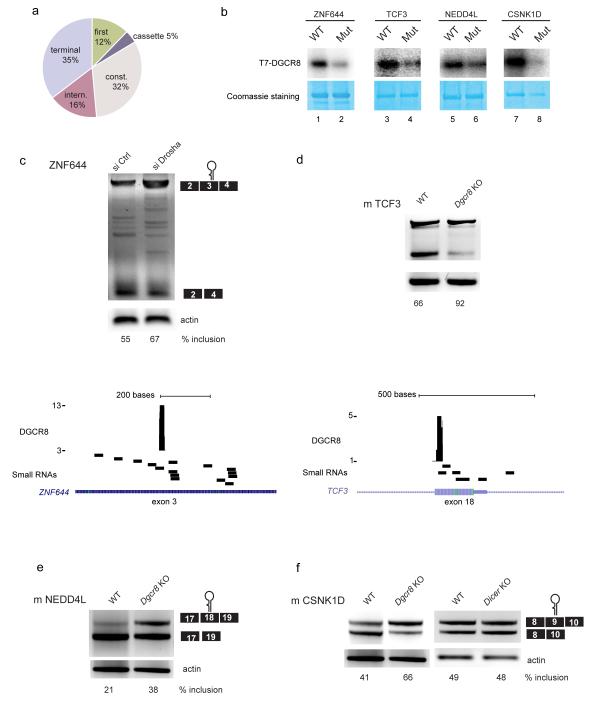
Binding of DGCR8 to cassette exons can modulate the relative abundance of alternatively spliced isoforms. (a) HITS-CLIP data revealed that 241 predicted cassette exons are bound by DGCR8. (b) UV cross-linking assay determines binding of DGCR8 to cassette exons that were identified by CLIP (upper panels). WT indicates intact RNA secondary structure, whereas Mut, indicates mutations introduced to disrupt this structure (lanes 2, 4, 6 and 8). Lower panels show Coomassie blue stainings of the same gels to show equal presence of T7-DGCR8 protein (c) The ZNF644 isoform including exon 3 is upregulated in human cells depleted of Drosha. The lower panel illustrates the presence of small RNAs identified in this region32. (d) The mouse TCF3 isoform including exon 18 is upregulated in mouse cells lacking DGCR8 or Drosha but not Dicer, indicating that is a miRNA-independent effect (Supplementary Fig. 5 f,g). The lower panel illustrates the presence of small RNAs identified in this region32 in human transcripts and the presence of DGCR8 binding sites by CLIP. (e) The mouse NEDD4L isoform including exon 18 is upregulated in mouse cells lacking DGCR8. (f) The mouse CSNK1D isoform including exon 9 is upregulated in the absence of DGCR8 (left) but not Dicer (right).
We first focused on four cassette exons shown to be bound by DGCR8 in the CLIP experiments that also display an RNA secondary structure resembling pri-miRNAs (Supplementary Fig. 5b-e). An UV-cross-linking assay revealed that DGCR8 binding to each of these cassette exons was affected when the predicted secondary structures were disrupted by mutation (Fig. 5b). Importantly, those isoforms bound by DGCR8 were stabilized in human cells depleted of Drosha or in mouse cells lacking DGCR8 (Fig. 5c-f and Supplementary Fig. 5f), strongly suggesting that the Microprocessor specifically cleaves and destabilizes mRNA isoforms containing DGCR8 binding sites leading to an altered ratio of the alternatively spliced isoforms. Accordingly, we also observed an overlap of these DGCR8 alternatively spliced exons with small RNA libraries (Fig 5c, d, lower panels), confirming that cleavage can occur at these locations, which could help to explain the change in the relative abundance of those isoforms containing the DGCR8 bound exon. In addition, in those cases that we tested, we could confirm that no changes in alternative splicing were observed in the absence of Dicer, indicating that these effects are independent from the presence of miRNAs (TCF3 and CSNK1D) (Fig 5f and Supplementary Fig. 5g). Altogether these data suggest that DGCR8 binding to cassette exons may act to influence the relative abundance of alternatively spliced isoforms.
DISCUSSION
In this study, we present a comprehensive strategy aimed at the identification of endogenous RNA targets for the Microprocessor. We found that DGCR8 binds to many different types of RNAs involved in different cellular pathways. We identified most of the miRNAs known to be expressed in these cells, which was expected. Other DGCR8 targets comprise mRNAs, non-coding RNAs including snoRNAs, long non-coding RNAs and retrotransposons.
Our analysis also revealed that DGCR8 binds to more than 2,000 mRNAs and in many cases depletion of Microprocessor components leads to an up-regulation of these mRNAs, suggesting that they are direct targets of the Microprocessor. For some of these mRNAs that were up-regulated upon depletion of Drosha or DGCR8, we could indeed confirm Drosha-dependent cleavage in vitro (Fig. 4). This extends a previous observation showing that a hairpin localized in the 5′ UTR of the DGCR8 pre-mRNA is targeted by the Microprocessor27-29. Nevertheless, it remains elusive whether any of these cleavage products are also Dicer substrates that would lead to the generation of miRNA-like small RNAs. However, we observed that a large proportion of DGCR8 binding sites also overlap with small RNAs harboring a 5′phosphorylated end, which have been proposed to be produced by the action of endonucleases. The fact that the Microprocessor destabilizes mRNAs containing DGCR8 binding sites in cassette exons suggested the interesting possibility that the Microprocessor might have a role in the processing of these alternative exons, which would impact on alternative splicing regulation. We confirmed that DGCR8 can modulate the relative abundance of alternatively spliced isoforms and that binding to these sequences depends on RNA secondary structure (Fig. 5 and Supplementary Fig. 5)
Functional studies of the miRNA processing machinery have shown that Drosha, DGCR8 and Dicer deficiencies generate similar phenotypes, resulting in early embryonic lethality, strongly suggesting that miRNAs are essential for normal development38,39. Furthermore, several lines of evidence indicated additional functions for Drosha and/or Dicer, although initial global studies suggested DGCR8 mRNA as the only non-canonical substrate for the Microprocessor40. First, microarray profiling of MEFs derived from Drosha and Dicer knock-out mice showed poor correlation between the populations of mRNAs affected by these two endonucleases. Secondly, Drosha was shown to recognize and cleave many protein-coding messenger RNAs (mRNAs) with secondary stem-loop structures in early-stage thymocytes41, as well as viral RNAs to directly regulate viral expression42. Finally, Drosha-dependent mRNA cleavage events that functionally regulate mRNA levels in mESCs, were recently described32. A simple explanation for these observations is that non-canonical functions for the Microprocessor and/or Dicer could be responsible for these effects. The comparison of these reports with our data only resulted in a partial overlap, probably due to the use of different cell lines for these studies (T cells versus HEK 293T cells). It also remains possible that binding of DGCR8 might not always lead to changes in relative mRNA expression.
What we provide here is a genome-wide view of DGCR8 binding sites in HEK 293T cells. Importantly, our studies show more alternative functions for the Microprocessor component DGCR8, implicating this protein in the control of non-coding RNAs stability (lncRNAs and snoRNAs) and on the relative abundance of alternatively spliced isoforms.. In vitro processing assays with immunoprecipitated DGCR8 showed cleavage of the mature snoRNA species in a Drosha-independent manner, suggesting the involvement of DGCR8 in cellular complexes with other endonucleases (Fig. 3 and Supplementary Fig. 2-3). A DGCR8-independent role for Drosha in pre-rRNA processing was also suggested by recent evidence. While Drosha is required for the processing of 12S pre-rRNA43,44, Dgcr8 KO mouse ES cells did not display evident defects in ribosomal RNA processing39, suggesting alternative complexes to the canonical Microprocessor. The presence of miRNA-like molecules encoded by H/ACA snoRNAs independent of Drosha function but involving a Dicer-dependent cleavage activity was previously reported37. Bioinformatics analyses of small RNAs encoded from snoRNAs showed that are dependent on both, DGCR8 and Dicer36, although the role of Drosha was not assessed. From an evolutionary point of view, some miRNAs have been shown to evolve from snoRNAs. Their structure conservation as well as the retained capacity to bind dyskerin and fibrillarin, two of the core components from snoRNP particles, suggests a common origin45-48. Interestingly, DGCR8 has been reported to localize to the nucleolus46. Our work proposes a new complex between DGCR8 and another endonuclease/s that would specifically cleave snoRNAs and probably other classes of cellular RNAs. Although it is well known how snoRNAs are synthesized, very little is known about their turnover and decay, and it would be of great interest to further study the implication of DGCR8 in this process. These findings raise the interesting possibility that there could be alternative DGCR8 complex/es using different nucleases to process a variety of cellular RNAs.
Significantly, all the new functions described here for the Microprocessor complex could be useful to reinterpret the phenotypes observed for Drosha or DGCR8 deficient cells that were mainly attributed to the deficiency of miRNAs. Importantly, the DGCR8 gene is present in the deleted genomic region (22q11.2) in DiGeorge syndrome patients that expands around 30 genes11,12. Mouse models for this deletion showed a minor alteration of miRNAs, but still exhibit behavioral and cognitive deficits and cardiac abnormalities that resemble the human condition47. Interestingly, the Dgcr8 heterozygous mouse still showed some of the traits present in DiGeorge patients, such as altered short-term plasticity that has been linked to schizophrenia48. Although it is clear that in this situation the miRNA biogenesis pathway is affected, it would be of great interest to determine how important are the new alternative DGCR8 functions for the origin and development of the disease.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Sara Heras (Medical Research Council Human Genetics Unit, Edinburgh) for discussions and critical reading of the manuscript; Narry Kim (Seoul National University) for the Flag-Drosha, Dominant negative DGCR8 and Drosha expression vectors; Bertrand Seraphin (IGBMC, Strasbourg) for the Dcp1 antibody and Robert Blelloch (University of California, San Francisco) for Dicer KO and Dicer flox/flox cell lines. This work was supported by the Medical Research Council and by the Wellcome Trust (WT084057MA) (A.S., G.M., S.M. and J.F.C.). E.E and M.P. were supported by grants from the Spanish Ministry of Science and by the Sandra Ibarra Foundation (BIO2008-01091, BIO2011-23920 and CSD2009-00080). S.M. was the recipient of an EMBO long-term postdoctoral fellowship. M.P. is supported by the Novo Nordisk Foundation. J.F.C is recipient of a Wellcome Trust Senior Investigator Award (Grant 095518/Z/11/Z).
Footnotes
AUTHOR CONTRIBUTIONS S.M. and J.F.C. conceived, designed, and interpreted the experiments. S.M., G.M. and A.S. performed the experiments and data analysis. M.P. and E.E. provided all the bioinformatics analysis, including mapping of the CLIP-tags to the genome and statistical analysis. J.F.C. supervised the whole project. The manuscript was co-written by all authors.
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
ACCESSION CODES Sequencing raw data for endogenous and overexpressed DGCR8 HITS-CLIP can be found at the Gene Expression Omnibus Database, under accession code (GSE xxx)
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morlando M, et al. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008 doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 8.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 11.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem. Biophys. Res. Commun. 2003;304:184–190. doi: 10.1016/s0006-291x(03)00554-0. [DOI] [PubMed] [Google Scholar]
- 12.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 14.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 15.Davis BN, Hata A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun. Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 17.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 18.Han J, et al. Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 19.Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat. Struct. Mol. Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- 20.Faller M, et al. DGCR8 recognizes primary transcripts of microRNAs through highly cooperative binding and formation of higher-order structures. RNA. 2010;16:1570–1583. doi: 10.1261/rna.2111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanford JR, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadener S, et al. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Triboulet R, Chang HM, LaPierre RJ, Gregory RI. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ule J, Jensen K, Mele A, Darnell RB. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karginov FV, et al. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 34.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 35.Kiss T. SnoRNP biogenesis meets Pre-mRNA splicing. Mol. Cell. 2006;23:775–776. doi: 10.1016/j.molcel.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 36.Taft RJ, et al. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ender C, et al. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein E, et al. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shenoy A, Blelloch R. Genomic analysis suggests that mRNA destabilization by the microprocessor is specialized for the auto-regulation of Dgcr8. PLoS. ONE. 2009;4:e6971. doi: 10.1371/journal.pone.0006971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chong MM, et al. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin YT, Sullivan CS. Expanding the role of Drosha to the regulation of viral gene expression. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11229–11234. doi: 10.1073/pnas.1105799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 44.Liang TJ, Qin CY. The emerging role of microRNAs in immune cell development and differentiation. APMIS. 2009;117:635–643. doi: 10.1111/j.1600-0463.2009.02520.x. [DOI] [PubMed] [Google Scholar]
- 45.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS. Comput. Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp. Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 47.Stark KL, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 48.Fenelon K, et al. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.