Abstract
γ-Glutamyltranspeptidase (EC 2.3.2.2) of Bacillus subtilis, which is an extracellular enzyme, hydrolyzes the γ-glutamyl linkage of glutathione. YwrD, which is homologous to γ-glutamyltranspeptidase, was speculated to have a similar physiological role. It was shown that γ-glutamyltranspeptidase, but not YwrD, is important in utilizing glutathione as the sole sulfur source in Bacillus subtilis.
γ-Glutamyltranspeptidase (GGT) (EC 2.3.2.2), which is widely distributed in living organisms (12), catalyzes the hydrolysis of γ-glutamyl compounds, such as glutathione (GSH), and the transfer of γ-glutamyl moieties to amino acids and peptides. Recently we constructed a Bacillus subtilis strain producing large quantities of GGT and characterized the enzymatic properties of the purified enzyme (6). The nucleotide sequence of the whole genome of B. subtilis 168 has been determined previously (5). From the deduced amino acid sequence of YwrD, Grundy and Henkin speculated that ywrD might encode a second GGT (3). YwrD exhibits 31 and 27% identity with the amino acid sequences of the Escherichia coli K-12 and B. subtilis 168 GGTs, respectively (11, 15). There is no signal sequence in ywrD, so YwrD may be a cytoplasmic enzyme (http://bacillus.genome.ad.jp). However, YwrD has never been characterized.
GSH is the most abundant thiol compound in the cell. It plays important roles in the cell, such as protection from oxidative stress and maintenance of the redox environment. It has been reported that GGT catalyzes the initial step in the cleavage of GSH as a source of cysteine in E. coli (9) and also in some mammals (4). Although B. subtilis GGT is able to catalyze the hydrolysis of GSH (6), it has been reported that Cys− or Gly− mutants of B. subtilis cannot utilize GSH as a source of these amino acids (15). On the other hand, GGT and YwrD were speculated to catalyze the initial step of the cleavage of GSH as a sulfur source in B. subtilis (3), although no experimental evidence has been reported. Although sulfur metabolism has been studied in B. subtilis (1, 8, 13, 14), GSH metabolism has not been clarified (2), and the roles of GGT and YwrD in GSH metabolism have remained to be elucidated. In this study, we analyzed GSH metabolism in ggt and ywrD mutants of B. subtilis.
Construction of ggt and ywrD mutants of B. subtilis.
To investigate the roles of GGT and YwrD, the chromosomal region in B. subtilis corresponding to the ggt or ywrD gene was deleted and replaced with the chloramphenicol acetyltransferase gene (cat) or the tetracycline resistance gene (tet) through a homologous recombination event. The plasmid containing the Δggt::cat or ΔywrD::cat gene was constructed as follows. The plasmid containing the ggt gene, pMH2279, was obtained in our previous study (6). Approximately 800 bp of the C-terminal region of GGT (from the AatI to the BglII site) was removed from this plasmid, and the cat gene from pPL623 (7) (BglII-HaeIII fragment) was inserted in this place.
The plasmid containing the ywrD gene was constructed by a method described previously (6). Chromosomal DNA from B. subtilis 168 was digested with HindIII. DNA fragments (approximately 2.9 kb in size) separated by 0.8% agarose gel electrophoresis were inserted into pMW119 (Nippon Gene Co.) cleaved with HindIII. The 2.9-kb HindIII fragment containing the ywrD gene was screened by colony PCR with two oligonucleotides, BF2 (5′-GACTCAATTGCTCCACCTGCTTTC-3′) and BR2 (5′-GCCTCCACAAGCACGTGATAATAC-3′), complementary to the 1.1-kb nucleotides containing the ywrD sequence deduced from the B. subtilis genome sequence (5). The middle part of YwrD, about 400 bp (from the BbrPI to the NaeI site), was removed from this plasmid, and the cat gene, on the BglII (end-filled)-HaeIII fragment from pPL623, was inserted in this place. As a result, a plasmid in which the ggt or ywrD gene was deleted and replaced with the cat gene was constructed.
The ggt mutant (trpC2 Δggt::cat+) and the ywrD mutant (trpC2 ΔywrD::cat+) were constructed by introducing the linear FspI fragments of these plasmids into strain 168 Marburg (trpC2) by electroporation. For the construction of ggt and ywrD double mutants (trpC2 Δggt::cat+ ΔywrD::tet+), ywrD was deleted as described above and was replaced with the tet gene from pHY300PLK (Takara Shuzo Co.) (DraI-SmaI fragment). The double mutant was constructed by introducing the linear FspI fragment of this plasmid into the ggt mutant. These mutants were viable and had the same phenotype as the wild-type strain in Luria broth and minimal medium (6 mM K2HPO4, 4.5 mM KH2PO4, 0.3 mM trisodium citrate, 0.02% MgSO4 · 7H2O, 0.2% NH4Cl, 0.2% sucrose, 22 mg of FeCl2 · 4H2O/liter, 50 mg of Trp/liter). Because the expression of B. subtilis GGT is repressed by glucose (15), sucrose was used as a carbon source, which did not repress the expression of GGT (data not shown). When GSH was used as a sulfur source, it was added to a final concentration of 250 μM and MgSO4 was replaced with MgCl2.
Growth of B. subtilis in minimal medium with GSH.
GSH can be utilized as a sulfur source in B. subtilis (2, 14). The growth of the wild type and the ggt, ywrD, and double mutants was analyzed in minimal medium with sulfate or GSH as a sulfur source. For analysis of growth, exponentially growing cells were used as the inoculum and various culture conditions were examined. The cells were grown in minimal medium with sulfate, washed twice with sterile water, and reinoculated into minimal medium with sulfate or GSH. Growth was monitored by measuring the absorbance at 600 nm.
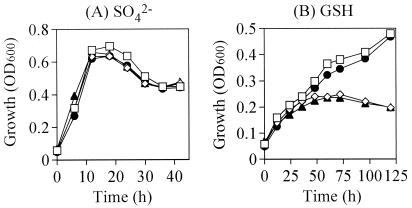
With sulfate as a sulfur source, the growth of the mutants was the same as that of the wild type (Fig. 1A). With GSH as a sulfur source, the growth of ggt and double mutants was dramatically reduced compared to that of the wild type and ywrD mutant (Fig. 1B). Our findings suggested that extracellular GGT catalyzes the hydrolysis of the γ-glutamyl linkage of exogenous GSH and then cysteinylglycine is utilized as a sulfur source in the cell. The growth of the wild type and ywrD mutant on GSH was slower than that on sulfate (Fig. 1). It was caused by the poor hydrolysis of GSH in exponential phase, because GGT of B. subtilis is synthesized during the mid-stationary phase (15).
FIG. 1.
The growth profiles of B. subtilis, the parental strain (open squares), ggt mutant (open diamonds), ywrD mutant (filled circles), and double mutant (filled triangles) in minimal medium with sulfate (A) or GSH (B) as a sulfur source. The growth was monitored by measuring the optical density at 600 nm (OD600). The experiments were repeated three times with essentially the same results, and the data for a representative experiment is shown.
ggt mutants grew very poorly in the minimal medium containing GSH (Fig. 1B). It was possible that exogenous GSH underwent natural decomposition or was cleaved and transported into the cell by tripeptidases or tripeptide transporters with an affinity for GSH, although such tripeptidases or tripeptide transporters have not yet been identified in B. subtilis. Even if there were such tripeptidases or tripeptide transporters, they would have little effect on GSH metabolism. Without GGT, B. subtilis was not able to utilize GSH as a sulfur source effectively or to grow fully in minimal medium with GSH.
On the other hand, the growth of the ywrD mutant was the same as that of the wild type, and growth of the ggt mutant and that of the ggt ywrD double mutant were the same (Fig. 1B). These findings suggested that YwrD is not involved in the metabolism of GSH, which is consistent with our finding that the cell extract of the ggt mutant of B. subtilis expressing YwrD from the ywrD gene on the pHY300PLK plasmid had no detectable GSH hydrolysis activity (data not shown). To concentrate and amplify the activity of YwrD, YwrD with a histidine tag was purified. The maltose binding protein tag on the N-terminal amino acid of the large subunit had no effect on the activity of E. coli GGT (10). The histidine tag was attached to the N-terminal amino acid of YwrD. The purified YwrD with histidine tag, which consisted of a 57-kDa subunit, could not hydrolyze GSH (data not shown).
When glucose was used as the carbon source, the growth of the wild type and ywrD mutant was reduced compared to that seen when sucrose was used as the carbon source (data not shown). This may be because the expression of B. subtilis GGT is repressed by glucose (15). The various culture conditions, poor or rich nutrient state, were examined. Although the level of growth was different in each culture condition, the growth of ggt and double mutants was reduced compared to that of the wild type and ywrD mutant with GSH as a sulfur source (data not shown).
It can thus be concluded that GGT, but not YwrD, is important in the utilization of extracellular glutathione as a sulfur source in B. subtilis.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research, grant 15580061 to H.S. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by research funds to H.S. from Nagase Science and Technology Foundation.
REFERENCES
- 1.Auger, S., A. Danchin, and I. Martin-Verstraete. 2002. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coppée, J.-Y., S. Auger, E. Turlin, A. Sekowska, J. P. Le Caer, V. Labas, V. Vagner, A. Danchin, and I. Martin-Verstraete. 2001. Sulfur-limitation-regulated proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 147:1631-1640. [DOI] [PubMed] [Google Scholar]
- 3.Grundy, F. J., and T. M. Henkin. 2002. Synthesis of serine, glycine, cysteine, and methionine, p. 245-254. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. American Society for Microbiology, Washington, D.C.
- 4.Hanigen, M. H., and W. A. Ricketts. 1993. Extracellular glutathione is a source of cysteine for cells that express γ-glutamyl transpeptidase. Biochemistry 32:6302-6306. [DOI] [PubMed] [Google Scholar]
- 5.Kunst, F., N. Ogasawara, I. Moszer, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 6.Minami, H., H. Suzuki, and H. Kumagai. 2003. Salt-tolerant γ-glutamyltranspeptidase from Bacillus subtilis 168 with glutaminase activity. Enzyme Microb. Technol. 32:431-438. [Google Scholar]
- 7.Schoner, R. G., D. M. Williams, and P. S. Lovett. 1983. Enhanced expression of mouse dihydrofolate reductase in Bacillus subtilis. Gene 22:47-57. [DOI] [PubMed] [Google Scholar]
- 8.Sekowska, A., S. Robin, J.-J. Daudin, A. Hénaut, and A. Danchin. 2001. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2:0019.1-0019.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki, H., W. Hashimoto, and H. Kumagai. 1993. Escherichia coli K-12 can utilize an exogenous γ-glutamyl peptide as an amino acid source, for which γ-glutamyltranspeptidase is essential. J. Bacteriol. 175:6038-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki, H., and H. Kumagai. 2002. Autocatalytic processing of γ-glutamyltranspeptidase. J. Biol. Chem. 277:43536-43543. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki, H., H. Kumagai, T. Echigo, and T. Tochikura. 1989. DNA sequence of the Escherichia coli K-12 γ-glutamyltranspeptidase gene, ggt. J. Bacteriol. 171:5169-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tate, S. S., and A. Meister. 1981. γ-Glutamyl transpeptidase: catalytic, structural and functional aspects. Mol. Cell. Biochem. 39:357-368. [DOI] [PubMed] [Google Scholar]
- 13.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Expression of the Bacillus subtilis sulphonate-sulphur utilization genes is regulated at the levels of transcription initiation and termination. Mol. Microbiol. 39:1356-1365. [PubMed] [Google Scholar]
- 14.van der Ploeg, J. R., N. J. Cummings, T. Leisinger, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]
- 15.Xu, K., and M. A. Strauch. 1996. Identification, sequence, and expression of the gene encoding γ-glutamyltranspeptidase in Bacillus subtilis. J. Bacteriol. 178:4319-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]