Abstract
Shigella are human-adapted Escherichia coli that have gained the ability to invade the human gut mucosa and cause dysentery1,2, spreading efficiently via low-dose fecal-oral transmission3,4. Historically, S. sonnei has been predominantly responsible for dysentery in developed countries, but is now emerging as a problem in the developing world, apparently replacing the more diverse S. flexneri in areas undergoing economic development and improvements in water quality4-6. Classical approaches have shown S. sonnei is genetically conserved and clonal7. We report here whole-genome sequencing of 132 globally-distributed isolates. Our phylogenetic analysis shows that the current S. sonnei population descends from a common ancestor that existed less than 500 years ago and has diversified into several distinct lineages with unique characteristics. Our analysis suggests the majority of this diversification occurred in Europe, followed by more recent establishment of local pathogen populations in other continents predominantly due to the pandemic spread of a single, rapidly-evolving, multidrug resistant lineage.
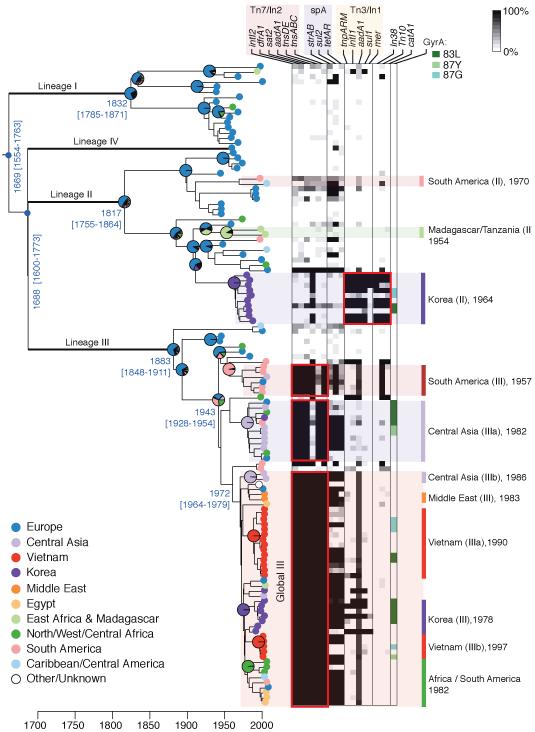
To establish an accurate population framework we sequenced the whole genomes of 132 S. sonnei isolated between 1943 and 2008, spanning four continents (Supplementary Table 1). We detected 10,111 chromosomal single nucleotide polymorphisms (SNPs) randomly distributed around the S. sonnei chromosome, approximately one per 430 bp (0.23% nucleotide divergence) (Supplementary Fig. 1). To investigate the population structure of S. sonnei, we analysed these chromosomal SNPs using multiple phylogenetic methods. Maximum likelihood (ML) phylogenetic analysis (Supplementary Fig. 2) revealed a strong correlation between root-to-tip branch lengths and the known dates of isolation for the sequenced S. sonnei, indicative of rapid, clock-like evolution (Supplementary Fig. 3). There appears to be some rate variation between lineages, possibly associated with differences in effective population size or in the mean number of generations per year (replication rate), which may in turn be associated with different lifestyles or niches. We used a Bayesian approach (BEAST8) to infer the evolutionary dynamics of the global S. sonnei population as a whole. Importantly, this yielded the same tree topology as the ML analysis, while also providing estimates of nucleotide substitution rates and divergence times for key S. sonnei lineages (Fig. 1). Interestingly, the phylogenies identified four distinct S. sonnei lineages, three encompassing isolates spanning the 1940s through the 2000s and another comprising a single isolate from France (Fig. 1). These lineages each had 100% ML bootstrap support, 100% Bayesian posterior support (BEAST) and were also recovered using a Bayesian clustering analysis (see Online Methods). Whilst these lineages are uniquely characterized by hundreds of SNPs they display only minor differences in gene content and were correlated with traditional typing methods used to subdivide S. sonnei (biotypes a-g9 and CRISPR types10) (Supplementary Note, Supplementary Fig. 2, Supplementary Table 3). We estimated a mean substitution rate of 2.0 × 10−4 site−1 year−1 among the 10,111 chromosomal SNP loci [95% Highest Posterior Density (HPD) 1.6 × 10−4 – 2.3 × 10−4], corresponding to the accumulation of approximately 2.2 SNPs chromosome−1 year−1 ([95% HPD 1.8 – 2.6], excluding repeated and phage regions). This scales to a genome-wide substitution rate of 6.0 × 10−7 substitutions site−1 year−1 [95% HPD = 5.2 × 10−7 - 6.7 × 10−7], which likely represents the upper bound of the true genome-wide substitution rate and is similar to that calculated for the enteric pathogen Vibrio cholerae (8 × 10−7 site−1 year−1)11 but lies between the rates estimated for Yersinia pestis (2 × 10−8)12 and Staphylococcus aureus (3 × 10−6)13. From BEAST analysis, we estimated the most recent common ancestor (MRCA) of all contemporary S. sonnei existed less than 500 years ago [median calendar year for divergence date, 1669; 95% HPD, 1554 - 1763] (Fig. 1). Similarly, we estimate the MRCA for each of Lineages I and II existed in the early 19th century and that all Lineage III isolates descend from a hypothetical ancestor that existed around the turn of the 20th century (Fig. 1). Critically, these data indicate that though the extant S. sonnei population descends from a single ancestor existing in the 17th century, by the late 19th century S. sonnei had become segregated into at least four distinct lineages that still persist today.
Figure 1. Bayesian maximum clade credibility phylogeny for S. sonnei.
Branches defining major lineages in bold (each had 100% posterior support); pie charts indicate ML estimates for geographic origin of major nodes, according to inset legend (lower left). Time (x-axis) is relative to the Common Era; divergence dates (median estimate and 95% HPD) are given in blue for major nodes. Distribution of antimicrobial resistance determinants is indicated in the heatmap according to the legends provided, which reflect percentage of bases in each gene sequence that are covered by reads from each isolate (top right). Geographically localised clonal expansions are highlighted on the right, labeled with their median estimated divergence date.
There was strong evidence for regional clustering of S. sonnei within the phylogenetic tree (Fig. 1), indicating significant geographic structure in the global bacterial population (p <1×10−5 for association between phylogeny and geographic region14). Interestingly, the European population shows the richest diversity, with isolates distributed across all four lineages (31% lineage I, 35% lineage II, 31% lineage III, sole lineage IV isolate) and occupying basal branches in each lineage (Fig. 1). In contrast, S. sonnei isolates from Asia, Africa and America were mainly from lineage III (67-77%) with fewer lineage II representatives (22-26%) and just two from Lineage I. Furthermore, ancestral state reconstruction analysis indicated a >50% likelihood of a European common ancestor for each of the lineages I, II and III (Fig. 1). The data also indicate Lineage III has been more successful at global dispersal than other lineages, with only low numbers of Lineage I or II detected outside Europe (Fig. 1). In particular, a recently derived clade within Lineage III (Global III, MRCA = 1972 [95% HPD = 1964-1979 C.E.]) has been particularly successful at global dissemination, comprising 49% of all isolates sampled since 1995 and detected in all regions represented in our collection (Fig. 1). Unlike the European isolates, isolates from non-European countries form tight shallow-rooted phylogenetic clusters, consistent with and suggestive of contemporary dispersal (Fig. 1). In many cases, these clusters contain multiple isolates from the same country, indicating localized clonal expansions (Fig. 1). For example, isolates from Korea formed two subclades within lineages II and III that likely represent separate introductions of S. sonnei into Korea during the 1960s and 1970s, each followed by local clonal expansions (Fig. 1). Similarly, isolates originating in Vietnam form two subclades, indicating the local establishment of Lineage III clones in Vietnam in the 1990s (Fig. 1). At a regional level, there appears to have been an establishment of a Lineage III subclade in South America during the 1950s to which isolates from Brazil and Peru could be traced, followed by dissemination of the Global III clade into Africa and America in the early 1980s (Fig. 1).
Critically, the phylogeographic analysis indicates that all contemporary S. sonnei infections are caused by a small number of clones that have recently become globally dispersed (Fig. 1). The distribution of antimicrobial resistance genes and mutations within the S. sonnei phylogeny suggest that selection for multiple drug resistance (MDR) played a pivotal role in driving this global dissemination (Fig. 1, Supplementary Fig. 2, Supplementary Table 1). In particular, the establishment of local S. sonnei Lineage III populations outside Europe is intimately associated with the carriage of transposon Tn7 and class II integrons (In2) encoding resistance to multiple antimicrobials (Fig. 1). All three major Lineage III subgroups carry a distinct In2 variant, which is either plasmid-encoded (South America III) or integrated into the chromosome adjacent to glmS (Central Asia IIIa, Global III), suggesting independent acquisitions of the integron in each group during the 1960s-1970s followed by clonal expansion and subsequent international spread (Fig. 1). Studies from Europe, Asia, Africa, South America and Australia have reported a high prevalence of In2-bearing, MDR, biotype g S. sonnei, often associated with local epidemics15. Our data demonstrate biotype g is a marker for Lineage III due to a conserved nonsense mutation in rhamnose regulatory gene rhaR (Supplementary Fig. 2) and indicate that the global distribution of MDR biotype g/In2 S. sonnei is the result of global dissemination of multiple In2-bearing subclades of Lineage III S. sonnei. Half of the In2-bearing Lineage III isolates also harboured the small MDR plasmid spA2 containing tetAR, strAB and sul2 genes, which confer additional resistance to tetracycline, streptomycin and sulfonamides (Fig. 1). All quinolone resistant isolates harboured one of three point mutations in the chromosomal DNA gyrase gene, gyrA, known to confer quinolone resistance (Fig. 1, Supplementary Table 1; we detected no plasmid-mediated quinolone resistance genes). The distribution of gyrA mutations within the phylogeny shows these resistance mutations have arisen independently on at least nine occasions among our S. sonnei collection, including two separate mutations within the clonal group Korea II, indicative of surprisingly strong selection for quinolone resistance even among MDR isolates (Fig. 1). To investigate other signals of selection, we examined the clustering of SNPs within genes and chromosomal regions (Supplementary Note). We found evidence of phage and transposase insertions and a single case of homologous recombination affecting the sitABCD operon in isolate 31382, but identified only two genes displaying amino acid variation significantly higher than expected under a random distribution of SNPs. Neither of these genes (rpoS and mreB) encodes an extracellular protein, suggesting a lack of immune selection, in common with another human restricted pathogen Salmonella Typhi (typhoid fever)16. However, we detected a large number of nonsynonymous SNPs (nsSNPs) and a high rate of nonsyonymous to synonymous substitutions per site (dN/dS) in the drug efflux pump component genes acrD (8 nsSNPs, dN/dS =2.5) and acrB (12 nsSNPs, dN/dS =1.8). Currently, antimicrobial treatment is recommended for the management of dysentery17, but may not significantly impact the resolution of S. sonnei or S. flexneri infections18,19. However, there is evidence such treatment can prevent shedding of S. sonnei after the resolution of symptoms20. Thus, while antimicrobial resistance may have only minor implications for dysentery treatment, this phenotype may be important in sustaining S. sonnei transmission within human populations and our data indicates there is a strong selective pressure for its maintenance. It has been hypothesized that free-living amoebae may represent an environmental reservoir for Shigella, which are able to survive intracellularly within Acanthamoeba21,22. This could potentially provide another niche in which selective pressure for antibiotic resistance may be exerted, although intracellular Shigella are likely to be protected from most antibiotics by their amoebae hosts23,24.
Previous studies have proposed that the acquisition of virulence plasmid pINV B, encoding the Plesiomonas shigelloides related O antigen, was the defining event in the emergence of S. sonnei25. Unfortunately, the S. sonnei virulence plasmid is highly unstable on laboratory media and is commonly lost on sub-culturing26 and, as a consequence, less than half of our isolates yielded sufficient virulence plasmid sequence data for analysis (46 isolates with >10x read depth). Phylogenetic analysis of the available virulence plasmid sequences (which contained 84 SNPs) identified three distinct lineages (Supplementary Fig. 4). There was a parallel relationship between chromosomal and plasmid lineages, consistent with co-evolution of the plasmid and host chromosome, stable maintenance of the plasmid in the natural environment and no transfer of plasmid variants among host bacteria. It has also been proposed that exposure to P. shigelloides via contaminated water protects humans from S. sonnei infection5 as the O antigens are indistinguishable and cross-react27,28. This may explain increases in S. sonnei incidence following economic development and water quality improvements, as the result of a decline in passive cross-protection by environmental immunization with P. shigelloides. If this cross-protection acts as a barrier to the establishment of S. sonnei in human populations, one would predict that S. sonnei infections would gradually increase following improvements in water quality, and that the geographical expansion of S. sonnei will be characterized by the introduction and expansion of novel clones moving into human populations with falling natural immunity previously obtained from exposure to P. shigelloides. Our model of recent dissemination out of Europe is remarkably consistent with these hypotheses. Transmission of S. sonnei into other continents has likely occurred sporadically over centuries through human migration, trade and travel; however the establishment of local S. sonnei populations – which we would observe as geographically clustered clonal groups outside Europe – is not evident until the last few decades.
Our findings have major implications for global public health and diarrheal infections. Improvement of drinking water, one of the Millenium Development Goals, is an undeniably important aim and is expected to reduce morbidity and mortality due to a diverse array of waterborne diseases. However, we predict that fulfilling this aim will produce a concurrent increase in S. sonnei dysentery incidence in transitional countries. The combination of increased incidence and excessive antimicrobial resistance among globally disseminated S. sonnei indicates an anti-S. sonnei vaccine will be increasingly important for the control and long-term prevention of dysentery and associated morbidity and mortality. A suitable vaccine is an achievable goal, since all S. sonnei share a single O antigen that has proven to be a successful vaccine target29. Interestingly, the success of S. sonnei in the face of diminishing S. flexneri incidence suggests important epidemiological distinctions in transmission of the two pathogens. S. sonnei outbreaks have been associated with schools, care facilities, contaminated food and insects moving between fecal waste and food preparation areas30-32. These modes of transmission are considerably more direct than waterborne transmission and may explain the persistence of S. sonnei even when water infrastructure is improved, implying that vaccination and improved hygiene standards will be pivotal in eliminating S. sonnei infections in industrializing countries.
Online Methods
Bacterial isolates and sequencing
Bacterial isolates analysed in this study are detailed in Supplementary Table 1. DNA was prepared using the Wizard Genomic DNA Kit (Promega, Madison, WI) or phenol extraction. Index-tagged paired end Illumina sequencing libraries were prepared using one of 12 unique indexing tags as previously described13. These were combined into pools each containing 11-12 uniquely tagged libraries and sequenced on the Illumina Genome Analyzer GAII according to manufacturer’s protocols to generate tagged 54 bp paired-end reads.
Read alignment and SNP detection
Reads from each isolate were mapped to the S. sonnei reference genome (strain Ss046 chromosome, NC_007384; strain Ss046 plasmids, NC_007385, NC_009347, NC_009346, NC_009345; plasmid pEG356, NC_013727) using BWA33 with default parameters. Average read depths are given in Supplementary Table 1. SNPs were identified using SamTools34. SNPs in the previously sequenced S. sonnei strain 53G were identified using the same mapping procedure to analyse reads simulated from the finished genome (chromosome: HE616528; plasmids: HE616529, HE616530, HE616531 and HE616532) using SamTools’ wgsim algorithm. SNPs called in phage regions or repetitive sequences (10.2% of bases and 15.5% of genes in the Ss046 reference chromosome) were excluded16, resulting in a final set of 10,111 chromosomal SNP loci. The allele at each locus in each isolate was determined by reference to the consensus base in that genome (using SamTools pileup and removing low confidence alleles with consensus base quality ≤20, read depth ≤5 or a heterozygous base call).
The SNP calling procedure was repeated using S. sonnei 53G (Lineage II) as the reference for mapping. This resulted in an identical tree topology with near-identical branch lengths (Pearson correlation coefficient = 0.995, p<1×10−15), demonstrating the robustness of the method and its independence from the choice of reference genome. The Ss046-mapped data was used for all analyses reported, since the Ss046 genome has been widely used in previous comparative studies while the 53G genome is reported here for the first time.
The same procedures were followed to identify SNPs in the invasion plasmid. The analysis was restricted to strains with a mean plasmid read depth of ≥10x and the 137 kbp of non-repetitive plasmid sequence (63% of the S. sonnei pSs046 reference plasmid sequence).
Alleles in outgroup genomes were determined using the same approach to analyse reads simulated from other Shigella and E. coli reference genomes (Supplementary Table 2) using wgsim (distributed with SamTools).
Phylogenetic and temporal analyses
Chromosomal SNP alleles were concatenated for each strain to generate a multiple alignment of all SNPs (where high confidence base calls could not be determined, the allele was recorded as a gap character). Clusters of SNPs introduced via horizontal transfer (see SNP distribution section below) were removed from the alignment. The resulting alignment was further filtered to remove loci at which alleles were unknown for >40% of isolates (indicating the site is not conserved) and an ML phylogeny was estimated using RAxML35. The BEAST package8 was utilized for the Bayesian inference of phylogeny and divergence dates. Additionally, we used the BAPS program (Bayesian Analysis of Population Structure)36 to examine clustering of isolates based on SNP data.
For ML analysis, RAxML was run ten times using the generalized time-reversible model with a Γ distribution to model site-specific rate variation (i.e., the GTR+Γ substitution model; GTRGAMMA in RAxML). 1000 bootstrap pseudo-replicate analyses were performed to assess support for the ML phylogeny. The final result (Supplementary Fig. 2) is the tree with the highest likelihood across all ten runs, with ML estimates of branch length and confidence in major bipartitions calculated using the bootstrap values across all runs. This phylogeny was rooted using E. coli and Shigella outgroups (Supplementary Table 2).
Root-to-tip branches were extracted from the ML tree using the program TreeStat (see URLs). The relationship between root-to-tip distances, year of isolation and lineage were analysed using linear regression. Plots and regression lines are shown in Supplementary Figure 3, along with Pearson correlation coefficients.
For BEAST analysis, we also used the GTR+Γ substitution model and defined tip dates as the year of isolation (restricting the analysis to those sequences with recorded dates). We performed multiple analyses using both constant size and Bayesian skyline demographic models, in combination with either a strict molecular clock or a relaxed clock (uncorrelated lognormal distribution). BEAST (v1.6) uses a Markov chain Monte Carlo (MCMC) method for sampling the posterior probability distributions. Analyses of all model combinations (demographic and clock) were performed using ten chains of 100 million generations each to ensure convergence, with samples taken every 1,000 MCMC generations. Parameters were estimated after combining all replicate analyses, totaling 900 million MCMC generations post-burnin, with all reported parameter estimates (i.e., medians and 95% Highest Probability Densities – HPDs) calculated using the program Tracer v1.5. The relaxed clock models provided much better fit to the data (Bayes Factor > 100; using the harmonic mean estimator of the marginal likelihood) and the standard deviation of inferred substitution rates across branches was 0.45 [95% HPD = 0.38 - 0.52], providing additional strong support for a relaxed molecular clock. Bayesian skyline plots indicated a constant population size through time and estimates under a constant population model yielded very similar results to that under a Bayesian skyline model. Therefore, all parameter estimates quoted are from analyses using relaxed clock and Bayesian skyline demographic models. To test the validity of the temporal signal in the data, we performed 20 additional BEAST runs (of 200 million MCMC generations each) with identical substitution (GTR+Γ), clock (relaxed), and demographic (Bayesian skyline) models, but with randomized tip dates (Supplementary Fig. 5). This randomization procedure produces a null set of tipdate and sequence correlations that may be analysed to produce null substitution rate distributions, which can then be compared with empirical rate estimates.
Phylogeographic analysis
The geographic region of isolation of each S. sonnei was analysed as a discrete character trait using two complementary methods. Phylogeographic analyses were performed using the 126 isolates which had complete information on both year and geographic region of isolation (see Supplementary Table 1). First, the association between the phylogenetic relationships of S. sonnei isolates (inferred by BEAST) and their geographic region of isolation was tested using the Bayesian Tip-Significance software (BaTS14). A random selection of 50,000 trees sampled during the Bayesian phylogenetic analysis described above were used as input, and 1,000 randomizations were used to generate a null distribution for significance testing. Second, ancestral state reconstruction of the geographic origin of hypothetical common ancestors (i.e., internal nodes in the phylogeny) was performed using the ‘ace’ function implemented in the ‘ape’ package for R37. The percent probability estimates quoted, and illustrated by pie charts in Figure 1, are scaled likelihoods for the discrete character trait (i.e., region of isolation) at each node.
Gene content analysis
Each read set was assembled using the de novo short read assembler Velvet38 and Velvet Optimiser (see URLs). Contigs less than 100 bp in size were excluded from further analysis. The S. sonnei 53G genome (chromosome: HE616528; plasmids: HE616529, HE616530, HE616531 and HE616532) and de novo assembled contig sets were mapped iteratively to the pan-genome reference set (initialized as the concatenation of S. sonnei Ss046 chromosome, NC_007384; Ss046 plasmids, NC_007385, NC_009347, NC_009346, NC_009345; plasmid pEG356, NC_013727) using MUMmer (nucmer algorithm)39. At each iteration i, sequences not aligning to the current pan-genome Pi-1 set were incorporated into an extended pan-genome, Pi. The final pan-genome, P, was annotated using a combination of annotation transfer (for S. sonnei reference sequences) and de novo annotation using the RAST annotation server40 for novel sequences assembled from reads. The latter included 1.67 Mbp of sequence in 862 contigs, in which 2,422 genes were annotated (incorporating 80.5% of bases), resulting in a total of 6,852 genes.
S. sonnei read sets were then aligned to the pan-genome using BWA27 with default mapping parameters. A pileup was generated for each aligned read set using SamTools28 and used to summarize, for each annotated gene in the pan-genome P, the coverage (% of bases covered) and presence of inactivating mutations (nonsense SNPs or non-triplet indels resulting in frameshifts) in each genome. The results were used to identify genes whose presence or inactivation was associated with specific lineages (Supplementary Note, Supplementary Fig. 6).
Resistance gene analysis
The presence of resistance genes was initially determined from mapping data described above. The genetic context of resistance genes was examined by blastn search of each contig set with known resistance, transposase or integrase genes as query sequences. The resulting contigs were compared to the NCBI non-redundant nucleotide database to annotate the resistance genes and mobile elements. Mapping was then repeated using annotated mobile elements to generate the gene coverage maps shown in Figure 1 and Supplementary Figure 2, which indicate the proportion of bases in each gene sequence that are covered by reads from each isolate (reference sequences are provided in Supplementary Fig. 2).
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust (#0689); and a Victorian Life Sciences Computation Initiative (VLSCI) grant (#VR0082) on its Peak Computing Facility at the University of Melbourne, an initiative of the Victorian Government, Australia. KEH was supported by a Fellowship from the NHMRC of Australia (#628930); SB is supported by an OAK Foundation Fellowship through Oxford University (#OAKF9) and the Li Ka Shing foundation (#LG13); FXW was partially funded by the Institut de Veille Sanitaire; JY was supported by a MRC grant (#G0800173); DWK was partially supported by grant RTI05-01-01 from the Ministry of Knowledge and Economy (MKE). We thank Myron Levine (University of Maryland School of Medicine, Center for Vaccine Development, USA) and Christoph Tang (University of Oxford, UK) for their kind gift of Shigella sonnei strain 53G.
Footnotes
URLs
Illumina sequence data provided at http://www.ebi.ac.uk/ena/data/view/ERP000182
TreeStat: http://tree.bio.ed.ac.uk/software/treestat/
Velvet Optimiser: http://www.ebi.ac.uk/~zerbino/velvet/
Author Contributions
KEH, NRT, ECH and AK analysed the data and performed phylogenetic analysis. NRT, GD, JY, SB, JJF, KEH and JP were involved in the study design. FXW, DJB, JEC, JY, VS, DWK, SYC, SHK, WDS and DJP were involved in isolate collection, DNA analysis and resistance phenotyping. KEH, SB, NRT, GD, AK, ECH and FXW contributed to the manuscript writing.
Accession Numbers
The finished genome of S. sonnei 53G is available under EMBL accessions HE616528 (chromosome) and HE616529, HE616530, HE616531 and HE616532 (plasmids). Sequence reads for the 132 Illumina-sequenced S. sonnei are deposited in the European Nucleotide Archive under accession ERP000182.
The authors declare no competing financial interests.
References
- 1.Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA. 2000;97:10567–72. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang F, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–58. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DuPont HL, Levine MM, Hornick RB, Formal SB. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 1989;159:1126–8. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 4.Kotloff KL, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651–66. [PMC free article] [PubMed] [Google Scholar]
- 5.Sack DA, Hoque AT, Huq A, Etheridge M. Is protection against shigellosis induced by natural infection with Plesiomonas shigelloides? Lancet. 1994;343:1413–5. doi: 10.1016/s0140-6736(94)92531-3. [DOI] [PubMed] [Google Scholar]
- 6.Vinh H, et al. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis. 2009;9:204. doi: 10.1186/1471-2334-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karaolis DK, Lan R, Reeves PR. Sequence variation in Shigella sonnei (Sonnei), a pathogenic clone of Escherichia coli, over four continents and 41 years. J Clin Microbiol. 1994;32:796–802. doi: 10.1128/jcm.32.3.796-802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nastasi A, Pignato S, Mammina C, Giammanco G. rRNA gene restriction patterns and biotypes of Shigella sonnei. Epidemiol Infect. 1993;110:23–30. doi: 10.1017/s0950268800050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Touchon M, et al. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J Bacteriol. 2011;193:2460–7. doi: 10.1128/JB.01307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–5. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morelli G, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42:1140–3. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol. 2008;8:239–46. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Ranjbar R, et al. Genetic relatedness among isolates of Shigella sonnei carrying class 2 integrons in Tehran, Iran, 2002-2003. BMC Infect Dis. 2007;7:62. doi: 10.1186/1471-2334-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–93. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae 1. WHO Document Production Services; Geneva, Switzerland: 2005. [Google Scholar]
- 18.Christopher PR, David KV, John SM, Sankarapandian V. Antibiotic therapy for Shigella dysentery. Cochrane Database of Systematic Reviews. 2010 doi: 10.1002/14651858.CD006784.pub3. CD006784. [DOI] [PubMed] [Google Scholar]
- 19.Vinh H, et al. A multi-center randomized trial to assess the efficacy of gatifloxacin versus ciprofloxacin for the treatment of shigellosis in Vietnamese children. PLoS Negl Trop Dis. 2011;5:e1264. doi: 10.1371/journal.pntd.0001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vinh H, et al. Treatment of bacillary dysentery in Vietnamese children: two doses of ofloxacin versus 5-days nalidixic acid. Trans Royal Soc Trop Med Hyg. 2000;94:323–6. doi: 10.1016/s0035-9203(00)90343-2. [DOI] [PubMed] [Google Scholar]
- 21.Jeong HJ, et al. Acanthamoeba: could it be an environmental host of Shigella? Exp Parasitol. 2007;115:181–6. doi: 10.1016/j.exppara.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Saeed A, Abd H, Edvinsson B, Sandstrom G. Acanthamoeba castellanii an environmental host for Shigella dysenteriae and Shigella sonnei. Arch Microbiol. 2009;191:83–8. doi: 10.1007/s00203-008-0422-2. [DOI] [PubMed] [Google Scholar]
- 23.Winiecka-Krusnell J, Linder E. Free-living amoebae protecting Legionella in water: the tip of an iceberg? Scand J Infect Dis. 1999;31:383–5. doi: 10.1080/00365549950163833. [DOI] [PubMed] [Google Scholar]
- 24.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–33. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shepherd JG, Wang L, Reeves PR. Comparison of O-antigen gene clusters of Escherichia coli (Shigella) sonnei and Plesiomonas shigelloides O17: sonnei gained its current plasmid-borne O-antigen genes from P. shigelloides in a recent event. Infect Immunity. 2000;68:6056–61. doi: 10.1128/iai.68.10.6056-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sansonetti PJ, Kopecko DJ, Formal SB. Shigella sonnei plasmids: evidence that a large plasmid is necessary for virulence. Infect Immunity. 1981;34:75–83. doi: 10.1128/iai.34.1.75-83.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Verg LL, Herrington DA, Boslego J, Lindberg AA, Levine MM. Age-specific prevalence of serum antibodies to the invasion plasmid and lipopolysaccharide antigens of Shigella species in Chilean and North American populations. J Infect Dis. 1992;166:158–61. doi: 10.1093/infdis/166.1.158. [DOI] [PubMed] [Google Scholar]
- 28.Shimada T, Sakazaki R. On the serology of Plesiomonas shigelloides. Jap J Med Science Biol. 1978;31:135–42. doi: 10.7883/yoken1952.31.135. [DOI] [PubMed] [Google Scholar]
- 29.Kaminski RW, Oaks EV. Inactivated and subunit vaccines to prevent shigellosis. Exp Review Vaccines. 2009;8:1693–704. doi: 10.1586/erv.09.127. [DOI] [PubMed] [Google Scholar]
- 30.Genobile D, et al. An outbreak of shigellosis in a child care centre. Communicable Dis Intell. 2004;28:225–9. [PubMed] [Google Scholar]
- 31.Lewis HC, et al. Outbreaks of Shigella sonnei infections in Denmark and Australia linked to consumption of imported raw baby corn. Epidemiol Infect. 2009;137:326–34. doi: 10.1017/S0950268808001829. [DOI] [PubMed] [Google Scholar]
- 32.Cohen D, et al. Reduction of transmission of shigellosis by control of houseflies (Musca domestica) Lancet. 1991;337:993–7. doi: 10.1016/0140-6736(91)92657-n. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 36.Tang J, Hanage WP, Fraser C, Corander J. Identifying currents in the gene pool for bacterial populations using an integrative approach. PLoS Comp Biol. 2009;5:e1000455. doi: 10.1371/journal.pcbi.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 38.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 42.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–4. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.