Abstract
Non-coding RNAs (ncRNAs) are involved in an increasing number of cellular events1. Some ncRNAs are processed by DICER and DROSHA ribonucleases to give rise to small double-stranded RNAs involved in RNA interference (RNAi)2. The DNA-damage response (DDR) is a signaling pathway that originates from the DNA lesion and arrests cell proliferation3. So far, DICER or DROSHA RNA products have not been reported to control DDR activation. Here we show that DICER and DROSHA, but not downstream elements of the RNAi pathway, are necessary to activate DDR upon oncogene-induced genotoxic stress and exogenous DNA damage, as studied also by DDR foci formation in mammalian cells and zebrafish and by checkpoint assays. DDR foci are sensitive to RNase A treatment, and DICER- and DROSHA-dependent RNA products are required to restore DDR foci in treated cells. Through RNA deep sequencing and studies of DDR activation at an inducible unique DNA double-strand break (DSB), we demonstrate that DDR foci formation requires site-specific DICER- and DROSHA-dependent small RNAs, named DDRNAs, which act in a MRE11-RAD50-NBS1 (MRN) complex-dependent manner. Chemically synthesized or in vitro-generated by DICER cleavage, DDRNAs are sufficient to restore DDR in RNase A-treated cells, also in the absence of other cellular RNAs. Our results describe an unanticipated direct role of a novel class of ncRNAs in the control of DDR activation at sites of DNA damage.
Keywords: DICER, DROSHA, small non coding RNAs, DNA damage response (DDR), ATM, cellular senescence, zebrafish
Mammalian genomes are pervasively transcribed with most transcripts apparently not associated with coding functions4,5. An increasing number of ncRNAs have been shown to play a variety of relevant cellular functions, often with estimated very low expression levels6,7,8. DICER and DROSHA are two RNase type III enzymes that process ncRNAs hairpin structures to generate small double-stranded RNAs9 (see Introduction in SI).
Detection of a DSB triggers the kinase activity of ATM, which initiates a cascade by phosphorylating the histone variant H2AX (γH2AX) at the DNA damage site and recruiting additional DDR factors. This establishes a local self-feeding loop that leads to accumulation of upstream DDR factors in the form of cytologically-detectable foci at damaged DNA sites3,10. DDR has been considered exclusively a protein-made signaling cascade, with no direct contributions of RNA species to its activation. Oncogene-induced senescence (OIS) is a non-proliferative state characterized by a sustained DDR11 and senescence-associated heterochromatic foci (SAHF)12. Since ncRNAs participate in heterochromatin formation13, we investigated whether they could control SAHF and OIS. We used small interfering RNAs (siRNA) to knock down DICER or DROSHA in OIS cells and monitored SAHF and cell-cycle progression. Knockdown of either DICER or DROSHA, as well as ATM as control14, restored DNA replication and entry into mitosis (Figures S1, S2); we did not detect overt SAHF changes, however (Figure S3a, b). Instead, we observed that DICER or DROSHA inactivation significantly reduces the number of cells positive for DDR foci containing 53BP1, the autophosphorylated form of ATM (pATM) and the phosphorylated substrates of ATM and ATR (pS/TQ), but not γH2AX, without decreasing DDR proteins expression (Figure S3c). Importantly, the simultaneous inactivation of all three GW182-like proteins, TNRC6A, B and C, essential for the translational inhibition mediated by microRNAs (canonical DICER and DROSHA products involved in RNAi)15, does not impact DDR foci formation (Figure S4).
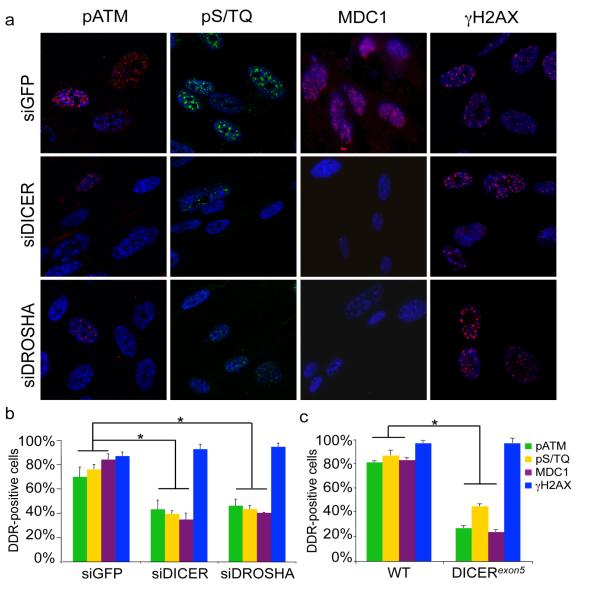
We next asked whether DICER or DROSHA inactivation also affects ionizing radiation (IR)-induced DDR activation. We transiently inactivated DICER or DROSHA by siRNA in human normal fibroblasts (HNF), exposed cells to IR and monitored DDR foci. We observed that few hours post IR DICER or DROSHA inactivation impairs the formation of pATM, pS/TQ, and MDC1, but not γH2AX, foci without decreasing their protein levels (Figures 1a,b and S5). At an earlier time point (10′) after IR, 53BP1 foci were significantly reduced also (Figure S6a). Using an RNAi-resistant form of DICER in DICER knock down cells, we observed that re-expression of wild-type DICER, but not a DICER endonuclease mutant (DICER44ab)16, rescues DDR foci formation (Figure S6b-d). The simultaneous knock down of TNRC6A/B/C, or DICER shows a comparable impact on a reporter system specific for miRNA-dependent translational repression17, but only DICER inactivation reduces DDR foci formation (Figure S7). To further confirm the involvement of DICER in DDR activation, we used a cell line carrying a hypomorphic allele of DICER (DICERexon5) defective in microRNA maturation18. In DICERexon5-irradiated cells, pATM, pS/TQ and MDC1, but not γH2AX, foci formation is impaired without a decrease in their protein levels, and 53BP1 foci formation is delayed compared to the DICER wild-type parental cell line (Figure S8). These defects could be reversed by the re-expression of wild-type DICER but not the mutant allele DICER44ab (Figure S9). We confirmed that ATM autophosphorylation is reduced in DICER- or DROSHA-knocked down HNF and in DICERexon5 cell lines by immunoblotting (Figure S10). These results indicate that DICER and DROSHA RNA products control DDR activation and act independently from canonical microRNA-mediated translational repression mechanisms.
Figure 1. DICER or DROSHA inactivation impairs DDR foci formation in irradiated cells.
a. DICER or DROSHA knock down WI38 cells were irradiated (10Gy) and fixed 7 h later. b. Histogram shows the percentage of cells positive for pATM, pS/TQ, MDC1 and γH2AX foci. c. WT and DICERexon5 cells were irradiated (2 Gy) and fixed 2 h later. Histogram shows the percentage of cells positive for pATM, pS/TQ and MDC1 and γH2AX, foci. Error bars indicate s.e.m. (n ≥3). Differences are statistically significant (*p-value <0.01).
DDR signaling enforces cell-cycle arrest at G1/S and G2/M checkpoints3. We observed that DNA damage-induced checkpoints were impaired in DICER- or DROSHA-inactivated cells and that wild-type DICER re-expression in DICERexon5 cells restores checkpoint functions while two independent mutant alleles of DICER fail to do so (Figures S11-13). Thus, DICER and DROSHA are required for DNA damage-induced checkpoint enforcement.
To test the role of DICER in DDR activation in a living organism, we inactivated it by morpholino antisense oligonucleotide injection in Danio rerio (zebrafish) larvae19. Such Dicer inactivation results in a dramatic impairment of pATM and γH2AX accumulation in irradiated zebrafish larvae as detected both by immunostaining and immunoblotting of untreated or Dicer morpholino-injected larvae and of chimeric animals (Figures S14, 15).
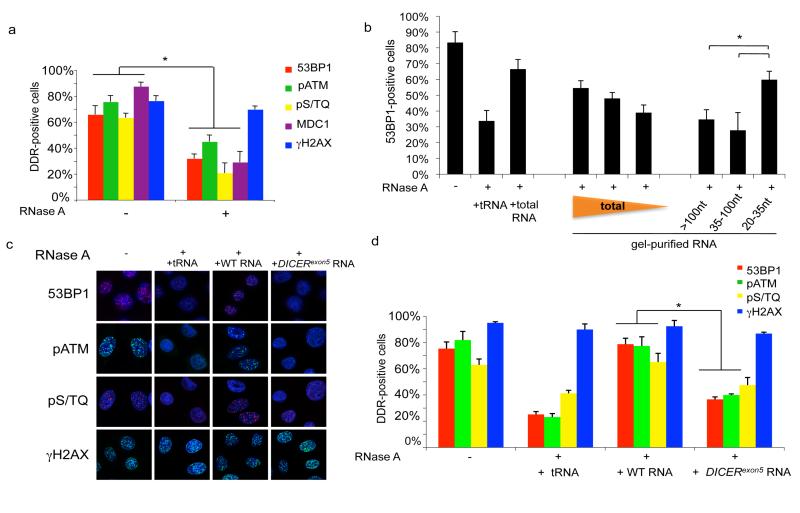
Previous reports have shown that mammalian cells can withstand a transient membrane permeabilization and RNase A treatment, enabling the study of the contribution of RNA to heterochromatin organization and 53BP1 association to chromatin20,21. We used this approach to address the direct contribution of DICER and DROSHA RNA products in DDR activation. Irradiated HeLa cells were permeabilized and treated with RNase A, leading to degradation of all RNAs, without significantly affecting protein levels (Figure S16a). We observed that 53BP1, pATM, pS/TQ and MDC1 foci become markedly reduced in number and intensity upon RNA degradation while, similarly to DICER- or DROSHA-inactivated cells, γH2AX is unaffected (Figures 2a and S16b). Notably, 53BP1, MDC1 and γH2AX triple staining shows that RNA degradation reduces 53BP1 and MDC1 accumulation at unperturbed γH2AX-foci (Figure S16c). When RNase A is inhibited, DDR foci progressively reappear within minutes and α-amanitin prevents this (Figure S17a, b), suggesting that DDR foci stability is RNA polymerase II-dependent.
Figure 2. Irradiation-induced DDR foci are sensitive to RNase A treatment and are restored by small and DICER dependent RNAs.
a. Irradiated HeLa cells (2 Gy) were treated with PBS (-) or RNase A (+) and probed for 53BP1, pATM, pS/TQ, MDC1 and γH2AX foci. Histogram shows the percentage of cells positive for DDR foci. b. 100, 50 or 20 ng of gel-extracted total RNA and 50 ng of RNA extracted from each gel fraction (>100, 35-100 and 20-35 nt) were used for DDR foci reconstitution after RNase treatment. c. 53BP1, pS/TQ and pATM foci are restored in RNase-treated cells when incubated with RNA of wild-type cells but not with RNA of DICERexon5 cells or tRNA. e. Histogram shows the percentage of cells positive for DDR foci. Error bars indicate s.e.m. (n ≥3). Differences are statistically significant (*p-value <0.01).
We tested if DDR foci can reform upon addition of exogenous RNA to RNase A-treated cells. We observed that DDR foci robustly reform in RNase A-treated cells following their incubation with total RNA purified from the same cells, but not with tRNA control (Figure 2b-d). Similar conclusions were reached using an inducible form of PpoI and AsiSI site-specific endonucleases22,23(data not shown).
Next, we attempted to characterize the length of the RNA species involved in DDR foci reformation, which we refer to as DDRNAs. We observed that an RNA fraction enriched by chromatography for species <200 nt was sufficient to restore DDR foci (Figure S17c-e). To attain better size separation, we resolved total RNA on a polyacrylamide gel and recovered RNA fractions of different lengths (Figure S17f,g). Using equal amounts of each fraction, we observed that only the 20-35 nt fraction could restore DDR foci (Figure 2b), consistent with the size range of DICER and DROSHA RNA products.
To test the hypothesis that DDRNAs are DICER and DROSHA products, we attempted DDR foci restoration with total RNA extracted from wild-type or DICERexon5 cells. While RNA extracted from wild-type cells restores pATM, pS/TQ and 53BP1 foci, RNA from DICERexon5 cells does not (Figure 2c,d). Importantly, RNA from DICERexon5 cells re-expressing wild-type, but not endonuclease mutant, DICER allows DDR foci reformation (Figure S18a,b). These results were reproduced using RNA extracted from cells transiently knocked-down for DICER or DROSHA (Figure S18c-f).
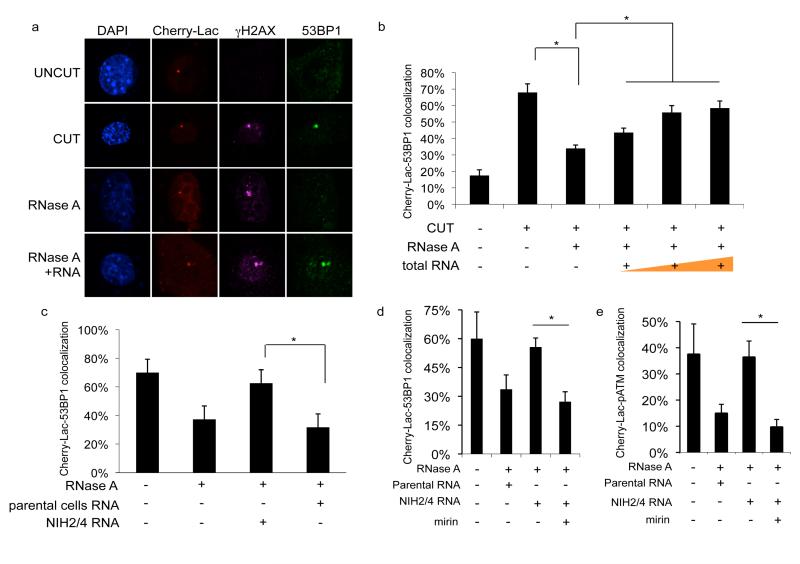
IR induces DNA lesions that are heterogeneous in nature and random in their genomic location. To reduce this complexity, we studied a single DSB at a defined and traceable genomic locus. We therefore took advantage of NIH2/4 mouse cells carrying an integrated copy of the I-SceI restriction site flanked by arrays of Lac- or Tet-operator repeats at either sites24. In this cell line, the expression of the I-SceI restriction enzyme together with the fluorescent protein Cherry-Lac-repressor allows the visualization of a site-specific DDR focus that overlaps with a focal Cherry-Lac signal (cut NIH2/4 cells). No DDR focus formation is observed on Cherry-Lac signal in the absence of I-SceI expression (uncut NIH2/4 cells). Also in this system, RNase-A treatment causes the disappearance of the 53BP1, but not the γH2AX, focus at the I-SceI-induced DSB; total RNA addition from cut cells restores 53BP1 focus formation in a dose-dependent manner (Figure 3a,b). Therefore, a DDR focus generated on a defined DSB can disassemble and reassemble in a RNA-dependent manner.
Figure 3. Site-specific DDR focus formation is RNase A-sensitive and can be restored by site-specific RNA in a MRN-dependent manner.
a. Cut NIH2/4 cells display a 53BP1 and γH2AX focus colocalizing with Cherry-Lac focus. 53BP1, but not γH2AX, focus is sensitive to RNase A and is restored by incubation with total RNA. b. Histogram shows the percentage of cells in which 53BP1 and Cherry-Lac foci co-localize. Addition of 50, 200, or 800 ng of RNA purified from cut NIH2/4 rescues 53BP1 foci formation in a dose-dependent manner. c. RNA purified from cut NIH2/4 restores 53BP1 focus while RNA from parental cells expressing I-SceI does not. d, e. RNase A-treated cut NIH2/4 cells were incubated with RNA from cut NIH2/4 cells, or parental ones, to test 53BP1 or pATM focus reformation in the presence of the MRN inhibitor mirin (100 μM). Histogram shows the percentage of cells positive for DDR focus. Error bars indicate s.e.m. (n ≥3). Differences are statistically significant (*p-value <0.05).
To discriminate whether DDRNAs are generated at the damaged locus or elsewhere in the genome, we took advantage of the fact that the I-SceI-induced DSB is generated within an integrated exogenous sequence, which is not present in the parental cell line. Since RNAs extracted from NIH2/4 or parental cells are expected to differ only in the potential presence of RNA transcripts generated at the locus, we used these two RNA preparations to attempt to restore 53BP1 focus formation at the I-SceI-induced DSB in RNase A-treated cells. The 53BP1 focus formation was efficiently recovered only by RNA purified from NIH2/4 cells and not from parental cells (Figure 3c), indicating that DDRNAs originate from the damaged genomic locus.
The MRE11/RAD50/NBS1 (MRN) complex is necessary for ATM activation25, and pATM and MRE11 foci formation is sensitive to RNase A treatment in the NIH2/4 cell system (Figure S19a,b). To probe the molecular mechanisms by which RNA modulates DDR focus formation, we used a specific MRN inhibitor26, mirin, that prevents ATM activation also in the NIH2/4 system (Figure S19d). In the presence of mirin, NIH2/4 RNA is unable to restore 53BP1 or pATM focus formation (Figure 3d, e), indicating that DDRNAs act in a MRN-dependent manner.
To detect potential short RNAs originating from the integrated locus, we deep-sequenced libraries generated from short (<200 nt) nuclear RNAs of cut or uncut NIH2/4 cells, as well as from parental cells expressing I-SceI as negative control. Sequencing revealed short transcripts arising from the exogenous locus (Figure S20a-e), 47 reads in cut cells, 20 reads in uncut cells and none in parental cells, indicating that even an exogenous integrated locus lacking mammalian transcriptional regulatory elements is transcribed and can generate small RNAs.
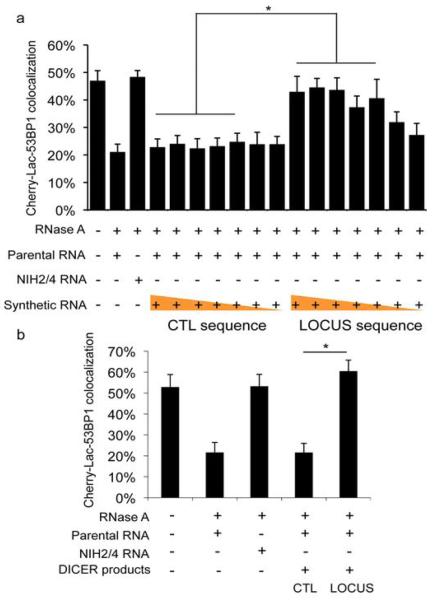
To test whether the identified locus-specific small RNAs are biologically active and have a causal role in DDR activation, we chemically synthesized four potential pairs among the sequences obtained and used them to attempt to restore DDR focus in RNase A-treated cells. Strikingly, we observed that addition of locus-specific synthetic RNAs, but not equal amounts of control RNAs, triggers site-specific 53BP1 focus reformation over a large range of concentrations both in the presence or absence of parental cells total RNA (Figures 4a and S20f). To additionally prove the biological activity of RNAs processed by DICER, we in vitro transcribed both strands of the sequence spanning the locus, or a control one, and processed the resulting RNAs with recombinant DICER. In vitro-generated locus-specific DICER RNA products, but not control RNAs, allowed DDR focus reformation in RNase A-treated cells (Figures 4b and S20g,h). Overall, these results indicate that DDRNAs are small RNAs with the sequence of the damaged locus, which play a direct role in DDR activation.
Figure 4. Chemically synthesized small RNAs and in vitro generated DICER RNA products are sufficient to restore DDR focus formation in RNase A-treated cells in a sequence-specific manner.
a. Chemically synthesized oligonucleotides were annealed and were tested to restore DDR focus formation in RNase A-treated cut NIH2/4 cells. Mixed with a constant amount (800 ng) of parental cells RNA, a concentrations range (1 ng/μl – 1 fg/μl, ten-fold dilution steps) of locus-specific or GFP RNAs was used. Locus-specific synthetic RNAs (down to 100 fg/μl) allow site-specific DDR activation. b. Short double-stranded RNAs generated by recombinant DICER were tested to restore DDR focus formation in RNase A-treated cut NIH2/4 cells. 1 ng/μl RNAs were tested mixed with 800 ng of parental cells RNA. Locus-specific DICER RNAs, but not control RNAs, allow site-specific DDR activation. Histograms show the percentage of cells positive for DDR focus. Error bars indicate s.e.m. (n ≥3). Differences are statistically significant (*p-value <0.05).
To investigate the biogenesis of such RNAs in vivo, we performed deeper sequencing of small nuclear RNAs from cut and uncut wild-type as well as Dicer or Drosha knock down NIH2/4 cells (Figure S21). As expected, Dicer or Drosha knockdown significantly reduced reads mapping to the known miRNAs (Figure S22). Our statistical analyses reveal that the percentage of 22-23 nt RNAs arising from the locus significantly increases in wild-type cut sample compared to the uncut one and that Dicer inactivation significantly reduces it (Figure S23a,b); the decrease in Drosha-inactivated cells did not reach statistical significance. Since the fraction of 22-23 nt RNAs from the locus is significantly higher respect to that of non-miRNA genomic loci, the RNAs detected are very unlikely to be random degradation products (Figure S23c). Finally, 22-23 nt RNAs at the locus tend to have an A/U at their 5′and a G at their 3′ end (Figure S23d), a nucleotide bias significantly different from the originating locus and from the rest of the genome.
In summary here we demonstrate that different sources of DNA damage, including oncogenic stress, ionizing irradiation and site-specific endonucleases, activate the DDR in a manner dependent on DDRNAs, which are DICER- and DROSHA-dependent RNA products with the sequence of the damaged site. DDRNAs control DDR foci formation and maintenance, checkpoint enforcement and cellular senescence in cultured human and mouse cells and in different cell types in living zebrafish larvae. They act differently from canonical microRNAs, as inferred by their demonstrated biological activity independent of other RNAs and of GW proteins.
Methods
Cultured cells
Early passage WI38 cells (ATCC) were grown under standard tissue culture conditions (37°C, 5% CO2) in MEM supplemented with 10% fetal bovine serum, 1% L-glutamine, 1% non-essential aminoacids, 1% Na Pyruvate. HeLa, Phoenix ecotrophic and HEK293T cell lines were grown under standard tissue culture conditions (37°C, 5% CO2) in DMEM, supplemented with 10% fetal bovine serum, 1% glutamine, 1% penicillin/streptomycin. DICERExon5 colon cancer cell lines18 were cultured in Mc’Coy 5A medium +10% fetal calf serum, 1% penicillin/streptomycin. NIH2/4 24 where grown in DMEM, supplemented with 10% fetal bovine serum, 1% glutamine, gentamicine (40μg/ml), and hygromycin (400μg/ml).
H-RasV12 overexpressing senescent BJ cells were generated as in14. BrdU incorporation assays were carried at least a week after cultures had fully entered the senescent state, as determined by ceased proliferation, DDR activation and SAHF formation. Ionizing radiation (IR) was induced by a high-voltage X-rays generator tube (Faxitron X-Ray Corporation). In general, WI38 cells were exposed to 5Gy and transformed cell (RKO, HCT116 and HeLa) to 2Gy for the DDR foci formation studies. We used 5Gy for the G2/M checkpoint assays and 10Gy for the G1/S checkpoint assays.
Cherry-Lac and I-Sce I-restriction endonuclease expressing vectors were transfected by lipofectamine 2000 (Invitrogen) in a ratio of 3:1. 16h post transfection around 70% of the cells were scored positive for DDR markers at the Lac array. For generation of Dicer and Drosha knocked down, NIH2/4 cells were infected with lentiviral particles carrying pLKO.1, shDicer or shDrosha vectors. After 48h cells were superinfected with Adeno Empty Vector (kind gift of Elisabetta Dejana, IFOM, Milan, Italy) or Adeno I-Sce I (kind gift of Philip Ng, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas). Nuclei were isolated the day after the adenoviral infection.
Antibodies
Mouse anti-γH2AX, anti-H3K9me3, rabbit polyclonal anti-pH3 (Upstate Biotechnology); anti-pS/TQ (Cell Signaling Technology); anti-H2AX, anti-H3 and anti DICER (13D6) (Abcam); rabbit polyclonal anti-53BP1 (Novus Biological) and mouse monoclonal anti-53BP1 (a gift from Thanos Halazonetis, University of Geneva); anti-MRE11 (a gift from SP. Jackson, Gurdon Institute, Cambridge, UK); anti-BrdU (Becton Dickinson); rabbit polyclonal anti-MCM2 (a gift of Marine Melixetian, IEO, Milan Italy); anti MRE11 rabbit polyclonal raised against recombinant MRE11; anti-pATM (Rockland); mouse monoclonal anti-ATM and anti-MDC1 (SIGMA); anti-Lamin A/C (Santa Cruz), anti-vinculin (clone hVIN-1), anti-□-tubulin (clone AA2) and anti-Flag M2 monoclonal antibodies (SIGMA).
Indirect immunofluorescence
Cells were grown on poly-D-lysinated coverslips (poly-D-lysine was used at 50μg/ml final concentration) and plated (15-20×103 cells/cover) one day before staining. DDR and BrdU staining was performed as in 14. Cells were fixed in 4% paraformaldehyde or methanol:acetone 1:1. NIH2/4 mouse cells were fixed by 4% paraformaldehyde as in 24. Images were acquired using a wide field Olympus Biosystems Microscope BX71 and the analySIS or the MetaMorph software (Soft Imaging System GmbH). Comparative immunofluorescence analyses were performed in parallel with identical acquisition parameters; at least 100 cells were screened for each antigen. Cells with more than two DDR foci were scored positive. Confocal sections were obtained with a Leica TCS SP2 or AOBS confocal laser microscope by sequential scanning.
Plasmids
DICER-flag, DICER44ab-flag and DICER110ab-flag were a kind gift of R. Shiekhattar, the Wistar Institute, Philadelphia, USA. DICER110ab-flag and DICER44ab-flag double mutants carry two amino acid substitution in the RNase III domains of Dicer (Asp1320->Ala and Asp1709->Ala for 44ab and Glu1652->Ala and Glu1813->Ala for 110ab mutant; both mutants were reported to be deficient in endonuclease activity16). pLKO.1 shDICER expressing vector was a kind gift of WC. Hahn. Short hairpin sequence for DICER is: CCG GCC ACA CAT CTT CAA GAC TTA ACT CGA GTT AAG TCT TGA AGA TGT GTG GTT TTT G. pRETROSUPER shp53 as in14. Short hairpin sequence for p53 was: AGT AGA TTA CCA CTG GAG TCT T. Cherry-Lac-repressor and I-Sce I-restriction endonuclease expressing vectors were kind gifts of E. Soutoglou, National Cancer Institute, NIH, Bethesda, USA24. shRNA against mouse Dicer and Drosha expressing vectors were a kind gift of W.C. Hahn, Dana-Farber Cancer Institute, Boston, USA. shRNA for mouse Dicer: CCG GGC CTC ACT TGA CCT GAA GTA TCT CGA GAT ACT TCA GGTCAA GTG AGG CTT TTT. shRNA for mouse Drosha: CCG GCC TGG AAT ATG TCC ACA CTT TCT CGA GAA AGT GTG GAC ATA TTC CAG GTT TTT G.
siRNA
The DHARMACON siGENOME SMARTpool siRNA oligonucleotide sequences for human 53BP1, ATM, DICER, DROSHA were: 53BP1: GAG AGC AGA UGA UCC UUU A; GGA CAA GUC UCU CAG CUA U; GAU AUC AGC UUA GAC AAU U; GGA CAG AAC CCG CAG AUU U.
ATM: GAA UGU UGC UUU CUG AAU U; AGA CAG AAU UCC CAA AUA A; UAU AUC ACC UGU UUG UUA G; AGG AGG AGC UUG GGC CUU U.
DICER: UAA AGU AGC UGG AAU GAU G; GGA AGA GGC UGA CUA UGA A; GAA UAU CGA UCC UAU GUU C; GAU CCU AUG UUC AAU CUA A.
DROSHA: CAA CAU AGA CUA CAC GAU U; CCA ACU CCC UCG AGG AUU A; GGC CAA CUG UUA UAG AAU A; GAG UAG GCU UCG UGA CUU A.
The DHARMACON siGENOME si RNA sequences for Human TNRC6A, B and C were: GW182/TNRC6A: GAA AUG CUC UGG UCC GCU A; GCC UAA AUA UUG GUG AUU A. TNRC6B: GCA CUG CCC UGA UCC GAU A; GGA AUU AAG UCG UCG UCA U. TNRC6C: CUA UUA ACC UCG CCA AUU A; GGU AAG UCC UCC AUU GAU G.
siRNA against human DICER 3′ UTR: CCG UGA AAG UUU AAC GUU U.
siRNA against GFP: AAC ACU UGU CAC UAC UUU CUC.
siRNA against Luciferase: CAU UCU AUC CUC UAG AGG AUG dTdT; dTdT GUA AGA UAG GAG AUC UCC UAC.
siRNAs were transfected by Oligofectamine or Lipofectamine RNAi Max (Invitrogen) at a final concentration of 200nM in OIS cells and 100nM in HNF. In the siRNA titration experiment we transfected OIS cells in parallel with 20nM and 200nM siRNA oligos. For siRNA transfection with deconvolved siRNA oligos we used 50nM for smart pools and 12.5nM for deconvolved siRNAs.
Real-time quantitative PCR (qRT-PCR)
Total RNA was isolated from cells using TRIzol (Invitrogen) or RNAeasy kit (Qiagen) according to the manufacturer’s instructions, and treated with DNase before reverse transcription. For small RNA isolation we used mirVana™ miRNA Isolation Kit (Ambion). cDNA was generated using the Superscript II Reverse Transcriptase (Invitrogen) and used as template in real-time quantitative PCR analysis. TaqMan® MicroRNA Assays (Applied Biosystems) were used for the evaluation of mature miR-21 and rnu44 and rnu19 expression levels (Assay ID: 000397, 001094 and 001003). 18S or □-actin was used as a control gene for normalization. Real-time quantitative PCR reactions were performed on an Applied Biosystems ABI Prism 7900HT Sequence Detection System or on a Roche LightCycler 480 Sequence Detection System. The reactions were prepared using SyBR Green reaction mix from Roche. Ribosomal protein P0 (RPP0) was used as a human and mouse control gene for normalization.
Primer sequences for real-time quantitative PCR were: RPP0: TTCATTGTGGGAGCAGAC (Forward), CAGCAGTTTCTCCAGAGC (Reverse); human endogenous DICER: AGCAACACAGAGATCTCAAACATT (Forward), GCAAAGCAGGGCTTTTCAT (Reverse); human endogenous and overexpressed DICER: TGTTCCAGGAAGACCAGGTT (Forward), ACTATCCCTCAAACACTCTGGAA (Reverse); human DROSHA: GGCCCGAGAGCCTTTTATAG (Forward), TGCACACGTCTAACTCTTCCAC (Reverse); human GW182: CAGCCAGTCAGAAAGCAGTG (Forward), TGTGAGTCCAGGATCTGCTACTT (Reverse); mouse Dicer: GCAAGGAATGGACTCTGAGC (Forward), GGGGACTTCGATATCCTCTTC (Reverse); mouse Drosha: CGTCTCTAGAAAGGTCCTACAAGAA (Forward), GGCTCAGGAGCAACTGGTAA (Reverse).
RNase A treatment and RNA complementation experiments
Cells were plated on poly-D-lysinated coverslips and irradiated with 2Gy of IR. 1h later HeLa cells were permeabilized with 2% Tween 20 in PBS for 10 minutes at RT while I-Sce I-transfected NIH2/4 cells were permeabilized in 0,5% Tween 20 in PBS for 10 minutes at RT. RNase A treatment was carried out in 1ml of 1mg/ml Ribonuclease A from bovine pancreas (Sigma-Aldrich cat n: R5503) in PBS for 25 minutes at room temperature. After RNase A digestion, samples were washed with PBS, treated with 80 units of RNase inhibitor (RNaseOUT Invitrogen 40units/□l) and □0μg/ml of αamanitin (SIGMA) for 15 minutes in a total volume of 70μl. For experiments with mirin, NIH2/4 cells were incubated at this step also with 100μM mirin (SIGMA) or DMSO for 15 minutes. Then, RNase A-treated cells were incubated with total, small or gel extracted RNA, or the same amount of tRNA, for additional 15 minutes at room temperature. If using mirin, NIH2/4 cells were incubated with total RNA in the presence of 100μM mirin or DMSO for 25 minutes at room temperature. Cell were then fixed with 4% paraformaldehyde or methanol:acetone 1:1.
In complementation experiments with synthetic RNA oligonucleotides, eight RNA oligonucleotides with the potential to form four pairs were chosen among the sequences that map at the integrated locus in NIH2/4 cells, obtained by deep sequencing. Synthetic RNA oligonucleotides were generated by SIGMA with a monophosphate modification at the 5′ end. Sequences map to different regions of the integrated locus: two pairs map to a unique sequence flanking the I-Sce I restriction site, one to the Lac-operator and one to the Tet-operator repetitive sequences. Two paired RNA oligonucleotides with the sequences of GFP were used as negative control. Sequences are reported below.
Oligo 1: 5′-AUA ACA AUU UGU GGA AUU CGG CGC-3′, oligo 2: 5′-CGA AUU CCA CAA AUU GUU AUC C-3′, oligo 3: 5′-AUU UGU GGA AUU CGG CGC CUC UAG AGU CGA GG-3′, oligo 4: 5′-CCU CGA CUC UAG AGG CG-3′, oligo 5: 5′-AGC GGA UAA CAA UUU GUG GCC ACA UGU GGA-3′, oligo 6: 5′-UGUGGC CAC AAA UUG UU-3′, oligo 7: 5′-ACU CCC UAU CAG UGA UAG AGA AAA GUG AAA GU-3′, oligo 8: 5′-CUU UCA CUU UUC UCU AUC ACU GAU AGG GAG UG-3′
GFP 1: 5′-GUU CAG CGU GUC CGG CGA GUU-3′, GFP 2: 5′-CUC GCC GGA CAC GCU GAA CUU-3′
RNAs were resuspended in 60mM KCl, 6mM HEPES-pH 7.5, 0.2mM MgCl2, at the stock concentration of 12□M, denatured at 95°C for 5 minutes and annealed for 10 minutes at room temperature.
DICER RNA products were generated as follows. A 550bp DNA fragment carrying the central portion of the genomic locus studied (three Lac repeats, the I-Sce I site and two Tet repeats) was flanked by T7 promoters at both ends and was used as a template for in vitro transcription with the TurboScript T7 transcription kit (AMSBIO). The 500nt long RNAs obtained were purified and incubated with human recombinant DICER enzyme (AMSBIO) to generate 22-23nt RNAs. RNA products were purified, quantified and checked on gel. As a control, the same procedure was followed with a 700bp construct containing the RFP DNA sequence. Equal amounts of DICER RNA products generated in this way were used in complementation experiment in NIH2/4 cells following RNase A treatment.
Small RNA preparation
Total RNA was isolated from cells using TRIzol (Invitrogen) according to the manufacturer’s instructions. To generate small RNA-enriched fraction and small RNA-devoid fraction we used mirVana™ microRNA Isolation Kit (Ambion) according to the manufacturer’s instructions. The mirVana microRNA isolation kit employs an organic extraction followed by immobilization of RNA on glass-fiber (silica-fibers) filters to purify either total RNA, or RNA enriched for small species. For total RNA extraction ethanol is added to samples, and they are passed through a Filter Cartridge containing a glass-fiber filter, which immobilizes the RNA. The filter is then washed a few times, and finally the RNA is eluted with a low ionic-strength solution. To isolate RNA that is highly enriched for small RNA species, ethanol is added to bring the samples to 25% ethanol. When this lysate/ethanol mixture is passed through a glass-fiber filter, large RNAs are immobilized, and the small RNA species are collected in the filtrate. The ethanol concentration of the filtrate is then increased to 55%, and it is passed through a second glass-fiber filter where the small RNAs become immobilized. This RNA is washed a few times, and eluted in a low ionic strength solution. Using this approach consisting of two sequential filtrations with different ethanol concentrations, an RNA fraction highly enriched in RNA species ≤200nt can be obtained 18,27.
RNA extraction from gel
Total RNA samples (15ng) were heat-denatured, loaded and resolved on a 15% denaturing acrylamide gel [1X TBE, 7 M urea, 15% acrylamide (29:1 acryl:bis-acryl)]. Gel was run for 1h at 180 V and stained in GelRed solution. Gel slices were excised according to the RNA molecular weight marker, moved to a 2 ml clean tube, smashed and RNA was eluted in 2 ml of ammonium acetate 0.5 M, EDTA 0.1 M in RNase-free water, rocking overnight at 4°C. Tubes were then centrifuged 5 minutes at top speed, the acqueous phase was recovered and RNA was precipitated and resuspended in RNase free water.
G1/S checkpoint assay
WI38 cells were irradiated with 10Gy and 1h afterwards incubated with BrdU (10μg/ml) for 7h; HCT116 cells were irradiated 2Gy and incubated with BrdU for 2h. Cells were fixed with 4% paraformaldehyde and probed for BrdU immunostaining. At least 100 cells per condition were analyzed.
G2/M checkpoint assay
HEK 293 calcium phosphate transfected cells were irradiated with 5Gy and allowed to respond to IR-induced DNA damage in a cell culture incubator for 12, 24 or 36h. Then, at these three time points post irradiation, together with not irradiated cells, 1×106 cells were collected for Fluorescence Activated Cell Sorting (FACS) analysis, fixed in 75% ethanol in PBS, 30 minutes on ice. Afterwards, cells were treated 12h with 250μg/ml of RNase A and incubated at least 1h with propidium iodide (PI). FACS profiles were obtained by the analysis of at least 5×105 cells. In the complementation experiments HEK 293 were transfected using Lipofectamine RNAi Max (Invitrogen) and 48h later irradiated with 5Gy. Cells were then treated as explained above.
Immunoblotting
Cells were lysed in sample buffer and 50-100μg of whole cell lysate were resolved by SDS-PAGE, transferred to nitrocellulose and probed as in 14. For zebrafish immunoblotting protein analysis, 72h post fertilization (hpf) larvae were deyolked in Krebs Ringer’s solution containing 1mM EDTA, 3mM PMSF and proteases inhibitor (Roche complete protease inhibitor cocktail). Embryos were then homogenized in SDS sample buffer containing 1mM EDTA with a pestle, boiled 5 minutes and centrifuged 13000 rpm for 1 minute. Protein concentration was measured with the BCA method (Pierce) and proteins (50μg-900μg) were loaded in an SDS-12% (for γH2AX and H3) and SDS-6% polyacrylamide gel (for pATM and ATM), transferred to a nitrocellulose membrane, and incubated with anti-γH2AX (1:2000, a gift from J. Amatruda28), H3 (1:10000, Abcam), pATM (1:1000, Rockland), ATM (1:1000, Sigma). Immunoreactive bands were detected with horseradish peroxidaseconjugated anti-rabbit or anti-mouse IgG and an ECL detection kit (Pierce, Springfield, IL, USA). Protein loading was normalized to equal amounts of total ATM and H3.
Zebrafish embryo injection, cell transplantation and staining
Zebrafish embryos at the stage of 1-2 cells were injected with a morpholino against Dicer1 29 diluted in Danieau buffer. The morpholino oligonucleotide was injected at a concentration of 5ng/nl, and a volume of 2nl/embryo. To assess the efficiency of the morpholino to block microRNA maturation, we co-injected the morpholino with in vitro synthesized mRNA, encoding for red fluorescent protein (RFP) and carrying 3 binding site for miR126 in the 3′ UTR 17. The oligonucleotides carrying the binding sites for miR126 used for construction of pCS2:RFPmiR126 sensor are: 5′GCATTATTACTCACGGTACGAATAAGGCATTATTACTCACGGTACGAAT AAGGCATTATTACTCACGGTACGA 3′ and 5′CGTAATAATGAGTGCCATGCTTATTCCGTAATAATGAGTGCCATGCTTAT TCCGTAATAATGAGTGCCATGCT 3′. The construct was verified by sequencing and used to synthesize mRNA in vitro using the mMessage Kit (Ambion). Messenger RNA encoding for RFPmiR126 sensor was injected alone or in combination with Dicer1 morpholino at a concentration of 10pg/nl. For cell transplantation experiments, we injected donor embryos with a mixture of dicer1 morpholino and mRNA encoding for GFP (5pg/nl). Approximately 20 cells were transplanted from donor embryos at dome stage (5 hpf) to uninjected host at the same stage. Successfully transplanted larvae (displaying GFP+ cells) were irradiated as described below. Mature miRNA were reverse transcribed to produce 6 different cDNAs for TaqMan® MicroRNA assay (30ng of total mRNA for each reaction; Applied Biosystems). Real-time PCR reactions based on TaqMan reagent chemistry were performed in duplicate on ABI PRISM® 7900HT Fast Real-Time PCR System (Applied Biosystems). The level of miRNA expression was measured using CT (threshold cycle). Fold change was generated using the equation 2-CT.
For immunofluorescence in zebrafish larvae: 72 hpf larvae were irradiated with 12Gy, fixed in 2% paraformaldehyde for 2h at room temperature. After equilibration in 10 and 15% sucrose in PBS, larvae were frozen in OCT compound on coverslips on dry ice. Sections were cut with a cryostat at a nominal thickness of 14□m and collected on Superfrost slides (BDH). Antisera used were zebrafish γH2AX – a kind gift of J. Amatruda28 – and pATM (Rockland). GFP fluorescence in transplanted embryos was still easily visible in fixed embryos. Images were acquired with a confocal (Leica SP2) microscope and 63X oil immersion lens.
RNA sequencing
Nuclear RNA shorter than 200nt was purified using mirVana™ microRNA Isolation Kit. RNA quality was checked on a small RNA chip (Agilent) before library preparation. For Illumina hi Seq Version3 sequencing, spike RNA was added to each RNA sample in the RNA: spike ratio of 10000:1 before library preparation and libraries for Illumina GA IIX were prepared without spike. An improved small RNA library preparation protocol was used to prepare libraries 30. In brief, adenylated 3′ adapters were ligated to 3′ ends of 3′-OH small RNAs using a truncated RNA ligase enzyme followed by 5′ adapter ligation to 5′-monophosphate ends using RNA ligase enzyme, ensuring specific ligation of non degraded small RNAs. cDNA was prepared using a primer specific to the 3′adapter in the presence of Dimer eliminator and amplified for 12-15 PCR cycles using a special forward primer targeting the 5′ adapter containing additional sequence for sequencing and a reverse primer targeting the 3′ adapter. The amplified cDNA library was run on a 6% polyacrylamide gel and the 100bp band containing cDNAs up to 33nt was extracted using standard extraction protocols. Libraries were sequenced after quality check on a DNA high sensitivity chip (Agilent). Multiplexed barcode sequencing was performed on Illumina GA-IIX (35bp Single end reads) and Illumina Hi seq version3 (51bp single end reads). Sequence data have been deposited in the DNA Data Bank of Japan under accession code DRA000540.
Statistical analyses
Results are shown as means plus/minus standard error (s.e.m.). p-value was calculated by Chi-squared test. qRT-PCR results are shown as means of a triplicate plus/minus standard deviation (s.d.) and p-value was calculated by Student’s t-test as indicated. * indicates p-value<0.05. n stands for number of independent biological experiments.
Statistical analysis of small RNA sequencing data
Statistical significance of downregulation of normalized miRNAs in Dicer and Drosha knockdown samples were calculated using the Wilcoxon signed-rank test.
The differences in the fraction of 22-23nt vs total small RNAs at the locus between the wildtype, Dicer knockdown, and Drosha knockdown before and after cut was calculated by fitting a negative binomial model to the small RNAs count data and performing a likelihood ratio test, keeping the fraction of 22-23nt vs total small RNAs at the locus fixed across conditions under the null hypothesis and allowing it to vary between conditions under the alternative hypothesis.
Supplementary Material
Acknowledgments
We thank E. Soutoglou, W.C. Hahn, M. Kastan, V. Orlando, R. Shiekhattar, J. Amatruda, T. Halazonetis, E. Dejana, P. Ng and F. Nicassio for sharing reagents, M. Fumagalli and Francesca Rossiello for reading the manuscript, M. Dobreva, V. Matti and F. Pezzimenti for technical support, G. D’Ario for help with statistical analyses, B. Amati, M. Foiani, V. Costanzo and FdAdF group members for help and discussions. FdAdF laboratory was supported by AIRC, European Community’s 7th Framework Programme (FP7/2007-2013) under grant agreement n° 202230, acronym “GENINCA”, HFSP, AICR, the EMBO Young Investigator Program. The initial part of this project was supported by Telethon. PC was supported by 7th Framework of the European Union commission to the Dopaminet consortium, a Grant-in-Aids for Scientific Research (A) No.20241047, Funding Program for the Next Generation World-Leading Researchers (NEXT Program) to PC and a Research Grant to RIKEN Omics Science Center from MEXT. SF is supported by Center for Genomic Science of IIT@SEMM and AIRC. MM was supported by Cariplo (grant no. 2007-5500) and AIRC. AS is supported by a JSPS fellowship P09745 and grant in aid by JSPS and DT is supported by the European Union Seventh Framework Programme under grant agreement FP7-People-ITN-2008-238055 (“BrainTrain” project) to PC.
Footnotes
Author Contributions A.S., D.T. and P.C. planned, generated and analysed the genomics data as in S20 a-e, S21, S22b, S23. M. d. H. performed statistical analysis of the genomics data. A.S., P.C. also edited the manuscript. M.M. and V.A. generated data in S14 and S15. F.M. generated data shown in 2b; 3d, e; 4b; S14 d, f; S2 b, e; S3 e; S4b; S5 f, g; S6 b-d; S7 d; S9; S13 d-f; S17 f, g; S18 a,b; S19; S20 g,h; S22a and generated RNA for deep sequencing; contributed to: S16a; S5 d, e; S11 c, d and edited the manuscript. S.F. generated the data shown in remaining figures, contributed to experimental design and edited the manuscript. F.d.A.d.F. conceived the study, designed the experiments and wrote the manuscript.
Sequence data have been deposited in the DNA Data Bank of Japan under accession code DRA000540.
References
- 1.Esteller M. Non-coding RNAs in human disease. Nature reviews. Genetics. 2011;12:861–874. doi: 10.1038/nrg3074. doi:10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews. Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. doi:10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. doi:10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark MB, et al. The reality of pervasive transcription. PLoS biology. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102, doi:10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. doi:10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. doi:10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. doi:10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 9.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. doi:10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 10.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13:1161–1169. doi: 10.1038/ncb2344. doi:10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 11.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. doi:10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 12.Narita M, et al. Rb-Mediated Heterochromatin Formation and Silencing of E2F Target Genes during Cellular Senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 13.White SA, Allshire RC. RNAi-mediated chromatin silencing in fission yeast. Curr Top Microbiol Immunol. 2008;320:157–183. doi: 10.1007/978-3-540-75157-1_8. [DOI] [PubMed] [Google Scholar]
- 14.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 15.Tritschler F, Huntzinger E, Izaurralde E. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of deja vu. Nature reviews. Molecular cell biology. 2010;11:379–384. doi: 10.1038/nrm2885. doi:10.1038/nrm2885. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 17.Nicoli S, et al. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. doi:10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummins JM, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. doi:10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 20.Maison C, et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 21.Pryde F, et al. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J Cell Sci. 2005;118:2043–2055. doi: 10.1242/jcs.02336. [DOI] [PubMed] [Google Scholar]
- 22.Berkovich E, Monnat RJ, Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. doi:10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 23.Iacovoni JS, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. The EMBO journal. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. doi:10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. doi:10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nature reviews. Molecular cell biology. 2011;12:90–103. doi: 10.1038/nrm3047. doi:10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dupre A, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nature chemical biology. 2008;4:119–125. doi: 10.1038/nchembio.63. doi:10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. doi:10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 28.Sidi S, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. doi:10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 30.Kawano M, et al. Reduction of non-insert sequence reads by dimer eliminator LNA oligonucleotide for small RNA deep sequencing. BioTechniques. 2010;49:751–755. doi: 10.2144/000113516. doi:10.2144/000113516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.