Abstract
Alternating hemiplegia of childhood (AHC) is a rare, severe neurodevelopmental syndrome characterized by recurrent hemiplegic episodes and distinct neurologic manifestations. AHC is usually a sporadic disorder with unknown etiology. Using exome sequencing of seven patients with AHC, and their unaffected parents, we identified de novo nonsynonymous mutations in ATP1A3 in all seven AHC patients. Subsequent sequence analysis of ATP1A3 in 98 additional patients revealed that 78% of AHC cases have a likely causal ATP1A3 mutation, including one inherited mutation in a familial case of AHC. Remarkably, six ATP1A3 mutations explain the majority of patients, including one observed in 36 patients. Unlike ATP1A3 mutations that cause rapid-onset-dystonia-parkinsonism, AHC-causing mutations revealed consistent reductions in ATPase activity without effects on protein expression. This work identifies de novo ATP1A3 mutations as the primary cause of AHC, and offers insight into disease pathophysiology by expanding the spectrum of phenotypes associated with mutations in this gene.
Alternating hemiplegia of childhood (AHC) was first characterized as a distinct syndrome in 1971 with a report describing eight patients with episodes of intermittent hemiplegia on alternating sides of the body, developmental delay, dystonia, and choreoathetosis beginning in infancy1. Since that time, specific diagnostic criteria have more clearly defined the classic paroxysmal and interictal neurologic manifestations associated with this disease2-6. AHC is estimated to affect approximately one in one million individuals7, with most cases occurring sporadically5,8-10. While the etiology of AHC is usually unknown, a missense mutation in ATP1A2 was reported in one case of atypical familial alternating hemiplegia9,10; however the clinical presentation of some of the family members with the ATP1A2 mutation was more consistent with familial hemiplegic migraine9, which is caused by mutations in ATP1A211,12. To date, no cases of sporadic AHC have been attributed to ATP1A2 mutations.
In this study, we used next-generation sequencing (NGS) to exome or whole-genome sequence ten AHC probands and their unaffected parents where possible. We identified and confirmed rare (MAF <0.01%) mutations in ATP1A3 in eight of the 10 probands; for all seven patients where parental DNA was available we could demonstrate that the mutations had occurred de novo. The ATP1A3 mutations included five distinct nonsynonymous mutations, one of which was found in four AHC patients (Supplementary Table 1). ATP1A3 was then further interrogated in the two unexplained AHC probands for structural variants in the whole genome sequence data, and for overlooked single nucleotide and insertion-deletion variants by Sanger sequencing of the protein coding exons; neither analysis identified candidate causal ATP1A3 mutations in these individuals. Given the rarity of functional de novo mutations, the occurrence of seven de novo mutations in the same gene in seven AHC patients, provides definitive genetic evidence that mutations in ATP1A3 cause sporadic AHC.
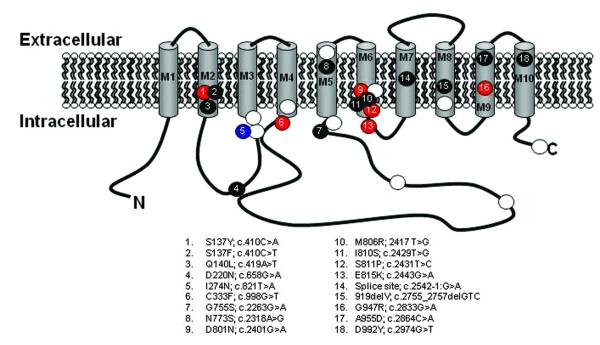
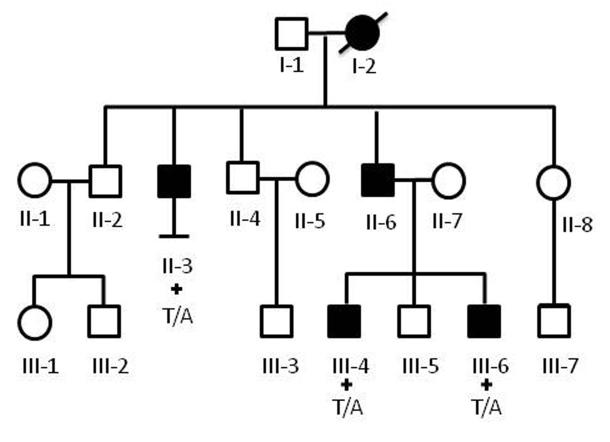
We then Sanger-sequenced the protein-coding exons of ATP1A3 in an additional cohort of 95 AHC patients. In these 95 subjects, we identified rare (MAF <0.01%) ATP1A3 mutations in 74 patients (Table 1), all of which were found to be de novo in the 59 sporadic AHC patients with parental DNA available. Including samples sequenced with NGS, we, in total, identified 18 different ATP1A3 mutations in 82 out of 105 (78%) patients studied. The majority of these mutations fell in or near transmembrane domains of ATP1A3 (Fig. 1). Six of the mutations were identified in multiple AHC cases, including D801N and E815K that were identified in 36 (34%) patients and 19 (18%) patients, respectively (Table 1). One of the 95 AHC patients evaluated was a case of autosomal dominant alternating hemiplegia, first described in 19928. In this familial case of AHC, we identified a rare ATP1A3 mutation (I274N) in the cytoplasmic domain that co-segregates with the AHC phenotype (Fig. 2).
Table 1.
ATP1A3 mutations identified in AHC patients.
| ATP1A3 mutationa |
Nucleotide changea |
Number of AHC probands with the mutation |
|---|---|---|
| S137Y | c.410C>A | 2 |
| S137F | c.410C>T | 1 |
| Q140L | c.419A>T | 1 |
| D220N | c.658G>A | 1 |
| I274N | c.821T>A | 1 |
| C333F | c.998G>T | 2 |
| G755S | c.2263G>A | 1 |
| N773S | c.2318A>G | 1 |
| D801N | c.2401G>A | 36 |
| M806R | c.2417T>G | 1 |
| I810S | c.2429T>G | 1 |
| S811P | c.2431T>C | 4 |
| E815K | c.2443G>A | 19 |
| splice site | c.2542-1:G>A | 1 |
| V919del | c.2755_2757delGTC | 1 |
| G947R | c.2839G>A | 7 |
| A955D | c.2864C>A | 1 |
| D992Y | c.2974G>T | 1 |
ATP1A3 mutation coordinates are defined based on UniProt ID P1363724 and Consensus CDS ID CCDS12594.125.
Figure 1. Diagram of the ATP1A3 protein showing positions of AHC-causing and DYT12-causing mutation.
AHC-causing mutations seen in a single case, in multiple cases and in a familial case are represented by black, red and blue dots, respectively. The white dots represent DYT12 mutations compiled from the HGMD database23. ATP1A3 mutation coordinates are defined based on UniProt ID P1363724 and Consensus CDS ID CCDS12594.125.
Figure 2. Pedigree of family with autosomal dominant AHC.
Black shading indicates affectation status. An ATP1A3 mutation (c.821T>A;I274N) was identified in patients II-3, III-4, and III-6 where DNA was available (indicated by a plus sign). DNA was unavailable in the father and grandmother of patients III-4 and III-6. Details of the family were reported previously1. Phenotypic details of the affected patients are provided in Supplementary Note.
Thirteen of the 17 ATP1A3 mutations seen in sporadic AHC cases were confirmed to be de novo (Supplementary Table 1). We also observed, however, 15 sporadic patients with rare (MAF <0.01%) ATP1A3 variants where parents were not available. This raises the possibility that some of these are inherited benign polymorphisms. While this is unlikely given the rarity of functional variants in ATP1A3, we can conservatively estimate the number of patients with pathogenic ATP1A3 mutations by only considering those mutations observed as de novo in at least one patient as pathogenic. Under this assumption, 11 of the 15 patients have a pathogenic ATP1A3 mutation. We can therefore conclude that at least 74% of sporadic patients with typical presentation of AHC studied here harbor disease-causing mutations in ATP1A3.
ATP1A3 encodes an alpha-subunit of the Na+/K+-ATPase pump that is partly responsible for establishing and maintaining electrochemical gradients of sodium and potassium ions across the plasma membrane of neurons13. Mutations in ATP1A3 have been shown to cause rapid-onset-dystonia-parkinsonism (DYT12)14-18. None of the mutations known to cause DYT12 was found in AHC patients. Two AHC mutations (D801N and I274N), however, affect the same amino acids as the DYT12 mutations.
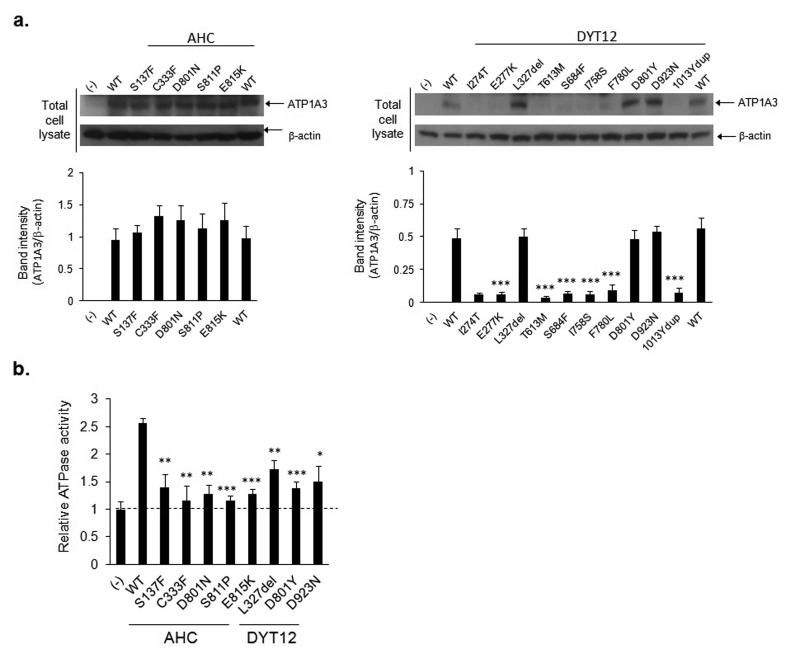
To better understand how ATP1A3 mutations cause two clinically distinct disorders, the functional consequences of the five mutations identified in the NGS screens of AHC patients and ten mutations that cause DYT12 were studied in vitro. All fifteen mutations were introduced in expression constructs and assessed for ATP1A3 expression and function. None of the mutations was found to affect ATP1A3 mRNA expression (Supplementary Fig. 1). However, seven out of ten mutations that cause DYT12 reduced ATP1A3 protein expression to undetectable levels, while none of the AHC-causing mutations reduced protein expression compared to wild-type (Fig. 3 and Supplementary Fig. 2). Despite different effects on protein expression both the mutations that cause AHC and those that cause DYT12 reduced ATPase activity in vitro by 54-90% (Fig. 3). We note that the previous study of ATP1A3 protein expression in DYT12 reported that D801Y attenuates protein expression and I274T does not affect protein expression14, whereas we observe opposite effects in our study. This discrepancy may be attributed to different sensitivities of the assay in specific cell lines. Despite this inconsistency, both studies show that DYT12 mutations typically reduce ATP1A3 protein expression, whereas in our study no AHC mutation reduces protein levels. These data suggest that ATP1A3 mutations causing DYT12 do so through hypomorphic effects on the Na+/K+-ATPase pump, while AHC-causing mutations modulate the activity of the pump. The reason for this inference is that if hypomorphic mutations could cause AHC we would expect some of the mutations to reduce protein level, and that the mutations would be distributed through the protein rather than concentrated in transmembrane domains. Further supporting the hypothesis, evaluation of the crystal structure of the Na+/K+-ATPase19 predicts that a change from aspartic acid at position 801 to asparagine (i.e. D801N) in AHC will prevent the binding of potassium ions. One possible exception to this pattern is the de novo splice-site mutation in an AHC patient that may result in protein elimination by a frame-shift, although it could result in a protein with altered activity.
Figure 3. Effects of disease-causing mutations on protein expression and enzyme activity in COS-7 cells.
Panel a. shows the effects of AHC-causing mutations (left) to DYT12 (right) on ATP1A3 protein abundance compared to WT. Panel b. compares the ATPase activity of disease-causing mutations (AHC and DYT12, mean ± s.d.) to that of WT.
*P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test (uncorrected for multiple testing), compared to WT
As noted previously, six of the AHC mutations result in the same amino acid substitution in multiple patients. A recurrent mutation in FGFR3 was previously reported to cause the majority of cases of achondroplasia20, suggesting the presence of hypermutable sequence in that gene. In AHC, the recurrence of de novo mutations may be due to hypermutable sequences in ATP1A3, the ascertainment effect of only a specific subset of mutations causing AHC, or both. Some contribution of hypermutability is indicated by a simple analysis. We observe 13 sites that carry de novo mutations in ATP1A3. Under the null hypothesis that the mutation rate is equal amongst these sites, the chance that any single site would have 36 or more mutations (as observed for D801N) out of the total 77 observations (patients studied with a “pathogenic” mutation) is low (P < 0.0001). Furthermore, three of six of these sites recurrently mutated in AHC are G>A substitutions occurring at hypermutable methylated CpG-dinucleotide sequences21 (D801N, E815K, G947R). It also appears, however, that only a specific subset of ATP1A3 mutations produce AHC since nearly all identified AHC mutations fall in or near transmembrane domains of the protein, while the DYT12 mutations are more evenly distributed (Fig. 1). Collectively, this suggests that the observed patterns of AHC-causing mutations across ATP1A3 results from both hypermutable sequence and that only a small set of specific mutations can cause AHC. This is consistent with the functional analyses suggesting that AHC mutations may have specific effects on protein function as opposed to simply reducing activity. While not evaluated in this study, functional evaluation of the inherited ATP1A3 mutation (I124N) may reveal distinct effects on the activity of the Na+/K+-ATPase that may help explain the atypical familial AHC phenotype.
We also evaluated whether patients with and without ATP1A3 mutations have different clinical presentations. Myshkin mice that are heterozygous for missense Atp1a3 mutation (I801N) that inactivates Na+/K+-ATPase are predisposed to seizure activity that is rescued by replacement with functional Atp1a322. We therefore first evaluated whether patients with ATP1A3 mutations were more likely to have seizures than those without, and found a minor but significant effect (54% versus 29% respectively, P = 0.01, binomial probability calculation). For one patient cohort studied here (n = 30) with consistent phenotyping5 we also compared age at first paroxystic event, age at first hemiplegic attack, and a series of disability indices, but found no statistically significant differences. More detailed investigations are needed to characterize the phenotypic spectrum associated with ATP1A3 mutations, and to compare phenotypes among patients with different ATP1A3 mutations.
In conclusion, mutations in ATP1A3 likely account for at least 74% of patients with a diagnosis of typical sporadic AHC. Since the present study only assessed patients with typical AHC further work will be needed to assess whether ATP1A3 mutations cause distinct but related conditions. In addition to identifying the cause of the majority of AHC patients, our results now implicate another clinically-distinct disease linked to ATP1A3, and open the door to detailed functional characterization of mutations that cause the different diseases. Having the ability to test the functional consequences of two groups of mutations that lead to clinically-distinct phenotypes offers unique insight into the pathophysiologic processes unique to each disease and will likely facilitate drug discovery for these and related conditions.
Online Methods
Study population
All sequenced patients met the diagnostic criteria for typical AHC4. Exome and genome sequenced patients were recruited to take part in this study through the Genetics of Epilepsy study at Duke University Medical Center, the Genetics of Epilepsy study at University College London, and at the University of Utah. Blood samples for DNA extraction were collected from each affected child and from unaffected parents when possible. Patients comprising the follow-up cohort were obtained from Duke University Medical Center (n = 2), the I.B.AHC Biobank and Clinical Registry for Alternating Hemiplegia (n = 34), the European AHC Genetics Consortium (n = 16), the University of Melbourne (n = 7), the University of Utah (n = 1), the French DNA and Cell Biobank for AHC (n = 30), Our Lady’s Children’s Hospital (Dublin, Ireland, n = 4), and The Children’s University Hospital (Dublin, Ireland, n = 1). All patients were recruited and consented based on the standards set forth by the ethics boards at the patient collection sites. Detailed phenotypic and demographic information of AHC patients with ATP1A3 mutations are provided in Supplementary Table 1.
Next-generation sequencing
Samples were either exome sequenced using Agilent’s All Exon (50 MB) capture, or whole-genome sequenced. All exome and whole genome sequencing was performed on either Illumina GAIIx or HiSeq 2000 machines in the Genomic Analysis Facility within the Center for Human Genome Variation (Duke University). Sequence data from 484 controls non-enriched for neuropsychiatric diseases that were sequenced as part of other in-house studies (e.g. genetics of cognition, birth weight, and HIV resistance) were used to ascertain candidate variant genotype frequencies. Sequencing was performed using standard protocols. The targeted exonic regions of all exome sequenced AHC samples and their parents were sequenced to an average coverage of 90-fold (minimum 65-fold), with at least 95 percent of the captured region having >5-fold coverage. Whole genome sequenced patient samples were sequenced to an average coverage of greater than 25-fold.
Paired-end reads were aligned to the Human Reference Genome (NCBI Build 36) using BWA software26. Variant calling to detect single nucleotide variants and indels was performed using SAMtools software27. Structural variation was called from whole-genome sequenced samples using software developed in the Center for Human Genome Variation, ERDS28 and SV-Finder. SequenceVariantAnalyzer (SVA)29 was used to annotate variants identified from the sequence data (Ensembl 50_36l). This software provides each variant with a genomic context (nonsynonymous coding vs. splice site, gene name, transcript, associated GO term, etc)29. Identity-by-descent calculations were used to confirm paternity and maternity.
Pictures of aligned NGS fragments covering the de novo ATP1A3 variants in the seven sequenced AHC trios (parents and affected child) are provided in Supplementary Fig. 3.
Prioritization of candidate disease-causing mutations
Genotypes of variants identified in each of the seven patients were evaluated for presence in their unaffected parents and also in a set of 484 population controls. Any genotype identified in either the unaffected parent or a population control was assumed to be non-causal. Genotypes present in the patients and absent in the unaffected parents, 484 sequenced controls, and 5400 individuals sequenced as part of the NHLBI Exome Sequencing Project (ESP) Exome Variant Server (v.0.0.8, release ESP5400) were considered as possibly causal.
Candidate disease-causing mutations were confirmed to be both present and de novo using Sanger sequencing.
Sanger sequencing of the protein-coding exons of ATP1A3
Some or all of the protein-coding exons of ATP1A3 were Sanger sequenced in a follow-up cohort of 95 AHC patients using standard methods. When parental DNA samples were not available, heterozygous mutations absent in any publically available database were presumed to be causal (confirmed de novo status is noted in Supplementary Table 1). Sequencing primers are available upon request. We compared the sample sources between clinical sites to ensure no overlap. For 59 of the 70 AHC patients with recurrent ATP1A3 mutations, we genotyped ten common polymorphisms to confirm that samples harboring the same ATP1A3 mutation and each were unique.
Functional characterization of disease-causing mutations
Pathogenic mutations in ATP1A3 identified in this study and a series of mutations previously implicated in DYT1214-18 were evaluated for effects on protein expression and overall ATPase activity in vitro.
Plasmids
Human APT1A3 cDNA samples were amplified in four fragments from first-strand cDNA derived from total RNA extracted from human neuroblastoma A172. Each part of ATP1A3 cDNA was then subcloned into the pCR-Blunt II-TOPO vector (Invitrogen-Life Technologies, Carlsbad, CA, USA). Fifteen different mutant alleles were produced by PCR-directed mutagenesis and the sequences were confirmed. The wild-type or mutant cDNA parts were then subcloned into pCR-Blunt II to generate full length cDNAs. Each full length ATP1A3 cDNA (wild-type and 15 mutants) was then subcloned into the expression vector, pcDNA3.1 (+). The sequences of the constructs were confirmed by sequence analysis. Primer sequences used for the generation of the plasmids are available upon request.
Quantitative RT-PCR
Empty pcDNA3.1(+) vector, pcDNA3.1(+)-ATP1A3-wildtype vector, and each allele of pcDNA3.1(+)-ATP1A3 vectors were transfected into human epithelial carcinoma cell line HeLa by lipofection using Lipofectamine 2000 (Invitrogen-Life Technologies). Total RNA was extracted from transfectant, and cDNAs were synthesized. ATP1A3 and GAPDH mRNA expression was estimated using RT–PCR (primer sequences available upon request).
Western Blotting
Empty pcDNA3.1 (+) vector, pcDNA3.1(+)-ATP1A3-wildtype vector, and each of the pcDNA3.1(+)-ATP1A3-mutant allele vectors were transfected into human epithelial carcinoma cell line (HeLa) and monkey kidney cell line (COS-7) by lipofection. After 48h of transfection, the cell lysates were subjected to SDS–PAGE gel and transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA). The membranes were then incubated with anti-human-ATP1A3 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), or anti-beta-actin (Cell Signaling Technology, Beverly, MA). Proteins were visualized with the ECL plus western blotting detection system (GE Healthcare, Piscataway, NJ). The effects of the mutations were quantified using Image J software.
ATPase assay
COS-7 cells expressing wild-type or mutant alleles of ATP1A3 were lysed gently by Mammalian Protein Extraction Buffer (GE Healthcare) and protease inhibitor cocktail (Sigma-Aldrich). Only mutant alleles with detectable ATP1A3 protein levels in the Western blot assay were assessed in the ATPase assay. COS-7 cells were chosen for the ATPase analysis because HeLa cells do not express ATP1A2, an essential subunit of the Na+/K+-ATPase. The lysates were then incubated with 250 μM of ATP, 40mM of NaCl, 25mM of KCl, 3mM of MgCl2 and 1mM EGTA in 37 degree for 1h. Synthesized ADP by ATPase reaction was detected by ADP-Glo™ Kinase Assay (Promega). ATPase activity was estimated by subtracting the luminescence signal with the addition of 100 μM of Ouabain from the luminescence reading without Ouabain.
Supplementary Material
Web box.
David Goldstein and Mohamad Mikati report identification of de novo mutations in ATP1A3 in alternating hemiplegia of childhood, which is a rare neurodevelopmental syndrome characterized by recurrent hemiplegic episodes and distinct neurologic manifestations.
Acknowledgements
We are deeply indebted to all the AHC families for their participation in this study.
We would like to thank the Alternating Hemiplegia of Childhood Foundation for their efforts in coordinating the collection of US samples, the financial support of Kathryn Swoboda, MD, Sandra Reyna, MD, & Tara Newcomb, MS in the form of grants, and the facilitation of US research collaboration. We also would like to thank the French Family Foundation (Dominique Poncelin) and the Italian AHC Family Foundation (Rosaria Vavassori) for facilitating the international collaboration.
We thank A.I.S.EA Onlus, the Italian Patient Association for Alternating Hemiplegia, for coordinating and funding the project I.B.AHC Biobank and Clinical Registry for Alternating Hemiplegia. Specifically, we thank Maria Teresa Bassi and Erika Tenderini for preparing all of the AHC samples for analysis. Many thanks also to the Scientific Institute E. Medea, Lecco, Italy, that hosts the I.B.AHC Biobank, according to the I.B.AHC protocol.
We also thank the ENRAH for SMEs Consortium, the ENRAH validation committee and all collaborating physicians for the data collection5. We are also grateful to the DNA and cell bank of Genethon for the processing of French blood samples.
We would like to acknowledge the following individuals who contributed next-generation sequenced control samples to this study: D. Attix, E. Behr, R.Brown, J. Burke, D. Daskalakis, V. Dixon, Farfel, R.Gbadegesin, A. Holden, E. Holtzman, J. Hoover-Fong, C. Hulette, S. Kerns, D. Lancet, W. Lowe, P. Lugar, D. Marchuk, J. McEvoy, J. Milner, H. Oster, R. Ottman, S. Palmer, E. Pras, V. Shashi, N. Sobriera, D. Valle, K. Welsh-Bohmer, and M. Winn, as well as the MURDOCK study community registry. Funding for the collection of control samples was funded in whole or part with federal funds by the Center for HIV/AIDS Vaccine Immunology (“CHAVI”) under a grant from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Grant Number UO1AIO67854, Bryan ADRC NIA P30 AG028377, NINDS RC2NS070344, NINDS 1RC2NS070342-01, and the Division of Intramural Research, NIAID, NIH.
This study was funded in part by the ENRAH for SMEs Consortium grant (LSSM-CT-2005-516513 ENRAH for SMEs) of the European Commission Research Programme FP6, Inserm, CNRS, UPMC Univ Paris 06, Association Française contre les myopathies (SN), Association Française de l’Hémiplégie Alternante (SN, AMvdM, BdV), A.I.S.EA Onlus (FG, GN), the Center for Human Genome Variation, the Wellcome Trust (084730, SMS), UL1RR025764 to the University of Utah, Center for Clinical and Translational Sciences (KJS), NIH grant 1T32HL105321-01 (CH), The University of Luxembourg–Institute for Systems Biology Program (CH), and the Center for Medical Systems Biology established in the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (project nr. 050-060-409 from AMvdM/MDF). SN is a recipient of a Contrat d’Interface from Assistance Publique-Hôpitaux de Paris.
Footnotes
Competing Financial Interests DBG, ELH, KVS, MAM, and Duke University are named on a patent application filed by Duke University based on this work.
Author Contributions MAM, SMS, and DBG jointly supervised this research. ELH, YH, SMS, MAM, DBG conceived and designed the study. Genetic data were generated and analyzed by ELH, KJS, YH, FG, SN, BdV, FDT, CH, LBJ, KVS, CG, LL, GN, AA, and AMJMvdM. Patient DNA samples and phenotypic information of the AHC patients were collected, compiled, and analyzed by KJS, FG, SN, NMW, BdV, FDT, BF, SH, EP, MTS, TMN, LV, SPR, KJM, KS, LJP, JH, MDF, AMB, GKH, CMW, DW, BJL, PU, MDK, IES, GN, AA, SMS, MAM, the European AHC Genetics Consortium, IBAHC Consortium, and the ENRAH for SME Consortium. ELH, AMJMvdM, SMS, MAM, and DBG wrote the paper. All authors reviewed the compiled manuscript.
URLs Ensembl database, www.ensembl.org
Human Gene Mutation Database (HGMD), www.hgmd.org
I.B.AHC Biobank and Clinical Registry for Alternating Hemiplegia, http://en.ibahc.org
Image J software, http://rsbweb.nih.gov/ij/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://snp.gs.washington.edu/EVS
SequenceVariantAnalyzer (SVA), http://www.svaproject.org
References
- 1.Verret S, Steele JC. Alternating hemiplegia in childhood: a report of eight patients with complicated migraine beginning in infancy. Pediatrics. 1971;47:675–680. [PubMed] [Google Scholar]
- 2.Bourgeois M, Aicardi J, Goutieres F. Alternating hemiplegia of childhood. J. Pediatr. 1993;122:673–679. doi: 10.1016/s0022-3476(06)80003-x. [DOI] [PubMed] [Google Scholar]
- 3.Mikati MA, Kramer U, Zupanc ML, Shanahan RJ. Alternating hemiplegia of childhood: clinical manifestations and long-term outcome. Pediatr. Neurol. 2000;23:134–141. doi: 10.1016/s0887-8994(00)00157-0. [DOI] [PubMed] [Google Scholar]
- 4.Sweney MT, et al. Alternating hemiplegia of childhood: early characteristics and evolution of a neurodevelopmental syndrome. Pediatrics. 2009;123:e534–541. doi: 10.1542/peds.2008-2027. [DOI] [PubMed] [Google Scholar]
- 5.Panagiotakaki E, et al. Evidence of a non-progressive course of alternating hemiplegia of childhood: study of a large cohort of children and adults. Brain. 2010;133:3598–3610. doi: 10.1093/brain/awq295. [DOI] [PubMed] [Google Scholar]
- 6.Rho JM, Chugani HT. Alternating hemiplegia of childhood: insights into its pathophysiology. J. Child Neurol. 1998;13:39–45. doi: 10.1177/088307389801300107. [DOI] [PubMed] [Google Scholar]
- 7.Neville BG, Ninan M. The treatment and management of alternating hemiplegia of childhood. Dev. Med. Child Neurol. 2007;49:777–780. doi: 10.1111/j.1469-8749.2007.00777.x. [DOI] [PubMed] [Google Scholar]
- 8.Mikati MA, et al. A syndrome of autosomal dominant alternating hemiplegia: clinical presentation mimicking intractable epilepsy; chromosomal studies; and physiologic investigations. Neurology. 1992;42:2251–2257. doi: 10.1212/wnl.42.12.2251. [DOI] [PubMed] [Google Scholar]
- 9.Swoboda KJ, et al. Alternating hemiplegia of childhood or familial hemiplegic migraine? A novel ATP1A2 mutation. Ann. Neurol. 2004;55:884–887. doi: 10.1002/ana.20134. [DOI] [PubMed] [Google Scholar]
- 10.Bassi MT, et al. A novel mutation in the ATP1A2 gene causes alternating hemiplegia of childhood. J. Med. Genet. 2004;41:621–628. doi: 10.1136/jmg.2003.017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanmolkot KR, et al. Novel mutations in the Na+, K+-ATPase pump gene ATP1A2 associated with familial hemiplegic migraine and benign familial infantile convulsions. Ann. Neurol. 2003;54:360–366. doi: 10.1002/ana.10674. [DOI] [PubMed] [Google Scholar]
- 12.De Fusco M, et al. Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat. Genet. 2003;33:192–196. doi: 10.1038/ng1081. [DOI] [PubMed] [Google Scholar]
- 13.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13:163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 14.de Carvalho Aguiar P, et al. Mutations in the Na+/K+ -ATPase alpha3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron. 2004;43:169–175. doi: 10.1016/j.neuron.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 15.Anselm IA, Sweadner KJ, Gollamudi S, Ozelius LJ, Darras BT. Rapid-onset dystonia-parkinsonism in a child with a novel ATP1A3 gene mutation. Neurology. 2009;73:400–401. doi: 10.1212/WNL.0b013e3181b04acd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svetel M, et al. Rapid-onset dystonia-parkinsonism: case report. J. Neurol. 2010;257:472–474. doi: 10.1007/s00415-009-5385-y. [DOI] [PubMed] [Google Scholar]
- 17.Kamm C, et al. Novel ATP1A3 mutation in a sporadic RDP patient with minimal benefit from deep brain stimulation. Neurology. 2008;70:1501–1503. doi: 10.1212/01.wnl.0000310431.41036.e0. [DOI] [PubMed] [Google Scholar]
- 18.Blanco-Arias P, et al. A C-terminal mutation of ATP1A3 underscores the crucial role of sodium affinity in the pathophysiology of rapid-onset dystonia-parkinsonism. Hum. Mol. Genet. 2009;18:2370–2377. doi: 10.1093/hmg/ddp170. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa H, Shinoda T, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump (Na+,K+-ATPase) with bound potassium and ouabain. Proc. Natl. Acad. Sci. USA. 2009;106:13742–13747. doi: 10.1073/pnas.0907054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellus GA, et al. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am. J. Hum. Genet. 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum. Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- 22.Clapcote SJ, et al. Mutation I810N in the alpha3 isoform of Na+,K+-ATPase causes impairments in the sodium pump and hyperexcitability in the CNS. Proc. Natl. Acad. Sci. USA. 2009;106:14085–14090. doi: 10.1073/pnas.0904817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenson PD, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 24.Jain E, et al. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruitt KD, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu M, et al. Inferring copy number variants in high-coverage genomes using ERDS. Am. J. Hum. Genet. doi: 10.1016/j.ajhg.2012.07.004. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge D, et al. SVA: software for annotating and visualizing sequenced human genomes. Bioinformatics. 2011;27:1998–2000. doi: 10.1093/bioinformatics/btr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.