SUMMARY
Ubiquitin modification is mediated by a large family of specificity determining ubiquitin E3 ligases. To facilitate ubiquitin transfer, RING E3 ligases bind both substrate and a ubiquitin E2 conjugating enzyme linked to ubiquitin via a thioester bond, but the mechanism of transfer has remained elusive. Here we report the crystal structure of the dimeric RING of RNF4 in complex with E2 (UbcH5a) linked by an isopeptide bond to ubiquitin. While the E2 contacts a single protomer of the RING, ubiquitin is folded back onto the E2 by contacts from both RING protomers. The C-terminal tail of ubiquitin is locked into an active site groove on the E2 by an intricate network of interactions, resulting in changes at the E2 active site. This arrangement is primed for catalysis as it can deprotonate the incoming substrate lysine residue and stabilise the consequent tetrahedral transition state intermediate.
By altering the fate of modified proteins, conjugation with ubiquitin and its homologues plays a central role in eukaryotic biology underpinning cell signaling, protein degradation and stress responses. In most cases ubiquitin is transferred to its target proteins from a thioester complex with a ubiquitin conjugating enzyme (E2) by a large family of ubiquitin E3 ligases (E3)1. The RING family of E3s, of which over 600 are encoded in the human genome, possess a conserved arrangement of cysteine and histidine residues that co-ordinate two zinc atoms2. RING E3 ligases bind both substrate and E2~ubiquitin (E2~Ub) thioester, but the molecular basis by which the RING activates the E2~Ub bond for transfer of ubiquitin to substrate has remained elusive.
RNF4 is a SUMO-targeted ubiquitin ligase3 that plays a key role in the DNA damage response4-6 and in arsenic therapy for Acute Promyelocytic Leukaemia7,8. RNF4 contains multiple SUMO-interaction motifs, allowing it to engage polySUMO modified substrates, and a RING domain2 that is responsible for dimerization and catalysis of ubiquitin transfer3,9. Our understanding of RING-catalysed ubiquitylation has been hindered by the lack of structures of the key intermediate: a RING bound to E2~Ub. Obtaining this key complex is difficult, since the thioester (or engineered oxyester) bond linking E2 and ubiquitin is highly activated and unstable in the presence of an E3.
Structure of the RING–UbcH5a~Ub complex
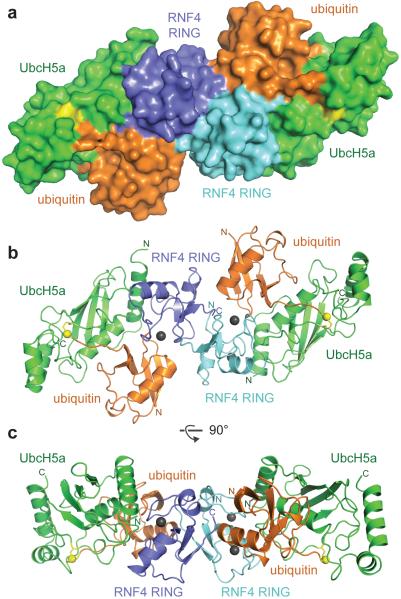
We have engineered a mimic of the E2~Ub thioester bond by replacing the active site cysteine of the E2 UbcH5a with a lysine to generate an isopeptide (amide) bond between the C-terminus of ubiquitin and the ε-amino group of the introduced lysine (Supplementary Fig. 1, 2). Isopeptide-linked UbcH5a~Ub bound selectively to the RNF4 RING and acted as a potent inhibitor of RNF4 mediated substrate ubiquitylation confirming it is indeed an excellent mimic, but crucially, it is stable in the presence of RNF4 (Supplementary Fig. 3). The E2~Ub mimic was mixed in a 2:1 ratio with a fused RNF4 RING dimer3 and crystallised. A 2.2 Å structure of the resulting complex was determined (Supplementary Table 1). The asymmetric unit contains the central RNF4 RING dimer, two UbcH5a molecules and two ubiquitin molecules related by a two-fold axis (Fig. 1). Each UbcH5a molecule contacts a single RING domain and is linked by an isopeptide bond to ubiquitin (Supplementary Fig. 4) that sits at the RING dimer interface. The complex can be envisaged as a dimer of heterotrimers (RING monomer, UbcH5a and ubiquitin).
Figure 1. Structure of the RNF4 RING bound to ubiquitin-loaded UbcH5a.
a, Surface representation of the complex. Individual RING protomers are coloured cyan and blue, UbcH5a is green, ubiquitin is orange and the isopeptide linkage between the C-terminus of ubiquitin and K85 of UbcH5a is shown in yellow. b, Ribbon diagram of the complex with the same orientation and colour scheme as in a. Zinc atoms are indicated as grey spheres. c, As in b, but the complex is rotated by 90o as indicated.
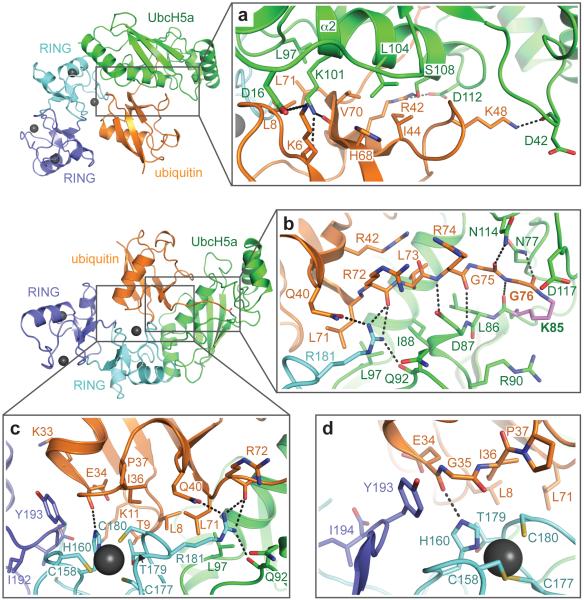
Strikingly, ubiquitin is folded back on to E2, creating an interface that buries approximately 1800 Å2, has 15 hydrogen bonds and 4 salt bridges. L8 of ubiquitin interacts with L97 and K101 of UbcH5a, whereas I44, H68 and V70 in ubiquitin are close to L104, S105 and S108 on the α2 helix of the E2 (Fig 2a). Extensive contacts are evident between the C-terminal 6 residues of ubiquitin and loops surrounding the active site of UbcH5a, particularly residues L86, D87, Q92 and N114, while the side chain of N77 in UbcH5a hydrogen bonds to the isopeptide carbonyl (Fig. 2b). Mapping conserved E2 residues (Supplementary Fig. 5) shows that highly conserved residues surround the active site and the shallow groove that accommodates the C-terminal region of the linked ubiquitin (Supplementary Fig. 6). The other conserved cluster of E2 residues constitutes the binding site for the E3 ligase.
Figure 2. Molecular interfaces in the RNF4 RING–UbcH5a~Ub complex.
a, Detail of the interaction between ubiquitin (orange) and the α2 helix of UbcH5a (green). b, Detail of the interaction interface between ubiquitin (orange) and UbcH5a (green) in the E2 active site groove. The side chain of K85 in UbcH5a that forms the isopeptide bond with ubiquitin is coloured violet. c, The hydrophobic cluster at the centre of the ubiquitin (orange), UbcH5a (green), RING (cyan) heterotrimer. d, Stacking interaction between the main chain of ubiquitin (orange) in one heterotrimer and Y193 of the RING (blue) from the other heterotrimer.
UbcH5a contacts a single protomer of the RING (Supplementary Fig. 7) and the interface is very similar to that previously described for RING–E2 complexes10,11. At the junction of the three molecules in the heterotrimer is a hydrophobic cluster formed by L8, T9 and L71 of ubiquitin, A96 and L97 of UbcH5a, and P137, P178 and R181 of the RING (Fig. 2c and Supplementary Fig. 7). Ubiquitin contacts both protomers of the RING dimer and the interface buries 940 Å2 (Fig. 2c). L8 to K11 and L71 with R72 of ubiquitin contact RING residues T179 to R181 within the same heterotrimer, while the Q31 to Q40 loop of ubiquitin contacts both protomers of the RING dimer. The backbone carbonyl of ubiquitin E34 makes a hydrogen bond with RING residue H160 (zinc ligand) and the main chain E34 to G35 of ubiquitin stacks with the side chain of Y193 of the RING domain from the other heterotrimer (Fig. 2d). These interfaces explain why dimerization of the RNF4 RING is required for activity3,9. Phylogenetic analysis of RNF4 from a wide range of species and sequence comparison of RNF4 with other dimeric RING and U-box E3 ligases indicates that the bound ubiquitin interacts with conserved features of the RING (Supplementary Fig. 8).
The RING domain does not undergo any major structural change as a result of complex formation (Supplementary Fig. 9a). Ubiquitin shows little change in overall structure up to R72, the remaining five residues are however positioned differently as a consequence of being held in the active site groove of UbcH5a. The loop at L8 in ubiquitin has moved over 4 Å to form the hydrophobic cluster with UbcH5a and the RING (Supplementary Fig. 9b). Superposition of the coordinates of unconjugated UbcH5a either free (PDB ID 2C4P), or in a variety of non-covalent complexes12-14, and UbcH5a in the present structure reveals a clear shift at D117. The unconjugated structures all have the side chain of D117 pointing towards C85 in a position that would clash with the isopeptide (thioester) bond (Supplementary Fig. 9c,d).
E2 and Ub residues required for activity
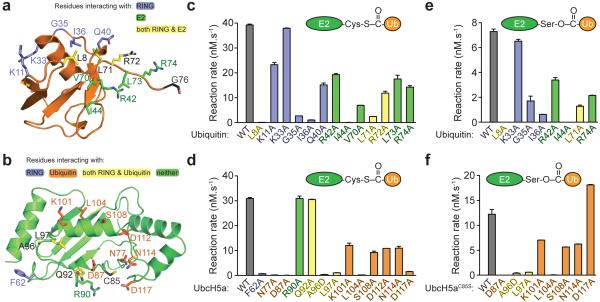
Previous mutational analysis revealed the importance of the RING residue R181, which contacts both E2 and ubiquitin in the present structure (Fig. 2), in ubiquitylation activity of RNF43. Moreover, Y193 in the RING plus L8 and I44 in ubiquitin were shown to be required for activity3. While it was thought that these residues might interact directly, the present structure emphasizes their importance but shows that they are not in direct contact. To further validate our structure, we introduced mutations into ubiquitin and UbcH5a (Fig. 3a,b) and tested these in a single-turnover substrate ubiquitylation assay. Mutations of hydrophobic residues I44 (ubiquitin) and L104 (UbcH5a) at the interface between ubiquitin and UbcH5a abolished ubiquitylation activity, while mutations K101A, S108A and D112A in UbcH5a and R42A in ubiquitin reduced activity modestly (Fig. 3c,d and Supplementary Fig. 10-13). Ubiquitin mutations G35A and I36A at the RING interface substantially (>10x) reduced activity. This was also observed for mutations of hydrophobic residues L8 and L71 in ubiquitin and L97 in UbcH5a at the junction of all the three molecules in the heterotrimer. In the E2 active site groove, mutations N77A and D87A in UbcH5a abolished activity, while D117A severely compromised activity. N114A UbcH5a and R72A, L73A, and R74A ubiquitin all displayed modestly reduced activity (Fig. 3c,d and Supplementary Fig. 10-13).
Figure 3. Mutational analysis of the RNF4 RING–UbcH5a~Ub complex.
a, Side chains of altered residues in ubiquitin contacting RNF4 (blue), UbcH5a (green), or both RNF4 and UbcH5a (yellow). b, Side chains of altered residues in UbcH5a contacting RNF4 (blue), ubiquitin (orange), both RNF4 and ubiquitin (yellow), or neither (green). c, Reaction rates were determined (mean ± s.d. of duplicates) for single-turnover, RNF4-dependent substrate ubiquitylation assays with mutant forms of ubiquitin. Wild-type ubiquitin is in grey and mutants are colour coded as in a. d, Assays with UbcH5a mutants quantified as in c and colour coded as in b. e, RNF4-mediated hydrolysis of UbcH5aC85S~Ub oxyesters with mutations in ubiquitin. Rates are mean ± s.d. of duplicates. f, As in e, with mutations in UbcH5a.
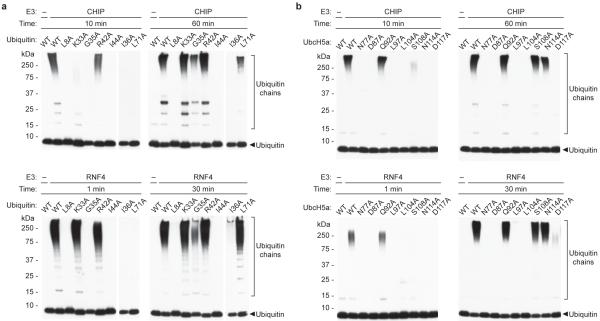
To discriminate between residues in ubiquitin and E2 that influence the ability of the substrate lysine to carry out nucleophilic attack on the E2~Ub thioester and those residues involved in activating the E2~Ub bond, we carried out substrate-independent assays that measure the ability of the RNF4 RING to catalyse hydrolysis of an E2~Ub oxyester bond3 (Fig. 3e,f and Supplementary Fig. 14,15). Mutations in ubiquitin and UbcH5a that reduced substrate-dependent ubiquitylation also reduced oxyester hydrolysis, with the important exception of D117A which was defective in substrate ubiquitylation, but displayed wild-type levels of oxyester hydrolysis (Fig. 3d,f). We investigated whether residues in ubiquitin and UbcH5a that are important for RNF4-mediated ubiquitylation have a more general role in E3 catalyzed transfer. The ubiquitin and UbcH5a mutants were tested in combination with the unrelated U-box E3 ligase CHIP (C-terminus of Hsp70-Interacting Protein) using an autoubiquitylation assay. Although there are relatively modest quantitative differences in ubiquitylation, the effect of the mutations on CHIP and RNF4 activity is very similar (Fig. 4). Thus, it is likely that a conserved mechanism is employed by CHIP and RNF4 to catalyze ubiquitin transfer.
Figure 4. The same interfaces in E2 and ubiquitin are important for CHIP and RNF4 activity.
a, Autoubiquitylation activity of CHIP (upper panel) and RNF4 (lower panel) with ubiquitin mutants. Western blots probed with anti-ubiquitin antibody are shown. Longer exposure is shown for I36A and L71A ubiquitin, as binding of the antibody is affected by these mutations. b, Autoubiquitylation activity as in a, but with UbcH5a mutants.
Mechanism of RING mediated ubiquitylation
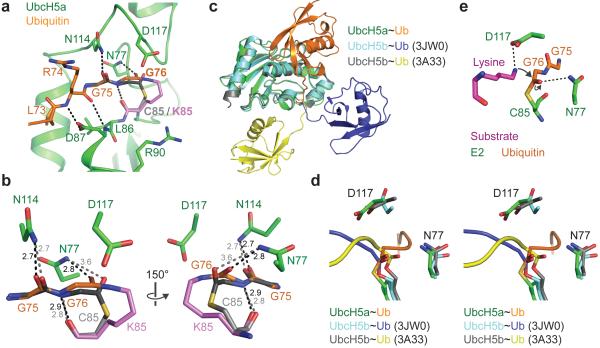
Using the isopeptide-linked E2~Ub in our crystal structure we constructed a model of the E2~Ub thioester by replacing K85 in UbcH5a with a cysteine and minimizing the geometry. The resulting model shows very minor changes; the Sγ and Cα atoms in C85 are shifted 1.0 Å and 0.2 Å from Cγ and Cα atoms of K85, with smaller changes in I84 and L86. In ubiquitin the Cα atoms of G76 and G75 have moved 0.5 Å and 0.2 Å, respectively. The carbonyl group of the thioester at G76 has moved 0.6 Å and rotated around 45 °, resulting in the hydrogen bond with N77 being extended to 3.6 Å (Fig. 5a,b). Coupled with the mutational analysis and evidence that the isopeptide-linked E2~Ub is a competitive inhibitor of ubiquitylation, we conclude that the crystal structure is a relevant model for the key E2~Ub–RING heterotrimeric intermediate. In the absence of an E3 ligase, the ubiquitin thioester-linked to the E2 can adopt a wide range of different conformations that also include a “folded-back” conformation15-17. As free ubiquitin has no detectable affinity for the RNF4 RING we suggest the initial interaction will be between E2 and the RING. In this encounter, with the E2 bound to one RING protomer, the thioester-linked ubiquitin would be engaged by Y193 of the other RING protomer and “folded back” to contact the α2 helix of UbcH5a, while its C-terminus is extended and locked in the active site groove of the E2. This orientates the planar thioester bond such that the ubiquitin G76 thioester carbonyl is in the optimal arrangement for nucleophilic attack by the incoming substrate lysine. This arrangement of the E2 active site was not observed in a UbcH5b~ubiquitin oxyester alone18 or when a UbcH5b~ubiquitin oxyester is bound to a HECT E3 ligase19 (Fig. 5c,d). The nucleophilic attack by the substrate lysine would result in formation of a tetrahedral intermediate on the G76 carbonyl carbon. The G76 carbonyl oxygen, with its developing negative charge, would move down below the plane of the original thioester bond and hydrogen bond to N77, stabilizing the tetrahedral intermediate. In fact the atoms would move towards the experimental orientation of the carbonyl in the isopeptide bond that makes a hydrogen bond with N77 of 2.8 Å. The role of UbcH5a D117, which sits above the thioester and is re-positioned by ubiquitin binding, has been clarified by analysis of the D117A mutant. Of the mutants which are defective in the ubiquitylation assay, only D117A retains wild-type levels of oxyester hydrolysis (Fig. 3f). Since the E2~Ub oxyester bond is hydrolyzed in the presence of E3 (no transfer to substrate)3, only a residue whose sole function is to position and/or activate the incoming lysine nucleophile should possess activity in oxyester hydrolysis assays but be inactive in ubiquitylation.
Figure 5. E3-mediated structural changes associated with the catalytically primed form of UbcH5a~Ub.
a, Model of UbcH5a~ubiquitin thioester (grey) with UbcH5a K85 (violet). b, Comparison of modeled thioester with isopeptide linkage. Hydrogen bonds are black (isopeptide) or grey (modeled thioester) dashes. c, Comparison of position of ubiquitin relative to E2 in UbcH5a~ubiquitin–RING complex reported here with UbcH5b~ubiquitin–HECT(NEDD4L) complex (PDB 3JW0)19, and UbcH5b~Ub oxyester (PDB 3A33)18. d, RING mediated remodeling of UbcH5a active site. The position of the C-terminus of ubiquitin linked to the active site cysteine/serine of the E2 is shown relative to residues N77 and D117 in the three structures shown in c. e, Model for nucleophilic attack by substrate lysine (pink) on the E2~Ub thioester bond, based on the SUMO-RanGAP1–Ubc9–RanBP2 structure25.
Implications for transfer of Ub and Ubls
This is the first structure of a RING E3 ligase bound to a ubiquitin-loaded E2, but the mechanism proposed here for ubiquitin transfer to substrate is consistent with previous work. The key roles played by residues N7720 and D11721 in E2 catalytic activity have been suggested. Evidence that activation of the thioester bond requires both ubiquitin/ubiquitin-like modifier (Ubl) and E2 to be bound by the E3 comes from previous work on RNF43, the Siz122 and RanBP223 SUMO E3 ligases, and the NEDD4L HECT E3 ligase19. The “folded-back” conformation where the I44 hydrophobic patch of ubiquitin (or equivalent region of SUMO) engages the α2 helix of the E2 has been suggested as an intermediate in ubiquitin/Ubl transfer based on NMR models15,17, mutagenesis coupled with modeling22,24, and from the structure of a SUMO-substrate, E2, E3 product complex23,25. Comparing the NMR model of Ubc1~Ub thioester15 with the present structure shows that although ubiquitin in the Ubc1~Ub thioester is in the “folded-back” conformation, it is different from the present structure where interactions between ubiquitin and the RING extend and tension the ubiquitin C-terminus, locking it down into the E2 active site groove. In the absence of its cognate E3 the ubiquitin C-terminal tail in the Ubc1~Ub complex is not locked down in the Ubc1 active site groove and the thioester is thus not activated (Supplementary Fig. 16).
The “folded-back” conformation was also observed in the structure of SUMO-modified RanGAP1 in complex with Ubc9 and the SUMO E3 ligase RanBP225 (trapped product complex). Position of the SUMO C-terminal tail and hydrogen bonding interactions within the active site groove of Ubc9 are remarkably similar to those seen for UbcH5a~Ub bound to the RNF4 RING (Supplementary Fig. 17). Although both RNF4 and RanBP2 interact with ubiquitin/SUMO to lock it into this conformation, molecular details of these contacts are rather different. While the RING domain interacts with a hydrophobic patch in ubiquitin containing L8, I36, and L71, RanBP2 holds SUMO using a SUMO-interaction motif. Superimposing Ubc9 from the RanGAP1-SUMO–Ubc9–RanBP2 complex with UbcH5a from the RNF4 RING–UbcH5a~Ub structure allows a model of the catalytic transfer complex to be constructed (E2~Ub thioester, E3 and substrate) (Fig. 5e, Supplementary Fig. 17d,e). This model both unifies and provides clear molecular rational for a body of existing data on ubiquitylation.
Although RNF4 is a rather simple E3 ligase it seems likely that similar principles of E2~Ub activation will be employed by structurally more complex ubiquitin ligases such as the Cullin based ligases26 and the Anaphase Promoting Complex/Cyclosome27 that are also RING dependent. E3 ligases for other Ubls are also likely to use a similar catalytic mechanism23,25. The unifying concept is that the E3 activates E2~Ub/Ubl thioester by holding the Ub/Ubl in the “folded-back” position, extending its C-terminal tail. This is akin to tensioning a spring that would be released by cleavage of the thioester and formation of the isopeptide bond. Although details of the molecular contacts that fold back the Ub/Ubl will vary, it is the position of the C-terminal tail of the Ub/Ubl in the active site groove of the E2 that is key to the process.
Methods
Cloning, expression and purification of recombinant proteins
Expression and purification of Rattus norvegicus RNF4, human UbcH5a, and His6-tagged linear fusion of four SUMO-2 molecules (4 × SUMO-2) has been described previously3. Mutations S22R28 and C85K were introduced into UbcH5a using PCR-based site-directed mutagenesis and the mutant protein was expressed and purified as described for wild-type UbcH5a. A linear fusion of two RNF4 RING domains was generated by sub-cloning the first RING domain (RING1, residues 131-194 of R. norvegicus RNF4) into pLou3 vector8 using NcoI and BamHI restriction sites. The second RING domain (RING2, residues 131-194) was inserted using BamHI and HindIII restriction sites with a single glycine residue as a linker between the two RINGs. The RING1–RING2 linear fusion was expressed and purified as described for wild-type RNF43. Human ubiquitin (residues 1-76) was sub-cloned into pHIS-TEV-30a vector29 and expressed in BL21(DE3) E. coli cells at 37°C for 4 hours after induction with 1 mM IPTG. His6-tagged ubiquitin was purified by Ni-NTA (Qiagen) affinity chromatography and dialyzed overnight into 20 mM Tris, 150 mM NaCl, pH 8.0. To cleave off the His6-tag, ubiquitin was incubated with TEV protease, followed by Ni-NTA affinity chromatography to remove any uncleaved His6-tagged ubiquitin, the free His6-tag and the TEV protease (also His6-tagged). Purified untagged ubiquitin was then dialyzed against 50 mM Tris, pH 7.5. As a result of cloning, the ubiquitin construct contains four extra residues at the N-terminus (Gly-Ala-Met-Gly) after cleavage with TEV protease.
Preparation UbcH5a~Ub connected with an isopeptide bond
To generate the UbcH5aS22R,C85K~Ub conjugate, UbcH5a S22R,C85K (200 μM) was incubated with His6-tagged ubiquitin (200 μM) and His6-Ube1 (1 μM) at 35°C for 26 hours in a buffer containing 3 mM ATP, 5 mM MgCl2, 50 mM Tris pH 10.0, 150 mM NaCl, and 0.8 mM TCEP. Subsequently, imidazole was added to a final concentration of 20 mM and the sample was applied onto a Ni-NTA column pre-equilibrated with binding buffer (50 mM Tris, 150 mM NaCl, 20 mM imidazole, 0.5 mM TCEP, pH 7.5). The column was washed with binding buffer and the E2~Ub conjugate was eluted with elution buffer (50 mM Tris, 150 mM NaCl, 150 mM imidazole, 0.5 mM TCEP, pH 7.5). Elution fractions containing the E2~Ub conjugate were pooled and TEV protease was added to the sample to cleave off the His6-tag from ubiquitin, followed by overnight dialysis at 4°C against 50 mM Tris, 150 mM NaCl, 0.5 mM TCEP, pH 7.5. Subsequently, the sample was passed through a Ni-NTA column pre-equilibrated in binding buffer to remove any uncleaved E2~HisUb conjugate and the TEV protease (also His6-tagged). A flow-through fraction was concentrated and applied onto a HiLoad 16/60 Superdex 75 gel filtration column (GE Healthcare) pre-equilibrated in 20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH 7.0. The purified UbcH5aS22R,C85K~Ub conjugate was concentrated to 5 mg.ml−1, flash-frozen in liquid nitrogen and stored at −80°C.
Crystallization of the RNF4 RING–UbcH5aS22R,C85K~Ub complex
The UbcH5aS22R,C85K~Ub conjugate was mixed with the linear fusion of two RNF4 RING domains in a 2:1 molar ratio and the complex was concentrated to 17 mg.ml−1. Proteins were buffer-exchanged into 20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH 7.0 during the concentration step. Crystals were grown at 20 °C using the sitting-drop vapour diffusion method by mixing 1 μl of protein complex with 1 μl of reservoir solution (18% (w/v) PEG 3,000, 0.1 M Tris pH 7.2, 0.2 M calcium acetate). Crystals appeared after one or two days and grew to their final size within ~5-7 days. Crystals were briefly soaked in a cryprotectant solution (10% (v/v) ethyleneglycol, 18% (w/v) PEG 3,000, 0.1 M Tris pH 7.2, 0.2 M calcium acetate) before flash-freezing in liquid nitrogen.
Data collection and structure determination
Diffraction data were recorded on a Rigaku Saturn CCD with X-rays generated from a Rigaku 007 HF generator. Resolution of the crystals was limited by our ability resolve the long cell edge due to high mosaic spread (approx 1°) and orientation of the crystal. The structure was solved by molecular replacement using PHASER30 as implemented in the CCP4 package31. A lower resolution (3 Å) data set for the heterotrimer was solved by finding a single RNF4 RING domain (PDB 2XEU)3, followed by E2 UbcH5a (2YHO)14 and ubiquitin (1UBQ, truncated at residue R72)32. Interestingly, searching for a second copy of each domain alone did not produce a clear solution. Instead searches using the RING dimer, followed by E2, ubiquitin and then the E2~ubiquitin conjugate, or RING monomer, then E2, then ubiquitin, followed by RING–E2~ubiquitin heterotrimer gave solutions. When a higher resolution data set (2.2 Å) was obtained, the heterotrimer from the low resolution structure was used to solve this data. The models were adjusted manually using COOT33, the isopeptide bond and the missing ubiquitin residues were clearly visible and built into the model. The model was refined using REFMAC534, NCS restrains were used throughout. MolProbity35 was used to correct side chain conformations and as a guide to manual building. The final model has good geometry with MolProbity score of 1.42 (99th percentile). 98.6% of residues are in the favoured regions of Ramachandran plot and no residues are in the disallowed regions. Molecular interfaces were analyzed using the PISA server36. The two RING molecules in the crystal are fused together into a single protomer but comparison with the native (unfused) dimeric RING domain structure3 shows that the arrangement of the domains relative to each other and the contacts between them are very similar. For clarity we therefore discuss the dimeric RING domain structure in this crystal as if it were formed by two proteins.
UbcH5a~Ub thioester model
The UbcH5a~Ub thioester model was generated from the crystal structure by replacing K85 in UbcH5a with a cysteine using COOT33. The N-Cα-Cβ-Sγ dihedral angle was set to 180 ° (the same conformer as in 3PTF12). The geometry of the model was then minimized by REFMAC34 for 10 cycles, adding hydrogens at expected positions. Restraints for the thioester linkage were generated using JLigand37.
Ubiquitylation assays
A single-turnover substrate ubiquitylation assay for RNF4 has been described previously3. Briefly, UbcH5a~Ub thioester was first prepared in the absence of RNF4 and a substrate. The charging reaction contained 100 μM UbcH5a, 120 μM ubiquitin, 0.2 μM His-Ube1 (E1), 3 mM ATP, 5 mM MgCl2, 50 mM Tris, 150 mM NaCl, 0.5 mM TCEP, pH 7.5. Apyrase (4.5 U.ml−1, New England BioLabs) was then added to deplete ATP and thus to stop the charging reaction. The UbcH5a~Ub thioester (~20 μM) was then mixed with RNF4 (0.275 μM) and a substrate (5.5 μM) buffered with 50 mM Tris, 150 mM NaCl, 0.5 mM TCEP, 0.1% (v/v) NP40, pH 7.5. A linear fusion of four SUMO-2s (4 × SUMO-2), labelled with iodine-125, was used as a substrate for RNF4. Reactions were incubated at room temperature, stopped by SDS-PAGE loading buffer and analysed by SDS-PAGE, followed by phosphorimaging. Reactions were performed in duplicate and reaction rates are shown as mean ± standard deviation. In assays comparing mutant forms of ubiquitin, untagged UbcH5a and untagged ubiquitin (the construct described above) were used. His6-tagged UbcH5a and untagged ubiquitin (obtained from Sigma) were used in assays comparing UbcH5a mutants.
Single-turnover autoubiquitylation assays contained ~20 μM UbcH5a~Ub thioester and either 0.55 μM RNF4 or 1.1 μM CHIP38 buffered with 50 mM Tris, 150 mM NaCl, 0.5 mM TCEP, 0.1% (v/v) NP40, pH 7.5. Reactions were incubated at room temperature, stopped by SDS-PAGE loading buffer and analysed by Western blotting with anti-ubiquitin antibody (Dako).
UbcH5aC85S~Ub oxyester hydrolysis assay
UbcH5aC85S~Ub oxyesters were prepared by incubating UbcH5a C85S (100 μM) with ubiquitin (120 μM) and His-Ube1 (1 μM) in buffer containing 3 mM ATP, 5 mM MgCl2, 50 mM Tris, 150 mM NaCl, 0.5 mM TCEP, pH 7.5 for ~14 hours at 37°C. Apyrase (4.5 U.ml−1) was then added to deplete ATP. UbcH5aC85S~Ub oxyesters were mixed with RNF4 (8.8 μM), followed by incubation at room temperature. Reactions were stopped by SDS-PAGE loading buffer and analysed by SDS-PAGE. Gels were stained with Coomassie blue, scanned using the Odyssey CLx Infrared Imaging System (LI-COR Biosciences) and quantified using the LI-COR software. Reactions were performed in duplicate and reaction rates are shown as mean ± standard deviation.
Pull-down assay
Binding between MBP-tagged RNF4 and ubiquitin-loaded UbcH5a was analyzed by a pull-down assay as described previously3.
Mass spectrometry
UbcH5a S22R,C85K and the UbcH5aS22R,C85K~Ub conjugate (both 5 μg) were fractionated by 10% SDS-PAGE. Coomassie stained bands were excised and tryptic peptides extracted as described previously39, substituting iodoacetamide for chloroacetamide to limit false identifications of ubiquitylation sites40. Peptide samples were analysed by LC-MS/MS using a Q Exactive mass spectrometer (Thermo Scientific) employing high resolution HCD fragmentation. Peptides were identified and quantified by MaxQuant (v 1.2.2.5) running the Andromeda search engine41 using both a human proteome (Human IPI v3.68) and the recombinant protein sequence databases. Both Gly-Gly and Leu-Arg-Gly-Gly variable modifications to lysine were included in the search to detect ubiquitylation by two methods.
Supplementary Material
Acknowledgements
We thank Magnus Alphey and Emma Branigan for assistance with data collection. His-Ube1 was a kind gift from the Division of Signal Transduction Therapy, University of Dundee. CHIP was a kind gift from Axel Knebel and Philip Cohen (University of Dundee). A.P. was funded by the Wellcome Trust. This work was supported by a grant to R.T.H. from Cancer Research UK. Structural biology was supported by Scottish Funding Council (ref SULSA) and Wellcome Trust (program grant JHN).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
A.P. cloned, expressed and purified proteins, carried out structural analysis, conducted biochemical experiments and interpreted the data. E.G.J. purified recombinant proteins and carried out biochemical analysis. M.H.T. carried out mass spectrometry analysis. J.H.N. contributed to structural analysis and data analysis. A.P., J.H.N. and R.T.H. wrote the paper. R.T.H conceived the project and contributed to data analysis.
Author Information
Coordinates and structure factors of the RNF4 RING–UbcH5aS22R,C85K~Ub complex were deposited in the Protein Data Bank under accession code 4AP4. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J. Cell Sci. 2012;125:539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 2.Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biochem. Sci. 2011 doi: 10.1016/j.tibs.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Plechanovová A, et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nature Struct. Mol. Biol. 2011;18:1052–1059. doi: 10.1038/nsmb.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012;26:1179–1195. doi: 10.1101/gad.188284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo K, Zhang H, Wang L, Yuan J, Lou Z. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 2012 doi: 10.1038/emboj.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin Y, et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012;26:1196–1208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lallemand-Breitenbach V, et al. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nature Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 8.Tatham MH, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nature Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 9.Liew CW, Sun H, Hunter T, Day CL. RING domain dimerization is essential for RNF4 function. Biochem. J. 2010;431:23–29. doi: 10.1042/BJ20100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mace PD, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 11.Bentley ML, et al. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosanac I, et al. Modulation of K11-linkage formation by variable loop residues within UbcH5A. J. Mol. Biol. 2011;408:420–431. doi: 10.1016/j.jmb.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Bosanac I, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol. Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, et al. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 2011;25:1262–1274. doi: 10.1101/gad.2056211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton KS, et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 16.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme~ubiquitin conjugate. Biochemistry. 2011;50:1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakata E, et al. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure. 2010;18:138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B~ubiquitin-HECT(NEDD4L) complex. Mol. Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu PY, et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nature Struct. Mol. Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 22.Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell. 2009;35:669–682. doi: 10.1016/j.molcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gareau JR, Reverter D, Lima CD. Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2. J. Biol. Chem. 2012;287:4740–4751. doi: 10.1074/jbc.M111.321141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calabrese MF, et al. A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nature Struct. Mol. Biol. 2011;18:947–949. doi: 10.1038/nsmb.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber A, et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 28.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Martin SF, Hattersley N, Samuel ID, Hay RT, Tatham MH. A fluorescence-resonance-energy-transfer-based protease activity assay and its use to monitor paralog-specific small ubiquitin-like modifier processing. Anal. Biochem. 2007;363:83–90. doi: 10.1016/j.ab.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 30.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Lebedev AA, et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M, et al. Chaperoned ubiquitylation-crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 39.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature protocols. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen ML, et al. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nature methods. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 41.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.