Abstract
Sialoadhesin (Sn) is a macrophage (Mϕ)-restricted receptor that recognizes sialylated ligands on host cells and pathogens. Although Sn is thought to be important in cellular interactions of Mϕs with cells of the immune system, the functional consequences of pathogen engagement by Sn are unclear. As a model system, we have investigated the role of Sn in Mϕ interactions with heat-killed Campylobacter jejuni expressing a GD1a-like, sialylated glycan. Compared to Sn-expressing bone marrow-derived macrophages (BMDM) from wild-type mice, BMDM from mice either deficient in Sn or expressing a non-glycan–binding form of Sn showed greatly reduced phagocytosis of sialylated C. jejuni. This was accompanied by a strong reduction in MyD88-dependent secretion of TNF-α, IL-6, IL-12, and IL-10. In vivo studies demonstrated that functional Sn was required for rapid TNF-α and IFN-β responses to i.v.-injected sialylated C. jejuni. Bacteria were captured within minutes after i.v. injection and were associated with Mϕs in both liver and spleen. In the spleen, IFN-β–reactive cells were localized to Sn+ Mϕs and other cells in the red pulp and marginal zone. Together, these studies demonstrate that Sn plays a key role in capturing sialylated pathogens and promoting rapid proinflammatory cytokine and type I IFN responses.
Introduction
Sialic acids are a large family of nine-carbon sugars that are normally found at the terminal, exposed positions of glycans at the cell surface and on secreted proteins. Sialic acid can also be expressed on the surface of several medically important human pathogens such as Campylobacter jejuni, Neisseria meningitidis, Group B Streptococcus, and Trypanosoma cruzi (reviewed in Ref. 1), as well as on enveloped viruses including HIV (2). The presence of sialic acid on pathogens may contribute to molecular mimicry in which pathogens disguise themselves as host cells and thus circumvent and/or counteract the host's immune responses (reviewed in Ref. 3).
The major group of host sialic acid binding proteins are the siglecs, which are type I transmembrane proteins mostly expressed in the immune system (reviewed in Ref. 1). Sialoadhesin (Sn) is one of the most prominent siglecs with 17 Ig-like extracellular domains. It is well conserved between mice and humans where it is expressed by macrophage (Mϕ) subsets both under resting and inflammatory conditions (4, 5). Sn is distinct from other siglecs in having an unusually large number of Ig domains and in not having cytoplasmic or transmembrane signaling motifs (1, 6, 7). These features are more consistent with a role in cell–cell recognition functions as opposed to intrinsic cell signaling. Recent findings have shown that Sn mediates cross talk between inflammatory Mϕs and T cell subsets that express Sn ligands (8), thereby promoting inflammatory responses during certain autoimmune diseases (9–11). Sn is constitutively expressed at high levels on subsets of Mϕs that are strategically positioned to encounter pathogens in plasma and lymph, namely the marginal zone of the spleen and the subcapsular sinus of the lymph nodes (4). These Sn-expressing Mϕs have been shown to play important roles in Ag capture and transfer to B cells (12–14) and dendritic cells (15), as well as directly presenting glycolipid Ags to invariant NKT cells (16).
To investigate the role of Sn in Mϕ phagocytosis and cytokine responses to a sialylated pathogen, we have used heat-killed C. jejuni as a model system. C. jejuni is a cause of human gastroenteritis and is able to synthesize sialic acid-containing, ganglioside-like mimics on its surface-exposed lipooligosaccharide (LOS) core (1, 17). A complication of C. jejuni infection in a small proportion of infected individuals is the triggering of an acute postinfectious neuroinflammatory disease, Guillain–Barré syndrome, which can develop, generally, 2–3 wk after the initial C. jejuni infection (18). This autoimmune disease is thought to be mediated by structural similarities between peripheral nerve gangliosides and ganglioside-like carbohydrates expressed on the surface LOS of C. jejuni. These LOS glycans induce autoantibody responses that subsequently bind nerve gangliosides and damage tissue (19). Previous studies have demonstrated that sialylation can enhance pathogenicity by increasing invasiveness in intestinal epithelial cells (20). Sialylated C. jejuni can also be recognized by siglecs depending on the type of oligosaccharide presented on the LOS (17, 21). Gangliosides GT1b, GD1a, and GM3, which express terminal α2→3 linked sialic acid, are recognized by Sn, whereas binding is weak to the gangliosides GM1 and GM2, which only express an “internal” sialic acid residue (22). In the case of C. jejuni displaying GD1a-like structures, Sn expressed on CHO cells has been shown to enhance the binding of bacteria (21). This suggests that Sn may be targeted by bacteria in vivo and play a role in modulating immune responses to C. jejuni, but direct evidence for this is lacking.
In the current study, we have examined the in vitro and in vivo role of Sn in recognition and uptake of C. jejuni. Using mice that either lack Sn or express a mutant form of Sn that is unable to bind sialic acid, we demonstrate that Sn is required for rapid uptake of the GB11 strain of C. jejuni displaying GD1a- and GM1-like structures, leading to triggering of cytokine responses. Remarkably, when sialylated bacteria were injected i.v., they were rapidly localized within the spleen and liver and triggered high-level production of TNF-α and IFN-β in an Sn-dependent manner, as measured by cytokine levels in serum, spleen mRNA responses, and immunohistochemistry. This indicates that Sn is a potentially important pathogen recognition molecule for systemically introduced sialylated bacteria and is required for rapid production of proinflammatory and type I IFN responses, which are likely to play an important role in host defense.
Materials and Methods
Mouse strains used and generation of SnW2QR97A mice carrying targeted mutations in the sialic acid binding site of Sn
The generation of Sn-deficient (Sn−/−) mice has previously been described (23). MyD88-deficient (MyD88−/−) mice (24) were kindly provided by Prof. Shizuo Akira (Osaka University). To study mice expressing a non-sialic acid binding form of Sn, we generated a “knockin” mouse strain designated SnW2QR97A. This mouse was engineered to carry two mutations in the Sn gene resulting in the conversion of 2 aa required for sialic acid recognition (25), namely Trp2 and Arg97, which were changed to Gln and Ala, respectively. To produce a targeting construct, a BAC clone containing the Sn gene was isolated from a 129Sv mouse BAC library and used as a template to prepare 5′ and 3′ homology arms as depicted in Supplemental Fig. 1. A neo-pA cassette flanked by loxP sites was inserted between exons 2 and 3 in the 5′ arm. Sequences in exon 2 contained within the 5′ arm were mutated to code for Gln2 in place of Trp2 and Ala97 in place of Arg97. The PGK-TK-pA cassette was cloned into the end of the 5′ homology arm to aid negative selection. Targeting of the Sn gene to generate Sn+/W2QR97A ES cells (Supplemental Fig. 1) was performed by electroporation of E14 mouse embryonic stem cells, followed by positive selection with G418 and negative selection with ganciclovir. Targeted cell lines were identified by PCR and Southern blotting and the neo cassette removed by transfection with pMC-CrePuro as described (26). Primer P1 (5′-CACCACGGTCACTGTGACAA-3′) and primer P2 (5′-GGCCATATGTAGGGTCGTCT-3′) were used to identify clones containing the deleted neo cassette as well as to distinguish the wild-type allele (471 bp) from the targeted allele (635 bp) (Supplemental Fig. 1). Heterozygous Sn+/W2QR97A ES cell clones were microinjected into C57BL/6 × BALB/c blastocysts, which were then reimplanted into recipient female mice. Chimeric mice that had a high degree of ES cell contribution were identified by coat color and crossed to BALB/c mice to allow germline transmission of the mutant allele. Genotyping of Sn+/W2QR97A mice was carried out by PCR of genomic DNA using primers P1 and P2 described earlier (Supplemental Fig. 1).
All wild-type (WT), Sn−/−, and SnW2QR97A mice used in experiments were derived from heterozygotes back-crossed for eight generations onto a C57BL/6 background. Mice were age- and sex-matched and used at 8–15 wk of age. Animals were housed under specific pathogen-free conditions. The animal protocols used in this study were approved by the Ethical Review Committee of the University of Dundee. All procedures involving living animals were conducted according to the requirements of the United Kingdom Home Office Animals Scientific Procedures Act, 1986, under PPL 60/3856.
Isolation and culture of bone marrow-derived macrophages
Femurs and tibias were dissected from euthanized mice and sterilized by incubation in 70% ethanol for 1 min. Bone ends were removed, and bone marrow plugs were flushed with DMEM (Life Technologies, Invitrogen) using a syringe and 25-gauge needle. These plugs were mechanically disrupted by pipetting vigorously with a 5-ml pipette and finally passed through a 70-μm cell strainer prior to centrifugation at 400 × g for 5 min. The bone marrow cells from each mouse were resuspended in 15 ml complete growth medium: DMEM supplemented with 10% heat-inactivated FBS (PAA Laboratories), 2 mM l-glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin (Life Technologies, Invitrogen), and containing 25 ng/ml murine M-CSF (PeproTech). The bone marrow-derived macrophages (BMDM) were expanded on 9-cm bacterial plastic Petri dishes (BD Biosciences) for 7 d and then lifted using 4 mg/ml lidocaine-HCl (Sigma-Aldrich) and 5 mM EDTA in PBS (Ca2+-free and Mg2+-free; Invitrogen). The BMDM were centrifuged at 400 × g for 5 min and cultured for 3 d on bacterial Petri dishes (BD Biosciences) with 250 U/ml IFN-α (PBL IFNSource) to stimulate Sn expression.
Bacterial culture and fluorescent labeling
The WT GB11 strain of C. jejuni expresses a LOS carrying a mixture of GD1a and GM1-like oligosaccharides. The Campylobacter sialic acid transferase II (cst-II) knockout mutant (19) derived from GB11 C. jejuni expresses a truncated LOS outer core due to the absence of sialic acids and therefore lacks ganglioside-like structures. The GB11 WT and GB11 cst-II mutant strains are referred to hereafter as sialylated and non-sialylated C. jejuni, respectively. CFSE-labeled sialylated and non-sialylated C. jejuni were obtained by resuspending the stock at OD600 to OD = 1 and adding an equal volume of 2 μM CFSE (Sigma-Aldrich) for 30 min at 37°C. Bacteria were then inactivated at 65°C for 1 h, and equal mean fluorescence of CFSE was confirmed by flow cytometry. Bacteria were washed twice with PBS and stored at –80°C in PBS with 10% glycerol, and all subsequent studies were conducted on heat-killed organisms. PKH67-labeled bacteria were obtained by labeling a heat-inactivated GB11 WT stock with PKH67 Green Fluorescent cell linker kit (Sigma-Aldrich) according to the manufacturer’s instructions.
Flow cytometry to assay BMDM association with bacteria
BMDM from age- and sex-matched WT, Sn−/−, SnW2QR97A, or MyD88−/− mice were prepared and stimulated with IFN-α as described earlier. BMDM were then lifted using 4 mg/ml lidocaine-HCl and 5 mM EDTA in PBS (Ca2+-free and Mg2+-free), washed, and incubated in microcentrifuge tubes in the dark, rotating at 37°C for 90 min with CFSE-labeled sialylated or non-sialylated C. jejuni (ratio of 1 Mϕ/20 bacteria). Unattached bacteria were then removed by washing. To discriminate between bacteria bound to the cell surface and bacteria that had been phagocytosed, BMDM from half of each replicate were incubated for 15 min with 0.04% trypan blue (Sigma-Aldrich) in PBS to quench extracellular fluorescence as described previously (27). The Escherichia coli–Alexa 488 bacteria were IgG-opsonized for 1 h at 37°C using E. coli opsonizing reagent (both Invitrogen) according to the manufacturer’s recommendations. After extensive washing, bacteria were added to BMDM at a ratio of 10:1 in complete growth media for 20 min on ice and the cells then transferred to a 37°C incubator for 1 h. To remove unbound bacteria, the cells were washed with PBS five times and fixed with 1% formaldehyde. Bacterial uptake and association with BMDM were analyzed by FACSCalibur flow cytometer (BD Biosciences), and the data were processed using FlowJo software 7.6.4 (Tree Star).
Flow cytometry of BMDM cell-surface Ags
BMDM from WT and age- and-sex matched Sn−/− or SnW2QR97A mice were prepared and stimulated with IFN-α as described earlier. BMDM were then lifted using 4 mg/ml lidocaine-HCl and 5 mM EDTA in PBS (Ca2+-free and Mg2+-free) and washed. Ab staining was performed according to conventional protocols at 4°C. For Sn staining on BMDM, cells were incubated at 4°C for 30 min in 2.4G2 hybridoma supernatant. Biotin-conjugated rat anti-mouse Sn mAbs 3D6 and SER-4 were added and incubated for another 1 h. Cells were washed three times with FACS wash buffer (0.5% BSA, 5 mM EDTA, and 2 mM NaN3) and incubated with streptavidin–allophycocyanin (BD Biosciences) for 30 min. Finally, cells were washed three times and fixed with 1% formaldehyde. Fixed cells were analyzed on a FACSCalibur (BD Biosciences) using FlowJo software 7.6.4 (Tree Star).
Live cell video microscopy phagocytosis assays
WT and Sn−/− BMDM (1 × 106) in 1 ml supplemented DMEM medium were seeded onto glass-bottomed Iwaki dishes (VWR) and cultured for at least 2 h at 37°C with 5% CO2. Immediately prior to experiments, DMEM medium was replaced with 0.5 ml prewarmed supplemented CO2-independent medium (Life Technologies, Invitrogen) containing 1 μl LysoTracker Red DND-99 (Invitrogen). PKH-stained C. jejuni (20 × 106) in 0.5 ml prewarmed supplemented CO2-independent medium were added to Mϕs immediately prior to imaging. Video microscopy experiments were performed using a DeltaVision Core microscope (Applied Precision) with an environmental control chamber set at 37°C and oil immersion ×60 objective with a 1.4 numerical aperture. Images were captured at 1-min intervals for 2 h using a CoolSNAP HQ camera (Photometrics) and SoftWoRx Explorer acquisition software (Applied Precision). DeltaVision movies were processed using Volocity 5.0 image analysis software (PerkinElmer).
Cytokine analysis from BMDM supernatants by ELISA
Sialylated and non-sialylated bacteria were either untreated or opsonized with IgG by incubation for 1 h with antiserum from C. jejuni immunized mice. Bacteria were incubated with 106 IFN-α–induced BMDM in microcentrifuge tubes on a rotating wheel at 37°C for 90 min in 0.4 ml DMEM plus 0.1% BSA at a ratio of 20 bacteria per Mϕ. After the incubation, the unbound bacteria were removed by washing BMDM with PBS, and the cells were reseeded in triplicate at 2.5 × 105 cells per well in 0.5 ml complete growth medium on 24-well plates (TPP) for 24 h. Concentrations of TNF-α, IL-6, IL-10, and IL-12 in cell culture supernatants were determined by ELISA Development Kits (PeproTech) according to the manufacturer’s instructions. Supernatants from BMDM that were not incubated with bacteria but were subjected to the same experimental procedures were used as negative controls. Supernatants from BMDM incubated with heat-inactivated Salmonella enterica serovar Typhimurium M525P were used as positive controls.
In vivo cytokine responses to C. jejuni
Mice were injected i.v. with a 100-μl suspension of sialylated C. jejuni in PBS at 109 bacteria/ml or 100 μl PBS as a control. In some experiments, mice were also injected i.v. with 100 μl of polyinosinic:polycytidylic acid [poly(I:C)] solution (1.0 mg/ml) in PBS (Sigma-Aldrich). At the indicated time points, mice were euthanized and blood collected via heart puncture. Sera were separated by Microtainer serum separator tubes (BD Diagnostics) and stored at −20°C prior to measurement of TNF-α, IL-6, IL-10, and IL-12 by ELISA according to the manufacturer’s guidelines (PeproTech). Spleens and livers were immediately snap frozen in liquid nitrogen for RNA extraction and immunohistochemistry.
The localization of sialylated C. jejuni in spleen sections after an i.v. challenge
Mice were injected i.v. with 100 μl PKH67-labeled sialylated C. jejuni diluted in PBS at a concentration of 109 bacteria/ml. After 5 and 20 min, mice were euthanized, exsanguinated, and the spleens collected and frozen in OCT solution (Sakura Finetek). Eight-micrometer cryostat sections were mounted on Superfrost Plus slides (VWR) and stored at −20°C. Prior to immunofluorescence staining, the slides were thawed, fixed for 15 min at −20°C with precooled methanol, washed, and then blocked for 30 min with 5% goat serum (Sigma-Aldrich) in PBS. Sections were stained with rat anti-mouse Sn mAbs 3D6 and SER-4, B220-PE (BioLegend), CD68 (AbD Serotec), polyclonal rabbit anti-mouse IFN-β (PBL IFNSource), and/or F4/80–allophycocyanin (eBioscience), and where necessary with goat anti-rat IgG Alexa 488, goat anti-rat IgG Alexa 647, goat anti-rabbit IgG Alexa 488, and goat anti-rabbit IgG Alexa 555 (all Invitrogen). All sections were counterstained with DNA dye DAPI (Sigma-Aldrich) before mounting slides on coverslips with fluorescent mounting medium (DAKO). A minimum of 10 random images of each biological sample were taken using an LSM 700 laser-scanning confocal microscope (Carl Zeiss) equipped with EC Plan-Neofluar 40×/1.30 Oil DIC M27 objective. Images were acquired with ZEN 2009 software (Carl Zeiss) and processed with ZEN 2009 Light Edition software (Carl Zeiss). Bacterial cell surface area on tissue sections was quantified by Volocity 5.0 imaging software (PerkinElmer).
RNA isolation and quantitative real-time PCR
Total RNA was extracted from cells using RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions and reverse transcribed using the QuantiTect Rev. Transcription kit (Qiagen). cDNA was quantified with a StepOne Plus sequence detection system (Applied Biosystems) using specific primers for each cytokine (Supplemental Table I) and an SYBR Green-based detection system (Applied Biosystems). Input RNA was normalized between samples using endogenous control GAPDH. Fold upregulation of the target gene was calculated based on the relative quantification method as ratio target gene expression (experimental/control).
Statistical analyses
Data were analyzed for statistical significance with GraphPad Prism 5.0 software (GraphPad Software).
Results
Sn is required for efficient uptake of sialylated C. jejuni by BMDM
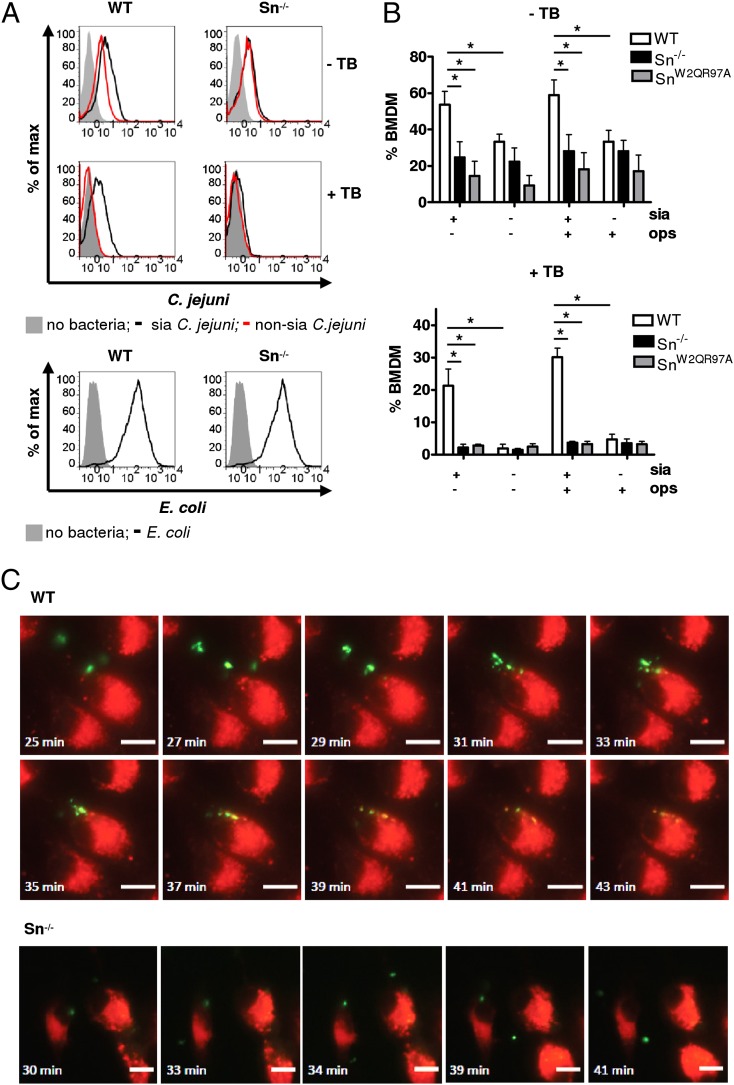
To investigate the potential role of Sn in Mϕ binding and uptake of sialylated bacteria, we used BMDM cultured with IFN-α to upregulate Sn expression (Supplemental Fig. 2A). BMDM prepared from WT mice, Sn−/− mice, and “knockin” mice expressing a non-sialic-acid–binding mutant form of Sn (SnW2QR97A) were incubated with fluorescently labeled sialylated or non-sialylated C. jejuni and analyzed by flow cytometry. A significantly greater percentage of WT BMDM was associated with the sialylated strain of C. jejuni compared with Sn−/− or SnW2QR97A BMDM, whereas non-sialylated bacteria bound at low levels to all BMDM populations (Fig. 1A, 1B, upper panels). After trypan blue quenching of extracellular bacteria (Supplemental Fig. 2B), it was found that only WT BMDM contained intracellular fluorescent sialylated bacteria (Fig. 1B, lower panel). Internalization of the bacteria into WT BMDM phagolysosomes was confirmed by live cell video microscopy (Fig. 1C). Sn−/− BMDM typically showed reduced binding and internalization over the same time frame (Fig. 1C). When BMDM were incubated with IgG-opsonized E. coli, similarly high levels of phagocytosis were seen with both WT and Sn-deficient Mϕs (Fig. 1A, lower panel), showing that there is no intrinsic defect of Sn-deficient Mϕs in phagocytosis.
FIGURE 1.
Binding and phagocytosis of GB11 C. jejuni by WT and Sn−/− BMDM in vitro. (A and B) IFN-α–stimulated BMDM were incubated with CFSE-labeled heat-killed sialylated (sia) or non-sialylated GB11 C. jejuni and analyzed by flow cytometry. To quench the fluorescence of extracellular bound but not phagocytosed bacteria, trypan blue (TB) was added for 15 min and washed out before the analysis. Opsonized E. coli were used as a positive control. One representative experiment (A) and data pooled from three independent experiments (B) are shown. Bars represent the average ± SD. Two-way ANOVA tests were used to assess differences and interactions between groups. *p < 0.01. (C) Live cell video microscopy images showing binding and engulfment of C. jejuni by WT and Sn−/− BMDM. In the upper panels, the first image shows initial contact between the WT Mϕs (red) and C. jejuni (green). Subsequent images show C. jejuni being engulfed by the Mϕs. In the later images, colocalization of C. jejuni with LysoTracker Red DND-99 can be seen (yellow) indicating phagolysosomal fusion. The lower panel shows Sn−/− Mϕ binding to two C. jejuni bacteria. The bacteria remain associated with the Mϕ for 10 min and then dissociate. The time points at which images were taken are displayed; images are representative from two independent experiments. Scale bars, 10 μm.
Sn-dependent phagocytosis triggers elevated cytokine levels
Sn-dependent uptake of sialylated bacteria was shown to result in higher production of cytokines TNF-α, IL-6, IL-12, and IL-10 by IFN-α–induced BMDM after 24-h incubation (Fig. 2A). Non-sialylated bacteria stimulated much lower levels of cytokine production in both WT and Sn-deficient Mϕs, but this was increased in the cases of TNF-α and IL-12 after IgG opsonization of the bacteria (Fig. 2A). When BMDM were exposed to heat-killed Salmonella, high-level production of cytokines was seen with both WT and Sn-deficient Mϕs, showing that there was no intrinsic defect of Sn-deficient Mϕs in cytokine production (Fig. 2A). This Sn-dependent cytokine response is likely due to activation of TLRs by C. jejuni LOS and MyD88-dependent signaling (28, 29), rather than reflecting intrinsic signaling by Sn. To demonstrate this directly, BMDM from MyD88−/− mice were prepared and shown to express normal levels of Sn after induction by IFN-α (Fig. 2B) and phagocytose sialylated bacteria to the same extent as WT BMDM (Fig. 2C). However, MyD88−/− BMDM failed completely to produce TNF-α or IL-6 after phagocytosis of sialylated C. jejuni (Fig. 2D). Therefore, Sn-dependent production of proinflammatory cytokines is strictly MyD88 dependent, supporting a nonsignaling role for Sn in this response.
FIGURE 2.
In vitro cytokine production by BMDM in response to C. jejuni. (A) Heat-killed sialylated (sia) or non-sialylated GB11 C. jejuni were either untreated or opsonized (ops) with mouse anti-C. jejuni antiserum and then incubated with BMDM from WT and Sn−/− mice for 90 min. After removal of unbound bacteria, BMDM were cultured for a further 24 h. Concentrations of TNF-α, IL-6, IL-12, and IL-10 in cell culture supernatants were measured by ELISA. Bars show the average of three independent experiments (three replicates) ± SD. Media from unstimulated cells were analyzed as a negative control. Heat-killed Salmonella were used as a positive control. (B) Sn expression on IFN-α–stimulated WT, Sn−/−, and MyD88−/− BMDM. (C) IFN-α–stimulated WT, Sn−/−, and MyD88−/− BMDM were incubated with CFSE-labeled heat-killed sialylated C. jejuni and binding and phagocytosis analyzed by flow cytometry. To quench the fluorescence of extracellular bound but not phagocytosed bacteria, trypan blue (TB) was added for 15 min and washed out before the analysis. (D) BMDM from WT, Sn−/−, and MyD88−/− mice were incubated for 90 min with heat-killed sialylated C. jejuni, and after removal of unbound bacteria, cells were cultured for a further 24 h. Concentrations of TNF-α and IL-6 in cell culture supernatants were measured by ELISA. Bars show the average of two independent experiments with three replicates ± SD. Media from unstimulated cells were analyzed as a negative control. Two-way ANOVA tests were used to assess differences and interactions between groups. *p < 0.01. US, Unstimulated cells.
Cytokine responses to C. jejuni in vivo
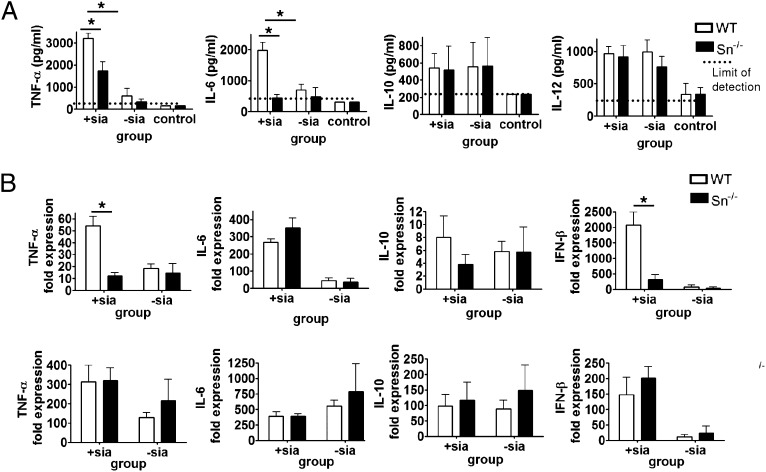
As Sn is expressed differentially on tissue Mϕ populations in vivo, we asked whether i.v.-injected sialylated C. jejuni would be targeted to certain subsets of Mϕs in the spleen and liver via Sn recognition and trigger an Sn-dependent inflammatory response. Cytokine responses were initially evaluated at 1 h after i.v. injection, both by measurement of protein concentrations in the sera as well as mRNA changes in livers and spleens, using the same groups of WT or Sn−/− mice (Fig. 3). Although the mice responded to both sialylated and non-sialylated strains of bacteria, a significantly higher Sn-dependent upregulation of TNF-α and IL-6 levels in serum was observed in response to sialylated C. jejuni, but not to non-sialylated C. jejuni. In contrast, no differences were seen for IL-10 and IL-12 (Fig. 3A). TNF-α mRNA was similarly upregulated in spleens of WT mice, but not in livers (Fig. 3B). There were no significant differences in mRNA levels in spleen and liver comparing WT and Sn−/− mice for IL-6, IL-10 (Fig. 4B), or IL-12 (data not shown). We also analyzed IFN-β mRNA as a readout for TIR domain-containing adapter inducing IFN-β–dependent activation (28) and observed a strong Sn-dependent induction with sialylated bacteria in spleens of WT mice (Fig. 3B).
FIGURE 3.
In vivo cytokine responses to C. jejuni in Sn−/− mice. Groups of WT and Sn−/− mice were injected i.v. with 108 heat-killed sialylated (+sia) or non-sialylated (−sia) C. jejuni or PBS (control) and cytokine responses assessed after 1 h by (A) ELISA measurements of serum cytokine concentrations or (B) changes in mRNA levels in spleen (top panels) or liver (bottom panels) by real-time quantitative PCR. Data are presented as means ± SD and are shown for one representative experiment out of two, with five to six mice per group injected with C. jejuni and three mice per group injected with PBS as a control. *p < 0.01 (Mann–Whitney U test).
FIGURE 4.
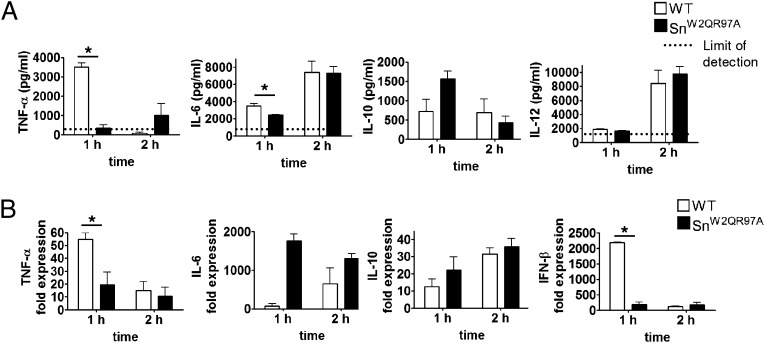
In vivo cytokine responses to C. jejuni in SnW2QR97A mice. Groups of WT and SnW2QR97A mice were injected i.v. with 108 heat-killed sialylated C. jejuni and cytokine responses measured at 1 h and 2 h by (A) ELISA measurements of serum cytokine concentrations or (B) changes in mRNA levels in spleen by real-time quantitative PCR. Data are presented as means ± SD and are shown for one experiment with three mice per group. *p < 0.05 (Mann–Whitney U test).
Similar results were obtained with SnW2QR97A mice, which were examined at 1 and 2 h after injection of sialylated C. jejuni (Fig. 4A, 4B). Serum levels of TNF-α, however, were greatly reduced at 2 h and were not significantly different between WT and SnW2QR97A mice at this time point. Similarly, splenic mRNA levels of TNF-α and IFN-β mRNA were greatly reduced at 2 h, whereas other cytokine mRNAs remained elevated at the later time point. Taken together, these experiments clearly demonstrated that sialylated C. jejuni can trigger a rapid Sn-dependent production of proinflammatory cytokines and type I IFN.
The capture and distribution of sialylated C. jejuni in vivo
To determine the in vivo distribution of the sialylated C. jejuni, fluorescently labeled bacteria were localized in spleen and liver sections by confocal microscopy at 5 and 20 min after i.v. injection (Figs. 5, 6). For these experiments, we compared WT mice with SnW2QR97A mice to visualize Sn+ Mϕs in both groups. Within 5 min of i.v. injection, bacteria in WT animals were observed in both the white pulp and red pulp of the spleen, in the red pulp being in close proximity to F4/80+ Mϕs that express low levels of Sn (Fig. 5A). In SnW2QR97A animals, however, there appeared to be little association of the bacteria with the red pulp and most were found in the white pulp of the spleen where they were specifically associated with CD68+ Mϕs within B220+ B cell zones (Fig. 5A). Similar observations were made after 20 min in both mouse strains (data not shown). In liver sections, bacteria were mostly localized with the Sn+ Kupffer cells in both WT and SnW2QR97A mice (Fig. 5B). Quantification of bacteria on tissue sections showed higher numbers in WT mice at 5 min compared with SnW2QR97A mice both in the spleen and liver, but numbers in both tissues were similar after 20 min (Fig. 6).
FIGURE 5.
Localization of C. jejuni in spleen and liver after i.v. injection. Groups of WT and SnW2QR97A mice were injected i.v. with 108 PKH67-labeled heat-killed sialylated C. jejuni. Spleens and livers were collected at 5 min or 20 min after the injection. (A) Representative images of spleen sections 5 min after injection are shown after staining with Abs to F4/80, Sn, CD68, and B220. Red pulp (RP) and white pulp (WP) areas are indicated. Scale bars, 100 μm. (B) Representative images of liver sections are shown after staining with anti-Sn Abs and DAPI. Scale bars, 100 μm. Data shown are representative of one experiment of two performed, each with four mice per group.
FIGURE 6.
Quantification of C. jejuni in spleen and liver after i.v. injection. Groups of WT and SnW2QR97A mice were injected i.v. with 108 PKH67-labeled heat-killed sialylated C. jejuni, and tissues were collected at 5 min or 20 min after the injection. (A) Spleen, average area (μm2) of C. jejuni per field of view. (B) Liver, average area (μm2) of C. jejuni per field of view. At least 20 images per sample were analyzed for each of the 5-min and 20-min time points. Data are shown are averages from two experiments with four mice per group. Data are presented as means ± SD. *p < 0.01 (paired t test).
The localization of IFN-β–producing cells in situ
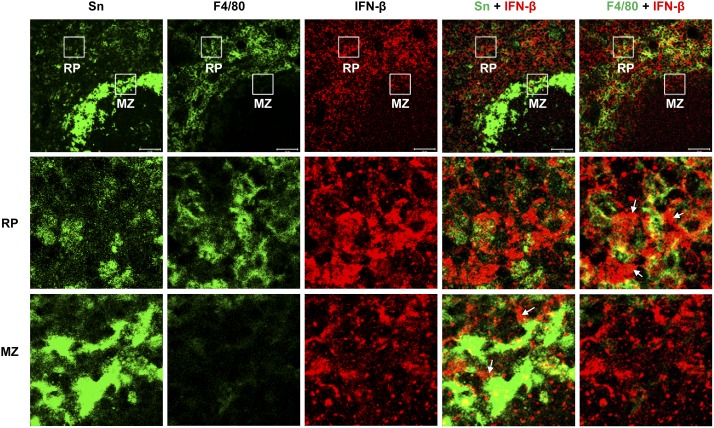
To gain insight into the nature of the cells in the spleen that produce rapid Sn-dependent cytokine responses, we stained tissue sections with Abs to IFN-β after injection of sialylated C. jejuni (Figs. 7, 8; Supplemental Fig. 3). Consistent with the strong Sn-dependent upregulation of IFN-β mRNA, IFN-β staining in WT spleens was markedly upregulated compared with both SnW2QR97A mice and PBS-treated control mice (Fig. 7). Similar findings were observed when comparing WT mice with Sn−/− mice (Supplemental Fig. 3), with IFN-β being seen mostly in the red pulp and marginal zone of the spleen, with less in the white pulp (Fig. 7). Poly(I:C) was used as a positive control for IFN-β induction. After i.v. injection, a similar strong staining for IFN-β was observed in spleens from WT and SnW2QR97A mice (Fig. 7). This demonstrates that Mϕs lacking functional Sn are not defective in their IFN-β response and that the failure to respond to sialylated C. jejuni is likely due to defective targeting of bacteria to these Mϕ subsets. Examination of red pulp and marginal zone areas of WT spleens showed that both Mϕs and non-Mϕs are associated with IFN-β production and that there was no obvious correlation at a cellular level between Sn expression and immunoreactivity for IFN-β (Fig. 8).
FIGURE 7.
Sn-dependent IFN-β production in spleen. WT and SnW2QR97A mice were injected i.v. with 108 sialylated C. jejuni, 100 μg poly(I:C), or PBS as a control. Spleens were collected after 1 h and cryostat sections stained with Abs to Sn, F4/80, and IFN-β and analyzed by confocal microscopy. Scale bars, 50 μm. Data are shown for one representative experiment of two with five to six mice per experimental group and three PBS control mice per group.
FIGURE 8.
Localization of IFN-β–associated cells in the red pulp and marginal zone of WT spleen. A WT mouse was injected with sialylated C. jejuni and the spleen collected after 1 h. Cryostat sections were stained with Abs to Sn, F4/80, and IFN-β. White boxes show areas selected for higher-power images of red pulp (RP) (middle panels) and marginal zone (MZ) (bottom panels). White arrows point to examples of IFN-β–associated, non-Mϕ cells. Scale bars, 50 μm.
Discussion
Several medically important pathogens including C. jejuni are thought to display sialic acid on their surface to circumvent and/or counteract the host's immune responses through a strategy of “molecular mimicry.” This has led to growing interest in the question of whether pathogen cell surface sialylation can modulate the host’s innate immune responses. In this study using a sialylated strain of C. jejuni as a model system, we provide the first functional evidence, to our knowledge, that Sn plays a potentially important role in this process. Using BMDM from mice either lacking Sn or expressing a nonbinding form of the receptor, we show that Sn is required for efficient sialic acid-dependent uptake of a sialylated strain of C. jejuni in vitro and that this interaction triggers enhanced cytokine responses. Crucially, this study has also demonstrated an in vivo requirement of Sn for rapid proinflammatory and type I IFN responses after i.v. injection of a sialylated strain of C. jejuni.
Sn was originally discovered as a nonphagocytic sialic acid binding receptor for erythrocytes and other hemopoietic cells (30–32). Recent studies on models of autoimmune disease have substantiated the view that its primary regulatory role in the host is in mediating sialic acid-dependent cell–cell interactions between Mϕ subsets and other cells of the immune system, for example by modulating the expansion of regulatory T cell subsets (8). A secondary role for Sn is as a receptor for “non-self” recognition of sialylated pathogens, including enveloped viruses (33–35), bacteria (21, 36), and protozoa (37), but the consequences of this for host immune function in vivo had not been examined prior to this study. Although previous studies showed that Sn does not function as a phagocytic receptor for sialylated RBC (32), with sialylated viruses and bacteria it appears to be capable of mediating efficient phagocytosis. Sn may function directly as a phagocytic receptor for some pathogens or alternatively it may synergize with other receptors to mediate efficient phagocytosis. The extended length of Sn, comprising 17 repeated Ig-like domains, may be important for mediating initial contacts with bacteria, thereby promoting interactions with other less accessible phagocytic receptors on the Mϕ surface. It has been proposed that porcine Sn can directly mediate endocytosis of sialylated enveloped Arterivirus (38) as well as F(ab′)2 fragments of an anti-Sn Ab (39). Consistent with a direct endocytic function for Sn, a recent study showed that liposomes displaying a high-affinity sialic acid-based ligand for Sn, but no other ligands, were rapidly cleared in mouse liver in an Sn-dependent manner (40). Therefore, under some circumstances, such as when particles are of a certain size or contain a high density of sialylated ligands, Sn may be capable of directly mediating uptake of particles into Mϕs.
The in vitro results showed clearly that Sn-dependent phagocytosis of sialylated C. jejuni by BMDM resulted in strong triggering of all cytokines tested, namely TNF-α, IL-6, IL-10, and IL-12, whereas non-sialylated cst-II mutants induced a much weaker response. Sn is unlikely to signal directly to trigger cytokine responses as its cytoplasmic tail lacks known signaling motifs and is poorly conserved in sequence and length (41). We therefore hypothesized that the Sn-dependent production of cytokines is likely due to synergy with TLRs, leading to MyD88-dependent signal transduction (24). For example, TLR-4 is strongly activated by C. jejuni LOS (28, 29), which also carries the GD1a-like structure recognized by Sn (21). We demonstrated in this study that MyD88 is required for Sn-dependent production of proinflammatory cytokines, but not for Sn-dependent binding or phagocytosis. Further studies are required to investigate the mechanisms involved, but one possibility is that Sn is required for effective delivery of bacterial products to TLR–MyD88 signaling compartments in macrophages.
When the same set of cytokines was analyzed in serum at 1 h after i.v. injection of bacteria, Sn-dependent upregulation of cytokine production was observed with TNF-α and IL-6 but no differences were seen for IL-10 and IL-12. Therefore, Sn-dependent responses in vivo are more restricted than those seen in vitro. This is likely to reflect the nature of the Mϕ subsets and other cell populations that are targeted in vivo. In WT mice, bacteria were seen to be associated with Sn+ Mϕs in the red pulp and marginal zone within 5 min of injection, whereas in mice lacking functional Sn, bacteria were found mainly in the white pulp, localizing to CD68+ Mϕs. Examination of cell subsets in the red pulp and marginal zone areas of spleens of WT mice showed that immunoreactive IFN-β was associated with Sn+ Mϕs, Sn− Mϕs, and neighboring cells. The latter did not appear to be Mϕs as they did not stain with either anti-Sn mAb or F4/80 mAb. The latter is thought to recognize all Mϕs in the red pulp of the spleen (42). IFN-β can strongly amplify its own production via a positive feedback loop involving both autocrine and paracrine signaling through the type I IFN receptor (43). Therefore, it is likely that the strong Sn-dependent IFN-β response observed throughout the red pulp and marginal zone is due to an initial secretion of IFN-β by Sn+ macrophages, followed by a secondary wave of production by other cells expressing type I IFN receptors.
The raised TNF-α levels in serum were paralleled by increased mRNA in the spleen at 1 h, but by 2 h this response was greatly attenuated, presumably due to the well-known feedback pathways in which TNF-α mRNA is rapidly degraded (44). In contrast, IL-6 mRNA levels were similar between WT mice and mice lacking functional Sn, despite WT mice having higher levels of this cytokine in the serum. This apparent discrepancy may be related to both the kinetics of IL-6 production and localization of bacteria in different tissues. Although the liver showed no differences in cytokine mRNA production between WT and Sn-deficient mice, there was a more rapid localization of bacteria to WT liver Kupffer cells within 5 min of injection, but this did not impact on cytokine mRNA levels measured at 1 h, probably because bacterial numbers in the liver were already similar by 20 min. The correlation between splenic TNF-α mRNA responses and TNF-α levels measured in serum suggests that the spleen is the major producer of TNF-α.
In conclusion, the data presented in this study support the idea that sialylation of C. jejuni LOS is an important factor for their uptake by Mϕ subsets in the spleen, which results in Sn-dependent cytokine production. Although this study focused on responses to sialylated C. jejuni as a model system, a similar Sn-dependent modulation of cytokine responses may occur with other sialylated bacteria such as N. meningitidis and Group B Streptococcus where Sn may similarly synergize with TLRs in promoting cytokine responses. In most cases, production of TNF-α and IFN-β are important for host resistance to infection by bacteria, although there are several examples of bacterial pathogens that exploit type I IFN responses for their own benefit (43). In the case of C. jejuni, future studies will be required to determine how these Sn-dependent cytokine responses contribute to host resistance versus bacterial survival and to the development of postinfectious Guillain–Barré syndrome.
Supplementary Material
Acknowledgments
We thank Drs. Hannah Richards and Sarah McMillan (University of Dundee) for useful discussions.
This work was supported by a studentship from the Biotechnology and Biological Sciences Research Council (to M.K.), a Pathological Society of Great Britain Fellowship (to A.E.), a Wellcome Trust Senior Fellowship (081882MA to P.R.C.), and Wellcome Trust Grants 089930 and 075470 (to L.P.E.).
The online version of this article contains supplemental material.
- BMDM
- bone marrow derived macrophage
- cst-II
- Campylobacter sialic acid transferase II
- LOS
- lipooligosaccharide
- Mϕ
- macrophage
- poly(I:C)
- polyinosinic:polycytidylic acid
- Sn
- sialoadhesin
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Crocker P. R., Paulson J. C., Varki A. 2007. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7: 255–266 [DOI] [PubMed] [Google Scholar]
- 2.Hart M. L., Saifuddin M., Spear G. T. 2003. Glycosylation inhibitors and neuraminidase enhance human immunodeficiency virus type 1 binding and neutralization by mannose-binding lectin. J. Gen. Virol. 84: 353–360 [DOI] [PubMed] [Google Scholar]
- 3.Hajishengallis G., Lambris J. D. 2011. Microbial manipulation of receptor crosstalk in innate immunity. Nat. Rev. Immunol. 11: 187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crocker P. R., Gordon S. 1989. Mouse macrophage hemagglutinin (sheep erythrocyte receptor) with specificity for sialylated glycoconjugates characterized by a monoclonal antibody. J. Exp. Med. 169: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartnell A., Steel J., Turley H., Jones M., Jackson D. G., Crocker P. R. 2001. Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations. Blood 97: 288–296 [DOI] [PubMed] [Google Scholar]
- 6.Crocker P. R., Gordon S. 1985. Isolation and characterization of resident stromal macrophages and hematopoietic cell clusters from mouse bone marrow. J. Exp. Med. 162: 993–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abram M., Vu ković D., Wraber B., Dorić M. 2000. Plasma cytokine response in mice with bacterial infection. Mediators Inflamm. 9: 229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C., Rauch U., Korpos E., Song J., Loser K., Crocker P. R., Sorokin L. M. 2009. Sialoadhesin-positive macrophages bind regulatory T cells, negatively controlling their expansion and autoimmune disease progression. J. Immunol. 182: 6508–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ip C. W., Kroner A., Crocker P. R., Nave K. A., Martini R. 2007. Sialoadhesin deficiency ameliorates myelin degeneration and axonopathic changes in the CNS of PLP overexpressing mice. Neurobiol. Dis. 25: 105–111 [DOI] [PubMed] [Google Scholar]
- 10.Jiang H. R., Hwenda L., Makinen K., Oetke C., Crocker P. R., Forrester J. V. 2006. Sialoadhesin promotes the inflammatory response in experimental autoimmune uveoretinitis. J. Immunol. 177: 2258–2264 [DOI] [PubMed] [Google Scholar]
- 11.Kobsar I., Oetke C., Kroner A., Wessig C., Crocker P., Martini R. 2006. Attenuated demyelination in the absence of the macrophage-restricted adhesion molecule sialoadhesin (Siglec-1) in mice heterozygously deficient in P0. Mol. Cell. Neurosci. 31: 685–691 [DOI] [PubMed] [Google Scholar]
- 12.Carrasco Y. R., Batista F. D. 2007. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity 27: 160–171 [DOI] [PubMed] [Google Scholar]
- 13.Junt T., Moseman E. A., Iannacone M., Massberg S., Lang P. A., Boes M., Fink K., Henrickson S. E., Shayakhmetov D. M., Di Paolo N. C., et al. 2007. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature 450: 110–114 [DOI] [PubMed] [Google Scholar]
- 14.Phan T. G., Grigorova I., Okada T., Cyster J. G. 2007. Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nat. Immunol. 8: 992–1000 [DOI] [PubMed] [Google Scholar]
- 15.Backer R., Schwandt T., Greuter M., Oosting M., Jüngerkes F., Tüting T., Boon L., O’Toole T., Kraal G., Limmer A., den Haan J. M. 2010. Effective collaboration between marginal metallophilic macrophages and CD8+ dendritic cells in the generation of cytotoxic T cells. Proc. Natl. Acad. Sci. USA 107: 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barral P., Polzella P., Bruckbauer A., van Rooijen N., Besra G. S., Cerundolo V., Batista F. D. 2010. CD169(+) macrophages present lipid antigens to mediate early activation of iNKT cells in lymph nodes. Nat. Immunol. 11: 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avril T., Wagner E. R., Willison H. J., Crocker P. R. 2006. Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect. Immun. 74: 4133–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes R. A., Cornblath D. R. 2005. Guillain-Barré syndrome. Lancet 366: 1653–1666 [DOI] [PubMed] [Google Scholar]
- 19.Godschalk P. C., Heikema A. P., Gilbert M., Komagamine T., Ang C. W., Glerum J., Brochu D., Li J., Yuki N., Jacobs B. C., et al. 2004. The crucial role of Campylobacter jejuni genes in anti-ganglioside antibody induction in Guillain-Barre syndrome. J. Clin. Invest. 114: 1659–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louwen R., Heikema A., van Belkum A., Ott A., Gilbert M., Ang W., Endtz H. P., Bergman M. P., Nieuwenhuis E. E. 2008. The sialylated lipooligosaccharide outer core in Campylobacter jejuni is an important determinant for epithelial cell invasion. Infect. Immun. 76: 4431–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heikema A. P., Bergman M. P., Richards H., Crocker P. R., Gilbert M., Samsom J. N., van Wamel W. J., Endtz H. P., van Belkum A. 2010. Characterization of the specific interaction between sialoadhesin and sialylated Campylobacter jejuni lipooligosaccharides. Infect. Immun. 78: 3237–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crocker P. R., Kelm S., Dubois C., Martin B., McWilliam A. S., Shotton D. M., Paulson J. C., Gordon S. 1991. Purification and properties of sialoadhesin, a sialic acid-binding receptor of murine tissue macrophages. EMBO J. 10: 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oetke C., Vinson M. C., Jones C., Crocker P. R. 2006. Sialoadhesin-deficient mice exhibit subtle changes in B- and T-cell populations and reduced immunoglobulin M levels. Mol. Cell. Biol. 26: 1549–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. 1999. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11: 115–122 [DOI] [PubMed] [Google Scholar]
- 25.Vinson M., van der Merwe P. A., Kelm S., May A., Jones E. Y., Crocker P. R. 1996. Characterization of the sialic acid-binding site in sialoadhesin by site-directed mutagenesis. J. Biol. Chem. 271: 9267–9272 [DOI] [PubMed] [Google Scholar]
- 26.McManus E. J., Collins B. J., Ashby P. R., Prescott A. R., Murray-Tait V., Armit L. J., Arthur J. S., Alessi D. R. 2004. The in vivo role of PtdIns(3,4,5)P3 binding to PDK1 PH domain defined by knockin mutation. EMBO J. 23: 2071–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persson T., Andersson P., Bodelsson M., Laurell M., Malm J., Egesten A. 2001. Bactericidal activity of human eosinophilic granulocytes against Escherichia coli. Infect. Immun. 69: 3591–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rathinam V. A., Appledorn D. M., Hoag K. A., Amalfitano A., Mansfield L. S. 2009. Campylobacter jejuni-induced activation of dendritic cells involves cooperative signaling through Toll-like receptor 4 (TLR4)-MyD88 and TLR4-TRIF axes. Infect. Immun. 77: 2499–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuijf M. L., Samsom J. N., van Rijs W., Bax M., Huizinga R., Heikema A. P., van Doorn P. A., van Belkum A., van Kooyk Y., Burgers P. C., et al. 2010. TLR4-mediated sensing of Campylobacter jejuni by dendritic cells is determined by sialylation. J. Immunol. 185: 748–755 [DOI] [PubMed] [Google Scholar]
- 30.Crocker P. R., Gordon S. 1986. Properties and distribution of a lectin-like hemagglutinin differentially expressed by murine stromal tissue macrophages. J. Exp. Med. 164: 1862–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Berg T. K., Brevé J. J., Damoiseaux J. G., Döpp E. A., Kelm S., Crocker P. R., Dijkstra C. D., Kraal G. 1992. Sialoadhesin on macrophages: its identification as a lymphocyte adhesion molecule. J. Exp. Med. 176: 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crocker P. R., Freeman S., Gordon S., Kelm S. 1995. Sialoadhesin binds preferentially to cells of the granulocytic lineage. J. Clin. Invest. 95: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanderheijden N., Delputte P. L., Favoreel H. W., Vandekerckhove J., Van Damme J., van Woensel P. A., Nauwynck H. J. 2003. Involvement of sialoadhesin in entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages. J. Virol. 77: 8207–8215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou Z., Chastain A., Moir S., Ford J., Trandem K., Martinelli E., Cicala C., Crocker P., Arthos J., Sun P. D. 2011. Siglecs facilitate HIV-1 infection of macrophages through adhesion with viral sialic acids. PLoS ONE 6: e24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rempel H., Calosing C., Sun B., Pulliam L. 2008. Sialoadhesin expressed on IFN-induced monocytes binds HIV-1 and enhances infectivity. PLoS ONE 3: e1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones C., Virji M., Crocker P. R. 2003. Recognition of sialylated meningococcal lipopolysaccharide by siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Mol. Microbiol. 49: 1213–1225 [DOI] [PubMed] [Google Scholar]
- 37.Monteiro V. G., Lobato C. S., Silva A. R., Medina D. V., de Oliveira M. A., Seabra S. H., de Souza W., DaMatta R. A. 2005. Increased association of Trypanosoma cruzi with sialoadhesin positive mice macrophages. Parasitol. Res. 97: 380–385 [DOI] [PubMed] [Google Scholar]
- 38.Delputte P. L., Costers S., Nauwynck H. J. 2005. Analysis of porcine reproductive and respiratory syndrome virus attachment and internalization: distinctive roles for heparan sulphate and sialoadhesin. J. Gen. Virol. 86: 1441–1445 [DOI] [PubMed] [Google Scholar]
- 39.Delputte P. L., Van Gorp H., Favoreel H. W., Hoebeke I., Delrue I., Dewerchin H., Verdonck F., Verhasselt B., Cox E., Nauwynck H. J. 2011. Porcine sialoadhesin (CD169/Siglec-1) is an endocytic receptor that allows targeted delivery of toxins and antigens to macrophages. PLoS ONE 6: e16827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W. C., Completo G. C., Sigal D. S., Crocker P. R., Saven A., Paulson J. C. 2010. In vivo targeting of B-cell lymphoma with glycan ligands of CD22. Blood 115: 4778–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klaas M., Crocker P. R. 2012. Sialoadhesin in recognition of self and non-self. Semin. Immunopathol. 34: 353–364 [DOI] [PubMed] [Google Scholar]
- 42.Hume D. A., Robinson A. P., MacPherson G. G., Gordon S. 1983. The mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Relationship between macrophages, Langerhans cells, reticular cells, and dendritic cells in lymphoid and hematopoietic organs. J. Exp. Med. 158: 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trinchieri G. 2010. Type I interferon: friend or foe? J. Exp. Med. 207: 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kontoyiannis D., Pasparakis M., Pizarro T. T., Cominelli F., Kollias G. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10: 387–398 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.