Abstract
DNA replication is spatially and temporally regulated during S-phase. DNA replication timing is established in early-G1-phase at a point referred to as timing decision point. However, how the genome-wide replication timing domains are established is unknown. Here, we show that Rif1 (Rap1-interacting-factor-1), originally identified as a telomere-binding factor in yeast, is a critical determinant of the replication timing programme in human cells. Depletion of Rif1 results in specific loss of mid-S replication foci profiles, stimulation of initiation events in early-S-phase and changes in long-range replication timing domain structures. Analyses of replication timing show replication of sequences normally replicating early is delayed, whereas that normally replicating late is advanced, suggesting that replication timing regulation is abrogated in the absence of Rif1. Rif1 tightly binds to nuclear-insoluble structures at late-M-to-early-G1 and regulates chromatin-loop sizes. Furthermore, Rif1 colocalizes specifically with the mid-S replication foci. Thus, Rif1 establishes the mid-S replication domains that are restrained from being activated at early-S-phase. Our results indicate that Rif1 plays crucial roles in determining the replication timing domain structures in human cells through regulating higher-order chromatin architecture.
Keywords: chromatin loop, replication foci, replication timing domains, S-phase, TDP (timing decision point)
Introduction
In eukaryotic cells, the initiation of DNA replication starts at multiple sites along the genome. Firing of replication origins are temporally and spatially regulated. Some origins are fired at the beginning of S-phase and others later during S-phase. This timing of replication may be coordinated with transcription and chromatin structures, ensuring that the entire genome replicates once during every cell cycle (Arias and Walter, 2007; Farkash-Amar et al, 2008; Karnani et al, 2007, 2009; Hiratani et al, 2009; Masai et al, 2010). Replication timing programme appears to be established in early-G1-phase through chromatin regulation (Dimitrova and Gilbert, 1999). The decision was proposed to take place at the TDP (timing decision point) concomitant with the repositioning of chromatin in the nucleus after mitosis (Dimitrova and Gilbert, 1999; Li et al, 2001; Gilbert, 2010; Lu et al, 2010). Studies in budding yeast also indicated a critical role of early G1 events in replication timing determination (Heun et al, 2001a, 2001b). Interaction of late-replicating loci with nuclear periphery at early G1 may play a crucial role in setting up the late-replicating domains in yeast.
Correlation of chromosome banding with replication timing as well as the S-phase-timing-specific replication foci pattern have suggested that various segments of chromosomes replicate with distinct timing in S-phase. Notably, mid-S replication domains are coincident with a major change of the replication profile including changes of nuclear replication foci pattern and the R/G banding pattern and reduced fork rate, leading to the prediction that instalment of mid-S replication domains would play a critical role in determination of replication timing (Takebayashi et al, 2005; Hiratani et al, 2009). More recently, the genome-wide replication profile has been determined in various cell lines through enrichment of S-phase-timing-specific nascent DNA and the use of genomic tiling arrays or deep sequencing (Karnani et al, 2007, 2009; Hiratani et al, 2008; Gilbert, 2010; Hansen et al, 2010; Ryba et al, 2010; Yaffe et al, 2010). These experiments led to the identification of the chromosome segments, called ‘replication domains’ that replicate simultaneously at specific times. The sizes of the replication timing domains were proposed to range from a few hundred kilobases to megabases, depending on the averaging window size adopted (Karnani et al, 2007, 2009; Hiratani et al, 2008). In experiments using undifferentiated mouse embryonic stem cells, the distribution of replication domains extensively changes after induction of differentiation (Hiratani et al, 2008; Ryba et al, 2010). Furthermore, extensive differences were detected in replication domain structures between different cell types (Hansen et al, 2010). Thus, replication timing domain structures can be regarded as a novel epigenetic mark which is specific to each cell type (Hiratani and Gilbert, 2009). However, the mechanisms by which the replication domains are determined are totally unknown.
In yeasts, late origin firing is inhibited by checkpoint pathways (Santocanale and Diffley, 1998; Shirahige et al, 1998). Dbf4, the Cdc7 (cell division cycle 7) activation subunit, and Sld3, a factor essential for initiation complex formation, have been shown to be critical targets of checkpoint inhibition in budding yeast, although precise mechanisms of inhibition are unknown (Lopez-Mosqueda et al, 2010; Zegerman and Diffley, 2010). Epigenetic modifications of histones have also been shown to regulate the timing of origin firing at some selective locations (Vogelauer et al, 2002; Aparicio et al, 2004; Goren et al, 2008; Hiratani et al, 2009). We recently have shown that Rif1 (Rap1-interacting-factor-1) may play an important role in determination of replication timing of early and late origins in fission yeast (Hayano et al, 2012). However, mechanisms with which the genome-wide replication timing domain is installed in mammalian cells have been elusive. In this manuscript, we will present evidence that the human homologue of Rif1 plays a major role in determination of the replication timing domain structures of the human genome.
Results
Depletion of human Rif1 protein leads to increased Cdc7-dependent phosphorylation and slight increase of DNA synthesis at the early-S-phase
It was recently reported in budding yeast that loss of rif1, a telomere-binding protein, resulted in early-S-phase replication of the late-replicating telomere segment (Lian et al, 2011). However, it is not known whether Rif1 regulates origin firing in other species. We recently discovered that loss of rif1 restores the growth of a null mutant of hsk1, the fission yeast homologue of Cdc7 kinase, by changing the origin firing programme (Hayano et al, 2012). This finding led us to examine the human homologue of Rif1 (hereafter, will be referred to as Rif1) for its involvement in regulation of the replication programme in mammalian cells. Rif1, which does not bind to telomeres, has been suggested to have a role in DNA damage responses (Silverman et al, 2004; Buonomo et al, 2009; Wang et al, 2009; Xu et al, 2010), but its roles in DNA replication have not been explored.
Depletion of Rif1 in HeLa cells synchronously released from the G1/S boundary did not significantly affect the overall S-phase progression as detected by FACS analyses of DNA contents (Figure 1A–C). However, we noted a slight but reproducible increase in BrdU (Bromodeoxyuridine) or [3H] thymidine incorporation at early-S-phase in Rif1-depleted cells (Supplementary Figure S1). The phosphorylation levels of minichromosome maintenance (MCM) proteins, targets of the Cdc7 kinase (Masai et al, 2006; Montagnoli et al, 2006), increased in Rif1-depleted cells (Figure 1D and E), and this increased phosphorylation was indeed dependent on Cdc7 kinase (Figure 1F). Analyses using the synchronized cell population indicated that the increased phosphorylation of MCM occurs specifically during early-S-phase (Figure 1E; Supplementary Figure S2). This was also confirmed by analyses of early- and late-S-phase cell populations fractionated and separated by FACS from asynchronously growing cells (Supplementary Figure S3). We then examined the chromatin binding of Cdc45 and PCNA (proliferating cell nuclear antigen), the proteins required for replication after the action of Cdc7. The chromatin binding of Cdc45 and PCNA during early-S-phase increased in Rif1-depleted cells (Figure 1G), consistent with enhanced action of Cdc7 kinase. This is not due to the enhanced Cdc7 kinase activity in Rif1-depleted cells since we measured the Cdc7 kinase activity in control and Rif1-depleted HeLa cells, and showed that it is not affected or slightly reduced in Rif1-depleted cells (Supplementary Figure S4). These results indicate that Rif1 depletion somehow alters the structures of pre-replicative complexes (pre-RCs) in that they can be more readily accessible for recognition by Cdc7. The measurement of DNA synthesis at known replication origins in early- or late-S-phase indicated that DNA replication at mid/late origins is enhanced in early-S-phase, showing that the sequences normally replicated late is replicated early in Rif1-depleted cells (Supplementary Figure S5). It should be noted that HU-induced checkpoint activation is not affected at all by Rif1 depletion, and Rif1 depletion does not activate checkpoint, either (Supplementary Figure S6). Thus, involvement of the checkpoint pathway in regulation of replication timing by Rif1 is unlikely. Previous report also states that depletion of Rif1 does not affect mitotic index (Xu and Blackburn, 2004).
Figure 1.
Depletion of Rif1 protein enhances initiation events in early-S-phase. (A) HeLa cells, transfected with Rif1 siRNA for 48 and 72 h, were harvested and the whole-cell extracts were immunoblotted with antibodies against Rif1. The Rif1 protein level was reduced by >90% at 48 h compared with control. Tubulin is shown as a loading control (C). (B) Asynchronous cells transfected with control or Rif1 siRNA for 48 h were stained with PI and DNA contents were examined by FACS. (C) HeLa cells treated with control or Rif1 siRNA were synchronized at the G1/S-phase by double-thymidine block. At indicated times after release, cell-cycle progression was examined by FACS. (D) HeLa and NHDF cells were harvested at 48 h after control (−) or Rif1 (+) siRNA transfection, and the cells were fractionated into Triton-soluble and -insoluble fractions by CSK buffer (containing 0.1% Triton X-100). The latter fractions (enriched in chromatin-bound proteins) were immunoblotted with the antibodies indicated. (E) HeLa cells treated with control (−) or Rif1 (+) siRNA were synchronized at the G1/S boundary, released into cell cycle and harvested at indicated times after release. The whole-cell extracts were immunoblotted with the antibodies indicated. (F) The chromatin-enriched fractions (insoluble fractions in CSK containing 0.1% Triton X-100) from HeLa cells, transfected with Rif1 and/or Cdc7 siRNAs for 48 h, were immunoblotted with the antibodies indicated. (G) HeLa cells treated with control or Rif1 siRNA were synchronized at the G1/S boundary by double-thymidine block. At the times indicated after release, cells were harvested and fractionated into Triton-soluble and Triton-insoluble chromatin-enriched fractions in CSK buffer (containing 0.1% Triton X-100). The latter fractions were immunoblotted with the antibodies indicated. In all the experiments, Rif1 siRNA #4 was used.
Depletion of Rif1 leads to specific loss of mid-S replication foci and major change in replication timing domain structures
We next examined the replication foci pattern in Rif1-depleted HeLa cells. It is well established that replication foci pattern changes during S-phase (Dimitrova and Berezney, 2002). Indeed, distinct early-, mid- and late-S-phase replication foci patterns are observed in control human cells during progression of S-phase. In contrast, Rif1-depleted cells exhibited predominantly early-S-phase-like characteristics almost throughout the S-phase (Figure 2A and B). At the end of the S-phase, late-S pattern appeared, indicating that the mid-S-phase pattern characterized by large nucleoli-associated foci and nuclear periphery foci selectively disappeared in the absence of Rif1. To further analyse changes of replication foci patterns in single cells, cells synchronously proceeding S-phase were sequentially labelled by EdU and Biotin-dUTP at early- and mid-/late-S-phase, respectively (Supplementary Figure S7A). In control cells, pattern A dominated, indicative of distinct labelling at early-S-replicating and mid-S-replicating foci by EdU and Biotin-dUTP, respectively, In contrast, in Rif1-depleted cells, pattern B dominated, where both labelings occur in the early-S-replicating foci pattern (Supplementary Figure S7B; Figure 2C and D). This further demonstrates that mid-S-replicating pattern is lost and DNA synthesis proceeds in the early-S pattern in the absence of Rif1.
Figure 2.
Mid-S-phase replication foci pattern is selectively lost in Rif1-depleted cells. (A) HeLa cells, transfected with control or Rif1 siRNA, were arrested at the G1/S boundary by double-thymidine block and were released into cell cycle. After treatment with BrdU for 30 min at the indicated times, cells were fixed, treated with 2.0 N HCl to denature DNA, and then stained with anti-BrdU antibody to detect DNA replication sites. Representative BrdU images of nuclei in cells at each stage of S-phase are shown. (B) Cells were analysed under fluorescence microscopy (Olympus FV1000 microscopy) and the numbers of the cells with each foci pattern (representing early-S-, mid-S- or late-S-phase cells) were counted at each time point after release. The averages of three independent experiments (N=100 for each) are presented. (C) Enlarged foci patterns of the cells from Supplementary Figure S7A. (D) The fractions of the two replication foci patterns were counted in control and Rif1-depleted cells under microscopy. The averages of three independent experiments (N=100 for each) are presented. (E) The cell-cycle progression of the cells used in (C) monitored by FACS. In all the experiments, Rif1 siRNA #4 was used.
These results indicate that replication timing profile may change in response to Rif1 depletion. Therefore, we analysed the replication timing domains on the 42-Mb genome segment of the human chromosome V. The control cells showed the replication domain pattern characteristic to HeLa cells. In contrast, the patterns were substantially different in Rif1-depleted cells (Figure 3A–C). The differences included conversion of late to early or early to late domains (reversion), the conversion of small domains into larger ones (consolidation) and loss of clear replication domains (homogenization) (Supplementary Figure S8A–C). These changes were validated by the quantitative PCR analyses of replication timing at each location where the alteration was observed in the tiling array analyses (Figure 3C). They occur on several hundred kilobases to megabase scale, suggesting that Rif1 affects the replication timing on a long-chromosome level. We have presented the early/late ratio of replication timing as a histogram on the entire 42 Mb in control and Rif1-depleted cells (Supplementary Figure S9A, upper). The histogram shows two early and late peaks in the control cells, whereas the peak is centred (at mid-S) in the Rif1-depleted cells in both 10 and 100 kb window analyses, indicating that early- and late-replicating segments shifted to mid-S. The histogram showing the ratio of Rif1-depleted cells to control indicated the almost equal occurrence of both late-to-mid-S and early-to-mid-S changes (Supplementary Figure S9A, lower). Investigation of replication timing change in selected segments in the 42-Mb region also indicated those shifting towards early as well as towards late (Supplementary Figure S9B and C). We also noted the presence of the genome segments the replication timing of which is not much affected by depletion of Rif1 (e.g., the segments C and E shown in Supplementary Figure S9B). This could be because the effect of Rif1 could be more enhanced in some regions than in others, maybe reflecting the binding profiles of Rif1 protein on the genome. Alternatively, replication timing of these segments may be determined predominantly by local chromatin structures, which may be independent of Rif1. We also analysed the replication timing at selective loci in ATR (ATM and Rad3-related)-depleted HeLa cells and found that the replication timing was largely same as the control cells (Supplementary Figure S10). This suggests that genome-wide structures of replication timing domains may not be affected by checkpoint functions as much as by Rif1, although some individual late-firing origins may be prematurely activated by loss of a replication checkpoint (Karnani and Dutta, 2011).
Figure 3.
Rif1 depletion causes major changes in replication timing domains on a 42-Mb segment of the human chromosome 5. (A) Asynchronously growing HeLa cells, treated with control or Rif1 siRNA #4, were pulse labelled for 90 min with BrdU and were fractionated into early- and late-S-phase fractions by FACS cell sorter (see Supplementary Figure S15). Nascent BrdU-substituted DNA, immunoprecipitated with anti-BrdU antibody, in each fraction was hybridized with oligonucleotide tiling arrays containing one probe every 1.0 kb, as described in ‘Materials and methods’. Data, processed in Genomic workbench software, generated the timing pattern on the 42-Mb segment on the chromosome 5. In the scatter plots, red and green indicate early- and late-replicating regions, respectively. The smoothed curve shows replication timing domains. The R/G band pattern of this segment is shown below. (B) Overlay of the results of the control (red) and Rif1-depleted (blue) cells. (C) Validation of the timing array results by quantitative PCR. Asynchronously growing HeLa cells, treated with control or Rif1 siRNA #2, were pulse labelled for 90 min with BrdU and were fractionated into early- and late-S-phase fractions by FACS cell sorter. BrdU-labelled nascent DNA was immunoprecipitated in each fraction. The nascent DNAs at the indicated loci (A–G; left) on the chromosome 5 were amplified by quantitative PCR to determine replication timing. The values presented are the ratios to the level of amplified mtDNA (right). Data means were calculated from at least three independent experiments. The data are in excellent agreement with those of tiling array analyses.
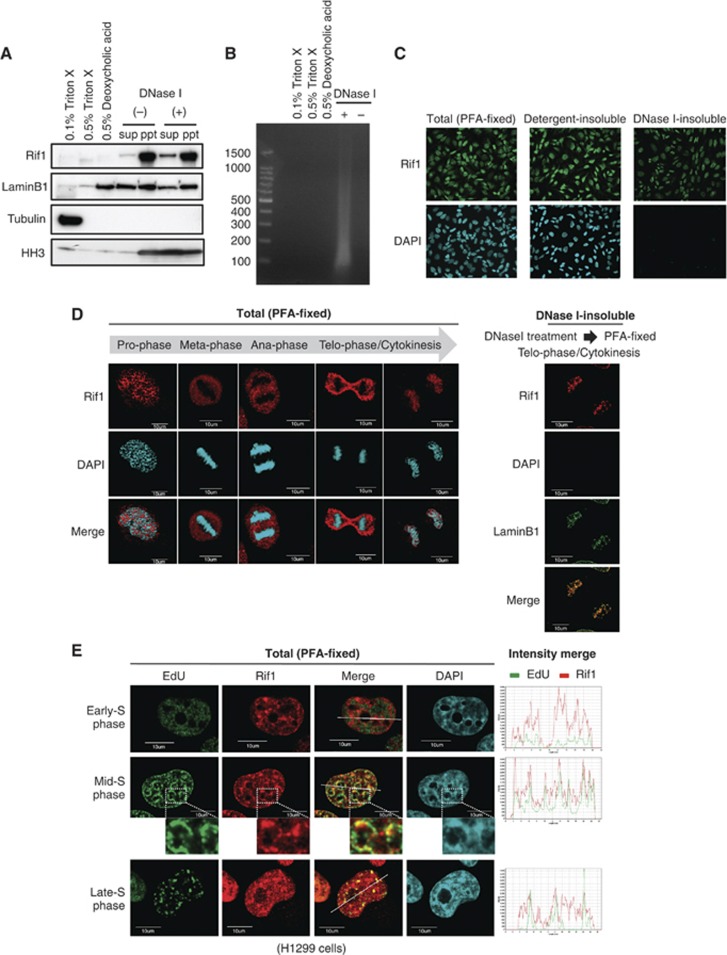
Rif1 binds to the DNase I-insoluble nuclear structures at late-M/early-G1
Above results indicate that the loss of Rif1 protein in human cultured cells leads to (1) stimulation of replication initiation events during early-S-phase, (2) loss of mid-S replication foci pattern and prevalence of early-S-phase-like foci pattern, and (3) alteration of replication timing domains on a large-genome basis. How can one account for these aberrations by the loss of Rif1, a single factor? In an attempt to answer this question, we first analysed the precise cellular localization of Rif1 protein. Rif1 is not solubilized by detergents such as 0.5% Triton X-100 or deoxycholic acid. A small fraction of Rif1 was solubilized after DNase I treatment in the presence of 0.25 M ammonium sulphate, but most remained in the pellet (Figure 4A and B). This suggests that the majority of Rif1 is present in the nuclear-insoluble fractions. We then conducted immunostaining of Rif1 protein (Supplementary Figure S11A). As expected from the fractionation study, the Rif1 signal did not change by prior wash with detergent. The significant Rif1 signals remained, even after the cells on the glass slides were pretreated with DNase I, while DAPI-stained DNA signals were largely gone (Figure 4C). The Rif1 signals in the nuclease-treated cells overlapped with those of Lamin B1, known to be associated with nuclear-insoluble structures (Supplementary Figure S11B and C). This confirms the results of the fractionation study and indicates that Rif1 is present in the DNase I-insoluble, matrix-associated structures. We then examined these nuclear-insoluble Rif1 signals during cell-cycle progression. Since Rif1 signals can be detected throughout the interphase, we examined the mitotic cells. In the prophase cells, the nuclear Rif1 signals did not overlap with the condensing chromatin, suggesting dissociation of Rif1 from DNA at this stage. As reported before (Xu and Blackburn, 2004), Rif1 did not localize on mitotic chromosomes. Rif1 rebound to the chromatin at late telophase, and this binding was resistant to DNase I (Figure 4D). The timing of chromatin binding of Rif1 is close to that of pre-RC formation. Therefore, we examined effect of Rif1 depletion on the chromatin binding of MCM. First, we show that depletion of Cdt1 significantly reduced the amount of MCM on the detergent-prewashed nuclei by immunostaining (Supplementary Figure S12A and B). The level of MCM in the chromatin-enriched fraction also significantly reduced by Cdt1 depletion (Supplementary Figure S12E). On the other hand, MCM staining and its level in the chromatin-enriched fraction was not affected by Rif1 depletion (Supplementary Figure S12C and D). These results indicate that Rif1 is not required for pre-RC formation.
Figure 4.
Rif1 tightly binds to DNase I-insoluble nuclear structures in nuclei at late M/early G1 and colocalizes with mid-S replication foci. (A, B) Asynchronously growing HeLa cells were fractionated as described in ‘Materials and methods’. (A) Each fraction was immunoblotted with indicated antibodies. (B) The soluble fractions were run on 2% agarose gel to examine the presence of DNA. (C) Immunostaining of Rif1 protein (green) after various treatment. Total, fixed with 4% paraformaldehyde; detergent-insoluble, pretreated with 0.5% Triton X-100 before fixation; DNase I-insoluble, pretreated with DNase I before fixation. (D) (Left) Mitotic HeLa cells as shown were fixed with 4% paraformaldehyde and immunostained with anti-Rif1 antibody. Rif1 dissociates from DNA in prophase and is not present on mitotic chromosomes. Rif1 binds to chromatin at late telophase. (Right) Rif1 staining on late telophase chromosome is resistant to prior DNase I treatment. (E) H1299 cells in early-, mid- and late-S-phase were labelled with EdU. The sites of the EdU signals (green) and Rif1 localization (red) were compared in each stage of S-phase. The insets show the enlarged images of the segments indicated. The intensities of both green and red signals on the white line are shown in the right graph (Olympus FV1000 microscopy). Blue, DNA.
Rif1 colocalizes with the mid-S replication foci
We then examined the correlation with the sites of DNA replication. The Rif1 foci did not overlap with early-S or late-S replication foci. However, we found that Rif1 protein colocalizes with the sites of DNA replication during mid-S-phase (Figure 4E). Rif1 localizes at the nuclear periphery as well as at the periphery of nucleoli, where mid-S replication foci are observed. Partial overlap of Rif1 with mid-S replication foci was previously reported in mouse cells as well (Buonomo et al, 2009). These results are consistent with the idea that Rif1 somehow directs the formation of nuclear structures that are prerequisite for the mid-S replication domain. The DNA damage-induced foci of Rif1 protein, that have been reported to colocalize with γ-H2AX signals (Silverman et al, 2004; Wang et al, 2009), are largely eliminated by prior DNase I treatment (Supplementary Figure S13), suggesting that the mechanisms of Rif1 recruitment to the sites of damage and its role in damage responses may be distinct from those at the nuclear-insoluble structures during the normal S-phase progression.
Rif1 regulates chromatin-loop structures
The above results suggest that Rif1 regulates chromatin architecture by binding to nuclear-insoluble structures and generating specific chromatin-loop structures. Therefore, we conducted nuclear halo assays that may indicate the sizes of the chromatin loops (Gerdes et al, 1994). We have shown that the halo size increased in HeLa cells treated with two different siRNAs #2 and #4. The extent of the inhibition of the Rif1 expression was better with #4, which gave rise to cells with larger halo than #2 did (Figure 5A and B; Supplementary Figure S2). These results strongly indicate that Rif1 regulates the chromatin-loop sizes. DNA fibre analyses on the DNA labelled with halogenated nucleotide derivatives indicated that replication fork rate increased in both early- and mid-S-phase. Increase is more striking in mid-S-phase, since fork speed is generally slower in mid-S replication domains than in early-S-phase in untreated cells. Disintegration of mid-S replication domains apparently results in conversion of replication modes from ‘mid-S’ type to ‘early-S’ type. Concomitantly, average inter-origin distances are longer in Rif1-depleted cells in both early- and mid-S (Supplementary Figure S14), consistent with the longer chromatin loops indicative of larger replicon sizes (Buongiorno-Nardelli et al, 1982; Courbet et al, 2008; Guillou et al, 2010). In contrast, the fork rate and inter-origin distances did not change in late-S-phase, consistent with the fact that late-replicating foci patterns are not affected by Rif1 depletion.
Figure 5.
Rif1 depletion causes increase of chromatin-loop sizes. (A, B) Control and Rif1-depleted HeLa cells were synchronized at G1/S-phase and released into cell cycle for 1 h. Cells were harvested and DNA halo assays were conducted (N=50). (A) Phase-contrast images of representative cells. (B) Chromatin-loop sizes were calculated according to the formula described in ‘Materials and methods’ (Guillou et al, 2010). The data with two representative siRNAs (#2 and #4) are presented.
Discussion
Rif1, originally identified as a telomere-binding factor in yeast, does not play significant roles in telomere maintenance in human cells (Hardy et al, 1992; Kanoh and Ishikawa, 2001; Silverman et al, 2004; Xu and Blackburn, 2004). Although deregulation of silent origins near telomere in rif1Δ cells was reported in budding yeast (Lian et al, 2011) and more recently fission yeast Rif1 was reported to regulate origin firing timing, it was not known whether Rif1 regulates DNA replication in mammalian cells. In this report, we have presented evidence that shows the crucial roles of Rif1 protein in setting up the replication timing domain structures in human cells.
The strong association of Rif1 with nuclear-insoluble structures suggests a possibility that Rif1 may be a crucial factor for generating higher-order chromatin architecture including special organization of chromatin loops (Figure 6). Rif1 may tether the chromatin fibres at the nuclear-insoluble structures to assemble proper chromatin architecture that may specify mid-S replication domains. Oligomerization and DNA-binding activity of Rif1 may facilitate this process (Xu et al, 2010). The nuclear replication foci surrounding nucleoli or those at nuclear periphery that are observed in mid-S depend on Rif1 and overlap with Rif1 localization. The Rif1-mediated chromatin structures are formed during late M-to-early-G1-phase, and are maintained throughout G1-to-S-phase. This mid-S chromatin architecture is somehow sequestered from the activation by Cdc7 kinase in early-S-phase, but this restraint is relieved at mid-S-phase to permit the action of Cdc7 kinase at the mid-S origins.
Figure 6.
A model for Rif1-mediated determination of replication timing domains. (Left) Normally, Rif1 binds to nuclear-insoluble structures at late-M to early-G1, generating mid-S replication domains some of which are clustered at nuclear periphery as well as around nucleoli. This could be related to TDP known to occur at early-G1. The origins associated with the mid-S domains are sequestered from activation until mid-S-phase (shown with dotted grey arrow emanating from Cdc7). (Right) In Rif1-depleted cells, mid-S replication domains are not generated and the origins normally associated with mid-S domains are scattered throughout the nuclei. This permits access of Cdc7 (shown with solid arrow) and other replication factors to mid-S origins throughout early- to mid-S-phase, resulting in stimulation of initiation events (Cdc7-mediated phosphorylation of MCM and chromatin loading of Cdc45 and PCNA, etc.) at early-S-phase. Portions of this figure were adopted from Gilbert (2010).
In the absence of Rif1, this mid-S-phase architecture is not formed, allowing Cdc7 kinase to gain access in early-S-phase to the origins that would normally not be activated until mid-S-phase. This would explain the increased Cdc7-dependent phosphorylation of the target proteins and hyper-loading of Cdc45 and PCNA onto chromatin in Rif1-depleted cells at early-S-phase (Figure 1). As a result of loss of mid-S replication domains, replication timing domain structures may undergo genome-wide alteration. It should be noted that replication timing is generally more centred to mid-S in the absence of Rif1, although mid-S replication foci pattern is lost and all the replication foci appear to adopt the early-S-like pattern (Supplementary Figure S9). This effect of Rif1 on replication timing is similar to what we observed in fission yeast rif1Δ mutant, where some early-firing origins are delayed and late-firing/dormant origins are fired earlier (Hayano et al, 2012). Changes of replication timing domain structures in Rif1-depleted cells are mostly consistent with this observation. The fusion of small replication domains into larger replication domains may be consistent with the extended chromatin-loop sizes. However, changes of replication domains are less dramatic in some genome regions. The replication timing in the absence of Rif1 may be determined directly by local chromatin structure or epigenetic state. It could also be due to the segment-specific association of Rif1 with chromatin/nuclear structures. Alternatively, other proteins (e.g., cohesin; Guillou et al, 2010), or other nuclear skelton-associating proteins may exert chromatin-loop regulation and/or local epigenetic modifications might affect replication timing domains (Hiratani et al, 2009).
All these data are consistent with the hypothesis that Rif1 plays a crucial role at or near TDP when the replication timing programme is determined. The timing of association of Rif1 with chromatin appears to be very close to that of pre-RC formation. This timing may be slightly earlier than the proposed timing of TDP which is early-G1-phase (Dimitrova and Gilbert, 1999), suggesting the requirement for another event(s) for execution of TDP. We show that Rif1 is not required for the pre-RC formation (Supplementary Figure S12) and think it is unlikely that Rif1 regulates replication timing through regulation of timing of pre-RC assembly (Wu and Nurse, 2009). However, the current result does not completely rule out this possibility.
In spite of rather dramatic effect of Rif1 depletion on replication timing during S-phase, effect of overall S-phase progression is surprisingly subtle in the experimental setting conducted. S-phase is apparently completed without delay, and cell viability is not much affected. This is in sharp contrast to the depletion of Cdc7 or other replication factors under the identical condition which disturbs S-phase progression and often induces strong cell death in cancer cells (Feng et al, 2003; Montagnoli et al, 2004; Ito et al, 2012). Although initiation events are stimulated at early-S-phase in Rif1-depleted cells (Figure 1), this occurs only in a limited window of cell cycle. We speculate this is because the amount of Cdc7-ASK is limited and is not able to activate all the early-/mid-S origins at one time. We show that fork speed increases in the absence of Rif1 in early- and mid-S-phases. This appears to be compensated by longer inter-origin distances. Thus, overall S-phase length stays unchanged. It should be noted that S-phase delay was observed in Rif1 knockout MEF cells (Buonomo et al, 2009). We speculate that acute depletion of Rif1 by siRNA may cause deregulation of replication programme but may not cause much effect on S-phase progression. It would be interesting to examine the effect of a long-term depletion of Rif1 on progression of S-phase.
Rif1 is overexpressed in embryonic stem cells and is rapidly downregulated upon induction of differentiation (our unpublished data and Loh et al, 2006). It was reported that replication domains are fused together (consolidated) after differentiation (Hiratani et al, 2008), which may be consistent with the reduction of Rif1 expression. Thus, the Rif1 protein level could be crucial for determining the chromatin higher-order structures specific for each cell type. We observed that >600 genes are either upregulated or downregulated in Rif1-depleted HeLa cells by >1.5 fold (unpublished observation). Thus, chromatin structures regulated by Rif1 obviously affect transcription activity as well. It was reported that replication timing domain structures exhibit striking similarity to the spatial proximity of chromatin as measured by Hi-C analyses (Ryba et al, 2010). This is also consistent with the idea that Rif1 may determine higher-order chromatin architecture that may regulate not only DNA replication but also transcription.
In conclusion, we identified Rif1 as a crucial determinant for replication timing domains in human cells. Rif1 regulates higher-order chromatin architecture by facilitating the chromatin-loop organization through binding at nuclear-insoluble structures. Rif1 may regulate TDP by setting up special chromatin architecture at late-M/early-G1-phase that specifies mid-S replication domains on the human chromosomes.
Materials and methods
Cell culture and cell-cycle synchronization
HeLa (human cervical cancer) and H1299 (human non-small cell lung carcinoma) cells were maintained in Dulbecco’s modified Eagle’s Medium supplemented with 10% fetal bovine serum (Hana-Nesco Bio) at 37°C. Normal human dermal fibroblasts (NHDF) cells were maintained in fibroblast basal medium (TaKaRa). For synchronization, cells were arrested at the G1/S boundary by two consecutive incubation in the presence of 2.5 mM thymidine for 14–16 h each with a 9-h interval of growth without thymidine. Cells were then released into cell cycle and harvested at the indicated time points after release. Cells were transfected with siRNA at 4 h before the first thymidine incubation.
Visualization of BrdU incorporating sites in nuclei
Asynchronous or synchronized cells were pulse labelled for 30 min with 20 μM BrdU before fixation with 4% paraformaldehyde. BrdU detection was performed as previously described (Dimitrova and Berezney, 2002).
Dual labelling of replication foci patterns
Cells arrested at the G1/S boundary were released into cell cycle for 1.5 h and pulse labelled with EdU (20 μM) for 20 min. After the first labelling, the cells, incubated in new medium for 4.5 h, were prewashed with KH buffer (10 mM HEPES (pH 7.4) and 50 mM KCl) as a hypotonic shift step (Koberna et al, 1999), were pulse labelled with Biotin-dUTP (100 μM) for 10 min, incubated in new medium for 30 min, and then fixed with 100% methanol.
Cell fractionation and immunofluorescence
Cell fractionation and immunofluorescence were performed as previously described (Gregson et al, 2001; Guillou et al, 2010). Immunofluorescence images at each extraction step were collected with the same exposure time. As a control for staining of nuclear matrix scaffold (DNase I-insoluble structures), cells were costained for Lamin B1.
Cell-cycle fractionation and isolation of BrdU-labelled DNA
HeLa cells were pulse labelled for 90 min with BrdU. After fixation in 70% ethanol at −20°C, cells were stained with PI solution. Sorting windows within S-phase were defined on the basis of DNA content. Cells were collected with two gates, corresponding to early- and late-S-phase, by FACS Aria III. The proper fractionation of S-phase cells has been validated by reanalysis of the DNA contents of the sorted cells (Supplementary Figure S15). Isolation of BrdU-labelled DNA with immunoprecipitation has been described (Ryba et al, 2011).
Replication timing array
The replication profiling protocol has been described (Ryba et al, 2011). Two independent biological replicates were analysed, in which early- and late-replicating DNA were labelled with Cy3 and Cy5 (with dye switch). Nascent DNA was amplified in the linear range (20 cycles) by using a WGA2 kit (Sigma-Aldrich) according to the protocol provided by the supplier and the amplified products were then purified with a PCR cleanup kit (Qiagen). The amplified samples were loaded onto gel to determine size range and reverified by PCR to confirm that no bias was introduced during amplification. Amplified DNA was labelled using the Genomic DNA Enzymatic Labelling kit (Agilent) and hybridized with custom microarrays according to the manufacturer’s instructions (Agilent: 4 × 44K: G4497A#022461). The oligonucleotide microarrays used contain one probe every 1.0 kb. The array slides were scanned with a Laser Scanner GenePix 4000B (Molecular Devices) using appropriate gains on the photomultiplier (PMT) to obtain the highest intensity without saturation. Scanned images were processed using Feature Extraction software (Agilent). Data analyses were done using Genomic workbench software. Smoothed replication timing profiles are shown with a moving average of 100 kb windows. The raw and processed microarray data are accessible through GEO Datasets under accession numbers GSE37971.
Replication timing analyses using real-time PCR
Immunoprecipitated nascent DNA was subjected to quantitative PCR with the Light Cycler 480 (Roche) using SYBR Premix Ex Taq™ (Perfect Real Time; TaKaRa) to quantify the amount of the replicating DNA at a given locus in each fraction (early- and late-S-phase). Primers used for PCR are listed in Supplementary Table S1. Mitochondrial DNA (mtDNA) known to replicate throughout cell cycle and to be equally represented in early- and late-S-phase fractions was used as control. The amount of each DNA relative to that of mtDNA is presented.
Halo assays
Control and Rif1-depleted HeLa cells were synchronized at early-S-phase by double-thymidine block. Harvested cells were washed twice in PBS and halo assays were preformed as previously described (Gerdes et al, 1994). The halo radius of each nucleus was determined by measuring the total areas of the nucleus and the central area, heavily stained with DAPI, and using the following formula. The halo radius  ; A, total area of nucleus; B, central area.
; A, total area of nucleus; B, central area.
Antibodies
Antibodies used in this study were anti-Rif1 antibody (1:1000 for western blotting and 1:50 for immunofluorescence; Bethyl Laboratories, Inc.), anti-phosphoMCM2 Ser53 (1:1000; Bethyl Laboratories, Inc.), anti-MCM2 (1:200; Santa Cruz Biotechnology, Inc.), anti-phosphoMCM4 Ser6Thr7 (1:1000), anti-MCM4 (1:1000) (Masai et al, 2006), anti-ATR (1:200; Santa Cruz Biotechnology, Inc.), anti-phosphoHistoneH3 Ser10 (1:200; Upstate), anti-PCNA (1:200; Santa Cruz Biotechnology, Inc.), anti-HistoneH3 (1:200; Santa Cruz Biotechnology, Inc.) anti-Cdc45 (1:200; Masai et al, 2006), anti-Lamin B1 (1:200; Santa Cruz Biotechnology, Inc.), anti-αTubulin (1:1000; Sigma), anti-Cdc7 (1:1000; MBL), and anti-BrdU antibody (MBL), and anti-Cdt1 antibody (1:200; Santa Cruz Biotechnology, Inc.).
RNAi experiments
HeLa and NHDF cells were transfected with siRNA for 48 or 72 h according to the Invitrogen protocol. In double gene silencing, both siRNAs were mixed and added to the cells for 48 h. siRNA#2, #3, #4 and #6 were designed as published previously (Silverman et al, 2004). Human Rif1 siRNA: #2, AAUGAGACUUACGUGUUAAAAdTdT; #3, AAGAGAAACCAGGUUCUGAAGdTdT; #4, AAGAAUGAGCCCCUAGGGAAAdTdT; #6, AAGAGCAUCUCAGGGUUUGCUdTdT; ATR siRNA, CCUCCGUGAUGUUGCUUGAdTdT; Cdt1 siRNA, GACAUGAUGCGUAGGCGUUdTdT. The sense oligonucleotide for each pair was used for control transfection.
Supplementary Material
Acknowledgments
We thank Dr Dave Gilbert for helpful discussion, advice and critical reading of the manuscript. We thank Naoko Kakusho for technical assistance and the members of our laboratory for helpful discussion. We thank Mr Daniel Jeffery for comments on the manuscript. We also thank Drs Hiroki Ashida and Yoshikazu Furuta for their help in analyses of replication timing data. This work was supported in part by Grants-in-Aid for Basic Scientific Research (A) and Grant-in-Aid for Scientific Research on Priority Area ‘Chromosome Cycle’ from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by Takeda Science Foundation and by Astellas Foundation for Research on Metabolic Disorders (to HM).
Author contributions: SY performed the majority of the experiments, analysed the data and prepared the figures. HM designed the experiments, directed, supervised the study and provided financial support. SY and HM wrote the paper. AI performed some experiments. YK and MO gave technical advice on BrdU ChIP and timing array analyses. YN conducted hybridization assay of timing arrays.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM (2004) The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 24: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC (2007) Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Buongiorno-Nardelli M, Micheli G, Carri MT, Marilley M (1982) A relationship between replicon size and supercoiled loop domains in the eukaryotic genome. Nature 298: 100–102 [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Wu Y, Ferguson D, de Lange T (2009) Mammalian Rif1 contributes to replication stress survival and homology-directed repair. J Cell Biol 187: 385–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M (2008) Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Berezney R (2002) The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J Cell Sci 115: 4037–4051 [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM (1999) The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993 [DOI] [PubMed] [Google Scholar]
- Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I (2008) Global organization of replication time zones of the mouse genome. Genome Res 18: 1562–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Tu Z, Wu W, Liang C (2003) Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Res 63: 7356–7364 [PubMed] [Google Scholar]
- Gerdes MG, Carter KC, Moen PT Jr, Lawrence JB (1994) Dynamic changes in the higher-level chromatin organization of specific sequences revealed by in situ hybridization to nuclear halos. J Cell Biol 126: 289–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM (2010) Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM (2010) Cell fate transitions and the replication timing decision point. J Cell Biol 191: 899–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, Cedar H (2008) DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev 22: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson HC, Schmiesing JA, Kim JS, Kobayashi T, Zhou S, Yokomori K (2001) A potential role for human cohesin in mitotic spindle aster assembly. J Biol Chem 276: 47575–47582 [DOI] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Mendez J (2010) Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 24: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA (2010) Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci 107: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D (1992) A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, Masai H (2012) Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev 26: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, Gasser SM (2001a) The positioning and dynamics of origins of replication in the budding yeast nucleus. J Cell Biol 152: 385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM (2001b) Chromosome dynamics in the yeast interphase nucleus. Science 294: 2181–2186 [DOI] [PubMed] [Google Scholar]
- Hiratani I, Gilbert DM (2009) Replication timing as an epigenetic mark. Epigenetics 4: 93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM (2008) Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi S, Lu J, Gilbert DM (2009) Replication timing and transcriptional control: beyond cause and effect--part II. Curr Opin Genet Dev 19: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Ishii A, Kakusho N, Taniyama C, Yamazaki S, Fukatsu R, Sakaue-Sawano A, Miyawaki A, Masai H (2012) Mechanism of cancer cell death induced by depletion of an essential replication regulator. PLoS One 7: e36372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630 [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A (2007) Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res 17: 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A (2009) Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell 21: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Dutta A (2011) The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev 25: 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna K, Stanek D, Malinsky J, Eltsov M, Pliss A, Ctrnacta V, Cermanova S, Raska I (1999) Nuclear organization studied with the help of a hypotonic shift: its use permits hydrophilic molecules to enter into living cells. Chromosoma 108: 325–335 [DOI] [PubMed] [Google Scholar]
- Li F, Chen J, Izumi M, Butler MC, Keezer SM, Gilbert DM (2001) The replication timing program of the Chinese hamster beta-globin locus is established coincident with its repositioning near peripheral heterochromatin in early G1 phase. J Cell Biol 154: 283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HY, Robertson ED, Hiraga S, Alvino GM, Collingwood D, McCune HJ, Sridhar A, Brewer BJ, Raghuraman MK, Donaldson AD (2011) The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol Biol Cell 22: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL et al. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maas NL, Jonsson ZO, Defazio-Eli LG, Wohlschlegel J, Toczyski DP (2010) Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li F, Murphy CS, Davidson MW, Gilbert DM (2010) G2 phase chromatin lacks determinants of replication timing. J Cell Biol 189: 967–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem 79: 89–130 [DOI] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, Tamai K, Ohtani K, Arai K (2006) Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem 281: 39249–39261 [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Tenca P, Sola F, Carpani D, Brotherton D, Albanese C, Santocanale C (2004) Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res 64: 7110–7116 [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Brotherton D, Troiani S, Rainoldi S, Tenca P, Molinari A, Santocanale C (2006) Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J Biol Chem 281: 10281–10290 [DOI] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM (2011) Genome-scale analysis of replication timing: from bench to bioinformatics. Nat Protoc 6: 870–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM (2010) Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H (1998) Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T (2004) Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev 18: 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi S, Sugimura K, Saito T, Sato C, Fukushima Y, Taguchi H, Okumura K (2005) Regulation of replication at the R/G chromosomal band boundary and pericentromeric heterochromatin of mammalian cells. Exp Cell Res 304: 162–1674 [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M (2002) Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhao A, Chen L, Zhong X, Liao J, Gao M, Cai M, Lee DH, Li J, Chowdhury D, Yang YG, Pfeifer GP, Yen Y, Xu X (2009) Human RIF1 encodes an anti-apoptotic factor required for DNA repair. Carcinogenesis 30: 1314–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Nurse P (2009) Establishing the program of origin firing during S phase in fission yeast. Cell 136: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Muniandy P, Leo E, Yin J, Thangavel S, Shen X, Ii M, Agama K, Guo R, Fox D 3rd, Meetei AR, Wilson L, Nguyen H, Weng NP, Brill SJ, Li L, Vindigni A, Pommier Y, Seidman M, Wang W (2010) Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J 29: 3140–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Blackburn EH (2004) Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol 167: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I (2010) Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2010) Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.