Abstract
Most common genetic factors known to cause intellectual disability are Down syndrome and Fragile X syndrome. However, the underlying cellular and molecular mechanisms of intellectual disability remain unclear. Recently, dendritic spine dysmorphogenesis and impaired local protein synthesis are posited to contribute to the cellular mechanisms of intellectual disability. Here, we show that Down syndrome critical region1 (DSCR1) interacts with Fragile X mental retardation protein (FMRP) and regulates both dendritic spine morphogenesis and local protein synthesis. Interestingly, decreasing the level of FMRP restores the DSCR1-induced changes in dendritic spine morphology. Our results imply that DSCR1 is a novel regulator of FMRP and that Fragile X syndrome and Down syndrome may share disturbances in common pathways that regulate dendritic spine morphology and local protein synthesis.
Keywords: down syndrome, DSCR1, FMRP, Fragile X syndrome, local protein synthesis
Introduction
Dendritic spines, which receive and integrate presynaptic signals, play critical roles in synaptic function and plasticity in the brain. Abnormal dendritic spine morphology thus hinders the cellular and molecular mechanisms of neuronal activity network, consequently affecting learning and memory. Dendritic pathology also appears to be a common feature of intellectual disability regardless of its origin, whether due to environmental or genetic factors (Dierssen and Ramakers, 2006). Both in Down syndrome, the most common genetic cause of intellectual disability caused by trisomy 21, and in Fragile X syndrome, the most prevalent form of intellectual disability resulting from a single gene mutation, there are reports of abnormal spine morphology (Belichenko et al, 2004, 2009; Dierssen and Ramakers, 2006; Bassell and Warren, 2008; Waung and Huber, 2009). However, the molecular mechanisms regulating dendritic spine morphogenesis are not well understood and it is not known whether the two disorders share common disturbances in molecular pathways.
Recent studies propose that in addition to spine dysmorphogenesis, defects in local protein synthesis at dendritic spines may also contribute to the cellular and molecular mechanisms of intellectual disability. One protein implicated in spine morphogenesis and intellectual disability is the Fragile X mental retardation protein (FMRP) (Dierssen and Ramakers, 2006; Bassell and Warren, 2008; Waung and Huber, 2009). The absence of the mRNA-binding protein FMRP, encoded by the fmr1 gene, causes mental impairment in Fragile X syndrome patients (Bagni and Greenough, 2005; Bassell and Warren, 2008; Garber et al, 2008; Waung and Huber, 2009). The fmr1 knockout mice have deficits in learning behaviour, altered synaptic activity, as well as abnormal spine morphology characterized by a high density of thin and elongated spines (Comery et al, 1997; Penagarikano et al, 2007). FMRP is located in the cell body, dendrites, and dendritic spines in neurons and emerging evidence suggest that FMRP is involved in local protein synthesis in dendritic spines, although its regulatory mechanism is not well understood (Bagni and Greenough, 2005; Bassell and Warren, 2008; Muddashetty et al, 2011; Nalavadi et al, 2012; Niere et al, 2012).
Down syndrome critical region1 (DSCR1, also called RCAN1 or regulator of calcineurin) is located on human chromosome 21 and highly expressed in the brain, especially enriched in hippocampal neurons (Fuentes et al, 1995). DSCR1 belongs to a conserved family of calcineurin (CaN) inhibitors called calcipressins, which includes RCN1P in yeast (Kingsbury and Cunningham, 2000), CBP1 in fungus (Görlach et al, 2000), nebula in Drosophila (Chang et al, 2003), as well as DSCR1 in mouse and human (Casas et al, 2001; Arron et al, 2006). DSCR1 knockout mice show learning deficits and impaired late-phase long-term potentiation (L-LTP), which requires new gene expression (Hoeffer et al, 2007). By blocking translation and transcription surgically or using inhibitors, several reports have shown that L-LTP requires local protein synthesis, while somatic transcription and translation may be dispensable (Sutton and Schuman, 2006; Costa-Mattioli et al, 2009). Together, it implies that DSCR1 is involved in local protein synthesis. Furthermore, a transgenic mouse overexpressing DSCR1 in the brain show significant defects in learning (Dierssen et al, 2011), suggesting that DSCR1 may play an important role in intellectual disability in Down syndrome. However, the roles of DSCR1 in dendritic spine morphogenesis or local protein synthesis at dendritic spines are not studied.
Here, we show that DSCR1 interacts with FMRP and regulates both dendritic spine morphogenesis and local protein synthesis. DSCR1 knockout mice exhibit reduced number and size of dendritic spines in hippocampal CA1 region, while DSCR1 transgenic mice have enlarged spines. Interestingly, reducing the level of FMRP restores the DSCR1-induced changes in dendritic spine morphology. DSCR1 specifically interacts with phosphorylated FMRP that suppresses translation. Using the photoswitchable fluorescent dendra2 protein fused with 5′UTR and 3′UTR of αCaMKII, we show that DSCR1 is required for local protein synthesis at dendritic spines following BDNF stimulation. We also find that neuronal BDNF stimulation induces phosphorylation of DSCR1, which may activate calcineurin to dephosphorylate phosphorylated FMRP, thus allowing protein synthesis to ensue at the dendritic spines.
Results
DSCR1 is involved in dendritic spine morphogenesis
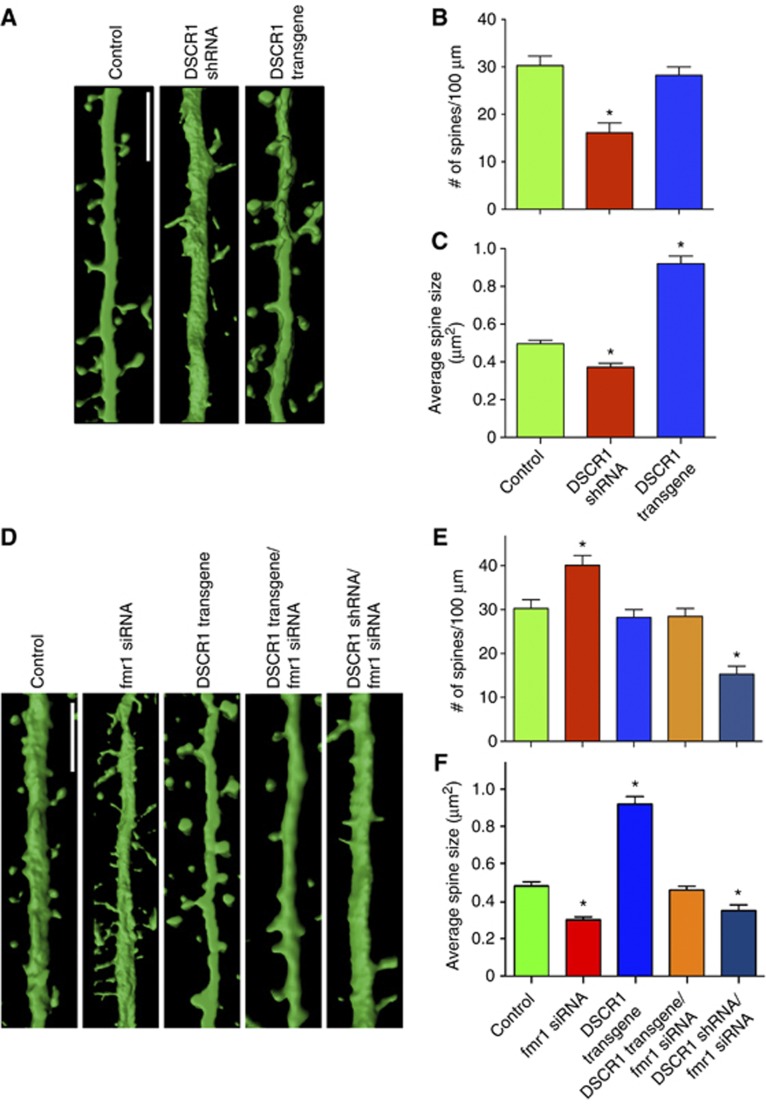
DSCR1 is highly expressed in the hippocampus (Fuentes et al, 1995), but its subcellular distribution within hippocampal neurons is unknown. Immunostaining revealed that DSCR1 is present in the cell bodies, dendrites, and dendritic spines, but not detected in the presynaptic terminals of axons (Supplementary Figure S1). As DSCR1 is highly expressed in the postsynaptic sites, we first investigated the role of DSCR1 in spine morphogenesis by manipulating the level of DSCR1 within hippocampal neurons. DSCR1 shRNA transfection reduced the level of DSCR1 protein to 50% while overexpression of DSCR1 increased DSCR1 in dendrites (Supplementary Figure S2). Reduction of the DSCR1 protein caused a significant decrease in dendritic spine density as well as the size of spine heads (Figures 1A–C; Supplementary Figure S3). On the contrary, overexpression of DSCR1 profoundly increased the size of dendritic spine heads (Figure 1A–C; Supplementary Figure S3). Together, these results suggest that DSCR1 is important for dendritic spine morphogenesis.
Figure 1.
DSCR1 and FMRP regulate spine morphogenesis. (A) 3D reconstruction images showing phenotypes of pyramidal hippocampal neurons transfected with DSCR1 shRNA or DSCR1 transgene. Scale bar, 5 μm. (B) Number of spines per 100 μm of dendrites and (C) size of spine head. (D) The increase in spine head size caused by DSCR1 overexpression is restored by FMRP reduction. Phenotypes of neurons having both DSCR1 shRNA and fmr1 siRNA are similar to DSCR1 reduction alone. Scale bar, 5 μm. (E) Dendritic spine density and (F) area of spine head. Eight to thirteen neurons for each condition from four independent experiments. In all, 100–150 spines were measured for each condition. Neurons at DIV 21 were used for the analyses. All values are shown as mean±s.e.m., and statistical significance determined by one-way ANOVA analysis. *P<0.005.
DSCR1 genetically interacts with fmr1 in regulating spine morphogenesis
Based on similar subcellular localization and role in spine morphogenesis, we hypothesized that DSCR1 and FMRP may participate in the same pathway to regulate synaptic morphology. To test the hypothesis, we first investigated phenotypes of hippocampal primary neurons containing DSCR1 shRNA or DSCR1 transgene with fmr1 siRNA (Figure 1D–F). Expression levels of DSCR1 and FMRP in neurons transfected with various expression vectors were confirmed by comparing the relative staining intensities of transfected versus untransfected primary hippocampal neurons within the same field of view (Supplementary Figure S4). Reduction of FMRP by fmr1 siRNA caused increased spine density and decreased spine size as reported previously (Comery et al, 1997; Penagarikano et al, 2007). Interestingly, a 50% reduction of FMRP by fmr1 siRNA restored the phenotypes caused by DSCR1 transgene expression, whereas reduction of both DSCR1 and FMRP resulted in decreased spine density and size similar to that seen in DSCR1 reduction only (Figure 1D–F; Supplementary Figure S4). Together, the results suggest that DSCR1 and FMRP are involved in the same biological pathway.
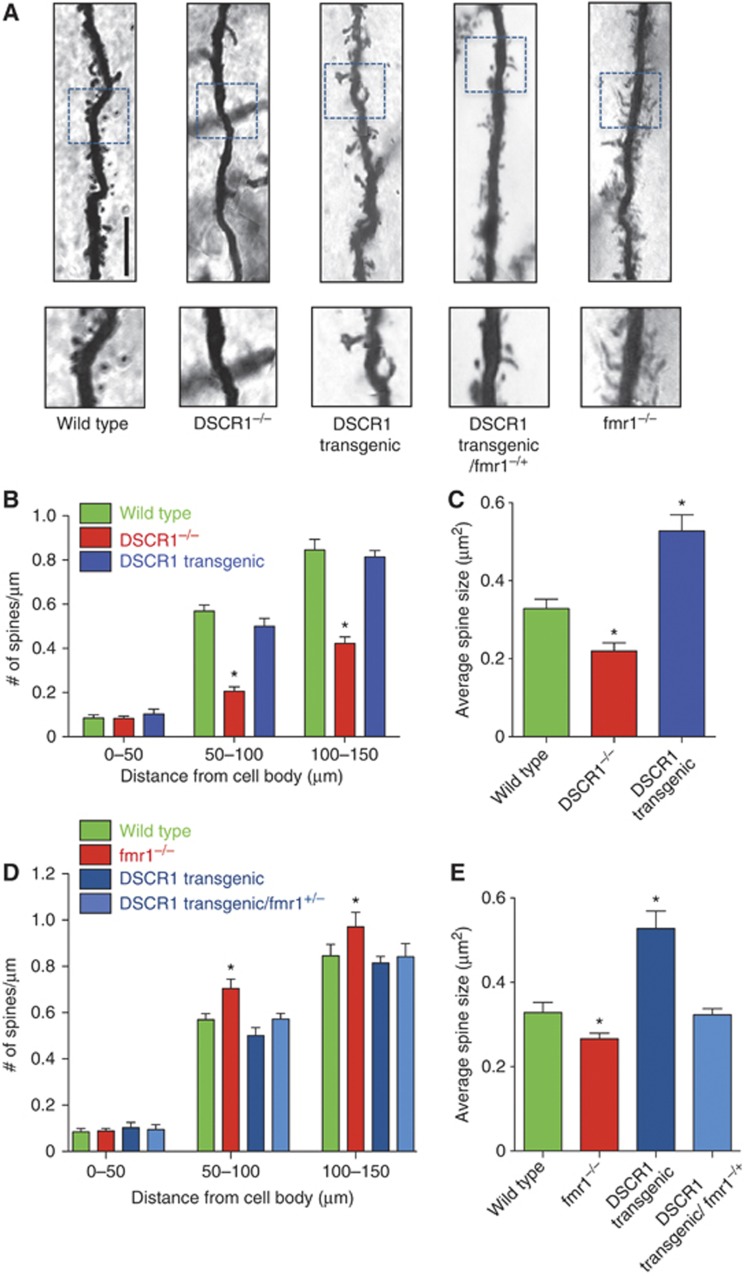
To confirm that DSCR1 regulates dendritic spine morphology and that DSCR1 and FMRP do indeed interact in vivo, we first investigated the role of DSCR1 in spine morphogenesis within hippocampal neurons in DSCR1−/− as well as in DSCR1 transgenic mice. Expression levels of DSCR1 in the brains of mutant mice were confirmed by western blot analyses using an antibody specific for DSCR1 (Supplementary Figure S5A). To assess the involvement of DSCR1 in the regulation of spine morphogenesis in vivo, we performed Golgi staining on P21 brains of DSCR1−/−, DSCR1 transgenic, and wild-type mice. Consistent with the aberrant dendritic phenotypes found in dendritic spines of primary hippocampal neurons transfected with DSCR1 shRNA and DSCR1 transgene (Figure 1), we also observed similar morphological changes in the pyramidal neurons of DSCR1 mutant mice (Figure 2A–C). Spine density and spine head size in the pyramidal neurons in hippocampal CA1 of DSCR1−/− mutant were decreased significantly (63 and 34% reduction in spine density and spine size, respectively, in the region 50–100 μm from the cell body), while DSCR1 transgenic mouse showed a 60% increase in the size of spine heads in the neurons without decreasing spine density (Figure 2B and C). The cortical layer V of DSCR1 mutant brains also showed similar phenotypes in spine density and size (Supplementary Figure S6). Together, these results confirm that DSCR1 is important for dendritic spine morphogenesis in vivo.
Figure 2.
DSCR1 and fmr1 interact genetically in spine morphogenesis. (A) Golgi staining of dendritic spines in hippocampal CA1 pyramidal neurons in wild-type, DSCR1−/−, DSCR1 transgenic, DSCR1 transgenic/fmr1−/+, and fmr1−/− mice. DSCR1−/− generated reduced number of spines, while DSCR1 transgenic mouse produced enlarged spine head. Consistent with the primary hippocampal neuron culture results, DSCR1 transgenic/fmr1−/+ restored the increase in the size of spine head observed in the brains of DSCR1 transgenic mice to normal. Inset box shows different sizes of spine heads. Each image is composed from an average of 30 focused Z-stacks. Scale bar, 10 μm. (B–E) Quantification of dendritic spines from the cell body on the CA1 pyramidal neurons. Eight neurons from brains of each genotype were used for Golgi staining. (B, D) Number of spines grouped by distance from the cell body and (C, E) average spine head area of CA1 pyramidal neurons. In all, 50–60 spines were used to measure the size. Postnatal 21 days of two mice brains of each strain were used for the analyses. Values represent mean±s.e.m., *indicates P<0.0001 as determined by ANOVA analysis.
To test whether DSCR1 and FMRP do indeed interact to regulate dendritic spine morphology in vivo, we next generated mutant mice containing the DSCR1 transgene but with reduced level of fmr1 (fmr1−/+) (Supplementary Figure S5B). Consistent with the primary hippocampal neuron culture results, removing one copy of fmr1 was sufficient to restore the average size of spine heads observed in the brains of DSCR1 transgenic mice (Figure 2A, D and E). Taken together, our results suggest that DSCR1 genetically interacts with fmr1 in regulating spine morphogenesis.
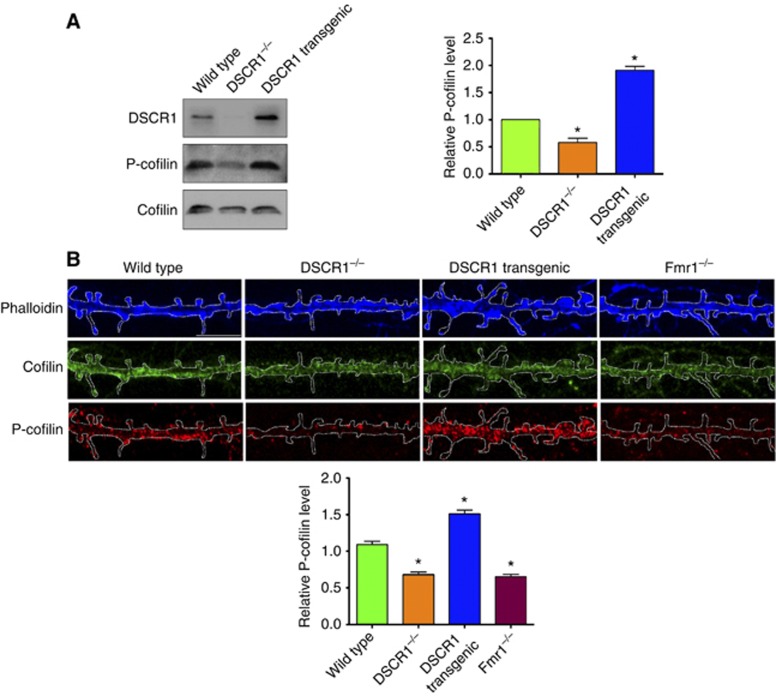
Studies have shown that phosphorylation of cofilin plays an important role in regulating spine morphology. Phosphorylated cofilin facilitates actin polymerization and produces enlarged spine heads, while dephosphorylated cofilin severs actin filaments, thus resulting in small and thin head spines (Cingolani and Goda, 2008). Hence, we investigated the phosphorylation status of cofilin in HEK cells, hippocampal primary neurons with DSCR1 overexpression or reduction, as well as hippocampal neurons from wild-type, DSCR1−/−, and DSCR1 transgenic mice. Figure 3 and Supplementary Figure S7 show that the phosphorylation level of cofilin in the cells is increased when DSCR1 is overexpressed, while reduction of DSCR1 decreased the amount of phospho-cofilin. These results are consistent with the enlarged and reduced spine head size observed in DSCR1 transgenic and DSCR1−/− neurons, respectively. Interestingly, while reducing the level of FMRP decreased the level of phospho-cofilin, reducing FMRP level in DSCR1 overexpression background restored the level of phospho-cofilin, which is accompanied by normal spine size (Figures 1F, 2E and 3; Supplementary Figure S7). Since DSCR1 inhibits calcineurin activity (Fuentes et al, 2000) and calcineurin can dephosphorylate phospho-cofilin (Zhou et al, 2004), it is plausible that DSCR1 plays a key role in spine morphogenesis by modulating calcineurin activity, which in turn changes the level of phospho-cofilin.
Figure 3.
DSCR1 regulates the phosphorylation level of cofilin. (A) Twenty-one days old postnatal hippocampi or (B) primary hippocampal neurons at DIV 21 prepared from wild-type, DSCR1−/−, and DSCR1 transgenic mice were examined for the level of phospho-cofilin by western blotting (A) and immunocytochemistry (B). Relative levels of phospho-cofilin are normalized to the level of cofilin. Decreasing DSCR1 reduced phospho-cofilin level, while overexpression of DSCR1 increased phospho-cofilin. Primary hippocampal neurons from fmr1−/− also showed decreased level of phosphorylated cofilin. n=3 per condition, values were shown as mean±s.e.m., and tested for statistical significance by ANOVA analysis. *Indicates P<0.001.
DSCR1 and phosphorylated FMRP form a complex
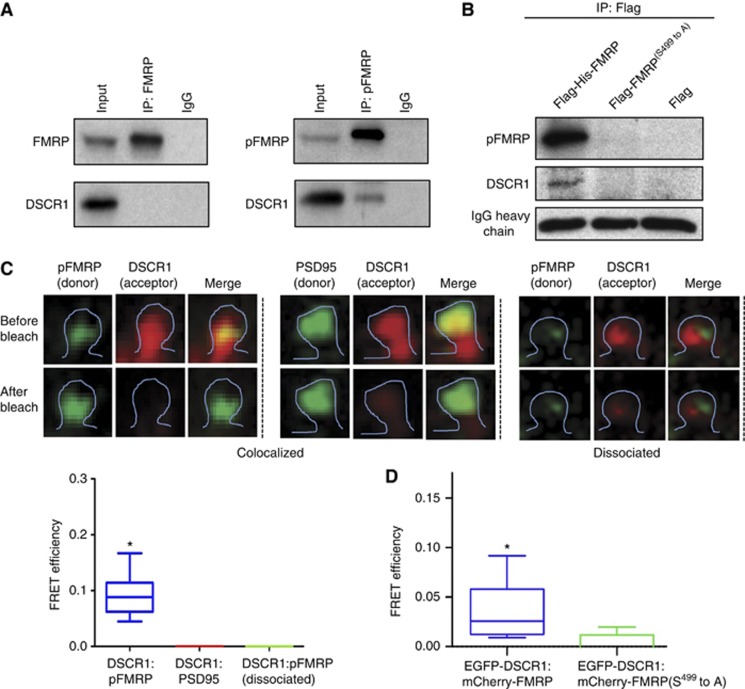
Having established that DSCR1 and fmr1 can interact genetically in the same pathway to modulate spine morphologies, we next sought to determine if DSCR1 and FMRP also interact physically. To examine whether DSCR1 binds to FMRP, we immunoprecipitated FMRP with an FMRP-specific antibody and checked for the presence of DSCR1 via western blot with a DSCR1-specific antibody. We found no interaction when the immunoprecipitation was performed with the FMRP antibody (Figure 4A). Since FMRP is also present as phosphorylated protein in neurons (Ceman et al, 2003; Narayanan et al, 2007), we next checked to see if DSCR1 can interact with phospho-FMRP. Figure 4A shows that DSCR1 is brought down by an antibody specific for phospho-FMRP. Note that we were not able to detect DSCR1 interaction when performing immunoprecipitation with the FMRP antibody because the FMRP antibody preferentially binds to non-phosphorylated FMRP, and with significantly less binding to phosphorylated FMRP (Supplementary Figures S8A and B). Next, to verify the interaction between DSCR1 and phospho-FMRP, we mutated the phosphorylation site on FMRP (serine residue at 499 to alanine). HEK cells transfected with either Flag-His-FMRP or Flag-His-FMRP(S499 to A) were used to perform immunoprecipitation with antibody against the Flag tag. We found that in cells transfected with Flag-His-FMRP, phosphorylated FMRP and DSCR1 were detected in the immuoprecipitates, but not in cells transfected with Flag-His-FMRP(S499 to A) (Figure 4B; Supplementary Figure S8C). These results confirm that DSCR1 specifically interacts with phosphorylated FMRP.
Figure 4.
DSCR1 and phospho-FMRP interact in vitro and in cells. (A) HEK cells transfected with DSCR1 transgene were used for immunoprecipitation. Immunoprecipitation with FMRP or phospho-FMPP antibodies was followed by blotting with DSCR1 antibody. The phospho-FMRP immunoprecipitates brought down DSCR1, while FMRP immunoprecipitates do not contain DSCR1. Input: HEK cell extracts. (B) Immunoprecipitation with Flag antibody done using HEK cell lysates transfected with Flag-His-FMRP or Flag-His-FMRP(S499 to A) and DSCR1. Flag-His-FMRP showed interaction with DSCR1, while Flag-His-FMRP(S499 to A) did not bind to DSCR1. (C) FRET analysis of DSCR1 and phospho-FMRP interaction using acceptor photo-bleaching method. Colocalization between DSCR1 and phospho-FMRP in spine shows strong FRET signals, while no FRET signal is found between DSCR1 and PSD95 although these two proteins are colocalized. DSCR1 and phospho-FMRP that are dissociated also do not show FRET signals. Primary hippocampal neurons at DIV 21 were used. n=27 spines from three independent experiments, *P<0.0001. Student’s t-test was used. (D) EGFP-DSCR1 and mCherry-FMRP show significant FRET, while there is negligible FRET signals between EGFP-DSCR1 and mCherry-FMRP(S499 to A). Live HEK cells were used to perform FRET analysis. Nine cells from three independent experiments were used. *P<0.01. Student’s t-test was used.
To further confirm whether DSCR1 and phospho-FMRP forms a complex in hippocampal primary neurons, we applied FRET detection method between DSCR1 and phospho-FMRP (Figure 4C). Since expression of fluorescent-labelled FMRP form aggregates in primary neurons and severely affect the health of the cells, we performed immuno-FRET using double-labelling immunofluorescence with fixed hippocampal neurons (König et al, 2006; David Gerecht et al, 2010; Ebrahimi et al, 2010). We used phospho-FMRP antibody detected with secondary antibody conjugated with Alexa 488 as donor and DSCR1 antibody detected with a secondary antibody conjugated with Alexa 555 as acceptor. Following photo-bleaching of the FRET acceptor, protein interaction is indicated by an increase in donor fluorescence. We found strong FRET signals when DSCR1 and phospho-FMRP are colocalized in dendritic spines. We also measured FRET signals between DSCR1 and PSD95 in spines as controls. No FRET signal was observed although DSCR1 and PSD95 colocalize, suggesting that DSCR1 and PSD95 do not interact (Figure 4C). Next, to confirm the interaction between DSCR1 and phospho-FMRP in live cells, we performed FRET experiment using live HEK cells containing EGFP-DSCR1 and mCherry-FMRP. There is significant FRET signal between the two proteins (Figure 4D). To verify the result, we have also mutated the phosphorylation site on FMRP (serine residue at 499 to alanine) and repeated the FRET experiment between EGFP-DSCR1 and mCherry-FMRP(S499 to A). We found negligible FRET signals between these two proteins in the cells, which is consistent with the IP and the FRET results showing that DSCR1 interacts with phosphorylated FMRP (Figure 4D). Collectively, these results imply that phosphorylated FMRP forms a complex with DSCR1 in vitro as well as in dendritic spines of hippocampal neurons.
DSCR1 is involved in local protein synthesis at the dendritic spines
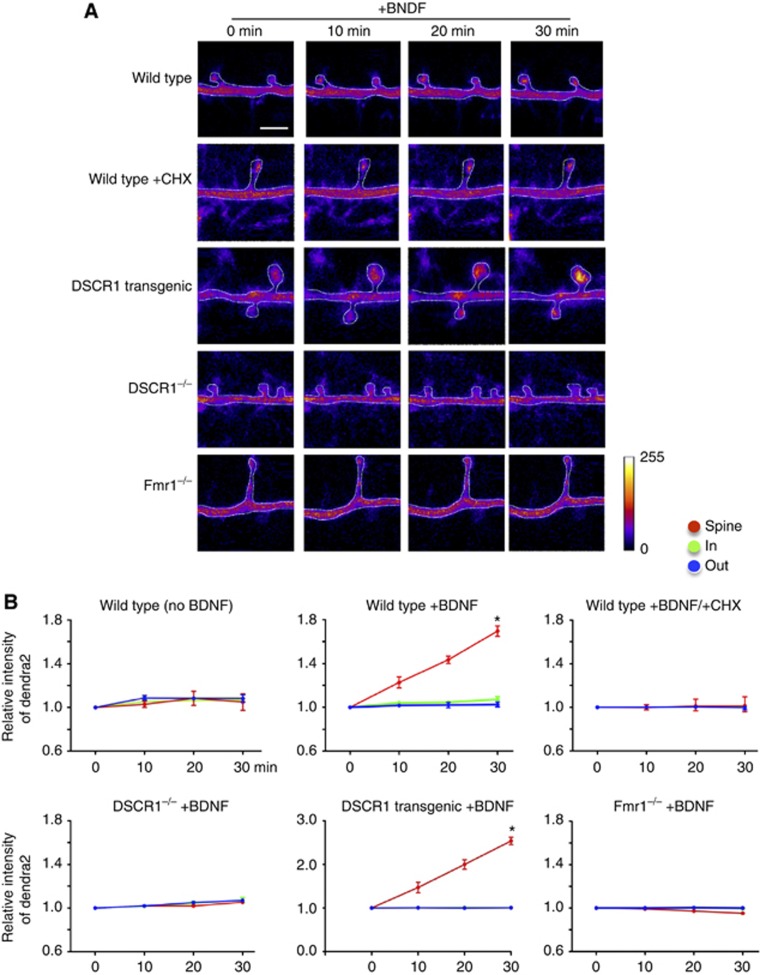
It has been suggested that phospho-FMRP associates with stalled polyribosome, and that dephosphorylation of the phospho-FMRP following neuronal stimulation leads to activation of mRNA translation (Ceman et al, 2003; Narayanan et al, 2007). It has also been shown that local protein synthesis in dendrites can be activated by exposure to the growth factor BDNF; BDNF stimulation triggers local protein synthesis of the FMRP target mRNAs encoding αCaMKII, Arc/Arg3.1, Map1B, and APP (Napoli et al, 2008). Based on report that DSCR1 knockout mice have learning deficits and impaired L-LTP (Hoeffer et al, 2007), which requires new gene expression, as well as our finding that DSCR1 interacts with phospho-FMRP (Figure 4), we hypothesized that DSCR1 is involved in regulation of local protein synthesis. To test this hypothesis in live neurons, we first constructed a vector containing a photoswitchable dendra2 protein fused with the 5′UTR and 3′UTR of αCaMKII mRNA (5′UTR-dendra2-3′UTRαCaMKII). The 5′UTR and 3′UTR of αCaMKII transcript are sufficient to direct its dendritic localization and translation of the reporter protein (Rook et al, 2000; Aakalu et al, 2001), and BDNF stimulation induces αCaMKII mRNA translation in dendritic spines (Aakalu et al, 2001; Smith et al, 2001). Dendra2 protein is normally a green fluorescent protein, but can be irreversibly converted to red fluorescence following photo-activation (Gurskaya et al, 2006). Using this construct, we are able to directly distinguish if the dendra2 protein is synthesized newly at the spines or is transported to the spines from the soma. We prepared hippocampal neurons from wild-type, DSCR1−/−, DSCR1 transgenic, and fmr1−/− mice that were transfected with 5′UTR-dendra2-3′UTRαCaMKII. A small dendritic region including spines is activated with the UV light, which converts the green fluorescent dendra protein in the region to red fluorescence (Supplementary Figure S9A). Hence, if additional green fluorescent dendra proteins are observed in the spine after photo-activation, then the proteins are either newly synthesized locally or transported into the spine from cell soma. Immediately after activation of a dendritic region, we applied BDNF to neurons and took time-lapse images every 10 min for 30 min (Figure 5). To assess local protein synthesis, we measured the intensity of newly synthesized green fluorescent dendra proteins in three different regions: spine, and dendritic regions proximal to and distal to the soma (Figure 5A; Supplementary Figure S9B and C). Without BDNF stimulation, no newly synthesized dendra protein was observed in the photo-activated spines or surrounding regions, suggesting that new protein synthesis does not occur at dendritic spines without synaptic stimulation (Figure 5B). After BDNF stimulation, however, the intensity of green fluorescent dendra within the dendritic spine increased significantly, while the surrounding dendritic regions of the spine showed no detectable increase in the green dendra protein. This result suggests that the green dendra proteins are indeed newly synthesized at the spines, and are not transported down from the soma (Figure 5B; Supplementary Figure S9). It is unlikely that the increase in green fluorescence is due to differences in spine size or dendra2 distribution, since the intensity of green fluorescence was not altered before BDNF treatment in all conditions and the conversion from green to red fluorescence is irreversible (Figure 5; Supplementary Figures S9 and S10). To further verify that the increased signals are not due to diffusion, we measured local proteins synthesis in neurons stimulated with BDNF in the presence of translational inhibitor, cycloheximide or anisomycin. The result clearly shows that cycloheximide and anisomycin treatment inhibited local proteins synthesis (Figure 5B; Supplementary Figure S10B), confirming that increased dendra signals due to local protein synthesis at spines. Next, we asked whether DSCR1 modulates local protein synthesis. The same experimental conditions as used for control neurons were applied to hippocampal neurons from DSCR1−/−, DSCR1 transgenic, and fmr1−/− mice. BDNF treatment failed to induce new protein synthesis in spines of primary hippocampal neurons from DSCR1−/−. In contrast, BDNF activation of neurons from DSCR1 transgenic mouse significantly enhanced local protein synthesis of dendra at spines compared to that of control. Fmr1−/− did not exhibit increased protein synthesis at dendritic spines. Also, no local protein synthesis was observed without BDNF stimulation in the mutant mice (Supplementary Figure S10A). These results with primary neurons from mutant mice are consistent with results using DSCR1 shRNA, DSCR1 transgene, and fmr1 siRNA (Supplementary Figure S9). We also tested whether the increase in local protein synthesis by DSCR1 overexpression is restored to normal if the level of FMRP is reduced. After 10 min of BDNF treatment, the amount of local protein synthesis in the spines is increased to the same extent as that of DSCR1 overexpression, however, in contrast to control or DSCR1 overexpression, local protein synthesis attenuated after 20 or 30 min of BDNF activation (Supplementary Figure S9). Taken together, these results indicate that DSCR1 is necessary for local protein synthesis in dendritic spines.
Figure 5.
DSCR1 is required for local protein synthesis. (A) Pseudo-coloured images of the green fluorescent dendra2 proteins in dendritic spines. An increase in intensity indicates newly synthesized dendra2 protein in the spines at various time points following BDNF treatment. Reduction of DSCR1 inhibited local protein synthesis in spine upon BDNF stimulation, while DSCR1 overexpression caused increased synthesis of new dendra2 protein in spines. FMRP reduction led to no local protein synthesis upon BDNF stimulation. Scale bar, 2 μm. (B) Change in fluorescence intensity was analysed in three different regions: near to soma (in), spine, and away from soma (out) for 30 min with BDNF stimulation. Protein synthesis occurs only after BDNF stimulation. Addition of translation inhibitor, cycloheximide (CHX), in the presence of BDNF, prevents local protein synthesis. Note the scale change in the relative intensity of dendra2 in the neurons from DSCR1 transgenic mouse. Neurons at DIV 14 were used. n=24 spines in each condition from three independent experiments, *P<0.0001. Values were shown as mean±s.e.m., and tested for statistical significance by ANOVA analysis.
BDNF stimulation phosphorylates DSCR1 that activates calcineurin followed by dephosphorylation of phospho-FMRP
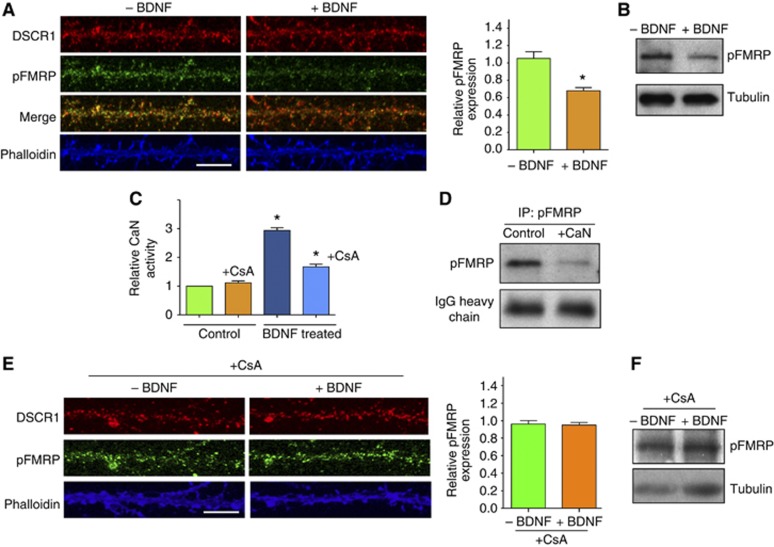
We next investigated the underlying molecular basis of how DSCR1 regulates local protein synthesis. Hilioti et al (2004) suggested that non-phosphorylated Rcn1, a yeast homologue of DSCR1, inhibits calcineurin, while phosphorylation of Rcn1 by GSK-3 may activate calcineurin. It is plausible that BDNF stimulation on dendritic spines triggers phosphorylation of DSCR1, thus DSCR1 no longer functions as an inhibitor of calcineurin. Subsequently, active calcineurin dephosphorylates phospho-FMRP, and in turn FMRP dissociates from target mRNAs thus allowing local mRNA translation to occur. To test this model, we first determined if BDNF treatment of hippocampal neurons alters FMRP phosphorylation. After BDNF treatment, the intensity of phospho-FMRP reduced significantly while the level of DSCR1 and the amount of FMRP remain mostly unaltered, suggesting that phospho-FMRP is dephosphorylated following BDNF treatment (Figure 6A and B; Supplementary Figure S11A). Next, we tested if BDNF treatment of hippocampal neurons elevates calcineurin activity. As shown in Figure 6C, calcineurin activity is indeed increased in BDNF-treated hippocampal neurons. These results suggest that calcineurin may act on phospho-FMRP after BDNF stimulation. To verify if calcineurin can dephosphorylate phospho-FMRP, we immunoprecipitated phospho-FMRP from HEK cells with phospho-FMRP-specific antibody, then added purified calcineurin to the immunoprecipitates. Calcineurin indeed dephosphorylated phospho-FMRP (Figure 6D). In addition, we examined the level of phospho-FMRP in control and DSCR1−/− transgenic mice. We found that the level of pFMRP is decreased in DSCR1−/− mice (Supplementary Figure S11B), suggesting that the ability of DSCR1 to regulate calcineurin leads to modulation of pFMRP. We also treated primary hippocampal neurons with BDNF in the presence of calcineurin inhibitor, cyclosporin A, and measured the level of phospho-FMRP. We found that in the presence of calcineurin inhibitor, BDNF no longer triggered dephosphorylation of pFMRP, confirming that BDNF induced dephosphorylation of phospho-FMRP acts through calcineurin.
Figure 6.
BDNF activates calcineurin to dephosphorylate phospho-FMRP. (A) BDNF stimulation decreased the level of phospho-FMRP in hippocampal dendrites. (B) Phospho-FMRP level is significantly reduced in hippocampal primary neurons treated with BDNF. (C) Calcineurin activity is increased in primary hippocampal neurons after BDNF treatment, while adding 5 μM of cyclosporin A (CsA) inhibits calcineurin. Note that basal calcineurin activity in untreated neurons is low. (D) Phospho-FMRP was brought down by immunoprecipitation with phospho-FMRP antibody, and then calcineurin was added to the immunoprecipitates. (E, F) Primary neurons from wild type were treated with BDNF in the presence of CsA and levels of phospho-FMRP were measured by immunoimaging (E) and western blotting (F). There is no difference in phospho-FMRP level after BDNF treatment, if CsA is present. Three independent experiments performed per condition and values represent mean±s.e.m., *P<0.001. Student’s t-test was used. Scale bar, 10 μm.
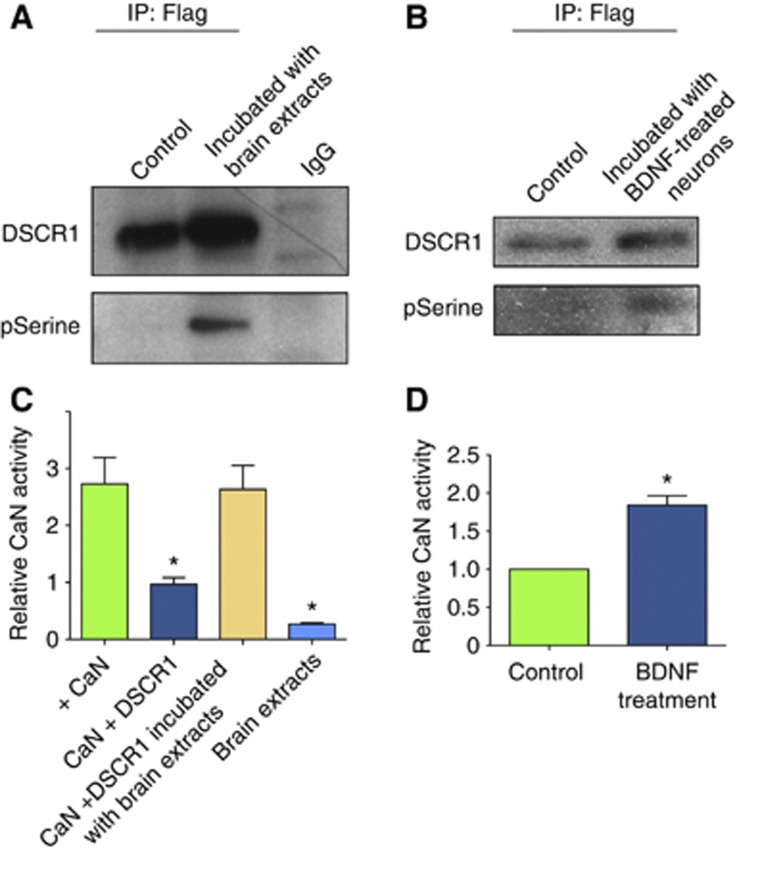
According to our hypothesis, BDNF treatment triggers phosphorylation of DSCR1, which may activate calcineurin and lead to increased dephosphorylation of phospho-FMRP. To test this model, we determined if DSCR1 could be phosphorylated by endogenous kinase(s) found in the mouse brain. This was achieved by incubating immunoprecipitated Flag-DSCR1 from HEK cells with extracts of mouse brain or hippocampal neurons treated with or without BDNF. The immunoprecipitated Flag-DSCR1 from HEK cells is not phosphorylated, however, Figure 7A and B showed that Flag-DSCR1 becomes phosphorylated after incubation with brain extracts or BDNF-treated hippocampal neuronal extracts, as revealed by western blotting using phosphorylated serine-specific antibody. We then determined whether phosphorylated DSCR1 would cause active calcineurin. Calcineurin activity assays revealed that Flag-DSCR1 inhibits calcineurin activity, but Flag-DSCR1 treated with brain extracts before the assay showed higher calcineurin activity (Figure 7C). Also, Flag-DSCR1 incubated with extracts of hippocampal primary neurons stimulated with BDNF showed more calcineurin activity compared to that of neurons without BDNF treatment (Figure 7D). Taken together, these data show that BDNF stimulation increases DSCR1 phosphorylation, which results in active calcineurin, thus leading to increased dephosphorylation of phospho-FMRP.
Figure 7.
Phosphorylated DSCR1 activates calcineurin. Flag-DSCR1 incubated with brain extracts (A) or neurons treated with BDNF (B) are phosphorylated on serine residues. (C) Flag-DSCR1 inhibits calcineurin activity. However, Flag-DSCR1 incubated with brain extracts prior to calcineurin activity measurement does not inhibit calcineurin, while brain extracts do show minimum calcineurin activity. (D) Flag-DSCR1 was incubated with extract of primary hippocampal neurons treated with or without BDNF before determination of calcineurin activity. (C, D) n=4–6 independent experiments and values represent mean±s.e.m., *P<0.001. Student’s t-test was used.
Discussion
Intellectual disability is a developmental brain disorder that is characterized by significantly impaired cognitive performance, which can be caused by various aetiological factors including prenatal fetus exposure to toxic materials, intrauterine virus infection, maternal malnutrition, premature birth, and other unknown reasons. However, it is also known that intellectual disability arises from genetic factors, and two of the most common genetic causes of intellectual disability are Fragile X syndrome and Down syndrome. Recent evidence indicate that spine dysmorphology and abnormal local protein synthesis are connected to intellectual disability. In this study, we find that DSCR1 is an important regulator of spine morphogenesis and local protein synthesis. DSCR1 interacts with phospho-FMRP and modulates BDNF induced local mRNA translation in the dendritic spines. This work for the first time demonstrates that proteins altered in Fragile X syndrome and Down syndrome share common signalling pathways.
Roles of DSCR1 in spine morphogenesis
During neuronal development, dendritic spines become stabilized once in contact with axon terminals, which require actin polymerization and organization. Modulation of actin dynamics by regulation of cofilin phosphorylation is important for determining and maintaining dendritic spine morphology, since cofilin can sever actin filaments to cause spine shrinkage while phospho-cofilin polymerizes actin filaments to promote spine growth (Cingolani and Goda, 2008). Hence, kinases and phosphatases that can alter the phosphorylation status of cofilin will play a key role in spine head formation (Calabrese et al, 2006). Calcineurin is known to dephosphorylate phospho-cofilin (Zhou et al, 2004; Cingolani and Goda, 2008), and we found that DSCR1, an inhibitor of calcineurin, functions in spine morphogenesis (Figure 1). Reduction of DSCR1 leads to more cofilin, lower number of dendritic spines, as well as smaller spine head size. In contrast, overexpression of DSCR1 increases the level of phospho-cofilin as well as increases the size of spine heads (Figures 1, 2, 3; Supplementary Figure S7). Together, our results indicate that DSCR1 inhibits calcineurin to modulate the level of phospho-cofilin, which in turn affect spine morphology. Another phosphatase known to control dephosphorylation of phospho-cofilin is the catalytic subunit of protein phosphatase 2A (PP2AC). It is reported that FMRP binds to the 5′UTR of pp2ac mRNA to negatively regulate translation of pp2ac mRNA (Castets et al, 2005), suggesting that an abundance of the PP2AC protein due to lack of FMRP may contribute to the abnormal dendritic spine morphology seen in Fragile X syndrome or fmr1 knockout mouse. We found that the enlarged spine head phenotype caused by DSCR1 overexpression or the elongated spine phenotype due to FMRP reduction is restored to normal by reducing FMRP level in DSCR1 overexpression background (Figures 1 and 2). This indicates that DSCR1 and FMRP act in the same spine morphogenesis pathway by regulating the level of phospho-cofilin to modulate actin dynamics at dendritic spines during neuronal development (Supplementary Figure S12A). It is interesting to note that two Down syndrome mouse models, Ts65Dn and Ts1Cje, show significantly enlarged dendritic spines in the hippocampus (Belichenko et al, 2004), while a third Down syndrome mouse model, Ts1Rhr, shows abnormal spines but to a lesser degree (Belichenko et al, 2009). Importantly, DSCR1 is triplicated in both Ts65Dn and Ts1Cje, but not in Ts1Rhr. Hence, it is plausible that DSCR1 is a candidate gene that contributes to enlarged spines as seen in the DS mouse models. Furthermore, postmortem brains of Down syndrome patients show various dendritic pathologies including enlarged but less abundant dendritic spines, as seen in Ts65Dn (Fiala et al, 2002; Belichenko et al, 2004). This suggests that DSCR1 may also play an important role in spine morphogenesis in human Down syndrome.
Roles of DSCR1 in local protein synthesis
Local protein synthesis at dendritic spines upon synaptic stimulation is required for long-term synaptic plasticity and memory. Studies have shown that mRNAs and translational machinery are present at dendritic spines and protein synthesis occurs when neurons are activated; however, mechanisms regulating this process are not well understood. It has also been challenging to prove that mRNAs are translated into new protein locally, and not due to transport from the cell body. In this study, we used in-vivo imaging assay that enables us to evaluate local protein synthesis at dendritic spines of live neurons. We constructed photoswitchable dendra2 protein with 5′UTR and 3′UTR of αCaMKII containing dendritic targeting element and monitored local protein synthesis at dendritic spines. We showed that mRNA transcripts of 5′UTR-dendra2-3′UTRαCaMKII are indeed translated at dendritic spines and not transported from cell soma when BDNF stimulation is applied to neurons. Using this assay, we demonstrate that reducing DSCR1 level prevented local mRNA translation upon BDNF stimulation (Figure 5; Supplementary Figures S9 and S10). Consistent with this, DSCR1 knockout mouse shows impaired L-LTP that requires local protein synthesis and deficits in learning and memory (Hoeffer et al, 2007). In contrast, we show that DSCR1 overexpression significantly increases local protein synthesis at dendritic spines. A recent report shows that DSCR1 transgenic mouse exhibits spatial learning defects similar to that of DSCR1 knockout mouse (Dierssen et al, 2011). We previously showed that overexpression or reduction of nebula, Drosophila homologue of DSCR1, generates disruption in learning and memory process in Drosophila (Chang et al, 2003). It is also known that both overexpression and deletion of αCaMKII show impaired spatial learning and LTP, suggesting that optimum amount of αCaMKII is necessary for proper learning and memory (Mayford et al, 1996; Silva, 2003). Adjusting threshold for synaptic plasticity is suggested as an important mechanism for learning and memory, and it is plausible that an imbalance in local protein synthesis at dendritic spines due to alteration of DSCR1 level may change homeostatic synaptic plasticity, resulting in deficits in learning and memory. Although our results are consistent with local protein synthesis in the spine, further confirmation with a diffusion restricted reporter would be desirable.
In our study, neurons from fmr1 knockout mouse or containing fmr1 siRNA did not exhibit increased protein synthesis at dendritic spines, although FMRP is known to suppress mRNA translation and basal protein synthesis has been shown to be increased in hippocampal slices of fmr1 knockout mouse (Osterweil et al, 2010). Consistent with our findings, however, a recent report shows that the level of αCaMKII at dendritic spines in cultured hippocampal neurons of wild-type mouse is significantly increased after DHPG treatment, while no elevation in the level of αCaMKII is found in fmr1 knockout mouse (Kao et al, 2010). The amount of αCaMKII mRNAs is not increased at dendritic spines of fmr1 knockout mouse after DHPG stimulation, because FMRP also delivers αCaMKII mRNAs to dendritic spines after stimulation (Kao et al, 2010). Hence, it is likely that BDNF stimulation has no measurable effect on translation of 5′UTR-dendra2-3′UTRαCaMKII transcripts due to insufficient amount of 5′UTR-dendra2-3′UTRαCaMKII transcripts in the spines of hippocampal neurons containing fmr1 siRNA. It is interesting to note that DSCR1 transgene/fmr1 siRNA combination failed to show local protein synthesis after 20 or 30 min of BDNF treatment; however, it showed local translation after 10 min of BDNF activation to the same extent as DSCR1 overexpression (Supplementary Figure S9). It suggests that DSCR1 overexpression could increase local protein synthesis even with low amount of 5′UTR-dendra2-3′UTRαCaMKII transcripts in the first 10 min of BDNF stimulation, but it cannot sustain local protein synthesis for the next 10–20 min during BDNF treatment because mRNA templates are depleted.
In resting neurons, DSCR1 prevents calcineurin from dephosphorylating phospho-FMRP, thus allowing phospho-FMRP to bind to and suppress translation of target mRNAs at dendritic spines. However, when neurons are stimulated by BDNF, DSCR1 becomes phosphorylated by currently unidentified kinase(s) that are downstream of the tyrosine receptor kinases B pathway. Phosphorylation of DSCR1 does not inhibit but rather stimulates or no longer inhibits calcineurin, thus allowing calcineurin to subsequently dephosphorylate phospho-FMRP. This leads to release of FMRP from target mRNAs, resulting in new local protein synthesis at dendritic spines. Activation of local protein synthesis may also trigger expression of proteins that influence local actin dynamics, thus leading to modulation of spine morphologies. We also expect that DSCR1 will become dephosphorylated, likely by calcineurin as suggested by Hilioti et al (2004), thus resetting the pathway. Spine morphology in mature spines will thus likely be regulated by a delicate balance of cofilin phosphorylation status and downstream effectors of FMRP translation pathway such as αCaMKII, Arc/Arg3.1, Map1B, and APP, both of which are modulated by DSCR1 (Supplementary Figure S12B). It is interesting to note that Arc synthesis in the dendritic spines is necessary for stabilization of polymerized actin and consolidation of LTP, suggesting that local protein translation and changes in spine morphology are linked during LTP (Bramham, 2008; Hotulainen and Hoogenraad, 2010).
In conclusion, we find that DSCR1 is a novel regulator of both dendritic spine morphogenesis and local protein synthesis. Our results suggest that DSCR1, calcineurin, and FMRP together act to regulate spine morphogenesis during neuronal development and local protein synthesis during neuronal stimulation. During early spine morphogenesis, DSCR1 and FMRP regulate the level of phospho-cofilin that controls spine morphology through actin dynamics. However, DSCR1 regulates local protein synthesis by directly binding to phospho-FMRP at mature dendritic spines. It is tantalizing to speculate that altered expression of DSCR1 may trigger dendritic spine dysmorphology and altered synaptic activity, thus leading to intellectual disability seen in Down syndrome. It will be interesting to test in the future if reduction of DSCR1 or fmr1 can alleviate the phenotypes seen in Down syndrome mouse model, or if overexpression of DSCR1 can ameliorate the phenotypes observed in Fragile X syndrome mouse model. We believe this work provides an important step towards understanding the multiple roles of DSCR1 in neurons, and will help to shed light on mechanisms underlying intellectual disability seen in Down syndrome and Fragile X syndrome.
Materials and methods
Cell culture
Hippocampi were dissected from E18 mouse embryos, and then treated with trypsin. Dissociated neurons were seeded in 24-well plate containing 12 mm glass coverslips or 6-well plate without glass coverslips both coated with poly-D-lysine (50 μg/ml). Neurons were plated at 15 000–20 000/cm2 for immunoimaging, and at 35 000–40 000/cm2 for transfection. Neurons were co-cultured with glial cell in neurobasal medium containing B27 supplement up to DIV 24. Neuronal cultures at DIV5 were transfected using Lipofectamine™ 2000 (Invitrogen) following manufacturer’s protocol. HEK293 cells were grown on 6-well plates and transfected using Lipofectamine™ 2000. The cells were harvested 24 h after transfection for biochemical analyses. BDNF (Alomone Labs) was added to hippocampal primary neurons at 30 ng/ml for 30 min. Cells were washed with PBS and then either fixed for immunostaining or used for biochemical analyses.
Animals
Animals were used in accordance with protocols approved by the Animal Care and Use Committees of Indiana University, Bloomington. DSCR1−/− and DSCR1 transgenic mice were obtained from Dr S Ryeom at University of Pennsylvania. C57BL/6 mouse strain and fmr1−/− were purchased from Jackson Labs. All these mice used in this paper are females and have the C57BL/6 strain background.
Immunocytochemistry
Hippocampal primary neurons (DIV 21) were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature (RT), then permeabilized with 0.2% Triton X-100 in PBS for 10 min, and blocked for 1 h at RT with PBS containing 1% BSA (Sigma-Aldrich). Neurons were incubated with primary antibodies overnight at 4°C. After several washes, Alexa Fluor-conjugated secondary antibodies were applied for 1 h at RT. Images were taken using Leica SP5 confocal microscope (Leica Microsystems Ltd). We used several antibodies to detect proteins in the neuronal cell: DSCR1 polyclonal antibody (Abgent), DSCR1 mouse monoclonal antibody (Abnova), FMRP (Millipore), phospho-FMRP, cofilin and phospho-cofilin (Abcam).
Analysis of dendritic spines
For image processing and analysis of spine morphology, length, and quantification of proteins, we used NIH ImageJ software. Fifteen Z-stack fluorescent images with 0.25 μm thickness were acquired with Leica SP5 confocal microscope, and 3D reconstruction was prepared using Imaris 7.0 software (Bitplane). Density and head size of dendritic spines in the primary hippocampal cultures or CA1 neurons in mouse was based on 2D images. Using ImageJ each dendritic spine head was outlined, then the measured area of each spine head was analysed and the number of spines was also determined. Spine density in CA1 hippocampal region of mouse brain was measured from three different distances from cell body: 0–50 μm, 50–100 μm and 100–150 μm.
Plasmids constructions
Flag-DSCR1 and EGFP-DSCR1 were prepared by cloning the mouse DSCR1-1L to the pFlag-CMV-6c vector and to pEGFP-C2, respectively. We constructed the DSCR1 shRNA vector with the target sequence of 5′-TCC ATG TAT GTG AGA GTG ATC-3′ in DSCR1 by using Block-iT™ pcDNA6.2-GW/EmGFP-miR vector system (Invitrogen). Dendra2-3′UTR of CaMKII was constructed by subcloning the 3′UTR of CaMKII (a gift from Dr Kosik) to pDendra2-C1. Fmr1 siRNA was obtained from Santa Cruz.
Site-directed mutagenesis
A mutant of FMRP was produced using QuickChange site-directed mutagenesis kit (Stratagene). FMRP S499A was produced using the primer 5′-gcatcaaatgctgctgaaacagaatctgacc-3′ and its reverse compliment 5′-ggtcagattctgtttcagcagcatttgatgc-3′.
Golgi staining
We used FD Rapid Golgi staining kit (FD Neuro Technologies). Freshly dissected P21 brains were immersed in solutions A and B for a week at RT, then transferred to solution C for 24 h. The brains were blocked and embedded in TFM tissue freezing medium (TBS) and sectioned using Leica CM1850 cryostat with thickness of 100 μm. Staining of the sections was performed according to the kit. Bright field microscope (Nikon E800) was used to take pictures of pyramidal neurons of CA1 and layer V region. ImageJ was applied to construct Z-stacks with a thickness of 1 μm, and 0.2 μm overlap between each Z-stack. The final image was composed of an average of 30 focused Z-stacks.
Immunoprecipitation
HEK cells containing Flag-DSCR1 were lysated in fresh lysis buffer, containing 25 mM Tris, pH 8.0, 100 mM NaCl, 1 mM MgCl2 and 1 mM CaCl2. The lysate was then incubated with either phospho-FMRP- or FMRP-specific antibodies at 4°C overnight. Then, phospho-FMRP or FMRP was pull down by A/G agarose beads and analysed by western blotting with the DSCR1 antibody.
Supplementary Material
Acknowledgments
We thank Dr S Ryeom for DSCR1 knockout and transgenic mice, Dr K Kosik for CaMKII 3′UTR, F Kennedy for assistance with preparation of hippocampal cell cultures, and the light microscopy imaging center at IUB. This work was supported by Indiana University, Ulsan National Institute of Science and Technology, Foundation Jerôme Lejeune, and KTC is supported by the R00 Grant NS052524 from the National Institutes of Health.
Author contributions: WW and JZZ performed experiments. WW, JZZ, KTC, and K-TM analysed the data. KTC and K-TM designed the experiments and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM (2001) Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30: 489–502 [DOI] [PubMed] [Google Scholar]
- Arron J, Winslow MM, Polleri A, Chang C, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, Yamasaki N, Miyakawa T, Francke U, Graef IA, Crabtree GR (2006) NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441: 595–600 [DOI] [PubMed] [Google Scholar]
- Bagni C, Greenough WT (2005) From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci 6: 376–387 [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST (2008) Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 60: 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belichenko NP, Belichenko PV, Kleschevnikov AM, Salehi A, Reeves RH, Mobley WC (2009) The ‘Down Syndrome Critical Region’ is sufficient in the mouse model to confer behavioral, neurophysiological, and synaptic phenotypes characteristic of down syndrome. J Neurosci 29: 5938–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belichenko PV, Masliah E, Kleschevnikov AM, Villar AJ, Epstein CJ, Salehi A, Mobley WC (2004) Synaptic structural abnormalities in the Ts65Dn mouse model of Down Syndrome. J Comp Neurol 480: 281–298 [DOI] [PubMed] [Google Scholar]
- Bramham CR (2008) Local protein synthesis, actin dynamics, and LTP consolidation. Curr Opin Neurobiol 18: 524–531 [DOI] [PubMed] [Google Scholar]
- Calabrese B, Wilson MS, Halpain S (2006) Development and regulation of dendritic spine synapses. Physiology (Bethesda, Md) 21: 38–47 [DOI] [PubMed] [Google Scholar]
- Casas C, Martínez S, Pritchard MA, Fuentes JJ, Nadal M, Guimerà J, Arbonés M, Flórez J, Soriano E, Estivill X, Alcántara S (2001) Dscr1, a novel endogenous inhibitor of calcineurin signaling, is expressed in the primitive ventricle of the heart and during neurogenesis. Mech Dev 101: 289–292 [DOI] [PubMed] [Google Scholar]
- Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel J-L, Bardoni B (2005) FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum Mol Genet 14: 835–844 [DOI] [PubMed] [Google Scholar]
- Ceman S, O’Donnell WT, Reed M, Patton S, Pohl J, Warren ST (2003) Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet 12: 3295–3305 [DOI] [PubMed] [Google Scholar]
- Chang KT, Shi Y-J, Min K-T (2003) The Drosophila homolog of Down’s syndrome critical region 1 gene regulates learning: implications for mental retardation. Proc Natl Acad Sci USA 100: 15794–15799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y (2008) Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci 9: 344–356 [DOI] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT (1997) Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci USA 94: 5401–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N (2009) Translational control of long-lasting synaptic plasticity and memory. Neuron 61: 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David Gerecht PS, Taylor MA, Port JD (2010) Intracellular localization and interaction of mRNA binding proteins as detected by FRET. BMC Cell Biol 11: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierssen M, Arqué G, Mcdonald J, Andreu N, Martínez-Cué C, Flórez J, Fillat C (2011) Behavioral characterization of a mouse model overexpressing DSCR1/RCAN1. PLoS ONE 6: e17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierssen M, Ramakers GJA (2006) Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes Brain Behav 5(Suppl 2): 48–60 [DOI] [PubMed] [Google Scholar]
- Ebrahimi S, Fraval H, Murray M, Saint R, Gregory SL (2010) Polo kinase interacts with RacGAP50C and is required to localize the cytokinesis initiation complex. J Biol Chem 285: 28667–28673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harris KM (2002) Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res Rev 1: 29–54 [DOI] [PubMed] [Google Scholar]
- Fuentes JJ, Genescà L, Kingsbury TJ, Cunningham KW, Pérez-Riba M, Estivill X, de la Luna S (2000) DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum Mol Genet 9: 1681–1690 [DOI] [PubMed] [Google Scholar]
- Fuentes JJ, Pritchard MA, Planas AM, Bosch A, Ferrer I, Estivill X (1995) A new human gene from the Down syndrome critical region encodes a proline-rich protein highly expressed in fetal brain and heart. Hum Mol Genet 4: 1935–1944 [DOI] [PubMed] [Google Scholar]
- Garber KB, Visootsak J, Warren ST (2008) Fragile X syndrome. Eur J Hum Genet 16: 666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach J, Fox DS, Cutler NS, Cox GM, Perfect JR, Heitman J (2000) Identification and characterization of a highly conserved calcineurin binding protein, CBP1/calcipressin, in Cryptococcus neoformans. EMBO J 19: 3618–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA (2006) Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 24: 461–465 [DOI] [PubMed] [Google Scholar]
- Hilioti Z, Gallagher DA, Low-Nam ST, Ramaswamy P, Gajer P, Kingsbury TJ, Birchwood CJ, Levchenko A, Cunningham KW (2004) GSK-3 kinases enhance calcineurin signaling by phosphorylation of RCNs. Genes Dev 18: 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Dey A, Sachan N, Wong H, Patterson RJ, Shelton JM, Richardson JA, Klann E, Rothermel BA (2007) The Down syndrome critical region protein RCAN1 regulates long-term potentiation and memory via inhibition of phosphatase signaling. J Neurosci 27: 13161–13172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC (2010) Actin in dendritic spines: connecting dynamics to function. J Cell Biol 189: 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao D-I, Aldridge GM, Weiler IJ, Greenough WT (2010) Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Natl Acad Sci USA 107: 15601–15606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury TJ, Cunningham KW (2000) A conserved family of calcineurin regulators. Genes Dev 14: 1595–1604 [PMC free article] [PubMed] [Google Scholar]
- König P, Krasteva G, Tag C, König IR, Arens C, Kummer W (2006) FRET–CLSM and double-labeling indirect immunofluorescence to detect close association of proteins in tissue sections. Lab Invest 86: 1–12 [DOI] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER (1996) Control of memory formation through regulated expression of a CaMKII transgene. Science 274: 1678–1683 [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ (2011) Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell 42: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ (2012) Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci 32: 2582–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C (2008) The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134: 1042–1054 [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST (2007) FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci 27: 14349–14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niere F, Wilkerson JR, Huber KM (2012) Evidence for a fragile x mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered arc translation and long-term depression. J Neurosci 32: 5924–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterweil EK, Krueger DD, Reinhold K, Bear MF (2010) Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of Fragile X syndrome. J Neurosci 30: 15616–15627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Mulle JG, Warren ST (2007) The pathophysiology of Fragile X syndrome. Annu Rev Genomics Hum Genet 8: 109–129 [DOI] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS (2000) CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci 20: 6385–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ (2003) Molecular and cellular cognitive studies of the role of synaptic plasticity in memory. J Neurobiol 54: 224–237 [DOI] [PubMed] [Google Scholar]
- Smith WB, Aakalu G, Schuman EM (2001) Local protein synthesis in neurons. Curr Biol 11: R901–R903 [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM (2006) Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127: 49–58 [DOI] [PubMed] [Google Scholar]
- Waung MW, Huber KM (2009) Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr Opin Neurobiol 19: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo MM (2004) Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron 44: 749–757 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.