Abstract
The eukaryotic genome is replicated according to a specific spatio-temporal programme. However, little is known about both its molecular control and biological significance. Here, we identify mouse Rif1 as a key player in the regulation of DNA replication timing. We show that Rif1 deficiency in primary cells results in an unprecedented global alteration of the temporal order of replication. This effect takes place already in the first S-phase after Rif1 deletion and is neither accompanied by alterations in the transcriptional landscape nor by major changes in the biochemical identity of constitutive heterochromatin. In addition, Rif1 deficiency leads to both defective G1/S transition and chromatin re-organization after DNA replication. Together, these data offer a novel insight into the global regulation and biological significance of the replication-timing programme in mammalian cells.
Keywords: chromatin organization, DNA replication, genome stability, replication timing, Rif1
Introduction
Replication of the mammalian genome is organized in both space and time. Large segments of the genome, called replication domains, are coordinately replicated through the nearly synchronous firing of clusters of replication origins. Replication domains are visualized as foci and their position in the nucleus as well as temporal order of activation are inherited throughout cell cycles (Jackson and Pombo, 1998; Ma et al, 1998; Dimitrova and Gilbert, 1999b; Sadoni et al, 2004). The molecular and genetic mechanisms underlying such highly orchestrated coordination and inheritance processes are still largely unknown. Transcriptional activity, epigenetic marks and tri-dimensional (3D) chromatin organization have all been proposed to play a role in defining the identity of replication domains and their order of activation (Gilbert and Gasser, 2006; Gondor and Ohlsson, 2009; Hiratani et al, 2009). Yet, the relationships between these inter-dependent processes are poorly understood. A particularly intriguing relationship is between replication timing and transcription, because of the strong correlation between early replication and active gene expression. A long-standing question has been whether transcriptional activity and replication timing are inter-dependently regulated. In some cases, changes in transcriptional activity, for example, induced by developmental transitions, have been shown to correlate with changes in replication timing (Hiratani et al, 2008) but recent genome-wide techniques have revealed that their relationship is not direct (Hiratani et al, 2009). Moreover, interfering with the activity of chromatin modifying enzymes can affect replication timing, but the effect is generally of modest extent and local (Aparicio et al, 2004; Li et al, 2005; Wu et al, 2006; Jorgensen et al, 2007; Goren et al, 2008; Yokochi et al, 2009). To date, no global determinants of the spatio-temporal organization of mammalian DNA replication have been identified, since all the mutations analysed influence either few loci or repetitive sequences such as rDNA or pericentric regions. The most dramatic rearrangements of replication timing thus far reported take place during embryonic stem cells (ESCs) differentiation (Hiratani et al, 2008).
The strongest correlation of chromatin properties to replication timing so far observed is to 3D chromatin organization. Indeed, both chromatin interactions and subnuclear position of different replication domains display a high degree of correspondence with their timing of replication (Gilbert et al, 2010; Ryba et al, 2010; Yaffe et al, 2010). After metaphase, chromatin is highly mobile in the nucleus. During G1, upon re-assembly of the nuclear envelope and lamina, chromatin is anchored and assumes defined positions. This moment coincides with the identification and segregation of the replication domains whose origins fire at different times and is therefore called ‘timing decision point’ (TDP) (Dimitrova and Gilbert, 1999a; Leonhardt et al, 2000; Chubb et al, 2002). One plausible theory about the mechanism of timing control proposes the existence of a protein or protein complex that is rate limiting for origin firing as recently identified in budding yeast (Mantiero et al, 2011). In this scheme, at the beginning of S-phase, origins with the highest probability to fire would be the ones with the highest affinity/accessibility for the limiting factor(s). Only upon release of the limiting factor(s) from the early domains, origins with lower affinity could then fire (Rhind, 2006). In line with the idea of regulated access to limiting initiation factors being a key mechanism of replication-timing regulation, it was recently shown that the clustering of origins in subnuclear domains mediated by Fkh1 and 2 is indeed essential for the origin-firing programme in budding yeast (Knott et al, 2012). The anchorage of chromosome domains to the nuclear periphery during G1 could be a means to spatially segregate different genomic compartments either with different protein content and/or to spatially define the accessibility of a limiting factor required for origin firing to a given replication domain. Thus, by regulating interaction with the limiting factor(s), chromatin anchoring could control the temporal order of origin activation.
The biological significance of spatial compartmentalization of DNA replication also remains unclear. It may be that this organization reflects the necessity of re-assembling different types of chromatin structures in the wake of the replication fork, creating a temporal and local concentration of specific factors for each chromatin type (Gilbert, 2001). For example, temporal segregation of replication of pericentric heterochromatin (pHC) to mid S-phase could serve the purpose of guaranteeing the availability/concentration of specialized chromatin remodelers and histone chaperones, especially required for assisting fork progression through highly compacted chromatin and for re-establishing the appropriate epigenetic features (Quivy et al, 2008; Loyola et al, 2009; Maison et al, 2010).
In this work, we identify Rif1 as the first key component of the molecular machinery that determines the replication-timing programme in mammalian cells. Rif1 was originally identified in budding yeast as a Rap1 interacting protein (Hardy et al, 1992). As for Rap1, Rif1 deletion causes telomere hyper-elongation, suggesting that Rif1 is a telomerase negative regulator. The mechanism by which Rif1 participates in telomere homoeostasis remains unclear. However, it was recently published that budding yeast Rif1 regulates telomere replication timing by temporally restricting telomerase access (Gallardo et al, 2011; Lian et al, 2011). In fission yeast, Rif1 does not bind Rap1, but still interacts with telomeres and participates in their length regulation (Kanoh and Ishikawa, 2001). In mammalian cells, others and we identified Rif1 as a double-strand break (DSBs) response factor (Silverman et al, 2004; Xu and Blackburn, 2004). In addition, we found that Rif1 had a very important role during fork re-start downstream of ATR (Buonomo et al, 2009; Xu et al, 2010). We therefore attributed the effects of Rif1 deletion on S-phase progression to its activity during fork re-start (Buonomo et al, 2009). Both during DSBs response and upon fork stalling, localization of Rif1 at the sites of DNA damage required 53BP1 (Silverman et al, 2004; Buonomo et al, 2009). However, Rif1 is an essential gene (Buonomo et al, 2009), while 53BP1 knockout mice are viable (Ward et al, 2003), prompting our investigation of non-DNA-damage related, 53BP1-independent functions of Rif1. Our work presented here shows that the most rapid and widespread effect of deleting Rif1 is a genome-wide alteration of the temporal order of origin firing. We also describe the consequences of Rif1 deficiency on chromatin organization and cell-cycle progression. Through studying this key regulator, we can therefore provide an insight into the links between replication-timing control, chromatin organization and genome stability.
Results
Rif1 shows a dynamic S-phase-specific pattern
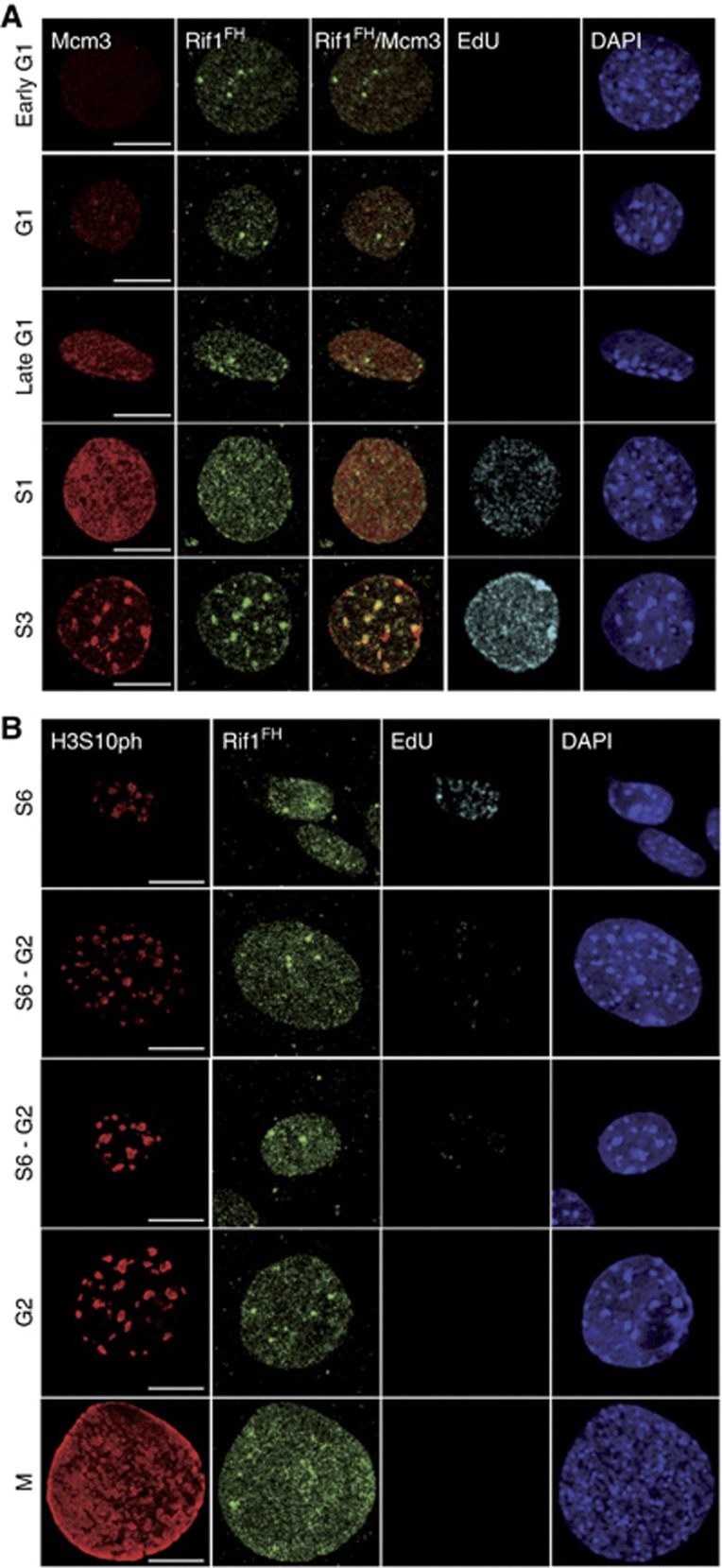
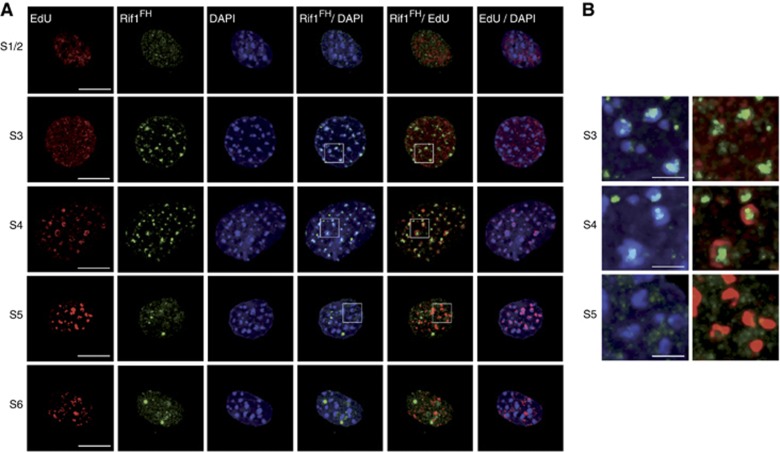
To gain molecular insight into Rif1 function during S-phase, we have generated a knockin mouse replacing the starting Rif1 ATG codon with the coding sequence of the FLAG–HA2 tag, resulting in the Rif1FH allele (Supplementary Figure S1A and B). The functionality of this allele was confirmed by the birth of Mendelian numbers of Rif1FH/FH mice (Supplementary Figure S1C) and their normal health status. Immunofluorescence using an anti-Rif1 antibody in heterozygous Rif1FH/+ mouse embryonic fibroblasts (MEFs) confirmed that the tagged protein fully recapitulated the dynamics of the untagged Rif1 (Supplementary Figure S1D). We therefore employed this endogenously epitope-tagged Rif1 protein to study in detail Rif1 localization during cell cycle by immunofluorescence. We performed our immunofluorescence studies in cells that were pre-extracted with a buffer containing 0.5% Triton X-100. This allowed us to concentrate on the fraction of the protein that is either chromatin-bound or present in the insoluble nuclear fraction. During G1 as well as G2, apart from a variable number of unclassified brighter foci, Rif1 was distributed diffusely through the nucleus (Figure 1A and B). S-phase can be divided in six different stages, each characterized by a specific spatial EdU pattern (Supplementary Figure S2) (Dimitrova and Berezney, 2002; Quivy et al, 2004). S1–3, S4–5 and S6 represent early, mid and late S-phase, respectively. In S1/2, Rif1 was present throughout the nucleus, although its distribution was not homogeneous and it never co-localized with the replication fork (Figure 2A, S1/2) or with the replicative helicase MCM3 (Figure 1A, S1). In S3, the latter part of early S and prior to the appearance of the replication forks within pHC, Rif1 was specifically enriched at chromocenters (Figure 2A), where it co-localized with MCM3 (Figure 1A, S3). As previously described (Quivy et al, 2004), replication of pHC occurs at the outer edges of the chromocenters and once replicated, the DNA moves to the interior of the chromocenter. During the initial steps of pHC replication (S4), Rif1 localization was restricted to the innermost of this structure, which is yet to be replicated, appearing always closely juxtaposed to the EdU but never overlapping with it (Figure 2B). In S5, Rif1 no longer co-localized with the fully replicated chromocenters (Figure 2A and B). In S6, Rif1 signal had again become more diffuse. These data show that Rif1 displays a highly dynamic and specific S-phase behaviour. Interestingly, Rif1 never co-localizes with the replication fork, but on the contrary, clearly precedes it, at least at pHC during mid S-phase.
Figure 1.
Rif1 localization during G1 and G2. (A) Confocal microscope images of cells in different stages of G1 and early S identified by the intensity and distribution of MCM3 signal and EdU pattern. During G1, Rif1FH (anti-HA, green) shows a diffuse nuclear staining (DAPI, blue). It co-localizes with MCM3 (red) only during the latter part of early S (S3). EdU is shown in Cyan. Scale bar: 10 μm. (B) Cells in late S-phase and different G2/M stages were identified by staining for phosphorylated Ser10 histone H3 (H3S10ph, red). Rif1FH (anti-HA, green) returns to its diffuse localization during G2 (DAPI, blue). EdU is shown in Cyan. Scale bar: 10 μm.
Figure 2.
Dynamic Rif1 localization during S-phase. (A) Confocal microscope images of fluorescently stained newly replicated DNA (EdU, red), immunofluorescence for Rif1FH (anti-HA, green) and DAPI (blue) in Rif1FH/FH MEFs. Scale bar: 10 μm. (B) Rif1 precedes replication at chromocenters. Magnifications of insets highlighted from (A). Scale bar: 2 μm.
Rif1 is required for the regulation of replication timing
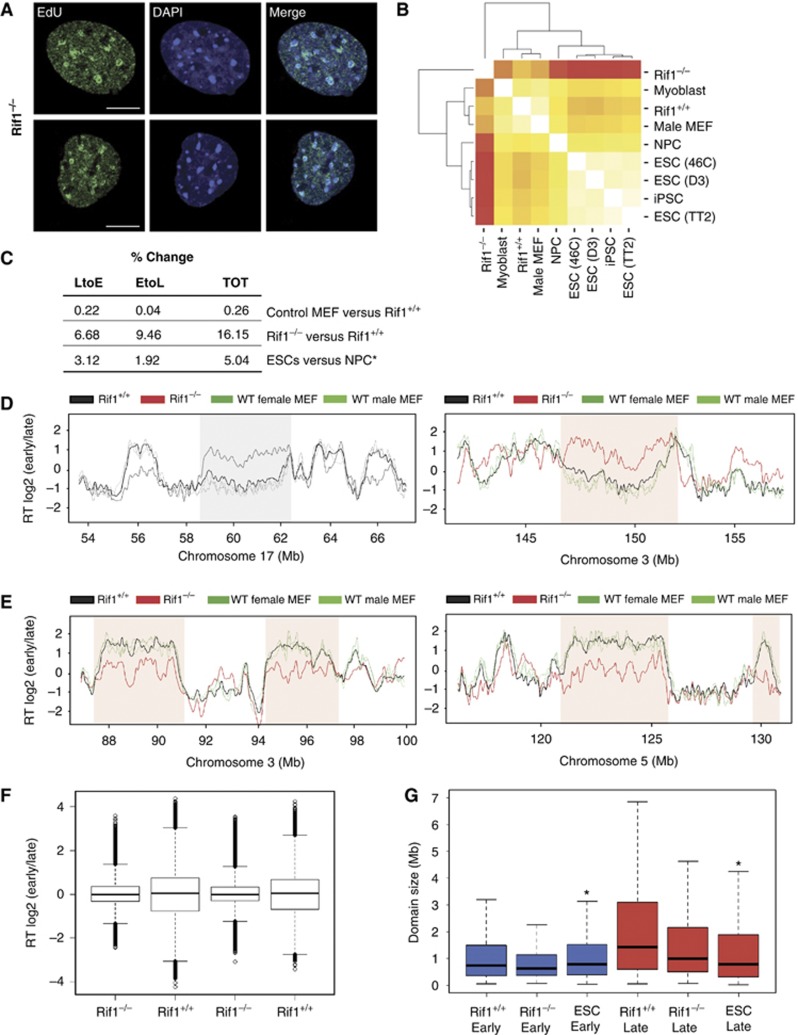
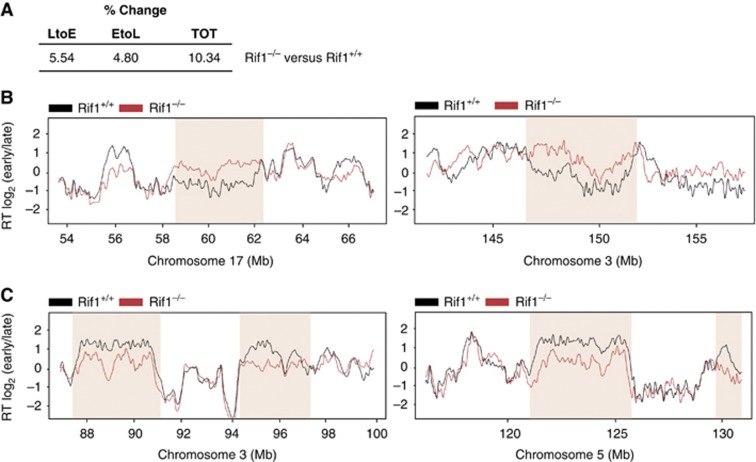
Rif1’s localization ahead of the replication fork, marking domains whose origins have yet to be activated, prompted us to investigate spatio-temporal replication patterns in Rif1 null cells. To this end, we have taken advantage of the Rif1 conditional allele (Rif1F) (Buonomo et al, 2009) to induce acute Rif1 deletion in logarithmically growing early passage Rif1F/F pMEFs (Supplementary Figure S3A). Upon CRE infection of Rif1F/F pMEFs, cells were pulse-labelled with EdU and the six replication patterns typically observed in mouse cells were identified. A low but reproducible percentage of Rif1−/− cells displayed an aberrant EdU pattern that we classified as mixed S2–S4 (Figure 3A). A similar observation was reported in Suv39h1/2 double-null MEFs (Wu et al, 2006), where it was shown that the appearance of this aberrant pattern coincided with advanced replication timing of the major satellites. We next performed genome-wide profiling (Hiratani et al, 2008) of replication timing in Rif1+/+ and Rif1−/− pMEFs. Several cell cycles following CRE-mediated deletion of Rif1, the replication-timing programme was dramatically re-organized (Figure 3B and C), to an extent substantially greater than that seen during ESCs differentiation (Figure 3C) (Hiratani et al, 2008), the most dramatic re-organization observed to date. The correlation between Rif1+/+ and Rif1−/− pMEFs was reduced to an average of 0.63, yet the correlation of replicates between two independent lines was 0.90 (Supplementary Figure S3B and C), demonstrating that the disrupted pattern upon Rif1 loss was reproducible. Rif1 deletion caused both late-to-early (LtoE) and early-to-late (EtoL) switches of replication timing (Figure 3D and E), with 40% more domains switching from EtoL than LtoE timing (Figure 3C). The distribution of replication-timing values in Rif1 null cells clustered near the middle of S-phase (Figure 3F), but due to the nature of our genome-wide analysis, we cannot distinguish whether these regions are replicating in mid-S, replicating randomly throughout S-phase, or whether the two homologues are replicating at specific but asynchronous times.
Figure 3.
Rif1 deficiency deregulates the replication-timing programme. (A) EdU staining (green) in Rif1−/− pMEFs reveals a mixed S2–S4 pattern as compared with wild-type cells (Figure 2, S2). A pan-nuclear EdU signal excluding nucleoli typical of S2 co-exists with the characteristic S4 pattern, identified by the typical EdU rings around the chromocenters. DAPI (blue). Scale bar: 10 μm. (B) Hierarchical clustering and correlation matrix heat map comparing replication-timing data (each an average of two bio-replicates) from pMEF Rif1+/+ and Rif1−/− to different cell types (previously published ESC, neural progenitor cells-NPC, Myoblasts, induced pluripotent Stem cells-iPSC) along with published wild-type MEF and two replicates of pMEF Rif1+/+ and Rif1−/−. (C) Table summarizing the percentages of changes comparing previously published wild-type MEFs with Rif1+/+ pMEFs and the latter with littermate Rif1−/− pMEFs. *As a reference, the % changes during ESCs differentiation into NPCs from Hiratani et al (2008) is shown. The percentage of changes is calculated as the number of probes on the array that change by a factor of more than 1 versus the total number of probes. (D) Exemplary regions (shaded) in chromosome 17 (left) and 3 (right) whose replication-timing switches from LtoE in Rif1−/− (red) compared with Rif1+/+ (black) pMEFs, shown by Loess smoothed replication-timing profiles. Profiles for the same region from male and female published wild-type MEFs are shown in green. (E) Exemplary regions (shaded) in chromosome 3 (left) and 5 (right) whose replication-timing switches from EtoL, depicted as in (D). (F) Distribution of replication-timing values in Rif1−/− versus Rif1+/+ cells. Two independent lines are shown for each genotype. (G) Comparison of early (blue) and late (red) domain size distribution between Rif1+/+, Rif1−/− and ESCs (*ESCs from Hiratani et al, 2008).
Interestingly, one striking effect of Rif1 deficiency on replication-domain structure was their fragmentation in size (Figure 3G). ESCs have the smallest replication domains observed among all cell types that have been analysed thus far, which consolidate into larger domains during differentiation (Hiratani et al, 2008; Ryba et al, 2010). Rif1 deletion in pMEFs induced fragmentation of replication-domain sizes into a total number even higher than in ESCs (1789 domains in ESCs versus 1425 in Rif1+/+ and 1956 in Rif1−/−pMEFs).
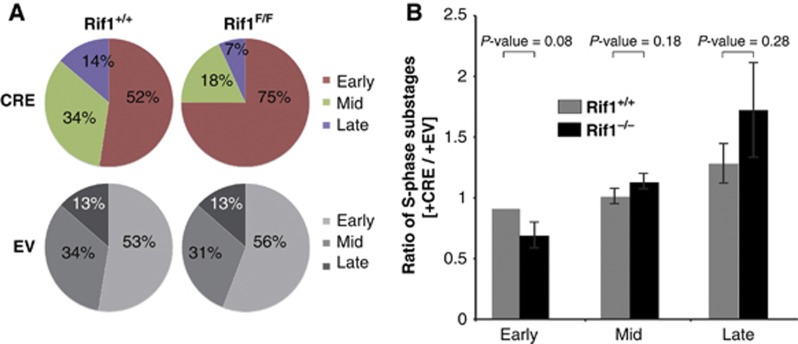
These data show that Rif1 deficiency induces dramatic changes in the temporal replication programme, to an extent not matched by the disruption of 3D spatial replication patterns. However, during the analysis of the spatial organization of DNA replication by EdU staining, we observed that there were about 50% more Rif1 null cells displaying an early S-phase pattern (Figure 4A). This result could be interpreted as an actual accumulation of cells in the early stages of S-phase, reflecting a temporal delay in the early-to-mid S-phase transition. Alternatively, in many cells the pattern that we had identified as early S could result from a loss of spatial organization as well as a fragmentation of the replication domains. The early S-phase pattern is indeed characterized by a diffuse localization and by the presence of numerous small replication foci. To distinguish between these two possibilities, we quantified the distribution of Rif1−/− cells among different S-phase stages by flow cytometry, relying on DNA content, rather than visual inspection (Figure 4B) to assess whether the cells were delayed in early S-phase. We subdivided S-phase into three equal fractions according to DNA content and determined the percentage of total 5′-bromo-2′-deoxyuridine (BrdU)-positive S-phase cells found in each fraction for wild-type versus Rif1−/− pMEFs. This analysis revealed comparable progression through S-phase for cells of both genotypes. We can therefore conclude that the visual accumulation of cells with a diffuse early-S-like EdU pattern in the Rif1−/− cells results from spatially and temporally disorganized replication, potentially due to fragmentation of replication domains.
Figure 4.
Consequences of Rif1 deletion on spatial replication-foci distribution. (A) In the absence of Rif1, cells with early replication spatial patterns accumulate. Upon 30′ EdU pulse, cells were fixed and stained for EdU. S-phase substages were evaluated by visual inspection of the cycling population. Pie charts show the relative proportion (percentage of total S) of early, mid and late S-phase. Cells were scored blinded for two independent Rif1+/+ and four Rif1F/F treated with CRE or EV pMEF clones. In all, 200 EdU+ cells were counted for each clone. Averages are shown. P-values early P<0.0001, mid=P<0.0001, late P<0.01. (B) Six independent Rif1+/+ and Rif1F/F pMEF clones treated with CRE or EV were pulse-labelled with BrdU and the percentage of positive cells was evaluated by FACS analysis. S-phase was subdivided into three equal fractions of increasing propidium iodide content to define early, mid and late S-phase. The ratio of the percentage of total BrdU-positive cells found in each S-phase fraction between EV and CRE treated was plotted.
Surprisingly, we found that Rif1 deletion had no major effect on the cell’s transcriptome (Supplementary Figure S3D), suggesting that the effect of Rif1 deletion on replication timing is neither mediated by nor causing gross changes in the transcriptional landscape. However, our transcriptome analysis has been performed on the population of cycling Rif1−/− cells. Therefore, we cannot formally exclude that a small number of cell-cycle specific genes could be deregulated as a consequence of Rif1 deletion. The effect of Rif1 deficiency on the mammalian replication-timing profile that we report here is the strongest observed to date, indicating that Rif1 plays a key role in the control of the spatio-temporal organization of the replication origin-firing programme.
Rif1 deficiency disrupts repackaging of newly replicated chromatin
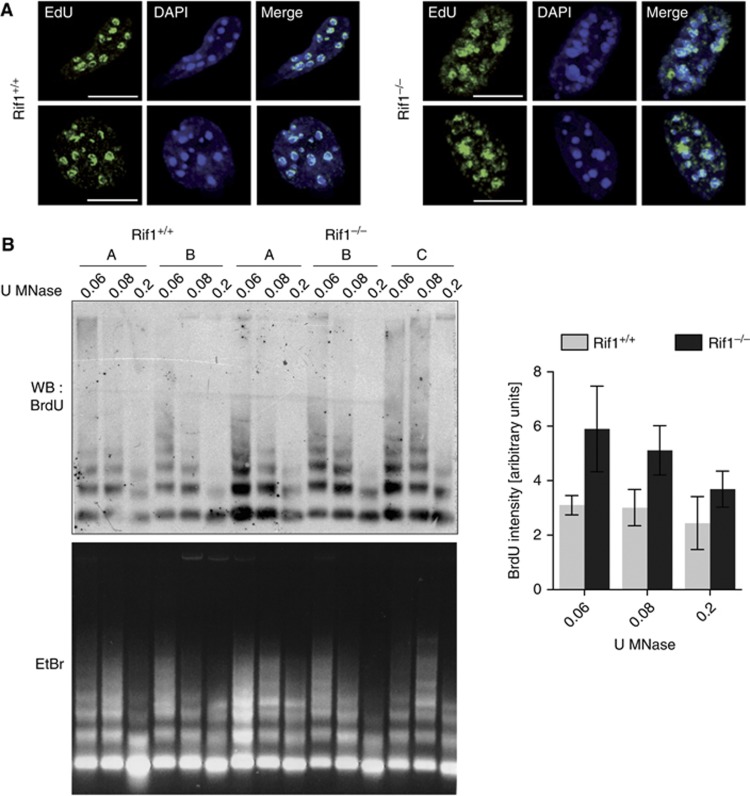
It has been speculated that the spatio-temporal organization of DNA replication could facilitate the repackaging of different types of chromatin after fork passage. Thus, one possible consequence of replication-timing deregulation could be disorganized chromatin assembly (Quivy et al, 2008; Maison et al, 2010). In agreement with this prediction, one of the most striking phenotypes associated with Rif1 deficiency is the presence of what we have defined ‘fluffy’ newly replicated chromocenters. In Rif1 null cells, the EdU-labelled DNA around the chromocenters appeared highly disorganized (Figure 5A) in comparison to wild-type controls, suggesting a defective pHC repackaging/assembly. However, none of the known post-translational histone modifications associated with heterochromatin analysed showed abnormal localization on newly replicated pHC by immunofluorescence, or overall levels by western blotting (Supplementary Figure S4A and B). We also analysed the general organization of newly replicated DNA by examining its accessibility to Micrococcal nuclease (MNase). For the same extent of total DNA digestion (visualized by ethidium bromide staining), a stronger BrdU signal in Rif1−/− pMEFs is detected (Figure 5B; Supplementary Figure S4C), suggesting increased accessibility of newly replicated DNA in Rif1−/− pMEFs. In summary, Rif1 deletion causes both an alteration of the replication-timing programme and the disorganization of chromatin re-assembly after the passage of the replication fork.
Figure 5.
Rif1 deficiency affects re-organization of newly replicated chromatin. (A) Rif1 deletion impairs post-replicative 3D re-organization of pHC. EdU staining (green) of cells in mid S-phase shows disorganization of newly replicated pHC in Rif1−/− cells. Scale bar: 10 μm. (B) BrdU pulse-labelled chromatin from two independent Rif1+/+ and three Rif1F/F; Rosa26CreERT2/+ (Rif1−/−) pMEFs treated with 4-hydroxytamoxifen was digested with different dilutions of MNase. DNA was extracted and used for Southern–Western with an anti-BrdU antibody (upper panel). The lower panel shows ethidium bromide staining of the agarose gel with the total DNA from MNase digested chromatin used for the Southern–Western. All the pMEFs lines showed a BrdU+ population around 4%, as quantified by FACS (not shown). The right panel shows the quantification of the BrdU immunoblot (WB) normalized for the major satellite Southern signal (SB) (Supplementary Figure S4C). U=enzymatic units.
Replication-timing deregulation does not affect inter-origin distances or fork progression
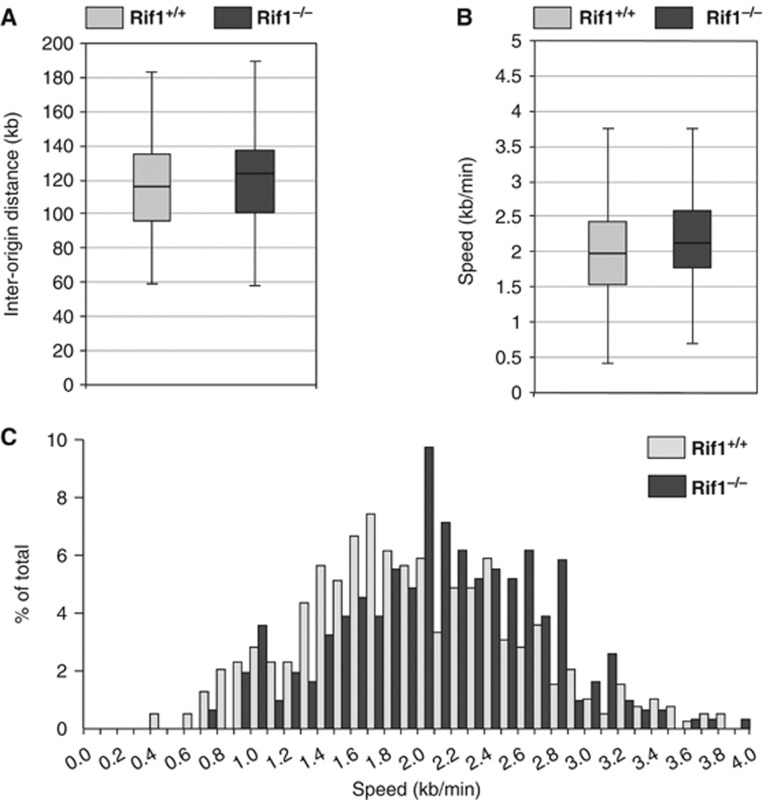
The profound effect of Rif1 deletion on replication timing offered the opportunity to investigate how alteration of this programme is related to DNA replication dynamics, namely the frequency of origin firing and fork progression speed. Upon CRE infection, we labelled Rif1-deficient and control pMEFs with consecutive pulses of IdU followed by CldU to visualize individual replication tracks by DNA Dynamic Molecular Combing (Michalet et al, 1997). Inter-origin distances were not significantly altered (Figure 6A) while replication fork speeds showed a minor increase in Rif1 null cells (Rif1+/+ 1.98 kb/min and Rif1−/− 2.16 kb/min), mostly resulting from a reduction in the abundance of slower replicating forks (Figure 6B and C). Moreover, we could not detect any difference in the number of collapsed or uni-directional forks, nor, by flow cytometry, in the amount of BrdU incorporated per cell (data not shown). In DT40 cells, it has been shown that Rif1 knockout causes an increase in the number of uni-directional replication forks (Xu et al, 2010). However differences both in cell type and system used could account for this discrepancy. Our data indicate that the correct establishment of the replication-timing programme is independent from and has no major impact on fork progression and frequency of origin firing.
Figure 6.
Rif1 deficiency does not affect the frequency of origin-firing or replication fork speed. (A) Boxplot diagram of inter-origin distance measured on individual DNA fibres in Rif1−/− (n=38) and Rif1+/+ cells (n=53). (B) Boxplot diagram of replication fork speed measured on individual DNA fibres in Rif1−/− (n=309) and Rif1+/+(n=392). P-value=3,75 10−5. (A, B) Bottom and top of the box indicate the upper and lower quartiles, respectively, the line in the box the median and the whiskers the 1.5 IQR of the lower and upper quartile. (C) Fork speed distribution in Rif1−/− and Rif1+/+ cells.
Replication-timing deregulation is a primary consequence of Rif1 deficiency
In order to determine whether replication-timing deregulation was upstream or downstream of the chromatin-repackaging defect, we examined replication timing in the first S-phase following Rif1 deletion. Rif1F/F pMEFs were arrested in G0, deletion was induced (Supplementary Figure S4D) and then cells were released synchronously into S-phase. Genome-wide analysis revealed that replication timing was substantially deregulated already in the first S-phase after Rif1 deletion, albeit to a lesser extent than after several cell cycles (correlation between Rif1+/+ and Rif1−/− 0.75, Figure 7A–C). This result is consistent with Rif1 playing a direct role in replication-timing determination. Interestingly, during the first cell cycle after the induction of Rif1 deficiency, the ratio between the percentages of EtoL and LtoE switching regions is nearly equal (Figure 7A), unlike after multiple cell cycles in the absence of Rif1, when a bias towards EtoL emerges (1.4, Figure 3C). Hence deregulation of the replication-timing programme is a primary effect of Rif1 deficiency and affects both early and late replicating domains equally.
Figure 7.
Deregulation of replication timing is an immediate consequence of Rif1 deletion. pMEFs Rif1+/+ and Rif1F/F; Rosa26CreERT2/+ (Rif1−/−) were arrested in G0, 4-hydroxytamoxifen was administered in order to induce CRE-mediated deletion and the cells were released into a synchronous cell cycle. (A) Table summarizing changes in replication timing in the first cell cycle after Rif1 deletion, compared with Rif1+/+, as in Figure 3C (B) Loess smoothed replication-timing profile of the same regions (shaded) in chromosome 17 (left) and 3 (right) shown in Figure 3D whose replication-timing switches from LtoE in Rif1−/− (red) compared with Rif1+/+ (black) pMEFs during the first S-phase (first) upon Rif1 deletion. (C) Replication-timing profile of the same regions (shaded) in chromosome 3 (left) and 5 (right) shown in Figure 3E whose replication-timing switches from EtoL during the first S-phase (first) upon Rif1 deletion, depicted as in (B).
Rif1 deficiency affects entry into the first S-phase upon deletion
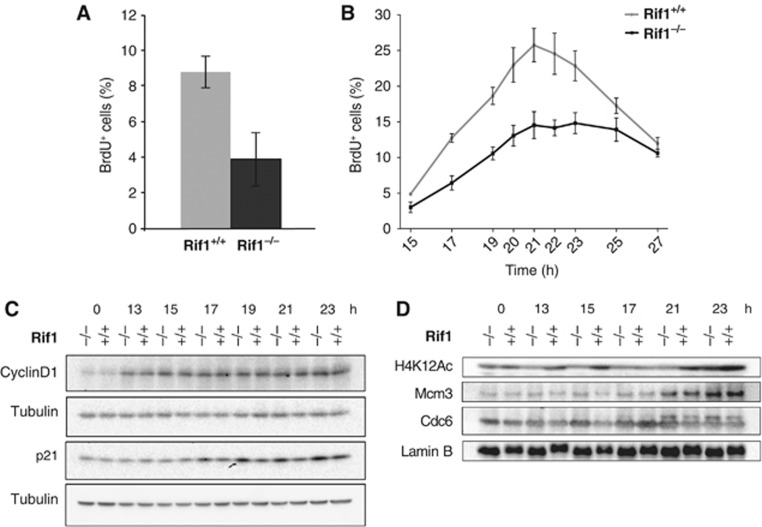
CRE-mediated deletion of Rif1 in logarithmically growing Rif1F/F pMEFs reduced the percentage of cells actively incorporating BrdU by about half (Figure 8A; Supplementary Figure S5A). This could be an indirect consequence of DNA damage progressively accumulated during multiple cell cycles in the absence of Rif1. Alternatively, entry into S-phase could be impeded by a more direct effect of Rif1 loss. In order to distinguish between these possibilities, we have analysed S-phase progression of Rif1F/F pMEFs released in a synchronous cell cycle after inducing the CRE-mediated deletion in G0. As in the case of deletion in logarithmically growing cells, the total number of cells incorporating BrdU was reduced by about half (Figure 8B). This was not due to a defective release from G0 to G1, as revealed by the comparable kinetics of accumulation of cyclin D1 (Figure 8C). The appearance of a defective BrdU incorporation already in the first cell cycle after Rif1 deletion excludes that this phenotype originates from accumulation of DNA damage over multiple cell cycles. Instead, these data suggest that the defect due to Rif1 deficiency is already sensed prior to the entry into S-phase and affects the G1/S transition (Supplementary Figure S5B and C). To better characterize the nature of the cell-cycle block, we analysed the behaviour of some of the crucial regulators of the G1/S transition, such as chromatin binding of key effectors of the pre-replication complex and p21. We detected no difference in the kinetics of chromatin binding for Cdc6 or Mcm3 between Rif1-proficient and -deficient cells (Figure 8D). However, Rif1 null cells accumulated at higher levels of p21 (Figure 8C). This is even more evident when Rif1 deletion is induced in cycling instead of contact-inhibited cells (Supplementary Figure S5D), in agreement with the fact that replication-timing deregulation becomes also progressively more pronounced with increasing cell cycles after deletion. p21 inhibits DNA replication both by interfering with PCNA function (Flores-Rozas et al, 1994; Waga et al, 1994; Cazzalini et al, 2003) and by silencing Cyclin-dependent kinases (Cdks) essential for S-phase (Sherr and Roberts, 1999; Vogelstein et al, 2000). In agreement with a reduced S-phase population, the amount of chromatin-bound newly incorporated histone H4 (acetylated Lys 12 histone H4, as an indicator for newly synthesized histones that display on H4 the double acetylation at K5 and K12) (Figure 8D) was also decreased (Sobel et al, 1995; Loyola et al, 2006). These results indicate that approximately half of the Rif1 null cells become blocked at the G1/S transition. However, half of the cells proceed into S, fire origins at a normal frequency and show no defect in fork progression or BrdU incorporation. At the moment we do not know what differentiates the null cells that arrest and the ones that don’t. Also, we don’t know if and how the arrested cells escape the checkpoint at later stages. However, the progressive quality of the phenotype suggests that they do. The fact that the effect of Rif1 deficiency is already sensed prior to the beginning of DNA replication is very interesting, suggesting the possibility that a defective replication domain definition could be sensed by the G1/S checkpoint.
Figure 8.
The absence of Rif1 impairs entry into the first S-phase after deletion. (A) Prolonged Rif1 deletion decreases the percentage of BrdU+ cells in an asynchronous population. A 30' BrdU pulse was given to three independent Rif1+/+ or Rif1F/F pMEF lines infected with a retrovirus encoding CRE recombinase (CRE). Bars show the average percentage of BrdU+ cells. (B) Rif1 deletion was induced as in Figure 7. S-phase progression was monitored by FACS analysis of BrdU content and the percentage of BrdU+ cells over the entire population was plotted versus time. An average of three independent pMEF lines for each genotype is presented. (C) Western blotting of Cyclin D1 and p21. Loading control: α-tubulin. h=hours. (D) Western blot of chromatin-bound Cdc6, MCM3, acetylated Lys12 histone H4 (H4K12-Ac). Loading control: lamin B. (C, D) One representative pair of Rif1+/+; Rosa26CreERT2/+ and Rif1F/F; Rosa26CreERT2/+(Rif1−/−), both+4-hydroxytamoxifen treated are shown.
Rif1 is associated to the nuclear matrix
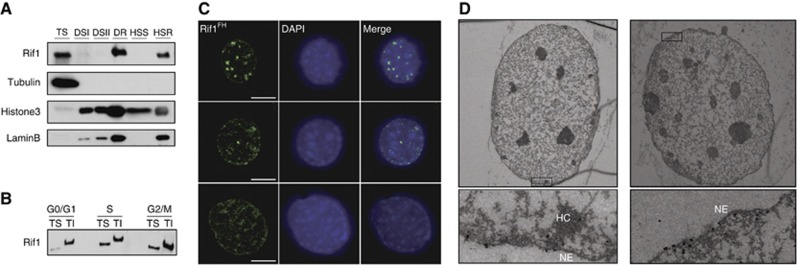
Recently, budding yeast Rif1 was found to be palmitoylated and it was shown that this modification contributes to its association with the nuclear membrane (Park et al, 2011). Chromatin anchoring to the nuclear membrane is a driver of spatial organization of the chromatin in the nucleus. Since chromatin anchoring to the nuclear periphery coincides with the establishment of the replication-timing programme, we determined whether mouse Rif1 was associated with the nuclear matrix. Approximately half of total Rif1 was indeed recovered in the lamin B-enriched fraction (Figure 9A), at least in G1 and S-phase (Figure 9B; Supplementary Figure S5E). Accordingly, Rif1 could also be detected in the nuclear interior of halo preparations, displaying various distribution patterns (Figure 9C) whose significance has yet to be defined. To examine at a higher resolution the distribution of the insoluble fraction of Rif1 in the cell, we performed electron microscopy. We could detect Rif1FH signal in ultra-thin sections mostly associated to the nuclear periphery and heterochromatin (Figure 9D). These data demonstrate that at least a portion of Rif1 is constitutively associated with the nuclear periphery. Given the fundamental role of this structure in multiple aspects of DNA metabolism, such as DNA replication and repair, Rif1 association to the nuclear matrix could potentially explain its involvement in both DNA repair and replication-timing regulation. In addition, Rif1 localization is intriguing in the light of the proposed role of 3D chromatin organization in the definition of the temporal order of replication domains activation.
Figure 9.
Rif1 is associated with the nuclear matrix. (A) Western blotting on extracts from MEFs fractionated for nuclear matrix isolation. Anti-Rif1 antibody used was 1240. TS=triton soluble; DS=DNase soluble; DR=DNase resistant. The DR fraction was further divided in HSS=high salt soluble and HSR=high salt resistant. (B) Live MEFs were FACS sorted in three fractions (G1, S and G2) on the basis of their DNA content using Hoechst 33342. The cells collected were fractionated in triton-soluble (TS) and -insoluble fraction (TI). Western blot was performed on extracts from equal number of cells for each fraction using anti-Rif1 antibody 1240. The migration shift of Rif1 TI fraction band is probably caused by the presence of UREA 8 M (see Materials and methods). (C) Nuclear halos were prepared from Rif1FH/FH MEFs. Immunofluorescence for Rif1FH (anti-HA, green) and DAPI (blue). Scale bar: 10 μm. (D) Electron microscopy visualization of Rif1FH in triton pre-extracted MEFs. One representative ultra-thin section per each cell and magnification of insets highlighted are shown. The black dots correspond to the Nanogold particles associated to the secondary antibody. HC, heterochromatin; NE, nuclear envelope.
Discussion
Although it has been recognized for a long time that the mammalian genome replicates according to a precise spatio-temporal programme, its determinants and its biological significance have remained elusive. The first case of a genome-wide precocious firing of late origins in early S was very recently reported in budding yeast (Mantiero et al, 2011). This study showed that deregulation of replication timing causes activation of the DNA damage checkpoint and decreases cell viability. Here, we identify Rif1 as a key regulator of the temporal DNA replication programme in mammalian cells. In the absence of Rif1, both the physical definition of replication domains as well as their temporal order of activation is altered. The effect of Rif1 deficiency extends throughout the genome, involving both early and late replication domains. Currently, we do not know what distinguishes the affected domains from the ones that are Rif1 insensitive. We also found that Rif1 displays cell cycle-dependent dynamic localization patterns. At least for mid S-phase, Rif1 accumulates at sites and is cleared from chromatin prior to its replication. It is therefore tempting to speculate that Rif1 could be part of the mechanisms establishing accessibility of different origin clusters for limiting replication factors. By doing so, Rif1 would therefore control the temporal sequence of firing of different origin clusters. Rif1 association with the nucleoskeleton is also compatible with this hypothesis. Intriguingly, it was recently shown that in budding yeast, Rif1 plays a similar role at telomeres. While telomeres are clustered and tethered to the nuclear membrane, Rif1 imposes a time constraint (late S-phase) for the accessibility of telomerase to the shortest telomeres (Gallardo et al, 2011). While our work was in preparation, it was also demonstrated that fission yeast Rif1 has a role in regulating the replication-timing programme (Hayano et al, 2012). As we find for mammalian cells, Schizosaccharomyces pombe rif1 deletion affects replication timing of both early and late replication domains. This role seems therefore to be evolutionarily conserved.
Our work also offers for the first time an insight on the possible consequences of replication-timing deregulation on multiple aspects of cell-cycle progression and chromatin organization in mammalian cells. Should replication timing serve the purpose of propagating distinct types of chromatin, the predicted consequences of its perturbation match the results of Rif1 deletion. We find that within the first few cell cycles in the absence of Rif1 there are problems packaging newly replicated chromatin, particularly evident at pHC. In addition, cell-cycle progression is affected during the first cycle after deletion. The fact that Rif1 deficiency affects the G1/S transition is intriguing. Also, a defect preceding DNA replication is consistent with the localization of Rif1 ahead of the replication fork. These results are important, since they show that deregulation of replication timing has an effect immediately prior to the initiation of DNA replication, but not on its progression. The frequency of origin-firing and fork-progression parameters are indeed mostly unaffected by Rif1 deficiency. We have observed that Rif1 deletion reproducibly decreases the abundance of the slower replication forks. However, we do not know if this is a direct consequence of Rif1 deficiency, or, alternatively, the result of altered chromatin status or a form of cell adaptation.
Previous studies with immortalized Rif1 null mouse and chicken cells detected activation of the DNA replication checkpoint, accumulation of chromatid breaks (Buonomo et al, 2009) and problems in fork progression (Xu et al, 2010). Comparison among these different data sets suggests that the immortalization process (perhaps loss of the G1/S checkpoint) allows cells a certain degree of tolerance to Rif1 deficiency (our unpublished data) with the consequent accumulation of replication stress and genomic instability (Buonomo et al, 2009). Mutations found in the Rif1 gene in breast cancer cell lines (Sjoblom et al, 2006; Howarth et al, 2008) have raised the possibility that deregulation of Rif1 function could be involved in cellular transformation. Our data provide potential mechanisms, by illustrating how loss of Rif1 function determines profound changes of the replication-timing programme, activation of the G1/S checkpoint and chromatin-repackaging defects. Each of these events on its own has been shown to either identify cancer cells (De and Michor, 2011; Ryba et al, 2012) or to contribute directly to cell transformation (Bartkova et al, 2005; Gorgoulis et al, 2005; Halazonetis et al, 2008; Zhu et al, 2011). In conclusion, the work we report here identifies Rif1 as one of the first key mammalian factors discovered in the yet unknown network that temporally controls DNA replication and, as a consequence, genome stability.
Materials and methods
Mice generation
The Rif1F mouse allele has been described previously (Buonomo et al, 2009). For the generation of the mouse Rif1FH allele, the second exon, containing the starting ATG, was replaced with FLAG–HA2 tag containing an ATG. The targeting vector was generated by recombineering (Lee et al, 2001). We inserted a FRT-NeomycinR(NeoR)-FRT cassette downstream of the second exon. The first FRT site is associated to a diagnostic NdeI restriction site, the second to BamHI. The construct was subcloned into the pDTA-TKIII vector that allows double-negative selection (DTA and TK genes), linearized with NotI and electroporated into Bruce4 C57BL/6 ES cells. ES cells colonies were screened for homologous recombination by Southern blot, digesting the DNA with BamHI and using the R5 probe. Positive clones were further confirmed by NdeI digest and injected by standard techniques into C57BL/6J blastocysts. Chimeras were evaluated based on tail genotyping. Chimeric founders were crossed to C57BL/6J females and the mice were kept in a pure C57BL/6J background. The NeoR cassette was removed by crossing the F1 mice with a FLPe deleter mouse strain (Jackson Labs). Genotyping of mice and derived cells was performed by PCR with the following primers: f1: GCGAACCTCGGACGCCGTGG; r1: GCACCTGTAATCTCAACCACTC. PCR generates a 146-bp product for the wild-type allele and a 248-bp product for the Rif1FH allele. The Southern probes R5 (848 bp) was generated by PCR from genomic DNA with the following primers: Rif1FH5′fw 5′-GTGTCACTACTCTCACATTT and Rif1FH5′rev 5′-TGTTTTTCCATTTAGAAGCCAG.
Cell manipulation
pMEFs were generated from E12.5 embryos according to standard protocols and propagated in DMEM high Glucose, 15% heat-inactivated fetal bovine serum (PAA), 100 U/ml penicillin, 0.1 mg/ml streptomycin, 0.2 mM L-glutamine, 0.1 mM non-essential amino acids and 50 μM β-mercaptoethanol. pMEFs at passage 2 were infected as described in Buonomo et al (2009), except that we performed 5 instead of 4 infections, spaced 12–6–6–6–12 h. After 4 days of selection in Hygromycin B (Sigma H3274) at 90 μg/ml cells were processed without further passaging.
Cell synchronization was performed by driving early passage pMEFs into contact inhibition and then inducing CREERT2 translocation into the nucleus by treating the cells with 600 nM 4-hydroxytamoxifen (Sigma H7904). During the last 2 days of treatment, the cells were also serum-starved.
Immunofluorescence
Cells were pulsed with EdU for 30′, and then processed as described in Buonomo et al (2009). EdU staining was performed according to the manufacturer’s instructions (Invitrogen Click-iT kit C10350). Evaluation of cells in different S-phase substages for the different lines was performed blind. Images were acquired using a Leica confocal TCS SP5 microscope with a × 63 1.4 NA oil objective and run by LAS AF Software (Leica). Contrast adjustment and cropping were performed in Photoshop (Adobe) and ImageJ. Figures were composed in Illustrator (Adobe).
Preparation of halos
Halos were prepared according to Guillou et al (2010). Anti-HA immunofluorescence was performed as above.
Micrococcal nuclease treatment and Southern–Western
Cells were pulsed for 1 h with 10 μM BrdU and collected. Nuclei were prepared by dounce homogenization in lysis buffer (20 mM Tris–HCl pH 7.5, 4 mM MgCl2, 250 mM sucrose, 0.1% Triton X-100 and protease inhibitors). After pelleting them 5′ at 2000 g at 4°C, nuclei were resuspended in lysis buffer and chromatin was isolated over a sucrose cushion (Lysis buffer with additional 30% sucrose) at 2400 g 15′ 4°C. The isolated chromatin was resuspended in MNase buffer (60 mM KCl, 15 mM NaCl, 15 mM Tri–HCl pH 7.4, 0.25 M sucrose, 1 mM CaCl2) and digested for 2 min with the indicated amounts of Micrococcal nuclease (Sigma N3755). The reaction was stopped with stop buffer (50 mM EDTA, 2.5% sarcosyl, 1% SDS) and proteins digested with Proteinase K 10 mg/ml. Equal amount of DNA were loaded on a 1.3% agarose gel prepared in Tris-Glycine buffer (5 × : 144 g Glycine, 30 g Trisbase per liter) and used for Southern blotting according to standard protocols. The membrane was crosslinked twice at 0.15 J/cm2 and washed in 0.1 M NaOH. After 1 h blocking in 3% BSA in PBS-T (Tween 0.1%), the membrane was incubated in anti-BrdU antibody for 1 h. The same membrane was probed for Southern with the major satellite probe from Lehnertz et al (2003).
Western blotting, chromatin fractionation and nuclear matrix isolation
Total protein extracts and western blotting were performed as described in Buonomo et al (2009).
Chromatin fractionation was performed by incubating cells in CSK buffer (0.5% Triton X-100, 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 1 mM EGTA, 10 mM PIPES pH 6.8) 10' on ice then spinning 4 at 500 g. Supernatants were recovered and represent the Triton-soluble (TS) fraction. Pellets were washed twice in CSK buffer, resuspended in 8 M Urea and represent the Triton-insoluble (TI) fraction.
Nuclear matrix isolation was performed as in Mladenov et al (2006).
Antibodies
Anti-HA (Covance HA.11 Clone 16B12) was used at 1:3000; Anti-BrdU (BD 347580) 1:2000 for western blotting 1:7 for FACS; Anti-Cdc6 (Cell Signaling 3387 Clone C42F7) 1:1000; Anti-p21 (Santa Cruz sc-6246 1:200); Anti-CyclinD1 (Santa Cruz M-20, sc-718) 1:200; Anti-MCM3 (Santa Cruz N-19, sc-9850) 1:200 for western blotting, 1:50 for immunofluorescence; Anti-H3 (Cell Signaling 9681) 1:1000; Anti-H3K9me3 for western blotting (Novus Biologicals 6F12H4) 1:1000; Anti-H3K9me3 (Abcam, ab8898) for immunofluorescence 1:500; Anti-HP1α for western blotting (Upstate 07-346) 1:1000; Anti-HP1α for immunofluorescence (Euromedex 2HP-1H5-AS) 1:500; Anti-H3K27me3 (Abcam, ab6147) 1:1000; Anti-H4K20me3 (Abcam ab9053); Anti-H4K12Ac (Cell Signaling 2591) 1:1000; Anti-Lamin B (Abcam, ab16048) 1:1000; Anti-MeCP2 (Abcam, ab2828) 1:1000; Anti-phospho-Histone H3 (Ser10) (Millipore 06-570) 1:200; Anti-Rif1 (Buonomo et al, 2009) 1:8000 for western blotting and 1:3000 for immunofluorescence; Anti-Smc1 (Bethyl Laboratories A300-055A) 1:5000; Anti-α-Tubulin (Sigma T9026) 1:10 000; Anti-γ-Tubulin (Sigma T6557) 1:5000.
FACS analysis and sorting
BrdU labelling, staining and FACS analysis were performed as in Buonomo et al (2009).
For FACS sorting logarithmically growing MEFs were stained with Hoechst 33342 and sorted in the three fractions according to their DNA content on a 5-laser ARIA SORP (BD Biosciences). Once collected, the cells were pelleted and the triton-soluble/insoluble fractions were obtained as described.
DNA combing
Neo-synthesized DNA was labelled with an IdU pulse followed by a CldU pulse as described in Anglana et al (2003). Genomic DNA was extracted and combed on silanized slides as in Letessier et al (2011). Immunofluorescence detection of neo-synthesized DNA and DNA was performed by successive incubations and washes as in Anglana et al (2003) with the following reagents: (1) mouse anti-bromodeoxyuridine (BrdU) (BD Biosciences) at 1:5 dilution and rat anti-BrdU (AbD Serotec) at 1:10 dilution; (2) Alexa-488-conjugated goat anti-mouse (Invitrogen) and Alexa-594-conjugated goat anti-rat (Invitrogen) both at 1:50 dilution; (3) mouse anti-single-stranded DNA (Chemicon) at 1:25 dilution; (4) Alexa-594-conjugated goat anti-rat (Invitrogen) and Alexa-647-conjugated goat anti-mouse (Invitrogen) both at 1:50 dilution. Data were generated with two independent Rif1+/+ and Rif1−/− pMEF preparations.
A Leica DMR600 epifluorescence microscope equipped a CoolSNAP HQ2 CCD camera and run by Metamorph software (Molecular Devices) was used for image acquisition with a × 40 objective. Overlays of IdU/CldU/DNA signals performed in Metamorph were used to determine CldU replication-track length and inter-origin distances. Only CldU signal (second pulse) juxtaposed to IdU signal was used to determine replication speed. Replication tracks with interruption of the DNA signal or with a length identical to the DNA signal were not considered since they could reflect broken fibres. Replication fork speed was calculated by dividing the length of a CldU track (in kb) with pulsing time (20′). Statistical analysis was performed with a Mann–Whitney test.
Transcriptome analysis
Total RNA was isolated using the TRIzol reagent (Invitrogen) following the manufacturer’s instructions and treated with DNAse (Promega). RNA quality was checked using a bioanalyzer (Agilent 2100; Agilent Technologies), and RNA quantity was measured with ND-1000 Nanodrop spectrophotometer. In all, 1 μg of RNA sample was used for microarray analysis Affymetrix Mouse Gene 1.0ST array (Affymetrix). Robust multi-array average (RMA) normalization was applied. Normalized data were then filtered based on the Affymetrix detection call so that only probes that had a Present call in at least one of the arrays were retained. CEL files were then imported in GeneSpringGX 11.5 software and statistical analysis was performed to detect significantly differentially expressed genes. A fold change cutoff of 2 was applied.
Replication profiling
Newly synthesized DNA was pulse-labelled with (BrdU) while the cells were in the exponential phase. Cells were then dissociated into a single-cell suspension and nuclei were isolated. DNA was subsequently stained with propidium idodide and cells were FACS sorted into early and late S-phase fractions based on their increasing DNA content during S-phase. The DNA that was synthesized either early or late during S-phase was then purified by immunoprecipitation of the BrdU-substituted nascent DNA (BrdU-IP). For the analysis of replication timing in the synchronous cell cycle, cells were collected at each time point after 2 h BrdU pulse. In this way, we ensured that only cells in the first S-phase after Rif1 deletion were analysed. After fixation and before FACS sorting, the different time points were pooled. Thus, the definition of early and late S-phase was based on the DNA content and not on the time of collection. A series of 10 DNA sites that are known to replicate at specific times were then analysed by PCR to verify the quality of the nascent strands, which was then subjected to whole-genome amplification. The amplified early and late fraction were differentially labelled and hybridized to a CGH whole-genome tiling array. The replication timing of each probe along the genome was then calculated as the log2 enrichment of early fraction over late fraction. Genome-wide replication-timing profiles were constructed as described (Hiratani et al, 2008) with exception that Rif1−/− pMEF data set was not scaled to the standard Inter-quartile range (IQR) of other data sets. This was due to the unusually narrow distribution of the majority of data points, which caused the outlining data points to go beyond scale during standard scaling. Since the scale of Rif1−/− data was significantly different from Rif1+/+ in both replicates, this points to a biological cause. Two replicates of Rif1−/− and Rif1+/+ pMEFs were averaged and used to create the replication-timing profiles. The replicates of Rif1+/+ data and Rif1−/− data were normalized separately using Limma package in R. Then, the data were Loess smoothed using 300 Kb window. After smoothing, the 2–98 percentile range of Rif1−/− was scaled to 2–98 percentile range of Rif1+/+. This scaled data were used for further analysis.
Electron microscopy
Rif1FH/FH MEFs were plated to 80% confluency on gridded MatTek dishes (MatTek, Ashland, USA). Cells were then rinsed with PBS, pre-extracted with Triton X-100 buffer pH 7.9 (0.5% Triton, 20 mM Hepes-KOH, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) for 10 min at 4°C. They were then rinsed with PBS and fixed with 3% paraformaldehyde and 2% sucrose in PBS for 10' at room temperature. Cells were then rinsed with 50 mM NH4Cl, extracted with 0.5% saponin and incubated with the EdU reaction mix for 30'. After the incubation, they were rinsed with PBS and imaged on a Zeiss AxioObserver epifluorescence microscope and S-phase nuclei were imaged. Cells were then blocked, incubated with an anti-HA antibody (Covance, Princeton, USA) for 1 h, rinsed and then incubated with an anti-mouse IgG Nanogold antibody overnight. They were then further processed for electron microscopy as described in Bahtz et al (2012). Thin sections (60 nm) were imaged on formvar-coated slot grids and stained with 2% uranyl acetate in 70% methanol and 1.5% lead citrate. Imaging was done on a CM120 Phillips microscope.
Supplementary Material
Acknowledgments
We gratefully acknowledge Violetta Parimbeni for mouse husbandry, Claudia Valeri for genotyping and Melanie Leuener for technical help. This study was technically supported by EMBL Monterotondo’s FACS and microscopy and EMBL Heidelberg Genomics Core Facility. We also acknowledge the services of J Rientjes from Monash University’s Gene Recombineering Facility and Patrick Varga-Weisz (Babraham Institute, Cambridge, UK) for the Micrococcal assay protocol and useful discussions. We greatly appreciate for helpful discussions Dònal O’Carroll (EMBL Monterotondo). We are especially grateful to Nadia Rosenthal. GA and JPQ received support from la Ligue Nationale contre le Cancer (Equipe labellisée Ligue 2010), the European Commission Network of Excellence EpiGeneSys (HEALTH-F4-2010-257082), ERC Advanced Grant 2009-AdG_20090506 ‘Eccentric’, ANR ‘ECenS’ ANR-09-BLAN-0257-01. RF was supported by EMBL Interdisciplinary Postdoc (EIPOD) under Marie Curie Actions (COFUND); SCBB thanks the EpiGeneSys Network of Excellence.
Author contributions: DC performed the majority of the experiments. VD performed the replication-timing experiment starting from the FACS sorting of the cells and analysed the data. JPQ performed the DNA combing and its analysis. RF did the transcriptome analysis. FT helped in several experiments and performed the BrdU FACSes. RM and CA have carried on the electron microscopy analysis. GA and DG have given scientific feedback on both the experiments and the manuscript preparation. SCBB designed the experiments and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anglana M, Apiou F, Bensimon A, Debatisse M (2003) Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 [DOI] [PubMed] [Google Scholar]
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM (2004) The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 24: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahtz R, Seidler J, Arnold M, Haselmann-Weiss U, Antony C, Lehmann WD, Hoffmann I (2012) GCP6 is a substrate of Plk4 and required for centriole duplication. J Cell Sci 125(Pt 2): 486–496 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870 [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Wu Y, Ferguson D, de Lange T (2009) Mammalian Rif1 contributes to replication stress survival and homology-directed repair. J Cell Biol 187: 385–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzalini O, Perucca P, Riva F, Stivala LA, Bianchi L, Vannini V, Ducommun B, Prosperi E (2003) p21CDKN1A does not interfere with loading of PCNA at DNA replication sites, but inhibits subsequent binding of DNA polymerase delta at the G1/S phase transition. Cell Cycle (Georgetown, TX) 2: 596–603 [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA (2002) Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12: 439–445 [DOI] [PubMed] [Google Scholar]
- De S, Michor F (2011) DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat Biotechnol 29: 1103–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Berezney R (2002) The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J Cell Sci 115(Pt 21): 4037–4051 [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM (1999a) DNA replication and nuclear organization: prospects for a soluble in vitro system. Critical Rev Eukaryot Gene Expression 9: 353–361 [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM (1999b) The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993 [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H, Kelman Z, Dean FB, Pan ZQ, Harper JW, Elledge SJ, O'Donnell M, Hurwitz J (1994) Cdk-interacting protein 1 directly binds with proliferating cell nuclear antigen and inhibits DNA replication catalyzed by the DNA polymerase delta holoenzyme. Proc Natl Acad Sci USA 91: 8655–8659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Laterreur N, Cusanelli E, Ouenzar F, Querido E, Wellinger RJ, Chartrand P (2011) Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol Cell 44: 819–827 [DOI] [PubMed] [Google Scholar]
- Gilbert DM (2001) Nuclear position leaves its mark on replication timing. J Cell Biol 152: F11–F15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Gasser SM (2006) Nuclear Structure and DNA Replication. InDNA Replication and Human Desease DePamphilis ML (ed). Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, [Google Scholar]
- Gilbert DM, Takebayashi SI, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I (2010) Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol 75: 143–153 [DOI] [PubMed] [Google Scholar]
- Gondor A, Ohlsson R (2009) Replication timing and epigenetic reprogramming of gene expression: a two-way relationship? Nat Rev Genet 10: 269–276 [DOI] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, Cedar H (2008) DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev 22: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD (2005) Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913 [DOI] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Mendez J (2010) Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 24: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science (New York, NY) 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D (1992) A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, Masai H (2012) Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev 26: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM (2008) Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi S, Lu J, Gilbert DM (2009) Replication timing and transcriptional control: beyond cause and effect--part II. Currt Opin Genet Dev 19: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth KD, Blood KA, Ng BL, Beavis JC, Chua Y, Cooke SL, Raby S, Ichimura K, Collins VP, Carter NP, Edwards PA (2008) Array painting reveals a high frequency of balanced translocations in breast cancer cell lines that break in cancer-relevant genes. Oncogene 27: 3345–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen HF, Azuara V, Amoils S, Spivakov M, Terry A, Nesterova T, Cobb BS, Ramsahoye B, Merkenschlager M, Fisher AG (2007) The impact of chromatin modifiers on the timing of locus replication in mouse embryonic stem cells. Genome Biol 8: R169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630 [DOI] [PubMed] [Google Scholar]
- Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM (2012) Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG (2001) A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73: 56–65 [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC (2000) Dynamics of DNA replication factories in living cells. J Cell Biol 149: 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M (2011) Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123 [DOI] [PubMed] [Google Scholar]
- Li J, Santoro R, Koberna K, Grummt I (2005) The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J 24: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HY, Robertson ED, Hiraga S, Alvino GM, Collingwood D, McCune HJ, Sridhar A, Brewer BJ, Raghuraman MK, Donaldson AD (2011) The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol Biol Cell 22: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G (2006) PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell 24: 309–316 [DOI] [PubMed] [Google Scholar]
- Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G (2009) The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep 10: 769–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R (1998) Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol 143: 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Quivy JP, Probst AV, Almouzni G (2010) Heterochromatin at mouse pericentromeres: a model for de novo heterochromatin formation and duplication during replication. Cold Spring Harb Symp Quant Biol 75: 155–165 [DOI] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P (2011) Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30: 4805–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, Bensimon A (1997) Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science (New York, NY) 277: 1518–1523 [DOI] [PubMed] [Google Scholar]
- Mladenov E, Anachkova B, Tsaneva I (2006) Sub-nuclear localization of Rad51 in response to DNA damage. Genes Cells 11: 513–524 [DOI] [PubMed] [Google Scholar]
- Park S, Patterson EE, Cobb J, Audhya A, Gartenberg MR, Fox CA (2011) Palmitoylation controls the dynamics of budding-yeast heterochromatin via the telomere-binding protein Rif1. Proc Natl Acad Sci USA 108: 14572–14577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G (2008) The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol 15: 972–979 [DOI] [PubMed] [Google Scholar]
- Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G (2004) A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J 23: 3516–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N (2006) DNA replication timing: random thoughts about origin firing. Nature Cell Biol 8: 1313–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Chang BH, Shirley JW, Buckley Q, Pope BD, Devidas M, Druker BJ, Gilbert DM (2012) Abnormal developmental control of replication timing domains in pediatric acute lymphoblastic leukemia. Genome Res (advance online publication, 24 May 2012; doi:; DOI: 10.1101/gr.138511.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM (2010) Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D (2004) Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci 117(Pt 22): 5353–5365 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512 [DOI] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T (2004) Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev 18: 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE (2006) The consensus coding sequences of human breast and colorectal cancers. Science (New York, NY) 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD (1995) Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA 92: 1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408: 307–310 [DOI] [PubMed] [Google Scholar]
- Waga S, Hannon GJ, Beach D, Stillman B (1994) The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 369: 574–578 [DOI] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, Chen J (2003) p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 23: 2556–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Singh PB, Gilbert DM (2006) Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J Cell Biol 174: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Muniandy P, Leo E, Yin J, Thangavel S, Shen X, Ii M, Agama K, Guo R, Fox D 3rd, Meetei AR, Wilson L, Nguyen H, Weng NP, Brill SJ, Li L, Vindigni A, Pommier Y, Seidman M, Wang W (2010) Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J 29: 3140–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Blackburn EH (2004) Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol 167: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I (2010) Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM (2009) G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci USA 106: 19363–19368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.