Abstract
Toxigenic conversion of Vibrio cholerae bacteria results from the integration of a filamentous phage, CTXφ. Integration is driven by the bacterial Xer recombinases, which catalyse the exchange of a single pair of strands between the phage single-stranded DNA and the host double-stranded DNA genomes; replication is thought to convert the resulting pseudo-Holliday junction (HJ) intermediate into the final recombination product. The natural tendency of the Xer recombinases to recycle HJ intermediates back into substrate should thwart this integration strategy, which prompted a search for additional co-factors aiding directionality of the process. Here, we show that Endo III, a ubiquitous base excision repair enzyme, facilitates CTXφ-integration in vivo. In vitro, we show that it prevents futile Xer recombination cycles by impeding new rounds of strand exchanges once the pseudo-HJ is formed. We further demonstrate that this activity relies on the unexpected ability of Endo III to bind to HJs even in the absence of the recombinases. These results explain how tandem copies of the phage genome can be created, which is crucial for subsequent virion production.
Keywords: cholera toxin, four-way DNA junctions, lysogenic conversion, Xer recombination
Introduction
Two related tyrosine recombinases, XerC and XerD, are encoded in the genome of most bacteria. They serve to resolve dimers of circular chromosomes by the addition of a crossover at a specific site, dif (Val et al, 2008). Many mobile elements exploit the existence of this recombination machinery. Indeed, XerC was initially discovered because of its role in the maintenance of the monomeric state of plasmids (Colloms et al, 1990). Several Integrative Mobile Elements exploiting Xer (IMEXs) were subsequently found in bacterial genomes (Das et al, 2011a), including phages (Huber and Waldor, 2002; Campos et al, 2003), cryptic elements related to phages or plasmids (Hassan et al, 2010) and genomic islands (Dillard and Seifert, 2001).
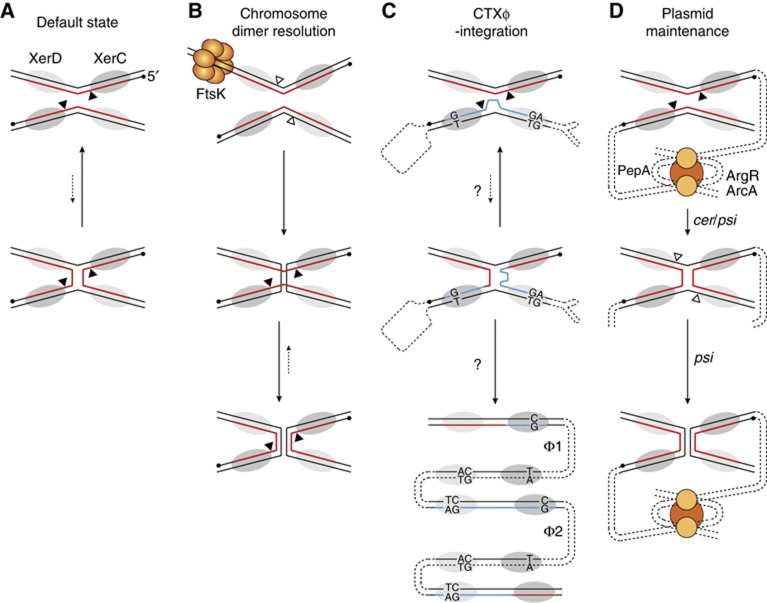
Exploitation of Xer is likely to be advantageous to mobile elements because the wide distribution of the Xer recombinases in bacteria and the high sequence conservation of their target sites on bacterial chromosomes should permit a quite easy extension of their host range (Val et al, 2008; Kono et al, 2011). However, it requires means to overcome the cellular mechanisms that normally restrict recombination to dif sites harboured by a chromosome dimer: Xer recombination occurs within synaptic complexes containing two molecules of each of XerC and XerD, each pair of recombinases catalysing the exchange of a specific pair of DNA strands (Figure 1A). Coordinated reciprocal switches in recombinase activity ensure that only one pair of recombinases is active at a time (Hallet et al, 1999). As a result, the reaction proceeds sequentially, with a necessary Holliday junction (HJ) intermediate. By default, the XerD recombinases are inactive and the XerC recombinases promote a low level of HJ formation (Figure 1A; Barre et al, 2000). However, it is not followed by a switch in the catalytic state of the two pair of recombinases (Arciszewska et al, 2000). As a result, HJs resulting from XerC-catalysis are rapidly eliminated by new rounds of XerC-strand exchanges (Figure 1A; Barre et al, 2000; Aussel et al, 2002). During chromosome dimer resolution, this futile recombination cycle is broken by a cell division protein, FtsK, which triggers the exchange of a first pair of strands by XerD-catalysis (Figure 1B; Aussel et al, 2002).
Figure 1.
Scheme of the different pathways of Xer recombination and their control. (A) Default catalytic state of the Xer recombinases. XerC and XerD are depicted by dark and light-shaded ovals, respectively. The nucleotidic positions attacked by XerC and XerD are indicated by black and white triangles, respectively. They border the overlap region of the sites, where strand exchanges occur. The 5′-end of the strands recombined by the XerC are shown as a black dot. In plasmid and dimer resolution sites, the strand recombined by XerC is shown in red. (B) Chromosome dimer resolution pathway. According to current models for the tyrosine recombinases pathway, alternative orders of strand exchange are dictated by different arrangements of the synapse. This is indicated in the figure by a different angle between the XerC- and XerD-binding sites in the two recombination substrates. (C) CTXϕ-integration pathway. In CTXϕ attP(+), the strand recombined by XerC is shown in blue to indicate the imperfect homology with dif. A loop depicts the unpaired bases in the overlap region of CTXϕ attP(+) and the unpaired bases in the XerC- and XerD-binding arms are indicated. Two phage DNA copies integrated in tandem are depicted in the bottom scheme, as observed in most Cholera epidemic strains (Φ1 and Φ2). A new dif and inverted attP2 and attP1 dsDNA sites flank the first copy whereas both sides of the second copy are flanked by attP2 and attP1. (D) Plasmid dimer resolution pathway.
The cholera toxin genes—the principal virulence factors of the deadly diarrhoea-causing bacterium Vibrio cholerae—are encoded in the genome of an IMEX, CTXϕ (Val et al, 2005; Das et al, 2010, 2011b). CTXϕ is a filamentous bacteriophage (Waldor and Mekalanos, 1996). The dsDNA replicative form of its genome contains two pairs of binding sites for XerC and XerD in inverted orientations, attP1 and attP2. In the (+) ssDNA form of its genome, the one that is packaged in viral particles, they fold back to form the stem of a double hairpin structure, attP(+) (Figure 1C; Val et al, 2005). Lysogenic conversion results from the exchange of a single pair of strands between attP(+) and the dif site of one or the other of the two circular chromosomes of V. cholerae, which is catalysed by the XerC recombinases (Val et al, 2008). Resolution of the resulting pseudo-HJ by a second pair of strand exchanges would lead to the formation of a lethal linear covalently closed dsDNA chromosome (Bouvier et al, 2005). However, the lack of homology in the overlap regions of attP(+) and dif next to the XerD cleavage sites prevents any potential XerD-mediated strand exchange (Das et al, 2010, 2011b). Correspondingly, CTXϕ-integration does not depend on the catalytic activity of the XerD recombinases, which only play a structural role in the formation of the synaptic complex (Val et al, 2008). The pseudo-HJ is thought to be converted into product by replication (Figure 1C; Val et al, 2005; Das et al, 2010). Another important feature of the integration process of CTXϕ is its irreversibility, the active form of the phage attachment site being masked in the dsDNA prophage (Val et al, 2005). This prevents production of new extrachromosomal copies of the phage genome by Xer recombination. However, CTXϕ (+) ssDNA can be produced by a process analogous to rolling circle replication (RCR) in cells carrying tandemly arranged prophages (Moyer et al, 2001). Such phage arrays are found in most V. cholerae toxigenic strains (Chun et al, 2009; Mutreja et al, 2011; Das et al, 2011a, 2011b), which suggested that their formation could be inherent to the mechanism of integration of the phage. However, a molecular explanation was still lacking (Figure 1C). In addition, it was intriguing to observe that XerC catalysed the formation of the pseudo-HJ integration intermediate of CTXϕ (Val et al, 2005; Das et al, 2010) since its propensity to recycle back HJs to substrate (see Hallet et al, 1999; Arciszewska et al, 2000; Barre et al, 2000 for dif/dif HJs and Figure 4E for attP(+)/dif pseudo-HJs) should severely decrease the chances for replication to finalize the integration process (Figures 1A and C). Recombination events are also initiated by XerC during plasmid dimer resolution (Colloms et al, 1996). In this case, however, the catalytic state of the Xer recombinases is determined by accessory host proteins and accessory sequences flanking the recombination sites that direct the formation of a synapse with a specific architecture (Figure 1D; Colloms et al, 1997; Bregu et al, 2002), which supported the possibility that additional host factors than XerCD might be implicated in the formation and/or resolution of the pseudo-HJ integration intermediate of CTXϕ (Figure 1C).
Here, we took a direct genetic approach to uncover host factors that might participate in the toxigenic conversion of V. cholerae by CTXϕ. We found that nth disruption considerably reduced the efficiency of integration of the phage. In addition, we found that the number of integrated copies of the phage genome decreased. The nth gene codes for Endo III, a ubiquitous DNA glycosylase/lyase of the base excision repair (BER) pathway (Aspinwall et al, 1997; Denver et al, 2003). However, the catalytic activity of Endo III was not required for CTXϕ-integration, nor were other BER enzymes. In vitro analysis revealed that Endo III had the so far unexpected ability to bind to HJs in general and that it displaced XerC from the pseudo-HJ integration intermediate of the phage when it bound to it, thereby impeding new rounds of XerC-strand exchanges. Taken together, these results suggest that Endo III increases the efficiency of integration of CTXϕ by stabilizing its pseudo-HJ integration intermediate until its conversion into product by replication and that this permits multiple successive integration events.
Results
A screen for host factors implicated in CTXϕ-integration
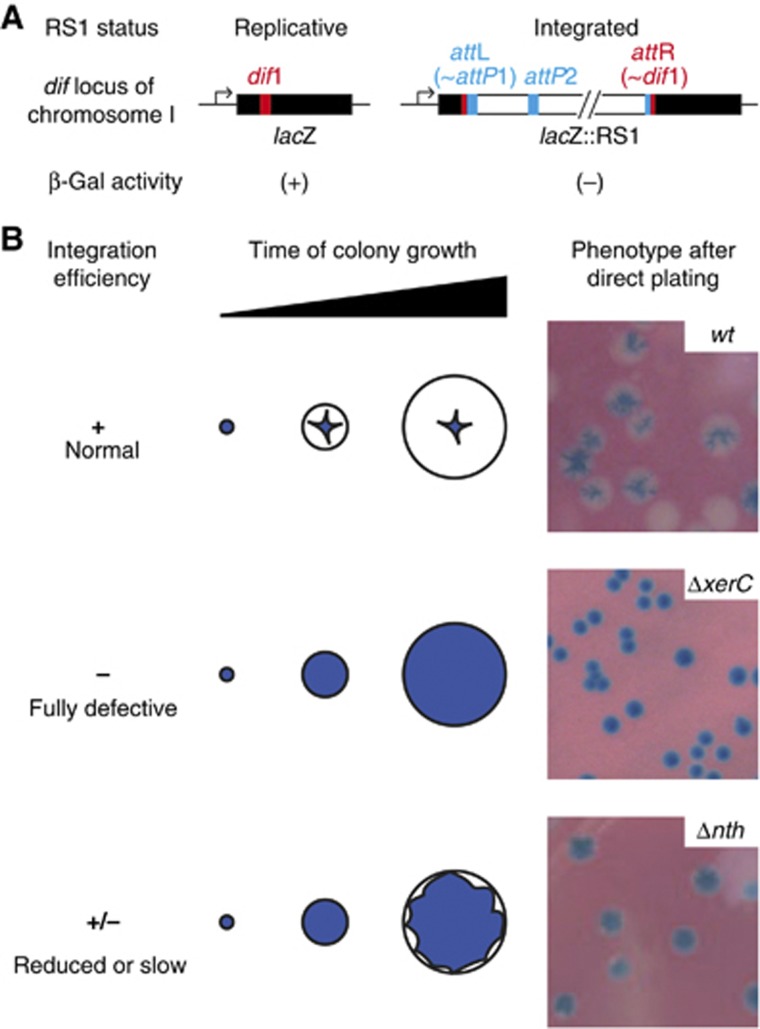
We previously described a colorimetric assay to monitor IMEX integration events in V. cholerae (Das et al, 2010). Briefly, the dif site of the largest of the two chromosomes harboured by the V. cholerae N16961 El Tor strain, dif1, was inserted in the coding region of the Escherichia coli lacZ gene in such a manner that the produced protein might retain its β-galactosidase activity (Figure 2A). The lacZ::dif1 was then inserted in place of the normal dif1 site in a N16961 El Tor strain in which the endogenous lacZ gene had been deleted. This reporter strain gives rise to blue colonies on X-gal media. Site-specific integration events into dif1 disrupt the lacZ::dif1 ORF (Figure 2A), leading to the appearance of white sectors (Figure 2B). Using this assay, we demonstrated that RS1, a truncated derivative of CTXϕ that lacks the phage morphogenesis genes and the cholera toxin genes, eventually integrated in 100% of the colonies (Das et al, 2010). This led to fully white colonies or colonies with large white sectors around a blue star-shaped centre on X-gal plates (Figure 2B, normal integration). We reasoned that introduction of RS1 in a reporter cell in which a host factor implicated in CTXϕ-integration was disrupted would lead to the formation of fully blue colonies or colonies with small white sectors at the periphery of a large blue heart, depending on whether the factor was absolutely necessary for integration (Figure 2B, fully defective integration) or not (Figure 2B, reduced or slow integration). In both cases, the colonies would be distinguishable from the normal integration pattern, which could serve as a screen for host factors implicated in CTXϕ-integration. In addition, the procedure could be used to check the homogeneity of the integration defect of potentially interesting clones by curing them of the replicative form of RS1 and subjecting them to a second round of conjugation.
Figure 2.
Screening for host factors implicated in CTXϕ-integration. (A) Scheme of the lacZ::dif1 allele and of its disruption upon RS1-integration. (B) Phenotypes of colonies in which RS1-integration was normal (top), fully defective (middle) and reduced or slow (bottom). Cells were plated immediately after conjugation to mimick the condition of the screen.
We implemented this screen in two independent mariner transposition libraries of the lacZ::dif1 reporter strain by conjugating a R6K suicide vector harbouring a chloramphenicol resistance gene and the RS1 replication and integration region (Das et al, 2010). Conjugants were selected on plates supplemented with chloramphenicol and X-gal. We thus screened over 24 000 clones, which allowed us to isolate four independent clones in which the xerC ORF was disrupted and two independent clones in which the mariner transposon had inserted within the xerC promoter, demonstrating the validity of the procedure (Supplementary Table S1).
Endo III facilitates RS1-integration
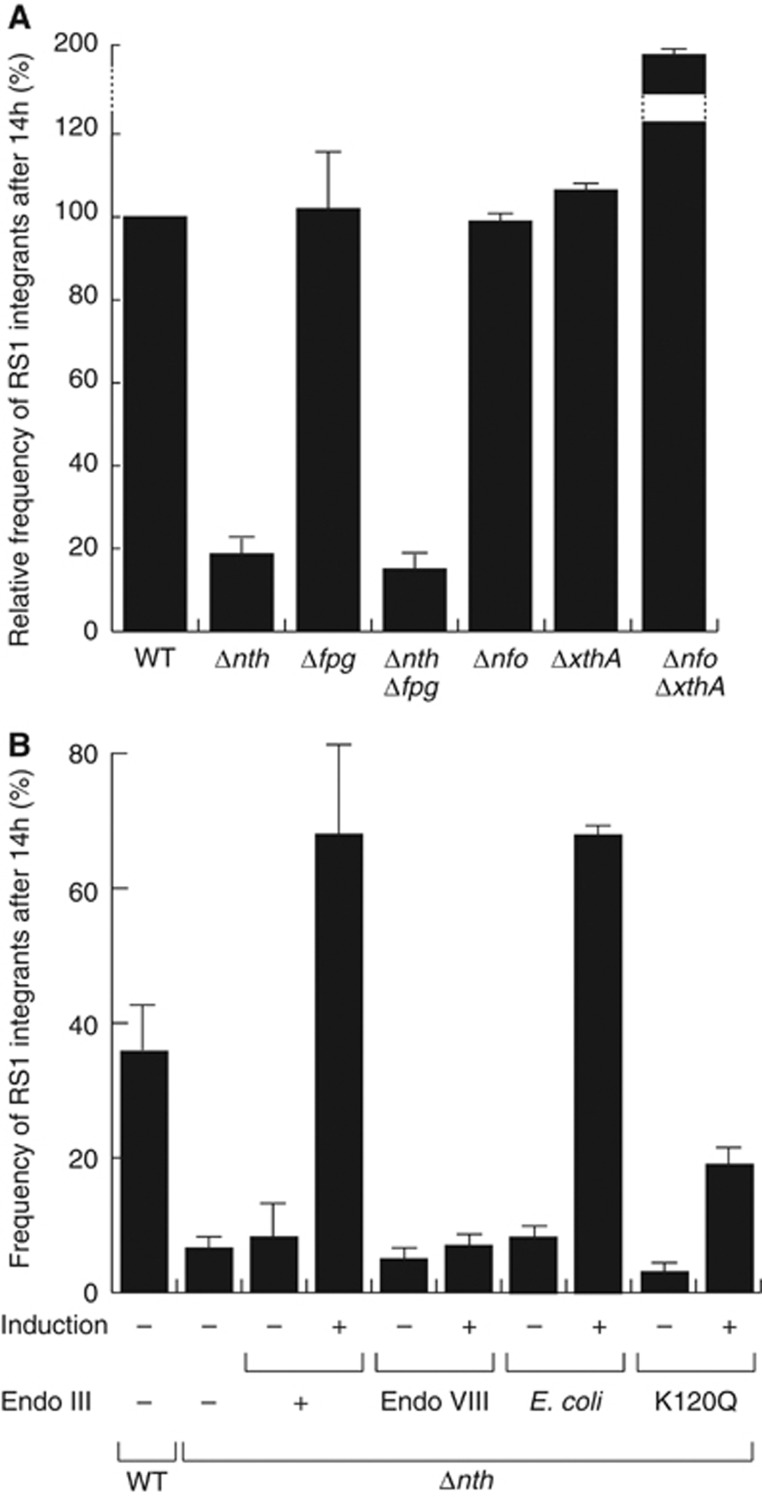
We isolated five independent clones in which transposition disrupted the nth ORF, the gene encoding for Endo III (Supplementary Table S1). To confirm this observation, we engineered a complete deletion of the nth ORF in the lacZ::dif1 reporter strain. We did not observe any defect of proliferation of the Δnth cells after conjugation of the R6K suicide vector harbouring the RS1 replication and integration region. However, the conjugants yielded colonies with small white sectors at the periphery of a large blue heart (Figure 2B). In addition, the number of integrated RS1 copies was smaller in the absence of Endo III: 36% of Δnth lysogens carried a single integrated RS1 copy whereas wild-type lysogens always harboured two or more copies of it (Supplementary Figure S1). In order to gain a quantitative measure of the integration defect of Δnth cells, we compared the number of integrants 14 h after conjugation (Figure 3). Deletion of nth led to a five-fold reduction in the integration efficiency of RS1 at this time (Figure 3A). Integration was fully restored when Endo III was produced from an ectopic plasmid under the control of the arabinose promoter, excluding any polar effect of the deletion (Figure 3B).
Figure 3.
Endo III facilitates V. cholerae toxigenic conversion. (A) Relative frequency of RS1 integrants in BER mutants. (B) Frequency of RS1 integrants in Δnth cells complemented with V. cholerae Endo III, its K120Q catalytically inactive mutant, E. coli Endo VIII or E. coli Endo III. Conjugants were grown overnight (14 h) at 37°C in rich media supplemented with chloramphenicol. The proportion of fully white colonies (lysogenics) over the total number of CmR clones was used to calculate the frequency of integrants. Mean and standard deviation of at least three independent experiments.
BER is not implicated in integration
Endo III is a DNA glycosylase/lyase of the BER pathway. No other mutation on the BER pathway was isolated in our screen. However, we did not isolate clones in which xerD was disrupted, suggesting that the screen might not have been exhaustive. Endo III serves to remove damaged pyrimidines. A second DNA glycosylase/lyase, Fpg, serves to remove damaged purines in V. cholerae. We reasoned that Fpg could be implicated in the remaining integration events observed in Δnth cells. However, deletion of fpg did not affect RS1-integration whether alone or in combination with the nth deletion, highlighting the specificity of action of Endo III (Figure 3A). Most bacteria encode for a third DNA glycosylase/lyase enzyme, Endo VIII, which is absent from V. cholerae. Endo VIII is structurally related to Fpg but acts on the same substrates as Endo III. Whereas production of E. coli Endo III restored a high rate of integration in V. cholerae Δnth cells (Figure 3B), production of E. coli Endo VIII did not (Figure 3B), suggesting that the role of Endo III was not linked to its action on damaged pyrimidines. Cleavage of damaged DNA by Endo III generates 3′ unsaturated aldehydic ends, which need to be recessed into 3′-hydroxyl ends by either Exo III or Endo IV, the respective products of the xthA and nfo genes. Neither xthA and/or nfo-deletions reduced the efficiency of CTXϕ-integration (Figure 3A), further suggesting the idea that the catalytic activity of Endo III was not required for integration. Correspondingly, production of a form of V. cholerae Endo III carrying the catalytically inactivating mutation K120Q (Thayer et al, 1995) partially complemented the nth deletion (Figure 3B). Taken together, these results indicate that Endo III facilitates RS1-integration independently of its normal BER function.
Endo III directly affects the integration reaction of CTXϕ
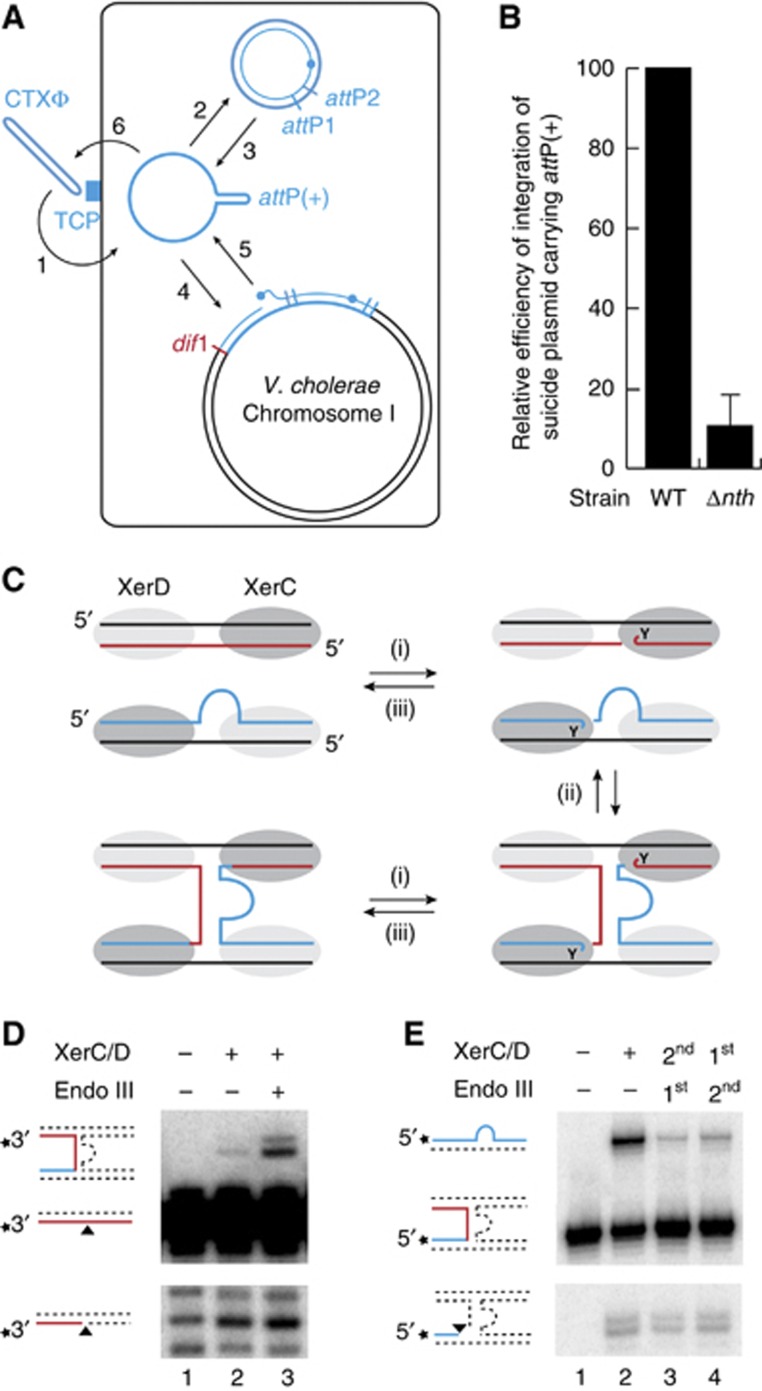
Because only RS1 was delivered in our screen, we could exclude that deletion of nth decreased the amount of ssDNA available for integration by influencing transduction, viral DNA packaging, phage particle production and/or subsequent re-infection of the cells during colony growth (Figure 4A). In addition, deletion of nth led to a 10 × decrease in the frequency of integration of non-replicative plasmids harbouring attP(+) (Figure 4B). In this context, the only available ssDNA substrate is the one transferred by conjugation, excluding the possibility that Endo III affected RS1-integration by interfering with the amount of ssDNA produced by RCR of RS1 (Figure 4A). Finally, deletion of nth also reduced the integration of the replication and integration region of two CTXϕ variants, El Tor and Classical, in which production of the RstA relaxase is governed by different regulators (data not shown; Safa et al, 2009). Taken together, these results suggested that Endo III facilitated toxigenic conversion of V. cholerae by a direct action on the integration process of CTXϕ.
Figure 4.
Endo III stabilizes the pseudo-HJ integration intermediate of CTXϕ. (A) Scheme of the life cycle of CTXϕ. 1: adsorption to the Toxin-Coregulated Pilus (TCP); 2: conversion of the circular (+) ssDNA phage genome into its dsDNA replicative form; 3: production of (+) ssDNA by RCR; 4: Integration of the ssDNA by Xer recombination; 5: production of new extrachromosomal copies of the phage genome by a process analogous to RCR between two tandem copies of the prophage; 6: encapsidation of the (+) ssDNA and production of new virions. (B) Efficiency of integration of a non-replicative vector harbouring RS1 attP. Integration was monitored directly after conjugation. Mean and standard deviation of at least three independent experiments. (C) Scheme of the three reversible steps of the recombination reaction leading to the formation of the attP/dif pseudo-HJ. Legend is as in Figure 1. (i) XerC-mediated cleavage; (ii) exchange of the liberated 5′-hydroxyl extremities; and (iii) self-re-ligation or re-ligation to the partner recombining strand. (D) In-vitro equilibrium reached after 4 h of incubation of a short radioactively labelled dif1 substrate and a longer cold attP(+) substrate with the Xer recombinases. The shorter migration product reveals cleavage of the recombining dif1 strand. The longer migration product reveals formation of the attP/dif pseudo-HJ formation. Unlabelled oligonucleotides cannot be directly visualized and do not interfere with the migration of the labelled fragments in the denaturing gel. Schemes of substrate and products are drawn on the left of the gels. A star indicates the position of the radioactive label. Unlabelled oligonucleotides are represented as dashed lines. (E) Cleaved and resolved products obtained after 1 h of incubation of labelled attP(+)/dif pseudo-HJs with the Xer recombinases. Samples were treated with proteinase K to avoid any interference of the covalent 3′-phosphotyrosyl bonds between XerC and the cleaved fragments in the oligonucleotide migration. First and second indicate the order of addition of Endo III and of the Xer recombinases.
Endo III stabilizes the pseudo-HJ integration intermediate of CTXϕ
The Xer recombination reaction leading to the formation of attP(+)/dif pseudo-HJs can be reconstituted in vitro using purified V. cholerae XerC and XerD recombinases and annealed oligonucleotides mimicking the attachment site of the different CTXϕ variants and their cognate chromosomal target (Das et al, 2010). Three steps can be defined in the process (Figure 4C): (i) a single strand in each of the two recombining sites is cleaved by the XerC recombinases engaged in the synaptic complex, which generates two 3′-phosphorotyrosyl recombinase/DNA covalent intermediates; (ii) the liberated 5′-hydroxyl extremities are exchanged; and (iii) they attack the phosphorotyrosyl bond of the partner site to form phosphodiester bonds. Each of these steps is reversible, leading to an equilibrium (Figure 4C).
Here, we used synthetic oligonucleotides that mimicked dif1 and the attachment site of RS1, attP(+), which is identical to the attachment site of the El Tor variant of CTXϕ. Cleavage of the dif1 recombining strand and its subsequent ligation to its partner strand in attP(+) were monitored by labelling its 3′ extremity: strand cleavage led to the appearance of a shorter migration product on a sequencing gel (Figure 4D, bottom gel, second lane); ligation to the partner strand, which was designed to have a longer extension on the 5′ side of the XerC-binding site, led to the appearance of a longer recombinant product (Figure 4D, top gel, second lane). As production of E. coli Endo III fully complemented the integration defect of V. cholerae Δnth cells (Figure 3B), it was initially used for our in vitro investigations.
Addition of Endo III significantly increased the amount of detected pseudo-HJ (Figure 4D, top gel, third lane) without affecting the formation of cleaved substrate (Figure 4D, bottom gel, third lane). This could result from either an increase in the rate of pseudo-HJ formation by XerC-catalysis or from a decrease in the rate of the reverse reaction, that is, the conversion of attP(+)/dif1 pseudo-HJs into attP(+) and dif1. To differentiate between these two possibilities, we reconstituted a synthetic attP(+)/dif1 pseudo-HJ by annealing four oligonucleotides. The oligonucleotide that mimicks the ligation of the 5′-side of the recombining strand in attP(+) to the 3′-end of the recombining strand in dif1 was labelled on its 5′ side: strand cleavage led to the appearance of a shorter migration product on a sequencing gel after proteinase K treatment (Figure 4E, bottom gel, second lane); ligation to its partner strand in the pseudo-HJ, which was designed to have a longer extension on the 3′ side of the XerC-binding site, led to the appearance of a longer recombinant product (Figure 4E, top gel, second lane). Addition of Endo III blocked the conversion of the pseudo-HJ back into substrate.
The same in-vitro activities were observed with purified V. cholerae Endo III (Supplementary Figure S2). Similarly, the K120Q mutant form of the V. cholerae protein was able to stabilize the pseudo-HJ integration intermediate of the phage in vitro, albeit with a lower efficiency (Supplementary Figure S2). Taken together, these results suggest that Endo III facilitates toxigenic conversion by stabilizing the pseudo-HJ integration intermediate of CTXϕ.
Endo III binds to HJs independently of XerC and XerD
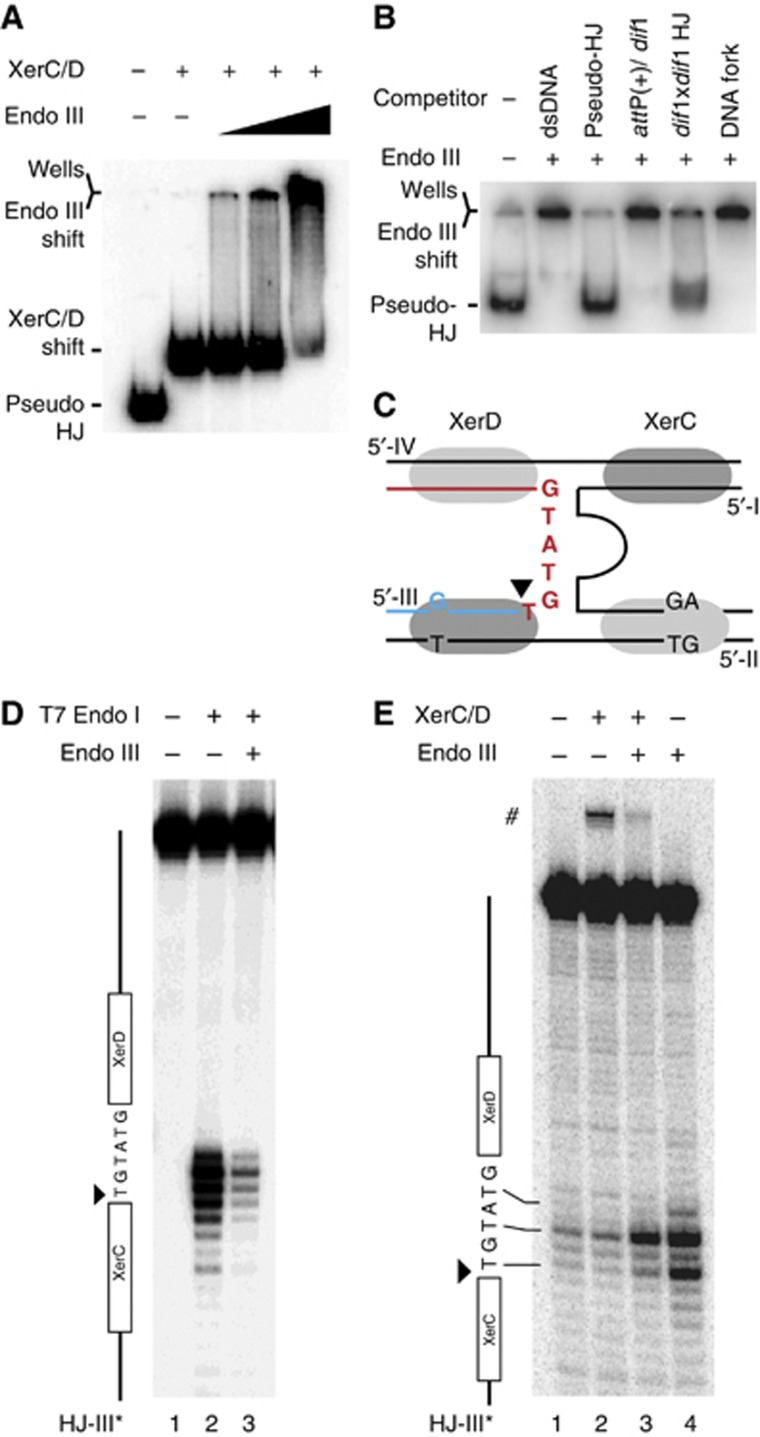
Stabilization of the attP/dif1 pseudo-HJ by Endo III suggested that the protein could either specifically interact with the Xer recombinases when engaged on a pseudo-HJ or with the pseudo-HJ itself, even if it should be partially masked by the recombinases. In both cases, we expected Endo III to be able to target the XerCD/pseudo-HJ complex. To test this possibility, we reconstituted XerCD/pseudo-HJ complexes by pre-incubating radioactively labelled pseudo-HJs with an excess amount of XerC and XerD. Conditions for full coverage of the pseudo-HJs were determined based on the conditions required for full coverage of attP and dif1 dsDNA substrates (Supplementary Figure S3). The XerCD/pseudo-HJ complex was visualized as a single retarded band in a gel shift assay (Figure 5A, second lane). Addition of Endo III led to the loss of the XerCD/pseudo-HJ band and the progressive accumulation of a very slow migrating complex (Figure 5A, lanes 3–5). In contrast, Endo III did not affect the migration of XerCD/attP(+) or XerCD/dif1 nucleoprotein complexes (Supplementary Figure S4). Taken together, these results indicate that Endo III can specifically interact with the XerCD/pseudo-HJ complex.
Figure 5.
Endo III binds with a high affinity to HJs and to pseudo-HJs. (A) EMSA of attP(+)/dif1 pseudo-HJs in the presence of XerC (150 nM), XerD (100 nM) and increasing concentrations of Endo III (0 lane 2, 50 nM lane 3, 100 nM lane 4 and 200 nM lane 5). (B) Competition between attP/dif pseudo-HJs and dsDNA, a dif1-attP(+) mix, dif1/dif1 HJs and DNA forks for Endo III binding. The amount (in mass) of competitor DNA was left constant in each lane. (C) Scheme of the attP(+)/dif1 pseudo-HJ substrate indicating the sequence of the overlap region of the recombining strand that was analysed in the T7 Endo I protection assay (D) and in the KMnO4 sensitivity assay (E). The analysed strand is indicated on top of the gels and a scheme of the strand is drawn on their left. # indicates the XerC strand exchange product.
In the absence of XerCD, addition of Endo III also retarded the migration of attP(+)/dif1 pseudo-HJs, demonstrating direct recognition of the nucleotidic part of the XerCD/pseudo-HJ complexes (Figure 5B, second lane). Addition of an excess amount of unlabelled pseudo-HJs (30-fold excess in weight) fully displaced Endo III from radioactive pseudo-HJs (Figure 5B, third lane). The complexes resisted similar quantities of competitor dsDNA (Figure 5B, second lane), further indicating that Endo III recognized a peculiarity of the attP(+)/dif1 pseudo-HJ structure (Figure 5C). However, we could exclude that the single mismatch in the XerC-binding site, the two contiguous mismatches in the XerD-binding site and the small ssDNA loop of attP(+) participated in the binding of Endo III to the pseudo-HJ because excess amounts of dif1 and attP(+) dsDNA substrates were unable to compete against them for Endo III binding (Figure 5B, fourth lane). In contrast, addition of an excess amount of unlabelled dif1/dif1 HJs displaced Endo III from pseudo-HJs, demonstrating that it recognized branched DNA structure (Figure 5B, fourth lane). Finally, DNA forks did not compete against pseudo-HJs (Figure 5B, sixth lane), suggesting preferential association to four-way DNA junctions in general. Indeed, Endo III retarded the migration of a HJ with arms unrelated to dif1 or attP(+) (Supplementary Figure S5).
Binding of Endo III to four-way DNA junctions was confirmed by a protection experiment against T7 Endo I: incubation of attP(+)/dif1 pseudo-HJs with T7 Endo I resulted in specific cleavages at their branch point (Figure 5D, second lane), which was repressed in the presence of Endo III (Figure 5D, third lane).
Because the bases at the branch point of dif/dif HJs or attP(+)/dif pseudo-HJs are protected from nucleases by the binding of XerC and XerD (see Arciszewska et al, 1995, 1997 and Figure 6C, respectively), we wondered if the capacity of Endo III to bind four-way DNA junctions was sufficient to explain its ability to bind pseudo-HJs covered by excess amounts of XerC and XerD (Figure 5A). Addition of Endo III increased the exposure of the bases at the branch point of attP(+)/dif1 pseudo-HJs to KMnO4, a chemical agent that preferentially attacks unstacked thymines (Figure 5E, fourth lane). In contrast, binding of XerC and XerD to pseudo-HJs did not alter the chemical modification pattern of KMnO4 (Figure 5E, second lane). Even though the samples were incubated at 4°C, XerC-catalysis was observed (Figure 5E, #), demonstrating efficient binding of the recombinases. Nevertheless, XerCD binding did not prevent Endo III from unstacking the bases at the junction of the four arms of the pseudo-HJ (Figure 5E, third lane).
Figure 6.
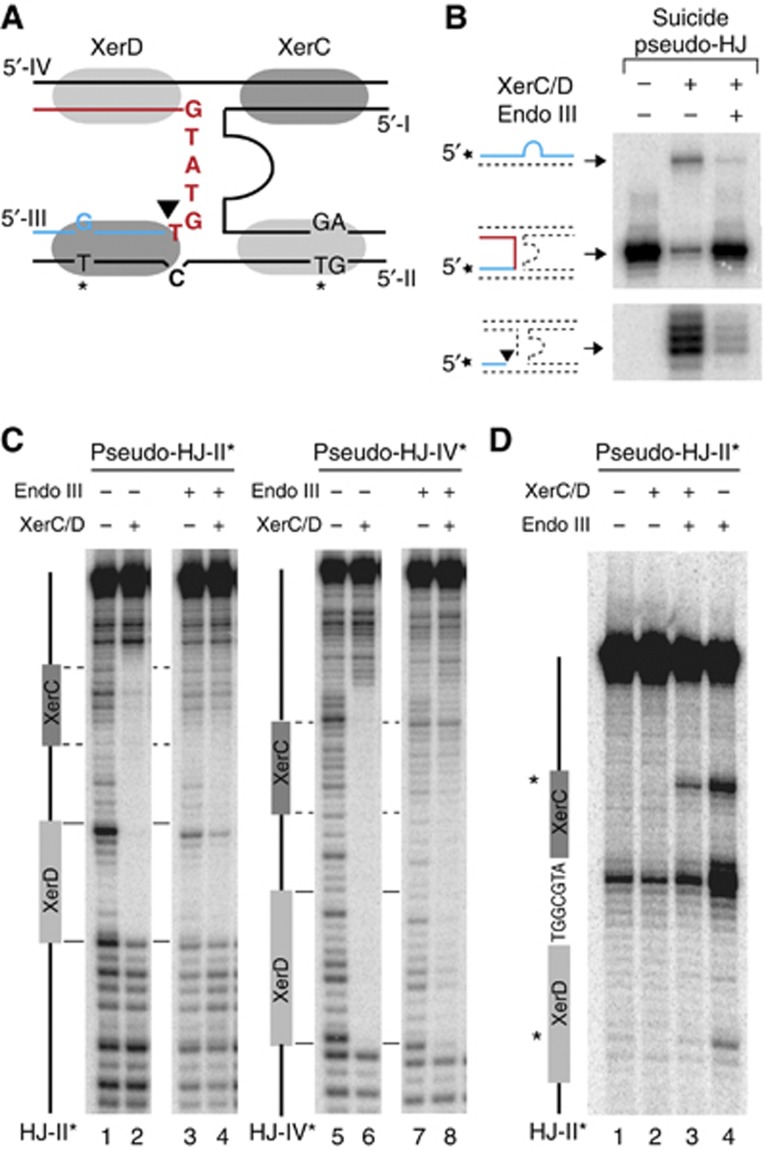
Endo III blocks XerC-catalysis on pseudo-HJ and displaces it from them. (A) Scheme of the suicide pseudo-HJ indicating the mismatch engineered to slow down re-ligation after XerC-cleavage. KMnO4 sensitive residues in the XerC- and XerD-binding attP(+) arms are indicated by a star. (B) Resolution of attP(+)/dif suicide pseudo-HJs. Legend as in Figure 4D. (C) DNase I protection and (D) KMnO4 sensitivity assays of the attP(+)/dif1 pseudo-HJ substrate. The analysed strand was labelled on its 5′ end. A scheme of the analysed strand is drawn on the left of the gels. KMnO4 sensitive residues in the XerC- and XerD-binding sites are indicated by a star.
Taken together, these results demonstrate that Endo III can bind four-way DNA junctions and modify the structure around their branch point, even if these junctions are initially covered by XerC and XerD.
Endo III binding blocks XerC-catalysis after HJ formation
We next investigated which step of the strand exchange reaction catalysed by XerC was prevented by the binding of Endo III to the XerCD/pseudo-HJ complex (Figure 4C): (i) XerC-mediated cleavage of the recombining strands of the pseudo-HJ (strand I and strand III of Figure 5C); (ii) exchange of the liberated 5′-hydroxyl extremities; and (iii) ;re-ligation to the partner recombining strand. By labelling one of the two recombining strands at its 5′ extremity, we can detect the amount of cleaved strand that is reached after a given time (Figure 4D). However, it only reflects the equilibrium between cleavage and re-ligation, whether strand exchange has occurred or not. Therefore, any decrease in the efficiency of cleavage (Figure 4C, (ii)) could be masked by a similar decrease in the efficiency of re-ligation (Figure 4C, (iii)). We circumvented this difficulty by using synthetic suicide pseudo-HJs that abolish any possibility for self-re-ligation and/or ligation to the partner recombining strand. As ligation events rely on the possibility to form stable base-pair interactions in the immediate vicinity of the cleavage site (Das et al, 2010), we reasoned that the introduction of a single base-pair mutation in the overlap region of the strand complementary to a particular recombining strand would be sufficient to achieve this (Figure 6A). Indeed, such a substrate considerably decreased XerC-mediated strand exchanges (Figure 6B, top gel, second lane) and led to the rapid accumulation of cleaved products (Figure 6B, bottom gel, second lane). Addition of Endo III led to a further diminution in the amount of detected strand exchanges, as expected (Figure 6B, top gel, third lane). Moreover, it almost completely blocked the accumulation of cleaved fragments (Figure 6B, bottom gel, second lane), indicating that Endo III inhibits the first step of the strand exchange catalysed by XerC. In agreement with this result, a lower amount of cleaved product was detected in the presence of Endo III at early time points, that is, at a time when the equilibrium between cleavage and re-ligation had less chances of being reach, with the non-suicide version of the attP(+)/dif pseudo-HJ (Supplementary Figure S6).
Finally, we investigated what modifications Endo III could operate on the structure of the XerCD/pseudo-HJ nucleoprotein complex to prevent XerC-catalysis. To this aim, we subjected attP(+)/dif1 pseudo-HJs to DNase I digestion (Figure 6C; Supplementary Figure S7). XerC and XerD almost completely blocked DNase I digestion along the four Xer-binding sites and at the branch point of the pseudo-HJ (Figure 6C, lanes 2 and 6). Addition of Endo III considerably decreased the protection of the two XerC-binding sites, suggesting that Endo III could displace the recombinase (Figure 6C, lanes 4 and 8). To further probe the changes operated in the XerCD/pseudo-HJ complex by the binding of Endo III, we made use of two mismatched thymines in the attP(+) arms of the pseudo-HJ substrate, one in the XerC-binding site and one in the XerD-binding site (Figure 6A): Endo III rendered both thymines sensitive to KMnO4 treatment in naked pseudo-HJs (Figure 6D, lane 4). However, only the thymine of the XerC-binding site remained hyper-reactive when pseudo-HJs were covered by excess amounts of XerC and XerD (Figure 6D, lane 3). Taken together, these results suggest that Endo III can penetrate the XerCD/pseudo-HJ complex and can dislodge XerC.
Discussion
Endo III is a co-factor required for efficient integration of CTXϕ
We previously demonstrated that XerC and XerD were sufficient to reconstitute the integration reaction between CTXϕ attP(+) sites and V. cholerae dif sites in vitro (Val et al, 2005; Das et al, 2010). However, addition of the recombinases only yielded very small amounts of pseudo-HJs (Figure 4D; Val et al, 2005; Das et al, 2010). The low efficiency of the reaction was not unexpected since it relied on XerC-catalysis, which leads to futile recombination cycles between dif sites (Figure 1A; Barre et al, 2000). In contrast, comparative genomics revealed a rapid and continuous drift in the arrangement and combination of the different CTXϕ variants harboured by epidemic strains of V. cholerae (Chun et al, 2009; Das et al, 2010; Mutreja et al, 2011), suggesting a very efficient integration strategy in the environment. This is partly explained by cooperative interactions between CTXϕ and several other IMEXs (Hassan et al, 2010; Das et al, 2011b). Nevertheless, introduction of CTXϕ-derivatives in cells devoid of IMEXs led to fully white colonies or colonies with large white sectors around a blue star-shaped centre on X-gal plates in our colorimetric integration assay (Figure 2B), indicating that the integration strategy of CTXϕ was intrinsically highly efficient. Therefore, we reasoned that additional host factors might facilitate the process. Correspondingly, we isolated several V. cholerae transposition insertion mutants in which CTXϕ-integration was reduced (Figure 2). The most promising mutants corresponded to insertions in the nth ORF, which codes for a ubiquitous enzyme of the BER pathway, Endo III. Subsequent analysis revealed that Endo III facilitated integration independently of its role in BER (Figure 3) and in vitro work demonstrated that it directly participated in the first step of the integration reaction of CTXϕ, which is mediated by XerC-catalysis (Figure 4).
A new mechanism to control site-specific recombination
The catalytic activity of Endo III was not required for CTXϕ-integration (Figure 2; Supplementary Figure S2), which could have suggested a role in the assembly of a synapse with a specific architecture via its binding to accessory sequences flanking the core recombination sites. Such a role is played by Fis and IHF during phage λ integration and by PepA and ArgR/ArcA during plasmid dimer resolution. However, Endo III does not bind to specific DNA motifs and the DNA molecules employed in our in vitro reactions lacked any homologies with the phage (+) ssDNA and the genomic DNA of V. cholerae apart from the attP(+) and dif1 sites. Indeed, our study unveiled a novel mechanism for the control of DNA site-specific recombination (Figure 7): we found that Endo III did not promote formation of the pseudo-HJ integration intermediate but rather stabilized it by impeding new rounds of strand exchanges by XerC-catalysis once it had been formed (Figure 4). Using a suicide pseudo-HJ DNA substrate, we further showed that Endo III inhibited pseudo-HJ resolution by blocking its cleavage by XerC (Figure 6). We observed that Endo III exclusively associated with the XerCD/pseudo-HJ nucleoprotein complex and not with the XerCD/attP(+) or XerCD/dif complex (Figure 5; Supplementary Figure S4), which explains why the XerC-strand exchange leading to the formation of the pseudo-HJ was left unaffected.
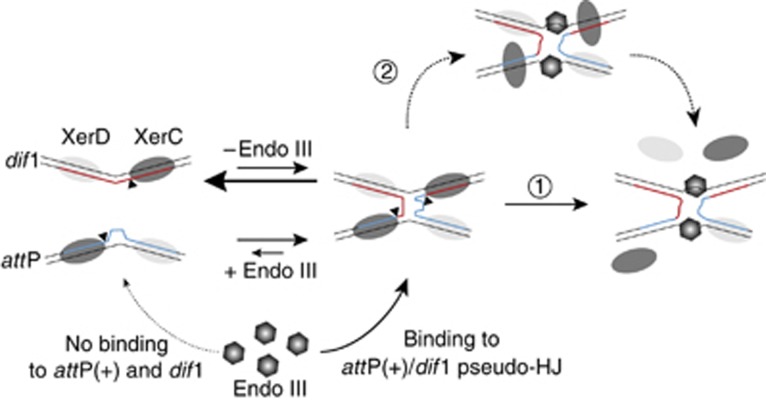
Figure 7.
A model for the role of Endo III in the stimulation of CTXΦ-integration. CTXΦ-integration result from the reversible formation of a pseudo-HJ intermediate between dif and attP(+) by XerC-catalysis. The length and width of the arrows indicate which of the two opposite reactions is favoured in the absence or presence of Endo III. Endo III (hexagons) inhibits XerC-cleavage after pseudo-HJ formation because it specifically recognizes and binds to four-way DNA junctions. Endo III binding results in the final displacement of the XerC recombinases. This might be due to a competition between XerC and Endo III for binding to the pseudo-HJ (arrow 1). Our results are in favour of a second pathway in which Endo III binding to the pseudo-HJ induces structural changes that perturb interactions between XerC and the DNA or XerC and XerD, leading to its destabilization (arrow 2).
This mechanism is reminiscent of the mode of action of small peptide inhibitors of phage λ site-specific recombination, which were isolated by screening synthetic combinatorial peptide libraries (Klemm et al, 2000). These peptides trap HJs by binding in the centre of the recombinase-bound DNA junction and interacting with solvent-exposed bases near the junction branch point. Correspondingly, we observed that Endo III recognized four-way DNA junctions independently of XerC and XerD and of any specific sequence (Figure 5B; Supplementary Figure S5). Endo III is monomeric whereas proteins binding to HJs are generally multimeric (Declais and Lilley, 2008; Vitoc and Mukerji, 2011). Future work will be needed to solve the exact mechanism of recognition of four-way junctions by Endo III.
How Endo III achieves XerC-cleavage inhibition also remains to be more extensively explored. Enzymatic DNA footprints suggest displacement of the XerC recombinases from their specific DNA-binding sequences on the pseudo-HJ (Figures 5 and 6). However, pre-incubation of pseudo-HJs with Endo III before the addition of the Xer recombinases did not potentiate resolution inhibition (Figure 4E, lanes 3 and 4), indicating that Endo III does not directly compete with the recombinases for DNA binding (Figure 7, arrow 1). Therefore, we favour the idea (Figure 7, arrow 2) that Endo III can bind to pseudo-HJs covered by the Xer recombinases (as suggested in Figure 5A), induce DNA structure changes in the central region of the pseudo-HJs (as suggested in Figure 5E), which eventually inhibits the catalysis of the recombinases and promotes their dissociation. As XerC displays a much weaker affinity for dif and attP than XerD (Supplementary Figure S3), the primary effect of Endo III binding would be the loss of XerC-catalysis (as suggested in Figure 4) and its subsequent dissociation (as suggested in Figure 6).
Implications for our understanding of the life cycle of CTXϕ
The replicative form of CTXϕ is not stably propagated. In addition, CTXϕ attP(+) is a complex site, which is created by the folding of its (+) ssDNA genome. As such, it is expected to be a very transient structure in the cell: packaging of the (+) ssDNA genome of CTXϕ into new virus particles and conversion into dsDNA limit the amount of substrate that is available for integration, which probably explains the crucial importance of Endo III for CTXϕ-integration (Figure 3A). Correspondingly, we did not observe any effect of the nth deletion on the integration of VGJϕ, an IMEX that displays a very different life cycle than the one of CTXϕ, using our in vivo suicide integration assay. Similarly to CTXϕ, VGJϕ-integration results from a single pair of strand exchanges, which is catalysed by XerC, and the resulting pseudo-HJ needs to be converted into product by replication (Das et al, 2011b). However, the attachment site of VGJϕ is found on the dsDNA replicative form of the phage, which is stably maintained (Das et al, 2011b). This allows for repeated events of synapsis and strand exchange to occur, masking any possible effect of Endo III on the life time of the VGJϕ pseudo-HJ integration intermediate.
A drawback to the integration strategy of CTXϕ is that the replicative form of the phage cannot be generated by prophage excision, which would eventually stop virion production (Figure 4A; Val et al, 2005). However, most V. cholerae toxigenic strains harbour tandemly arranged copies of the prophage (Chun et al, 2009; Das et al, 2011a, 2011b; Mutreja et al, 2011), which permits production of new (+) ssDNA by a process analogous to RCR (Figure 4A; Moyer et al, 2001). To date, it was unknown how these tandem copies were generated. They could result from a single integration event, the phage DNA being duplicated by an uncharacterized DNA repair and/or replication event, or from multiple successive integration events (Figure 1C). Our observation that the number of integrated copies was reduced in Δnth cells compared with nth+ cells (Supplementary Figure S1) is in favour of the latter scenario since the average number of integrated copies should have been independent of the efficiency of integration if the former hypothesis had been correct. We conclude that Endo III permits the formation of tandem prophage arrays by increasing the chances for success of independent integration events, which is crucial for the dissemination of new phage particles in the environment.
Possible implications of Endo III in other recombination processes
Both in vivo and in vitro, E. coli Endo III had the same properties than its V. cholerae orthologue in terms of HJ-binding and Xer recombination regulation (Figure 3; Supplementary Figure S2). Therefore, it is very likely that Endo III also serves as a co-factor in the integration of enterobacterial IMEXs that are related to CTXϕ, such as CUS-1ϕ (Gonzalez et al, 2002) and Ypfϕ (Derbise et al, 2007). Endo III was also able to impede the resolution of dif1/dif1 HJs by XerC-catalysis in vitro (Supplementary Figure S8), raising the possibility that it could act on the integration intermediate of IMEXs that would be unrelated to CTXϕ. We observed that deletion of nth did not dramatically affect the integration of VGJϕ, a representative of a second category of IMEXs found in V. cholerae (Das et al, 2011a). Nevertheless, it will be important to address the possible implication of Endo III in the integration strategies and life cycles of the other classes of IMEXs that are yet to be explored, such as Neisseriales genomic islands (Dillard and Seifert, 2001) and cryptic elements related to phages or plasmids (Hassan et al, 2010). In addition to CTXϕ and CTXϕ-related phages, several mobile elements rely on a recombination event between ssDNA and dsDNA molecules. As Endo III targets four-way DNA junctions independently of the presence of XerC and XerD, it is tempting to speculate that it might participate in the processing of several such elements (Figure 5; Supplementary Figure S5). In particular, it would be interesting to study the possible implication of Endo III in the integration of integron cassettes, which is most similar to the integration strategy of CTXϕ since it relies on the formation of a pseudo-HJ between the ssDNA attachment sites of the cassettes and their dsDNA attachment site on the host genome by a tyrosine recombinase (Bouvier et al, 2005; MacDonald et al, 2006). Finally, our observation that Endo III, which is shared by most organisms, including humans (Aspinwall et al, 1997; Denver et al, 2003), binds to HJs in general (Supplementary Figure S5) provides a direct connection between BER and recombinational repair. Such a connection might not be fortuitous since homologous recombination and BER are both required for fork progression through alkylated DNA (Vazquez et al, 2008). Thus, it is tempting to propose that Endo III might participate in the balance between the different processes that take care of HJs in the cell (Chen et al, 2001; Wu and Hickson, 2003; Ip et al, 2008; Taylor and McGowan, 2008; Andersen et al, 2009; Fekairi et al, 2009; Svendsen et al, 2009; Wechsler et al, 2012).
Materials and methods
Strains, plasmids, oligonucleotides and proteins
Strains, plasmids and oligonucleotides are described in Supplementary Tables S2, S3 and S4, respectively. All V. cholerae mutants were constructed by allele exchange methods using derivatives of a suicide vector carrying rpsL as a counter selectable marker (Skorupski and Taylor, 1996; Philippe et al, 2004). Engineered strains were confirmed by PCR and sequencing. For ectopic complementation, the ORFs of E. coli nth, E. coli nei and V. cholerae nth were cloned downstream of the arabinose promoter on a pSC101 derivative. The K120Q mutation of Endo III was introduced by site-specific mutagenesis. All the oligonucleotides that served to create synthetic recombination substrates were purified by PAGE. T4 DNA polynucleotide kinase and [32P] γ-ATP were used for 5′-end labelling of oligonucleotides; Klenow exo- and [32P] α-dCTP were used for 3′-end labelling. XerC and XerD were prepared as previously described (Das et al, 2010, 2011b). E. coli Endo III, T7 endo I and Dnase I were purchased from New England Biolabs. V. cholerae Endo III and the K120Q mutant were produced in JM101 strains. Cells were grown at 37°C until OD600 nm 0.5 then induced with 0.5 mM IPTG for five additional hours. Cells were collected and re-suspended in 40 mM Hepes (pH 7.5), 250 mM NaCl, 1 mM EDTA, 1 mM DTT and 5% glycerol, treated with lysozyme for 45 min on ice and broken with a Carver press. Lysates were sonicated and cleared by centrifugation for 45′ at 25 000 g. The proteins were purified on a ÄKTA purifier system, using HITRAP MonoQ, Heparin HP MonoS 5/50 GL columns (GE Healthcare). They were stored in 25 mM Tris–HCl pH 7.4, 250 mM NaCl, 1 mM DTT, 0.1 mM EDTA and 20% glycerol at −20°C. Protein purification was estimated to be over 95% according to Coomassie blue staining (Supplementary Figure S9). Protein concentrations were evaluated by the Bradford methods using BSA as a standard.
Mariner transposon-mutagenesis, screening and mutant characterization
Over 162 000 clones were pooled in mariner transposon-mutagenesis bank of the V. cholerae reporter strain. Each bank was conjugated with a chloramphenicol resistant (CmR) derivative of RS1 and individual colonies were selected on X-Gal, IPTG and chloramphenicol plates after 24 h of growth at 37°C. Candidate clones were cured from RS1 by overnight growth in the absence of antibiotic and their integration defect was quantitatively monitored. The insertion was mapped by direct sequencing of the DNA flanking the point of insertion of the mariner transposons, which was amplified by arbitrary-random PCR (O’Toole and Kolter, 1998).
In-vivo integration assays
In order to compare the efficiency of integration of RS1 in different genetic backgrounds, we monitored the proportion of fully white colonies (lysogenics) over the total of number of CmR clones present in LB cultures that were grown at 37°C for 14 h after RS1 conjugation. The frequency of integration of a suicide CmR vector harbouring attP(+) was calculated as the number of CmR clones per number of conjugants. Co-conjugation of a replicative pSC101 vector carrying an ampicillin resistance gene was used to estimate the total number of conjugants.
In-vitro assays
The standard reaction buffer contained 25 mM Tris–HCl (pH 7.4), 100 mM NaCl, 1 mM EDTA, 0.1 μg/ml BSA, 150 nM of XerC and 100 nM of XerD. Recombination reactions were performed in the presence of 5 nM of each of the cold and radioactively labelled recombination substrates and 40% glycerol. They were incubated for 3 h at 37°C. HJ-resolution assays were performed in the presence of 10 nM of 5′-labelled HJs and 10% glycerol. They were incubated for 1 h, stopped by the addition of 0.1% SDS and 1 mg/ml Proteinase K and heated for 1 h at 55°C. Unless otherwise indicated, Endo III was added at 200 nM final concentration immediately after the addition of XerC and XerD. DNA-binding assays were performed in the presence of 10 nM of radio-labelled probe and 10% glycerol. XerC and XerD were used at 150 nM and 100 nM concentration, respectively. Complexes were assembled during 5 min at room temperature and analysed by electrophoresis on a 5% polyacrylamide, 1 × TBE gel at 4°C. Complexes for T7 Endo I and DNase I footprints and for KMnO4 assays were assembled in the presence of 100 nM of non-specific DNA competitor. DTT was omitted for KMnO4 treatment. Complexes were treated with either 1 unit of T7 Endo I and 5 mM MgCl2 for 2 min at 37°C, 0.1 unit of DNase I and 5 mM MgCl2 for 1 min at room temperature or 8 mM KMnO4 for 30 s at room temperature. The reactions were quenched with 200 mM NaCl, 0.1% SDS, 25 mM EDTA, 0.5 mg/ml Proteinase K and 100 μg/ml glycogen, and ethanol precipitated. For KMnO4 treatment, the DNA was further treated with 1 M piperidine for 30 min at 90°C and ethanol precipitated a second time. They were analysed on a 10% polyacrylamide DNA sequencing gel.
Supplementary Material
Acknowledgments
We thank S Boiteux, L Sperling, C Possoz and the members of the team for helpful discussions. This work was supported by the Fondation pour la Recherche Médicale [Equipe 2007] and the Agence Nationale pour la Recherche [ANR-09-BLAN-0258; ANR-11-BSV3-024].
Author contributions: JB and F-XB designed the research; JB and CS performed the research; JB and F-XB analysed the data and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J (2009) Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell 35: 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska L, Grainge I, Sherratt D (1995) Effects of Holliday junction position on Xer-mediated recombination in vitro. EMBO J 14: 2651–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska LK, Baker RA, Hallet B, Sherratt DJ (2000) Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J Mol Biol 299: 391–403 [DOI] [PubMed] [Google Scholar]
- Arciszewska LK, Grainge I, Sherratt DJ (1997) Action of site-specific recombinases XerC and XerD on tethered Holliday junctions. EMBO J 16: 3731–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinwall R, Rothwell DG, Roldan-Arjona T, Anselmino C, Ward CJ, Cheadle JP, Sampson JR, Lindahl T, Harris PC, Hickson ID (1997) Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc Natl Acad Sci USA 94: 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D (2002) FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108: 195–205 [DOI] [PubMed] [Google Scholar]
- Barre FX, Aroyo M, Colloms SD, Helfrich A, Cornet F, Sherratt DJ (2000) FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev 14: 2976–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M, Demarre G, Mazel D (2005) Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J 24: 4356–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregu M, Sherratt DJ, Colloms SD (2002) Accessory factors determine the order of strand exchange in Xer recombination at psi. EMBO J 21: 3888–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J, Martinez E, Suzarte E, Rodriguez BL, Marrero K, Silva Y, Ledon T, del Sol R, Fando R (2003) VGJ phi, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTX phi. J Bacteriol 185: 5685–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH (2001) Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell 8: 1117–1127 [DOI] [PubMed] [Google Scholar]
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Munk AC, Chertkov O, Meincke L, Saunders E, Walters RA, Huq A et al. (2009) Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA 106: 15442–15447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colloms SD, Bath J, Sherratt DJ (1997) Topological selectivity in Xer site-specific recombination. Cell 88: 855–864 [DOI] [PubMed] [Google Scholar]
- Colloms SD, McCulloch R, Grant K, Neilson L, Sherratt DJ (1996) Xer-mediated site-specific recombination in vitro. EMBO J 15: 1172–1181 [PMC free article] [PubMed] [Google Scholar]
- Colloms SD, Sykora P, Szatmari G, Sherratt DJ (1990) Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J Bacteriol 172: 6973–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Bischerour J, Barre FX (2011a) Molecular mechanism of acquisition of the cholera toxin genes. Indian J Med Res 133: 195–200 [PMC free article] [PubMed] [Google Scholar]
- Das B, Bischerour J, Barre FX (2011b) VGJφ-integration and excision mechanisms contribute to the genetic diversity of Vibrio cholerae epidemic strains. Proc Natl Acad Sci USA 108: 2516–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Bischerour J, Val M-E, Barre FX (2010) Molecular keys of the tropism of integration of the cholera toxin phage. Proc Natl Acad Sci USA 107: 4377–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declais AC, Lilley DM (2008) New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr Opin Struct Biol 18: 86–95 [DOI] [PubMed] [Google Scholar]
- Denver DR, Swenson SL, Lynch M (2003) An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol 20: 1603–1611 [DOI] [PubMed] [Google Scholar]
- Derbise A, Chenal-Francisque V, Pouillot F, Fayolle C, Prevost MC, Medigue C, Hinnebusch BJ, Carniel E (2007) A horizontally acquired filamentous phage contributes to the pathogenicity of the plague bacillus. Mol Microbiol 63: 1145–1157 [DOI] [PubMed] [Google Scholar]
- Dillard JP, Seifert HS (2001) A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol 41: 263–277 [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR 3rd, Russell P, Fuchs RP, McGowan CH, Gaillard PH (2009) Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MD, Lichtensteiger CA, Caughlan R, Vimr ER (2002) Conserved filamentous prophage in Escherichia coli O18:K1:H7 and Yersinia pestis biovar orientalis. J Bacteriol 184: 6050–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallet B, Arciszewska LK, Sherratt DJ (1999) Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol Cell 4: 949–959 [DOI] [PubMed] [Google Scholar]
- Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM (2010) Satellite phage TLCphi enables toxigenic conversion by CTX phage through dif site alteration. Nature 467: 982–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KE, Waldor MK (2002) Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417: 656–659 [DOI] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC (2008) Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361 [DOI] [PubMed] [Google Scholar]
- Klemm M, Cheng C, Cassell G, Shuman S, Segall AM (2000) Peptide inhibitors of DNA cleavage by tyrosine recombinases and topoisomerases. J Mol Biol 299: 1203–1216 [DOI] [PubMed] [Google Scholar]
- Kono N, Arakawa K, Tomita M (2011) Comprehensive prediction of chromosome dimer resolution sites in bacterial genomes. BMC Genomics 12: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN (2006) Structural basis for broad DNA-specificity in integron recombination. Nature 440: 1157–1162 [DOI] [PubMed] [Google Scholar]
- Moyer KE, Kimsey HH, Waldor MK (2001) Evidence for a rolling-circle mechanism of phage DNA synthesis from both replicative and integrated forms of CTXphi. Mol Microbiol 41: 311–323 [DOI] [PubMed] [Google Scholar]
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JL, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J et al. (2011) Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477: 462–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28: 449–461 [DOI] [PubMed] [Google Scholar]
- Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D (2004) Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51: 246–255 [DOI] [PubMed] [Google Scholar]
- Safa A, Nair GB, Kong RY (2009) Evolution of new variants of Vibrio cholerae O1. Trends Microbiol 18: 46–54 [DOI] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK (1996) Positive selection vectors for allelic exchange. Gene 169: 47–52 [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW (2009) Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ER, McGowan CH (2008) Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc Natl Acad Sci USA 105: 3757–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J 14: 4108–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val M-E, Bouvier M, Campos J, Sherratt DJ, Cornet F, Mazel D, Barre F-X (2005) The single stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol Cell 19: 559–566 [DOI] [PubMed] [Google Scholar]
- Val M-E, Kennedy SP, El karoui M, Bonné L, Chevalier F, Barre F-X (2008) FtsK-dependent dimer resolution on multiple chromosomes in the pathogen Vibrio cholerae. PLoS Genet 4: e1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez MV, Rojas V, Tercero JA (2008) Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair 7: 1693–1704 [DOI] [PubMed] [Google Scholar]
- Vitoc CI, Mukerji I (2011) HU binding to a DNA four-way junction probed by Forster resonance energy transfer. Biochemistry 50: 1432–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor MK, Mekalanos JJ (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914 [DOI] [PubMed] [Google Scholar]
- Wechsler T, Newman S, West SC (2012) Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature 471: 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID (2003) The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.