Abstract
Homologous DNA recombination is a fundamental, regenerative process within living organisms. However, in most organisms, homologous recombination is a rare event, requiring a complex set of reactions and extensive homology. We demonstrate in this paper that Beta protein of phage λ generates recombinants in chromosomal DNA by using synthetic single-stranded DNAs (ssDNA) as short as 30 bases long. This ssDNA recombination can be used to mutagenize or repair the chromosome with efficiencies that generate up to 6% recombinants among treated cells. Mechanistically, it appears that Beta protein, a Rad52-like protein, binds and anneals the ssDNA donor to a complementary single-strand near the DNA replication fork to generate the recombinant. This type of homologous recombination with ssDNA provides new avenues for studying and modifying genomes ranging from bacterial pathogens to eukaryotes. Beta protein and ssDNA may prove generally applicable for repairing DNA in many organisms.
Homologous recombination has been described most thoroughly in the bacterium Escherichia coli. Recombination with linear DNA in E. coli is limited because the transformed linear DNA is rapidly degraded by the bacterial RecBCD nuclease (1). Several strategies for recombination of linear double-stranded DNA (dsDNA) fragments in RecBCD-defective derivatives of E. coli have been described (2, 3). However, in these mutants, the frequency of recombination is very low and requires thousands of base pairs of homology. Newly developed phage-mediated recombination systems, like the bacteriophage λ Red system, permit efficient linear DNA recombination even in wild-type E. coli and can use short (50-bp) homologies to generate recombinants (4–7). This λ Red-mediated recombination requires the phage Gam, Exo, and Beta functions but does not require E. coli RecA function. RecA or a RecA-like function is normally central to all DNA recombination activities (8). The three λ recombination functions each contribute to provide homologous recombination activity with linear dsDNA. λ Gam inhibits activities of RecBCD, including the nuclease activity, thus protecting electroporated linear dsDNA from degradation (9, 10). λ Exo is a dsDNA-dependent exonuclease that digests in the 5′ to 3′ direction, leaving 3′ overhangs that act as substrates for recombination (11). λ Beta is a single-stranded DNA (ssDNA) binding protein that promotes annealing of complementary single strands and has been shown to mediate strand exchange in vitro (12, 13).
ssDNA tails are key intermediates in homologous recombination between dsDNAs (14, 15). We expected that in the λ Red system, Exo could generate 3′ ssDNA overhangs to which Beta could bind and finish the recombination process. Here, we show Beta can promote homologous recombination between the chromosome and a synthetic ssDNA donor.
Materials and Methods
Genotypes of Strains Used.
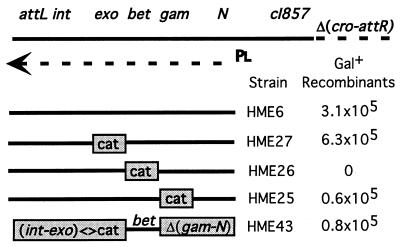
Strains were constructed either by λ Red recombination (4), or by standard P1 transduction. HME5 is W3110 Δ(argF-lac)U169 {λcI857 Δ(cro-bioA)} (see Fig. 2 for description of λ). HME5 is the parent of the following strains with additional markers shown: HME6, galKtyr145UAG; HME9, galKtyr145UAG ΔtyrTV<>cat; HME10, galKtyr145UAG ΔtyrTV<>cat Δ(srl-recA)301∷Tn10; HME25, galKtyr145UAG {λ Δgam<>cat}; HME26, galKtyr145UAG {λ Δbet<>cat}; HME27, galKtyr145UAG {λ Δexo<>cat}; HME31, galK<>catsacB; HME36, Δgal<>galETKM; HME41, INgal<>(galM+Ktyr145UAGT+E+); HME43, galKtyr145UAG {λ Δ(exo-int)<>cat Δ<>(gam-N)}. Note that we are using the symbol <> to indicate a replacement generated by homologous recombination technology.
Figure 2.
The defective λ prophage. The prophage contains λ genes from cI to int (4). Some λ genes are not shown for clarity. A deletion (Δ) removes cro through attR from the right side of the prophage. pL and the arrow indicate the early left promoter and transcript used to express Gam, Beta, and Exo. The CI857 repressor prevents pL expression at 32o but is inactive at 42o. attL and attR indicate the left and right junction of the prophage. Gene replacements with the cat cassette and a simple deletion Δ(gam-N) are shown below the prophage map and are located to the left of the strain name they represent. For the experiment, cells were induced for 15 min at 42°C and electroporated with 200 ng of the 70-base oligo of Fig. 1. The Gal+ recombinants were selected on minimal galactose medium, and the numbers are normalized per 108 cells surviving each electroporation.
Materials.
Oligonucleotides (oligos) were provided by Life Technologies (Frederick, MD) as salt free but were not purified in any special way. We note, however, that oligonucleotides obtained from some other sources were not as active as the ones we used here. The sequences of the most active of the two oligos used for curing each Tn10 insertion is indicated below: cysI(cw)GTGTCATTCCCGACTTGCCCGCTGGCGATGGCGGAAGCAGAGCGTTTCCTGCCGTCTTTTATCGACAACA; metC(cw)TACAACAAGCGATGTGTGAACTGGAAGGTGGCGCAGGCTGCGTGCTATTTCCCTGCGGGGCGGCAGCGG; thrA(cc)TTGGTGATTTTGGCGGGGGCAGAGAGGACGGTGGCCACCTGCCCCTGCCTGGCATTGCTTTCCAGAATATCGGCAACACG; trpC(cc)CAAAAGGCGTGATCCGTGATGATTTCGATCCAGCACGCATTGCCGCCATTTATAAACATTACGCTTCGGC; and nadA(cc)AATCTCACCCCAGCGACTAACAAAGTAGAAGCGGGATGCTTTGCACCGAAGCGCGCCATTTCCAGAGAAT.
Construction of HME31, a Genetic Insertion.
The cat-sacB cassette, carrying genes for chloramphenicol resistance and sucrose sensitivity, was amplified by PCR using oligos that contain homologies with the galK gene. The oligos are GACCGTTAAGCGCGATTTGTGCGCCGTCCAGCGGCAGATG/ATCAAAGGGAAAACTGTCCA and ACTGGAAGTCGCGGTCGGAACCGTATTGCAGCAGCTTTAT/AAAATGAGACGTTGATCGGCACG. The “/” indicates the junction between the galK homology on the 5′ end and the cat-sacB primers on the 3′end of each oligo. The PCR product was used for homologous recombination in the strain HME6. Recombinants that had inserted the cassette into galK were selected as chloramphenicol-resistant colonies and tested as described (4).
Construction of HME41, a Genetic Inversion.
To create an inversion of the gal operon, the operon was first deleted by recombination with the gal<>del-specific oligonucleotide (see below) into strain HME5 to create HME36 ΔgalETKM. Cells carrying this gal operon deletion were selected on minimal media with 0.1% glycerol, 0.2% 2-deoxygalactose with biotin (4). Presence of the deletion was verified by PCR with flanking primers. The gal<>del oligonucleotide includes 41 bases of sequence upstream of the gal operator, followed by 40 bases of sequence downstream of galM. The 81-base gal<>del sequence is 5′TCGCGCATAAAAAACGGCTAAATTCTTGTGTAAACGATTCC/GGCTTTATCTCATATTGTTCAAATCACCAGCAAACACCGA, where the slash indicates the deletion junction. To reinsert the gal operon in the inverted orientation, we created a PCR product of the gal operon in which homology segments found upstream (44 bases) and downstream (43 bases) flanked the complete operon, which was in inverted orientation with respect to these flanking chromosomal homology regions. PCR primers were 5′CATTCATAAACCCTCTGTTTTATAATCACTTAATCGCGCATAAAA/CTCATGACGAGGGCGTAAC and 5′TATGTCGGTGTTTGCTGGTGATTTGAACAATATGAGATAAAGCC/AACGGCTAAATTCTT GTGTAAAC, where slashes indicate the novel joints between the inversion and the chromosome. This PCR product was crossed into the Gal deletion strain HME36, generating Gal+ recombinants. The presence of the gal inversion was confirmed in four of four Gal+ isolates tested by PCR with primers specific to either the inverted or wild-type orientation of the gal operon. Finally, the galK amber mutation replacing codon tyr-145 was introduced by recombination as described previously (4). The experiments described here used the gal operon inversion strain HME41, which is Δ(argF-lac)U169 INgal(galM+Ktyr145UAGT+E+) {λ cI857 Δ(cro-bioA)}.
Construction of Strains Carrying Tn10.
DY374 is W3110 nadA∷Tn10 {λcI857 Δ(cro-bioA)}. Strains HME52 through HME55 were constructed by P1 transduction of the Tn10 insertions into a nadA+ derivative of DY374 from the following strains selecting for tetracycline resistance: CAG18442 thr34∷Tn10, CAG18455 trpB83∷Tn10, CAG12173 cysC95∷Tn10, CAG18475 metC162∷Tn10 (16).
Construction of HME43.
To create the “Beta only” strain HME43, int through exo were first replaced in HME6 by cat (4), and gam through N were subsequently deleted by using ssDNA recombination with the oligo ACATCAGCAGGACGCACTGACCACCATGAAGGTGA/TGATATTGATTCAGAGGTATAAAACGAATGAGTAC, in which the slash represents the deletion junction. In this case, deletion recombinants were selected as temperature-resistant survivors because the gam through N segment causes cell death when expressed constitutively.
Results
Homologous Recombination with ssDNA.
Activities of the λ Red proteins suggested that cells expressing the Red functions might be able to use a simple single-stranded oligo as substrate for recombination. To test this idea, we asked whether the λ Red system could use a 70-base single-stranded oligo to correct the amber mutation galKtyr145am (4) located on the bacterial chromosome. For this purpose, we synthesized an oligo corresponding to the non-template DNA strand of galK (see Fig. 1). Strains HME6 and HME9 carrying the amber mutation were induced for λ Red expression by shifting the culture to 42°C for 15 min and were prepared for electroporation as described by Yu et al. (4). These cells were then electroporated with the “wild-type” oligo and plated on minimal galactose plates to select for Gal+ recombinants. We obtained ≈2 × 105 Gal+ recombinants per 108 viable cells by using 10 ng of oligo. Another 70-base oligo encoding the same amino acids, but carrying changes at the third position of 13 codons, failed to generate any Gal+ recombinants, demonstrating that recombination depended upon homology between the oligo and galK.
Figure 1.
The galK DNA segment. At the top is a sequence from the galK gene illustrating an amber mutation galKam. The sequence shown encodes amino acids 135 through 156. The amber mutation is at tyr-145. Below is a 70-base oligo with a sequence identical, except for one base, to the non-template strand in galKam. It was used to replace the mutation with the wild-type allele.
To determine the minimum length of homology for recombination with a single-stranded oligo, we compared the ability of oligos from 20 to 70 bases long to generate Gal+ recombinants in the galKtyr145am strain HME9. Compared with the 70-base oligo, which gave 2.2 × 105 recombinants per 108 viable cells, the 60-, 50-, and 40-base oligos recombined less efficiently, yielding 4–5 × 104 recombinants per 108 cells. Recombination efficiency dropped to 5 × 103 recombinants per 108 viable cells with the 30-base oligo, and Gal+ colonies were near background levels with a 20-base oligo. A similar requirement for homology length was observed for recombination of linear dsDNA using the λ Red system (4). The drop in recombination efficiency from 40 to 30 bases is consistent with the observation that Beta binds poorly to oligos shorter than 36 bases (17).
Our demonstration that recombination with ssDNA could change a single base on the chromosome at galK led us to test whether deletions could also be engineered by using ssDNA. To compare the efficiency of these two types of changes using our protocol, we first inserted a 3.3-kbp chloramphenicol resistance cassette at the galK amber site in HME6, creating strain HME31. HME6 and HME31 were induced for Red functions and electroporated with the 70-base oligo described above. Gal+ recombinants are generated with the same frequency in both strains, demonstrating that single strand-mediated recombination can create a 3.3-kbp deletion as efficiently as a single base change.
ssDNA Recombination Is RecA Independent.
We reported that recombination of dsDNA substrates catalyzed by the λ Red system had a modest requirement for RecA (4), with recombination depressed less than 10-fold in RecA-defective strains. To determine whether RecA is required for λ Red-mediated recombination with ssDNA, we compared recombination efficiency of the 70-base oligo in recA+ or recA− strains. Recombination efficiency in the recA− strain was, if anything, slightly greater than that seen in the isogenic recA+ strain, with 2.6 × 105 versus 1.1 × 105 Gal+ recombinants, respectively, per 108 cells, indicating that λ Red-mediated recombination of ssDNA is completely RecA independent.
ssDNA Recombination Requires Only Beta.
To determine which λ functions are required for recombination with ssDNA, we created a series of replacements of λ genes by a cat cassette (Fig. 2). The physical structure of each of these replacements was verified by PCR. We then tested each of these mutant prophage strains for efficiency of gal recombination with the 70-base oligo. The results (Fig. 2) indicate that only λ Beta is absolutely required for ssDNA recombination with the chromosome. Deletion of exo had no effect, and deletion of gam caused a minor ≈5-fold reduction in efficiency, suggesting that Gam, although not required, enhances recombination efficiency. To confirm that Beta was the only λ function required for ssDNA recombination, we created a strain in which bet is the only λ gene expressed from the prophage. Recombination levels with this “Beta only” strain were similar to the gam deletion strain, confirming that induction of λ Beta was sufficient for recombination between ssDNA and the chromosome. This result is in contrast to recombination with dsDNA substrates that require expression of Exo, Beta, and Gam (4, 18).
Exo action on linear dsDNA produces 3′ overhangs that are thought to act as a substrate for Beta-mediated ssDNA annealing. Thus, Exo would not be expected to affect recombination of single-stranded linear DNA. Gam inhibits the activity of the RecBCD nuclease, as well as the hairpin nuclease SbcCD (19). Because RecBCD has single-stranded nuclease activity (20), the decreased recombination efficiency of ssDNA in a strain lacking Gam might be due to the RecBCD single-stranded nuclease activity, to the SbcCD hairpin nuclease activity, or perhaps to some unknown function normally inhibited by Gam. Beta bound to ssDNA protects it from nuclease attack (21); this may be another reason that ssDNA recombination is proficient even in the absence of Gam.
A Strand Bias in Recombination Levels When Comparing Two Complementary ssDNAs.
Although Beta can mediate strand exchange between ssDNA and a target DNA with single-stranded ends (13), it could not be shown to mediate strand invasion of a fully double-stranded DNA (22). Because there is no requirement for RecA, recombination of ssDNAs may not involve strand invasion. Instead, we believe that the recombination events described here involve pairing of the Beta-bound oligo at transiently single-stranded regions of the chromosome. Single-stranded regions are formed during replication of the chromosome, repair of DNA, and transcription-induced supercoiling.
The 70-base oligo corresponding in sequence to the non-template strand for transcription of galK generated ≈5-fold more Gal+ recombinants than the complementary oligo corresponding to the template strand (Table 1). A strand bias of 4- to 10-fold was seen in each of four independent experiments. This strand bias might result from differences in oligo sequence, or from inherently asymmetric processes in the cell, such as transcription. To test whether strand bias resulted from the direction of transcription at galK or some other asymmetry, an inversion of the entire gal operon containing the galK amber mutation was created (see Materials and Methods). If the bias results from the asymmetry of transcription, the non-template strand oligo should still recombine more efficiently than the template strand oligo within the gal inversion. Instead, in this case, recombination of the template strand oligo in the gal inversion strain was 14-fold more efficient than recombination of the non-template strand oligo (Table 1). Thus, the strand bias observed does not result from the asymmetry of transcription. The strand bias is also not a consequence of simple sequence differences between the two oligos because either one may be more recombinogenic depending on orientation of the operon within the chromosome. As shown below, our results are more consistent with a bias caused by the direction of replication through the region containing the gal operon.
Table 1.
Strand bias of recombination at galK
| Strain* | Oligo* | Gal+ recombinants† |
|---|---|---|
| HME6 | NT | 1.5 × 105 |
| HME6 | T | 0.3 × 105 |
| HME41 | NT | 0.1 × 105 |
| HME41 | T | 1.4 × 105 |
Cells were induced for 15 min at 42°C and electroporated with 200 ng of either the non-template (NT) or template (T) oligo. HME41 is HME6 with the gal operon inverted. By convention, the non-template DNA has the same sequence as the transcript.
Total recombinant colonies on minimal galactose medium per 108 viable cells following electroporation.
Strand Bias Correlates with Direction of Replication Through the Recombination Site.
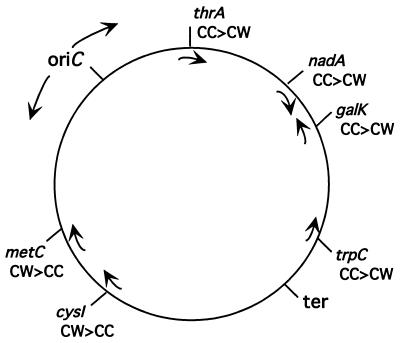
To determine whether strand bias is a general phenomenon or specific to galK, recombination at five additional genes on the chromosome was examined (Fig. 3). Each gene tested contained a Tn10 insertion causing an auxotrophic defect (16). For each mutant, two complementary 70-base oligos that included 35 bases of wild-type sequence on each side of the Tn10 insertion point were tested for efficiency of recombination (see Materials and Methods). We detected a strand bias at each of these sites, ranging from a 2- to 50-fold difference in recombination efficiency between the two complementary oligos used (Table 2). The strand bias was reproduced at each locus in three independent experiments (data not shown). As was the case for recombination at galK, the preferred strand at each of these genes had no correlation with the direction of transcription. Instead, strand preference correlated in every case with the direction of DNA replication through the gene being tested. Replication forks elongate in both directions from the origin (ori; Fig. 3), and the oligonucleotide that recombines more readily corresponds in sequence to the lagging strand DNA (Table 2 and Fig. 3).
Figure 3.
Correlation of single strand recombination with direction of replication. The bidirectionally replicated E. coli chromosome is illustrated as a circle with the directions of replication from the origin (oriC) toward the terminus (ter) indicated by arrows outside the circle. Arrows inside the circle indicate the direction of transcription of each gene. Mutations in each of these genes were corrected by oligo recombination (see Tables 1 and 2). The relative efficiency of two complementary oligos (cw or cc) was compared for each mutation (see Table 2). The oligo generating the greater (>) recombination is indicated.
Table 2.
Strand bias in recombination at several locations
| Strain and (relevant allele)* | Oligo† | Recombinants‡ |
|---|---|---|
| HME52(thrA∷Tn10) | cc | 3.8 × 105 |
| cw | 1.8 × 105 | |
| DY374 (nadA∷Tn10) | cc | 6.9 × 106 |
| cw | 3.8 × 106 | |
| HME53 (trpC∷Tn10) | cc | 1.0 × 105 |
| cw | 0.3 × 105 | |
| HME54 (cysI∷Tn10) | cc | 0.1 × 105 |
| cw | 5.1 × 105 | |
| HME55 (metC∷Tn10) | cc | 0.4 × 105 |
| cw | 1.3 × 105 |
Cells were induced for 15 min at 42°C and electroporated with 200 ng of the appropriate oligo containing 35 bases of wild-type sequence to each side of the indicated Tn10 element (16).
Oligos are labeled as clockwise (cw) referring to the DNA strand whose 5′ to 3′ sequence proceeds in the clockwise direction with respect to the genetic map of the E. coli chromosome, and counterclockwise (cc) for the opposite strand (see Fig. 3).
Total prototrophic recombinants per 108 viable cells following electroporation.
Discussion
Strand bias resulting from DNA replication suggests that ssDNA recombination may occur near the replication fork. The process of replication results in transient regions of ssDNA that may be accessible to Beta-mediated annealing of the introduced oligo. The increased recombination efficiency of the “lagging strand” oligos may reflect the increased frequency of single-stranded regions during lagging versus leading strand synthesis (23). DNA polymerase and DNA ligase may complete joining of the annealed strand to the chromosome. We observed rather substantial differences between the loci in both the degree of strand bias and the absolute number of recombinants (see Table 2). Our data do not provide an explanation of these differences; however, they may reflect changes in the structure or rate of replication of the chromosomal regions.
The result demonstrating that a single base change in galK is repaired at the same rate as a large insertion at the same position raises another important question that has not been resolved. How does the 70 base ssDNA used to repair these two mutations find its complementary sequence in these two examples? One might expect a gap created by discontinuous synthesis to reveal a 70-base segment to which Beta could anneal the complementary 70-base oligo. However, in the insertion mutant, the 35-base homology segments that flank the insertion DNA would be 3.3 kbp apart. How does Beta anneal to these separated 35-base targets at the same efficiency as it does to one 70-base target? Because the efficiency is the same, we must assume the two targets are not independent annealing events. Therefore, we assume either that the gap is very long, including the entire 3.3 kbp to allow direct annealing, or that there are multiple gaps spanning this distance and that the limiting event is finding the first complementary sequence. Most studies would support the latter of these two possibilities (23). Alternatively one can imagine that Beta does not depend on gaps at all and can stimulate strand invasion of the ssDNA into dsDNA. This model, however, ignores the in vitro result that strand invasion (or displacement) occurs only after the ssDNA has been first annealed to an adjacent gapped region (16, 23).
λ Red-mediated ssDNA recombination provides a rapid and highly efficient approach for generating recombinant DNA molecules. Because this process is highly efficient, one can directly screen for mutations in the absence of selection (24). The examples here involve chromosomal genes; however, it is also possible to create mutations in genes cloned on plasmids. Swaminathan et al. (24) have used this Red-mediated recombination system to create mutations within the murine Brca2 gene contained on a bacterial artificial chromosome (BAC). In this case, the high rate of recombination made it possible to use a PCR-based screen to identify recombinants.
Recombination with oligos can also be used to create deletions. Although we have not made extensive comparisons, deletion of an insertion cassette at galK by oligo recombination occurred with the same high efficiency as a single base change. Note that deletion of the different Tn10 insertion elements described here is also very efficient and again is high enough to permit screening for desired deletions in the absence of selection. For the nadA∷Tn10 insertion, recombinants that deleted the insertion made up 6% of cells that survive electroporation. The ability to delete individual genes or chromosome segments efficiently and precisely makes Red-mediated recombination with oligos a very useful tool for systematic deletion analysis of the E. coli genome. This system is useful for analysis of other bacterial genomes. λ Red recombination has already been adapted for use in Salmonella (N. P. Higgins, unpublished observations), and it seems reasonable to propose that expression of the λ Red functions, or related proteins, may mediate recombination by oligos in other bacteria. Note that, because bet is the only λ gene required for ssDNA recombination, appropriate shuttle vectors containing the bet gene could be used to export this recombination system to other species.
Thirty years ago the discovery of restriction enzymes led to revolutionary changes in molecular biology and to the advent of genetic engineering. However, homologous recombination is a more precise, efficient, and versatile means of engineering DNA molecules (4, 25). The precision by which recombinant DNA molecules can be created using restriction enzymes and DNA ligase is limited by the availability of unique and correctly positioned restriction sites. In vivo recombination can generate recombinant molecules without the requirements for unique or special sites. Moreover, the products engineered by recombination are generated directly in the chromosome or episomal plasmid without intermediate cloning steps. Engineering recombinant DNA molecules by in vivo homologous recombination, a method we will call “recombineering,” has been confined to yeast (25), and only recently has it been fully exploited in E. coli (4, 6). Here, we have described an extension of recombineering using ssDNA that we suggest can be adapted to modify the genomes of diverse organisms.
Recombination of oligos is not unique to E. coli. Short single-stranded oligos have been shown to recombine in yeast with the CYC1 gene. As we have observed for E. coli, oligo recombination in yeast demonstrated a strand bias that appeared unrelated to transcription (26, 27). Although the direction of replication through CYC1 was unknown, the authors speculated that this bias might reflect preferential annealing of the oligo to either the leading or lagging strand during replication. The yeast functions required for this recombination could not be determined. Although yeast Rad52 has been suggested to be functionally and structurally similar to λ Beta (28), Rad52 function was not required for oligo recombination (29), possibly because a second Beta-like function is present.
Oligo-mediated recombination occurs in both E. coli and yeast, suggesting that recombination via annealing of ssDNAs may occur in a wide range of organisms. Thus, recombineering with ssDNA may be applicable to higher eukaryotes as well, perhaps by a mechanism as simple as overexpressing Beta or a functionally similar ssDNA annealing protein, or more directly by introducing the protein itself into the eukaryotic cell during electroporation. We believe that homologous recombination with ssDNA will provide valuable technology for studying bacterial pathogens, and may open new avenues to engineer mutations or markers in the chromosomes of eukaryotic cells. Because oligos are so efficiently incorporated in vivo, the potential also exists to cross special chemical adducts and modifications from the oligos directly into the chromosome, thus providing special “tags” in the DNA of living cells.
Acknowledgments
We thank N. Costantino, E.-C. Lee, H. Peters, J. Sawitzke, K. Sergueev, S. K. Sharan, and H. Wilson for discussions and advice. We also thank T. Baker for technical help and M. Mills for preparing the manuscript and editorial comments. T. DiTizio is presently an undergraduate at Hood College in Frederick, MD.
Abbreviations
- oligo(s)
oligonucleotide(s)
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
References
- 1.Benzinger R, Enquist L W, Skalka A. J Virol. 1975;15:861–871. doi: 10.1128/jvi.15.4.861-871.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy K C. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jasin M, Schimmel P. J Bacteriol. 1984;159:783–786. doi: 10.1128/jb.159.2.783-786.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu D, Ellis H M, Lee E C, Jenkins N A, Copeland N G, Court D L. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. . (First Published May 16, 2000; 10.1073/pnas.100127597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Buchholz F, Muyrers J P, Stewart A F. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 6.Muyrers J P, Zhang Y, Testa G, Stewart A F. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee E-C, Yu D, de Velasco J M, Tessarollo L, Swing D A, Court D L, Jenkins N A, Copeland N G. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 8.Kuzminov A. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karu A E, Sakaki Y, Echols H, Linn S. J Biol Chem. 1975;250:7377–7387. [PubMed] [Google Scholar]
- 10.Murphy K C. J Bacteriol. 1991;173:5808–5821. doi: 10.1128/jb.173.18.5808-5821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Little J W. J Biol Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- 12.Kmiec E, Holloman W K. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- 13.Li Z, Karakousis G, Chiu S K, Reddy G, Radding C M. J Mol Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- 14.Van Dyck E, Stasiak A Z, Stasiak A, West S C. Nature (London) 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- 15.Zhou B B, Elledge S J. Nature (London) 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 16.Nichols B P, Shafiq O, Meiners V. J Bacteriol. 1998;180:6408–6411. doi: 10.1128/jb.180.23.6408-6411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mythili E, Kumar K A, Muniyappa K. Gene. 1996;182:81–87. doi: 10.1016/s0378-1119(96)00518-5. [DOI] [PubMed] [Google Scholar]
- 18.Muyrers J P, Zhang Y, Buchholz F, Stewart A F. Genes Dev. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni S K, Stahl F W. Genetics. 1989;123:249–253. doi: 10.1093/genetics/123.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Chen R, Julin D A. J Biol Chem. 2000;275:507–513. doi: 10.1074/jbc.275.1.507. [DOI] [PubMed] [Google Scholar]
- 21.Karakousis G, Ye N, Li Z, Chiu S K, Reddy G, Radding C M. J Mol Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 22.Muniyappa K, Radding C M. J Biol Chem. 1986;261:7472–7478. [PubMed] [Google Scholar]
- 23.Marians K J. In: Escherichia coli and Salmonella. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 749–63. [Google Scholar]
- 24.Swaminathan S, Ellis H M, Waters L S, Yu D, Lee E-C, Court D L, Sharan S K. Genesis. 2001;29:14–21. doi: 10.1002/1526-968x(200101)29:1<14::aid-gene1001>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 25.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moerschell R P, Tsunasawa S, Sherman F. Proc Natl Acad Sci USA. 1988;85:524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto T, Moerschell R P, Wakem L P, Komar-Panicucci S, Sherman F. Genetics. 1992;131:811–819. doi: 10.1093/genetics/131.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passy S I, Yu X, Li Z, Radding C M, Egelman E H. Proc Natl Acad Sci USA. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto T, Moerschell R P, Wakem L P, Ferguson D, Sherman F. Yeast. 1992;8:935–948. doi: 10.1002/yea.320081104. [DOI] [PubMed] [Google Scholar]