Abstract
The suf operon is composed of four genes (sufB, sufC, sufD, and sufS) and is highly conserved in the genomes of cyanobacteria. Open reading frame sll0088 in Synechocystis sp. strain PCC 6803 is located near the 5′ end of the suf operon but is transcribed in the direction opposite that of the suf operon. We previously reported the isolation of two independent suppressor strains of C14SPsaC that mapped to sll0088 and restored photoautotrophic growth. The protein encoded by sll0088 has two significant features: (i) a DNA-binding domain near the N terminus and (ii) four highly conserved cysteine residues near the C terminus. The protein has high sequence similarity to transcription regulatory proteins with a conserved DNA-binding domain and can be classified in the DeoR family of helix-loop-helix proteins. The protein falls into a further subclass that contains a C-X12-C-X13-C-X14-C motif near the C terminus, which may represent a metal-binding site. The expressed Sll0088 protein harbored an iron-sulfur cluster as shown by optical and electron paramagnetic resonance spectroscopy. Compared to the wild type, expression levels of the sufBCDS genes were elevated when cells were grown under conditions of oxidative and iron stress and were even higher in a null mutant of Synechococcus sp. strain PCC 7002 in which the sll0088 homolog was insertionally inactivated. In agreement with the proposed role of the sufBCDS genes in iron metabolism, the growth rate of the null mutant was significantly higher than that of the wild type under iron-limiting conditions. We propose that the protein encoded by sll0088 is a transcriptional repressor of the suf operon, and we name the gene sufR.
The biogenesis and assembly of fully functional photosystem I (PS I) require the assembly of FX, FA, and FB, which are [4Fe-4S] clusters, and soluble ferredoxin, which contains a [2Fe-2S] cluster. Iron-sulfur clusters can be assembled and inserted into proteins in vitro by incubating the apoprotein with iron, sulfide, and a thiol-containing reducing agent, such as 2-mercaptoethanol (22). This approach has been used in conjunction with bacterial expression systems to elucidate the structure and function of a variety of photosynthetic iron-sulfur proteins, including PsaC (reviewed in reference 11). In contrast, the in vivo biosynthesis of iron-sulfur clusters in photosynthetic complexes involves biochemical assembly processes that are poorly characterized. One known factor is a membrane-bound rubredoxin, which appears to be associated exclusively with the assembly of the FX iron-sulfur cluster (33, 34). Another known factor is the open reading frame sll0088 in Synechocystis sp. strain PCC 6803, which appears to have a role in regulating the biogenesis of PS I (43) and is the topic of this study.
In nonphotosynthetic organisms, iron-sulfur cluster assembly is known to be a multistep process that involves cluster biosynthesis, insertion, and stabilization (9). The isc operon is implicated in generalized iron-sulfur cluster assembly in many organisms, including Azotobacter vinelandii (45), Escherichia coli (25), and Saccharomyces cerevisiae (31). Homologs of several genes in the isc operon, including iscS and iscA, have been identified in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 (16, 17). In fact, three distinct iscS-like homologs have been identified in genomes of Synechocystis sp. strain PCC 6803 (16, 18), Anabaena sp. strain PCC 7120 (15, 26), and Synechococcus sp. strain PCC 7002 (J. Zhao, T. Li, J. Marquardt, and D. A. Bryant, unpublished data). The presence of multiple iscS homologs hints at different functions or regulatory mechanisms in the biosynthesis of iron-sulfur clusters. Two additional iron-sulfur cluster assembly systems that are more specialized in function, nif and suf, exist in bacteria. The nif operon in A. vinelandii is involved in the biosynthesis of the nitrogenase iron-molybdenum cofactor (8). The suf operon has been shown to function in the assembly of iron-sulfur clusters under conditions of oxidative stress (24, 41). It is known that at least two of these systems, isc and suf, exist in mitochondria (31, 39) and in the chloroplasts of higher plants (20, 28, 40), respectively. However, virtually nothing is known concerning their regulation.
As reported previously (43), a methodology of selecting suppressors to primary mutations in iron-sulfur proteins was used to isolate spontaneous suppressors from psaC site-directed mutants of Synechocystis sp. strain PCC 6803. The phenotypes of the C51DPsaC (FB) and C14SPsaC (FA) mutants of PsaC were such that the strains failed to grow photoautotrophically, yet electron throughput from cytochrome c6 to flavodoxin in isolated PS I complexes was similar to that of the wild type (14, 44). The mutants were sensitive to high light intensities and could not grow photomixotrophically under white-light intensities in the range from 20 to 60 μE m−2 s−1. The mutants had two additional phenotypes: the amount of PS I, but not PS II, was lower in the mutants than in the wild type on a per cell basis, and the mutants were able to grow photomixotrophically at a light intensity of 20 to 60 μE m−2 s−1 only in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), an inhibitor of PS II. Thus, the failure to grow photoautotrophically was apparently due to the lowered ratio of PS I to PS II in the mutant cells rather than an inefficiency in forward electron transfer in any individual PS I complex. Screening the C51DPsaC and C14SPsaC mutants under high-light intensity resulted in the appearance of suppressor mutants. Two of the intergenic suppressor strains, C14SPsaC-R18 and C14SPsaC-R62, which were capable of photoautotrophic growth at normal light intensities, were selected for further study. These two suppressor strains retained the primary mutation, which was verified through amplification of the psaC gene fragment by PCR and sequencing. The suppressor mutations were mapped to a specific gene, sll0088, by phenotypic complementation and identified by DNA sequencing (43).
To elucidate the function of the sll0088 gene homologs in cyanobacteria, we sought to characterize the physical properties of the protein encoded by this gene, to investigate the regulation of the sufBCDS operon, and to study the physiology of a sll0088 null mutant. We show that the protein encoded by sll0088 is an iron-sulfur protein that functions as a transcriptional repressor, regulating the expression of the sufBCDS operon. Thus, it is similar in function to IscR, which is an iron-sulfur protein that functions as a transcriptional repressor of the isc operon (32).
MATERIALS AND METHODS
Growth of the wild-type and mutant cells under optimal conditions.
Synechococcus sp. strain PCC 7002 was grown under photoautotrophic conditions in A medium supplemented with 1 mg of NaNO3 ml−1 (hereafter called A+ medium) (37). Solid medium for maintaining the mutant strains of Synechococcus sp. strain PCC 7002 was supplemented with 0.3% (wt/vol) sodium thiosulfate and 100 μg of kanamycin ml−1. Synechococcus sp. strain PCC 7002 cells were grown under photoheterotrophic conditions in A+ liquid medium containing 5 mM glycerol. Synechocystis sp. strain PCC 6803 cells were grown under photoautotrophic conditions in BG11 medium as described previously (36). All liquid cultures were bubbled with air supplemented with 1% (vol/vol) CO2. Fluorescent bulbs provided continuous white-light illumination. The temperature was maintained at 30°C for Synechocystis sp. strain PCC 6803 and 38°C for Synechococcus sp. strain PCC 7002 by a water bath. We monitored growth of the wild-type and mutant strains by measuring the optical density at 730 nm (OD730) with a Cary-14 spectrophotometer modified for computerized data acquisition by On-Line Instruments, Inc. (Bogart, Ga.).
Growth of the wild-type and mutant cells under stress conditions.
For growth studies under iron stress conditions, wild-type and mutant cells of Synechococcus sp. strain PCC 7002 were grown to mid-exponential phase, collected by centrifugation at 4,000 × g for 10 min, and resuspended to an OD730 of 0.1 in fresh medium containing 50 μM 2,2′-dipyridyl or 0.25 μM streptonigrin. For growth studies under iron starvation conditions, FeCl3 was eliminated from the A+ medium. For growth studies under oxidative stress conditions, wild-type and mutant cells of Synechococcus sp. strain PCC 7002 were grown to mid-exponential phase under standard growth condition with the addition of 5 mM glycerol in the medium. The cell cultures were diluted with fresh medium to an OD730 of 0.1 and grown to an OD730 of 0.5. A total of 0.5 μM DCMU (an inhibitor of PS II), 10 μM 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone (DBMIB) (an inhibitor of cytochrome b6 or f), or 150 μM hydrogen peroxide (H2O2) was then added to the medium. The cells were grown for 10 to 12 h, during which time they undergo no more than one division, and collected for total RNA isolation.
Protein quantitation and absorption spectroscopy.
Protein concentrations were measured by using the Coomassie Plus protein assay with bovine serum albumin as the standard (Pierce, Rockford, Ill.). Absorption spectra were recorded on a Cary-14 spectrophotometer modified for computerized data acquisition by On-Line Instruments, Inc.
DNA isolation and Southern blotting.
Chromosomal DNA from Synechococcus sp. strain PCC 7002 and Synechocystis sp. strain PCC 6803 was isolated as previously described (35). The DNA fragment containing the sll0088 gene in Synechocystis sp. strain PCC 6803 or its homolog in Synechococcus sp. strain PCC 7002 was cloned and sequenced. DNA sequencing was performed at the Penn State Nucleic Acid Facility. Southern blot and PCR analyses were used to verify whether the gene interruption was homozygous. For Southern hybridization, genomic DNA of the wild-type and mutant strains of Synechococcus sp. strain PCC 7002 was digested with appropriate restriction endonucleases and transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, N.H.). The DNA probe fragments were labeled with [α-32P]dATP (New England Nuclear, Boston, Mass.) using a random priming method.
Expression and purification of the protein encoded by sll0088.
The sll0088 gene from Synechocystis sp. strain PCC 6803 (17) was amplified by PCR. NdeI and HindIII restriction enzyme sites were incorporated via the Esll0088F and Esll0088R primers used for PCR as listed in Table 1. The amplified sll0088 gene fragment was digested with NdeI and HindIII and cloned into expression vector pET24a (Novagen, Madison, Wis.), resulting in the pET24a/sll0088 expression plasmid. The resulting construct was resequenced to ensure the correctness of the PCR amplification. Plasmid pET24a/sll0088 was transformed into E. coli strain BL21(DE3). Overproduction of the Sll0088 protein in E. coli cells was induced by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were harvested and washed once in TS buffer (10 mM Tris-HCl [pH 8.0], 50 mM NaCl). The cell pellets were resuspended in TS buffer with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 0.1 mg of DNase I (Sigma, St. Louis, Mo.) ml−1 and passed twice through a chilled French pressure cell. The overexpressed protein was present in inclusion bodies, which were pelleted by low-speed centrifugation at 7,000 × g for 20 min. The pellet was washed twice with TS buffer and solubilized in 7 M urea in TS buffer by addition of 2 mM dithiothreitol (DTT), 0.1% (wt/vol) Triton X-100, and 1 mM PMSF. After centrifugation at 12,000 × g to pellet the insoluble material, the supernatant was applied to a Sepharose G-100 column that was preequilibrated with TS buffer containing 5 mM DTT.
TABLE 1.
Sequences of oligonucleotides used for RT-PCR analysis and cloning the sll0088 gene
| Primer | Sequencea |
|---|---|
| sufRF | 5′-ACCCTTTCTGACACCCGCGCCGAAAC |
| sufRR | 5′-ATCTTTGGCTTGGATGATGTAGCCGC |
| sufBF | 5′-AGTGCGACCGTCAAAACCCTTGTCAA |
| sufBR | 5′-CAACAGTGCCTTCGAGTTTGAGGCTG |
| sufCF | 5′-GAGTGAAGTTATCTTAGCGATTA |
| sufCR | 5′-ACAACCCCCGCAGCCAGTTCTTG |
| sufDF | 5′-TCTAGCGGTACCATGACAGCGGCGATC |
| sufDR | 5′-GTCAACGGTGCGACAGGCAACACAAC |
| sufSF | 5′-GATCACAACCCAACCGAAACCCCTTG |
| sufSR | 5′-CTACCCATGCTGGTAAAGAAATCAATG |
| Esll0088F | 5′-CTTCTCCTGTTGTTGCATATGACCCTCAGTTCT |
| Esll0088R | 5′-CGTCAATTGATTAAAAAGCTTTCTGCCGCCCG |
Nucleotides of the restriction site sequence are underlined.
Reconstitution of iron-sulfur cluster in the Sll0088 protein and EPR spectroscopy.
Solubilization of the inclusion bodies with urea under aerobic conditions leads to disassembly of any iron-sulfur clusters that may be present in the Sll0088 protein. To reconstitute an anticipated iron-sulfur cluster, the apoprotein was treated with ferric chloride, sodium sulfide, and 2-mercaptoethanol (22). The reconstituted holoprotein was resuspended in 50 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) buffer (pH 10.5) and purified from the inorganic reconstitution agents by repeated ultrafiltration. Samples for electron paramagnetic resonance (EPR) spectroscopy contained 5 mg of protein ml−1 and 20% (vol/vol) glycerol and were reduced with 20 mM sodium dithionite in an anaerobic chamber (Coy Products, Grass Lake, Mich.). The EPR spectrum was recorded using a Bruker ECS106 EPR X-band (9.2 GHz) spectrometer operating with an ER 4012 ST resonator and an Oxford liquid helium cryostat. The microwave frequency was determined with a Hewlett-Packard 5340A frequency counter.
Production of antibodies.
To obtain polyclonal antibodies, the recombinant Sll0088 protein was partially purified by Sepharose G-100 chromatography and subjected to preparative polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE). An extra lane was loaded identically and used for Coomassie blue staining to locate the position of the protein band. The protein-containing region of the gel was excised, and the protein was eluted using a Bio-Rad electroelutor (Bio-Rad, Hercules, Calif.). Polyclonal antibodies against the SufR protein were generated in rabbits at the Centralized Biological Laboratory at Pennsylvania State University.
Protein electrophoresis and Western immunoblotting.
Methods used for SDS-PAGE and immunoblotting were identical to those described previously (35). Samples, including uninduced cells, induced cells, inclusion bodies, and purified proteins, were solubilized in loading buffer and boiled for 2 min before they were loaded onto an SDS-polyacrylamide gel. A 12% acrylamide gel was used to resolve proteins, which were stained with Coomassie blue. To prepare cyanobacterial extracts, cells were harvested by centrifugation, suspended in 50 mM Tris-HCl buffer (pH 8), and broken by sonication. For immunoblots, proteins separated by electrophoresis were transferred electrophoretically to nitrocellulose membranes (Schleicher & Schuell) by using the Semi-Dry system (Bio-Rad). The nitrocellulose membranes were subjected to immunoreaction with polyclonal antibodies and developed.
RNA isolation and RT-PCR analysis.
For RNA preparation, cells of the wild-type and mutant strains of Synechococcus sp. strain PCC 7002 were harvested during exponential growth. Total RNA was isolated using the High Pure RNA isolation kit according to the protocol provided by the manufacturer (Roche Diagnostics, Indianapolis, Ind.). Contaminating DNA was eliminated by incubating the sample with DNase I for 1 h at room temperature during the preparation procedure. The absence of DNA contamination in RNA samples was confirmed by PCR analysis. For Northern blot analysis, 10 μg of total RNA from the wild-type and mutant strains was loaded per lane, and the samples were electrophoresed in 1.3% (wt/vol) denaturing agarose-formaldehyde gels. The RNA was transferred to nylon membranes by capillary transfer, fixed by UV illumination for 1 min, and baked for 1 h at 80°C in a vacuum. The sll0088 homolog gene-specific probe was generated by PCR using Synechococcus sp. strain PCC 7002 genomic DNA as the template. The PCR product was purified by electrophoresis on an agarose gel, and the DNA was extracted and purified by using the QIAGEN Gel Extract kit (Valencia, Calif.). The probe was labeled by random priming as described previously (35). Hybridization with 32P-labeled DNA was performed overnight at 42°C. After the membranes were washed, they were incubated for fluorography at −80°C. RNA concentrations were determined by absorption spectroscopy. Reverse transcriptase PCR (RT-PCR) experiments were performed by using the QIAGEN OneStep RT-PCR kit. The primers were designed specifically to amplify the sufBCDS genes of Synechococcus sp. strain PCC 7002 (Table 1). Total RNA (2.4 ng) was used as the template for the wild-type and mutant strains. The RT-PCRs were performed as follows: a 30-min reverse-transcription reaction at 50°C, a 15-min initial heating step at 95°C, and 32 three-step cycles (1 min at 94°C, 1 min at 59°C, and 1.5 min at 72°C).
RESULTS
The sll0088 gene is adjacent to the suf operon in Synechocystis sp. strain PCC 6803.
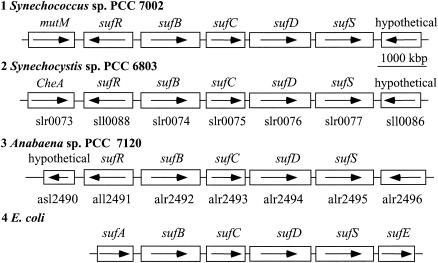
The majority of functional studies described in this paper were performed in Synechococcus sp. strain PCC 7002, because we were able to construct a fully segregated, null mutation in the sll0088 homolog of this cyanobacterium, which we could not do for Synechocystis sp. strain PCC 6803. Because the genome sequence is not yet available for Synechococcus sp. strain PCC 7002, we needed to determine the gene organization surrounding the sll0088 homolog. A DNA fragment containing the sll0088 homolog and its upstream flanking region were cloned and sequenced from the genome of Synechococcus sp. strain PCC 7002 (GenBank accession number AY375041). DNA sequence analysis of the sll0088 homolog and the open reading frames in the region upstream from this gene indicate that the gene organization is identical to that in Synechocystis sp. strain PCC 6803 (16, 17) and Anabaena sp. strain PCC 7120 (15). As shown in Fig. 1, sequence comparisons show that four conserved genes in cyanobacterial genomes, sufB, sufC, sufD, and sufS, have homologs in the suf operon of E. coli (4, 27). The genes in the suf operon have been shown to function in iron-sulfur cluster biogenesis under conditions of oxidative stress (24, 41). In cyanobacteria, the genes homologous to sll0088 are located near the 5′ end of the presumed suf operons but are transcribed in the direction opposite that of the sufBCDS genes. The sufA and sufE genes also exist in cyanobacteria (slr1417 and slr1419, respectively, in Synechocystis sp. strain PCC 6803), but they are not contiguous with the sufBCDS genes, as they are in E. coli and certain other bacteria.
FIG. 1.
Map of gene organization of sll0088 (sufR) and the sufBCDS operon of three cyanobacteria compared to the sufABCDSE operon in E. coli. Genomic sequences of DNA fragments containing the sufBCDS operon were compared for three available sequences of cyanobacteria, Synechococcus sp. strain PCC 7002, Synechocystis sp. strain PCC 6803 (17), and Anabaena sp. strain PCC 7120 (15), and the E. coli genome sequence (4). The direction of transcription is indicated by the arrows.
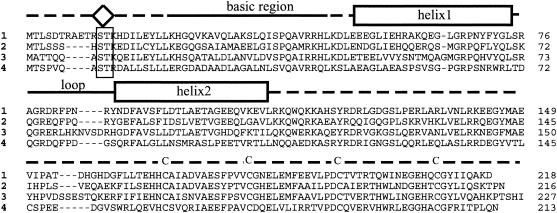
As shown in Fig. 2, the amino acid sequences of the proteins coded by the sll0088 homologs are highly conserved in Synechococcus sp. strain PCC 7002, Synechocystis sp. strain PCC 6803, Anabaena sp. strain PCC 7120, and Synechococcus WH8102. A BLAST search shows that highly similar proteins also exist in the photosynthetic prokaryotes Trichodesmium erythraeum IMS101, Nostoc punctiforme, Thermosynechococcus elongatus, and Prochlorococcus marinus strain MIT9313. Three features can be identified in these proteins using PROSITE program analysis: (i) near the N terminus, a basic region preceded by the elements of a putative protein kinase C phosphorylation site (Ser-Thr-Lys); (ii) a DNA-binding domain that contains a basic region and a putative helix-loop-helix motif whose sequence is very similar to the sequences of DNA-binding proteins from other organisms; and (iii) near the C terminus four highly conserved cysteine residues in a C-X12-C-X13-C-X14-C motif with a possible function in metal binding. Comparative analyses show that the sequences of the sll0088 homologs are very similar to the sequences of transcription regulatory proteins that bind DNA by a conserved helix-loop-helix motif near the N terminus. The C terminus, with similarly spaced cysteine residues, may confer functional specificity to this class of regulatory proteins.
FIG. 2.
Conserved domains in SufR proteins from four cyanobacterial strains. The sequences of four cyanobacterial strains are indicated in the figure as follows: 1, Synechococcus sp. strain PCC 7002; 2, Synechocystis sp. strain PCC 6803; 3, Anabaena sp. strain PCC 7120; and 4, Synechococcus sp. strain WH8102. Their amino acid sequences were aligned using the Clustal-W algorithm in the MacVector DNA analysis program (Accelerys, Madison, Wis.). The basic region, the helix-loop-helix domain, and the four conserved cysteines in the C-terminal region are indicated above the alignment. The diamond shape near the N terminus indicates the putative protein kinase C phosphorylation site. Gaps introduced to maximize alignment are indicated by dashes.
The purified Sll0088 protein binds an iron-sulfur cluster.
To characterize the properties of the protein coded by sll0088, the gene from Synechocystis sp. strain PCC 6803 was cloned into the expression vector pET24a and expressed in E. coli strain BL21(DE3). Very little Sll0088 protein was present in the soluble fraction; rather, the majority was found in inclusion bodies, which were light brown in color, indicating that this protein might harbor an iron-sulfur cluster. The inclusion bodies were solubilized in a solution containing 7 M urea, 4 mM DTT, and 0.1% (wt/vol) Triton X-100, and the Sll0088 protein was purified by gel exclusion chromatography. The Sll0088 protein had an apparent mass of 24 kDa as shown by gel exclusion chromatography and SDS-PAGE (data not shown). This measured mass is in close agreement with a calculated mass of 24.6 kDa determined from the DNA sequence. (A molecular mass of 27.4 kDa is calculated from the DNA sequence of the sll0088 open reading frame in CyanoBase (http://www.kazusa.or.jp/cyanobase/). However, due to the presence of two closely spaced ATG start codons near the beginning of the open reading frame, this mass may be higher than that of the actual protein. Based on the results of a comparison with Sll0088 sequences from other cyanobacteria [Fig. 1], it is likely that the real start is the second ATG codon. Therefore, the Sll0088 protein from Synechocystis sp. strain PCC 6803 is probably made up of 217 amino acids.)
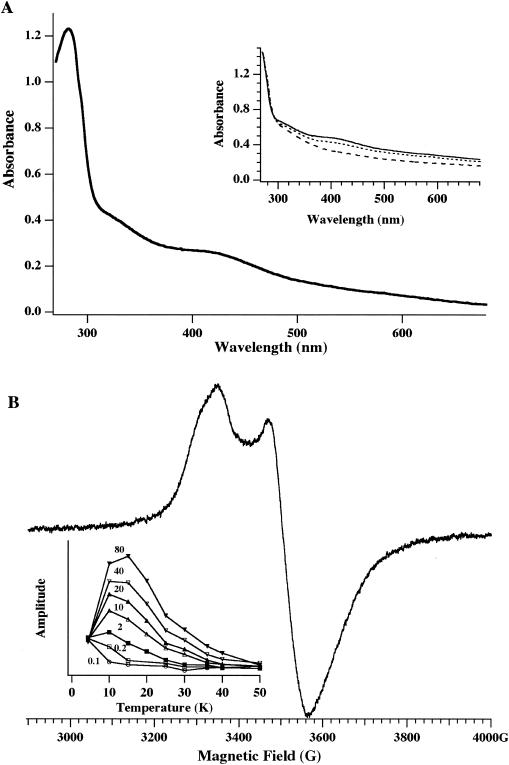
The presence of four highly conserved cysteines in the C-terminal region suggests that their side groups might play a role as metal-binding ligands. To test the possibility that they bind an iron-sulfur cluster, purified Sll0088 protein was incubated with ferric chloride, sodium sulfide, and 2-mercaptoethanol and repurified under anaerobic conditions by size exclusion chromatography (22, 42). The UV-visible spectrum of the reconstituted Sll0088 protein showed absorbance in the UV and visible regions, with maxima at wavelengths of 280, 320, and 411 nm (Fig. 3A). The peaks at 320 and 411 nm are typical of the broad S → Fe charge transfer transitions found in iron-sulfur proteins. The A411/A280 ratio of 0.29 is within the usual range for a typical iron-sulfur protein (2, 13). The absence of absorption peaks in the 450- and 550-nm-wavelength regions is more characteristic of a [4Fe-4S] cluster than a [2Fe-2S] cluster (5, 42). Similar to other iron-sulfur proteins, the spectrum lost about one-half of its amplitude after reduction of the protein with sodium hydrosulfite. When the protein was oxidized in air, the spectrum lost most of its absorbance in the 320- to 420-nm-wavelength region (Fig. 3A, inset), suggesting that the iron-sulfur cluster is oxygen sensitive. Figure 3B shows the low-temperature, X-band EPR spectrum of the reconstituted Sll0088 protein after chemical reduction with dithionite. The axial spectrum of the reduced protein, with a low-field peak at g = 2.02 and a high-field trough at g = 1.89 is characteristic of proteins that contain an iron-sulfur cluster. The spectrum and high spin relaxation rate implied from the microwave power dependence (Fig. 3B, inset) and the lower amplitude of the signal at temperatures above 20 K (Fig. 3B, inset) are consistent with the presence of a [4Fe-4S] cluster. The cluster is stable for days in either the oxidized or reduced state when the protein is maintained under anaerobic conditions.
FIG. 3.
UV-visible absorption spectrum (A) and EPR spectrum (B) of the recombinant Sll0088 protein after reconstitution with iron, sulfide, and 2-mercaptoethanol. The UV-visible absorption spectrum shows maxima at wavelengths of 280, 320, and 411 nm. The inset shows the loss of the absorbance of the oxidized protein upon exposure to air for 30 min (solid line), 60 min (dotted line), and 90 min (dashed line). The EPR spectrum shows the axial line shape of an iron-sulfur cluster, with a low-field peak at g = 2.02 and a high-field trough at g = 1.89. The inset shows the signal intensity as a function of temperature and microwave power (in milliwatts). EPR conditions follow: microwave frequency, 9.4709 GHz; microwave power, 40 mW; modulation amplitude, 10 G; receiver gain, 6.3 × 104; temperature, 14 K. The spectrum represents an average of four scans.
The sufBCDS genes compose an operon in cyanobacteria.
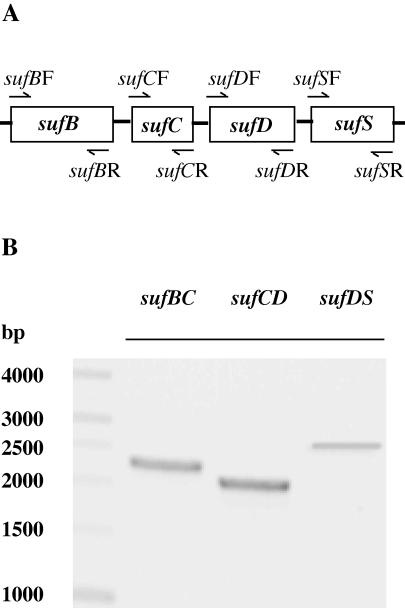
The sufABCDSE genes in E. coli (41) and Erwinia chrysanthemi (24) are organized into operons. In cyanobacteria, the adjacent sufBCDS genes suggest that they also compose an operon (Fig. 1). To determine whether sufB, sufC, sufD, and sufS are cotranscribed in Synechococcus sp. strain PCC 7002, the 5′ primers for sufB, sufC, and sufD were added to an RT-PCR mixture together with the 3′ primers for sufC, sufD, and sufS, respectively, to form primer pairs (Fig. 4A). As shown in Fig. 4B, three fragments were amplified with sizes corresponding to sufBC (2.2 kbp), sufCD (2.1 kbp), and sufDS (2.6 kbp). The cotranscription of the sufB and sufC, sufC and sufD, and sufD and sufS genes clearly indicates that in cyanobacteria the four genes also compose an operon.
FIG. 4.
The primer pairs used for the RT-PCR experiment (A) and the cotranscription of suf genes detected by RT-PCR (B). The sufBF and sufCR primers were used to amplify the sufBC fragment, the sufCF and sufDR primers were used to amplify the sufCD fragment, and the sufDF and sufSR primers were used to amplify the sufDS fragment. Products of the expected sizes for sufB and sufC, sufC and sufD, and sufD and sufS establish that the four genes were cotranscribed.
Increased sufBCDS mRNA levels in a null mutant of the sll0088 homolog.
The proximity of the sll0088 homologs to the sufBCDS operon in several cyanobacteria and the identification of the product of the sll0088 gene from Synechocystis sp. strain PCC 6803 as a possible transcription regulatory protein suggested that Sll0088 might play a role in regulation of expression of the sufBCDS operon. To test this idea, we compared the mRNA levels for the sufB, sufC, sufD, and sufS genes in the wild type and a null strain of the sll0088 homolog in Synechococcus sp. strain PCC 7002. As we have shown previously (43), null mutants were created by inserting the aphII gene into the StuI restriction site of a gene homologous to sll0088 in Synechococcus sp. strain PCC 7002 either parallel or antiparallel to the transcription direction of the sll0088 homolog. Both mutants were completely segregated, as verified by PCR and Southern blot analysis. Their growth rates under low (50 μmol · m−2 · s−1) and normal (250 μmol · m−2 · s−1) light intensities were very similar (data not shown), suggesting that the insertion direction of aphII gene has no effect on the mutant phenotype. The complete absence of the sll0088 homolog in the null strains was confirmed by immunoblot analysis (Fig. 5). Given these results, all further studies reported here in which the wild type and the null mutant were compared used only the strain in which the aphII gene was inserted into the sll0088 homolog parallel to its transcription orientation. We chose this orientation, because in principle, a parallel insertion is likely to have less effect on the other genes downstream of the sll0088 homolog than an antiparallel insertion.
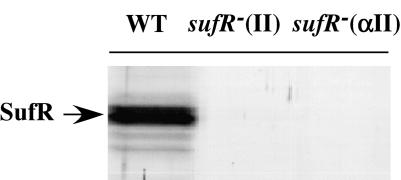
FIG. 5.
Immunodetection of the SufR protein in the cells of the wild type (WT) and two sufR null mutants of Synechococcus sp. strain PCC 7002 by use of antibodies against the recombinant SufR protein of Synechocystis sp. strain PCC 6803. Proteins corresponding to 10 μg of chlorophyll from the whole-cell lysates were loaded onto each lane. sufR−(II) and sufR−(αII) strains are mutants in which the inserted aphII gene is transcribed in the same direction (II) or in the direction opposite (αII) that of the sufR gene. Only the sufR−(II) strain was used for subsequent work; it is referred to in the text as the sufR null mutant and is indicated in the figures as the sufR− strain.
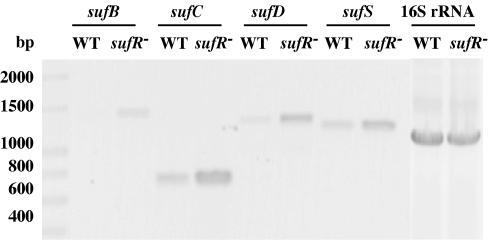
The transcript level for the sufS gene, probed by Northern blotting analysis, was found to be higher in the null strain than in the wild type (data not shown). The intensities of the hybridization signals for the sufS gene were quantified using the National Institutes of Health (NIH) Image Program, and the expression level was three to four times higher in the null strain. In addition, RT-PCR analysis was performed to probe more sensitively the transcript levels for the sufB, sufC, sufD, and sufS genes in cells of the wild-type and null strains. As shown in Fig. 6, mRNA levels for the sufB, sufC, sufD, and sufS genes were consistently higher in the sll0088 null strain. These results therefore implicate the sll0088 gene product in regulation of suf operon expression in Synechococcus sp. strain PCC 7002. We propose that the sll0088 homolog be named sufR and that the protein coded by this gene be named SufR.
FIG. 6.
RT-PCR analysis of the suf gene transcription levels in Synechococcus sp. strain PCC 7002 (wild type) and sufR null strain. Primers specific for the sufB, sufC, sufD, and sufS genes were used in RT-PCRs and are listed in Table 1. The 16S RNA gene was used as a control to assure that the RNAs of the wild type (WT) and sufR mutant strain were added to the RT-PCR mixtures at equal concentrations and that both were equally competent for the RT-PCR.
Transcript levels of sufB and sufS are elevated in the sufR null strain under stress conditions.
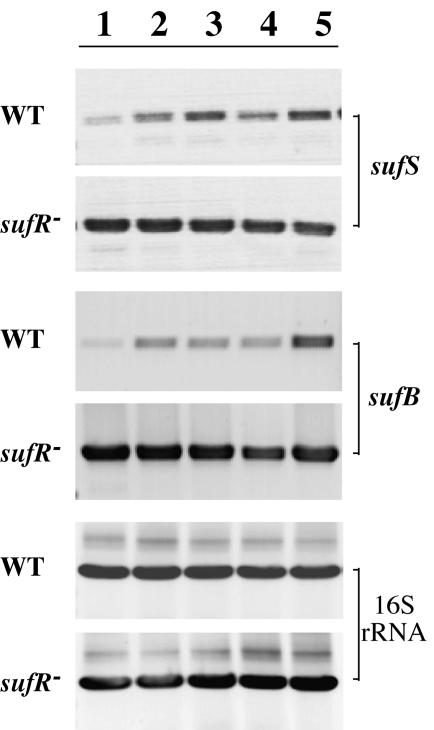
Proteins encoded by the suf operon in E. coli are thought to be involved in the assembly of iron-sulfur clusters, particularly under conditions of oxidative stress (24, 41). To probe expression of the suf genes in cyanobacteria under stress conditions, the wild type and the sufR null strain were grown in normal media and shocked with DCMU (an inhibitor of PS II), DBMIB (an inhibitor of cytochrome b6 or f), H2O2, and Fe deprivation for various lengths of time before being harvested and processed for RNA isolation. The transcription levels of the sufB and sufS genes under these conditions were compared with those under normal growth conditions. As shown in Fig. 7, the expression levels of sufS were higher in the presence of DCMU, DBMIB, and H2O2 and in the absence of iron in the medium than the level in the wild type. In the sufR null strain (Fig. 5), the expression levels of sufS were higher than the level in the wild type, regardless of the stress imposed. Thus, the genes in the sufBCDS operon are constitutively expressed when the SufR repressor is absent. The expression levels of sufB were also higher in the presence of DCMU, DBMIB, and H2O2 and in the absence of iron in the medium than in the wild type; in the sufR null strain, the sufB mRNA levels were even higher and independent of the stress imposed. Thus, the expression levels of the sufB and sufS genes in the sufR null mutant are constant and higher under all conditions than those in the wild type. This would be expected if the sufBCDS operon were no longer under SufR regulation. Therefore, these observations suggest that the iron-sulfur cluster assembly proteins encoded by the sufBCDS operon have increased functional importance under conditions of oxidative stress in cyanobacteria.
FIG. 7.
RT-PCR analysis of the expression of the sufS and sufB genes in the wild-type (WT) and sufR mutant strains of Synechococcus sp. strain PCC 7002. Total RNA was isolated from cells grown under the following conditions: standard growth conditions (lane 1), A+ medium supplemented with 5 mM glycerol and 0.5 μM DCMU (lane 2), A+ medium supplemented with 5 mM glycerol and 10 μM DBMIB (lane 3), A+ medium supplemented with 150 μM H2O2 (lane 4), and A+ medium with no iron salts added (lane 5). The gene for 16S RNA was used as a control to assure that the RNAs of the wild type and sufR mutant strain were added to the RT-PCRs at equal concentrations and that both were equally competent for the RT-PCR.
Growth rates of the sufR null strain under stress conditions are higher than that of the wild type.
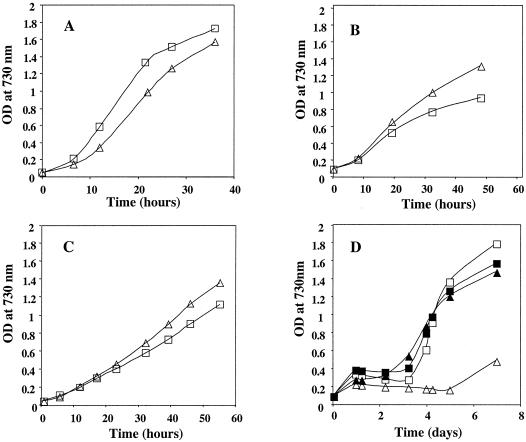
The increased mRNA levels of the sufBCDS operon in the sufR null strain would be expected to have physiological consequences if the proteins coded by these genes normally function in response to stress. As shown in Fig. 8A, the 4-h doubling time of the wild type at normal (250 μmol · m−2 · s−1) light intensities increased to 6.5 h in the sufR null strain, suggesting that the overexpression of the Suf proteins is mildly inhibitory in cells grown under near-optimal nutrient and illumination conditions. To probe the relationship between sufR and iron metabolism, cells were transferred to fresh growth medium containing 2,2′-dipyridyl (an iron chelator) or streptonigrin (an iron-activated antibiotic) during mid-exponential phase, and the growth rates were measured. In the presence of 2,2′-dipyridyl (Fig. 8B), the 15-h doubling time of the wild type decreased to 10.5 h in the sufR null strain. This might be expected if the iron-sulfur cluster biosynthetic enzymes in the sufR null mutant were up-regulated and were thereby able to scavenge low levels of iron more efficiently than the wild type. To test this idea further, growth rates of the wild type and sufR null strain of Synechococcus sp. strain PCC 7002 were compared when iron was eliminated from the growth medium. When the cells were transferred to fresh medium without added FeCl3 during mid-exponential phase, the 14-h doubling time of the wild type decreased to 11 h in the sufR null strain (Fig. 8C). When the cells were transferred to fresh growth medium in the presence of streptonigrin (Fig. 8D, open symbols), growth of the wild type and the mutant were inhibited for 3 and 7 days, respectively. The higher sensitivity of the mutant compared to the wild type could indicate that iron accumulated in the mutant cells, thereby providing the cells with a higher intracellular iron content than the wild type. The inhibiting effect of streptonigrin could be alleviated by iron starvation; the sufR mutant resumed growth after 2 days, and the wild type resumed growth after 3 days (Fig. 8D, filled symbols). Thus, the sufR null strain survived the presence of streptonigrin under iron starvation conditions better than in normal growth medium. This can be explained if most of the iron were tied up in iron-sulfur clusters due to the up-regulation of the sufBCDS operon. In contrast, growth of the wild type showed no significant differences under these conditions, probably because iron starvation exerted contradictory effects: on the one hand, it inhibited wild-type growth, and on the other hand, it alleviated the toxicity of streptonigrin.
FIG. 8.
Growth curves of Synechococcus sp. strain PCC 7002 under different conditions. The wild type (open square) and the sufR mutant (open triangle) under various conditions are shown. (A) Standard conditions (250 μE · m−2 · s−1, 38°C, A+ medium); (B) standard conditions in the presence of 2,2′-dipyridyl; (C) standard conditions in A+ medium with no FeCl3 added; (D) standard conditions in the presence of streptonigrin for the wild type with (open symbols) or without (filled symbols) addition of FeCl3 and for the sufR mutant strain with (open symbols) or without (filled symbols) addition of FeCl3. These experiments were repeated three times; all showed similar growth profiles. Data from only one experiment are depicted in this figure.
These growth studies clearly show that the increased transcript levels for the suf genes in the sufR null mutant allow cyanobacterial cells to grow more efficiently in a low-iron environment. This result is in clear contrast to the inhibitory effect exerted by increased suf gene expression level on the growth of sufR null mutants under standard growth conditions. The inhibitory effect of the high expression of the suf genes is more obvious under iron-replete conditions, as shown by the streptonigrin-containing cultures. As shown previously in Fig. 7, the expression level of suf genes in the sufR null mutant always exceeds that of the wild type in response to oxidative stresses. High expression levels of the Suf proteins are also toxic in E. coli (41). These studies show that increased mRNA levels for the suf genes in the sufR null mutant lead to the predicted physiological response of enhanced survivability under iron-limiting conditions.
DISCUSSION
The sufR gene is usually located adjacent to the sufBCDS operon in cyanobacteria.
The sll0088 (sufR) gene is immediately upstream of the sufBCDS operon in Synechocystis sp. strain PCC 6803, and homologs occupy a similar position in other cyanobacteria, including Synechococcus sp. strain PCC 7002, Anabaena sp. strain PCC 7120, Trichodesmium erythraeum IMS101, and Prochlorococcus marinus MIT9313. In Synechococcus sp. strain WH8102, the ferredoxin-thioredoxin reductase catalytic chain, a subunit of heterodimeric ferredoxin-thioredoxin reductase (ftrC, which corresponds to open reading frame sll0554 in Synechocystis sp. strain PCC 6803), is located between sufR and the sufBCDS operon. There is only a 7-bp-long spacer between ftrC and the sufBCDS operon, suggesting that the ftrC homolog is cotranscribed with the downstream sufBCDS genes. Ferredoxin-thioredoxin reductase is a [4Fe-4S] protein through which thioredoxin undergoes redox regulation of protein function and signaling via thiol redox control. Thioredoxin has additional functions in defense against oxidative stress and is required by a number of transcription factors for DNA binding (1). The organization of the sufR, ftrC, and sufBCDS genes in Synechococcus sp. strain WH8102 implies that SufR may also be involved in regulation of the ferredoxin-thioredoxin system. A more pronounced exception to the gene organization exists in Thermosynechococcus elongatus, in which sufB is located immediately upstream of the sll0088 homolog, and even though the sufC and sufD genes are adjacent to each other, they are located elsewhere in the genome, as is the sufS gene. This difference raises the possibility that SufR might act to control a regulon rather than simply the sufBCDS operon. An indication that this might be the case is that the mRNA level for the bacterioferritin gene of Synechococcus sp. strain PCC 7002 is also increased in the sufR mutant strain (data not shown).
Homologs of the sufR gene may exist in nonphotosynthetic pathogenic bacteria.
Nonphotosynthetic bacteria, particularly pathogens and organisms that live under extreme conditions, also contain genes with strong similarity to sufR. Bacterial pathogens have developed efficient iron acquisition systems to counteract the defensive sequestration of iron by their hosts (6). These organisms must deal with oxidative stress exerted by either the host defense response or the environment. Although all four cysteines are conserved in sufR homologs of cyanobacteria, only three cysteines (the first, second, and fourth) are conserved in sufR homologs of nonphotosynthetic bacteria. The third position in the bacterial sufR homologs is occupied by either Ala (in Bacillus anthracis and Thermoplasma acidophilum), Thr (in Mycobacterium leprae and Streptomyces coelicolor), or Val (in Yersinia pestis), just to name a few possibilities. This group of homologs also includes Chloroflexus aurantiacus, a thermophilic photosynthetic green nonsulfur bacterium in which the SufR homolog contains only three cysteine residues. It should be noted that certain nonphotosynthetic bacteria have SufR homologs with additional cysteine residues near the C terminus. For example, Vibrio cholerae SufR contains two cysteines in addition to the four conserved cysteine residues that are found in cyanobacterial SufR. The iron-sulfur cluster coordinated by the common cysteine motif near the C terminus of these proteins may sense oxidative stress and thus control the expression of their target genes. Because there are only three conserved cysteine residues in the SufR homologs of nonphotosynthetic organisms, a noncysteine residue may be required as the fourth ligand to the iron-sulfur cluster. Indeed, IscR, a transcriptional repressor of the iscSUA operon, contains only three conserved cysteines, but EPR studies show that it binds a stable [2Fe-2S] cluster (32). Among the bacteria that contain sufR homologs, it is only in Mycobacterium tuberculosis strain H37RV and Mycobacterium leprae that four suf genes and one iscU-like gene (sufBB′CS-iscU) are located adjacent to the sufR homolog. In Thermoplasma acidophilum, only three suf genes (sufCBD) are adjacent to the sufR homolog, and in the other organisms that bear a sufR homolog, sufR and the other suf genes are separated by significant distances on the genome. This strengthens the notion suggested above that SufR may act to control a regulon rather than only the sufBCDS operon. E. coli and Erwinia chrysanthemi also contain a suf operon that is composed of all six suf genes (sufABCDSE), but no sufR homolog could be discerned.
The SufR protein is a member of the DeoR family of helix-loop-helix proteins.
An amino acid alignment and BLAST search against the GenBank database shows that the proteins coded by the sll0088 homologs contain three putative motifs: a protein kinase C phosphorylation site near the N terminus, a helix-loop-helix DNA-binding domain, and a motif of four cysteine residues separated by 12, 13, and 14 amino acids near the C terminus. The DNA-binding motif is highly conserved among sufR homologs of cyanobacteria and can be classified in the DeoR family of helix-loop-helix proteins. These proteins are typically highly diverse. However, because the helix-loop-helix motif is situated near the N terminus, SufR falls into the DeoR subfamily, which includes DeoR (23), LacR (30), and FucR (21). SufR appears to be a member of yet another DeoR subclass that contains a highly conserved C-X12-C-X13-C-X14-C motif near the C terminus.
The SufR protein may utilize an iron-sulfur cluster to sense iron levels or stress.
The SufR protein harbors an iron-sulfur cluster, as shown by optical absorption and EPR spectroscopy, and the spectra and relaxation properties are consistent with its identification as a [4Fe-4S] cluster. In future work, we will verify its identity as a [4Fe-4S] cluster, determine which of the cysteines provide ligands to the cluster, and decide whether the reconstituted protein is a monomer or dimer. It should be kept in mind that the iron-sulfur cluster was inserted in vitro into the E. coli-expressed apoprotein; hence, it is possible that SufR does not harbor an iron-sulfur cluster in vivo or that it harbors a different type of cluster (i.e., a [2Fe-2S] cluster) when functioning within the cyanobacterium. Nevertheless, given the functions of the proteins encoded by the sufBCDS operon, it is reasonable that SufR would bind an iron-sulfur cluster and serve as a sensor of oxidative stress that is commonly encountered by this organism. We propose that SufR indirectly senses the levels of iron-sulfur clusters in the cells through its own unstable iron-sulfur cluster; when this cluster is present, SufR binds to its operator and functions as a regulatory repressor of the sufBCDS operon. The oxygen sensitivity of the iron-sulfur cluster on the SufR protein may limit the lifetime of the active state, thereby allowing the poise of a quasi-steady-state population that could respond rapidly as the level of oxidative stress rises or falls.
There are other regulatory proteins that are proposed to fill a similar role (3). In a study with Erwinia chrysanthemi, Nachin and colleagues found that the suf operon might participate in a SoxR-dependent response to oxidative stress (24). SoxR is a transcription activator and contains a helix-loop-helix motif that confers sequence-specific DNA-binding capability (reviewed in reference 29). SoxR in E. coli is a homodimer and contains one [2Fe-2S] cluster per monomer. It has been shown that the iron-sulfur cluster is not required for SoxR to bind to the promoter of the soxS gene and that only the form of SoxR that contains the 2+ state iron-sulfur cluster can lead to the initiation of transcription of the soxS gene (12). SoxR senses reversible oxidation/reduction of the iron-sulfur cluster, a one-electron redox system (10). A SoxR homolog, a MerR-like protein, can be identified from searches in genomic sequences of Synechocystis sp. strain PCC 6803 (17), Synechococcus sp. strain PCC 7002 (Zhao et al., unpublished) and Anabaena sp. strain PCC 7120 (15). Whether this merR-like gene participates in redox regulation of the suf operon in cyanobacteria will be investigated in future studies. The IscR protein of E. coli also functions as a repressor of the iscRSUA operon, because deletion strains of iscR exhibit increased expression of this operon. IscR isolated anaerobically from a bacterial expression system contains a [2Fe-2S]1+ cluster that appears to be important for IscR function. It has been proposed that IscR may function in a autoregulatory mechanism that senses the iron-sulfur cluster assembly status of cells (32). The presence of a [4Fe-4S] cluster in SufR and the presence of a [2Fe-2S] cluster in IscR constitute significant differences between these two proteins, even though both function as iron-sensing transcriptional regulators. If the [4Fe-4S] cluster were indeed present in vivo, SufR would represent an entirely new subclass of regulator.
The SufBCDS proteins function in iron-sulfur cluster biogenesis in cyanobacteria.
Proteins encoded by the sufABCDSE genes have been proposed to be involved in the biogenesis and assembly of iron-sulfur clusters in bacteria (24, 41). However, detailed functions of most of these proteins are unknown. SufC is known to play a key role in iron metabolism and oxidative stress response (24); it contains an ATP-binding domain and may function as a versatile orphan ATPase in the iron-sulfur cluster assembly machinery. Reverse genetics has shown that the sufB and sufC genes are important in iron metabolism and oxidative stress in Erwinia chrysanthemi (24). Homologs of sufB and sufC, named ycf24 and ycf16, respectively, have been located in the chloroplast of the red alga Porphyra purpurea (38) and the cyanelle of Cyanophora paradoxa (19). SufC is also encoded in the plastid genome of the cryptomonad Guillardia theta, whereas in Arabidopsis thaliana, the sufC gene is encoded in the nucleus. However, the existence of a transit peptide in A. thaliana SufC indicates that the SufC protein is targeted to plant chloroplasts. This suggests that SufC may play a role in the assembly of the photosynthetic apparatus in cyanobacteria and the chloroplasts of higher plants. SufB and SufD share a highly conserved UPF0051 domain, suggesting that they assume similar functions in iron metabolism. In a sufD deletion mutant in E. coli, no iron-sulfur clusters are assembled in the FhuF protein (27); therefore, SufD is thought to be necessary for the stability of the iron-sulfur cluster proteins. The sufS gene codes for a NifS homolog; NifS has been shown to be essential for the construction iron-sulfur clusters in the nitrogenase enzyme of A. vinelandii (46). In E. coli, the iscS gene codes for a NifS homolog that has been shown to be a pyridoxal phosphate-dependent cysteine desulfurase (7). The cyanobacterial sufS gene is most closely related to the chloroplast nifS-like gene of A. thaliana. The SufS homolog also has some similarities to a protein encoded by the at1g08490 gene in A. thaliana, named AtNFS2 (20) or AtCpNIFS (28), which is a cysteine desulfurase targeted to the chloroplast. Indeed, cysteine desulfurase activity has been detected in the chloroplasts of A. thaliana (28). The SufS homolog is less similar to a protein encoded by the at5g65720 gene in A. thaliana, which is a putative cysteine desulfurase targeted to the mitochondria (L. McIntosh, unpublished data). Both organelles contain high concentrations of iron-sulfur clusters due largely to the presence of NADH dehydrogenase and fumarase in the mitochondria and PS I in the chloroplast. On the basis of these proposed functions, it is reasonable to propose that the suf operon in cyanobacteria is also involved in iron-sulfur cluster assembly for the photosynthetic complexes. Hence, the SufB, SufC, SufD, and SufS proteins probably work together to form iron-sulfur clusters in cyanobacteria.
The C14PsaC suppressor mutations are functionally equivalent to sufR null mutants.
The relationship between sll0088 (renamed here sufR) and PS I biogenesis and assembly was established by our earlier finding that a secondary mutation to sufR in the C14SPsaC mutant of Synechocystis sp. strain PCC 6803 restored photoautotrophic growth (43). The C14PsaC-R62 and C14SPsaC-R18 suppressor mutants exhibits an increased number of PS I reaction centers per cell (43). C14SPsaC-R62 was found to substitute Pro for Arg at residue 161 as the result of a G482→C change, and C14SPsaC-R18 was found to have a 3-amino-acid insertion of Gly-Tyr-Phe following Cys231 as the result of a TGGTTATTT duplication at T690. These two mutations were proposed to disrupt the structure, and hence the function, of sufR in Synechocystis sp. strain PCC 6803. We propose that the C14PsaC-R62 and C14SPsaC-R18 suppressor mutants are phenotypically identical to the sufR null mutants in that both result in the up-regulation of the sufBCDS genes that encode proteins involved in the synthesis of iron-sulfur clusters. PS I is the most abundant iron-sulfur protein complex in the cyanobacterial cell. The primary C14SPsaC mutation results in the occurrence of a mixed-ligand (3 Cys · 1 Ser) [4Fe-4S] cluster in the FB site. We propose that these PS I complexes are targeted as faulty, resulting in the turnover of the entire PS I complex. The level of PS I in the C14SPsaC mutant would then represent a balance between the rate of synthesis and the rate of turnover. If the rate-limiting step in the biosynthesis of PS I is indeed the formation of iron-sulfur clusters, then the overexpression of enzymes involved in iron-sulfur cluster biosynthesis would explain why the C14SPsaC-R62 and C14SPsaC-R18 mutants are able to increase the steady-state population of PS I reaction centers per cell in the secondary mutants. The nearly normal ratio of PS I to PS II would support photoautotrophic growth, which was the criterion by which the secondary suppressor mutants were initially selected. Thus, the mixed-ligand iron-sulfur cluster in the FB site of the C14S mutant is fully functional in vivo, as was implied by the ability of isolated PS I complexes to support high rates of electron transfer from cytochrome c6 to flavodoxin in vitro (43, 44).
Conclusions.
Four suf genes (sufBCDS) in cyanobacteria constitute an operon, whose transcription is negatively regulated by the SufR protein. SufR is therefore similar in function to iscR, which is an iron-sulfur protein that functions as a transcriptional repressor of the isc operon. The transcript levels of the suf genes increase under conditions of oxidative stress and iron limitation. As an iron-sulfur protein, SufR may serve as a sensor of oxidative stress. As for the distribution of suf genes, all four suf gene homologs in A. thaliana contain a transit peptide sequence at the N terminus of the Suf proteins that targets them to the chloroplast. This makes it very likely that the Suf proteins are involved in iron-sulfur assembly and biogenesis in chloroplasts. We propose that SufR plays an indirect role in controlling PS I biogenesis by regulating suf gene expression for the control and synthesis of iron-sulfur clusters.
Acknowledgments
This work was supported in part by the U.S. Department of Agriculture award 2001-35318-10125 to J.H.G., by a Department of Energy contract to L.M., and by National Science Foundation grant MCB-0077586 and NIH grant GM-31625 to D.A.B.
We thank David Becker for critically reading the manuscript.
REFERENCES
- 1.Arner, E. S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267:6102-6109. [DOI] [PubMed] [Google Scholar]
- 2.Aspinwall, R., D. G. Rothwell, T. Roldan-Arjona, C. Anselmino, C. J. Ward, J. P. Cheadle, J. R. Sampson, T. Lindahl, P. C. Harris, and I. D. Hickson. 1997. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl. Acad. Sci. USA 94:109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beinert, H., and P. J. Kiley. 1999. Fe-S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol. 3:152-157. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Duin, E. C., M. E. Lafferty, B. R. Crouse, R. M. Allen, I. Sanyal, D. H. Flint, and M. K. Johnson. 1997. [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry 36:11811-11820. [DOI] [PubMed] [Google Scholar]
- 6.Dussurget, O., and I. Smith. 1998. Interdependence of mycobacterial iron regulation, oxidative-stress response and isoniazid resistance. Trends Microbiol. 6:354-358. [DOI] [PubMed] [Google Scholar]
- 7.Flint, D. H. 1996. Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J. Biol. Chem. 271:16068-16074. [PubMed] [Google Scholar]
- 8.Frazzon, J., and D. R. Dean. 2002. Biosynthesis of the nitrogenase iron-molybdenum-cofactor from Azotobacter vinelandii. Met. Ions Biol. Syst. 39:163-186. [PubMed] [Google Scholar]
- 9.Frazzon, J., J. R. Fick, and D. R. Dean. 2002. Biosynthesis of iron-sulphur clusters is a complex and highly conserved process. Biochem. Soc. Trans. 30:680-685. [DOI] [PubMed] [Google Scholar]
- 10.Gaudu, P., and B. Weiss. 1996. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc. Natl. Acad. Sci. USA 93:10094-10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golbeck, J. H. 1999. A comparative analysis of the spin state distribution of in vivo and in vitro mutants of PsaC. A biochemical argument for the sequence of electron transfer in Photosystem I as FX → FA → FB → ferredoxin/flavodoxin. Photosynth. Res. 61:107-149. [Google Scholar]
- 12.Hidalgo, E., H. Ding, and B. Demple. 1997. Redox signal transduction via iron-sulfur clusters in the SoxR transcription activator. Trends Biochem. Sci. 22:207-210. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda, S., T. Biswas, R. Roy, T. Izumi, I. Boldogh, A. Kurosky, A. H. Sarker, S. Seki, and S. Mitra. 1998. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem. 273:21585-21593. [DOI] [PubMed] [Google Scholar]
- 14.Jung, Y. S., I. R. Vassiliev, J. P. Yu, L. McIntosh, and J. H. Golbeck. 1997. Strains of Synechocystis sp. PCC 6803 with altered PsaC. 2. EPR and optical spectroscopic properties of FA and FB in aspartate, serine, and alanine replacements of cysteines 14 and 51. J. Biol. Chem. 272:8040-8049. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213, 227-253. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement). DNA Res. 3:185-209. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko, T., A. Tanaka, S. Sato, H. Kotani, T. Sazuka, N. Miyajima, M. Sugiura, and S. Tabata. 1995. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 2:153-166, 191-198. [DOI] [PubMed] [Google Scholar]
- 19.Lambert, D. H., D. A. Bryant, V. L. Stirewalt, J. M. Dubbs, S. E. Stevens, Jr., and R. D. Porter. 1985. Gene map for the Cyanophora paradoxa cyanelle genome. J. Bacteriol. 164:659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leon, S., B. Touraine, J. F. Briat, and S. Lobreaux. 2002. The AtNFS2 gene from Arabidopsis thaliana encodes a NifS-like plastidial cysteine desulphurase. Biochem. J. 366:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, Z., and E. C. Lin. 1989. The nucleotide sequence of Escherichia coli genes for L-fucose dissimilation. Nucleic Acids Res. 17:4883-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehari, T., K. G. Parrett, P. V. Warren, and J. H. Golbeck. 1991. Reconstitution of the iron-sulfur clusters in the isolated FA/FB protein: EPR spectral characterization of same-species and cross-species photosystem I complexes. Biochim. Biophys. Acta 1056:139-148. [Google Scholar]
- 23.Mortensen, L., G. Dandanell, and K. Hammer. 1989. Purification and characterization of the deoR repressor of Escherichia coli. EMBO J. 8:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nachin, L., M. El Hassouni, L. Loiseau, D. Expert, and F. Barras. 2001. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39:960-972. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura, M., K. Saeki, and Y. Takahashi. 1999. Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster. J. Biochem. (Tokyo) 126:10-18. [DOI] [PubMed] [Google Scholar]
- 26.Ohmori, M., M. Ikeuchi, N. Sato, P. Wolk, T. Kaneko, T. Ogawa, M. Kanehisa, S. Goto, S. Kawashima, S. Okamoto, H. Yoshimura, H. Katoh, T. Fujisawa, S. Ehira, A. Kamei, S. Yoshihara, R. Narikawa, and S. Tabat. 2001. Characterization of genes encoding multi-domain proteins in the genome of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:271-284. [DOI] [PubMed] [Google Scholar]
- 27.Patzer, S. I., and K. Hantke. 1999. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 181:3307-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilon-Smits, E. A., G. F. Garifullina, S. Abdel-Ghany, S. Kato, H. Mihara, K. L. Hale, J. L. Burkhead, N. Esaki, T. Kurihara, and M. Pilon. 2002. Characterization of a NifS-like chloroplast protein from Arabidopsis. Implications for its role in sulfur and selenium metabolism. Plant Physiol. 130:1309-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 30.Rosey, E. L., and G. C. Stewart. 1992. Nucleotide and deduced amino acid sequences of the lacR, lacABCD, and lacFE genes encoding the repressor, tagatose 6-phosphate gene cluster, and sugar-specific phosphotransferase system components of the lactose operon of Streptococcus mutans. J. Bacteriol. 174:6159-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schilke, B., C. Voisine, H. Beinert, and E. Craig. 1999. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:10206-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98:14895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen, G., M. L. Antonkine, A. van der Est, I. R. Vassiliev, K. Brettel, R. Bittl, S. G. Zech, J. Zhao, D. Stehlik, D. A. Bryant, and J. H. Golbeck. 2002. Assembly of photosystem I. II. Rubredoxin is required for the in vivo assembly of FX in Synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J. Biol. Chem. 277:20355-20366. [DOI] [PubMed] [Google Scholar]
- 34.Shen, G., J. Zhao, S. K. Reimer, M. L. Antonkine, Q. Cai, S. M. Weiland, J. H. Golbeck, and D. A. Bryant. 2002. Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of Photosystem I activity. J. Biol. Chem. 277:20343-20354. [DOI] [PubMed] [Google Scholar]
- 35.Shen, G. Z., and D. A. Bryant. 1995. Characterization of a Synechococcus sp. strain PCC 7002 mutant lacking Photosystem I. Protein assembly and energy distribution in the absence of the Photosystem I reaction center core complex. Photosynth. Res. 44:41-53. [DOI] [PubMed] [Google Scholar]
- 36.Shen, G. Z., and W. F. J. Vermaas. 1994. Mutation of chlorophyll ligands in the chlorophyll-binding CP47 protein as studied in a Synechocystis sp. PCC 6803 photosystem I-less background. Biochemistry 33:7379-7388. [DOI] [PubMed] [Google Scholar]
- 37.Stevens, D. R., and R. Porter. 1980. Transformation in Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. USA 77:6052-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugiura, M. 1995. The chloroplast genome. Essays Biochem. 30:49-57. [PubMed] [Google Scholar]
- 39.Tachezy, J., L. B. Sanchez, and M. Muller. 2001. Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol. Biol. Evol. 18:1919-1928. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi, Y., A. Mitsui, T. Hase, and H. Matsubara. 1986. Formation of the iron sulfur cluster of ferredoxin in isolated chloroplasts. Proc. Natl. Acad. Sci. USA 83:2434-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi, Y., and U. Tokumoto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277:28380-28383. [DOI] [PubMed] [Google Scholar]
- 42.Ugulava, N. B., C. J. Sacanell, and J. T. Jarrett. 2001. Spectroscopic changes during a single turnover of biotin synthase: destruction of a [2Fe-2S] cluster accompanies sulfur insertion. Biochemistry 40:8352-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, J., G. Shen, T. Wang, D. A. Bryant, J. H. Golbeck, and L. McIntosh. 2003. Suppressor mutations in the study of photosystem I biogenesis: sll0088 is a previously unidentified gene involved in reaction center accumulation in Synechocystis sp. strain PCC 6803. J. Bacteriol. 185:3878-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, J. P., I. R. Vassiliev, Y. S. Jung, J. H. Golbeck, and L. McIntosh. 1997. Strains of Synechocystis sp. PCC 6803 with altered PsaC. 1. Mutations incorporated in the cysteine ligands of the two [4Fe-4S] clusters FA and FB of Photosystem I. J. Biol. Chem. 272:8032-8039. [DOI] [PubMed] [Google Scholar]
- 45.Zheng, L., V. L. Cash, D. H. Flint, and D. R. Dean. 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273:13264-13272. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]