SUMMARY
Karyopherinβ (Kapβ) proteins bind nuclear localization and export signals (NLSs and NESs) to mediate nucleocytoplasmic trafficking, a process regulated by Ran GTPase through its nucleotide cycle. Diversity and complexity of signals recognized by Kapβs have prevented prediction of new Kapβ substrates. The structure of Kapβ2 (also known as Transportin) bound to one of its substrates, the NLS of hnRNP A1, that we report here explains the mechanism of substrate displacement byRanGTPase. Further analyses reveal three rules for NLS recognition by Kapβ2: NLSs are structurally disordered in free substrates, have overall basic character, and possess a central hydrophobic or basic motif followed by a C-terminal R/H/KX(2–5)PY consensus sequence. We demonstrate the predictive nature of these rules by identifying NLSs in seven previously known Kapβ2 substrates and uncovering 81 new candidate substrates, confirming five experimentally. These studies define and validate a new NLS that could not be predicted by primary sequence analysis alone.
INTRODUCTION
Karyopherinβ proteins (Kapßs; also known as Importins and Exportins) are responsible for the majority of nucleo-cytoplasmic transport in the cell. At least 20 members of the Kapβ family have been identified in humans. Kapßs bind specific sets of transport substrates and target them to the nuclear pore complex. The Ran GTPase regulates Kapβ-substrate interactions and transport directionality through its nucleotide cycle (Chook and Blobel, 2001; Conti and Izaurralde, 2001; Gorlich and Kutay, 1999; Weis, 2003). RanGTP is concentrated in the nucleus, while RanGDP is concentrated in the cytoplasm. In import pathways, RanGTP and substrates bind Kapßs competitively, allowing substrate binding in the cytoplasm and RanGTP-mediated release in the nucleus. In contrast, in export pathways, RanGTP, substrates, and Kapßs bind cooperatively, resulting in substrate binding in the nucleus and release in the cytoplasm as the Ran bound nucleotide is hydrolyzed.
In humans, ten import Kapßs have been shown to carry a diverse set of macromolecular substrates into the nucleus (Mosammaparast and Pemberton, 2004). Despite significant efforts, only a few substrates have been identified for most import Kapßs, and large panels of substrates have been identified for only two pathways: those of Kapβ1 and Kapβ2 (see below). Each import Kapβ appears to bind distinct sets of substrates, suggesting that each Kapβ recognizes a different nuclear localization signal(s) (NLS[s]). However, large sequence diversity among various substrates has prevented identification of NLSs for most Kapßs, and it remains extremely difficult to predict NLSs in candidate import substrates.
The classical NLSs are short, lysine-rich sequences that bind the adaptor protein Kapα, which forms a heterodimer with Kapβ1, which in turn mediates nuclear import (Conti and Izaurralde, 2001). Most other proteins imported into the nucleus do not utilize such an adaptor but rather bind directly to a Kapβ. The few characterized NLSs that bind directly to Kapßs are diverse, encompassing both structural domains and linear epitopes. For example, crystal structures of three Kapβ1-substrate complexes show structurally diverse substrates binding at different sites on the karyopherin (Cingolani et al., 1999, 2002; Lee et al., 2003). Furthermore, most proteins that bind Kapβ1 show little sequence or structural homology, and thus general features among substrates in this pathway cannot be inferred at this time.
In another import pathway, more than 20 mRNA processing proteins (including hnRNPs A1, D, F, M, HuR, DDX3, Y-box binding protein 1, and TAP) have been identified as import substrates of Kapβ2 (Bonifaci et al., 1997; Fan and Steitz, 1998; Guttinger et al., 2004; Kawamura et al., 2002; Pollard et al., 1996; Rebane et al., 2004;Siomi et al., 1997; Suzuki et al., 2005; Truant et al., 1999). Kapβ2 binds its best-characterized substrate, splicing factor hnRNP A1, through the 38 residue M9 sequence (Bonifaci et al., 1997; Pollard et al., 1996) that we will refer to as M9NLS. Many studies have shown that the M9NLS peptide is both necessary and sufficient for nuclear import mediated by Kapβ2 (Siomi and Dreyfuss, 1995; Weighardt et al., 1995). Other than hnRNP A1, only NLSs in HuR (Fan and Steitz, 1998), TAP (Truant et al., 1999), and hnRNP D and its homologs, the JKTPB proteins (Kawamura et al., 2002; Suzuki et al., 2005), have been characterized. The NLSs of hnRNP D and HuR show marginal sequence homology to M9NLS, that of TAP shares no sequence homology with M9NLS, and none of the other Kapβ2 substrates contain obviousM9NLS-like sequences. Like the Kapβ1 system, the diversity of substrates and known NLSs in Kapβ2 has also prevented prediction of NLSs in this pathway.
In the nucleus, RanGTP binds import Kapßs with high affinity and dissociates substrates (Chook et al., 2002;Floer and Blobel, 1996; Gorlich et al., 1996). The unique repertoire of substrates for individual Kapßs suggests significant differences in their mechanisms of substrate recognition and therefore also differences in their regulation by Ran. The latter is illustrated in two different models for Ran-mediated substrate dissociation in the Kapβ1 and Kapβ2 pathways. For structurally diverse Kapβ1 substrates that also bind at different sites on the karyopherin, Ran-mediated dissociation involves both a global conformational change that locks the Kapβ1 superhelix into a substrate-incompatible conformation and a direct displacement by Ran (Cingolani et al., 1999, 2002; Lee et al., 2003, 2005; Vetter et al., 1999). Alternatively, structural and biochemical analyses of the Kapβ2-RanGTP complex suggest that RanGTP and substrate binding sites do not overlap and that an internal loop of Kapβ2 is crucial for substrate dissociation in the presence of Ran (Chook and Blobel, 1999; Chook et al., 2002). Thus it appears that the two best-known nuclear import pathways may utilize Ran to dissociate substrates in different manners.
In order to understand the mechanism of substrate recognition and distill the critical elements for NLS recognition by Kapβ2, and to understand the mechanism of Ran-mediated substrate dissociation for this import pathway, we have determined the structure of Kapβ2 bound to the M9NLS of hnRNP A1. The structure and complementary biochemical studies reveal a set of rules for NLS recognition by Kapβ2: NLSs imported by Kapβ2 should occur within large (>30-residue) structurally disordered elements, have overall basic character, and contain a set of consensus sequences. These rules are predictive and have allowed us to identify and biochemically confirm NLSs in seven known Kapβ2 substrates. Most importantly, we used these NLS rules in a bioinformatics approach and identified 81 new candidate import substrates for Kapβ2. We have confirmed that five of these bind Kapβ2 through the predicted NLS in a Ran-dependent manner. Finally, comparison with the previously determined structure of the Kapβ2-Ran complex (Chook and Blobel, 1999) has revealed the mechanism of Ran-mediated substrate dissociation. M9NLS binds in the C-terminal arch of Kapβ2, in a site spatially distinct from the Ran binding site. However, in the Ran complex, the acidic loop of Kapβ2 occupies this substrate binding site. Thus, Ran binding induces structural changes in Kapβ2 that are incompatible with substrate binding.
RESULTS AND DISCUSSION
Kapβ2-M9NLS Complex: Structure Overview
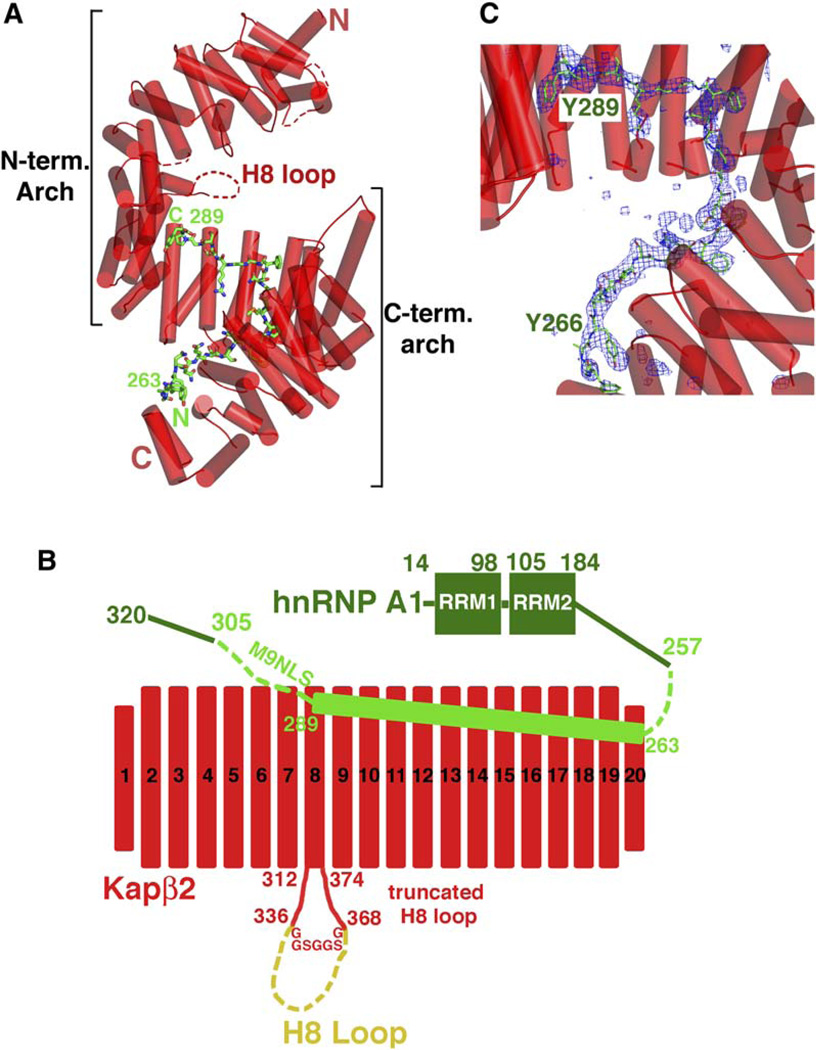
Kapβ2 is a superhelical protein with 20 HEAT repeats. It is almost exclusively α helical except for a 62 residue loop in repeat 8 (H8 loop, Figure 1A). Each repeat consists of two antiparallel helices, A and B, each lining the convex and concave sides of the superhelix, respectively (Chook and Blobel, 1999; Chook et al., 2002). Details of HEAT repeat nomenclature are described in the Supplemental Data. The Kapβ2-M9NLS crystals contain a Kapβ2 mutant with a truncated H8 loop bound to residues 257–305 of hnRNP A1 (Figure 1B). Biochemical studies showed that the loop neither hinders nor is necessary for substrate binding. However, it is sensitive to proteolytic degradation in substrate bound Kapβ2, suggesting structural flexibility (Chook et al., 2002). In the final Kapβ2 construct, the H8 loop was truncated (a GGSGGSG linker replaces residues 337–367) to minimize disorder in the crystal. The Kapβ2-M9NLS crystal structure was solved to 3.05 Å resolution (Table S1, PDB ID code 2H4M).
Figure 1. Crystal Structure of the Kapβ2-M9NLS Complex.
(A) Ribbon diagram of the Kapβ2-M9NLS complex with Kapβ2 in red (α helices represented as cylinders and structurally disordered loops as red dashes) and M9NLS shown as a stick figure (carbon: green, oxygen: red, nitrogen: blue, and sulfur: orange).
(B) The 20 HEAT repeats and H8 loop of Kapβ2 used in structural analyses (red) and M9NLS (light green) within hnRNP A1 (green). The deleted portion of the H8 loop is in yellow.
(C) The M9NLS binding site with Fo-Fc map (2.5 σ) calculated using Kapβ2 alone (blue mesh), drawn with PYMOL (DeLano, 2002).
The asymmetric unit of the crystal contains two Kapβ2-M9NLS complexes (I and II). All residues in both Kapβ2s are modeled except for three short loops at the N termini, H8 loop residues 320–337, and the engineered GGSG GSG H8 loop linker (disordered regions are indicated by dashes in Figures 1A, S1A, and S1B). Substrate residues 267–289 are observed in complex I, while additional substrate residues 263–266 are modeled in complex II (Figure 1C). Thus, the latter is used in structural analysis and discussion below. HEAT repeats 5–20 share similar conformations in both complexes (rmsd 1.7 Å). In contrast, HEAT repeats 1–4 diverge to a distance of 7Å at their N termini with high average B factors (93 Å2 for complex I and 118 Å2 for complex II), suggesting inherent conformational flexibility in this region of Kapβ2.
The 20 HEAT repeats of the Kapβ2-M9NLS complex form an almost perfect superhelix (pitch ~72 Å, diameter ~60 Å, and length ~111 Å; Figure 1A). The superhelix can also be described as two overlapping arches, with the N-terminal arch spanning HEAT repeats 1–13 and the C-terminal arch spanning repeats 8–20. In the Kapβ2-Ran complex, RanGTP binds in the N-terminal arch (Chook and Blobel, 1999). Here, we observe that M9NLS binds in the C-terminal arch (Figures 1A and 1C).
The Kapβ2-M9NLS Binding Interface
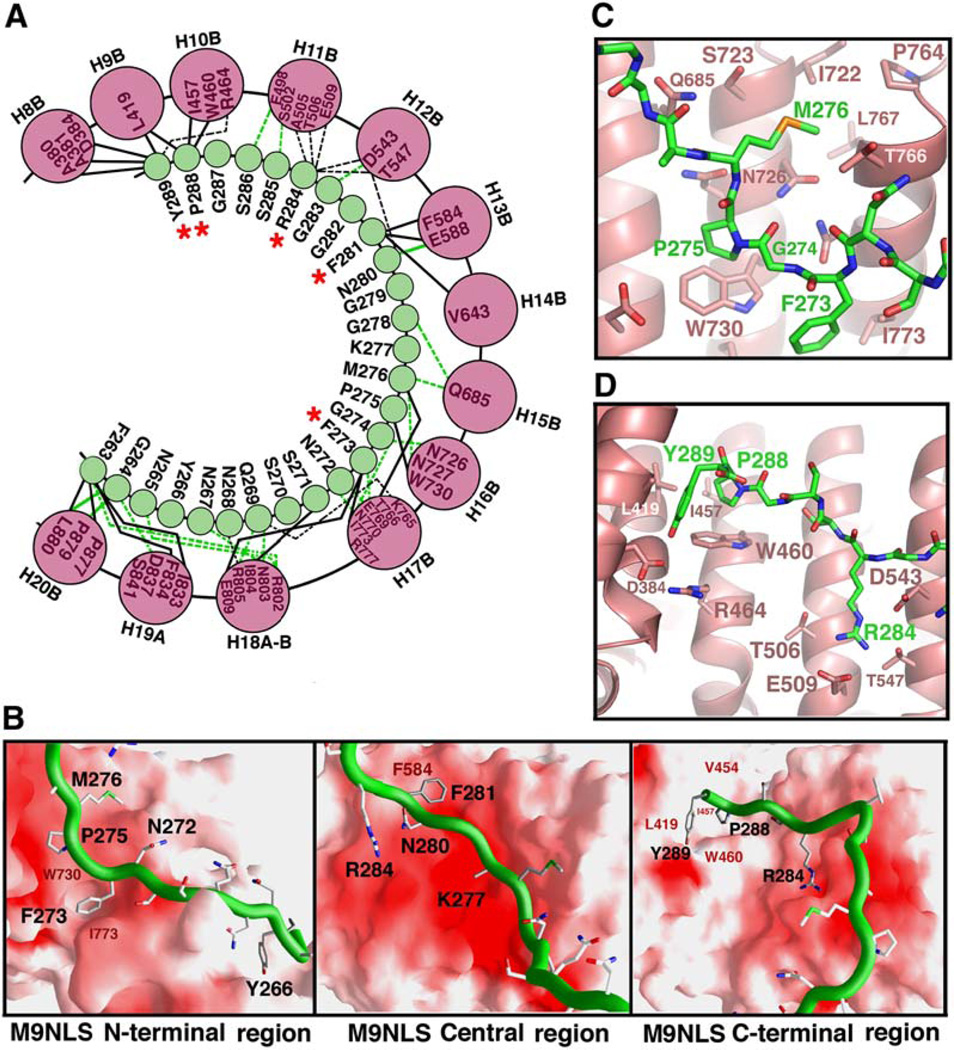
M9NLS binds in extended conformation to line the concave surface of the C-terminal arch of Kapβ2 (Figure 1A). Its peptide direction is antiparallel to that of the karyopherin superhelix, and substrate buries 3432 Å2 of surface area in both binding partners. Tracing M9NLS from N to C terminus, residues 263–266 interact with helices H18A, H19A, and H20B of Kapβ2, while residues 267–269 drape over the intra-HEAT 18 loop into the C-terminal arch of the karyopherin. The rest of M9NLS follows the curvature of the C-terminal arch to contact B helices of repeats 8–17 (Figures 1A and 2A). The substrate interface on Kapβ2 comprises ~30% of the concave surface of the C-terminal arch, which is relatively flat and devoid of deep pockets or grooves. Most of this surface, which includes the M9NLS interface, is also highly acidic (Figure 2B).
Figure 2. Kapβ2-M9NLS Interactions.
(A) Kapβ2-M9NLS contacts (<4.0 Å ) with M9NLS residues in green circles and Kapβ2 helices as pink circles. Contacts involving main chain atoms of M9NLS are shown with green lines. Contacts involving M9NLS side chains are shown with black lines. Solid lines are hydrophobic contacts and dashed lines are polar contacts. Red asterisks label M9NLS residues that make two or more side-chain contacts in both complexes in the asymmetric unit.
(B) The Kapβ2-M9NLS interface. The N-terminal third (left), the central region (middle), and the C-terminal third (right) of M9NLS. Substrate is shown as a green ribbon and the Kapβ2 electrostatic potential is mapped onto its surface, all drawn using GRASP (Nicholls et al., 1991). Red indicates negative electrostatic potential, white neutral, and blue positive. Residues in the hydrophobic patches of Kapβ2 are labeled in red and M9NLS residues labeled in black.
(C) Interactions between Kapβ2 (red) and substrate at M9NLS (green) residues 273FGPM276, drawn using PYMOL (DeLano, 2002).
(D) Interactions between Kapβ2 (red) and M9NLS (green) at the C terminus of the substrate, drawn using PYMOL (DeLano, 2002).
M9NLS forms an extensive network of polar and hydrophobic interactions with Kapβ2, involving both the main chain and side chains of the substrate (Figure 2A). Most of the substrate interface on Kapβ2 is acidic with the exception of several scattered hydrophobic patches. At the N terminus of M9NLS, residues 263–266 contact a hydrophobic patch on Kapβ2 helices H19A and H20B (Figure 2B, left). In the central region, a hydrophobic stretch 273FGPM276 contacts hydrophobic Kapβ2 residues I773 and W730 (Figures 2B and 2C). Further toward the C terminus, F281 binds near a hydrophobic patch formed by Kapβ2 residues F584 and V643 (Figure 2B, center), and finally, the C-terminal 288PY289 residues bind a large hydrophobic swath that includes Kapβ2 residues A380, A381, L419, I457, and W460 (Figures 2B, right and 2D). Despite the extensive acidic interface on Kapβ2, there are only two basic residues in M9NLS. R284 forms salt links with Kapβ2 residues E509 and D543, and the side chain of K277 is not observed.
Distribution of Binding Energy along M9NLS
In order to understand the distribution of binding energy along M9NLS, we measured dissociation constants (KDs) of a series of M9NLS mutants binding to Kapβ2 using isothermal titration calorimetry (ITC). The results of the binding studies using MBP-fusion proteins of M9NLS residues 257–305 and wild-type Kapβ2 are summarized in Table 1 and Figure S2. Wild-type M9NLS binds Kapβ2 with a KD of 42 nM. This ITC-measured affinity is somewhat lower than the previous KD of 2 nM measured by fluorescence titration but may be explained by the presence of both a covalently attached aromatic fluorophore and a significantly longer M9NLS spanning residues 238–320 in the earlier studies (Chook et al., 2002). Substrate residues that make two or more side-chain contacts with Kapβ2 (F273, F281, R284, P288, and Y289) were systematically mutated to alanines. Additional residues G274, P275, and M276 were also mutated given their implied importance in yeast-two-hybrid studies (Bogerd et al., 1999).
Table 1.
Kapβ2 Binding to M9NLS Mutants: Dissociation Constants by Isothermal Calorimetry
| MBP-M9NLS(257-305) proteins | KD |
|---|---|
| Wild type | 42 ± 2 nM |
| F273A | 61 ± 10 nM |
| G274A | 746 ± 63 nM |
| P275A | 74 ± 5 nM |
| M276A | 83 ± 17 nM |
| F281A | 56 ± 11 nM |
| R284A | 92 ± 9 nM |
| P288A | 158 ± 20 nM |
| Y289A | 133 ± 21 nM |
| P288A/Y289A | 136 ± 8 nM |
| R284A/P288A/Y289A | 461 ± 27 nM |
| G274A/P288A/Y289A | 5.9 ± 0.7 µM |
G274A is the only single mutant that shows significant (18-fold) decrease in Kapβ2 binding (Table 1). Single mutants of C-terminal residues P288 and Y289 follow with modest decreases of 3- to 4-fold. Thus, it appears that M9NLS binds Kapβ2 in a mostly distributive fashion, with a strict requirement for glycine at position 274 and modest though possibly important energetic contributions from C-terminal residues P288 and Y289. The importance of the PY motif is suggested in the R284/P288/Y289 and G274/P288/Y289 triple mutants, where 10-fold and 140-fold decreases were observed, respectively. Both triple mutants show nonadditivity in their binding energies when compared with single G274A and R284A and the double PY mutants, suggesting cooperativity between the C-terminal PY motif and both upstream binding sites at R284 and G274. The significance of the G274A mutation had previously been reported in both Kapβ2 binding and nuclear import assays (Fridell et al., 1997; Nakielny et al., 1996). The α carbon of G274 is in close proximity to neighboring substrate side chains F273 and P275 as well as Kapβ2 residue W730, such that a side chain in position 274 may result in a steric clash (Figure 2C).
The important energetic contributions of the substrate’s C-terminal PY motif and its central G274 residue are also supported by mutations of interacting residues in Kapβ2. Double and triple Kapβ2 mutants, W460A/W730A and I457A/W460A/W730A, both show significant decreases in Kapβ2 binding (Figure S1C). I457 and W460 interact with the substrate PY motif, while W730 makes a hydrophobic contact with substrate P275 and is also close to G274 (Figures 2C and 2D).
Rules for Substrate Recognition by Kapβ2
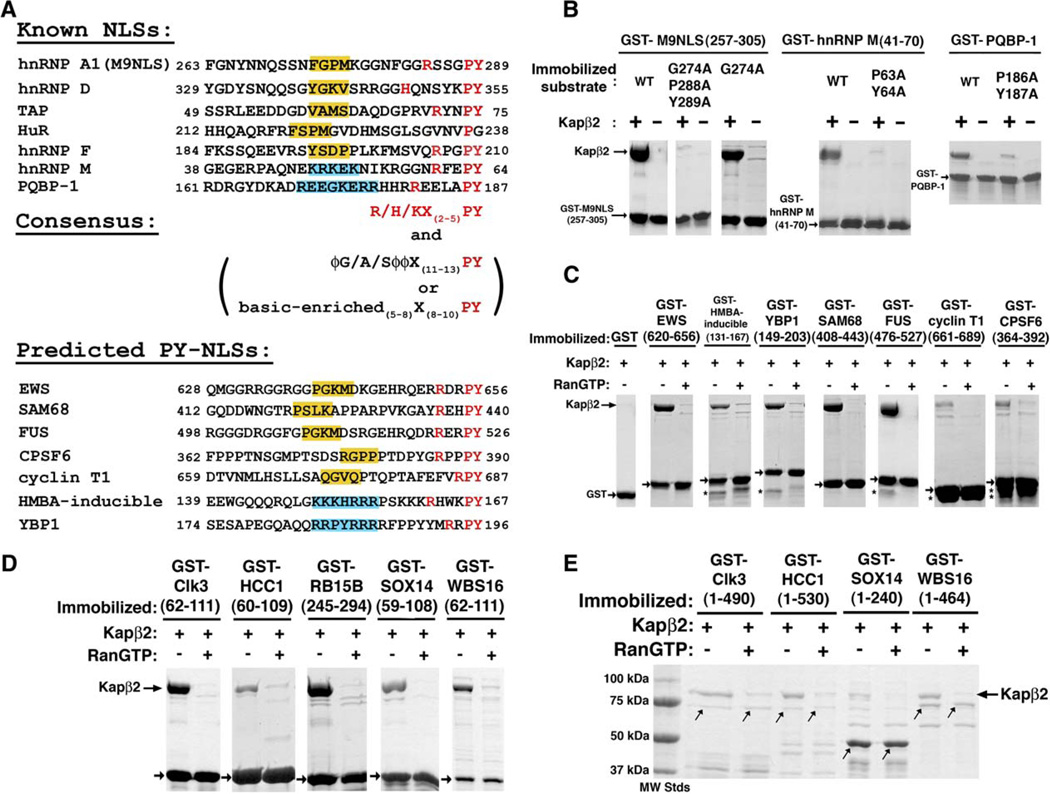
Prior to this study, among more than 20 known Kapβ2 substrates, only NLSs from hnRNP A1, D, HuR, TAP, and their homologs had been identified (Fan and Steitz, 1998; Kawamura et al., 2002; Siomi and Dreyfuss, 1995;Suzuki et al., 2005; Truant et al., 1999). All four NLSs span 30–40 residues, are rich in glycine and serine residues, and have overall basic character but share little sequence homology. To aid in assessment of the rules for NLS recognition by Kapβ2 suggested below, we constructed a series of deletion mutants to map three additional NLSs from hnRNP F, M, and PQBP-1. The results of in vitro binding assays map the NLSs to residues 151–190 in PQBP-1, residues 41–70 in hnRNP M, and residues 190–245 in hnRNP F (Figures S3A–S3C). Structural and mutagenesis analysis of the Kapβ2-M9NLS complex combined with sequence comparison and analysis of all seven NLSs reveals three rules for NLS recognition by Kapβ2.
Rule 1: NLS Is Structurally Disordered in Substrate
The extended conformation of the 26 residue M9NLS results in a linear epitope that traces a path of ~110 Å. The structure of the bound substrate suggests that an NLS recognized by Kapβ2 should exist within a stretch of at least 30 residues that lacks secondary structure in its native, unbound state. Thus, the NLS is most likely structurally disordered in the free substrate. The prediction of this NLS requirement is further supported by the fact that all seven known NLSs in Kapβ2 substrates occur within sequences with high probability of structural disorder (>0.7) calculated by the program DisEMBL (Linding et al., 2003). All seven NLSs are found either in loop regions between the RNA binding or other folded domains or at the termini of the substrates.
Rule 2: Overall Positive Charge for NLS Is Preferred
A second requirement for an NLS recognized by Kapβ2 emerges from the observation that Kapβ2’s substrate interface is highly negatively charged. An acidic peptide would likely not bind due to electrostatic repulsion, while an NLS with overall positive charge would most likely be favored. Examination of all known Kapβ2 NLSs indicates overall basic character spanning at least 30 residues in six of seven cases (Figure 3A). In addition, regions that flank the NLSs most likely also contribute favorably to electrostatics. For example, although the TAP-NLS sequence delineated in Figure 3A has slightly more acidic than basic residues, flanking regions are highly basic and may ultimately contribute to overall basic character to promote Kapβ2 binding. The importance of basic flanking regions is also observed in hnRNP A1. Here, the entire 135 residue C-terminal tail of the substrate has overall positive charge. A recent study showed that following osmotic shock stress in cells, four serine residues C-terminally adjacent to the M9NLS are phosphorylated, resulting in decreased binding to Kapβ2 and accumulation of hnRNP A1 in the cytoplasm (Allemand et al., 2005). Phosphorylation of the M9NLS-flanking serines may decrease the basic character of M9NLS and thus modulate interactions with Kapβ2.
Figure 3. Consensus Sequences of NLSs Recognized by Kapβ2.
(A) Alignment of all known (top) and predicted (bottom) NLSs recognized by Kapβ2 at conserved PY residues. NLSs in known Kapβ2 substrates are predicted by the presence of the R/K/H-X(2–5)-P-Y C-terminal motifs (red) within structurally disordered and positively charged regions of 30 amino acids. Central hydrophobic motifs ϕG/A/Sϕϕ (ϕ is a hydrophobic side chain) are shaded yellow. Central basic motifs are shaded blue.
(B) Binding assays of Kapβ2 and immobilized alanine mutants of M9NLS, PQBP-1, and NLS-containing fragments of hnRNP M. Bound proteins are visualized with Coomassie blue.
(C) Binding assays of predicted NLSs from known Kapβ2 substrates EWS, HMBA-inducible protein, YBP1, SAM68, FUS, Cyclin T1 and CPSF6. Kapβ2 is added to immobilized GST-NLSs (arrows) in the presence and absence of excess RanGTP, and bound proteins visualized with Coomassie blue. Asterisks label degraded fragments of substrates.
(D) Five predicted Kapβ2 substrates (Clk3, HCC1, RB15B, Sox14, and WBS16) are validated experimentally. GST NLSs (arrows) are immobilized on glutathione sepharose.
(E) Binding assays of full-length substrates Clk3,HCC1, Sox14, and WBS16 to Kapβ2. Expression of recombinant full-length RB15B was not successful. Coomasie-stained bands at the size of the GST substrates are labeled with arrows. Lower-molecular-weight proteins are likely degraded substrates.
Rule 3: Consensus Sequences for the NLS
All seven characterized NLSs recognized by Kapβ2 exist in structurally disordered regions, suggesting that this class of NLS is represented by linear epitopes and not folded domains. However, apparent sequence diversity among previously characterized NLSs from hnRNP A1, HuR, TAP, and JKTBP homologs had prevented delineation of a consensus sequence that could be used to identify new NLSs or substrates. However, despite apparent NLS diversity, mutagenesis, structural, and sequence analyses have resulted in the identification of two regions of conservation within the sequences.
The first region of conservation is found at the C terminus of the NLSs. Mutagenesis of M9NLS suggested the importance of its C-terminal PY motif (Table 1). Sequence examination of previously characterized NLSs from hnRNP D, HuR, and TAP, as well as the newly characterized NLSs of hnRNP F, M and PQBP-1, identified consecutive PY residues in six of the seven sequences (Figure 3A). Mutations of the PY residues in PQBP-1 and hnRNP M also decreased Kapβ2 binding, suggesting that they make energetically important contacts (Figure 3B). Mutations of the PY motif in JKTBP proteins and M9NLS were also previously shown to inhibit nuclear import (Iijima et al., 2006; Suzuki et al., 2005). In addition, we observe that a basic residue is always found several residues N-terminal of the PY sequence, consistent with an adjacent acidic surface on Kapβ2 (Figures 2B, 2D, and 3A). Based on these observations, we propose a C-terminal consensus sequence R/K/H-X(2–5)-P-Y (where X is any residue) for NLSs recognized by Kapβ2. We refer to this class of NLSs as PY-NLSs.
A second region of conservation within the PY-NLSs is found in the central region of the peptides. Examination of the central region divides the seven PY-NLSs into two subclasses. The first subclass includes M9NLS and NLSs of hnRNP D, F, TAP, and HuR, where four consecutive predominantly hydrophobic residues are located 11–13 residues N-terminal to the PY residues (Figure 3A). We refer to this subclass of sequences as hydrophobic PY-NLSs or hPY-NLSs. In contrast, the central regions of NLSs from hnRNP M and PQBP-1 are virtually devoid of hydrophobic residues but are instead enriched in basic residues. They appear to represent a distinct subclass of PY-NLSs that we call the basic PY-NLSs or bPY-NLSs.
The central hydrophobic motif in M9NLS spans residues 273FGPM276 previously found in yeast-two-hybrid studies and mutagenesis analysis to be important for import by Kapβ2, and a consensus sequence of Z-G-P/K-M/L/V-K/R (where Z is a hydrophobic residue) was previously suggested (Bogerd et al., 1999). The mutagenesis-derived consensus holds in the context of the M9NLS sequence but does not describe NLSs in other Kapβ2 substrates. A loose consensus of ϕ-G/A/S-ϕ-ϕ (where ϕ is a hydrophobic side chain) seems more appropriate upon comparison of the five central hydrophobic motifs in hnRNPs A1, D, F, TAP, and HuR (Figure 3A). The Kapβ2-M9NLS structure explains preferences for hydrophobic side chains in positions 1, 3, and 4, as well as small or no side chain in position 2. Position 1 in M9NLS is F273, which occupies a hydrophobic pocket formed by Kapβ2 residues W730 and I773 (Figure 2C). Position 3 is occupied by P275, which stacks on top of the indole ring of Kapβ2 W730, and M276 in position 4 binds a small hydrophobic patch on Kapβ2 formed by I722, P764, L766, and the Cβ of S767. Thus, hydrophobic or long aliphatic side chains at positions 1, 3, and 4 in other hydrophobic hPY-NLSs would provide energetically favorable hydrophobic contacts with Kapβ2. Mutagenesis of M9NLS suggests a strict requirement for glycine at position 2 (residue G274 in M9NLS) of the central hydrophobic motif. G274 is surrounded by adjacent substrate residues F273, P275, and Kapβ2 residue W730, suggesting that the strict requirement for glycine is likely heavily dependent on the identity of adjacent substrate residues. Nevertheless, hydrophobic neighbors, even those not as bulky as F273 and P275 in M9NLS, will likely still not accommodate large side chains in position 2.
The Kapβ2-M9NLS structure provides some suggestion for how the central basic motif in the bPY-NLSs could be accommodated. In the structure, the M9NLS hydrophobic motif interacts with Kapβ2 hydrophobic residues that are surrounded by numerous acidic residues (Figures 2B and 2C). Thus, the highly acidic substrate interface on Kapβ2 that contacts the central region of an NLS should also be able to interact favorably with numerous basic side chains. It is possible that the central basic and hydrophobic motifs in the two subclasses of PY-NLSs may take slightly different paths on Kapβ2. Structures of Kapβ2 bound to bPY-NLSs will be necessary to understand the difference between the two subclasses of PY-NLSs.
The NLS Rules Are Predictive
We have examined the sequences of the following eight recently identified Kapβ2 substrates: Ewing Sarcoma protein (EWS), hexamethylene bis acetamide (HMBA)-inducible protein, Y-box binding protein 1 (YBP1), SAM68, FUS, DDX3, CPSF6, and Cyclin T1 (Guttinger et al., 2004). We found the C-terminal R/K/H-X(2–5)-P-Y consensus within structurally disordered and positively charged regions of seven of them. The predicted NLSs for EWS, HMBA-inducible protein, YBP1, SAM68, FUS, CPSF6, and Cyclin T1 are listed in the bottom half of Figure 3A. The predicted signals in EWS, SAM68, FUS, CPSF6, and Cyclin T1 are hPY-NLSs, and those from HMBA-inducible protein and YBP1 are bPY-NLSs (Figure S3D). The easily detected PY motif is absent from DDX3, and we have not been able to show direct binding of DDX3 to Kapβ2 (data not shown). Thus, DDX3 may not be a substrate of Kapβ2 but may enter the nucleus by binding to a bona fide Kapβ2 substrate. All seven predicted NLSs bind Kapβ2 and are dissociated from the karyopherin by RanGTP, consistent with NLSs imported by Kapβ2 (Figure 3C). The NLSs of Cyclin T1 and CPSF6 bind Kapβ2 but more weakly than other substrates. It is not clear if this is due to proteolytic degradation of the substrates or to poor central hydrophobic motifs (Figures 3A, 3C, and S3D). Confirmation of these seven NLSs indicates that the three rules for NLS recognition by Kapβ2 described above are predictive.
We have also applied the NLS rules to human proteins in the SwissProt protein database (Bairoch et al., 2004) to identify potential Kapβ2 substrates. A search for proteins containing NLS-sequence motifs (Figures 3A and S3D) using the program ScanProsite (Gattiker et al., 2002) followed by filtering for structural disorder (DisEMBL) (Linding et al., 2003) and for overall positive charge in the NLS resulted in 81 new candidate Kapβ2 substrates (Tables 2 and 3). We chose five of these at random—protein kinase Clk3 (P49761), transcription factor HCC1 (Q14498), mRNA processing proteins RB15B (Q8NDT2) and Sox14 (O95416), and the Williams-Beuren syndrome chromosome region 16 protein/WBS16 (Q96I51)—and showed that both their predicted NLSs and the full-length proteins (except for RB15B, which could not be expressed in bacteria) bind Kapβ2 and can be dissociated by RanGTP (Figures 3D and 3E). Thus, the rules not only identify NLSs in known substrates but also are highly effective in predicting entirely new substrates.
Table 2.
Predicted Kapβ2 Substrates with Hydrophobic PY-NLSs
| Accession Number |
Name | Localizationa | N-Term. Residue |
Sequences for Candidate Hydrophobic PY-NLSb | C-Term. Residue |
|---|---|---|---|---|---|
| Q8IZP0 | Abl interactor 1 | C, N | 158 | KHGNNQPARTGTLSRTNPPTQKPPSPP MSGRGTLGRNTPYKTLEPVKPPT | 207 |
| Q9UKA4 | A-kinase anchor protein 11/AKAP 220 | C, Centrosome | 385 | QRKGHKHGKSCMNPQKFKFDRPALPA NVRKPTPRKPESPYGNLCDAPDSP | 434 |
| P50995 | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) |
C, N | 84 | PVPPGGFGQPPSAQQPVPPYGMYPP PGGNPPSRMPSYPPYPGAPVPGQPM | 133 |
| Q13625 | Apoptosis-stimulating of p53 protein 2 | C, N | 474 | TLRKNQSSEDILRDAQVANKNVAKVP PPVPTKPKQINLPYFGQTNQPPSD | 523 |
| Q9BXP5 | Arsenite-resistance protein 2c | not known | 53 | GEYRDYDRNRRERFSPPRHELSPP QKRMRRDWDEHSSDPYHSGYEMPYAG | 102 |
| Q92560 | Ubiquitin carboxyl-terminal hydrolase BAP1(BRCA1-associated protein 1)c |
N | 685 | EGMLANLVEQNISVRRRQGVSIGRL HKQRKPDRRKRSRPYKAKRQ | 729 |
| P48634 | Large proline-rich protein BAT2 (HLA-B-associated transcript 2) |
C, N | 690 | VPAPQAPPPPPKALYPGALGRPPPM PPMNFDPRWMMIPPYVDPRLLQGRP | 739 |
| O15178 | Brachyury protein | N | 251 | TSTLCPPANPHPQFGGALSLPSTHS CDRYPTLRSHRSSPYPSPYAHRNNS | 300 |
| O60885 | Bromodomain-containing protein 4 (HUNK1 protein) |
N | 1015 | QGQQPPHPPPGQQPPPPQPAKPQQV IQHHHSPRHHKSDPYSTGHLREAPSP | 1064 |
| Q14004 | Cell division cycle 2-like protein kinase 5 |
not known | 376 | YERGGDVSPSPYSSSSWRRSRSPYSPVLRRSGKSRSRSPYSSRHSRSRSR | 425 |
| Q9NYV4 | Cell division cycle 2-related protein kinase 7 |
N | 256 | SSNYDSYKKSPGSTSRRQSVSPPYK EPSAYQSSTRSPSPYSRRQRSVSPY | 305 |
| Q5TGI0 | Protein C6orf168 | not known | 94 | IDSKDAIILHQFARPNNGVPSLSPF CLKMETYLRMADLPYQNYFGGKLSA | 143 |
| P49761 | Dual specificity protein kinase CLK3 (CDC-like kinase 3/Clk3)c |
N | 18 | YRWKRRRSYSREHEGRLRYPSRREP PPRRSRSRSHDRLPYQRRYRERRDS | 67 |
| P05997 | Collagen alpha-2(V) chain precursor | not known | 611 | MGLPGPKGSNGDPGKPGEAGNPGVP GQRGAPGKDGKVGPYGPPGPPGLRG | 660 |
| Q03692 | Collagen alpha-1(X) chain precursor | not known | 84 | GYGSPGLQGEPGLPGPPGPSAVGKP GVPGLPGKPGERGPYGPKGDVGPAG | 133 |
| Q8TBR5 | Protein C19orf23c | not known | 70 | TWQTRNHTRTGHAYPRFTRPSFPSC NRNGKRRKLRLGLPY | 119 |
| Q96RT6 | Protein cTAGE-2 | not known | 692 | PPGTVFGASPDYFSPRDVPGPPRAP FAMRNVYLPRGFLPYRPPRPAFFPQ | 741 |
| Q9NSV4 | Protein diaphanous homolog 3 (Diaphanous-related formin-3) |
not known | 1070 | GAAFRDRRKRTPMPKDVRQSLSPM SQRPVLKVCNHGNKPYL | 1110 |
| P56177 | Homeobox protein DLX-1 | N | 44 | CLHSAGHSQPDGAYSSASSFSRPLG YPYVNSVSSHASSPYISSVQSYPGS | 93 |
| O95147 | Dual specificity protein phosphatase 14/MAP kinase phosphatase 6 |
not known | 156 | RQLIDYERQLFGKSTVKMVQTPYGIV PDVYEKESRHLMPYWGI | 200 |
| Q9BUP0 | EF-hand domain-containing protein 1 Swiprosin-2) |
not known | 42 | PPARAPTASADAELSAQLSRRLDINE GAARPRRCRVFNPYTEFPEFSRRL | 91 |
| Q6ZV73 | FYVE, RhoGEF and PH domain- containing protein 6 (Zinc finger FYVE domain-containing protein 24) |
C | 269 | SSELEALENGKRSTLISSDGVSKKSE VKDLGPLEIHLVPYTPKFPTPKPR | 318 |
| Q92837 | Proto-oncogene FRAT1 | N | 89 | PAVPLLLPPALAETVGPAPPGVLRCA LGDRGRVRGRAAPYCVAELATGPS | 138 |
| Q96AE4 | FUSE-binding protein 1/DNA helicase V |
N | 465 | PGPHGPPGPPGPGTPMGPYNPAPYNP GPPGPAPHGPPAPYAPQGWGNAYP | 514 |
| Q8NEA6 | Zinc finger protein GLIS3 | N | 601 | LTAVDAGAERFAPSAPSPHHISPRRV PAPSSILQRTQPPYTQQPSGSHLK | 650 |
| Q8TEK3 | Histone H3-K79 methyltransferase | N | 775 | SPAKIVLRRHLSQDHTVPGRPAASEL HSRAEHTKENGLPYQSPSVPGSMK | 824 |
| P35452 | Homeobox protein Hox-D12 (Hox-4H) |
N | 175 | AGVASCLRPSLPDGKRCPCSPGRPAVG GGPGEARKKRKPYTKQQIAELEN | 224 |
| Q13422 | DNA-binding protein Ikaros (Lymphoid transcription factor LyF-1) |
N | 254 | CKIGSERSLVLDRLASNVAKRKSSMPQ KFLGDKGLSDTPYDSSASYEKEN | 303 |
| O43474 | Kruppel-like factor 4 (Epithelial zinc-finger protein EZF) (Gut-enriched Krueppel-like factor) |
N | 218 | GKFVLKASLSAPGSEYGSPSVISVSKGS PDGSHPVVVAPYNGGPPRTCPK | 267 |
| Q8NEZ4 | Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3 |
N | 2427 | NVNQAFTRPPPPYPGNIRSPVAPPLGPR YAVFPKDQRGPYPPDVASMGMR | 2476 |
| Q96G25 | Mediator of RNA polymerase II transcription subunit 8 homolog (ARC32) |
N | 227 | GAPSQQQPMLSGVQMAQAGQPGKMPSG IKTNIKSASMHPYQR | 268 |
| Q93074 | Mediator of RNA polymerase II transcription subunit 12 |
N | 1854 | DLLHHPNPGSITHLNYRQGSIGLYTQN QPLPAGGPRVDPYRPVRLPMQKL | 1903 |
| O43312 | Metastasis suppressor protein 1 (Metastasis suppressor YGL-1) |
not known | 379 | LPRVTSVHLPDYAHYYTIGPGMFPSSQ IPSWKDWAKPGPYDQPLVNTLQR | 428 |
| Q13310 | Polyadenylate-binding protein 4 | C | 484 | GAAQQGLTDSCQSGGVPTAVQNLAPRA AVAAAAPRAVAPYKYASSVRSPH | 533 |
| Q9Y6V0 | Piccolo protein (Aczonin) | C | 2874 | VVYKLPFGRSCTAQQPATTLPEDRFGYR DDHYQYDRSGPYGYRGIGGMKP | 2923 |
| Q8NFH8 | RalBP1-associated Eps domain- containing protein 2 (RalBP1- interacting protein 2) |
C | 188 | PTMSPLASPPSSPPHYQRVPLSHGYSKL RSSAEQMHPAPYEARQPLVQPE | 237 |
| O75177 | SS18-like protein 1 (SYT homolog 1) | not known | 196 | SHYSSAQGGSQHYQGQSSIAMMGQGSQGSSMMGQRPMAPYRPSQQGSSQQ | 245 |
| Q92922 | SWI/SNF complex 155 kDa subunit (BRG1-associated factor 155) |
C, N | 960 | QQQHGQNPQQAHQHSGGPGLAPLGAAGHPGMMPHQQPPPYPLMHHQMPPP | 1009 |
| P09012 | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) |
N | 123 | AVQGGGATPVVGAVQGPVPGMPPMTQAPRIMHHMPGQPPYMPPPGMIPPP | 172 |
| P18583 | SON3/Negative regulatory element-binding protein/DBP-5 |
N | 945 | GQDPYRLGHDPYRLTPDPYRMSPRPYRI APRSYRIAPRPYRLAPRPLMLA | 994 |
| Q8IXZ3 | Transcription factor Sp8 (Specificity protein 8) |
N | 164 | GGSSAHSQDGSHQPVFISKVHTSVDGL QGIYPRVGMAHPYESWFKPSHPG | 213 |
| Q15532 | SSXT protein (SYT protein) | not known | 214 | QYNMPQGGGQHYQGQQPPMGMMGQVNQGNHMMGQRQIPPYRPPQQGPPQQ | 263 |
| Q9UMS6 | Synaptopodin-2 (Myopodin) (Genethonin 2)c |
C, N | 931 | PSYPLAALKSQPSAAQPSKMGKKKGKK PLNALDVMKHQPYQLNASLFTFQ | 980 |
| Q9Y5Q8 | General transcription factor 3C polypeptide 5 |
N | 31 | GVVRDVAKMLPTLGGEEGVSRIYADPT KRLELYFRPKDPYCHPVCANRFS | 80 |
| Q04206 | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit) |
C, N | 310 | KSIMKKSPFSGPTDPRPPPRRIAVPSR SSASVPKPAPQPYPFTSSLSTIN | 359 |
| Q9NRE2 | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer- related protein 10-2) |
N | 558 | LPMGSRVLQIRPNLTNKLRPIAPKWKV MPLVSMPTHLAPYTQVKKESEDK | 607 |
| Q9UJT2 | Testis-specific serine kinase substrate |
not known | 275 | PAATSQGCPGPPGSPDKPSRPHGLVPA GWGMGPRAGEGPYVSEQELQKLF | 324 |
| Q8TAP9 | TTD nonphotosensitive 1 protein | N | 15 | GPGGGGWGSGSSFRGTPGGGGPRPPSPRDGYGSPHHTPPYGPRSRPYGSS | 64 |
| Q96I51 | Williams-Beuren syndrome chromosome region 16 protein (WBS16) |
N | 62 | FVWGFSFSGALGVPSFVVPSSGPGPRAG ARPRRRIQPVPYRLELDQKISS | 111 |
| P19544 | Wilms’ tumor protein (WT33) | N | 94 | VHFSGQFTGTAGACRYGPFGPPPPSQAS SGQARMFPNAPYLPSCLESQPA | 143 |
| P17861 | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5) |
N | 202 | ISCWAFWTTWTQSCSSNALPQSLPAWRS SQRSTQKDPVPYQPPFLCQWGR | 251 |
| Q8NAP3 | Zinc finger and BTB domain- containing protein 38 |
N | 539 | HAIDHRLSISKKTANGGLKPSVYPYKLY RLLPMKCKRAPYKSYRNSSYEN | 588 |
| Q9C0A1 | Zinc finger homeobox protein 2 | N | 784 | VKPPATATPASLPKFNLLLGKVDDGTGR EAPKREAPAFPYPTATLASGPQ | 833 |
As annotated in the UniProtKB/Swiss-Prot entries. C represents cytoplasm and N represents nucleus.
Central hydrophobic motifs are underlined and the R/K/H-PY motifs are in bold.
Substrates also identified using bPY-NLS motif.
Table 3.
Predicted Kapβ2 Substrates with bPY-NLSs
| Accession Number |
Name | Localizationa | N-Term. Residue |
Sequences for Candidate Hydrophobic PY-NLSb | C-Term. Residue |
|---|---|---|---|---|---|
| Q13023 | A-kinase anchor protein 6 (AKAP 100) | not available | 1851 | GSVKRVSENNGNGKNSSHTHELGT KRENKKTIFKVNKDPYVADMENGNIE | 1900 |
| Q9BXP5 | Arsenite-resistance protein 2c | not available | 61 | NRRERFSPPRHELSPPQKRMRRDWD EHSSDPYHSGYEMPYAGGGGGPTYG | 110 |
| Q92560 | BRCA1-associated protein 1c | N | 685 | EGMLANLVEQNISVRRRQGVSIG RLHKQRKPDRRKRSRPYKAKRQ | 729 |
| Q9NYF8 | Bcl-2-associated transcription factor 1 | C, N | 32 | KRYSSRSRSRTYSRSRSRDRMYSRD YRRDYRNNRGMRRPYGYRGRGRGYY | 81 |
| Q9ULD4 | Bromodomain and PHD finger-containing protein 3 |
not available | 1 | MRKPRRKSRQNAEGRRSPSPYSLKC SPTRET | 31 |
| Q9UK58 | Cyclin-L1 | N | 337 | ASKPSSPREVKAEEKSPISINVKTVKK EPEDRQQASKSPYNGVRKDSKRS | 386 |
| Q9NYF5 | Protein C5orf5 (GAP-like protein N61) | not available | 531 | QRFLHDPEKLDSSSKALSFT RIRRSSFSSKDEKREDRTPYQLVKKLQKKI | 580 |
| P49761 | CDC-like kinase 3c | N | 62 | RERRDSDTYRCEERSPSFGEDYYGPS RSRHRRRSRERGPYRTRKHAHHCH | 111 |
| Q8TBR5 | Protein C19orf23c | not available | 70 | TWQTRNHTRTGHAYPRFTRPSFPS CNRNGKRRKLRLGLPY | 109 |
| Q92782 | Zinc-finger protein neuro-d4 | C, N | 156 | EDLEDDIPRRKNRAKGKAYGIGGLRKR QDTASLEDRDKPYVCDKFYKELA | 205 |
| O00358 | Forkhead box protein E1/Thyroid transcription factor 2 |
N | 17 | TVKEERGETAAGAGVPGEATGRGAGG RRRKRPLQRGKPPYSYIALIAMAI | 66 |
| Q13461 | Forkhead box protein E3 (FKHL12) (Forkhead-related transcription factor 8) |
N | 35 | AEPGREPEEAAAGRGEAAPTPAPGPG RRRRRPLQRGKPPYSYIALIAMAL | 84 |
| O75593 | Forkhead box protein F1 | N | 1 | MDPASSGPSKAKKTNAGIRRPEKPPYSYIALIVMAI | 36 |
| O75593 | Forkhead box protein H1/Forkhead activin signal transducer 1 |
N | 1 | MGPCSGSRLGPPEAESPSQPPKRRKKR YLRHDKPPYTYLAMIALVI | 46 |
| Q9UPW0 | Forkhead box protein J3 | N | 142 | SKDDPGKGSYWAIDTNPKEDALPT RPKKRARSVERASTPYSIDSDSLGME | 191 |
| P55317 | Hepatocyte nuclear factor 3-alpha (Forkhead box protein A1). |
N | 135 | MNPCMSPMAYAPSNLGRSRAGGGGDAKTFKRSYPHAKPPYSYISLITMAI | 184 |
| P55318 | Hepatocyte nuclear factor 3-gamma (Forkhead box protein A3) |
N | 81 | LGVSGGSSSSGYGAPGPGLVHGKEMP KGYRRPLAHAKPPYSYISLITMAI | 130 |
| Q9Y483 | Metal-response element-binding transcription factor 2 |
N | 370 | HEFKIKGRKASKPISDSREVSNGIE KKGKKKSVGRPPGPYTRKMIQKTAE | 419 |
| O95644 | NFAT transcription complex cytosolic Component |
C, N | 238 | PSTSPRASVTEESWLGARSSRPASP CNKRKYSLNGRQPPYSPHHSPTPSP | 287 |
| Q9ULL1 | Pleckstrin homology domain- containing family G member 1 |
not available | 1304 | SKFVDADFSDNVCSGNTLHSLNSP RTPKKPVNSKLGLSPYLTPYNDSDKL | 1353 |
| Q99575 | Ribonucleases P/MRP protein subunit POP1 |
N | 372 | QTELPDEKIGKKRKRKDDGENAKPIKK IIGDGTRDPCLPYSWISPTTGII | 421 |
| Q8NEY8 | Periphilin 1/Gastric cancer antigen Ga50 | C, N | 84 | YRWTRDDHSASRQPEYRDMRDGFRRKS FYSSHYARERSPYKRDNTFFRES | 133 |
| Q8NDT2 | RNA-binding protein 15B | N | 245 | SRSGERWGADGDRGLPKPWEERRKRR SLSSDRGRTTHSPYEERSRTKGSG | 294 |
| Q14498 | Splicing factor HCC1 | N | 60 | DRERKKSKSRERKRSRSKERRRSRSRSRDRRFRGRYRSPYSGPKFNSAIR | 109 |
| P62241 | 40S ribosomal protein S8 | N | 1 | GISRDNWHKRRKTGGKRKPYHKKRKYELGR | 30 |
| O95416 | Transcription factor SOX-14 | N | 59 | DEAKRLRAQHMKEHPDYKYRPRRKPKNLLKKDRYVFPLPYLGDTDPLKAA | 108 |
| Q9Y651 | Transcription factor SOX-21 (SOX-A) | N | 59 | DEAKRLRAMHMKEHPDYKYRPRRKPKTLLKKDKFAFPVPYGLGGVADAEH | 108 |
| O00267 | Transcription elongation factor SPT5 | N | 678 | GGQRGGFGSPGGGSGGMSRGRGRRDNELIGQTVRISQGPYKGYIGVVKDA | 727 |
| Q9UMS6 | Synaptopodin-2 (Myopodin) (Genethonin 2)c | C, N | 931 | PSYPLAALKSQPSAAQPSKMGKKKGKKPLNALDVMKHQPYQLNASLFTFQ | 980 |
| Q8IWR0 | Zinc finger CCCH-type domain-containing protein 7A |
N | 464 | ANIDHKCKKDILIGRIKNVEDKSWKKIRPRPTKTNYEGPYYICKDVAAEE | 513 |
| Q9H091 | Zinc finger MYND domain-containing protein 15 |
not available | 522 | RDSLEVSVRPGSGISARPSSGTKEKGGRRDLQIKVSARPYHLFQGPKPDL | 571 |
| Q9H116 | Zinc finger protein 336 | N | 177 | LTDSLDYPGERASNGMSSDLPP KKSKDKLDKKKEVVKPPYPKIRRASGRL | 226 |
| Q8N895 | Zinc finger protein 366 | N | 49 | RGPFSQFRYEPPPGDLDGFPGVFEGAGS RKRKSMPTKMPYNHPAEEVTLA | 98 |
As annotated in the UniProtKB/Swiss-Prot entries. C represents cytoplasm and N represents nucleus.
Central basic-enriched regions are underlined and the R/K/H-PY motifs are in bold.
Substrates also identified using hPY-NLS motif.
Of the 81 candidate Kapβ2 substrates, 48 contain hPY-NLSs (Table 2), 28 contain bPY-NLSs (Table 3), and 5 contain PY-NLSs with both basic and hydrophobic central motifs. Forty-nine of the new substrates (~60%) are involved in transcription or RNA processing, 18 have unknown cellular activity, and the rest are involved in signal transduction (8), cell-cycle regulation (3), and the cytoskeleton (3). Interestingly, information on subcellular localization is available for 62 of the predicted substrates, of which 57 (92%) are annotated to have nuclear localization. The SwissProt database used in the search is the most highly annotated and nonredundant protein database but it is still incomplete for human proteins (Apweiler et al., 2004). Thus, the number of new Kapβ2 substrates listed in Tables 2 and 3 is a lower limit of the complete set of Kapβ2 import substrates. The large number of Kapβ2 substrates currently predicted by our NLS rules already implies the generality and prevalence of PY-NLSs. Kapβ1 and Crm1 are also involved in mitosis and centrosome duplication (Ar-naoutov et al., 2005 and reviewed in Budhu and Wang, 2005; Harel and Forbes, 2004; Mosammaparast and Pemberton, 2004), suggesting that many other Kapβs may be similarly involved in multiple cellular functions in addition to nuclecytoplasmic transport. Thus, Kapβ2 substrates will likely include ligands responsible for other still unknown cellular functions of Kapβ2 as well as large numbers of cargoes for nuclear import.
Mechanism of Ran-Mediated Substrate Dissociation from Kapβ2
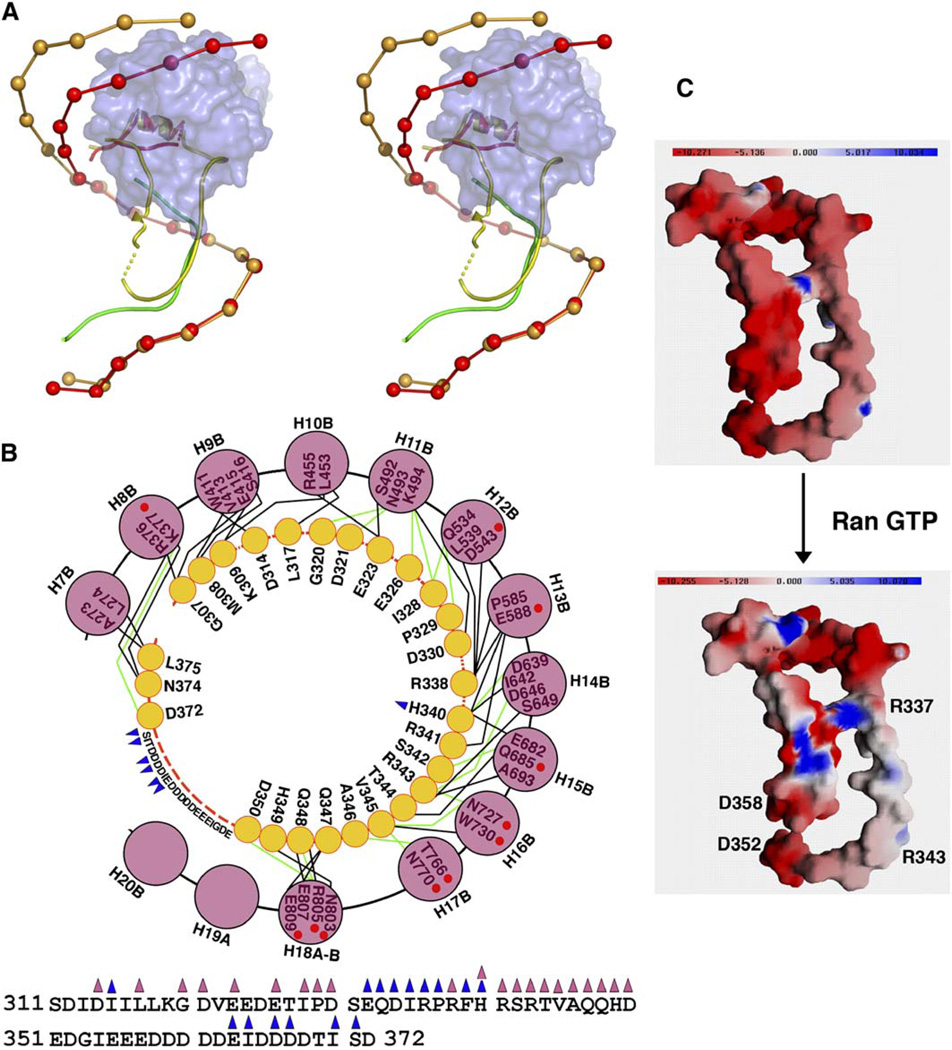
The interaction of RanGTP with Kapβ2 to dissociate substrates in the nucleus is a crucial step in nuclear import. Structural comparison of Kapβ2s in the M9NLS and RanGTP complexes (Chook and Blobel, 1999) shows large differences in their H8 loops (Figure 4A) and finally reveals the mechanism of Ran-mediated substrate dissociation. In the Kapβ2-Ran structure, the H8 loop makes extensive contacts with both Ran and the Kapβ2 C-terminal arch (Figures 4A and 4B) (Chook and Blobel, 1999). In fact, much of the H8 loop is sequestered in the C-terminal arch, such that loop residues 338–350 occupy the same binding site as M9NLS residues 268–281. In contrast, proteolysis studies have suggested that the loop is exposed when Ran is absent (Chook et al., 2002), and this is confirmed by the Kapβ2-M9NLS structure. Even though the H8 loop in the M9NLS complex is truncated, only 14 of its 32 residues are observed, indicating disorder in much of the loop. Ordered loop residues include 312–319 that emerge from helix H8A and residues 369–374 that precede helix H8B (Figures S1A and S1B). Residues 312–319 are in similar positions in both the Ran and substrate complexes, but residues 369–374 have shifted to direct the loop away from the arch in the substrate complex (Figures 4A and S1B). In summary, the concave surface of the C-terminal arch is free to bind substrate when Ran is absent, but the H8 loop occupies the substrate binding site when Ran is present. Interestingly, most of the substrate binding site remains unchanged in both ligand bound states with repeats 9–17 superimposing well at rmsd of 1.2 Å (Figure 4A). The mechanism of Ran-mediated substrate dissociation described here is a thermodynamic one. Ran may increase the dissociation rate of substrate, thus accelerating its release from Kapβ2. Alternatively, the system is limited by the intrinsic dissociation rate of the substrate, and Ran-induced changes in the loop prevent substrate rebinding once dissociation has occurred.
Figure 4. The Comparison of Kapβ2-M9NLS and Kapβ2-Ran Complexes.
(A) Stereo diagram of Kapβ2-M9NLS complex superimposed on the Ran complex. Kapβ2s are drawn as spheres at the geometric center of each HEAT repeat, and the H8 loops are drawn as ribbons. In the M9NLS complex, Kapβ2 is red and substrate green. In the Ran complex, Kapβ2 is light brown and its H8 loop is yellow. The molecular surface of RanGTP is shown in blue.
(B) Contacts (<4.0Å ) between the H8 loop and the C-terminal arch of Kapβ2 in the Ran state with the sequence of the H8 loop shown at bottom. Yellow circles are loop residues that contact the Kapβ2 arch and pink circles are Kapβ2 helices. Red dashed lines indicate intervening loop residues that do not contact the Kapβ2 arch. Blue triangles label residues that contact Ran, pink triangles label residues that contact the Kapβ2 arch, and red circles label Kapβ2 residues that also contact M9NLS. Polar contacts are shown with green lines and hydrophobic contacts with black lines.
(C) Electrostatic surface potential of the H8 loop in the presence and absence of RanGTP, drawn with GRASP (Nicholls et al., 1991). Top shows molecular surface of the isolated H8 loop (Kapβ2 residues 310–372, Kapβ2-Ran complex 1QBK). RanGTP and the Kapβ2 superhelix are omitted from the electrostatic calculation to approximate charges of the loop in the absence of Ran. Bottom shows molecular surface of the H8 loop with electrostatic surface potential calculated using both RanGTP and the H8 loop to represent the Ran bound state.
Despite extensive spatial overlap between the Ran bound H8 loop and M9NLS, no obvious sequence similarity is shared. This is not surprising since they bind in antiparallel directions to each other and their backbones deviate in path even where spatial overlap is greatest (loop residues 338–350 and M9NLS residues 268–281, Figure S4). However, the H8 loop obviously contains a linear epitope that binds Kapβ2 and raises the possible existence of a different class of NLSs.
Why does the H8 loop only bind the C-terminal arch in the presence of Ran? The calculated electrostatic surface potential of the H8 loop in the presence and absence of RanGTP is distinct (Figure 4C). The H8 loop contains many acidic residues, particularly through 351EDGIEEEDDDDDEIDDDD368 directly C-terminal to residues 338–350, which overlap with M9NLS. Negative charges here may prevent binding of the loop to the acidic C-terminal arch (Figure 4C, top). When Ran binds Kapβ2, its basic patch (K127, R129, K132, K134, R140, K141, and K159) inter-acts with H8 loop residues 332–340 and 363–371. Again, long-range electrostatic effects of the basic interface of Ran may substantially decrease the negative charge of the loop, converting residues 338–350 into a more suitable ligand for the Kapβ2 substrate binding site (Figure 4C, bottom). Ran probably also imparts conformational constraints to orient the H8 loop in the substrate site. The relative importance of electrostatic versus conformational effects of Ran binding is not known. Biophysical studies of H8 loop mutants with varying charge and H8 loop peptides in trans will be crucial to parse the different effects of Ran on the loop.
Another structural difference between the Kapβ2-M9NLS and Kapβ2-Ran complexes is found at the N-terminal arches (Figure 4A). Small changes in the orientation of α helices within and between HEAT repeats 1–10 result in a maximum displacement of over 23 Å at the N terminus. The M9NLS complex in the crystal cannot accommodate RanGTP, but biochemical studies had shown that Kapβ2 can adopt a Ran-competent conformation when bound to substrate in solution (Chook et al., 2002). The two Kapβ2-M9NLS complexes in the asymmetric unit also diverge structurally with high B factors at the N-terminal four repeats, suggesting inherent flexibility in that region. Many Kapßs have been shown to exhibit structural plasticity and adopt multiple conformations (Fukuhara et al., 2004). The Kapβ2-M9NLS crystals have trapped a conformation of the N-terminal arch that is incompetent for Ran binding.
Many other Kapßs contain large insertions like the Kapβ2 H8 loop. Kapβ1 has a short 15 residue acidic loop in repeat 8 (Cingolani et al., 1999; Lee et al., 2005), Cse1 has a 2 helix insertion in repeat 8 (Cook et al., 2005; Matsuura and Stewart, 2004), and Crm1, Kapβ3, Imp4, Imp7, Imp8, Imp9, and Imp11 are all predicted to have large insertions in their central repeats. Mutational studies of the predicted Crm1 insertion suggest that it also directly couples Ran and substrate binding (Petosa et al., 2004). However, in Kapβ1 and Cse1, the mechanisms of substrate dissociation appear distinct from those in Kapβ2 and Crm1. Kapβ1 binds three different substrates in three different binding sites, and RanGTP causes a drastic change in superhelical shape that distorts binding sites of substrates Kapα and SREBP-2 while directly displacing substrate PTHrP from the N-terminal arch (Cingolani et al., 1999, 2002; Lee et al., 2003, 2005). Similarly, the Cse1 insertion is a pivot point for global conformational change like that in Kapβ1 (Cook et al., 2005). Trends for coupling Ran and substrate binding in the Kapβ family are emerging. Kapβ2 and probably Crm1 employ a large insertion to directly couple the two ligands with little conformational change in the substrate binding site. In contrast, Kapβ1 and Cse1 use large-scale conformational changes to transition from closed substrate-free to open substrate bound conformations.
Conclusions
The crystal structure of Kapβ2 bound to its substrate M9NLS has revealed a set of rules that describe the recognition of a large class of nuclear import substrates. M9NLS adopts an extended conformation for 26 residues when bound to Kapβ2, leading to the first rule that NLSs recognized by Kapβ2 are structurally disordered in the free substrates. The structure also shows that the substrate binding site on Kapβ2 is highly acidic, leading to the second rule that NLSs will have an overall positive charge. Finally, biochemical analyses of Kapβ2-M9NLS interactions have mapped M9NLS residues that are important for Kapβ2 binding, and examination of other Kapβ2 substrates has revealed consensus motifs at these regions. The consensus motifs include a central hydrophobic or basic motif followed by a C-terminal R/K/HX(2–5)PY motif, leading to the name PY-NLSs for this class of signals. Although these rules are not strong filters individually or in pairs (not shown), together they provide substantial restrictions in sequence space. The three rules have been used to identify NLSs in seven previously identified Kapβ2 substrates and, more importantly, to predict 81 new candidate Kapβ2 substrates in our initial bioinformatics endeavor. Of the members of this predicted group with annotated subcellular localization, >90% are reported to be nuclear localized. We have experimentally validated all seven new NLSs of known Kapβ2 substrates and five new bioinformatics-predicted substrates for Kapβ2 recognition as well as Ran-mediated dissociation, demonstrating the predictive nature of the rules. The large number of predicted Kapβ2 substrates further suggests the prevalence of PY-NLSs in the genome. Finally, the fact that all 81 proteins likely use Kapβ2 suggests potential functional linkages in the group that may be revealed by comparison with other genome-wide analyses.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Complex Formation
In the crystallographic studies Kapβ2 residues 337–367 were replaced with a GGSGGSG linker. This protein was expressed in E. coli BL21 (DE3) as a GST fusion from pGEX-Tev vector and purified as previously reported (Chook and Blobel, 1999; Chook et al., 2002). M9NLS was expressed in E. coli as a GST fusion of hnRNP A1 residues 257–305 and purified as previously described (Chook et al., 2002). Two-fold molar excess of GST-M9NLS was added to purified Kapβ2, cleaved with Tev protease, and the complex purified by gel filtration chromatography. Selenomethionine-Kapβ2 and selenomethionine-M9NLS were purified and assembled as for the native proteins. All complexes were concentrated to 25 mg/ml for crystallization.
Crystallization, Data Collection, and Structure Determination
Native Kapβ2-M9NLS complex was crystallized by vapor diffusion (reservoir solution: 40 mM MES pH 6.5, 3M potassium formate, and 10% glycerol) and flash frozen in liquid propane. These crystals diffracted at best to 3.5Å. However, soaking the crystals in crystallization solution containing 0.7 mM of a 12 residue FXFG peptide (sequence: TGGFTFGTAKTA) improved diffraction to 3.05 Å . Data from an FXFG-soaked crystal were collected on the X-ray Operations and Research beamline 19-ID at the Advanced Photon Source, Argonne National Laboratory, and processed using HKL2000 (Otwinowski and Minor, 1997) (Table S1). Crystals of the selenomethionine complex were also obtained by vapor diffusion (reservoir solution: 0.1M Tris 8.0, 3M potassium formate, and 15% glycerol), soaked in FXFG peptide, and diffracted to 3.3 Å . Single-wavelength anomalous dispersion (SAD) data were collected on SBC-19-ID (Table S1) and processed with HKL2000 (Otwinowski and Minor, 1997).
Native Kapβ2-M9NLS crystals (space group C2, unit cell parameters of a = 152.0Å, b = 154.1 Å, c = 141.7 Å, and β = 91.7°) contain two complexes in the asymmetric unit. Selenomethionine Kapβ2-M9NLS also crystallized space group C2 but has a significantly different unit cell length in its a axis (unit cell parameters of a = 155.6 Å, b = 154.6 Å, c = 141.6 Å, and β = 91.6°; Table S1). Native Patterson maps indicate that the two complexes in the asymmetric unit are related by pseudo-translation along the crystallographic c axis. Molecular replacement trials using the Kapβ2-Ran structure were unsuccessful but SAD phasing followed by solvent flipping, both using the program CNS, produced interpretable electron density maps (Brunger et al., 1998). A model comprising 90% of Kapβ2 was built using O (Jones et al., 1991), but electron density for the substrate remained uninterpretable even though M9NLS residue M276 could be clearly placed using a selenium site. The partial SAD-phased model was used as a search model for molecular replacement using the program Phaser with the higher-resolution native dataset (McCoy et al., 2005). Positional refinement using REFMAC5 (CCP4, 1994), followed by solvent flipping using CNS (Brunger et al., 1998), yielded electron density maps that allowed 97% of Kapβ2 to be built. The density was further improved by rigid body, positional, and simulated annealing refinement of Kapβ2 alone, using the program CNS (Brunger et al., 1998). The Fo-Fc map plotted at 2.5 σ clearly showed strong density for M9NLS residues 267–289 in the complex I and residues 263–289 in complex II (Figure 1C). Even though soaking the crystals in FXFG peptide improved diffraction, no density was observed for the FXFG peptide. The final refined model shows good stereochemistry with R factor of 24.2% and Rfree of 27.2%.
NLS Mapping, Site-Directed Mutagenesis, and Kapβ2 Binding Assays
cDNA for hnRNPs F, M, PQBP-1, EWS, SAM68, HMBA-inducible protein, YBP1, FUS, DDX3, Clk3, Sox14, and WBS16 were obtained from Open Biosystems. cDNA for HCC1 and RB15B were obtained by PCR from a human fetal thymus cDNA library (Clontech). The full-length proteins as well as fragments listed in Figures 3C and S3B were sub-cloned using PCR into pGEX-Tev vector. Expression constructs for NLSs of Cyclin T1 and CPSF6 were generated using synthetic complementary oligonucleotides coding for the 28-mer peptides. Single, double, and triple mutations to alanine residues were performed using the Quickchange method (Stratagene), and all constructs were confirmed by nucleotide sequencing. Substrate proteins were expressed in E. coli BL21 (DE3) cells. GST-M9NLS was expressed at 37°C, GST-Kapβ2 was expressed at 30°C, and the other substrates were expressed at 25°C, and all were purified using glutathione sepharose (GE Healthcare).
In each binding reaction involving new NLSs, mutant NLSs, and new Kapβ2 substrates, approximately 18 µg of Kapβ2 were added to 5–10 µg of GST substrate immobilized on glutathione sepharose, followed by extensive washing of the beads with buffer containing 20 mM Hepes pH 7.3, 110 mM potassium acetate, 2 mM DTT, 1 mM EGTA, 2 mM Magnesium acetate, and 20% glycerol. Immobilized proteins were visualized using SDS-PAGE and Coomassie Blue staining. Three- to five-fold molar excess of RanGTP (compared to Kapβ2) is also used in some binding assays. Binding assays involving mutants of Kapβ2 were performed similarly, with each reaction using approximately 10 µg of MBP-M9NLS added to 5–10 µg of GST-Kapβ2.
Quantitation of Binding Affinity with ITC
Binding affinities of wild-type and mutant MBP-M9NLS to Kapβ2 were quantitated using ITC. The ITC experiments were done using a Micro-Cal Omega VP-ITC calorimeter (MicroCal Inc., Northampton, MA). Proteins were dialyzed against buffer containing 20 mM Tris pH 7.5, 100 mM NaCl, and 2 mM β-mercaptoethanol. 100–500 µM wild-type and mutant MBP-M9NLS proteins were titrated into a sample cell containing 10–100 µM full-length Kapβ2. Most ITC experiments were done at 20°C with 35 rounds of 8 µl injections. ITC experiments involving wild-type M9NLS were similar but with 56 rounds of 5 µl injections. Data were plotted and analyzed using MicroCal Origin software version 7.0, with a single binding site model.
Bioinformatics Search for New Kapβ2 Substrates
Candidate Kapβ2 substrates were identified by the program ScanProsite (Gattiker et al., 2002) using motifs ϕ1-G/A/S-ϕ3-ϕ4-X7–12-R/K/H-X2–5-P-Y (where ϕ1 is strictly hydrophobic, ϕ3 and ϕ4 are hydrophobic and also include long aliphatic side chains R and K) and K/R-X0–2-K/R-K/R-X3–10-R/K/H-X1–5-P-Y and human proteins in the UniProtKB/Swiss-Prot protein database (Bairoch et al., 2004). All resulting entries were filtered for structural disorder using the program DisEMBL (Linding et al., 2003) and for positively charged NLS segments of 50 amino acids (beginning 40 residues N terminus of the PY to 10 residues C terminus of that motif). Proteins with potential PY-NLSs that are found in transmembrane proteins and those that occur within identified domains were eliminated from the list even though some NLSs may occur in long loops within folded domains.
Supplementary Material
ACKNOWLEDGMENTS
We thank UT Southwestern SBL for technical advice and assistance in data collection; C. Thomas for advice on ITC; D. Schmidt, H. Gu, and L. Motta-Mena for assistance in protein expression and purification; N. Grishin for help with bioinformatics; G. Blobel for support of the initial stage of this project; and Z. Otwinowski, M. Phillips, S. Sprang, K. Gardner, R. Ranganathan, and M. Rosen for discussion. The U. S. Department of Energy, Office of Science, and Office of Basic Energy Sciences, under Contract No W-31-109-ENG-38, supported use of the Advanced Photon Source. NIH-R01 GM069909, Welch Foundation Grant I-1532, and the UT Southwestern Endowed Scholars Program support this work.
Footnotes
Supplemental Data
Supplemental Data include five figures and can be found with this article online at http://www.cell.com/cgi/content/full/126/3/543/DC1/.
Accession Numbers
The Kapβ2-M9NLS crystal structure has been deposited in the Protein Data Bank under ID code 2H4M.
References
- Allemand E, Guil S, Myers M, Moscat J, Caceres JF, Krainer AR. Regulation of heterogenous nuclear ribonucleo-protein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc. Natl. Acad. Sci. USA. 2005;102:3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH. Protein sequence data-bases. Curr. Opin. Chem. Biol. 2004;8:76–80. doi: 10.1016/j.cbpa.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Boeckmann B, Ferro S, Gasteiger E. Swiss-Prot: juggling between evolution and stability. Brief. Bioinform. 2004;5:39–55. doi: 10.1093/bib/5.1.39. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Benson RE, Truant R, Herold A, Phingbodhipakkiya M, Cullen BR. Definition of a consensus transportin-specific nucleocytoplasmic transport signal. J. Biol. Chem. 1999;274:9771–9777. doi: 10.1074/jbc.274.14.9771. [DOI] [PubMed] [Google Scholar]
- Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc. Natl. Acad. Sci. USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Rea RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project, Number 4) The CCP4 suite: programs for X-ray crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chook Y, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran.GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- Chook YM, Jung A, Rosen MK, Blobel G. Uncoupling Kapβ2 substrate dissociation and Ran binding. Biochemistry. 2002;41:6955–6966. doi: 10.1021/bi012122p. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Bednenko J, Gillespie MT, Gerace L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol. Cell. 2002;10:1345–1353. doi: 10.1016/s1097-2765(02)00727-x. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 2001;13:310–319. doi: 10.1016/s0955-0674(00)00213-1. [DOI] [PubMed] [Google Scholar]
- Cook A, Fernandez E, Lindner D, Ebert J, Schlenstedt G, Conti E. The structure of the nuclear export receptor Cse1 in its cytosolic state reveals a closed conformation incompatible with cargo binding. Mol. Cell. 2005;18:355–367. doi: 10.1016/j.molcel.2005.03.021. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL User’s Manual. San Carlos, CA: DeLano Scientific LLC).; 2002. . [Google Scholar]
- Fan XC, Steitz JA. HNS a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Blobel G. The nuclear transport factor karyo-pherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J. Biol. Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J. Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Fernandez E, Ebert J, Conti E, Svergun D. Conformational variability of nucleo-cytoplasmic transport factors. J. Biol. Chem. 2004;279:176–181. doi: 10.1074/jbc.M309112200. [DOI] [PubMed] [Google Scholar]
- Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Guttinger S, Muhlhausser P, Koller-Eichhorn R, Brennecke J, Kutay U. Transportin2 functions as importin and mediates nuclear import of HuR. Proc. Natl. Acad. Sci. USA. 2004;101:2918–2923. doi: 10.1073/pnas.0400342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M, Suzuki M, Tanabe A, Nishimura A, Yamada M. Two motifs essential for nuclear import of the hnRNP A1 nucle-ocytoplasmic shuttling sequence M9 core. FEBS Lett. 2006;580:1365–1370. doi: 10.1016/j.febslet.2006.01.058. [DOI] [PubMed] [Google Scholar]
- Jones TA, Cowan SW, Kjelgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kawamura H, Tomozoe Y, Akagi T, Kamei D, Ochiai M, Yamada M. Identification of the nucleocytoplasmic shuttling sequence of heterogeneous nuclear ribonucleoprotein D-like protein JKTBP and its interaction with mRNA. J. Biol. Chem. 2002;277:2732–2739. doi: 10.1074/jbc.M108477200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Sekimoto T, Yamashita E, Nagoshi E, Nakagawa A, Imamoto N, Yoshimura M, Sakai H, Chong KT, Tsukihara T, Yoneda Y. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science. 2003;302:1571–1575. doi: 10.1126/science.1088372. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Siomi MC, Siomi H, Michael WM, Pollard V, Dreyfuss G. Transportin: nuclear transport receptor of a novel nuclear protein import pathway. Exp. Cell Res. 1996;229:261–266. doi: 10.1006/excr.1996.0369. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Petosa C, Schoehn G, Askjaer P, Bauer U, Moulin M, Steuer-wald U, Soler-Lopez M, Baudin F, Mattaj IW, Muller CW. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol. Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Rebane A, Aab A, Steitz JA. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA. 2004;10:590–599. doi: 10.1261/rna.5224304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Iijima M, Nishimura A, Tomozoe Y, Kamei D, Yamada M. Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. FEBS J. 2005;272:3975–3987. doi: 10.1111/j.1742-4658.2005.04820.x. [DOI] [PubMed] [Google Scholar]
- Truant R, Kang Y, Cullen BR. The human tap nuclear RNA export factor contains a novel transportin-dependent nuclear localization signal that lacks nuclear export signal function. J. Biol. Chem. 1999;274:32167–32171. doi: 10.1074/jbc.274.45.32167. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- Weighardt F, Biamonti G, Riva S. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J. Cell Sci. 1995;108:545–555. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.