SUMMARY
The cytoplasmic polyadenylation element-binding protein 3 (CPEB3), a regulator of local protein synthesis, is the mouse homologue of ApCPEB, a functional prion protein in Aplysia. Here, we provide evidence that CPEB3 is activated by Neuralized1, an E3 ubiquitin ligase. In hippocampal cultures, CPEB3 activated by Neuralized1-mediated ubiquitination leads both to the growth of new dendritic spines and to an increase of the GluA1 and GluA2 subunits of AMPA receptors, two CPEB3 targets essential for synaptic plasticity. Conditional overexpression of Neuralized1 similarly increases GluA1 and GluA2 and the number of spines and functional synapses in the hippocampus, and is reflected in enhanced hippocampal-dependent memory and synaptic plasticity. By contrast, inhibition of Neuralized1 reduces GluA1 and GluA2 levels and impairs hippocampal-dependent memory and synaptic plasticity. These results suggest a model whereby Neuralized1-dependent ubiquitination facilitates hippocampal plasticity and hippocampal-dependent memory storage by modulating the activity of CPEB3 and CPEB3-dependent protein synthesis and synapse formation.
INTRODUCTION
Ubiquitin mediated proteolysis is an important post-translational modification for synaptic plasticity. Ubiquitination results from the covalent attachment to a protein substrate of the small protein ubiquitin, and can involve either single ubiquitin moieties or polymeric ubiquitin chains (DiAntonio and Hicke 2004). Ubiquitination requires the coordinated action of a ubiquitin-activating enzyme (E1), several ubiquitin conjugating enzymes (E2s) and numerous ubiquitin ligases (E3s) (Glickman and Ciechanover 2002).
Ubiquitination modulates neuronal plasticity by targeting of synaptic components for degradation and by regulating the trafficking of synaptic receptors (Mabb and Ehlers, 2010). In addition to triggering degradation, polyubiquitin chains formed through combinations of the lysine residues of ubiquitin can modify protein function and activity (Chen and Sun, 2009). One or more single ubiquitin moieties can modify protein-protein interactions, protein localization and protein activity (DiAntonio and Hicke 2004; Hicke, 2001). These non-degradative functions of ubiquitination have yet to be explored in the context of synaptic plasticity, learning and memory.
To better understand the non-degradative function of the ubiquitin system a number of studies have begun to identify the functional significance of the E3 ubiquitin ligases, the key regulatory determinants of the molecular targets of ubiquitination, and, together with the E2s, of the type of ubiquitination. Of the E3 ubiquitin ligases studied so far (Segref and Hoppe, 2009), Neuralized is of particular interest because it facilitates protein synthesis-dependent long-term memory in Drosophila (Pavlopoulos et al., 2008).
Based on the results in flies we have asked: does Neuralized also facilitate memory storage in mice? We find that Neuralized1 (Neurl1), the mouse orthologue of the fly protein, is expressed in the forebrain of the adult mouse, and that its level is elevated by synaptic activity. Experiments with dominant negative inhibition of Neurl1 in adult mouse forebrain indicate that it reduces the levels of GluA1 and GluA2 proteins and is critical for synaptic plasticity and hippocampal-dependent memory. Conversely, regulated overexpression of Neurl1 in the adult forebrain leads to an overall increase in the levels of GluA1 and GluA2 and an increase in the number of spines and facilitates hippocampal synaptic plasticity and enhances hippocampal-dependent learning and memory.
We next asked: how does Neurl1 mediate its actions? We find that Neurl1 directly interacts with and monoubiquitinates the Cytoplasmic Polyadenylation Element Binding Protein-3 (CPEB3), an RNA binding protein and translational regulator that is the mouse homologue of the functional prion protein in Aplysia ApCPEB (Theis et al., 2003; Huang et al., 2006). Monoubiquitination of CPEB3 leads to its activation, the production of its targets GluA1 and GluA2 and the formation of dendritic spines in cultured hippocampal neurons. Neurl1 overexpression increases the level of monoubiquitinated CPEB3 and the polyadenylation and translation of GluA1 and GluA2. Conversely, inhibition of Neurl1 reduces the translation of these targets. These results suggest a model whereby Neurl1-mediated ubiquitination facilitates hippocampal plasticity and hippocampal-dependent memory storage by modulating CPEB3 activity and CPEB3-dependent protein synthesis and synapse formation.
RESULTS
Neurl1 is Localized in Postsynaptic Sites in the Adult Hippocampus
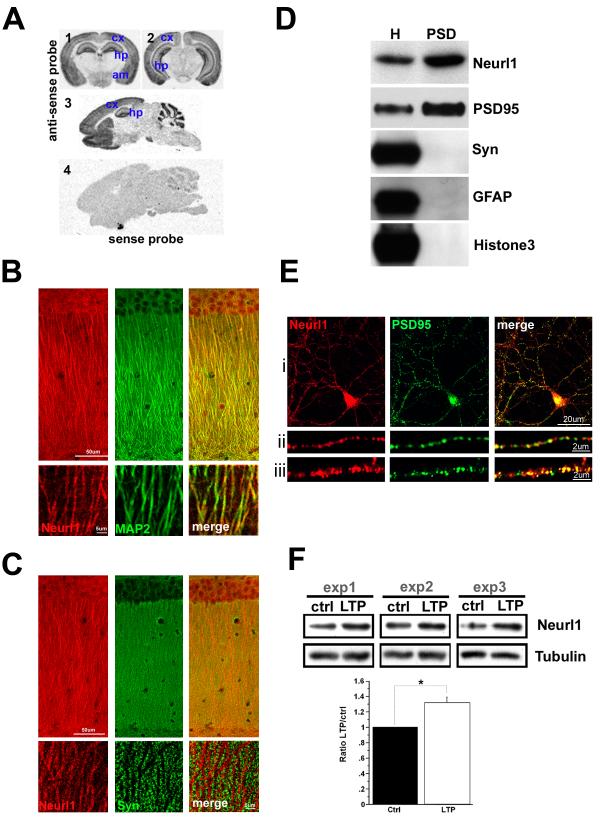
Neurl1 is expressed throughout the adult forebrain including the cerebral cortex, amygdala, striatum and hippocampus (Figure 1A). We also found Neurl1 protein in the CA1 area of the hippocampus (Figure 1B & S1). Within the CA1 region Neurl1 was detected in the cell bodies of the pyramidal neurons, and was distributed along their apical dendrites (Figure 1B). Consistent with its presence at postsynaptic sites, Neurl1 had a complementary immunostaining pattern with the presynaptic marker Synaptophysin (Figure 1C). The postsynaptic localization of Neurl1 was further documented by means of its presence in the Post-Synaptic Density fraction isolated from the hippocampus of adult mice and by colocalization with the postsynaptic protein PSD95 in cultured mouse hippocampal neurons (Figure 1D&E).
Figure 1. Neurl1 is expressed in the adult mouse forebrain and is localized in dendrites and at post-synaptic sites in hippocampal neurons.
(A) In situ hybridization analysis of Neurl1 mRNA expression on coronal (1&2) and sagittal (3) brain slices from adult (3 ½ months) wild type mice. cx: cerebral cortex, hp: hippocampus, st: striatum, am: amygdala.
(B & C) Confocal sections of adult CA1 pyramidal neurons. Coimmunostaining for Neurl1 and the dendritic marker MAP2 or the presynaptic marker Synaptophysin (Syn). Lower panels: high magnification of apical dendrites.
(D) Immunoblots of hippocampal homogenates (H) and PSD extracts. Neurl1 and PSD95 are enriched in the PSD fraction. Synaptophysin, the glial protein GFAP and histone 3 are controls.
(E) Confocal sections of cultured hippocampal neurons (16 DIV) showing colocalization of endogenous Neurl1 and PSD95. (ii &iii) high magnification of dendrites.
(F) Neurl1 protein level in the CA1 area is increased 30min after LTP induction at the Schaffer collateral pathway using 4 TBS. ctrl: unstimulated controls. Top: examples of western blots from 3 independent experiments. Bottom: Mean + SEM from 5 independent experiments (4 mice; *p<0.01). See also Figure S1.
Synaptic Activity Increases Neurl1 Levels
In the fly Neuralized overexpression facilitates memory formation. This finding led us ask whether Neurl1 protein level is upregulated by synaptic activity. We found that 30 minutes after the induction of LTP Neurl1 protein level was significantly increased (Figure 1F).
Regulated Inhibition of Neurl1 in the Adult Forebrain Impairs Hippocampal-Dependent Memory
Activity-dependent upregulation of Neurl1 in the hippocampus suggested its possible involvement in mechanisms underlying synaptic plasticity and memory storage. Consistent with this view, Neuralized was found to be limiting for memory formation in Drosophila (Pavlopoulos et al., 2008). We therefore asked: is the function of Neurl1 also critical for memory and synaptic plasticity in the mammalian brain? To address this question, we inhibited Neurl1 in the adult mouse forebrain by generating double transgenic (DT) mice using the tetO/tTA system (Mayford et al., 1996) that allowed us to regulate, reversibly, the expression of the transgene in the adult forebrain of DT mice (Figure S2A). We inhibited Neurl1 by expressing a dominant negative form (Neurl1DN) that lacks its RING Zn finger, the domain that possesses the ubiquitin ligase activity (Koutelou et al., 2008). A Flag epitope tag was fused at the C-terminus of Neurl1DN. Neurl1DN was expressed in the same CA1 neurons with Neurl1 and within those neurons in the same neuronal compartments (Figure S2C). To avoid developmental effects, Neurl1DN transcription was inhibited until day P40 (Figure S2B).
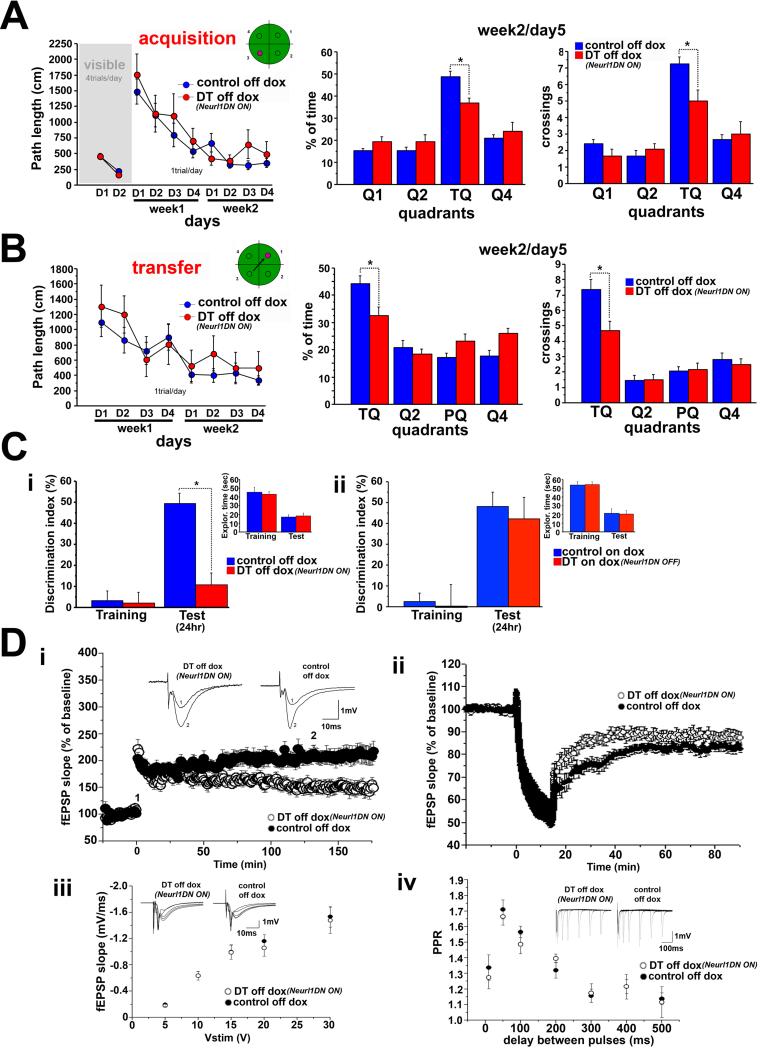
We first assessed memory in the Morris water maze (Morris et al., 1982). In the hidden version, which tests hippocampal-dependent spatial learning and memory, DT mice learned the task as well as their control littermates, but they had impaired memory of the platform position as evidenced by a probe trial one day after the end of training (Figure 2A & S2D). The mice were also impaired in forming a good memory of a new platform location (memory flexibility) and in novel object recognition, another hippocampal-dependent task (Figure 2B&C & S2D).
Figure 2. Inhibition of Neurl1 in the adult hippocampus impairs memory and synaptic plasticity.
(A) Averaged data (+SEM) from the Morris water maze of Neurl1DN expressing mice (DT off dox) and control siblings. The performance of DT was significantly lower in the probe trial (quadrant*genotype effect: p<0.04, t-test for training quadrant (TQ): *p<0.007).
(B) Transfer phase of the water maze. DT performed significantly worse than controls in the probe trial (quadrant*genotype effect: p<0.04, t test for TQ, *p<0.013). PQ: TQ in A.
(C) Novel object recognition (mean +SEM). (i) The discrimination index of DT was significantly lower in the 24hr test (*p=0.001). Total exploration times were similar (p>0.4). (ii) Mice tested on dox. No differences were observed (p>0.6).
(D) Expression of Neurl1DN in the hippocampus impairs the late phase of LTP and LTD at the Schaeffer collateral pathway. (i) LTP induction was not affected in DT (t-test 0-60min: p=0.17). L-LTP was impaired (t-test 60-120min & 120-180min; p<0.02) Insets: example of 10 averaged traces before (1) and after (2) LTP induction. (ii) LTD was impaired in DT (t-test 40-90 min: p<0.05). Basal transmission (iii) and paired-pulse facilitation (iv) were unaltered (ANOVA; p>0.63). Insets in (iii) & (iv): sample traces (PPR: traces for 10, 50, 100, 200 & 300 ms delays between pulses).
Non-cognitive parameters and anxiety levels were similar between all groups (Table S1), suggesting that the impaired performance of DT mice results from impaired hippocampal-dependent memory. Moreover, when we blocked the expression of Neurl1DN in the adult forebrain by administering doxycycline (Figure S2B), DT mice displayed cognitive performance similar to controls (Figure 2C & S2E). Thus, the expression of Neurl1DN in adult hippocampal neurons impairs hippocampal-dependent memory formation, indicating the requirement of Neurl1-mediated ubiquitination in this process.
Expression of Neurl1DN in The Adult Hippocampus Impairs Synaptic Plasticity at Schaffer Collateral Synapses
Is the hippocampal-dependent memory impairment observed in Neurl1DN expressing mice accompanied by alterations of hippocampal synaptic plasticity? We obtained extracellular recordings from hippocampal slices and induced LTP at the Schaffer collateral pathway using a single theta-burst stimulus (1TBS). We found that in Neurl1DN expressing mice the maintenance of L-LTP was impaired (Figure 2D). These effects were not related to a defect in basal synaptic properties, as both basal transmission and paired-pulse facilitation were unaltered (Figure 2D). LTP in these mice was restored to control levels when Neurl1DN expression was blocked by doxycycline (Figure S2F).
In addition to their impaired memory, Neurl1DN mice exhibited an impairment of memory flexibility, an impairment thought to require LTD (Nicholls et al., 2008; Malleret et al., 2010). Consistent with this idea, we also found LTD to be impaired in these mice (Figure 2D). Taken together, our data suggest that Neurl1 is required for hippocampal synaptic plasticity, memory and memory flexibility, consistent with the observed activity-dependent upregulation of Neurl1 in wild type mice.
Upregulation of Neurl1 in the Adult Forebrain Enhances Hippocampal-Dependent Memory
The requirement of Neurl1 function in synaptic plasticity and memory, as revealed in the Neurl1DN mice, and the activity-dependent upregulation of Neurl1 level suggest that Neurl1 is likely limiting for these processes. We therefore asked: will overexpression of Neurl1 in the adult mouse forebrain similarly enhance memory and synaptic plasticity? We now generated a second line of DT mice: tetO-Neurl1-Flag/CaMKIIa-tTA. Neurl1-Flag was expressed throughout the forebrain with a pattern similar to that of the endogenous Neurl1 (Figure S3A). Both the wild type and the transgenic proteins were expressed in the same CA1 hippocampal neurons and in the same neuronal compartments (Figure S3A). Neurl1 protein level in DT mice was 1.88 + 0.14 times more than controls (Figure S3A). Similar to Neurl1DN, we activated the expression of Neurl1-Flag at day P40.
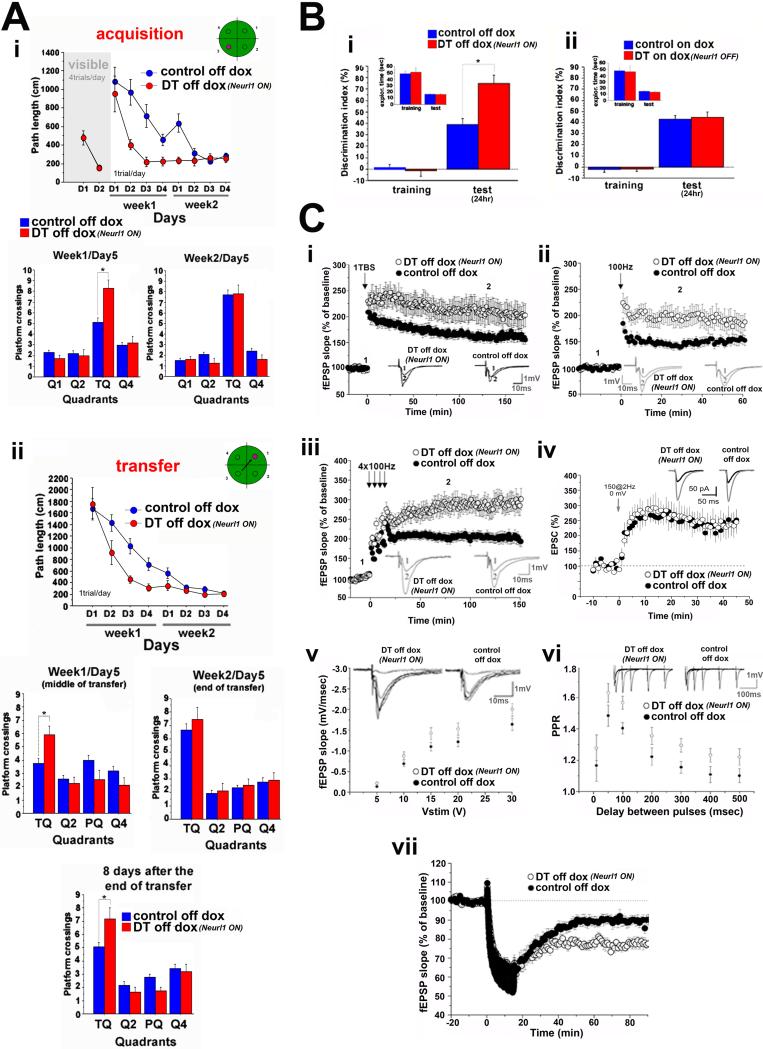
In marked contrast to Neurl1DN mice, Neurl1 overexpressing animals displayed an enhanced performance in learning and formed a good memory of the platform location in the hidden version of the Morris water maze. These mice also had significantly enhanced learning and memory flexibility (Figure 3A & S3B). Control mice needed almost twice as much training to reach a comparable level of performance. Moreover, even though by the end of training in the transfer phase both groups performed equally well, Neurl1 overexpressing mice again performed better in a probe trial eight days later (Figure 3A & S3B). Thus, Neurl1 overexpression also enhances the maintenance of long-term memory.
Figure 3. Overexpression of Neurl1 in the adult hippocampus results in enhanced learning and memory and increased synaptic plasticity.
(A) Data from the Morris water maze of Neurl1 overexpressing mice (DT off dox) and control siblings (mean +SEM). Acquisition and transfer: DT reached minimal path lengths faster than controls (*p<0.007) and performed better in the probe trial on week1/day5 (quadrant*genotype effect: p<0.003, t-test for training quadrant (TQ), p<0.007). In probe trial 8 days after the end of transfer, DT performed again better than controls (genotype*quadrant effect: p= 0.0047; t-test for TQ: p= 0.0059). PQ: TQ in (i).
(B) Novel object recognition (mean +SEM). (i) The discrimination index of DT was significantly higher in the 24hr test (*p=0.0005). (ii) Mice tested on dox. No differences were observed (p>0.6).
(C) (i) LTP induced by 1 TBS was enhanced in DT (mean % of baseline; ANOVA, p=0.01). Induction phase (0-60min) and maintenance (120-180min) were significantly enhanced in DT (mean % of baseline; unpaired t-test; induction: p<0.02; maintenance: p<0.02). LTP induced by 1 (ii) or 4 trains at 100Hz (iii) was also enhanced in DT (mean % of baseline; unpaired t-test; 1×100Hz: 1 hr recording, p<0.01; 4×100Hz: 15-60 & 60-120 min: p<0.01, 120-180 min: p=0.01). Insets: example of 10 averaged traces before (1) and after (2) LTP induction. (iv) Summary graph of LTP recorded in whole cell configuration. EPSCs were recorded at -73 mV in presence of GABAA blockers. LTP was evoked with a pairing protocol consisting of a presynaptic stimulation (150 pulses at 2Hz) combined with postsynaptic depolarization at 0 mV. LTP magnitude was similar between DT and interleaved control experiments (p=0.9). Inset: sample traces. (v) Basal synaptic transmission was slightly increased in DT (5V: p=0.01; 10V: p=0.04; 15V: p<0.05; 20 and 30V: p=0.1). PPR (vi) was increased (ANOVA, p=0.04). Insets in (v) & (vi): sample traces (PPR: traces for 10, 50, 100, 200 & 300 ms delays between pulses). (vii) LTD induced by one 15min train at 1 Hz was facilitated in DT (mean % of baseline 40-90 min: unpaired t-test p<0.001).
Neurl1 overexpressing mice also had enhanced memory for novel object recognition (Figure 3B).
Non-cognitive parameters and anxiety levels were similar between all groups (Table S2). Inhibition of Neurl1-Flag expression and return of Neurl1 level to the wild type state in the adult forebrain of DT mice reversed their enhanced performance in the cognitive tasks (Figure 3B & S3D; Table S2). Thus, consistent with the role of its Drosophila orthologue in flies, Neurl1 overexpression in adult hippocampal neurons facilitates hippocampal-dependent memory formation.
Neurl1 Upregulation in the Adult Hippocampus Enhances Synaptic Plasticity at Schaffer Collateral Synapses
Consistent with the results in Neurl1DN mice, L-LTP induced by 1TBS at the Schaeffer collateral pathway was increased in Neurl1 overexpressing animals (Figure 3C; Table S2). In addition to an increase in the late phase, the early phase of LTP also was increased, and was present even when we used a weaker LTP induction protocol (1 train at 100Hz; Figure 3D). This increase in the amplitude of E-LTP was likely due to a stronger depolarization produced by the tetanus. To test this idea more directly, we carried out recordings under whole-cell configuration using a pairing protocol consisting of a low-frequency presynaptic stimulation combined with sustained postsynaptic depolarization to 0 mV and found that indeed the magnitude of E-LTP was not different between Neurl1 overexpressing and control mice (Figure 3C). In further agreement with this idea, the basal synaptic transmission was slightly increased in Neurl1 overexpressing mice, which likely reflects a postsynaptic change as paired-pulse ratio was also slightly enhanced, consistent with a decrease in the probability of glutamate release (Figure 3C).
When we used 4 trains at 100Hz spaced by 5 minutes, which lead to the induction of L-LTP, we found in addition to the facilitation of the early phase a strong increase of the amplitude of L-LTP in Neurl1 overexpressing mice compared to controls (Figure 3C). The changes did not result from either impairment of inhibitory transmission or different basic membrane properties in DT mice (Figure S3C), and they were reversed when Neurl1 overexpression was blocked in the adult hippocampus (Figure S3E).
Consistent with the data obtained from Neurl1DN mice, the LTD also was significantly increased in Neurl1 overexpressing animals (Figure 3C).
Overexpression of Neurl1 Increases the Number of Synapses in the Adult Hippocampus
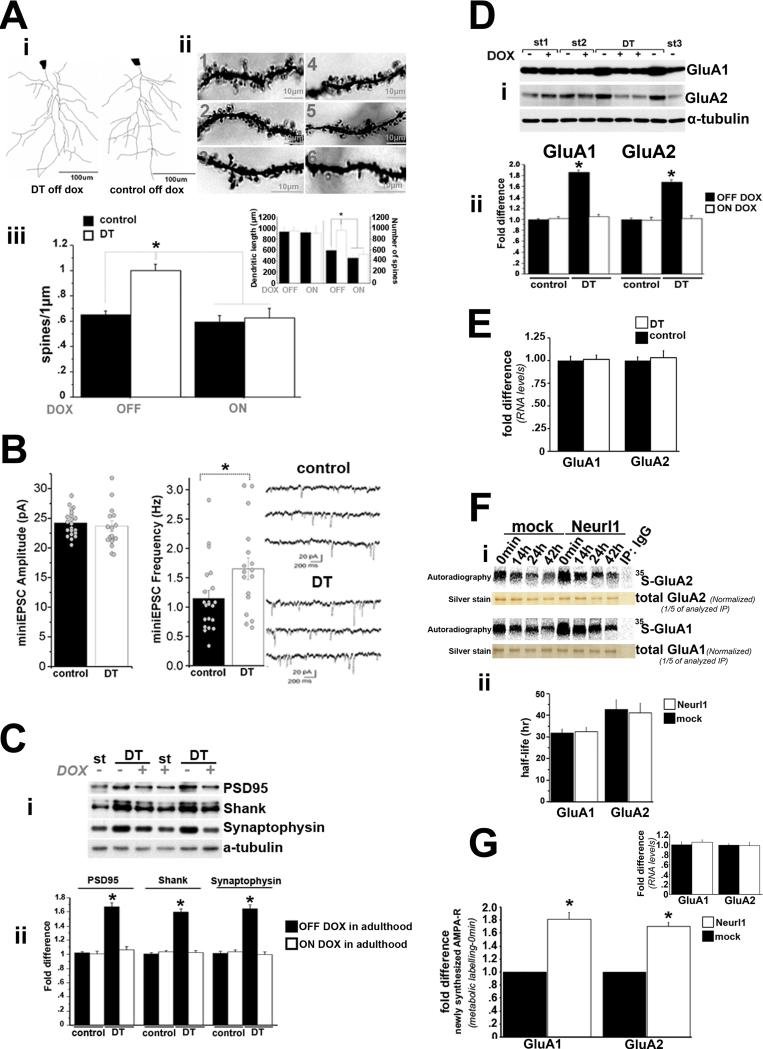
The increase in basal transmission accompanied by a decrease in glutamate release probability and the bidirectional facilitation of plasticity in Neurl1 overexpressing mice could reflect an increase of the number of functional synapses. To explore this possibility, we investigated the density of spines of the apical dendrites of CA1 pyramidal neurons in Neurl1 overexpressing mice and found it to be significantly increased (54.3%), and to return to control levels in DT mice on dox (Figure 4A & S4A).
Figure 4. Neurl1 overexpression in the hippocampus increases the number of spines and functional synapses and the number of AMPAR subunits GluA1 and GluA2.
(A) (i) Computer-assisted reconstructions of representative neurons in adult Neurl1 overexpressing (DT off dox) and control mice. (ii) Golgi stained apical dendrites from CA1 pyramidal neurons. 1-3: DT off dox. 4: DT on dox. 5: control on dox. 6: control off dox. (iii) Mean spine density (+ SEM). It was significantly increased in DT off dox. Inset: mean dendritic length and number of spines that were counted + SEM. *p<0.0001.
(B) Summary graph and sample traces of mEPSCs recorded in CA1 pyramidal neurons of Neurl1 overexpressing (DT) and control mice. Circles: individual experiments. Bars graph: mean + SEM. mEPSCs frequency was significantly increased in DT (*p = 0.036).
(C) Western blot of protein levels from adult hippocampal lysates (i) (different mouse/lane). St: tetO-Neurl1-Flag single transgenic mice. DT: double transgenic mice. (ii) mean fold difference + SEM (comparison with control off dox). Significant increase was observed only in DT off dox (overexpressed Neurl1; *p<0.0001).
(D) (i) Western blot of hippocampal lysates (different mouse/lane) from DT and controls (single transgenic; st1-st3). st1 & st3: tetO-Neurl1-Flag, st2: CaMKIIα-tTA. (ii) Mean fold difference + SEM of GluA1 and GluA2 protein levels (comparison with control off dox). DT off dox showed significant increase (*p<0.0001).
(E) Mean fold difference + SEM of mRNA levels in the hippocampus of Neurl1 overexpressing (DT) and control mice (real time qPCRs). No differences were found (p>0.3).
(F) Time course for 35S-GluA1 and 35S-GluA2 levels in control (mock; infection with control lentivirus) and Neurl1 overexpressing hippocampal cultures (16DIV) after a 1hr pulse of 35S-Met/35S-Cys. (i) Immunoprecipitated 35S-GluA1 and 35S-GluA2 were analyzed by SDS-PAGE and phosphoimager. (ii) Mean half-life + SEM from 3 independent experiments. No differences were found (p>0.8).
(G) Comparison of 35S-GluA1 and 35S-GluA2 at 0min. They were significantly increased in Neurl1 overexpressing neurons (*p<0.001). Real time qPCRs did not reveal differences at the mRNA level (inset; p>0.5).
To determine whether the increased spine number resulted in an increase in the number of functional synapses, we performed whole-cell recordings and monitored miniature EPSCs (mEPSCs) independent of action potentials. We found that the amplitude of mEPSCs was similar in controls and Neurl1 overexpressing mice, suggesting that the number of synaptic glutamate AMPA receptors (AMPAR) was likely not affected by Neurl1 overexpression (Figure 4B). However, the frequency of mEPSCs was significantly increased in the Neurl1 overexpressing animals (Figure 4B). Because release probability is likely not increased in these mice, these results suggest that Neurl1 overexpression in the adult hippocampus, leads to an increase in functional synapses, but not in the number of AMPAR per synapse.
In addition, the levels of three markers of synaptic density, the two post-synaptic markers PSD95 and Shank, and the pre-synaptic marker synaptophysin (Sala et al., 2001; Shimada et al., 2003), were significantly increased, and returned back to control levels when the overexpression of Neurl1 was blocked (Figure 4C).
The involvement of Neurl1 in the regulation of synapse formation is consistent with the role of Neurl1 in the maintenance of LTP and LTD (see discussion), and may also explain the facilitation of E-LTP in the Neurl1 overexpressing mice, as more synaptic contacts tend to increase the slope of EPSPs and facilitate the induction of LTP.
Neurl1 Overexpression Increases the Number of AMPA-Type Receptor Subunits GluA1 and GluA2
The increase of the number of functional synapses and the accompanying unaltered amplitude of mEPSCs compared to controls suggest that the overall synaptic population of AMPAR -critical components for synaptic plasticity- is likely increased in Neurl1 overexpressing hippocampal neurons. What is responsible for this increase? We found that the upregulation of Neurl1 in the adult hippocampus significantly increased both the GluA1 and the GluA2 subunits of AMPAR (Figure 4D). We confirmed these observations using cultured hippocampal neurons. We also found that Neurl1 overexpression does not affect the trafficking and endocytosis of AMPAR (Table S3; Figure S4B&C). Thus, Neurl1 is involved in regulating the overall protein levels of AMPAR. Perhaps most important, AMPAR levels and spine formation follow similar patterns of change, suggesting that Neurl1 may act on common or parallel molecular mechanisms to regulate these processes.
Neurl1 Facilitates Protein Synthesis of GluA1 and GluA2
In addition to not affecting the mRNA levels of GluA1 and GluA2 in the hippocampus (Figure 4E), Neurl1 overexpression did not affect the rate of degradation of GluA1 and GluA2 proteins. This was evident by: 1) the similar net decrease of GluA1 and GluA2 protein levels in the hippocampus of Neurl1 overexpressing and control mice one and a half hour after the administration of the protein synthesis inhibitor anisomycin, and 2) the similar rates of degradation and half-life of metabolically labelled (35S) GluA1 and GluA2 proteins in Neurl1 overexpressing and control cultured hippocampal neurons (Figure 4F & S4D). Importantly, the levels of newly synthesized 35S-GluA1 and 35S-GluA2 in neurons overexpressing Neurl1 were significantly higher than controls (Figure 4G). This increase was not due to differences of the mRNA levels of GluA1 and GluA2 (Figure 4G), suggesting that Neurl1 overexpression augments the protein levels of GluA1 and GluA2 by modulating their translation.
Consistent with the involvement of Neurl1 in the regulation of AMPAR protein synthesis, inhibiting Neurl1 reduced the synthesis of GluA1 and GluA2 proteins in cultured hippocampal neurons without affecting their rate of degradation. Similarly, the protein, but not the mRNA levels, of GluA1 and GluA2 were significantly reduced in the hippocampus of adult Neurl1DN expressing mice (Figure S4E&F). The fact that reduced basal levels of AMPAR in Neurl1DN mice do not alter basal transmission and the induction of LTP is not surprising as the synaptic population of AMPAR could remain unchanged due to insertion of extrasynaptic AMPAR in the synapse.
Neurl1 Interacts with and Ubiquitinates a Translational Regulator of GluA1 and GluA2: the Cytoplasmic Polyadenylation Element-Binding Protein 3
To investigate further how Neurl1 regulates GluA1 and GluA2 proteins, we applied Neurl1-specific immunoprecipitation and mass spectrometry to examine the proteins that interact with Neurl1 in the hippocampus of adult wild type mice (Extended Experimental Procedures). This analysis identified the Cytoplasmic Polyadenylation Element Binding Protein 3 (CPEB3; Theis et al., 2003) as one of the proteins interacting with Neurl1 (Figure 5A; Table S5&6). CPEB3 is an RNA binding protein that is a member of a protein family that controls polyadenylation of mRNAs in the cytoplasm, a widely used mechanism for the activation of the translation of dormant mRNAs critical for certain forms of synaptic plasticity in the mouse, in the fly, and in Aplysia (Richter, 2007). Of the seven Neurl1-interacting proteins that we identified, only CPEB3 was functionally related to the effect of Neurl1 overexpression in synapse formation and in causing an increase in GluA1 and GluA2 translation. Indeed, CPEB3 is a known translational regulator of GluA2 (Huang et al., 2006) while GluA1 has been recently identified in our laboratory as an additional target (Figure S5A). Moreover, CPEB3 is a potential functional homologue of CPEB in Aplysia (Theis et al., 2003), where it serves as a regulator of translation important for the maintenance of synaptic growth (Miniaci et al, 2008; Si et al., 2003, 2010). We, therefore, explored the possibility that CPEB3 may be the functional link between the increase of GluA1 and GluA2 translation and the formation of new synapses in Neurl1 overexpressing mice.
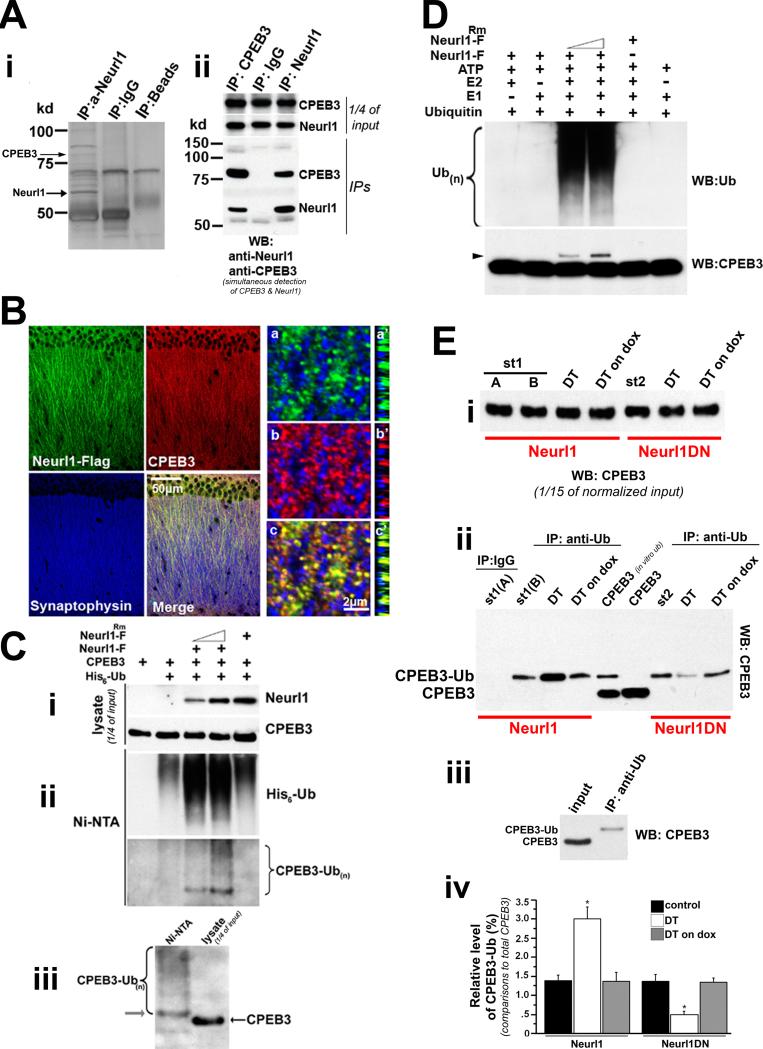
Figure 5. Neurl1 interacts with and ubiquitinates CPEB3.
(A) (i) Silver stain of hippocampal lysates from adult wild type mice after IP with anti-Neurl1 antibody. CPEB3 was detected as Neurl1-interacting protein. (ii) Co-IP from hippocampal lysates of adult wild type mice.
(B) Confocal images of CA1 pyramidal neurons from adult Neurl1 overexpressing mice. CPEB3 and Neurl1 proteins colocalize next to presynaptic sites (synaptophysin puncta). Because CPEB3 and Neurl1 antibodies were produced in rabbit, we used anti-Flag antibody to detect Neurl1-Flag, which exhibited similar subcellular distribution with endogenous Neurl1 and interacted with endogenous CPEB3 (Figure S3A & S5B). (a-c; a’-c’): high magnification of apical dendrites. (a’-c’): cross sections.
(C) Neurl1 promotes ubiquitination of CPEB3 in vivo. HEK293T cells were transiently expressing 6-his-tagged ubiquitin and the indicated proteins (i). Neurl1-F: Neurl1-Flag. Neurl1Rm-F: Neurl1-Flag without ubiquitin ligase activity. His-tagged ubiquitinated proteins were affinity-purified on nickel-agarose (Ni-NTA) under denaturing conditions and analyzed by immunoblotting with anti-CPEB3 antibody (ii). Anti-Ub antibody was used to test the recovery of ubiquitinated proteins. (iii) Immunoblot analysis of ubiquitinated species of CPEB3 next to lysate of cells transfected only with CPEB3. The most abundant ubiquitinated species of CPEB3 migrates in molecular size 8kD higher than the non-ubiquitinated protein, likely representing monoubiquitinated CPEB3 (arrow).
(D) In vitro ubiquitination assays of CPEB3. Monoubiquitinated CPEB3 (arrowhead) is detected only when Neurl1-F and the rest of the reaction components are present. Anti-Ub antibody was used to test Neurl1-F activity, as it is known to form poly-Ub chains.
(E) Isolation of ubiquitinated species of CPEB3 from adult hippocampal lysates using sequential IP with anti-CPEB3 and anti-Ub antibodies. (i) Normalized input of CPEB3 used in IPs. (ii) Detection of ubiquitinated CPEB3. CPEB3 (in vitro ub): Sample from in vitro ubiquitination of CPEB3 by Neurl1. CPEB3: non-ubiquitinated CPEB3 (in vitro assay without Neurl1). St1 & St2: single tetO-Neurl1-Flag and tetO-Neurl1DN-Flag mice, respectively. (iii) Western blot of monoubiquitinated CPEB3 isolated from hippocampal lysate (St1) next to the respective input. (iv) Mean + SEM from 3 independent experiments (1 mouse/genotype, treatment & experiment; controls: single tetO & tTA). Monoubiquitinated CPEB3 was increased in Neurl1 DT (*p<0.0016), while it was reduced in Neurl1DN DT (comparisons with respective control siblings and DT on dox; *p<0.0003).
See also Figure S5 & Table S4-6.
We first confirmed the interaction of Neurl1 with CPEB3. We immunoprecipitated CPEB3 from adult hippocampal lysates of wild type mice and detected Neurl1 in the same macromolecular complex (Figure 5A). We also found that CPEB3 and Neurl1 colocalize along the apical dendrites of adult CA1 neurons, in opposition to Synaptophysin (Figure 5B).
Using in vivo ubiquitination assays in HEK293T cells, we found that Neurl1 promotes ubiquitination of CPEB3 and that this depends on its ubiquitin ligase activity (Figure 5C). We also observed that the most abundant form of ubiquitinated CPEB3 migrated at a molecular size that was approximately 8kD higher than the non-ubiquitinated protein, which suggests that it may represent monoubiquitinated CPEB3.
Using in vitro ubiquitination assays, we found that Neurl1 targets CPEB3 directly for monoubiquitination (Figure 5D). Importantly, we detected monoubiquitinated CPEB3 in the adult hippocampus and found that it was significantly increased in Neurl1 overexpressing mice while it was significantly reduced in Neurl1DN animals (Neurl1: 3.01% + 0.31 of total CPEB3 vs 1.39% + 0.13 in controls; Neurl1DN: 0.48% +0.09 vs 1.37+0.15 in controls; p<0.0004; Figure 5E & S5C). When the expression of the transgenes in the adult hippocampus was blocked by doxycycline, monoubiquitinated CPEB3 returned back to wild type levels (Figure 5E & S5C). Consistent with our data, synaptic stimulation with glutamate in cultured hippocampal neurons led to upregulation of the protein levels of endogenous Neurl1 and increased monoubiquitination of CPEB3. Importantly, this activity-dependent increase of monoubiquitinated CPEB3 was blocked by lentiviral expression of Neurl1DN (Figure S5D&E).
Neurl1-dependent Ubiquitination Modulates the Activity of CPEB3 on its Translational Targets GluA1 and GluA2 and Increases their Protein Levels
A well-established function of ubiquitination is the degradation of proteins (DiAntonio and Hicke, 2004). In the basal state CPEB3 negatively regulates the translation of GluA1 and GluA2 (Huang et al., 2006; E.R.K. & L. F., unpublished data). Indeed, Neurl1-dependent degradation of CPEB3 might explain how overexpression of Neurl1 increases the levels of GluA1 and GluA2. However, we found this not to be case. In mice overexpressing Neurl1 the protein levels of CPEB3 in the hippocampus were not reduced; rather they were increased and this increase was not due to differences at the mRNA level (Figure S5F). Yet, in the hippocampus of Neurl1DN mice, we consistently found a reduction of CPEB3, without any alteration at the mRNA level (Figure S4E). Similarly, the activity-dependent upregulation of Neurl1 in cultured hippocampal neurons was accompanied by Neurl1-mediated increase of the overall protein levels of CPEB3 (Figure S5D). This paradox could be explained by a CPEB3-independent regulation of GluA1 and GluA2. Alternatively, Neurl1-dependent ubiquitination might affect CPEB3 translational activity by leading to the activation of CPEB3.
To distinguish between these alternatives, we overexpressed Neurl1 in cultured hippocampal neurons using lentiviral gene transfer. Neurl1 overexpression resulted in increased levels of GluA1 and GluA2 proteins (Figure 6A). CPEB3 levels were increased similar to what we observed in the adult hippocampus. But, when we overexpressed CPEB3 alone we found that the levels of GluA1 and GluA2 were dramatically reduced (Figure 6A). This effect of CPEB3 was rescued when Neurl1 was coexpressed with CPEB3 (Figure 6A). To rule out the possibility that Neurl1 affects GluA1 and GluA2 levels by a mechanism not related to the modulation of CPEB3 translational activity, we overexpressed Neurl1 and a truncated form of CPEB3 that retains its RNA binding properties but lacks the first 222 N-terminal aminoacids, a putative prion domain (CPEB3ΔNter; Huang et al., 2006). The deletion of this domain in Drosophila CPEB (orb2) resulted in impaired long-term memory formation, indicating a critical memory function of this protein region (Keleman et al., 2007). Importantly, we found that this domain is critical for both the interaction of CPEB3 with Neurl1 and for its ubiquitination (Figure S5G). If Neurl1 interacts and modulates the activity of CPEB3 and increases the translation of GluA1 and GluA2 mRNAs, this effect would be blocked if we abolished the interaction between CPEB3 and Neurl1. Indeed, CPEB3ΔNter was still capable of reducing the levels of GluA1 and GluA2 but this effect was not reversed by Neurl1 overexpression in contrast to full length CPEB3 (Figure 6A).
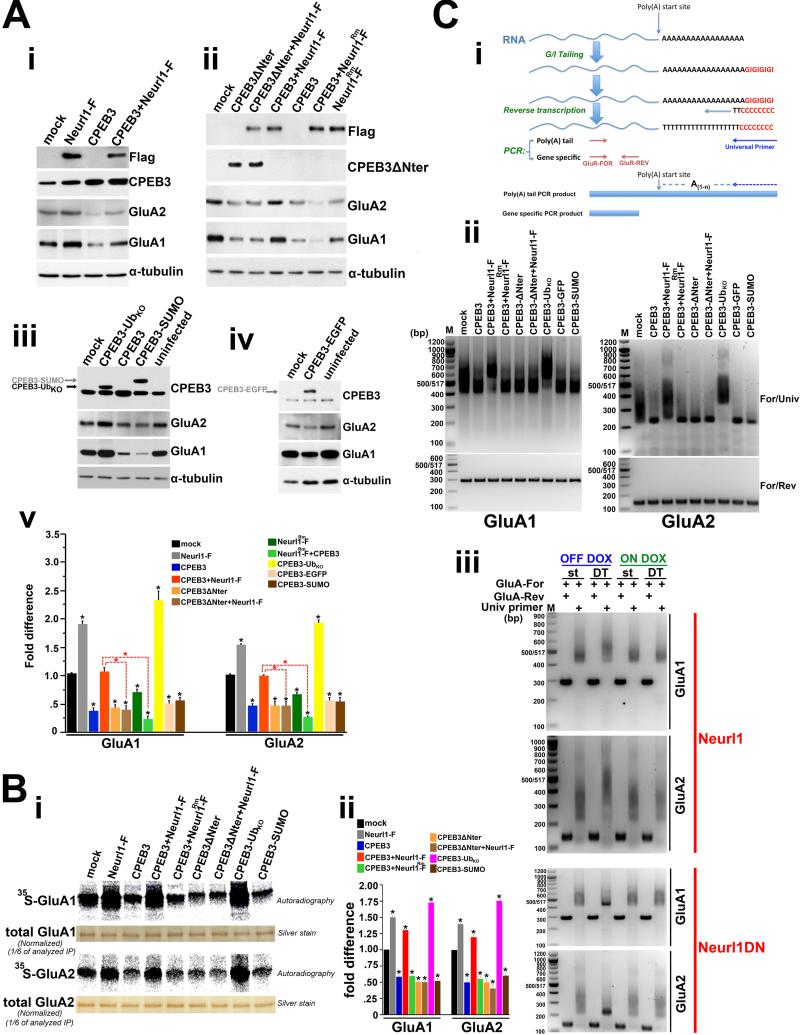
Figure 6. Neurl1-dependent ubiquitination and ubiquitin modulate the activity of CPEB3 and increase CPEB3-dependent polyadenylation and translation of GluA1 and GluA2 leading to an increase of their protein levels.
(A) Western blots (i-iv) and quantitative (v) analysis of GluA1 and GluA2 proteins in lysates of cultured hippocampal neurons. The neurons were infected with control lentivirus (mock) or lentiviruses expressing the indicated proteins. Anti-Flag antibody was used for the detection of Neurl1-Flag (Neurl1-F) and Neurl1Rm-Flag (Neurl1Rm-F). CPEB3-UbKO: chimeric CPEB3 having fused at its C-terminus single ubiquitin that cannot form poly-Ub chains. CPEB3-EGFP & CPEB3-SUMO: controls. (v) Averaged data from 4 independent experiments. Black asterisks: comparisons with mock. *p<0.0007.
(B) 35S-GluA1 and 35S-GluA2 immediately after a 1hr 35S-Met/35S-Cys pulse in cultured hippocampal neurons (16DIV) expressing the indicated proteins by viral gene transfer (i). (ii) Averaged data (+SEM) from 3 independent experiments. Compared to mock, 35S-GluA1 and 35S-GluA2 were significantly increased in neurons overexpressing Neurl1 (Neurl1-F expressing neurons), Neurl1 and CPEB3 together, and CPEB3-UbKO (p<0.008). 35S-GluA1 and 35S-GluA2 in the rest of neurons were similar and significantly lower compared to mock (*p<0.009).
(C) Poly(A) assay. (i) Schematic representation of the assay. Universal primer: poly(A)-specific primer. (ii) Assays in cultured hippocampal neurons expressing the indicated proteins. Mock: neurons expressing control lentivirus. (iii) Poly(A) tails of hippocampal GluA1 and GluA2 mRNAs from adult Neurl1 and Neurl1DN double transgenic mice (DT) and respective controls (single tetO; st) kept either off dox (transgene ON in DT) or on dox (transgene OFF in DT).
We also found that Neurl1 regulated CPEB3 activity via ubiquitination. Neurl1Rm-Flag, which does not ubiquitinate CPEB3 (Figure 5C), did not reverse the effect of CPEB3 overexpression on the levels of GluA1 and GluA2 (Figure 6A). Our results are consistent with the idea that Neurl1 ubiquitinates CPEB3 and that the ubiquitinated form of CPEB3 upregulates GluA1 and GluA2 protein synthesis.
Ubiquitin Modulates the Activity of CPEB3 on its Translational Targets GluA1 and GluA2
To test the idea that the addition of a single ubiquitin to CPEB3 is sufficient to modulate the activity of CPEB3 on GluA1 and GluA2 and mimic the effect of Neurl1 overexpression, we generated chimeric CPEB3 fused at its C-terminus to one moiety of ubiquitin with its seven lysines mutated to arginines and incapable to form polyubiquitin chains (CPEB3-UbKO). This approach has been used to study protein modification by ubiquitin as Ub fusion proteins have been shown to mimic ubiquitination in some instances (Qian et al. 2002; Li et al., 2003; Carter and Vousden, 2008). In contrast to wild type CPEB3, CPEB3-UbKO increased the protein levels of GluA1 and GluA2 in dissociated hippocampal neurons (Figure 6A). The effect of ubiquitin on CPEB3 activity was specific, and not an artifact of the fusion or a general effect of ubiquitin-like molecules, as chimeric CPEB3-EGFP and CPEB3-SUMO proteins reduced the levels of GluA1 and GluA2, similar to CPEB3 overexpression (Figure 6A). Thus, single ubiquitin is sufficient to activate CPEB3 and CPEB3-dependent translation of GluA1 and GluA2 and to increase their protein levels.
Similar to Neurl1, overexpression of CPEB3-Ub KO increased the surface population of AMPAR without affecting their trafficking and endocytosis (Table S3; Figure S4B).
Neurl1 and Ubiquitin Activate CPEB3-dependent Translation of GluA1 and GluA2 mRNAs
Our data suggest that Neurl1-dependent ubiquitination and ubiquitin modulate CPEB3 translational activity and activate CPEB3-dependent protein synthesis of GluA1 and GluA2. We tested this directly by using 35S-Methionine/35S-Cysteine metabolic labelling and examining the levels of newly synthesized GluA1 and GluA2. We obtained complementary results (Figure 6B). The observed effects were due to modulation of GluA1 and GluA2 translation, as we observed no differences at the level of mRNA (Figure S6A). The changes were specific. We tested the translation of actin and found it unaltered (Figure S6B). Using reporter assays in HEK293T cells we also found that the translational regulation of GluA1 and GluA2 mRNAs by Neurl1 and ubiquitin was mediated specifically by CPEB3 and required their 3′-UTRs (Table S3; Figure S6C), consistent with the role of CPEB3 as an RNA binding protein that binds the 3′-UTR of GluA1 and GluA2 mRNAs (Huang et al., 2006 & figure S5A). Taken together, these results directly demonstrate that Neurl1 and ubiquitin modulate CPEB3-dependent translation of GluA1 and GluA2.
Neurl1 and Ubiquitin Activate CPEB3-dependent Polyadenylation of GluA1 and GluA2 mRNAs
We next asked: Does ubiquitinated CPEB3 activity promote polyadenylation and increased translation of GluA1 and GluA2 mRNAs? Using polyadenylation assays in cultured hippocampal neurons, we found that consistent with its inhibitory role on GluA1 and GluA2 translation at the basal state overexpression of CPEB3 alone induced shortening of the poly(A) tails of GluA1 and GluA2 mRNAs (Figure 6C & S6D). This action of CPEB3 was reversed when Neurl1 was coexpressed. The effect of Neurl1 on CPEB3 activity was blocked when the N-terminal domain of CPEB3 was missing, and it was dependent on its ubiquitin ligase activity (Figure 6C). Expressing CPEB3 fused to single ubiquitin, but not SUMO and EGFP, also led to the elongation of the poly(A) tails of GluA1 and GluA2 mRNAs (Figure 6C).
GluA1 and GluA2 mRNA poly(A) tails were also elongated in the adult hippocampus of mice overexpressing Neurl1, while they were shortened in the hippocampus of mice expressing Neurl1DN (Figure 6C). The observed changes were specific, as the poly(A) length of actin mRNA was unaltered either in cultured neurons or the adult hippocampus (Figure S6E). Thus, Neurl1-mediated ubiquitination modulates CPEB3-dependent polyadenylation of GluA1 and GluA2 mRNAs.
Neurl1 and Ubiquitin Facilitate Spine Growth by Modulating CPEB3 Translational Activity
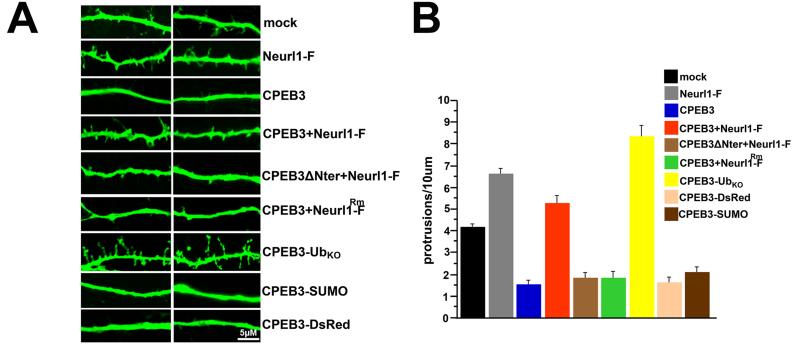
In addition to the increased protein levels of GluA1 and GluA2, the number of functional synapses increases in the Neurl1 overexpressing mice. Does Neurl1 also regulate spine formation by modulating CPEB3 translational activity? This modulation of translation would be consistent with a Neurl1-dependent molecular mechanism which links AMPA receptor translation and synapse formation to the role of local polyadenylation and protein synthesis in synaptic growth and plasticity (Sutton and Schuman, 2006). In agreement with and parallel to our observation in Neurl1 overexpressing mice, we found that Neurl1 overexpression in cultured hippocampal neurons resulted in significantly increased number of spines and filopodia-like protrusions (Figure 7). This number was reduced when CPEB3 was overexpressed alone. This inhibition was reversed when Neurl1 was coexpressed with CPEB3, similar to what we observed for Neurl1 enhancement of CPEB3 activity on GluA1 and GluA2 (Figure 7). Expression of CPEB3ΔNter in turn significantly reduced the number of spines and filopodia compared to control neurons and Neurl1 overexpression did not reverse this effect. Neurl1Rm-Flag was similarly not sufficient to reverse the inhibitory effect of CPEB3. However, expressing CPEB3-UbKO resulted in pronounced increase of number of spines and filopodia, whereas CPEB3 fused to either DsRed or SUMO resulted in a phenotype similar to wild type CPEB3 (Figure 7). Taken together, our data suggest that the ability of Neurl1 to ubiquitinate CPEB3 is a limiting factor and facilitates hippocampal synaptic plasticity and hippocampal-dependent memory by modulating CPEB3 translational activity through ubiquitination with consequent induction of an increase of GluA1 and GluA2 protein levels and the formation of new spines.
Figure 7. Neurl1 and ubiquitin increase spine number by modulating the translational activity of CPEB3.
(A) Dendrites of cultured hippocampal neurons (11DIV) expressing EGFP alone (mock) or EGFP and the indicated proteins. Branches of two neurons are shown. Neurl1-F: Neurons expressing Neurl1-F (Neurl1-Flag; Neurl1 overexpression). Neurl1Rm-F: mutant Neurl1-F. (B) Averaged density of dendritic protrusions + SEM [mock: 12 neurons, 2 branches/neuron (12×2×50μm); all the others: 8 neurons, 2 branches/neuron (8×2×50μm)]. The spine density of neurons overexpressing Neurl1, Neurl1+CPEB3, and CPEB3-UbKO were significantly increased compared to mock (CPEB3+Neurl1-F: p=0.041; all the others: p<0.0001). The difference between neurons overexpressing CPEB3 and neurons overexpressing CPEB3+Neurl1 was highly significant (p<0.0001). In the rest of the neurons, the density of protrusions was similar (p=0.6165), and significantly lower than mock (p<0.0003). See also Table S4 and Figure S7.
DISCUSSION
Neurl1-Mediated Ubiquitination Modulates Translational Activity of CPEB3
CPEB3 is a member of a group of cytoplasmic polyadenylation proteins that regulate local protein synthesis in Aplysia, Drosophila and mice. CPEB3 is a homologue of the Aplysia and Drosophila proteins that have been found to serve as functional prions, and to be essential for long-lasting changes of synaptic efficacy and long-term memory (Si et al., 2003, 2010; Keleman et al., 2007). One major question not been addressed in Aplysia and in Drosophila is: how are these proteins regulated? We find that CPEB3 directly interacts with Neurl1 and is monoubiquitinated by it. A single ubiquitin is sufficient to activate CPEB3 and the subsequent polyadenylation and translation of GluA1 and GluA2 mRNAs. Importantly, Neurl1 overexpression increases monoubiquitination and Neurl1 inhibition reduces monoubiquitination of CPEB3 in the adult hippocampus. These bidirectional changes are accompanied by parallel changes of the poly(A) tail lengths of GluA1 and GluA2 mRNAs and protein levels.
The proposed mechanism of Neurl1 in regulating translation is consistent with the proteasomeindependent action of ubiquitination in protein synthesis (Spence et al., 2000). In yeast, the ribosomal activity is regulated by the atypical Lys63 polyubiquitin chain. The current knowledge of the role of ubiquitination in regulating protein synthesis in mammals, however, is limited to proteasomal degradation of translational factors (Hitchcock et al., 2003; Peng et al., 2003; Hou et al., 2006). Our data, therefore, provide new evidence for a non-proteolytic function of ubiquitination in the control of protein synthesis in mammals in general and in neurons in particular.
A Novel Role for Ubiquitination in Synaptic Plasticity and Memory Storage
Hippocampal-dependent learning is correlated with increases in synaptic spines, and altered number of synaptic contacts is reflected in parallel changes in learning and memory in mice (Fischer et al., 2005; Guan et al., 2009). Consistent with these findings, Neurl1 overexpression and the consequent elevation of spine number and functional synapses is accompanied by hippocampal-dependent cognitive enhancements, whereas blockade of Neurl1 overexpression leads to restoration of spine number to wild type levels and to cognitive performances comparable to controls.
Using a reduced test system of hippocampal neurons in culture, we find that Neurl1 upregulation induces the formation of spines by modulating the translational activity of CPEB3. In the basal state, CPEB3 inhibits both spine formation and the translation of GluA1 and GluA2. By contrast, Neurl1 upregulation leads to monoubiquitination of CPEB3, which leads to increased translation of GluA1 and GluA2 accompanied by an increased number of spines. Consistently, increased monoubiquitination of CPEB3 and increased translation of GluA1 and GluA2 accompany the increased number of spines and functional synaptic contacts in the adult hippocampus of mice overexpressing Neurl1.
These findings are also in agreement with the role in synapse formation and synaptic plasticity of GluA1 and GluA2, the two known targets of CPEB3 (Nicoll, 2003; Kopec et al., 2007; Bassani et al., 2009). GluA2 is important for the formation of functional synapses. Its overexpression in hippocampal neurons increases the number of spines, the frequency of miniEPSCs and the protein levels of the postsynaptic proteins Shank and PSD95, (Passafaro et al., 2003; Saglietti et al., 2007). These effects are strikingly similar to those we observed in the adult hippocampus of the Neurl1 overexpressing mice. GluA1 also participates in the stabilization of spines and increases synaptic strength by means of its ligand-gated ion channel (Kopec et al., 2007).
The proposed mechanism of Neurl1 in synaptic plasticity is consistent with the requirement of Neurl1 activity for the maintenance of LTP and LTD, and with the activity-dependent upregulation of Neurl1 protein levels in the hippocampus of wild type mice. Protein synthesis-dependent increase in AMPAR expression has been described after LTP induction (Nayak et al., 1998) and LTP maintenance is believed to depend on both translation-dependent morphological changes (Tanaka et al., 2008) and GluA1 insertion-dependent spine alteration (Kopec et al., 2007). L-LTD also depends on new protein synthesis (Manahan-Vaughan et al., 2000; Kauderer and Kandel, 2000). In addition, decrease of spine density can induce LTD impairment (Brigman et al., 2006), whereas pharmacological or behavioral manipulations that increase spine density facilitate LTD (Mukai et al., 2007; Artola et al., 2006).
The model for Neurl1 function that we propose here is consistent with the role of local polyadenylation and translation in synaptic plasticity (Sutton and Schuman, 2006). The function of CPEB3 that we describe here is also consistent with the idea that CPEB3 is the mammalian functional homologue of ApCPEB (Theis et al., 2003). In Aplysia, CPEB regulates local protein synthesis and is required for the stabilization of synaptic growth and long-lasting changes of synaptic efficacy, much as we find for CPEB3 (Si et al., 2003, 2010; Miniaci et al., 2008).
EXPERIMENTAL PROCEDURES
Transgenic Mice
Neurl1 and Neurl1DN open reading frames (encoded amino acids: 1-574 & 1-520, respectively; Neurl1 Genebank accession numbers: Y15160 & BC099702) were cloned into the pMM400 vector. Mice were treated in compliance with the rules of IACUC.
Spine Counting
The FD Rapid GolgiStain kit was used. Neuronal tracing and spine counting were performed using the Neurolucida software (MicroBrightField). NeuroExplorer software (MicroBrightField) was used for the analysis. The experimenter was “blind” to the genotype/treatment. Standard criteria were followed for the selection of neurons for tracing (Extended Experimental Procedures).
In vivo Ubiquitination Assays
HEK293T cells transiently transfected with plasmids encoding 6xHis-Ub, CPEB3 and Neurl1-F or Neurl1Rm-F were lysed in 6M guanidine-HCl buffer. Lysates were sonicated and cleared by ultracentrifugation. His-Ub-conjugated proteins were purified on Ni-NTA-agarose under denaturing conditions, separated by SDS-PAGE, and analyzed by immunoblotting.
In vitro Ubiquitination Assays
CPEB3 produced by in vitro transcription/translation was incubated with Neurl1-F, Neurl1Rm-F and Ube1 (E1), UbcH5a (E2) and ubiquitin in the presence or absence of energy source (ATP). Reaction products were detected by immunoblotting.
Isolation of Hippocampal Ubiquitinated CPEB3
Adult hippocampal lysates from different genotypes were first normalized against CPEB3 by western blot and quantification. We next performed IP with anti-CPEB3 antibody. CPEB3 was eluted by competition with 100X of the respective peptide. Subsequent IP with anti-Ub antibody was performed. The final immunoprecipitants were analyzed for ubiquitinated species of CPEB3 by immunoblot using anti-CPEB3 antibody.
Metabolic Labelling of AMPAR
High density cultured hippocampal neurons (16DIV) were incubated for 1hr in 600 uCi/ml of 35S-Met/35S-Cys mix in serum and Met/Cys-free DMEM after an initial incubation for 90min at 37°C. Neurons were lysed in RIPA buffer.
Poly(A) Assays
Poly(A) tail-length assay kit was used (USB-Affymetrix).
Electrophysiology and Behavior
Adult littermate males (3 ½ months) were used. Single transgenic and wild type mice showed no differences and were pooled (control group). Statistical analyses used ANOVAs with genotype as the between-subject factor. The experimenter was “blind” to the genotype. Standard protocols were followed for electrophysiology (Extended Experimental Procedures).
Morris water maze
In the visible, hippocampal-independent, version of the task, the mice were trained with 4trials/day. One trial/day training protocol was used in the acquisition and the transfer phases of the hidden platform version of the maze. Escape latencies and path lengths were plotted against the day of the experiment. Probe trials, during which the platform was removed from the maze, lasted 1min. Platform crossings and exploration times were examined.
Novel object recognition
The mice first explored two identical novel objects for 15min. The 24hr memory test lasted 5min and a novel object had replaced one of the familiar objects. The discrimination index was determined by the difference in exploration time expressed as a ratio of the total time spent exploring the two objects.
The elevated plus maze and open field were used to measure anxiety.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Steven A. Siegelbaum for his comments and help with the mEPSC recordings. We are also grateful to Thomas Jessell for his reading of the manuscript and to Eleanor Simpson, Christoph Kellendonk, Kausik Si, Sathya Puthanveettil, Sebastien Thuault and Evangelia Koutelou for their critical comments. We also thank Kevin Karl for his technical help; and Joan Conaway and Nicholas Moschonas for the gift of the Neurl1Rm and Ub cDNAs. The work was supported by HHMI and the Broitman Program for Cognitive Disorders (to E.R.K). P.T. was supported by the Fondation pour la Recherche Medicale. E.R.K is a senior investigator of HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Artola A, von Frijtag JC, Fermont PC, Gispen WH, Schrama LH, Kamal A, Spruijt BM. Long-lasting modulation of the induction of LTD and LTP in rat hippocampal CA1 by behavioural stress and environmental enrichment. Eur J Neurosci. 2006;23:261–272. doi: 10.1111/j.1460-9568.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Bassani S, Valnegri P, Beretta F, Passafaro M. The GLUA2 subunit of AMPA receptors: synaptic role. Neuroscience. 2009;158:55–61. doi: 10.1016/j.neuroscience.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Brigman JL, Wright T, Talani G, Prasad-Mulcare S, Jinde S, Seabold GK, Mathur P, Davis MI, Bock R, Gustin RM, et al. Loss of GluN2B-containing NMDA receptors in CA1 hippocampus and cortex impairs long-term depression, reduces dendritic spine density, and disrupts learning. J Neurosci. 2006;30:4590–4600. doi: 10.1523/JNEUROSCI.0640-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S, Vousden KH. p53-Ubl fusions as models of ubiquitination, sumoylation and neddylation of p53. Cell Cycle. 2008;7:2519–2528. doi: 10.4161/cc.7.16.6422. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- DiAntonio A, Hicke L. Ubiquitin-dependent regulation of the synapse. Annu Rev Neurosci. 2004;27:223–246. doi: 10.1146/annurev.neuro.27.070203.144317. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Pang PT, Lu B, Tsai LH. Opposing roles of transient and prolonged expression of p25 in synaptic plasticity and hippocampus-dependent memory. Neuron. 2005;48:825–838. doi: 10.1016/j.neuron.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hitchcock AL, Auld K, Gygi SP, Silver PA. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc Natl Acad Sci U S A. 2003;100:12735–12740. doi: 10.1073/pnas.2135500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluA2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauderer BS, Kandel ER. Capture of a protein synthesis-dependent component of long-term depression. Proc Natl Acad Sci U S A. 2000;97:13342–13347. doi: 10.1073/pnas.97.24.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleman K, Krüttner SA, M., Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Kopec CD, Real E, Kessels HW, Malinow R. GluA1 links structural and functional plasticity at excitatory synapses. J Neurosci. 2007;27:13706–13718. doi: 10.1523/JNEUROSCI.3503-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E, Sato S, Tomomori-Sato C, Florens L, Swanson SK, Washburn MP, Kokkinaki M, Conaway RC, Conaway JW, Moschonas NK. Neuralized-like 1 (Neurl1) targeted to the plasma membrane by N-myristoylation regulates the Notch ligand Jagged1. J Biol Chem. 2008;283:3846–3853. doi: 10.1074/jbc.M706974200. [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer f., Chen D, Baer R, Gu W. Mono-Versus Polyubiquitination: Differential Control of p53 Fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malleret G, Alarcon JM, Martel G, Takizawa S, Vronskaya S, Yin D, Chen IZ, Kandel ER, Shumyatsky GP. Bidirectional regulation of hippocampal long-term synaptic plasticity and its influence on opposing forms of memory. J Neurosci. 2010;30(10):3813–3825. doi: 10.1523/JNEUROSCI.1330-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manahan-Vaughan D, Kulla A, Frey JU. Requirement of translation but not transcription for the maintenance of long-term depression in the CA1 region of freely moving rats. J Neurosci. 2000;20:8572–8576. doi: 10.1523/JNEUROSCI.20-22-08572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Miniaci MC, Kim JH, Puthanveettil SV, Si K, Zhu H, Kandel ER, Bailey CH. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59:1024–1036. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RGM, Garrud P, Rawlins JNP, O’Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- Mukai H, Tsurugizawa T, Murakami G, Kominami S, Ishii H, Ogiue-Ikeda M, Takata N, Tanabe N, Furukawa A, Hojo Y, et al. Rapid modulation of long-term depression and spinogenesis via synaptic estrogen receptors in hippocampal principal neurons. J Neurochem. 2007;100:950–967. doi: 10.1111/j.1471-4159.2006.04264.x. [DOI] [PubMed] [Google Scholar]
- Nayak A, Zastrow DJ, Lickteig R, Zahniser NR, Browning MD. Maintenance of late-phase LTP is accompanied by PKA-dependent increase in AMPA receptor synthesis. Nature. 1998;394:680–683. doi: 10.1038/29305. [DOI] [PubMed] [Google Scholar]
- Nicholls RE, Alarcon JM, Malleret G, Carroll RC, Grody M, Vronskaya S, Kandel ER. Transgenic mice lacking NMDAR-dependent LTD exhibit deficits in behavioral flexibility. Neuron. 2008;58(1):104–117. doi: 10.1016/j.neuron.2008.01.039. [DOI] [PubMed] [Google Scholar]
- Nicoll RA. Expression mechanisms underlying long-term potentiation: a postsynaptic view. Philos Trans R Soc Lond B Biol Sci. 2003;1432 doi: 10.1098/rstb.2002.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passafaro M, Nakagawa T, Sala C, Sheng M. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluA2. Nature. 2003;424:677–681. doi: 10.1038/nature01781. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Anezaki M, Skoulakis EM. Neuralized is expressed in the α/β lobes of adult Drosophila Mushroom Bodies and facilitates olfactory Long-Term Memory formation. Proc Natl Acad Sci U S A. 2008;105:14674–14679. doi: 10.1073/pnas.0801605105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Qian SB, Ott DE, Schubert U, Bennink JR, Yewdell JW. Fusion proteins with COOH-terminal ubiquitin are stable and maintain dual functionality in vivo. J Biol Chem. 2002;277:38818–38826. doi: 10.1074/jbc.M205547200. [DOI] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Saglietti L, Dequidt C, Kamieniarz K, Rousset MC, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, et al. Extracellular interactions between GluA2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- Segref A, Hoppe T. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Rep. 2009;10:44–50. doi: 10.1038/embor.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Keino H, Satoh M, Kishikawa M, Hosokawa M. Age-related loss of synapses in the frontal cortex of SAMP10 mouse: a model of cerebral degeneration. Synapse. 2003;48:198–204. doi: 10.1002/syn.10209. [DOI] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis M, Si K, Kandel ER. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc Natl Acad Sci U S A. 2003;100:9602–9607. doi: 10.1073/pnas.1133424100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.