Background: Rnds are constitutively active proteins whose regulation is poorly understood.
Results: Interaction with Syx or p190 RhoGAP stabilizes Rnds whereas ROCK activity stimulates Rnd3 turnover.
Conclusion: The steady-state level of Rnd proteins depends on the availability of cellular effectors that protect them from degradation.
Significance: This study uncovers a new mode of feedback regulation for Rnd proteins.
Keywords: Protein Degradation, Protein Stability, Protein Turnover, Protein-Protein Interactions, Ubiquitination, ROCK, Rnd3, Syx, p190 RhoGAP
Abstract
Rnd proteins are Rho family GTP-binding proteins with cellular functions that antagonize RhoA signaling. We recently described a new Rnd3 effector Syx, also named PLEKHG5, that interacts with Rnds via a Raf1-like “Ras-binding domain.” Syx is a multidomain RhoGEF that participates in early zebrafish development. Here we demonstrated that Rnd1, Rnd2, and Rnd3 stability is acutely dependent on interaction with their effectors such as Syx or p190 RhoGAP. Although Rnd3 turnover is blocked by treatment of cells with MG132, we provide evidence that such turnover is mediated indirectly by effects on the Rnd3 effectors, rather than on Rnd3 itself, which is not significantly ubiquitinated. The minimal regions of Syx and p190 RhoGAP that bind Rnd3 are not sequence-related but have similar effects. We have identified features that allow for Rnd3 turnover including a conserved Lys-45 close to the switch I region and the C-terminal membrane-binding domain of Rnd3, which cannot be substituted by the equivalent Cdc42 CAAX sequence. By contrast, an effector binding-defective mutant of Rnd3 when overexpressed undergoes turnover at normal rates. Interestingly the activity of the RhoA-regulated kinase ROCK stimulates Rnd3 turnover. This study suggests that Rnd proteins are regulated through feedback mechanisms in cells where the level of effectors and RhoA activity influence the stability of Rnd proteins. This effector feedback behavior is analogous to the ability of ACK1 and PAK1 to prolong the lifetime of the active GTP-bound state of Cdc42 and Rac1.
Introduction
The Rnd proteins are unusual in that they do not behave like conventional Rho proteins with respect to their mechanism of activation. Rnds are found in a GTP-bound state because they do not contain glycine at the RasG12 equivalent position (1–3) and thus do not catalyze GTP hydrolysis or require GDP-GTP exchange for activation. Consequently, Rnd proteins are instead regulated by mechanisms such as post-translational modification (4) and Rnd3/RhoE mRNA transcripts are up-regulated by diverse stimuli. For example, the oncogene B-Raf, genotoxic stress, and the mTOR pathway all promote increased Rnd3 expression (5–8). Rnd3 phosphorylation by RhoA-associated kinase (ROCK1)2 or protein kinase C (PKCα) have been reported to be important in regulating its localization and function (9, 10). Seven ROCK1 phosphorylation sites are identified in Rnd3, with Ser-7/Ser-11 reported to regulate Rnd3 stability (9). The structure and association of ROCK1 with Rnd3 suggest that this kinase can bind Rnd3 (11), but does not interfere with effector binding. It has also been suggested that PDK1 competition for Rnd3 binding prevents phosphorylation by ROCK1 (12).
Rnd1 expression is highest in adult brain and liver, Rnd2 is primarily expressed in the brain and testis, whereas Rnd3 is more ubiquitous. Nonetheless, loss of Rnd3 in mice causes severe abnormalities primarily in the nervous system, and these lead to death shortly after birth (13). We found that Rnd3 expression is strongly up-regulated during early vertebrate development (14). At the subcellular level, Rnd1 and Rnd3 are membrane-associated proteins that are concentrated at adherent junctions both in fibroblasts and in epithelial cells (15). The antagonistic effect of Rnd1/3 on RhoA signaling has been noted in terms of their effects on the actomyosin organization (2). Transient expression of Rnd1 or Rnd3 disassembles actin stress fibers and stimulates cell migration (15). Overexpression of Rnd3 can inhibit cell cycle progression and Ras-induced transformation, but this does not appear to involve RhoA, ERK, or PI3K/Akt pathways (16). Interestingly the micro-RNA associated with epithelial-mesenchymal transition, mir-200b, interacts with predicted target sites in the 3′-untranslated region of Rnd3 mRNA. In HeLa cells, mir-200b directly reduced Rnd3 mRNA and protein levels and promoted S phase entry (17). In Xenopus, Rnd1 and Rnd3 were found to be required for proper somite formation (18), and Rnd1 has also been demonstrated to regulate morphogenetic movements by modulating cell adhesion in early embryos (19).
Rnd1 and Rnd3 are closely related at the primary sequence level and appear to have very similar target proteins, including p190 RhoGAP and Socius, as reviewed in Ref. 4. Rnd1, Rnd2, and Rnd3 can stimulate the activity of the ubiquitous p190 RhoGAP to decrease RhoA GTP levels (20). It has been shown that myoblasts lacking Rnd3 show defects in p190 RhoGAP activation and impair down-regulation of RhoA at the onset of myoblast fusion and thus inhibit myotube formation (21). Recently, it has been reported that Rnd1 and Rnd3 but not Rnd2 have a basic-rich region in their N terminus which functions as lipid raft-targeting determinant, and this raft targeting is required for p190 RhoGAP-mediated RhoA inhibition (22). The single report of an interaction between Rnd3 and Socius (23) has not been followed up. We have demonstrated that Rnd3 binds a RhoGEF termed Syx and is required early in zebrafish development (14). A Rnd3 binding-defective mutant of Syx was more potent in rescuing the embryonic defects than WT Syx, indicating that Rnd negatively regulates Syx activity in vivo. In this context RhoA is required for tissue morphogenesis during early development, as exemplified by its role in the noncanonical Wnt signaling to activate DAAM1 and ROCK kinases (24, 25). Syx is also known as PLEKHG5/TECH in the context of the nervous system (26–28) and is most closely related to MyoGEF, although this protein does not contain the Rnd-binding domain (14). The ubiquitin-like “Rnd-binding domain” (RndBD) in Syx mediates an interaction with both Rnd1 and Rnd3. In this study, we demonstrate how such Rnd effectors can affect the stability of Rnd proteins. Specifically co-expression of Syx or p190 RhoGAP protects Rnd1 and Rnd3 from degradation, which does not occur with effector mutants of Rnd3. By contrast the more widely reported Rnd3 effector Plexin B1, which binds weakly to Rnd3, has no effect on Rnd3 stability. Our data also show that single substitution of Rnd3 K45R can largely prevent Rnd3 turnover, indicating that this represents a site that interacts with protein(s) modulating Rnd3 turnover.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK 293T (ATCC CRL-11268), HeLa (ATCC CCL-2), COS-7 (ATCC CRL-1651), and MDCK (ATCC CCL-34) cells were purchased from American Type Culture Collection. Human keratinocyte cells were obtained from National University Hospital and cultured in K-SFM medium supplemented with bovine pituitary extract and epidermal growth factor. Transient transfections were carried out with Genejuice (Novagen) for HEK 293T, COS-7, or HeLa cells and Lipofectamine 2000 (Invitrogen) for MDCK cells, according to the manufacturer's instructions.
Plasmid Constructions
The full-length and various truncated constructs of Syx, as well as Rnd1, Rnd2, and Rnd3, were obtained as described previously (14). Mouse RGS14(323–469) was cloned from mES cell line E14 by reverse transcription-PCR (RT-PCR) whereas human Plexin B1(1743–1869) was amplified from HeLa cells. Mouse p190B RhoGAP (amino acids 382–1007) in pCMV-myc was generous gift from Steen Hansen (Harvard Medical School), and the truncated constructs were obtained by PCR amplification and subcloned into N terminus fusion versions of HA or HA-GST-tagged pXJ40 vectors. Human ubiquitin in PXJ-HA vector was obtained from sGSK group (Astar Neuroscience Research Partnership). Mutants of Rnd3 (S11A, S11D, T37N, T55A, K45R, K115R, and C241S) were generated by PCR-mediated mutagenesis using the QuikChange II protocol (Stratagene) and sequenced. The chimeric Rnd3(C15) was obtained by PCR amplification of Rnd3(1–200) followed by the insertion of the last 15 C-terminal amino acid residues of Cdc42b.
Knockdown of Rnd3 Expression by SiRNA
COS cells were transfected with siRNA oligonucleotides by Lipofectamine 2000. Two sets of double-stranded siRNAs were used directed against Rnd3 (Thermo Scientific Dharmacon ON-TARGET plus SMARTpool L-064484-01-0005) and (Bioneer, 5′-GCACAUUAGUGGAACUCUCdTdT-3′). Control siRNA (5′-CCUACGCCACCAAU-UUCGUdTdT-3′) was from Bioneer. Knockdown of Rnd3 was determined by Western blotting at 24 h after transfection.
Proteasomal Degradation and Measurement of Rnd3 Turnover
HeLa, COS, or HEK 293T cells were treated with cycloheximide (20 μg/ml) (Sigma) in the presence of serum (unless otherwise indicated). MG132 was used at 10 μm final concentration (Sigma). The cells were lysed, and protein concentrations were determined using the Bradford assay. Equal amounts of proteins were resolved by SDS-PAGE and transferred to a PVDF membrane. Levels of Rnd3 proteins were determined by Western blotting and evaluated by densitometry of the film using ImageJ.
Streptavidin Pulldown
HEK 293T cells were transiently transfected with the indicated combination of plasmids and harvested at 24 h. When co-transfected with HA-tagged ubiquitin, MG132 (10 μm) was added to inhibit proteasome activity 24 h after transfection. The cells were treated for 6 h before harvesting. Harvested cells were washed in phosphate-buffered saline (PBS), lysed in buffer containing 10 mm Tris-HCl, pH 8.0, 250 mm NaCl, 0.1% Nonidet P-40, protease inhibitor mixture (Complete Mini EDTA-free; Roche Applied Science), and 2 mm sodium orthovanadate. The supernatants were incubated with streptavidin resin (50% slurry) (Amersham Biosciences) for 1 h at 4 °C, washed three times, and eluted with 2 mm biotin. The eluate containing the interacting complex was separated by SDS-PAGE, transferred to PVDF, and immunoblotted with the indicated antibodies.
Immunofluorescence Microscopy
MDCK cells seeded onto 13-mm glass coverslips were washed with cold PBS and fixed with 4% paraformaldehyde for 15 min at 37 °C. Fixed cells were permeabilized with 0.5% Triton X-100 for 15 min. Following this, the coverslips were incubated with primary antibodies at room temperature for 1 h. Coverslips were then washed three times in 0.5% Triton X-100 containing PBS before incubation with secondary fluorescent antibodies (Molecular Probes) coupled with Alexa Fluor 488. Images were viewed with an Olympus laser scanning microscope (FV-1000). All images were visualized with a 60× objective lens.
Antibodies
The following antibodies were used: monoclonal anti-FLAG (M2), anti-HA (HA-7), anti-Myc (all from Sigma), rabbit polyclonal HA (Santa Cruz Biotechnology), anti-Rnd3 (Upstate), and anti-β-actin (Millipore). For Western blotting, the primary antibodies were detected using horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) with GE Healthcare ECL or SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
RESULTS
Syx Interaction Stabilizes Rnd3
Rnd proteins are likely regulated by transcriptional, translational, or post-translational means because the proteins do not contain a GDP-bound “off” state. We observed that co-transfection of Syx(1–800) with Rnd was associated with significantly elevated levels of the G protein (Fig. 1B). Rho GTPases in their GTP-bound form are often targeted for ubiquitination and subsequent degradation (29, 30). To assess the stability of Rnd proteins versus other Rho GTPases (RhoA, Cdc42, and Rac1) we inhibited protein synthesis with cycloheximide and measured the levels of these proteins. As shown in Fig. 1C Rnds and other Rho proteins undergo turnover at similar rates. It has been shown that turnover of RhoA is driven by the E3 ligase Smurf-1 or the Cul3/BACURD ubiquitin ligase complex (31, 32) whereas (active) Rac1 degradation is apparently mediated by different complexes involving POSH and HACE (33, 34).
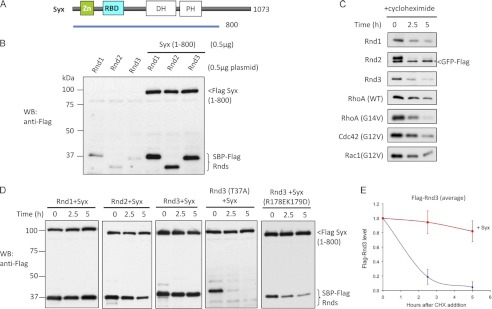
FIGURE 1.
Syx contributes to protein stability of Rnd3. A, schematic represents Syx construct used in 293T cell expression studies. B, 293T cells were transfected with Rnd plasmids in the absence or presence of Syx(1–800), lysed, and immunoblottted (WB). C, 293T cells were co-transfected with 0.5 μg of GFP-FLAG and 0.5 μg of the indicated plasmids. After 24 h, cells were incubated with cycloheximide (20 μg/ml) and then harvested at the indicated times with equal amounts of total protein loaded (40 μg/lane). Expression of GFP is marked by an asterisk (*). D, 293T cells were co-transfected with 0.5 μg of Rnd3 and 0.5 μg of Syx(1–800) plasmids and treated for the indicated times with cycloheximide, lysed, and immunoblottted. E, in the graphical representation, the y-axis corresponds to the ratio of Rnd3 relative to time 0. The average of three independent experiments is shown. Error bars, S.D.
The turnover of Rnd3 in 293T cells in the presence of transfected FLAG-Syx(1–800) is shown in Fig. 1D. After transient transfection, Rnd3 has a half-life of <2.5 h which can be increased to >5 h by the co-expression of Syx (Fig. 1E). Co-expression of full-length Syx similarly increased the half-life of Rnd3 (supplemental Fig. 1). This effect of Syx was also observed with Rnd1 and Rnd2 (Fig. 1D) although we found these bind more weakly to Syx (14). The Rnd3(T37A) mutant which cannot bind Syx (supplemental Fig. 2) undergoes turnover in the presence of Syx at a rate seen for Rnd3 alone. Similarly, the Syx mutant R178E/K179D that does not bind Rnd3 fails to stabilize Rnd3 (Fig. 1D). Taken together, we conclude that the stabilizing effect requires direct Syx interaction.
Evidence for Phosphorylation of Endogenous Rnd3
Rnd3 protein levels are specifically and transiently up-regulated upon keratinocyte differentiation (35). This up-regulation was found to require the activity of the RhoA target ROCK. Such Rnd3 activation in basal cells might facilitate detachment from the substratum and migration to form suprabasal layers (36). We found that Rnd3 has a half-life of <2.5 h in both nondifferentiated primary keratinocytes and HeLa cells (Fig. 2A), as assessed by Western blot analysis using an anti-Rnd3 Mab. Because Rnd3 is detected as a doublet (7, 10, 21, 35), we sought clarify whether both bands correspond to Rnd3. Antibody reactivity toward overexpressed Rnd1, Rnd2, and Rnd3 suggests that this antibody is specific for Rnd3 (supplemental Fig. 3A). Lysates from siRNA-treated COS-7 cells were probed with anti-Rnd3. Both bands were depleted in siRNA-treated cells, indicating that both correspond to Rnd3 (Fig. 2B). Calyculin treatment (10 min) resulted in a dramatic enrichment of the slower migrating Rnd3 band (Fig. 2C), likely representing a phosphorylated form of Rnd3. However, we could not cause an enrichment of the upper band by activating conventional PKCs with either bryostatin or PMA, as described previously (10), nor could this calyculin-induced size shift be blocked with Y-27632 (data not shown). We conclude that Rnd3 undergoes a rapid phosphorylation-dephosphorylation cycle in many cell types, as evidenced by the presence of a protein doublet, although this is less evident with overexpressed tagged Rnd3.
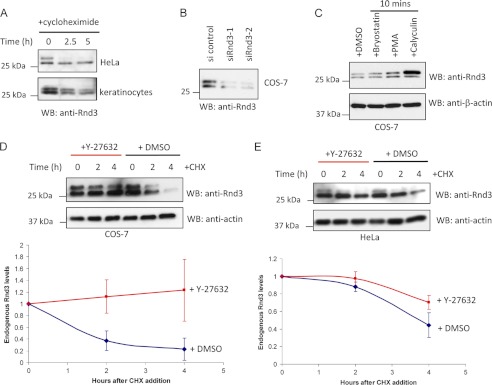
FIGURE 2.
ROCK activity stimulates Rnd3 turnover. A, HeLa cells and human keratinocytes treated for the indicated times with cycloheximide and then lysed and immunoblotted (WB) for endogenous Rnd3. B, Western blot analysis of Rnd3 levels in COS-7 cells transfected with control or two sets of Rnd3 siRNA duplexes for 24 h. C, COS cells treated with bryostatin (100 nm), phorbol 12-myristate 13-acetate (PMA) (100 nm), or calyculin (10 nm) for 10 min, lysed, and immunoblotted for endogenous Rnd3. D, Western blot analysis of endogenous Rnd3 protein in COS cells incubated in the presence or absence of 10 μm Y-27632 for 30 min before being treated with cycloheximide (CHX) for the indicated times. In the graphical representation, the y-axis corresponds to the ratio of Rnd3 relative to time 0. The average values derived from three independent experiments are shown with the level of the two bands normalized to time = 0 (using Image J). E, Western blot analysis of endogenous Rnd3 protein in HeLa cells treated and analyzed as in D. DMSO, dimethyl sulfoxide.
ROCK Stimulates Rnd3 Turnover
Rnd3 binds the N-terminal region of ROCK1, leading to its efficient phosphorylation predominantly at Ser-11 (9) which can stabilize (overexpressed) Rnd3. We therefore tested ROCK for its role in endogenous Rnd3 turnover. COS cells were incubated in the presence or absence of 10 μm Y-27632 to inhibit ROCK activity (added 30 min before addition of 20 μg/ml cycloheximide). In contrast to previous findings wherein long term incubation with Y-27632 decreased Rnd3 levels (9, 35, 37), we found that immediate inhibition of ROCK protected Rnd3 from degradation (Fig. 2D). The prolonged incubation with Y-27632 (i.e. >24 h) is likely to change the transcriptional profile of cells. In HeLa cells which have lower levels of Rnd3 (supplemental Fig. 3B), addition of Y-27632 also reduced the rate of Rnd3 degradation (Fig. 2E). We could not detect a difference in the rate of Rnd3(S11A) or Rnd3(S11D) turnover (supplemental Fig. 4), although other sites on Rnd3 are noted (9). Taken together, it appears that Rnd3 levels are down-regulated by a ROCK-sensitive mechanism in COS-7 cells, whereas in keratinocytes undergoing differentiation there is a ROCK-mediated up-regulation of Rnd3 levels probably through transcriptional mechanisms (35). These point to potentially complex regulation of Rnd3 mediated by phosphorylation at multiple sites (9, 11) as well as effector interaction.
Rnd3 Stabilization by Proteasome Inhibition Is Likely Indirect
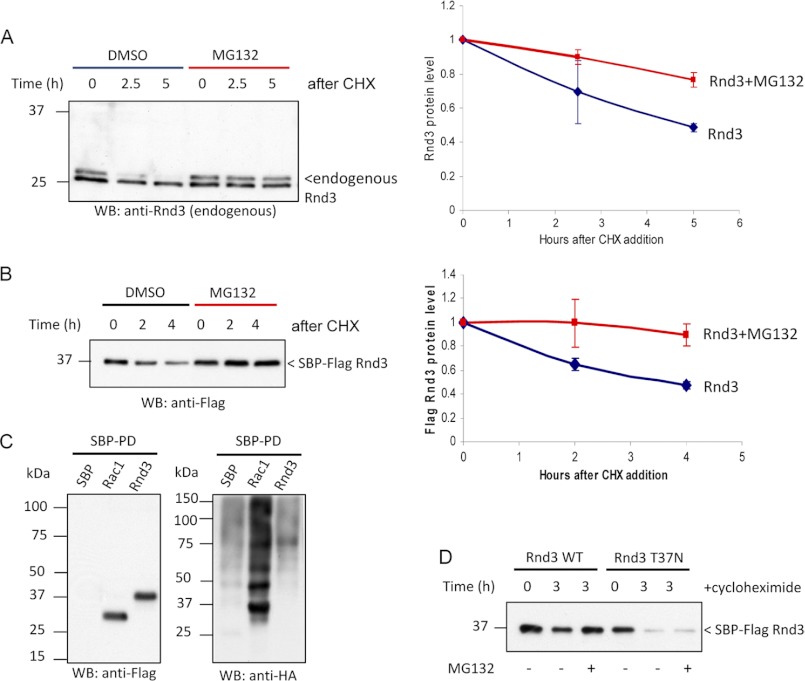
The turnover of endogenous as well as tagged Rnd3 was prevented by the proteasome inhibitor MG132 (Fig. 3, A and B), suggesting that Rnd3 turnover is proteasome-dependent. Most proteasome substrates are targeted for degradation by the addition of polyubiquitin chains. To determine whether Rnd3 is ubiquitinated, we transfected HeLa cells with plasmids encoding streptavidin-binding peptide (SBP)-Rnd3 (and HA-ubiquitin); lysates and purified proteins were probed with anti-HA to detect the ubiquitinated proteins. We were unable to detect any ubiquitinated species of Rnd3 whereas polyubiquitinated Rac1 (29, 38) was easily observed (Fig. 3C). To determine whether prolonged inhibition of proteasome function could stabilize putative ubiquitinated species of Rnd3, overnight treatment was performed, but again no ubiquitinated Rnd3 was detected (data not shown). To examine whether there are other post-translational modifications that underlie Rnd3 degradation, we probed for higher mobility species in the lysates of SBP-Rnd3 that were treated with dimethyl sulfoxide or MG132 (supplemental Fig. 5). These experiments suggest that any modified intermediates are unstable even with MG132 (i.e. not turned over by proteasome) or present in very low levels. Protein turnover via nonubiquitin pathways has been documented (39–42). To confirm that Rnd3 lifetime in cells is indeed dependent on effector binding we tested a nucleotide-binding deficient Rnd3(T37N) mutant, which undergoes normal rates of turnover but was not stabilized by adding MG132 (Fig. 3D). These experiments suggest that (i) the protein responsible for Rnd3 turnover can interact with Rnd3 independent of nucleotide state and (ii) MG132 stabilization requires the active form of Rnd3. This is consistent with Rnd3 effectors controlling endogenous Rnd3 levels. We sought to investigate these events further by making mutations to Rnd3 in regions known to be important for effector binding and protein localization.
FIGURE 3.
Rnd3 can be stabilized by proteasomal inhibition. A, HeLa cells were treated with cycloheximide (CHX) in the presence of 10 μm MG132 or dimethyl sulfoxide (DMSO), then lysed and immunoblotted (WB). In the graphical representation, the y-axis corresponds to the ratio of Rnd3 relative to time 0. The average of three independent experiments is shown. B, 293T cells overexpressing Rnd3 were treated for the indicated times with cycloheximide in the presence of 10 μm MG132 or dimethyl sulfoxide and analyzed as in A. C, HeLa cells were co-transfected with the indicated SBP-tagged plasmids and HA-ubiquitin. Cells were treated with 20 μm MG132 for 30 min prior to lysis. SBP pulldown was performed, and ubiquitinated proteins were visualized by immunoblotting using anti-HA. Conjugation of ubiquitin to Rnd3 was not detected. D, 293T cells overexpressing Rnd3(T37N) were treated for the indicated times with cycloheximide in the presence of 10 μm MG132 to assess whether the effect of proteasome inhibition is direct.
Identification of Rnd3 K45 as Important for Turnover
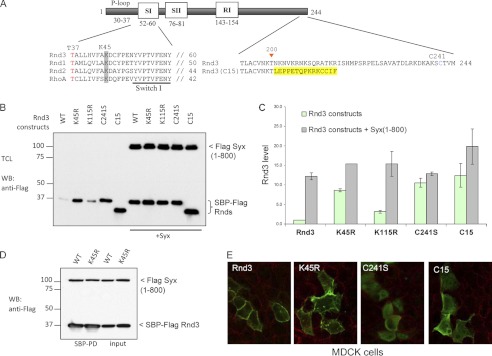
Our observations are consistent with protein(s) involved in this down-regulation of Rnd3 competing with effector binding around the Rnd3 switch 1–2 regions (43). We also considered modification of Rnd3 in this region and noted a conserved lysine (Lys-45 highlighted in Fig. 4A) in the vicinity of Rnd3 switch 1–2 regions which could represent a target for ubiquitin or sumoylation. The Rnd3 point substitutions K45R and K115R (the latter as a control, Fig. 4B) indicated that Rnd3(K45R) has a slower turnover than wild-type Rnd3 or the K115R mutant. Quantification of this effect indicated that at steady state Rnd3(K45R) is expressed ∼8-fold higher than wild-type Rnd3 (Fig. 4C) and has a much slower turnover rate (supplemental Fig. 6). In line with this finding, Syx co-expression has only a minor effect on Rnd3(K45R) levels whereas Rnd3(WT) was routinely increased >10-fold. The Rnd3(K115R) mutant was by contrast expressed at a lower level and significantly stabilized by Syx. Because Rnd3(K45R) binds normally to Syx (Fig. 4D) it seems that protein(s) interacting with residues including Lys-45 might contribute to its turnover. Note that the localization of Rnd3(K45R) was indistinguishable from wild-type Rnd3 (Fig. 4E). Both Rnd3 WT and Rnd3(K45R) lead to stress fiber loss, confirming that they interact normally with p190 RhoGAP (supplemental Fig. 7). However, under the same transfection conditions the K45R mutant is more potent in causing stress fibers disruption than wild-type Rnd3 (supplemental Fig. 7B). The GTP binding-deficient Rnd3(T37N) was used as an inactive control.
FIGURE 4.
Lys-45 and membrane localization are important for Rnd3 turnover. A, schematic shows Rnd3 construct and amino acid substitutions made in this study. The highly conserved Lys-45 is highlighted in gray. The Rnd3(C15) chimeric construct in which the last 44 C-terminal residues of Rnd3 is replaced by the last 15 C-terminal residues of Cdc42 is in yellow. B, 293T cells overexpressing the indicated plasmids were treated for 3 h with cycloheximide, lysed, and immunoblottted. C, graph represents the average of two independent experiments. D, 293T cells were co-transfected with SBP-FLAG Rnd3 or Rnd3(K45R) with Syx(1–800) constructs. The purified protein complex was immunoblotted to visualize bound Syx. E, localizations of the indicated constructs in MDCK cells were immunolabeled with anti-FLAG antibodies and examined by confocal microscopy.
It has recently been shown that the productive interaction of Rnd1 and Rnd3 with p190 RhoGAP is responsible for lipid raft targeting and activation of p190 RhoGAP (22). To address whether the correct subcellular localization in Rnd3 is required in its turnover, we first tested Rnd3(C241S) which is predominantly cytosolic (Fig. 4E), although the protein binds normally to Syx (supplemental Fig. 2). At steady state we observed a ∼10-fold higher level of Rnd3(C241S) compared with wild-type Rnd3, indicating that membrane localization is needed to promote turnover (Fig. 4, B and C). Co-expression of Syx did not significantly affect Rnd3(C241S) levels. Rnd3 has a stretch (∼40 residues) of nonconserved sequence (comparing Rnds) at its C terminus (Fig. 4A). When this region is removed and the Rnd3(1–200) fused to the last 15 residues of Cdc42 we found that the chimeric protein was also stabilized (Fig. 4, B and C). This chimera showed a >10-fold higher protein level compared with wild-type Rnd3 and is primarily localized to internal membranes (Fig. 4E) (44). We conclude that in part the correct localization of Rnd3 via its C terminus is required for its turnover.
Do Other Rnd Interacting Domains Stabilize Rnd3?
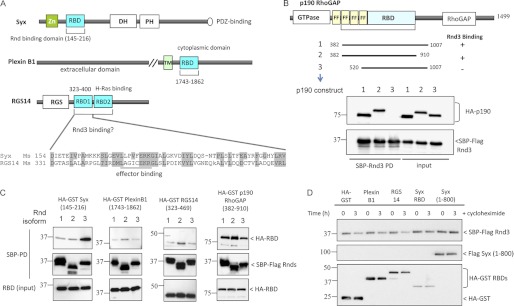
In contrast to conventional GTPases such as RhoA and Rac1, the mammalian Rnd proteins appear to have a more limited set of effectors that includes p190 RhoGAP (20), Plexin B1 (45–47), and Syx (14). The Ras-binding domains (RBD), such as that found in Raf1, is represented in approximately 50 human gene products (48). The Syx RndBD likely adopts the same type of effector complex as seen for the Raf1-Ras complex (49) judging by mutational analysis of the Syx RndBD (14). As illustrated in Fig. 5A, Rnd effector proteins have diverse domain organization. Although the RndBD of Syx can be mapped to a distinct small domain resembling the classic Raf1 RBD (49), the extent of p190 RhoGAP Rnd-binding domain has not been detailed (20) and was originally mapped to a 600-amino acid region (p190B RhoGAP, residues 382–1007) with no known similarity to other proteins. To assess whether the p190B RhoGAP RndBD sequence might contain a cryptic RndBD-like domain we first attempted to map a minimum productive binding region. Various p190B RhoGAP truncations (as illustrated in Fig. 5B) were tested for Rnd3 binding. Surprisingly, even removal of the N-terminal “FF” domains that flank the N-terminal GTPase domain of p190B RhoGAP abolished Rnd3 binding. We successfully deleted approximately 100 residues from the C-terminal side without compromising the interaction; but further deletions were not tolerated (data not shown). Thus, the p190B RhoGAP(382–910) appears to represent the minimal RndBD (Fig. 5B) and suggests a complex allosteric mode of binding. We were not able to detect primary sequence homology to the Syx RndBD within this region. The RndBD of Plexin B1 adopts a ubiquitin-like fold similar to the Syx RndBD but binds to Rnds (and many other Rho proteins) using the opposite face (50), suggesting that it is evolutionarily unrelated. We tested this, as well as the tandem RBDs of RGS14 (48) whose sequence (Fig. 5A) is quite similar to Syx RndBD. The three SBP-tagged Rnd isoforms were expressed in 293T cells, and their binding to putative RndBDs of Syx, p190B RhoGAP, Plexin B1, and RGS14 was measured by streptavidin pulldown. Rnd3 interacted strongly with Syx whereas Rnd1 and Rnd2 bind more weakly, at levels comparable with Plexin B1 and RGS14 sequences (Fig. 5C). All Rnds interacted equally with p190B RhoGAP(382–910), which is consistent with the observation that they activate p190B RhoGAP activity to the same extent (20). This is somewhat curious because Rnd2 is reported not to induce cell rounding (2, 15) and would suggest that p190 RhoGAP activation is not the key activity that underlies this phenotype.
FIGURE 5.
Rnd3 is not efficiently stabilized by all Rnd-interacting domains. A, schematic represents constructs used in 293T cell expression and Rnd interaction studies. Sequence alignment shows the highly conserved Raf1-like RBDs in mouse Syx (AAU04953) and RGS14 (NP_006471). B, p190B RhoGAP amino acid 382–910 is the minimum Rnd3-binding region. 293T cells were co-transfected with SBP-FLAG Rnd3 and HA or HA-GST p190B RhoGAP constructs. The purified protein complex was immunoblotted to visualize bound p190B RhoGAP. C, binding of Rnd isoforms to Plexin B1 was much weaker compared with Syx and p190 RhoGAP. 293T cells were transfected with expression vectors encoding SBP-FLAG Rnd1, Rnd2, or Rnd3 together with the indicated constructs and assessed for interaction. D, 293T cells overexpressing Rnd3 and the indicated plasmids were treated for 3 h with cycloheximide, lysed, and immunoblottted.
To determine whether RndBDs of the various proteins could stabilize Rnd3, cells were co-expressed with Rnd3 and HA-GST tagged Plexin B1(1743–1862), RGS14(323–469), or Syx(145–216). As controls, we also include HA-GST and Syx(1–800). Protein translation was inhibited for 3 h, and the levels of transfected proteins were determined by Western blotting (Fig. 5D). In keeping with their weak interaction, neither Plexin B1 nor RGS14 stabilizes Rnd3.
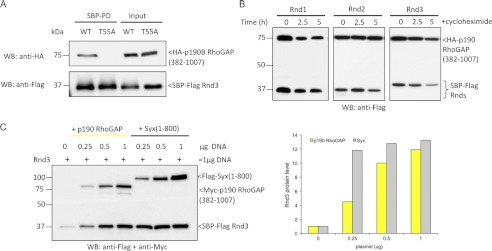
p190 RhoGAP Interaction with Rnd3 Is Effector-like and Stabilizes Rnd3
Syx and p190 RndBDs have equivalent “effector-like” behavior as seen by their inability to bind Rnd3(T55A), which cannot adopt a GTP-bound state (Fig. 6A). Thus, both Syx and p190 RhoGAP likely interact with Rnd3 via contacts typical of the switch I and II binders (51). We then examined whether co-expression of p190B RhoGAP(382–1007) affected the half-life of Rnd proteins. As shown in Fig. 6B, turnover of Rnds was markedly decreased in the presence of p190 RhoGAP. The effect of increasing levels of Syx(1–800) or p190B RhoGAP(382–1007) transfection (Fig. 6C) showed that Syx was somewhat more potent in stabilizing Rnd3. These experiments are consistent with a new form of regulation in which Rho effector proteins feed back to stabilize their partners and suggest that factors that disrupt this interaction would lead to Rnd turnover. Given the large number of RBD'-containing proteins (48), which are candidate Rnd3 binders in addition to p190 RhoGAP it is no surprise that the knockdown of Syx by itself has no obvious effect on Rnd3 levels (data not shown). We anticipate that in vivo Rnd3 protein levels are likely responsive to the availability of various multiple effector proteins (including Syx and p190 RhoGAP).
FIGURE 6.
p190 RhoGAP is a genuine Rnd3 effector that stabilizes Rnd3. A, 293T cells were transfected as indicated, and SBP pulldown was performed. Associated proteins were visualized by immunoblotting. No interaction between Rnd3 effector mutant (T55A) and p190 RhoGAP was observed. B, 293T cells were co-transfected with Rnd1, Rnd2, or Rnd3 together with p190 RhoGAP(382–1007) and treated for the indicated times with cycloheximide, lysed, and immunoblottted (WB). C, the stabilizing effect of Rnd3 by Syx and p190 RhoGAP is dose-dependent. 293T cells were transfected with 0.5 μg of Rnd3 and increasing amounts of Syx or p190 RhoGAP plasmids as indicated. The levels of Rnd3 were visualized by immunoblotting with anti-FLAG antibody. Graphical representation of the experiment is shown.
DISCUSSION
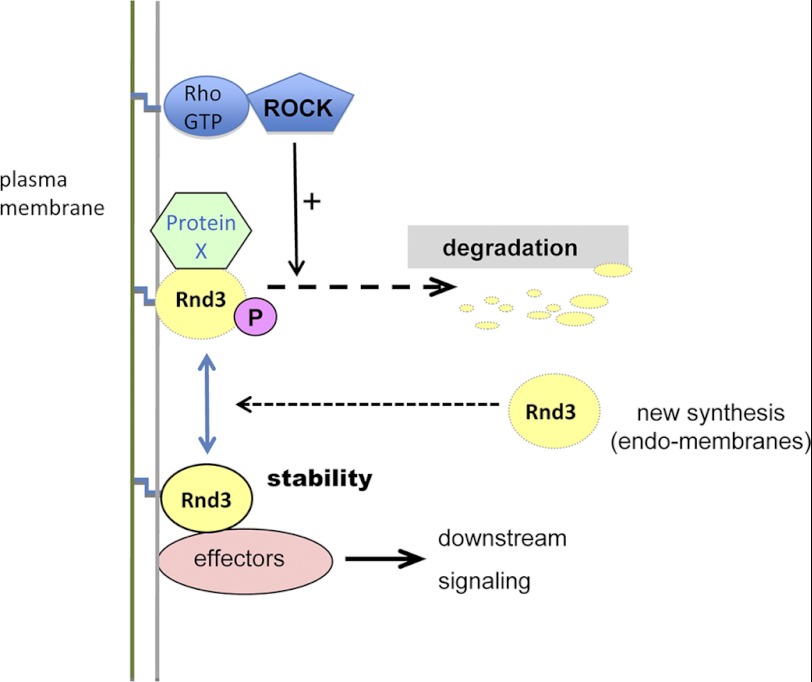
We have shown previously that Rnd3 binds to Syx via a Raf1-like RBD and refer to this related sequence as the RndBD. In this study, we show that this interaction is dependent on proper GTP loading of Rnd3. The observation that Syx protects Rnd3 from degradation has led us to a model (Fig. 7) in which effector binding impedes accessibility of protein(s) to access a region of Rnd3 that likely includes the conserved Lys-45. Such a mode of regulation has not been reported for other Rho GTPases.
FIGURE 7.
Model showing Rnd3 regulation by the level of effectors and ROCK activity. Rnd3 is targeted for degradation by binding of unknown protein (X) which is sensitive to both the Rnd3(K45R) mutation and the membrane localization of Rnd3. This turnover of Rnd3 is sensitive to Y-27632, indicating that ROCK stimulates such turnover. Effector interaction can prevent the turnover of Rnd3 probably by competing for binding of protein X.
We demonstrated that inhibition of ROCK kinase activity protects Rnd3 from turnover in COS-7 and HeLa cells. In contrast to previous studies (9, 35, 37) whereby ROCK stabilizes Rnd3, we demonstrate ROCK-dependent down-regulation of Rnd3. In this context, ROCK-mediated phosphorylation leading to Rnd3 turnover would serve as a feedback mechanism to modulate Rnd3 down-regulation of RhoA activity.
Rnd proteins contain both N- and C-terminal extensions; the C-terminal CAAM (methionine in last position) is farnesylated. Rnd3 exists largely in a GTP-bound state (1) and undergoes post-translational modifications such as phosphorylation (9, 10), which might affect its localization. We show here that the levels of Rnd3 are strongly influenced by co-expression of either Syx or p190B RhoGAP. The behaviors of nonfunctional Rnd3 mutant (T37A) and a Rnd3 binding defective Syx (Fig. 1D) are consistent with this notion. One can also observe that the expression level of a nonprenylated Rnd3 is severalfold higher than wild type, indicating that correct membrane targeting is required for degradation. Interestingly, when we target the protein using the Cdc42 C-terminal 15 residues, the localization is switched to intracellular membranes and protects the protein from turnover. Our data suggest that Rnd3 is not modified by ubiquitin but that the effects of MG132 might be mediated indirectly through the stabilization of its effectors. Thus, after cycloheximide treatment the turnover of Rnd effectors leads indirectly to their degradation. Alternatively, a specific endogenous component may be limiting in the ubiquitylation assay. Interestingly, Rnd3(K45R) is more stable and shows higher activity in disrupting stress fibers in cells (supplemental Fig. 7). This might provide a useful mutant for further studies because it is a more stable form of Rnd3. We hypothesize that effectors such as Syx compete out a protein that interacts with Rnd3 (in a nucleotide-independent manner) that is responsible for Rnd degradation. This is analogous to the ability of effectors such as ACK1 and PAK1 to prolong the lifetime of the active GTP-bound state of Cdc42 and Rac1 (52, 53). Thus, productive signaling by small G proteins results in increased or prolonged signaling.
In the current study, we have validated the interaction between Rnd3 and p190RhoGAP (Fig. 5), an abundant modulator of RhoA (20), and demonstrated that Syx and p190 RhoGAP bind with similar avidity to Rnd3 (Fig. 6A). The comparatively weak binding of Rnds to Plexin B1, which also binds diverse GTPases including Rac1 and RhoD (54), is insufficient to stabilize Rnd3 (Fig. 5D).The RndBD of p190B RhoGAP is a surprisingly large ∼500 residues (382–910), indicating perhaps that Rnd binding spans multiple domains (20). The contribution of the FF domains, also present within eukaryotic transcription factors such as CA150 (55), is curious. The first p190 RhoGAP FF(1) domain does not fold with the typical α-helical arrangement of other FF domains (56), in keeping with the low sequence homology in this domain. It has been suggested that the serum-responsive transcriptional regulator TFII-I interacts with the p190 RhoGAP tandem FF domains (57). Competition for binding could underlie an ability of Rnd proteins to affect growth factor-mediated entry into the cell cycle (58, 59), which is independent of RhoA.
In summary, we have shown that Syx and p190 RhoGAP bind with similar avidity to Rnd3 although their RndBDs are structurally unrelated. Both of these proteins can prevent Rnd1 and Rnd3 degradation (and to a lesser extend Rnd2), suggesting that a feedback loop exists to balance the activities of Rnd proteins and their targets. It is unlikely that individual Rnd3 effector serves as a critical regulator; we anticipate that the rate of Rnd turnover integrates across the whole spectrum of Rnd3 effectors in a given cell. In vivo this might be an important mechanism to down-regulate the level of Rnd proteins under conditions in which their effectors change concentration.
Supplementary Material

This article contains supplemental Figs. 1–7.
- ROCK
- RhoA-associated kinase
- MDCK
- Madin-Darby canine kidney
- RBD
- Ras-binding domain
- RndBD
- Rnd-binding domain
- SBP
- streptavidin-binding peptide.
REFERENCES
- 1. Foster R., Hu K. Q., Lu Y., Nolan K. M., Thissen J., Settleman J. (1996) Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol. Cell. Biol. 16, 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guasch R. M., Scambler P., Jones G. E., Ridley A. J. (1998) RhoE regulates actin cytoskeleton organization and cell migration. Mol. Cell. Biol. 18, 4761–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chardin P. (2006) Function and regulation of Rnd proteins. Nat. Rev. Mol. Cell Biol. 7, 54–62 [DOI] [PubMed] [Google Scholar]
- 4. Riou P., Villalonga P., Ridley A. J. (2010) Rnd proteins: multifunctional regulators of the cytoskeleton and cell cycle progression. Bioessays 32, 986–992 [DOI] [PubMed] [Google Scholar]
- 5. Hansen S. H., Zegers M. M., Woodrow M., Rodriguez-Viciana P., Chardin P., Mostov K. E., McMahon M. (2000) Induced expression of Rnd3 is associated with transformation of polarized epithelial cells by the Raf-MEK-extracellular signal-regulated kinase pathway. Mol. Cell. Biol. 20, 9364–9375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ongusaha P. P., Kim H. G., Boswell S. A., Ridley A. J., Der C. J., Dotto G. P., Kim Y. B., Aaronson S. A., Lee S. W. (2006) RhoE is a pro-survival p53 target gene that inhibits ROCK I-mediated apoptosis in response to genotoxic stress. Curr. Biol. 16, 2466–2472 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7. Klein R. M., Spofford L. S., Abel E. V., Ortiz A., Aplin A. E. (2008) B-RAF regulation of Rnd3 participates in actin cytoskeletal and focal adhesion organization. Mol. Biol. Cell 19, 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tyburczy M. E., Kotulska K., Pokarowski P., Mieczkowski J., Kucharska J., Grajkowska W., Roszkowski M., Jozwiak S., Kaminska B. (2010) Novel proteins regulated by mTOR in subependymal giant cell astrocytomas of patients with tuberous sclerosis complex and new therapeutic implications. Am. J. Pathol. 176, 1878–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Riento K., Totty N., Villalonga P., Garg R., Guasch R., Ridley A. J. (2005) RhoE function is regulated by ROCK I-mediated phosphorylation. EMBO J. 24, 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Madigan J. P., Bodemann B. O., Brady D. C., Dewar B. J., Keller P. J., Leitges M., Philips M. R., Ridley A. J., Der C. J., Cox A. D. (2009) Regulation of Rnd3 localization and function by protein kinase Cα-mediated phosphorylation. Biochem. J. 424, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Komander D., Garg R., Wan P. T., Ridley A. J., Barford D. (2008) Mechanism of multi-site phosphorylation from a ROCK-I:RhoE complex structure. EMBO J. 27, 3175–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pinner S., Sahai E. (2008) PDK1 regulates cancer cell motility by antagonising inhibition of ROCK1 by RhoE. Nat. Cell Biol. 10, 127–137 [DOI] [PubMed] [Google Scholar]
- 13. Mocholí E., Ballester-Lurbe B., Arqué G., Poch E., Peris B., Guerri C., Dierssen M., Guasch R. M., Terrado J., Pérez-Roger I. (2011) RhoE deficiency produces postnatal lethality, profound motor deficits and neurodevelopmental delay in mice. PLoS One 6, e19236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goh L. L., Manser E. (2010) The RhoA GEF Syx is a target of Rnd3 and regulated via a Raf1-like ubiquitin-related domain. PLoS One 5, e12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nobes C. D., Lauritzen I., Mattei M. G., Paris S., Hall A., Chardin P. (1998) A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J. Cell Biol. 141, 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Villalonga P., Fernández de Mattos S., Ridley A. J. (2009) RhoE inhibits 4E-BP1 phosphorylation and eIF4E function impairing cap-dependent translation. J. Biol. Chem. 284, 35287–35296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xia W., Li J., Chen L., Huang B., Li S., Yang G., Ding H., Wang F., Liu N., Zhao Q., Fang T., Song T., Wang T., Shao N. (2010) MicroRNA-200b regulates cyclin D1 expression and promotes S-phase entry by targeting RND3 in HeLa cells. Mol. Cell. Biochem. 344, 261–266 [DOI] [PubMed] [Google Scholar]
- 18. Goda T., Takagi C., Ueno N. (2009) Xenopus Rnd1 and Rnd3 GTP-binding proteins are expressed under the control of segmentation clock and required for somite formation. Dev. Dyn. 238, 2867–2876 [DOI] [PubMed] [Google Scholar]
- 19. Wünnenberg-Stapleton K., Blitz I. L., Hashimoto C., Cho K. W. (1999) Involvement of the small GTPases XRhoA and XRnd1 in cell adhesion and head formation in early Xenopus development. Development 126, 5339–5351 [DOI] [PubMed] [Google Scholar]
- 20. Wennerberg K., Forget M. A., Ellerbroek S. M., Arthur W. T., Burridge K., Settleman J., Der C. J., Hansen S. H. (2003) Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr. Biol. 13, 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fortier M., Comunale F., Kucharczak J., Blangy A., Charrasse S., Gauthier-Rouvière C. (2008) RhoE controls myoblast alignment prior fusion through RhoA and ROCK. Cell Death Differ. 15, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 22. Oinuma I., Kawada K., Tsukagoshi K., Negishi M. (2012) Rnd1 and Rnd3 targeting to lipid raft is required for p190 RhoGAP activation. Mol. Biol. Cell 23, 1593–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katoh H., Harada A., Mori K., Negishi M. (2002) Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol. Cell. Biol. 22, 2952–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Habas R., Kato Y., He X. (2001) Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell 107, 843–854 [DOI] [PubMed] [Google Scholar]
- 25. Tanegashima K., Zhao H., Dawid I. B. (2008) WGEF activates Rho in the Wnt-PCP pathway and controls convergent extension in Xenopus gastrulation. EMBO J. 27, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Toledo M., Coulon V., Schmidt S., Fort P., Blangy A. (2001) The gene for a new brain-specific RhoA exchange factor maps to the highly unstable chromosomal region 1p36.2–1p36.3. Oncogene 20, 7307–7317 [DOI] [PubMed] [Google Scholar]
- 27. Marx R., Henderson J., Wang J., Baraban J. M. (2005) Tech: a RhoA GEF selectively expressed in hippocampal and cortical neurons. J. Neurochem. 92, 850–858 [DOI] [PubMed] [Google Scholar]
- 28. Liu M., Horowitz A. (2006) A PDZ-binding motif as a critical determinant of Rho guanine exchange factor function and cell phenotype. Mol. Biol. Cell 17, 1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lerm M., Pop M., Fritz G., Aktories K., Schmidt G. (2002) Proteasomal degradation of cytotoxic necrotizing factor 1-activated rac. Infect. Immun. 70, 4053–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doye A., Mettouchi A., Bossis G., Clément R., Buisson-Touati C., Flatau G., Gagnoux L., Piechaczyk M., Boquet P., Lemichez E. (2002) CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell 111, 553–564 [DOI] [PubMed] [Google Scholar]
- 31. Boyer L., Turchi L., Desnues B., Doye A., Ponzio G., Mege J. L., Yamashita M., Zhang Y. E., Bertoglio J., Flatau G., Boquet P., Lemichez E. (2006) CNF1-induced ubiquitylation and proteasome destruction of activated RhoA is impaired in Smurf1−/− cells. Mol. Biol. Cell 17, 2489–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen Y., Yang Z., Meng M., Zhao Y., Dong N., Yan H., Liu L., Ding M., Peng H. B., Shao F. (2009) Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol. Cell 35, 841–855 [DOI] [PubMed] [Google Scholar]
- 33. Kärkkäinen S., van der Linden M., Renkema G. H. (2010) POSH2 is a RING finger E3 ligase with Rac1 binding activity through a partial CRIB domain. FEBS Lett. 584, 3867–3872 [DOI] [PubMed] [Google Scholar]
- 34. Torrino S., Visvikis O., Doye A., Boyer L., Stefani C., Munro P., Bertoglio J., Gacon G., Mettouchi A., Lemichez E. (2011) The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev. Cell 21, 959–965 [DOI] [PubMed] [Google Scholar]
- 35. Liebig T., Erasmus J., Kalaji R., Davies D., Loirand G., Ridley A., Braga V. M. (2009) RhoE Is required for keratinocyte differentiation and stratification. Mol. Biol. Cell 20, 452–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Watt F. M. (2002) Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J. 21, 3919–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ryan K. R., Lock F. E., Heath J. K., Hotchin N. A. (March 2012) Plakoglobin-dependent regulation of keratinocyte apoptosis by Rnd3. J. Cell Sci. 10.1242/jcs.101931 [DOI] [PubMed] [Google Scholar]
- 38. Visvikis O., Lorès P., Boyer L., Chardin P., Lemichez E., Gacon G. (2008) Activated Rac1, but not the tumorigenic variant Rac1b, is ubiquitinated on Lys-147 through a JNK-regulated process. FEBS J. 275, 386–396 [DOI] [PubMed] [Google Scholar]
- 39. Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. (1992) Ornithine decarboxylase is degraded by the 26 S proteasome without ubiquitination. Nature 360, 597–599 [DOI] [PubMed] [Google Scholar]
- 40. Glass J. R., Gerner E. W. (1987) Spermidine mediates degradation of ornithine decarboxylase by a non-lysosomal, ubiquitin-independent mechanism. J. Cell. Physiol. 130, 133–141 [DOI] [PubMed] [Google Scholar]
- 41. Asher G., Lotem J., Sachs L., Kahana C., Shaul Y. (2002) Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc. Natl. Acad. Sci. U.S.A. 99, 13125–13130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krappmann D., Wulczyn F. G., Scheidereit C. (1996) Different mechanisms control signal-induced degradation and basal turnover of the NF-κB inhibitor IκBα in vivo. EMBO J. 15, 6716–6726 [PMC free article] [PubMed] [Google Scholar]
- 43. Fiegen D., Blumenstein L., Stege P., Vetter I. R., Ahmadian M. R. (2002) Crystal structure of Rnd3/RhoE: functional implications. FEBS Lett. 525, 100–104 [DOI] [PubMed] [Google Scholar]
- 44. Xie C., Li N., Chen Z. J., Li B. L., Song B. L. (2011) The small GTPase Cdc42 interacts with Niemann-Pick C1-like 1 (NPC1L1) and controls its movement from endocytic recycling compartment to plasma membrane in a cholesterol-dependent manner. J. Biol. Chem. 286, 35933–35942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oinuma I., Katoh H., Harada A., Negishi M. (2003) Direct interaction of Rnd1 with Plexin B1 regulates PDZ-RhoGEF-mediated Rho activation by Plexin B1 and induces cell contraction in COS-7 cells. J. Biol. Chem. 278, 25671–25677 [DOI] [PubMed] [Google Scholar]
- 46. Oinuma I., Katoh H., Negishi M. (2004) Molecular dissection of the semaphorin 4D receptor Plexin B1-stimulated R-Ras GTPase-activating protein activity and neurite remodeling in hippocampal neurons. J. Neurosci. 24, 11473–11480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oinuma I., Ishikawa Y., Katoh H., Negishi M. (2004) The Semaphorin 4D receptor Plexin B1 is a GTPase-activating protein for R-Ras. Science 305, 862–865 [DOI] [PubMed] [Google Scholar]
- 48. Kiel C., Foglierini M., Kuemmerer N., Beltrao P., Serrano L. (2007) A genome-wide Ras-effector interaction network. J. Mol. Biol. 370, 1020–1032 [DOI] [PubMed] [Google Scholar]
- 49. Nassar N., Horn G., Herrmann C., Scherer A., McCormick F., Wittinghofer A. (1995) The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature 375, 554–560 [DOI] [PubMed] [Google Scholar]
- 50. Tong Y., Hota P. K., Penachioni J. Y., Hamaneh M. B., Kim S., Alviani R. S., Shen L., He H., Tempel W., Tamagnone L., Park H. W., Buck M. (2009) Structure and function of the intracellular region of the Plexin B1 transmembrane receptor. J. Biol. Chem. 284, 35962–35972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vetter I. R., Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 52. Manser E., Leung T., Salihuddin H., Tan L., Lim L. (1993) A non-receptor tyrosine kinase that inhibits the GTPase activity of p21cdc42. Nature 363, 364–367 [DOI] [PubMed] [Google Scholar]
- 53. Zhang B., Wang Z. X., Zheng Y. (1997) Characterization of the interactions between the small GTPase Cdc42 and its GTPase-activating proteins and putative effectors: comparison of kinetic properties of Cdc42 binding to the Cdc42-interactive domains. J. Biol. Chem. 272, 21999–22007 [DOI] [PubMed] [Google Scholar]
- 54. Tong Y., Chugha P., Hota P. K., Alviani R. S., Li M., Tempel W., Shen L., Park H. W., Buck M. (2007) Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the Plexin B1 effector domain. J. Biol. Chem. 282, 37215–37224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carty S. M., Goldstrohm A. C., Suñé C., Garcia-Blanco M. A., Greenleaf A. L. (2000) Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 97, 9015–9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bonet R., Ruiz L., Aragón E., Martín-Malpartida P., Macias M. J. (2009) NMR structural studies on human p190-A RhoGAPFF1 revealed that domain phosphorylation by the PDGF-receptor α requires its previous unfolding. J. Mol. Biol. 389, 230–237 [DOI] [PubMed] [Google Scholar]
- 57. Jiang W., Sordella R., Chen G. C., Hakre S., Roy A. L., Settleman J. (2005) An FF domain-dependent protein interaction mediates a signaling pathway for growth factor-induced gene expression. Mol. Cell 17, 23–35 [DOI] [PubMed] [Google Scholar]
- 58. Villalonga P., Guasch R. M., Riento K., Ridley A. J. (2004) RhoE inhibits cell cycle progression and Ras-induced transformation. Mol. Cell. Biol. 24, 7829–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Poch E., Miñambres R., Mocholí E., Ivorra C., Pérez-Aragó A., Guerri C., Pérez-Roger I., Guasch R. M. (2007) RhoE interferes with Rb inactivation and regulates the proliferation and survival of the U87 human glioblastoma cell line. Exp. Cell Res. 313, 719–731 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.