Abstract
The stem cell leukemia (SCL) gene encodes a tissue-specific basic helix–loop–helix (bHLH) protein with a pivotal role in hemopoiesis and vasculogenesis. Several enhancers have been identified within the murine SCL locus that direct reporter gene expression to subdomains of the normal SCL expression pattern, and long-range sequence comparisons of the human and murine SCL loci have identified additional candidate enhancers. To facilitate the characterization of regulatory elements, we have sequenced and analyzed 33 kb of the SCL genomic locus from the pufferfish Fugu rubripes, a species with a highly compact genome. Although the pattern of SCL expression is highly conserved from mammals to teleost fish, the genes flanking pufferfish SCL were unrelated to those known to flank both avian and mammalian SCL genes. These data suggest that SCL regulatory elements are confined to the region between the upstream and downstream flanking genes, a region of 65 kb in human and 8.5 kb in pufferfish. Consistent with this hypothesis, the entire 33-kb pufferfish SCL locus directed appropriate expression to hemopoietic and neural tissue in transgenic zebrafish embryos, as did a 10.4-kb fragment containing the SCL gene and extending to the 5′ and 3′ flanking genes. These results demonstrate the power of combining the compact genome of the pufferfish with the advantages that zebrafish provide for studies of gene regulation during development. Furthermore, the pufferfish SCL locus provides a powerful tool for the manipulation of hemopoiesis and vasculogenesis in vivo.
The stem cell leukemia (SCL) gene (also known as Tal-1) encodes a basic helix–loop–helix (bHLH) transcription factor that is a critical regulator of both hemopoiesis and vasculogenesis (1). SCL is normally expressed in hemopoietic stem cells, in committed cells of the erythroid, myeloid, and megakaryocytic lineages, in endothelium, and within specific regions of the central nervous system, a pattern of expression that is highly conserved across vertebrate species from mammals to teleost fish (2–6).
SCL is essential for the development of all hemopoietic lineages (7, 8). A role in early progenitors is also suggested by the observation that expression of anti-sense SCL suppressed the proliferation, cell cycle progression, and self renewal of a multipotent hemopoietic cell line (9). Distinct functions following lineage commitment are implied by the modulation of SCL mRNA and protein levels during erythroid and myeloid differentiation (10–14). Moreover, enforced SCL expression enhanced erythroid differentiation of hemopoietic cell lines (14, 15) and increased erythroid and megakaryocytic differentiation of normal CD34-positive progenitors (16, 17).
SCL also plays a crucial role in the formation of hemangioblasts, bipotent progenitors of both blood and endothelium. Ectopic expression of SCL mRNA in zebrafish embryos specifies hemangioblast formation from early mesoderm and results in excessive production of blood and endothelial progenitors (3). In keeping with this, SCL can partially rescue both the hemopoietic and vascular defects of the cloche zebrafish mutant (2), and SCL−/− murine embryonic stem cells fail to give rise to hemangioblasts during in vitro differentiation (18). A function for SCL later in endothelial development is also suggested by the observation that SCL−/− embryos exhibit defective yolk sac angiogenesis (19).
SCL expression is tightly regulated. Both human and mouse SCL are transcribed from two lineage-specific promoters (20–24). In addition, a panel of DNase I hypersensitive sites associated with enhancer or silencer activity have been identified within the murine SCL locus (25, 26). Studies using transgenic mice have subsequently defined a 30-kb region containing five distinct murine enhancers that are capable of directing reporter gene expression to hemopoietic progenitor cells, endothelium, and specific regions of the brain and spinal cord (6, 27, 28). However, long-range sequence comparisons of ≈200 kb from each of the human and murine SCL loci identified several peaks of homology that represent additional candidate regulatory elements (28).
Identification of the full complement of regulatory elements for a given gene amid the vast tracts of noncoding mammalian DNA presents a formidable challenge. One potentially powerful approach takes advantage of the observation that the pufferfish genome is ≈8-fold smaller than that of humans, yet contains a similar number of genes (29). Analysis of pufferfish DNA therefore provides an attractive strategy for identification of SCL regulatory elements, especially because the process of hemopoiesis (30, 31) and the pattern of SCL expression (2, 3, 6, 32) are both highly conserved from mammals to teleost fish. Moreover, enhancers conserved from fish to mammals have been identified in several other genes (33–40).
However, pufferfish are relatively large and difficult to maintain and so do not provide a tractable experimental system for in vivo studies. By contrast, the zebrafish is a powerful model organism for studies of vertebrate development (41, 42), including hemopoiesis (32, 43), and transgenic zebrafish carrying reporter constructs provide a potent approach to the characterization of regulatory elements in vivo. This strategy has been used to analyze enhancers and silencers from a number of zebrafish genes [sonic hedgehog (44), GATA-1 (45), GATA-2 (46), and six7 (47)] and from the mammalian GAP-34 (48) gene.
In this paper, we have sequenced and analyzed 33 kb of the SCL genomic locus from the pufferfish Fugu rubripes. Our data suggest that SCL regulatory elements responsible for its highly conserved pattern of expression are confined to the region between the flanking genes in different vertebrate species. Consistent with this concept, a 10.4-kb region encompassing the pufferfish SCL gene was expressed in the intermediate cell mass (ICM), head mesenchyme (HMC), and spinal cord in transgenic zebrafish, a pattern reflecting that of endogenous SCL (2, 3). These results demonstrate that combining the compact genome of pufferfish with the powerful developmental biology of zebrafish provides an effective strategy for dissecting gene regulation. Moreover, the pufferfish SCL locus represents a potent tool for the manipulation of hemopoiesis and vasculogenesis in vivo.
Materials and Methods
Southern Analysis.
Pufferfish DNA was isolated as described (49) and Southern analysis carried out by using standard procedures (50). The blot was probed with a bHLH probe from the pufferfish gene SLP-1 (49), generated by PCR by using the primers A and B, as described (3). The zebrafish SCL-specific C-terminal probe has been described (3).
Library Screening and DNA Sequence Analysis.
A Fugu cosmid library (CLONTECH QL1002 m) was screened by using a mouse SCL bHLH probe with standard protocols (50). Under high-stringency hybridization conditions (0.2 × SSC; 0.1% SDS; 65°C), 7 of 24 cosmids displayed the same restriction patterns as the putative SCL gene. One cosmid, in which the SCL gene was shown by Southern analysis to be centrally situated (Fr SCL1.311), was further characterized. Initially, cycle sequencing (AmpliCycle kit, Perkin–Elmer) and then direct sequencing of part of the cosmid insert revealed regions of homology to zebrafish SCL, both 5′ and 3′ to the bHLH. Primers are available on request. The entire insert of this cosmid was shotgun cloned into M13, fully sequenced, and analyzed as described (49).
Reverse Transcription–PCR (RT-PCR).
Total pufferfish RNA from heart, brain, and muscle were prepared as described (49). The remaining samples were kindly supplied by Gregg Elgar (HGMP Resource Centre, Hinxton, UK). RT-PCR was performed by using the Access RT-PCR kit (Promega). Specificity of RT-PCR products was confirmed by Southern hybridization and sequencing. Expression analysis of the pufferfish gene was performed by using the Calypso RT-PCR kit (DNAmp, Cambio, Cambridge, UK) and primers designed from sequence in exon 3 and 4 of pufferfish SCL and in exons 1 and 6 of the pufferfish p55 gene (GenBank accession no. X81359). Zebrafish RNA was prepared as above. RT-PCR was performed with primers in the first and last coding exons of zebrafish SCL cDNA and within zebrafish elongation factor 1α. All primer sequences are available on request.
DNA Microinjection and Analysis of Zebrafish Embryos.
FrSCL10.4 was generated by cloning a 10.4-kb XbaI/SalI fragment from cosmid FrSCL1.311 into pGEM11. DNA was prepared for microinjection by using a Qiagen maxiprep kit (Qiagen, Chatsworth, CA). An RNA probe for in situ hybridization was generated from a cloned 450-bp PCR product from the 3′ untranslated region of pufferfish SCL. This plasmid was linearized by digestion with HindIII and runoff transcripts generated by using T7 RNA polymerase (Promega) and digoxigenin-11-UTP (Boehringer Mannheim).
One-cell stage zebrafish embryos were injected with 105-106 molecules of supercoiled DNA per injection. Rhodamine dextran or green fluorescent protein mRNA was used as injection control. At least five independent experiments were performed for each construct and the results pooled. Embryos were analyzed at 22-h postfertilization (hpf) by RNA in situ hybridization and microtome sections, performed as described (51).
Results
Isolation of the Pufferfish SCL Gene.
We have previously characterized SLP-1, a pufferfish gene highly homologous to SCL within the bHLH domain (49) but which exhibits little sequence homology with the SCL genes of other vertebrate species outside of this region (see Fig. 2). No known mammalian orthologue of SLP-1 has been identified, and it is likely to represent an SCL paralogue. A pufferfish genomic Southern blot was hybridized with a SLP-1 bHLH probe at reduced stringency to reveal multiple bands in several digests. In each digest, one of these bands also hybridized to a zebrafish SCL cDNA fragment derived from a region downstream of the bHLH domain (Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org). These data demonstrate the presence of a single locus closely related to SCL in the pufferfish genome.
Figure 2.
Sequence comparisons of pufferfish SCL and SLP-1 with other vertebrate SCL proteins. The bHLH probe (black bar) and zebrafish SCL 3′ probe (gray bar) were used for library screening and Southern blotting.
Subsequent screening of a pufferfish (F. rubripes) genomic cosmid library led to the isolation of seven cosmids containing the putative pufferfish SCL gene. Cosmid FrSCL1.311, which had an insert size of ≈33 kb and in which the SCL gene was centrally situated (Fig. 1), was chosen for further analysis. The pufferfish SCL cosmid was completely sequenced (GenBank accession no. AJ131019) and annotated as described (49). Five new genes flanking the SCL gene were identified by using computer generated exon predictions, blast database searches and, where required, RT-PCR to confirm exon/intron structure. Details of these genes are available from the fully annotated sequence database entry and are published as supplemental data on the PNAS web site, www.pnas.org.
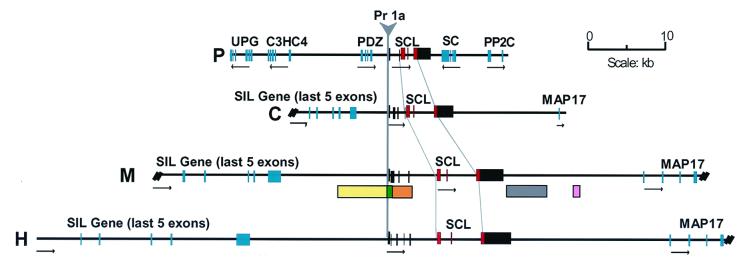
Figure 1.
Identification of the pufferfish SCL gene. Comparison of the genomic SCL loci of pufferfish (P), chicken (C), mouse (M), and human (H). Horizontal arrows under each gene indicate the direction of transcription. SCL coding and noncoding exons are shown as red and black boxes, respectively. The position of five enhancer elements that direct expression to endothelium (yellow box), hemopoietic progenitors (gray box), and neural tissue (green, orange and pink boxes) are indicated. SIL and MAP17 flank the SCL gene in human, mouse and chicken but are not found at the pufferfish SCL locus. UPG, unknown pufferfish gene; C3HC4, RING finger-like gene; PDZ, PDZ domain encoding gene; SC, saccular collagen gene; PP2C, protein phosphatase 2C gene; Pr1a, SCL promoter 1a.
Structure of the Pufferfish SCL Gene.
Comparison of the pufferfish SCL genomic sequence with genomic sequence for human, mouse, and chicken SCL (28) identified three coding exons that corresponded to exons 4, 5, and 6 of human SCL (Fig. 1) as well as a region of pufferfish sequence (18,531–18,697 bp) homologous to human, murine, and chicken SCL promoter 1a. To confirm the intron/exon structure of the pufferfish SCL gene, RT-PCR was performed by using a primer positioned immediately 3′ to the predicted transcription start site with a reverse primer in the predicted pufferfish exon homologous to exon 4 of human SCL. By using total RNA from heart and brain, an RT-PCR product was obtained which hybridized to an internal SCL probe. Direct sequencing of this product revealed the presence of a single upstream exon, with similarity to the first exon of human, mouse, and chicken SCL (data not shown), directly fused to the exon homologous to human exon 4. This suggests that, unlike the human, mouse, and chicken SCL genes, the pufferfish SCL gene lacks additional 5′ noncoding exons. RT-PCR with direct sequencing was also used to confirm the predicted coding exon boundaries. The positions of both the 2/3 and 3/4 exon junctions (homologous to human exon 4/5 and 5/6 junctions) were precisely conserved with those of higher vertebrates.
The size of the pufferfish SCL gene, from promoter to distal polyadenylation signal is 4.9 kb, ≈3.5-fold smaller than the human gene (16 kb). As with other pufferfish genes, this compression reflects a reduction in intron sizes. The predicted pufferfish SCL protein is larger (374 amino acids) than that of human, mouse, chicken, and zebrafish (331, 328, 311, and 324 amino acids, respectively). As shown in Fig. 2, the bHLH region (residues 208–259 of pufferfish SCL) exhibits 98% amino acid conservation between these species, and a number of additional domains of high homology can be identified in the N- and C-terminal regions.
Expression Pattern of Pufferfish SCL.
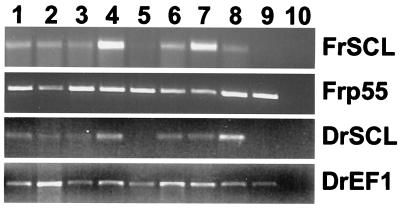
To determine the expression pattern of the pufferfish and zebrafish SCL genes, RT-PCR was performed by using total RNA from a number of tissues. Primers within the pufferfish homologue of the ubiquitously expressed gene p55 (52) or the zebrafish elongation factor α gene were used as loading controls for these experiments. In both pufferfish and zebrafish, SCL expression was observed in spleen, liver, kidney, brain, gill, and gonads but was not evident in gut and muscle (Fig. 3). These data are consistent with the highly conserved pattern of SCL expression previously reported in zebrafish (2, 3, 6) and in higher vertebrates (5, 6, 53). Although kidney and spleen are the main hemopoietic organs in fish, hemopoiesis has also been reported in both liver and heart (30), and the gills are rich in endothelial cells. Moreover, SCL is also expressed in murine testis and ovary (Fig. 6, which is published as supplemental data on the PNAS web site, www.pnas.org).
Figure 3.
Expression profile of SCL in pufferfish and zebrafish. RT-PCR analysis of pufferfish SCL (FrSCL) and zebrafish SCL (DrSCL). The pufferfish homologue of the ubiquitously expressed p55 gene (FRp55) and the zebrafish gene elongation factor α (DrEF1) were used as loading controls. 1, spleen; 2, liver; 3, gonads; 4, kidney; 5, gut; 6, gill; 7, brain; 8, heart; 9, muscle; 10, water control.
Expression of the Pufferfish SCL Gene in Transgenic Zebrafish Embryos.
The highly conserved pattern of SCL expression implies the existence of functionally homologous regulatory elements. In human, mouse, and chicken, the SCL gene is flanked upstream by the SCl interrupting locus (SIL) gene and downstream by the membrane-associated protein (MAP)17 gene (28, 54, 55). By contrast, no homologue of SIL or MAP17 was identified flanking the pufferfish SCL gene, which instead is flanked by a PDZ domain gene and a Saccular Collagen gene (Fig. 1). Furthermore, whereas human SCL maps to chromosome 1p (56), the likely human homologues of UPG and C3HC4 both map to chromosome 1q, and a human homologue of PP2C maps to chromosome 3. The simplest way to reconcile the loss of synteny at the pufferfish locus with the highly conserved pattern of SCL expression would be to postulate that the regulatory elements responsible for the conserved pattern of SCL expression are situated between the genes immediately upstream and downstream of SCL in each species, a region of only 8.5 kb in the pufferfish.
To test this hypothesis, we elected to study the expression of SCL from pufferfish genomic DNA in a transgenic zebrafish assay. The pattern of expression of the endogenous zebrafish SCL gene has been described in detail previously (2, 3). During zebrafish development, SCL is initially expressed in the lateral mesoderm in two stripes. The caudal portions of these stripes converge to form the midline ICM, from which hemopoietic cells develop. By 22 hpf, SCL is expressed in the ICM, in putative primary motorneurons in the ventrolateral regions of the spinal cord, and in the HMC (Fig. 4 b and c).
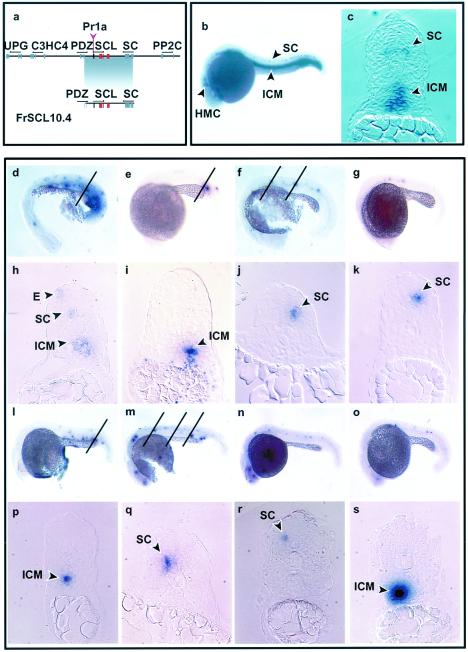
Figure 4.
Expression of the pufferfish SCL gene in transgenic zebrafish embryos. Views of embryos are lateral with anterior left and dorsal top. (a) Diagram of the pufferfish cosmid and the 10.4-kb fragment used to make transgenic zebrafish. (b) Whole-mount in situ analysis showing expression of endogenous zebrafish SCL at 22 hpf. Expression is seen in the HMC, in cell bodies within the ventrolateral region of the spinal cord in the position of motor neurons (SC), and in the cells of the ICM. The ICM consists of round cells that lie between the notochord and the endoderm above the yolk cell extension. (c) A section through the trunk shows expression of endogenous zebrafish SCL in the ICM and in the position of the motor neurons in the spinal cord. (d–k) Expression of the entire pufferfish SCL cosmid at 22 hpf. Varying numbers of cells expressing pufferfish SCL were seen in the ICM (d, e) and the spinal cord (f, g). The oblique lines in d indicates the plane of section corresponding to h, the line in e corresponds to i, and the two lines in f correspond to j and k. Sections confirmed expression in the ICM (h, i) and in the position of primary motor neurons in the spinal cord (h, j, and k). It should be noted that the ICM is formed by cells from the lateral mesoderm that migrate toward the midline, beginning anteriorly and progressing posteriorly. The width of the ICM therefore increases in more posterior regions of the trunk. E, area of ectopic expression. (l–s) Expression of the 10.4-kb construct in 22-hpf embryos. Expression of pufferfish SCL in the ICM (l, m) and the spinal cord (m, n, and o). The oblique line in l indicates the plane of section corresponding to p and the three lines in m correspond to q, r, and s. Expression in the ICM (p, s) and spinal cord (q, r).
To assess whether pufferfish regulatory elements would function appropriately in zebrafish, embryos at the single cell stage were injected with the entire pufferfish SCL cosmid. Expression of the pufferfish SCL gene was assessed by whole-mount in situ hybridization by using a 3′ untranslated region probe that lacked significant homology with the zebrafish SCL gene. Pufferfish SCL RNA was detected in the ICM, spinal cord, and HMC in 40, 18, and 15%, respectively, of green fluorescent protein (GFP)-positive embryos (Fig. 4 d–g; Table 1). No pufferfish SCL expression was detected in 31% of GFP-positive embryos, and ectopic expression was observed in the yolk cell and enveloping layer in 28 and 1% of embryos, respectively, both common sites of ectopic expression in similar studies (44, 46–48, 57, 58). To determine the exact location of the cells expressing the pufferfish SCL gene, transverse sections through the trunk of the embryos were performed. Sections confirmed that pufferfish SCL RNA was expressed within the ICM (Fig. 4 h and i) and in the position of primary motor neurons in the ventrolateral region of the spinal cord (Fig. 4 h, j, and k). The number of positive cells varied in different embryos. This has been observed previously after DNA injection (44, 47, 48, 57, 58) and is likely to reflect mosaic distribution of injected DNA together with delayed integration during the early rounds of cell division. These data therefore demonstrate that regulatory elements within a 33-kb region of the pufferfish SCL locus direct appropriate tissue specific expression within the ICM, spinal cord, and HMC in zebrafish embryos.
Table 1.
Expression of the FrSCL1.311 and FrSCL10.4 constructs in transgenic zebrafish embryos
| Construct | No. of embryos analyzed | Expression in ICM | Expression in spinal cord | Expression in HMC | Ectopic
|
||
|---|---|---|---|---|---|---|---|
| Yolk cell | Ectoderm | Other | |||||
| FrSCL-1.311 | 85 | 34 (40%) | 15 (18%) | 13 (15%) | 24 (28%) | 5 (1%) | 8 (1%) |
| FrSCL-10.4 | 158 | 46 (29%) | 54 (34%) | 41 (26%) | 67 (42%) | 57 (36%) | 48 (30%) |
To further localize the pufferfish SCL regulatory elements, a plasmid containing a 10.4-kb XbaI/SalI fragment (Fig. 4a) derived from the pufferfish cosmid was injected in a second series of experiments. This fragment contained the whole SCL gene and extended upstream to within the PDZ domain gene and downstream to within the Saccular Collagen gene. Whole-mount in situ analysis revealed SCL RNA expression in the position of the ICM, spinal cord, and HMC in 29, 34, and 26%, respectively, of GFP-positive embryos (Fig. 4 l–o; Table 1). Sections confirmed that the SCL mRNA was expressed within the ICM (Fig. 4 p and s) and in the position of primary motor neurons in the spinal cord (Fig. 4 q and r). Higher levels of ectopic expression were seen with the 10.4-kb fragment than with the entire cosmid (Table 1). Ectopic expression was particularly evident in the yolk cell and the enveloping layer (Fig. 4 l and m). These data are therefore consistent with the prediction that the regulatory elements responsible for the conserved pattern of SCL expression in the ICM, spinal cord, and HMC are located between the upstream and downstream flanking genes.
Discussion
We describe the isolation and characterization of the pufferfish SCL gene. Multiple lines of evidence argue that this pufferfish gene is indeed orthologous with the SCL genes of higher vertebrates. (i) Hybridization data demonstrated that the pufferfish genome contains a single SCL gene. (ii) Splice junctions between the coding exons were identical to those found in the SCL genes of higher vertebrates. (iii) Comparison of the predicted translation products revealed 98% conservation with the bHLH domain and, more importantly, a number of additional conserved domains were evident in the N- and C-terminal regions of the protein. (iv) In transgenic zebrafish embryos at 22 hpf, the pufferfish SCL locus recapitulated the expression pattern of the endogenous zebrafish SCL gene. (v) The region immediately upstream of the first exon of pufferfish SCL exhibits considerable homology with the human and murine SCL promoter 1a (unpublished data).
The extent to which synteny is conserved at the SCL locus in different species was particularly striking. The human SCL gene lies on the short arm of chromosome 1 and is flanked by SIL upstream and MAP17 downstream (28). Both mouse and chicken SCL genes are also flanked by SIL and MAP17. Present evidence demonstrates different expression patterns for SCL, SIL, and MAP17. SIL is widely expressed and plays a critical role in axial specification during embryonic development (59), and MAP17 is found in kidney tubular epithelial cells and is up-regulated in carcinomas (60). None of the five pufferfish genes identified here have human homologues that map to 1p. Instead, UPG and C3HC4 are highly homologous to genes that map to human 1q, and PP2C is highly related to a gene on chromosome 3 (Fig. 7, which is published as supplemental data on the PNAS web site, www.pnas.org). These data are consistent with previous observations that, in at least some regions of the genome, pufferfish speciation has been accompanied by particularly high rates of chromosomal rearrangements (61–64). It is possible that genome compression in the pufferfish has been achieved, at least in part, by clustering together genes with shared domains of expression. However, the Saccular Collagen gene (downstream of SCL) is specifically expressed in the inner ear (65), and the expression of the PDZ gene (upstream of SCL) is not known.
Genomic rearrangements represent an important substrate for natural selection during evolution and seem likely to at least partially underlie the enormous biological diversity of fish species (66). Such diversity could be generated not only by gene duplication and/or exon shuffling to create new coding entities but also by segregation of genes from their cognate regulatory elements, because altering the expression pattern of a gene can create or abolish biological functions associated with that gene. In light of these concepts, how can the marked lack of any conserved synteny around the SCL loci of fish and higher vertebrates be reconciled with the highly conserved pattern of SCL expression? At least three possible explanations can be envisaged. First, the elements responsible for directing SCL expression in teleosts and higher vertebrates may not be direct descendants of the same ancestral regulatory mechanisms, but instead represent convergent evolution. This seems unlikely, given the strong evidence presented in this study that the pufferfish SCL gene is orthologous to the SCL gene of higher vertebrates. Moreover, the midbrain-specific regulatory element upstream of mouse exon 1a has considerable sequence homology with the region upstream of pufferfish exon 1 (unpublished work). Second, some SCL elements may be capable of regulating the SCL gene over long distances. These may have been separated from the pufferfish SCL gene by genomic rearrangements associated with teleost speciation and as a result lie outside the FrSCL1.311 cosmid. However, the transgenic zebrafish data presented here argue against this possibility.
Instead, we favor the third explanation, that regulatory elements necessary for the conserved pattern of SCL expression in ICM, spinal cord, and HMC have remained closely linked to the SCL gene and lie between the immediate upstream and downstream flanking genes. This interpretation is consistent with our demonstration that a 10.4-kb fragment of the pufferfish SCL locus directed appropriate expression to these tissues in zebrafish embryos. Higher levels of ectopic expression were seen with the 10.4-kb fragment than with the entire cosmid. This might suggest that the 10.4-kb fragment lacks an insulator or boundary element. Alternatively, the larger cosmid may merely provide more flanking DNA which nonspecifically protects the SCL locus within it from cis-acting regulatory influences present at the sites of transgene integration.
The data presented here demonstrate that regulatory elements responsible for the conserved pattern of SCL expression in the ICM, spinal cord, and HMC all lie within a 10.4-kb fragment and probably within the 8.5-kb region between the upstream and downstream flanking genes. Several approaches can now be used to identify the pufferfish regulatory elements within this region. Pufferfish/mammalian sequence comparisons may prove informative, although the extent to which regulatory regions will be conserved remains unclear. Preliminary data suggest that such comparisons detect peaks of homology that correspond to only a minority of known murine SCL regulatory elements. However, this may reflect limitations of current computational approaches for long-range sequence comparisons. Several studies have used comparisons over short distances to demonstrate conservation of regulatory sequences between mammalian and pufferfish genes (33, 35, 37, 38). Sequence comparisons with zebrafish genomic DNA may be more informative. In addition, our results demonstrate that transgenic zebrafish reporter assays could be used to define the position of individual regulatory elements.
The pufferfish SCL locus also provides a valuable tool for several other lines of future work. It provides an expression cassette that can be used in vivo to target expression of exogenous genes to hemangioblasts, hemopoietic progenitors, endothelial progenitors, and spinal cord motor neurons. Furthermore, we predict that the pufferfish locus will direct expression to specific regions of the midbrain and hindbrain in which SCL is normally expressed (6) at later timepoints in zebrafish development. Finally, insertion of a reporter gene such as GFP into the pufferfish locus would provide a potent tool for lineage tracking experiments.
Supplementary Material
Acknowledgments
Work in our laboratories is supported by the Kay Kendall Leukaemia Fund (L.M.B.), the Wellcome Trust (B.G., J.G.R.G., D.G., J.R., R.P., and A.R.G.), and the Medical Research Council (M.G.). We thank sequencing team 47 at the Sanger Centre, Sarah J. Kinston for preparing zebrafish RNA and performing RT-PCR on these tissues, Dr. Adam Rodaway, Kings College, London, for helpful discussions, Dr. Sam Aparicio, Cambridge Institute for Medical Research, Cambridge, for the pufferfish cosmid library, and Dr. Greg Elgar, Human Genome Mapping Project, Cambridge, for pufferfish RNA.
Abbreviations
- SCL
stem cell leukemia
- bHLH
basic helix–loop–helix
- ICM
intermediate cell mass
- HMC
head mesenchyme
- RT-PCR
reverse transcription–PCR
- hpf
hours postfertilization
- GFP
green fluorescent protein
Footnotes
References
- 1.Begley C G, Green A R. Blood. 1999;93:2760–2770. [PubMed] [Google Scholar]
- 2.Liao E C, Paw B H, Oates A C, Pratt S J, Postlethwait J H, Zon L I. Genes Dev. 1998;12:621–626. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gering M, Rodaway A R F, Göttgens B, Patient R K, Green A R. EMBO J. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mead P E, Kelley C M, Hahn P S, Piedad O, Zon L I. Development (Cambridge, UK) 1998;125:2611–2620. doi: 10.1242/dev.125.14.2611. [DOI] [PubMed] [Google Scholar]
- 5.Drake C J, Brandt S J, Trusk T C, Little C D. Dev Biol. 1997;192:17–30. doi: 10.1006/dbio.1997.8751. [DOI] [PubMed] [Google Scholar]
- 6.Sinclair A M, Göttgens B, Barton L M, Stanley M L, Pardanaud L, Klaine M, Gering M, Bahn S, Sanchez M, Bench A J, et al. Dev Biol. 1999;209:128–142. doi: 10.1006/dbio.1999.9236. [DOI] [PubMed] [Google Scholar]
- 7.Robb L, Elwood N J, Elefanty A G, Köntgen F, Li R, Barnett L D, Begley C G. EMBO J. 1996;15:4123–4129. [PMC free article] [PubMed] [Google Scholar]
- 8.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt F W, Orkin S H. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 9.Green A R, De Luca E, Begley C G. EMBO J. 1991;10:4153–4158. doi: 10.1002/j.1460-2075.1991.tb04993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green A R, Visvader J, Lints T, Harvey R, Begley C G. Oncogene. 1992;7:653–660. [PubMed] [Google Scholar]
- 11.Murrell A M, Green A R. Oncogene. 1995;10:631–639. [PubMed] [Google Scholar]
- 12.Prasad K S, Jordan J E, Koury M J, Bondurant M C, Brandt S J. J Biol Chem. 1995;270:11603–11611. doi: 10.1074/jbc.270.19.11603. [DOI] [PubMed] [Google Scholar]
- 13.Cross M A, Heyworth C M, Murrell A M, Bockamp E-O, Cobley U T, Dexter T M, Green A R. Oncogene. 1994;9:3013–3016. [PubMed] [Google Scholar]
- 14.Hoang T, Paradis E, Brady G, Billia F, Nakahara K, Iscove N N, Kirsch I R. Blood. 1996;87:102–111. [PubMed] [Google Scholar]
- 15.Aplan P D, Nakahra K, Orkin S H, Kirsch I R. EMBO J. 1992;11:4073–4081. doi: 10.1002/j.1460-2075.1992.tb05500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elwood N J, Zogos H, Pereira D S, Dick J E, Begley C G. Blood. 1998;91:3756–3765. [PubMed] [Google Scholar]
- 17.Valtieri M, Tocci A, Gabbianelli M, Luchetti L, Masella B, Vitelli L, Botta R, Testa U, Condorelli G L, Peschle C. Cancer Res. 1998;58:562–569. [PubMed] [Google Scholar]
- 18.Robertson S M, Kennedy M, Shannon J M, Keller G. Development (Cambridge, UK) 2000;127:2447–2459. doi: 10.1242/dev.127.11.2447. [DOI] [PubMed] [Google Scholar]
- 19.Visvader J E, Fujiwara Y, Orkin S H. Genes Dev. 1998;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aplan P D, Begley C G, Bertness V, Nussmeier M A, Ezquerra A, Coligan J, Kirsch I R. Mol Cell Biol. 1990;10:6426–6435. doi: 10.1128/mcb.10.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Begley C G, Robb L, Rockman S, Visvader J, Bockamp E-O, Chan Y S, Green A R. Gene. 1994;138:93–99. doi: 10.1016/0378-1119(94)90787-0. [DOI] [PubMed] [Google Scholar]
- 22.Bockamp E-O, McLaughlin F, Murrell A M, Göttgens B, Robb L, Begley C G, Green A R. Blood. 1995;86:1502–1514. [PubMed] [Google Scholar]
- 23.Bockamp E O, McLaughlin F, Göttgens B, Murrell A M, Elefanty A G, Green A R. J Biol Chem. 1997;272:8781–8790. doi: 10.1074/jbc.272.13.8781. [DOI] [PubMed] [Google Scholar]
- 24.Lecointe N, Bernard O, Naert K, Joulin V, Larsen C J, Romeo P H, Mathieu-Mahul D. Oncogene. 1994;9:2623–2632. [PubMed] [Google Scholar]
- 25.Göttgens B, McLaughlin F, Bockamp E O, Fordham J L, Begley C G, Kosmopoulos K, Elefanty A G, Green A R. Oncogene. 1997;15:2419–2428. doi: 10.1038/sj.onc.1201426. [DOI] [PubMed] [Google Scholar]
- 26.Fordham J L, Gottgens B, McLaughlin F, Green A R. Leukemia. 1999;13:750–759. doi: 10.1038/sj.leu.2401420. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez M, Gottgens B, Sinclair A M, Stanley M, Begley C G, Hunter S, Green A R. Development (Cambridge, UK) 1999;126:3891–3904. doi: 10.1242/dev.126.17.3891. [DOI] [PubMed] [Google Scholar]
- 28.Göttgens B, Barton L M, Gilbert J G, Bench A J, Sanchez M J, Bahn S, Mistry S, Grafham D, McMurray A, Vaudin M, et al. Nat Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- 29.Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. Nature (London) 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- 30.Rowley A F, Hunt T C, Page M, Mainwaring G. Fish. Cambridge, U.K.: Cambridge Univ. Press; 1988. [Google Scholar]
- 31.Zon L I. Blood. 1995;86:2876–2891. [PubMed] [Google Scholar]
- 32.Hansen J D, Zapata A G. Immunol Rev. 1998;166:199–220. doi: 10.1111/j.1600-065x.1998.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 33.Marshall H, Studer M, Popperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. Nature (London) 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- 34.Popperl H, Bienz M, Studer M, Chan S K, Aparicio S, Brenner S, Mann R S, Krumland R. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 35.Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Proc Natl Acad Sci USA. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beckers J, Gerard M, Duboule D. Dev Biol. 1996;180:543–553. doi: 10.1006/dbio.1996.0327. [DOI] [PubMed] [Google Scholar]
- 37.Kimura C, Takeda N, Suzuki M, Oshimura M, Aizawa S, Matsuo I. Development (Cambridge, UK) 1997;124:3929–3941. doi: 10.1242/dev.124.20.3929. [DOI] [PubMed] [Google Scholar]
- 38.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 39.Armes N, Gilley J, Fried M. Genome Res. 1997;7:1138–1152. doi: 10.1101/gr.7.12.1138. [DOI] [PubMed] [Google Scholar]
- 40.Wentworth J M, Schoenfeld V, Meek S, Elgar G, Brenner S, Chatterjee V K. Gene. 1999;236:315–323. doi: 10.1016/s0378-1119(99)00265-6. [DOI] [PubMed] [Google Scholar]
- 41.Eisen J S. Cell. 1996;87:969–977. doi: 10.1016/s0092-8674(00)81792-4. [DOI] [PubMed] [Google Scholar]
- 42.Ingham P W. Hum Mol Genet. 1997;6:1755–1760. doi: 10.1093/hmg/6.10.1755. [DOI] [PubMed] [Google Scholar]
- 43.Amatruda J F, Zon L I. Dev Biol. 1999;216:1–15. doi: 10.1006/dbio.1999.9462. [DOI] [PubMed] [Google Scholar]
- 44.Muller F, Chang B, Albert S, Fischer N, Tora L, Strahle U. Development (Cambridge, UK) 1999;126:2103–2116. doi: 10.1242/dev.126.10.2103. [DOI] [PubMed] [Google Scholar]
- 45.Meng A, Tang H, Yuan B, Ong B A, Long Q, Lin S. Blood. 1999;93:500–508. [PubMed] [Google Scholar]
- 46.Jessen J R, Meng A, McFarlane R J, Paw B H, Zon L I, Smith G R, Lin S. Proc Natl Acad Sci USA. 1998;95:5121–5126. doi: 10.1073/pnas.95.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drivenes, Seo H, Fjose A. Biochim Biophys Acta. 2000;1491:240–247. doi: 10.1016/s0167-4781(00)00042-7. [DOI] [PubMed] [Google Scholar]
- 48.Reinhard E, Nedivi E, Wegner J, Skene J H, Westerfield M. Development (Cambridge, UK) 1994;120:1767–1775. doi: 10.1242/dev.120.7.1767. [DOI] [PubMed] [Google Scholar]
- 49.Göttgens B, Gilbert J G R, Barton L M, Aparicio S, Hawker K, Mistry S, Vaudin M, King A, Bentley D, Elgar G, Green A R. Genomics. 1998;48:52–62. doi: 10.1006/geno.1997.5162. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 51.Jowett T, Yan Y-L. Trends Genet. 1996;12:387–389. doi: 10.1016/s0168-9525(96)90091-8. [DOI] [PubMed] [Google Scholar]
- 52.Metzenberg A B, Gitschier J. Hum Mol Genet. 1992;1:97–101. doi: 10.1093/hmg/1.2.97. [DOI] [PubMed] [Google Scholar]
- 53.Drake C J, Fleming P A. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- 54.Aplan P D, Lombardi D P, Kirsch I R. Mol Cell Biol. 1991;11:5462–5469. doi: 10.1128/mcb.11.11.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gottgens B, Gilbert J G, Barton L M, Grafham D, Rogers J, Bentley D R, Green A R. Genome Res. 2001;11:87–97. doi: 10.1101/gr.153001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Begley C G, Aplan P D, Davey M P, Nakahara K, Tchorz K, Kurtzberg J, Hershfeld M S, Haynes B F, Cohen D I, Waldmann T A, Kirsch I R. Proc Natl Acad Sci USA. 1989;86:2031–2035. doi: 10.1073/pnas.86.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meng A, Tang H, Ong B A, Farrell M J, Lin S. Proc Natl Acad Sci USA. 1997;94:6267–6272. doi: 10.1073/pnas.94.12.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Long Q, Meng A, Wang H, Jessen J R, Farrell M J, Lin S. Development (Cambridge, UK) 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 59.Izraeli S, Colaizzo-Anas T, Bertness V L, Mani K, Aplan P D, Kirsch I R. Cell Growth Differ. 1997;8:1171–1179. [PubMed] [Google Scholar]
- 60.Kocher O, Cheresh P, Lee S W. Am J Pathol. 1996;149:493–500. [PMC free article] [PubMed] [Google Scholar]
- 61.Ohta Y, Okamura K, McKinney E C, Bartl S, Hashimoto K, Flajnik M F. Proc Natl Acad Sci USA. 2000;97:4712–4717. doi: 10.1073/pnas.97.9.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elgar G, Clark M S, Meek S, Smith S, Warner S, Edwards Y J, Bouchireb N, Cottage A, Yeo G S, Umrania Y, et al. Genome Res. 1999;9:960–971. doi: 10.1101/gr.9.10.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilley J, Fried M. Hum Mol Genet. 1999;8:1313–1320. doi: 10.1093/hmg/8.7.1313. [DOI] [PubMed] [Google Scholar]
- 64.Boeddrich A, Burgtorf C, Crollius H R, Hennig S, Bernot A, Clark M, Reinhardt R, Lehrach H, Francis F. Genomics. 1999;57:164–168. doi: 10.1006/geno.1998.5732. [DOI] [PubMed] [Google Scholar]
- 65.Davis J G, Oberholtzer J C, Burns F R, Greene M I. Science. 1995;267:1031–1034. doi: 10.1126/science.7863331. [DOI] [PubMed] [Google Scholar]
- 66.Wittbrodt J, Meyer A, Schartl M. BioEssays. 1998;20:511–515. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.