A live-imaging approach is used to demonstrate that nuclear proteins reorganize during mitosis to form a highly dynamic, viscous spindle matrix that embeds the microtubule spindle apparatus, stretching from pole to pole.
Abstract
The concept of a spindle matrix has long been proposed. Whether such a structure exists, however, and what its molecular and structural composition are have remained controversial. In this study, using a live-imaging approach in Drosophila syncytial embryos, we demonstrate that nuclear proteins reorganize during mitosis to form a highly dynamic, viscous spindle matrix that embeds the microtubule spindle apparatus, stretching from pole to pole. We show that this “internal” matrix is a distinct structure from the microtubule spindle and from a lamin B–containing spindle envelope. By injection of 2000-kDa dextran, we show that the disassembling nuclear envelope does not present a diffusion barrier. Furthermore, when microtubules are depolymerized with colchicine just before metaphase the spindle matrix contracts and coalesces around the chromosomes, suggesting that microtubules act as “struts” stretching the spindle matrix. In addition, we demonstrate that the spindle matrix protein Megator requires its coiled-coil amino-terminal domain for spindle matrix localization, suggesting that specific interactions between spindle matrix molecules are necessary for them to form a complex confined to the spindle region. The demonstration of an embedding spindle matrix lays the groundwork for a more complete understanding of microtubule dynamics and of the viscoelastic properties of the spindle during cell division.
INTRODUCTION
During cell division the entire nucleus undergoes a dramatic reorganization as the cell prepares to segregate its duplicated chromosomes. For many years the prevailing view on organisms possessing an open mitosis has held that the nucleus completely disassembled during early mitotic stages, thus enabling cytoplasmic microtubules emanating from the separated centrosomes to form a mitotic spindle. This cytocentric view largely discounted any nuclear contributions to the formation and/or function of the mitotic spindle (Johansen and Johansen, 2009; Simon and Wilson, 2011; Sandquist et al., 2011). However, in Drosophila we recently identified two nuclear proteins, Chromator (Rath et al., 2004; Ding et al., 2009; Yao et al., 2012) and Megator (Qi et al., 2004; Lince-Faria et al., 2009), from two different nuclear compartments that interact with each other and redistribute during prophase to form a molecular complex that persists in the absence of polymerized tubulin (Johansen et al., 2011). Chromator is localized to polytene chromosome interbands during interphase (Rath et al., 2004, 2006; Yao et al., 2012), whereas Megator occupies the nuclear rim and the intranuclear space surrounding the chromosomes (Zimowska et al., 1997; Qi et al., 2004). Chromator has no known orthologues in other species; however, Megator is the homologue of mammalian Tpr (Zimowska et al., 1997). The Megator/Tpr family of proteins is highly conserved through evolution, and structural homologues are present from yeast to humans (De Souza and Osmani, 2009). Moreover, in addition to Megator, the Aspergillus Mlp1 and human Tpr spindle matrix proteins have a shared function as spatial regulators of spindle assembly checkpoint proteins during metaphase (Lee et al., 2008; De Souza et al., 2009; Lince-Faria et al., 2009). Both Chromator and Megator are essential proteins required for normal mitosis to occur in Drosophila (Qi et al., 2004; Lince-Faria et al., 2009; Ding et al., 2009). These findings suggest that these proteins are molecular components of the hitherto-elusive spindle matrix that, based on theoretical considerations of the requirements for force production, has been proposed to help constrain and stabilize the microtubule-based spindle apparatus (Pickett-Heaps et al., 1982; Pickett-Heaps and Forer, 2009). Here we demonstrate that this nuclear-derived “internal” spindle matrix is a highly dynamic, self-contained structure that embeds the microtubule spindle apparatus from pole to pole. The findings further suggest that the spindle matrix may directly contribute to the viscoelastic micromechanical properties (Shimamoto et al., 2011) of the spindle.
RESULTS
The spindle matrix embeds the microtubule spindle apparatus
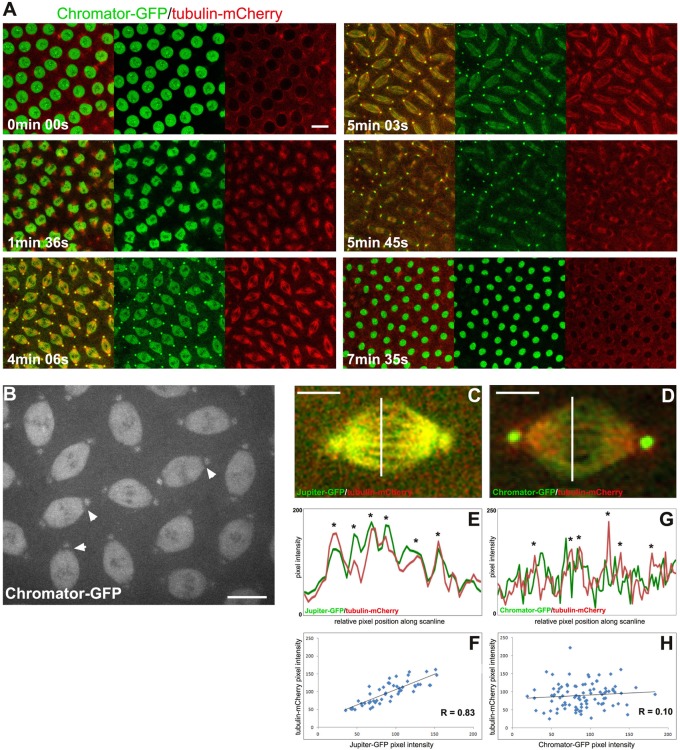
Figure 1 shows time-lapse imaging of Chromator–green fluorescent protein (GFP) and tubulin-mCherry during mitosis in syncytial Drosophila embryos. The results show that Chromator has reorganized away from the chromosomes as they begin to condense and fills the entire nuclear space before microtubule invasion (Figure 1A and Supplemental Movie S1; see also Supplemental Movie S5 for a clearer view of this transition). As spindle microtubules form, Chromator distribution attains a spindle-like morphology while also translocating to the centrosomes (Figure 1A). At anaphase and telophase Chromator dynamics closely mirror that of the microtubules before relocating back to the chromosomes in the forming daughter nuclei. This dynamic behavior of Chromator during mitosis is very different from microtubule-associated proteins (MAPs) such as Jupiter (Karpova et al., 2006; Supplemental Movie S2). Although Chromator is present throughout the spindle, its poleward boundary does not extend all the way to the centrosome (Figure 1B and Supplemental Movie S3), as also observed for the putative spindle pole matrix protein NuMA (Radulescu and Cleveland, 2010). Of interest, in line scans of pixel intensity across the spindle we found that peak intensities of the MAP Jupiter coincide with that of microtubules, indicating colocalization (Figure 1, C and E), whereas peak intensities of Chromator are notably distinct from those of microtubules and in many cases show an alternating pattern (Figure 1, D and G). Moreover, pixel intensities in line scans across the spindle for Jupiter-GFP and tubulin-mCherry were strongly correlated (r = 0.73 ± 0.10, n = 17; Figure 1F), whereas pixel intensities in line scans of Chromator-GFP and tubulin-mCherry showed little correlation (r = 0.32 ± 0.07, n = 17; Figure 1H). Taken together, these observations are consistent with the hypothesis that the Chromator-defined spindle matrix is part of a viscous, gel-like structure that embeds the microtubule-based spindle apparatus. Furthermore, the findings suggest that although this matrix forms independently of microtubules, its morphology and dynamic behavior during mitosis are governed by microtubule spindle dynamics.
FIGURE 1:
Confocal time-lapse analysis of Chromator-GFP during mitosis in syncytial Drosophila embryos. (A) Relative dynamics of Chromator-GFP (green) and tubulin-mCherry (red) during a complete mitotic cycle. Scale bar, 10 μm. (B) Chromator-GFP at metaphase. Arrowheads indicate the gap between Chromator-GFP's spindle matrix and centrosomal localization. Scale bar, 10 μm. (C) Relative localization of Jupiter-GFP (green) and tubulin-mCherry (red) at metaphase. Scale bar, 5 μm. (D) Relative localization of Chromator-GFP (green) and tubulin-mCherry (red) at metaphase. Scale bar, 5 μm. (E, G) Line-scan plots of pixel intensity across the spindle along the white lines in C and D for Jupiter-GFP/tubulin-mCherry and Chromator-GFP/tubulin-mCherry, respectively. The images in C and D are both from a single confocal optical plane. The asterisks indicate the likely position of microtubule K-fibers. (F, H) Plots of the correlation between pixel intensity between Jupiter-GFP/tubulin-mCherry and Chromator-GFP/tubulin-mCherry across the spindle along the white lines in C and D, respectively. The regression line and the value of Pearson's coefficient are indicated for each plot.
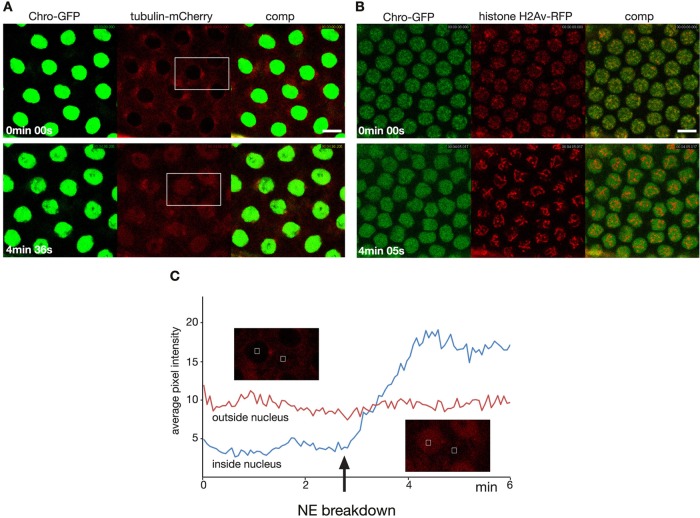
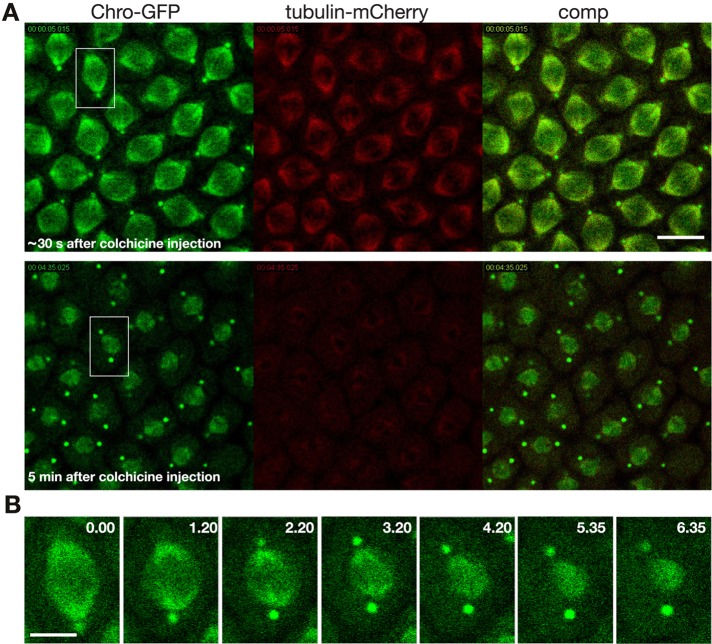
To further test this hypothesis, we depolymerized tubulin by injecting colchicine into embryos expressing GFP-Chromator and tubulin-mCherry or histone H2Av-RFP before prophase (Figure 2; Supplemental Movies S4 and S5). Under these conditions Chromator still relocates from the chromosomes to the matrix (Figure 2, A and B); however, in the absence of microtubule spindle formation the Chromator-defined matrix did not undergo any dynamic changes but instead statically embedded the condensed chromosomes for extended periods (>20 min). The movement observed within the matrix is caused by Brownian motion of the chromosomes. Of interest, Chromator under these conditions still relocated to the centrosomes, suggesting that this is a microtubule-independent process. Control embryos injected with vehicle only underwent normal mitosis indistinguishable from wild-type preparations (Supplemental Movie S6). Moreover, as illustrated in Figure 2C, unpolymerized tubulin accumulates within the nuclear space, as measured by relative average pixel intensity, to 1.6 ± 0.2 (n = 12, from five different preparations) times the levels outside the nuclear space in the colchicine-injected embryos (see also Figure 2, A and C, and Supplemental Movie S4). This finding suggests the presence of one or more tubulin-binding proteins within the spindle matrix.
FIGURE 2:
Spindle matrix dynamics after colchicine injection before nuclear envelope breakdown. (A) Two image panels from the beginning and end of a time-lapse sequence of Chromator-GFP (green) and tubulin-mCherry (red) after colchicine injection. (B) Two image panels from the beginning and end of a time-lapse sequence of Chromator-GFP (green) and histone H2Av-RFP (red). (C) Plot of the average pixel intensity in regions of interest (ROIs) outside the nucleus (red) and inside the nucleus (blue) as a function of time in a colchicine-injected embryo. The two image inserts correspond to the area outlined by a white boxes in A before and after NE breakdown, respectively. The ROIs are indicated by white squares. The difference in expression levels of Chromator-GFP in A and B is due to use of high- and low-expression driver lines, respectively.
The nuclear envelope and lamin B do not contribute to the internal spindle matrix
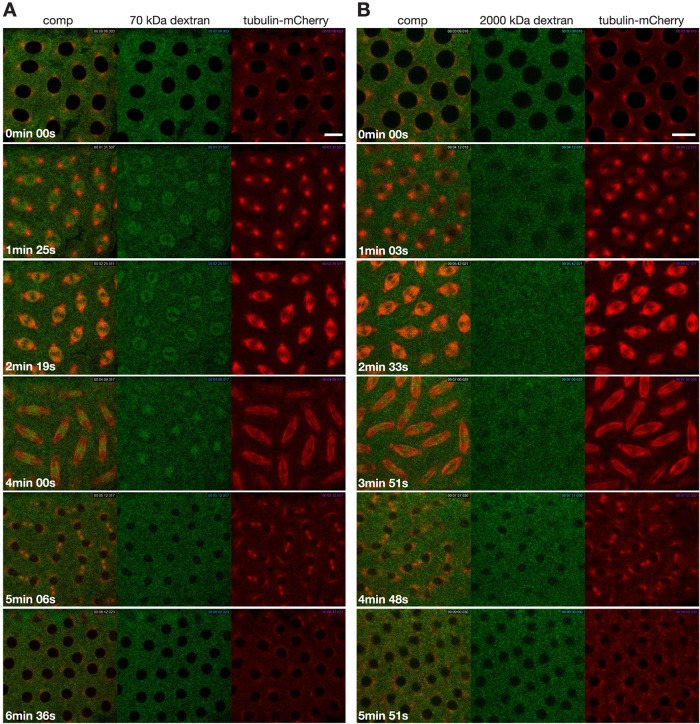
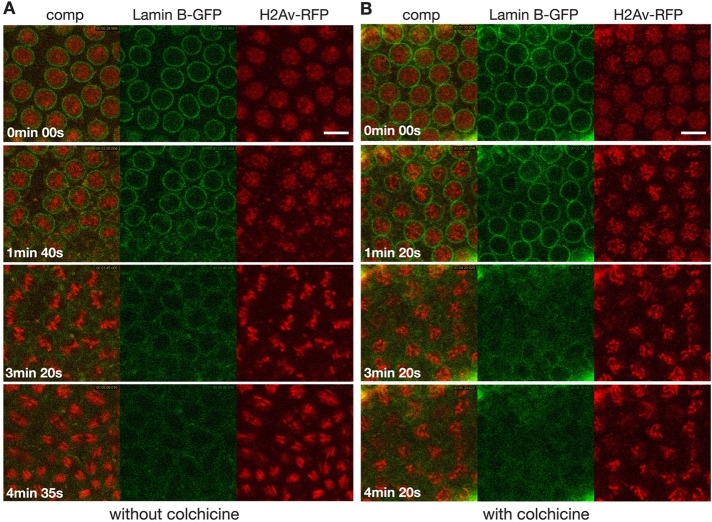
Drosophila embryos have semiopen mitosis in which the nuclear envelope (NE) initially breaks down only in the region of the centrosomes, and NE breakdown and dispersal of nuclear lamins such as lamin B (lamin Dm0 in Drosophila) is not completed until just before the end of metaphase (Stafstrom and Staehelin, 1984; Paddy et al., 1996; Civelekoglu-Scholey et al., 2010). This raises the question of whether the NE or the nuclear lamina presents a diffusion barrier during the early stages of mitosis and thus may contribute to the confinement of spindle matrix proteins. To test whether this is the case, we injected fluorescein-labeled dextrans of molecular mass 70, 500, or 2000 kDa, which are up to 10 times the molecular mass of the spindle matrix proteins Chromator and Megator, into tubulin-mCherry–expressing embryos treated with colchicine. The results showed that all three molecular-mass dextrans entered the nuclear space after NE breakdown on approximately the same timescale as tubulin-mCherry (Figures 3 and 4), indicating the absence of any significant diffusion barriers to spindle matrix proteins. Furthermore, in colchicine-injected embryos lamin B disperses within 2 min, on a timescale similar to that of uninjected embryos (Figure 5), and does not accumulate in the nuclear space. In contrast, the Chromator-defined matrix persists around the chromosomes for at least 10 times longer. Taken together, these findings suggest that the Chromator-defined “internal” spindle matrix is a distinct and independent structure from both the microtubule-based spindle apparatus and from the lamin B–containing spindle envelope previously described in Xenopus egg extracts (Zheng, 2010) and that the spindle matrix is held together by cohesive molecular interactions within the matrix.
FIGURE 3:
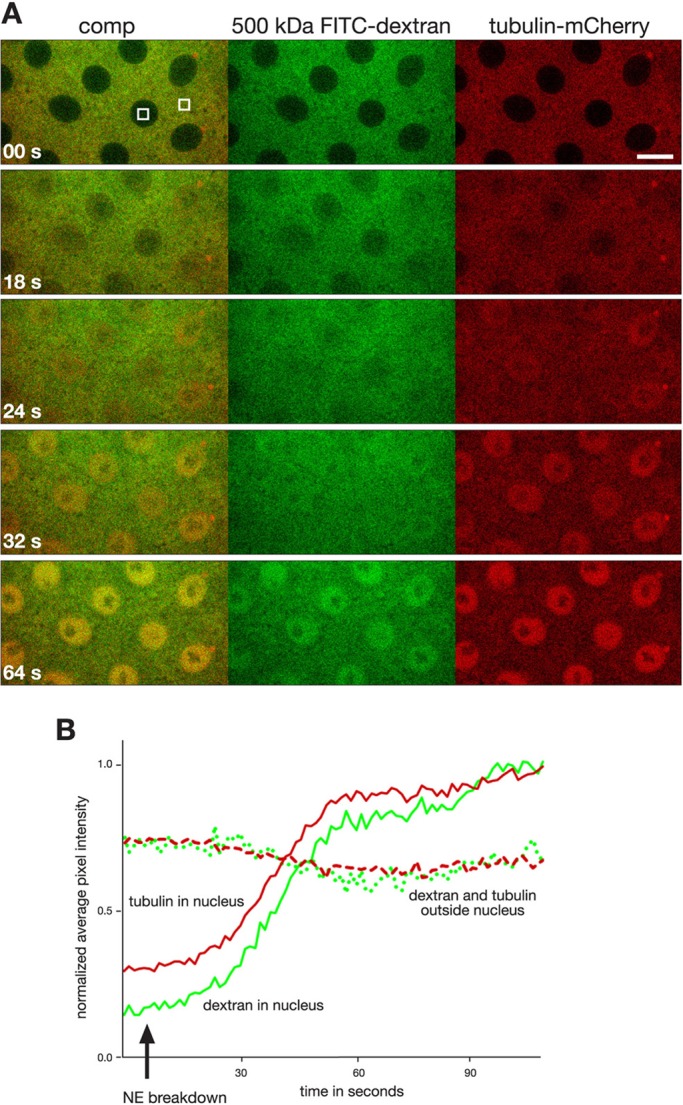
The 500-kDa dextran enters and accumulates in the nuclear space on the same timescale as tubulin in colchicine-injected embryos. (A) Image panels from a time-lapse sequence from a tubulin-mCherry (red)–expressing embryo coinjected with fluorescein-labeled dextran of molecular mass 500 kDa (green) and colchicine. Time is in seconds. Scale bar, 10 μm. (B) Plot of the normalized average pixel intensity in ROIs outside the nucleus and inside the nucleus of tubulin (red) and 500-kDa dextran (green) as a function of time in a colchicine-injected embryo. The solid and stippled lines correspond to areas inside and outside a nucleus, respectively, as outlined by the white boxes in A. The approximate time of NE breakdown is indicated by an arrow.
FIGURE 4:
The 70-kDa but not the 2000-kDa dextran incorporates into the spindle matrix during the cell cycle. (A) Image panels from a time-lapse sequence from a tubulin-mCherry (red)–expressing embryo injected with fluorescein-labeled dextran of molecular mass 70 kDa (green). (B) Image panels from a time-lapse sequence from a tubulin-mCherry (in red)–expressing embryo injected with fluorescein-labeled dextran of molecular mass 2000 kDa (green). Time is in minutes and seconds. Scale bars, 10 μm.
FIGURE 5:
Lamin B in colchicine-injected embryos disperses on a timescale similar to that of uninjected embryos during mitosis. (A) Image panels from a time-lapse sequence from a histone H2Av-RFP (red)– and lamin B-GFP (green)–expressing embryo. (B) Image panels from a time-lapse sequence from a histone H2Av-RFP (red)– and lamin B-GFP (in green)–expressing embryo injected with colchicine before nuclear envelope breakdown. Time is in minutes and seconds. Scale bars, 10 μm.
The 70- and 500-kDa dextrans incorporate into the spindle matrix
Of interest, we noted that 70- and 500-kDa dextrans accumulated within the nuclear space in a way similar to tubulin in colchicine-injected embryos, as illustrated in Figure 3 for 500-kDa dextran. This suggested that branched macromolecular polysaccharides can be incorporated into the spindle matrix. To further explore this possibility, we injected fluorescein-conjugated 70-, 500-, and 2000-kDa dextrans into tubulin-mCherry–expressing embryos without colchicine treatment. As exemplified in Figure 4A for 70-kDa dextran, both 70- and 500-kDa dextrans accumulate in the nuclear space before microtubule spindle formation, and its dynamics during mitosis until the end of telophase, when it gets excluded from the forming daughter nuclei (Supplemental Movie S7), closely resembles that of the spindle matrix proteins Chromator and Megator (Supplemental Movies S1 and S8). In contrast, although the 2000-kDa dextran did enter and equilibrate within the nuclear space at the time of NE breakdown, it did not show any enrichment within the spindle region (Figure 4B). We speculate that this difference between 70- and 2000-kDa dextrans is due to potential size exclusionary properties of the spindle matrix. These data provide additional support for the concept of a viscous matrix made up of macromolecules enriched in the spindle region by cohesive interactions.
The amino-terminal region of Megator is required for its spindle matrix localization
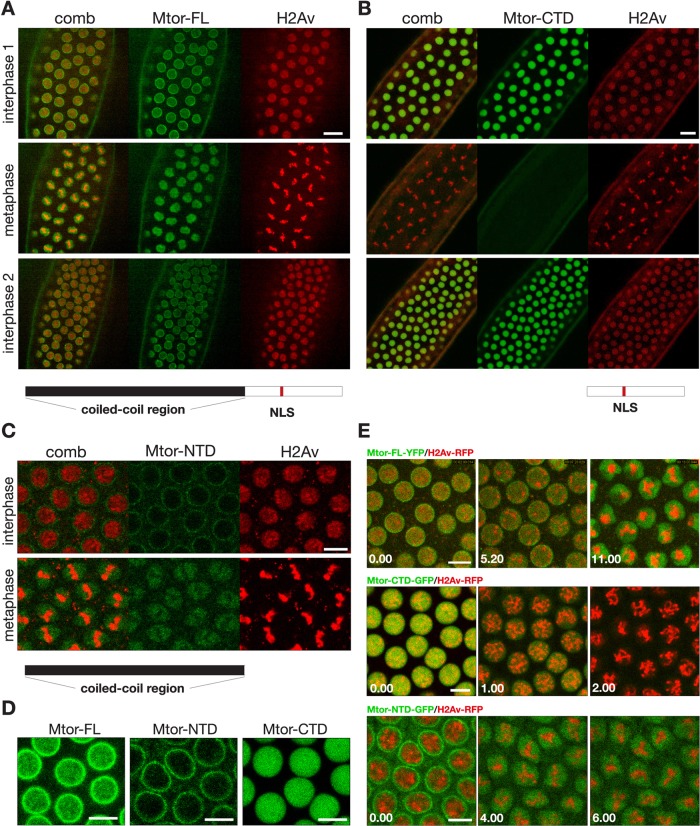
Megator is a large, 260-kDa protein (Mtor-FL) with an extended amino-terminal coiled-coil domain (Mtor-NTD) and an unstructured carboxy-terminal domain (Mtor-CTD). Coiled-coil domains are known protein interaction domains, as previously demonstrated for the spindle pole matrix protein NuMA (Radulescu and Cleveland, 2010). Therefore, to explore whether Megator's coiled-coil domain is required for Megator's spindle matrix localization, we conducted time-lapse imaging of full-length, yellow fluorescent protein (YFP)–tagged Megator (Mtor-FL), green fluorescent protein (GFP)–tagged Mtor-CTD, and GFP-tagged Mtor-NTD, together with histone H2Av-RFP in syncytial embryos (Figure 6). As illustrated in Figure 6A and Supplemental Movie S8, Mtor-FL localizes to the nuclear interior, as well as to the nuclear rim, at interphase and to the spindle matrix at metaphase. In contrast, Mtor-CTD, which contains the native nuclear localization signal (NLS), is diffusively present in the nucleoplasm without detectable nuclear rim localization at interphase and is absent from the spindle region at metaphase (Figure 6B and Supplemental Movie S9). Mtor-NTD is present at the nuclear rim with no or very little interior nuclear localization but relocalizes to the spindle matrix at metaphase (Figure 6C). The localization patterns of Mtor-FL, Mtor-NTD, and Mtor-CTD at interphase are illustrated at higher magnification in Figure 6D. These data suggest that the amino-terminal coiled-coil domain of Megator is required for localization to both nuclear pore complexes and to the spindle matrix, whereas Megator's carboxy-terminal domain facilitates Megator's interchromosomal localization during interphase. Furthermore, if microtubules are prevented from forming by colchicine injection before prophase, both Mtor-FL and Mtor-NTD still relocate to the spindle matrix and, as with the Chromator-defined matrix, do not undergo any dynamic changes but statically embed the condensed chromosomes (Figure 6E and Supplemental Movie S10). In contrast, under these conditions Mtor-CTD disperses on a rapid timescale in <2 min after NE breakdown (Figure 6E and Supplemental Movie S11). These findings provide further evidence that the cohesiveness of the spindle matrix depends on specific molecular interactions among the spindle matrix proteins.
FIGURE 6:
Time-lapse analysis of the spindle matrix protein Megator in syncytial embryos. (A) Relative dynamics of full-length Megator-YFP (Mtor-FL) and histone H2Av-RFP (H2Av) during a complete mitotic cycle. The images show their distribution at interphase 1, metaphase, and interphase 2, respectively. The diagram beneath the images shows the domain structure of Megator with the coiled-coil region in black, the CTD in white, and the endogenous NLS in red. Scale bar, 20 μm. (B) Relative dynamics of a truncated, GFP-tagged, carboxy-terminal construct of Megator (Mtor-CTD) and histone H2Av-RFP (H2Av) during a complete mitotic cycle. The images show their distribution at interphase 1, metaphase, and interphase 2, respectively. Mtor-CTD is diagrammed below the images. Scale bar, 20 μm. (C) Relative dynamics of a truncated, GFP-tagged, amino-terminal construct of Megator (Mtor-NTD) and histone H2Av-RFP (H2Av) during interphase and metaphase. Mtor-NTD is diagrammed below the images. Scale bar, 10 μm. (D) The localization patterns of Mtor-FL, Mtor-NTD, and Mtor-CTD at interphase. Mtor-FL localizes to the nuclear interior, as well as to the nuclear rim, Mtor-NTD is present at the nuclear rim with no or very little interior nuclear localization, and Mtor-CTD is diffusively present in the nucleoplasm without detectable nuclear rim localization. (E) Top, three images from a time-lapse sequence of Mtor-FL-YFP (green) and histone H2Av-RFP (red) after colchicine injection at interphase. Middle, three images from a time-lapse sequence of Mtor-CTD-GFP (green) and histone H2Av-RFP (red) after colchicine injection at interphase. Bottom, three images from a time-lapse sequence of Mtor-NTD-GFP (green) and histone H2Av-RFP (red) after colchicine injection at interphase. Time is in minutes and seconds. Scale bars, 10 μm.
Depolymerization of microtubules at metaphase collapses but does not disassemble the spindle matrix
To test the dependence of the spindle matrix on microtubule dynamics, we injected colchicine into Chromator-GFP– and tubulin-mCherry–expressing embryos during metaphase. As shown in the image sequence of Figure 7 and in Supplemental Movie S12, as the microtubules undergo depolymerization, the Chromator-defined matrix contracts and coalesces around the chromosomes. The reduction in the length of the spindle matrix was almost 60% from when the first image was obtained after colchicine injection to when microtubules were depolymerized (Figure 7B). This suggests that the spindle matrix is stretched by the microtubules. A similar result was obtained in S2 cells expressing the spindle matrix protein Megator (Lince-Faria et al., 2009), suggesting that the properties of the spindle matrix described here are a general feature of mitosis and not confined to only syncytial nuclei. Furthermore, the expectation would be that if microtubules were stabilized at metaphase instead of depolymerized, then the shape and form of the spindle matrix would not change. To test this prediction, we injected the microtubule-stabilizing agent Taxol into Mtor-FL– and tubulin-mCherry–expressing embryos during metaphase. As shown in Supplemental Movie S13, under these conditions both the spindle matrix and the microtubules do not undergo any dynamic changes but maintain their metaphase fusiform spindle morphology for extended time periods of >14 min.
FIGURE 7:
Depolymerization of microtubules at metaphase leads to contraction of the spindle matrix. (A) Two image panels from the beginning and end of a time-lapse sequence of Chromator-GFP (green) and tubulin-mCherry (red) after colchicine injection. The image sequence begins ∼30 s after colchicine injection. Scale bar, 10 μm. (B) Image sequence of Chromator-GFP after colchicine injection in the spindle outlined by white rectangles in A. Time is in minutes and seconds. Scale bar, 5 μm.
DISCUSSION
In this study we showed that at least two proteins from different nuclear compartments reorganize during mitosis to form a spindle matrix that embeds the microtubule spindle apparatus and that is likely to be part of a molecular complex stretching from pole to pole. As also indicated by previous experiments in S2 cells (Lince-Faria et al., 2009), the present observations are not compatible with a rigid matrix structure but instead with a highly dynamic viscous matrix made up of protein polymers forming a gel-like meshwork. For such a matrix to be stretched implies that components of the matrix physically be linked to microtubules and that changes to the shape and form of the matrix in turn are governed by microtubule dynamics. One possible mechanism to accomplish this is exemplified by NuMA, which, together with dynein, functions as a spindle pole matrix that tethers and focuses the majority of spindle microtubules to the poles largely independently of centrosomes (Dumont and Mitchison, 2009; Radulescu and Cleveland, 2010). Thus we propose that a spindle pole matrix may be a constituent of a larger pole-to-pole matrix that couples this matrix to microtubule dynamics.
In Xenopus egg extracts it was suggested that a membranous lamin B–containing envelope derived from the nuclear membrane could be part of the spindle matrix (Tsai et al., 2006; Zheng, 2010). However, our findings clearly demonstrate that the “internal” matrix as defined by the Chromator and Megator proteins is physically distinct from such a structure and that the internal matrix persists after dispersal of lamin B in nuclei arrested at metaphase. Nonetheless, the interplay between microtubules, the spindle matrix, and NE dynamics during mitosis is likely to be finely tuned and mutually dependent (Zheng, 2010). For example, evidence has been provided that the NE and lamin B in systems with semiopen mitosis may contribute to the robustness of spindle function and assembly during prometaphase and that the gradual disassembly of the lamin B envelope is coupled to proper spindle maturation during metaphase (Civelekoglu-Scholey et al., 2010).
In this study we present evidence by injection of high–molecular weight dextrans that the disassembling NE and nuclear lamina after their initial breakdown are not likely to present a diffusion barrier to most known proteins. Of interest, even in the absence of such a diffusion barrier we show that free tubulin (possibly as α/β-tubulin dimers) accumulates coextensively with the spindle matrix protein Chromator in colchicine-treated embryos independently of tubulin polymerization. We propose that this enrichment is dependent on one or more proteins within the spindle matrix with tubulin-binding activity. A similar enrichment within the nuclear region of free tubulin after NE breakdown has recently been reported in Caenorhabditis elegans embryos (Hayashi et al., 2012). The enhanced accumulation of free tubulin within the nascent spindle region may serve as a general mechanism to promote the efficient assembly of the microtubule-based spindle apparatus (Hayashi et al., 2012) and be mediated by spindle matrix constituents. The accumulation of tubulin in the nucleus under microtubule depolymerization conditions is not a general property of cytoplasmic proteins, as exemplified by the dynactin complex component DNC-1 in the nematode (Hayashi et al., 2012).
A surprising finding of the present study is that nonproteinaceous polysaccharide macromolecules such as dextrans have the ability to be incorporated into the spindle matrix. However, the results of previous studies showed that the spindle pole protein NuMA is highly poly(ADP-ribosyl)ated (Radulescu and Cleveland, 2010) and that poly(ADP-ribose) is required for spindle assembly and function in Xenopus (Chang et al., 2004). Thus it is possible that the size, branching, and charge distribution of such polymeric carbohydrate modifications of spindle matrix proteins might play a role in regulating its assembly and function. Furthermore, these modifications might contribute directly to the viscoelastic properties of the spindle and contribute to the modulation of microtubule dynamics and spindle stabilization.
An issue for the spindle matrix hypothesis has been to account for its molecular composition and structure, especially as the number and diversity of its possible constituents has grown (reviewed in Johansen et al., 2011). In Drosophila, in addition to Megator and Chromator, the nuclear proteins Skeletor, EAST, and Mad2 have been demonstrated to be associated with the spindle matrix (Walker et al., 2000; Qi et al., 2005; Katsani et al., 2008; Lince-Faria et al., 2009; Ding et al., 2009). Another candidate nuclear spindle matrix protein that relocates to the spindle region during mitosis in a microtubule-independent manner is the nucleoporin Nup107 (Katsani et al., 2008). Thus it is becoming clear that during mitosis many disassembled components of interphase nuclear structure do not simply disperse but rather reorganize, making important contributions to mitotic progression (De Souza and Osmani, 2009; Johansen and Johansen, 2007, 2009; Simon and Wilson, 2011). For example, many nuclear pore complex constituents in addition to Megator/Tpr and Nup107 have been demonstrated to relocate to the spindle region in both invertebrates and vertebrates (reviewed in De Souza and Osmani, 2009; Johansen et al., 2011). Of interest, certain nuclear pore proteins have been shown to form a three-dimensional polymer meshwork with hydrogel-like properties within the nuclear pore (Frey et al., 2006). If, as suggested here, the spindle matrix is a similar gel-like assembly of weakly associated protein polymers, its exact stoichiometry and composition may not be critical and it likely would be able to accommodate the inclusion of a wide array of proteins. However, it is important to note that not all nuclear proteins relocate to the spindle matrix during mitosis. For example, both lamin B and C (Paddy et al., 1996; Katsani et al., 2008) disperse, as does the nucleoporin Nup58 (Katsani et al., 2008). Furthermore, in this study we demonstrate that the amino-terminal coiled-coil region of Megator is required for its spindle matrix localization during mitosis, whereas the carboxy-terminal region disperses. In future experiments it will be of interest to determine the nature of the specific molecular interactions that govern which proteins are incorporated into the matrix.
Regardless of the exact composition and structure of the spindle matrix, the demonstration here of a self-contained macromolecular structure embedding the spindle apparatus during mitosis will have important implications for our understanding of microtubule dynamics (Dumont and Mitchison, 2009). Furthermore, in a recent study of the micromechanical properties of the metaphase spindle, the effective viscosity of the spindle region was measured to be ∼100 times higher than in the surrounding cytoplasm (Shimamoto et al., 2011). This difference was attributed largely to the actions of motor and nonmotor proteins cross-linking microtubules, with the assumption of negligible contributions from the spindle medium. However, the results of this study suggest that a gel-like spindle matrix is likely to directly contribute to the viscoelastic mechanical properties of the spindle.
MATERIALS AND METHODS
Drosophila melanogaster stocks and transgenic flies
Fly stocks were maintained according to standard protocols (Roberts, 1998), and Canton S was used for wild-type preparations. Full-length, GFP-tagged Chromator constructs under native or GAL-4 promoter control have been previously characterized (Ding et al., 2009). Tubulin-mCherry, Jupiter-GFP, and lamin-GFP fly stocks (stocks 25774, 6836, and 7378, respectively) and a tubulin-GAL-4 driver line (stock 7062) were obtained from the Bloomington Drosophila Stock Center, Indiana University (Bloomington, IN). The Megator YFP-trap fly line (w[1118]; PBac{602.P.SVS-1}Mtor[CPTI001044]) was obtained from the Drosophila Genetic Resource Center, Kyoto Institute of Technology (Kyoto, Japan; stock 115129). The H2AvDmRFP1 transgenic line was the gift of S. Heidmann and has been previously described (Deng et al., 2005). For the Megator-CTD construct under native promoter control a genomic region of 949 nucleotides upstream and 9 nucleotides downstream of the ATG start codon was PCR amplified and fused with an in-frame GFP tag, as well as with Megator carboxy-terminal coding sequence corresponding to residues 1758–2347, and inserted into the pUAST vector using standard techniques (Sambrook and Russell, 2001). For the Megator-NTD construct under native promoter control the same upstream region as for the Mtor-CTD construct was fused with an in-frame GFP tag, with Megator amino-terminal coding sequence corresponding to residues 1–1757, and with the NLS from the NLS-pECFP vector (Clontech, Mountain View, CA) and inserted into the pPFHW vector (Murphy, 2003) using standard techniques (Sambrook and Russell, 2001). Transgenic Mtor-CTD and Mtor-NTD fly lines were generated by P-element transformation by BestGene (Chino Hills, CA). Fly lines expressing combinations of transgenes were generated by standard genetic crosses.
Time-lapse confocal microscopy and injections
Time-lapse imaging of the fluorescently tagged constructs in live syncytial embryos were performed using a TCS SP5 tandem scanning microscope (Leica, Wetzlar, Germany) or an UltraView spinning-disk confocal system (PerkinElmer, Waltham, MA) as previously described (Ding et al., 2009). In brief, 0- to 1.5-h embryos were collected from apple juice plates and aged 1 h. The embryos were manually dechorinated, transferred onto a coverslip coated with a thin layer of heptane glue, and covered with a drop of halocarbon oil 700. Time-lapse image sequences of a single z-plane or of z-stacks covering the depth of the mitotic apparatus were obtained using a Plan-Apochromat 63×/1.4 numerical aperture objective. For colchicine injections, colchicine (Sigma-Aldrich, St. Louis, MO) was dissolved in dimethyl sulfoxide (DMSO) to a concentration of 100 mg/ml as a stock solution. The final concentration of colchicine for injection was 1 mg/ml by diluting the stock solution with PEM buffer (80 mM Na 1,4-piperazinediethanesulfonic acid, pH 6.9, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, 5% glycerol). Injections of ∼100–200 pl of 1 mg/ml colchicine into each embryo were performed with an IM-300 programmable microinjector system (Narishige, Tokyo, Japan) connected to the Leica confocal TCS SP5 microscope system, as previously described (Brust-Mascher and Scholey, 2009). For Taxol injections, ∼100–200 pl of 20 mg/ml Taxol (Sigma-Aldrich) in DMSO was injected into each embryo. Control injections were performed with DMSO alone or with PEM buffer with 1% DMSO. Fluorescein-labeled dextrans of molecular mass 70, 500, or 2000 kDa (Invitrogen, Carlsbad, CA) were injected into syncytial embryos using standard methods (Brust-Mascher and Scholey, 2009).
Image quantification and analysis
Image processing and quantification were carried out with the ImageJ 1.45 software (National Institutes of Health, Bethesda, MD) or with Photoshop (Adobe, San Jose, CA). QuickTime movies were generated with QuickTime Pro 7.6.6 (Apple, Cupertino, CA). Scatter plots, average pixel intensities of regions of interest, and determination of Pearson's correlation coefficient of the measured fluorescence intensity of line scans generated in ImageJ were performed and calculated using Excel (Microsoft, Redmond, CA).
Supplementary Material
Acknowledgments
We thank members of our laboratory for discussion, advice, and critical reading of the manuscript. We also acknowledge Atrez Norwood for technical assistance. We especially thank S. Heidmann, H. White-Cooper, and J. Roote for providing fly stocks. Work in the laboratory of J.J. and K.M.J. is supported by National Science Foundation Grant MCB0817107, and work in the laboratory of H.M. is supported by the Human Frontier Research Program.
Abbreviations used:
- DMSO
dimethyl sulfoxide
- GFP
green fluorescent protein
- MAP
microtubule-associated protein
- NE
nuclear envelope
- NLS
nuclear localization signal
- ROI
region of interest
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-06-0429) on August 1, 2012.
REFERENCES
- Brust-Mascher I, Scholey JM. Microinjection techniques for studying mitosis in the Drosophila melanogaster syncytial embryo. J Vis Exp. 2009;31:e1382.. doi: 10.3791/1382. Available at: http://www.jove.com/video/1382 (accessed 1 June 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Tao L, Brust-Mascher I, Wollman R, Scholey JM. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J Cell Biol. 2010;188:49–68. doi: 10.1083/jcb.200908150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell. 2009;20:2146–2159. doi: 10.1091/mbc.E08-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Osmani SA. Double duty for nuclear proteins—the price of more open forms of mitosis. Trends Genet. 2009;25:545–554. doi: 10.1016/j.tig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Zhang W, Bao X, Martin JN, Girton J, Johansen J, Johansen KM. The JIL-1 kinase regulates the structure of Drosophila polytene chromosomes. Chromosoma. 2005;114:173–182. doi: 10.1007/s00412-005-0006-8. [DOI] [PubMed] [Google Scholar]
- Ding Y, Yao C, Lince-Faria M, Rath U, Cai W, Maiato H, Girton J, Johansen KM, Johansen J. Chromator is required for proper microtubule spindle formation and mitosis in Drosophila. Dev Biol. 2009;334:253–263. doi: 10.1016/j.ydbio.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol. 2009;19:R749–R761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Richter RP, Görlich D. FG-repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Kimura K, Kimura A. Localized accumulation of tubulin during semi-open mitosis in the Caenorhabditis elegans embryo. Mol Biol Cell. 2012;23:1688–1699. doi: 10.1091/mbc.E11-09-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Johansen KM. The spindle matrix through the cell cycle in Drosophila. Fly. 2009;3:1–8. [PubMed] [Google Scholar]
- Johansen KM, Forer A, Yao C, Girton J, Johansen J. Do nuclear envelope and intranuclear proteins reorganize during mitosis to form an elastic, hydrogel-like spindle matrix? Chromosome Res. 2011;19:345–365. doi: 10.1007/s10577-011-9187-6. [DOI] [PubMed] [Google Scholar]
- Johansen KM, Johansen J. Cell and molecular biology of the spindle matrix. Int Rev Cytol. 2007;263:155–206. doi: 10.1016/S0074-7696(07)63004-6. [DOI] [PubMed] [Google Scholar]
- Karpova N, Bobinnec Y, Fouix S, Huitorel P, Depec A. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil Cytoskeleton. 2006;63:301–312. doi: 10.1002/cm.20124. [DOI] [PubMed] [Google Scholar]
- Katsani KR, Karess RE, Dostatni N, Doye V. In vivo dynamics of Drosophila nuclear envelope components. Mol Biol Cell. 2008;19:3652–3666. doi: 10.1091/mbc.E07-11-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M, Maffini S, Orr B, Ding Y, Florindo C, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J Cell Biol. 2009;184:647–657. doi: 10.1083/jcb.200811012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TD. 2003. The Drosophila Gateway™ Vector Collection. Available at: http://emb.carnegiescience.edu/labs/murphy/Gateway%20vectors.html (accessed 1 June 2012).
- Paddy MR, Saumweber H, Agard DA, Sedat JW. Time-resolved, in vivo studies of mitotic spindle formation and nuclear lamina breakdown in Drosophila early embryos. J Cell Sci. 1996;109:591–607. doi: 10.1242/jcs.109.3.591. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD, Forer A. Mitosis: spindle evolution and the matrix model. Protoplasma. 2009;235:91–99. doi: 10.1007/s00709-009-0030-2. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD, Tippit DH, Porter KR. Rethinking mitosis. Cell. 1982;29:729–744. doi: 10.1016/0092-8674(82)90435-4. [DOI] [PubMed] [Google Scholar]
- Qi H, Rath U, Ding Y, Ji Y, Blacketer MJ, Girton J, Johansen J, Johansen KM. EAST interacts with Megator and localizes to the putative spindle matrix during mitosis in Drosophila. J Cell Biochem. 2005;95:1284–1291. doi: 10.1002/jcb.20495. [DOI] [PubMed] [Google Scholar]
- Qi H, et al. Megator, an essential coiled-coil protein localizes to the putative spindle matrix during mitosis. Mol Biol Cell. 2004;15:4854–4865. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu AE, Cleveland DW. NuMA after 30 years: the matrix revisited. Trends Cell Biol. 2010;20:214–222. doi: 10.1016/j.tcb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath U, Ding Y, Deng H, Qi H, Bao X, Zhang W, Girton J, Johansen J, Johansen KM. The chromodomain protein, Chromator, interacts with JIL-1 kinase and regulates the structure of Drosophila polytene chromosomes. J Cell Sci. 2006;119:2332–2341. doi: 10.1242/jcs.02960. [DOI] [PubMed] [Google Scholar]
- Rath U, Wang D, Ding Y, Xu Y-Z, Qi H, Blacketer MJ, Girton J, Johansen J, Johansen KM. Chromator, a novel and essential chromodomain protein interacts directly with the putative spindle matrix protein Skeletor. J Cell Biochem. 2004;93:1033–1047. doi: 10.1002/jcb.20243. [DOI] [PubMed] [Google Scholar]
- Roberts DB. Drosophila: A Practical Approach. 2nd. Oxford: IRL; 1998. [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sandquist JG, Kita AM, Bement WM. And the dead shall rise: actin and myosin return to the spindle. Dev Cell. 2011;21:410–419. doi: 10.1016/j.devcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y, Maeda YT, Ishiwata S, Libchaber AJ, Kapoor TM. Insights into the micromechanical properties of the metaphase spindle. Cell. 2011;145:1062–107. doi: 10.1016/j.cell.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic “network of networks.”. Nat Rev Mol Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- Stafstrom JP, Staehelin LA. Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the Drosophila embryo. Eur J Cell Biol. 1984;34:179–189. [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Walker DL, Wang D, Jin Y, Rath U, Wang Y, Johansen J, Johansen KM. Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. J Cell Biol. 2000;151:1401–1411. doi: 10.1083/jcb.151.7.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Ding Y, Cai W, Wang C, Girton J, Johansen KM, Johansen J. The chromodomain-containing NH2-terminus of Chromator interacts with histone H1 and is required for correct targeting to chromatin. Chromosoma. 2012;121:209–220. doi: 10.1007/s00412-011-0355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. A membranous spindle matrix orchestrates cell division. Nat Rev Mol Cell Biol. 2010;11:529–535. doi: 10.1038/nrm2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska G, Aris JP, Paddy MR. A Drosophila Tpr homolog is localized both in the extrachromosomal channel network and to the nuclear pore complexes. J Cell Sci. 1997;110:927–944. doi: 10.1242/jcs.110.8.927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.