TBCB is a tubulin-binding cofactor required for tubulin heterodimerization. We show that in Drosophila, TBCB is required to sustain the levels of both α- and β-tubulins and for microtubule network formation in different tissues. Strikingly, TBCB is essential for cell polarity but not for cell division.
Abstract
Microtubules (MTs) are essential for cell division, shape, intracellular transport, and polarity. MT stability is regulated by many factors, including MT-associated proteins and proteins controlling the amount of free tubulin heterodimers available for polymerization. Tubulin-binding cofactors are potential key regulators of free tubulin concentration, since they are required for α-β–tubulin dimerization in vitro. In this paper, we show that mutation of the Drosophila tubulin-binding cofactor B (dTBCB) affects the levels of both α- and β-tubulins and dramatically destabilizes the MT network in different fly tissues. However, we find that dTBCB is dispensable for the early MT-dependent steps of oogenesis, including cell division, and that dTBCB is not required for mitosis in several tissues. In striking contrast, the absence of dTBCB during later stages of oogenesis causes major defects in cell polarity. We show that dTBCB is required for the polarized localization of the axis-determining mRNAs within the oocyte and for the apico-basal polarity of the surrounding follicle cells. These results establish a developmental function for the dTBCB gene that is essential for viability and MT-dependent cell polarity, but not cell division.
INTRODUCTION
Microtubules (MTs) are highly dynamic structures crucial for many cellular processes, such as cell division and cell polarity. MTs consist of α-β–tubulin heterodimer stacks (Wade, 2009) that generate a polarized structure. MT minus ends are usually stabilized at an MT-organizing center, whereas their plus ends are often highly dynamic, oscillating between phases of polymerization and depolymerization, a process known as dynamic instability (Mitchison and Kirschner, 1984; Desai and Mitchison, 1997). This instability triggers constant remodeling of the MT network in cells and is strictly regulated. An important mode of regulation involves MT-associated proteins (MAPs), which are distributed along the lattice (Amos and Schlieper, 2005) or restricted to growing MT plus ends (Akhmanova and Steinmetz, 2008, 2010). A second important mode of regulation of MT dynamics involves factors controlling the accessibility of free tubulin heterodimers. For instance, OP18/stathmin prevents MT growth by sequestering soluble heterodimers, thereby decreasing the concentration of tubulin molecules available for polymerization (Cassimeris, 2002; Holmfeldt et al., 2009).
Newly synthesized α- and β-tubulin monomers are labile molecules unable to fold and dimerize without the help of the cytosolic chaperonin and tubulin-binding cofactors (TBCs; Lopez-Fanarraga et al., 2001). In vitro, five TBCs (TBCA to TBCE) cooperate to trigger the release of functional α-β–tubulin dimers (Figure 1A; Tian et al., 1996, 1997). TBCs are thought to regulate MT dynamics by modulating the concentration of dimers available for polymerization (Lopez-Fanarraga et al., 2001). Although not essential in vitro, TBCB may enhance tubulin folding by transferring α-tubulin monomers to TBCE (Figure 1A; Tian et al., 1996, 1997).
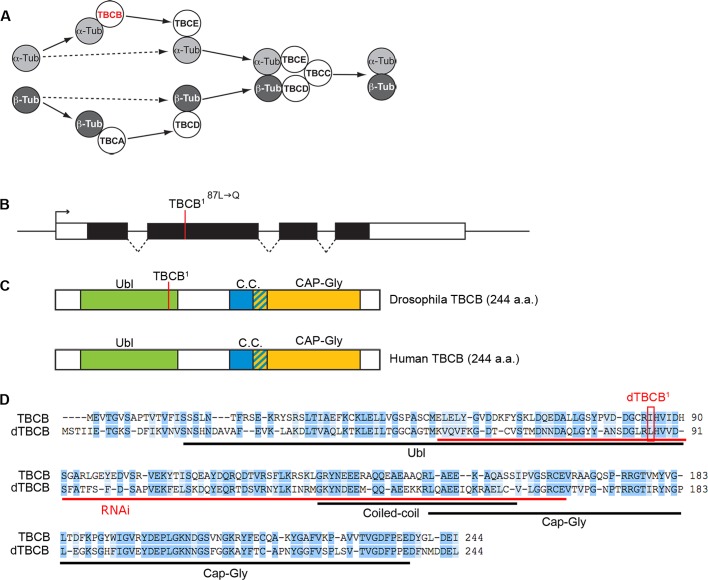
FIGURE 1:
Drosophila TBCB. (A) Schematic diagram of the conserved TBCs required for tubulin heterodimerization. TBCB is depicted in red. (B) Schematic diagram of the CG11242 locus, which encodes the Drosophila orthologue of dTBCB. The genomic dTBCB locus is shown with introns (dashed lines), exons (black boxes), and UTRs (open boxes). The dTBCB1 mutation corresponding to a T⇒A transition is located in the second exon. (C) The dTBCB protein, like its human orthologue, contains a ubiquitin-like domain (Ubl; green), a coiled-coil (C.C.; blue), and a CAP-Gly domain (orange). The L87-to-Q transition corresponding to the dTBCB1 mutation is located in the Ubl domain. (D) Alignment of Drosophila and human TBCB amino acid sequences. Identical amino acids are highlighted in dark blue and similar amino acids are highlighted in light blue. The dTBCB1 mutation is boxed in red. The sequence used as an inverted repeat for RNAi is highlighted in red.
Conversely, TBCB is also able to form a binary complex with TBCE that enhances the efficiency of TBCE to dissociate tubulin heterodimers in vitro. TBCB therefore has a potential role in the degradation or recycling of tubulin (Kortazar et al., 2007).
The function of TBCB in vivo is far less clear. In Schizosaccharomyces pombe, a knockout of TBCB results in a specific decrease of α-tubulin levels that correlates with an affected MT network and defects in cell division (Radcliffe et al., 1999; Radcliffe and Toda, 2000). In several mammalian cell types, however, TBCB knockdown does not affect tubulin levels nor does it destabilize the MT network (Vadlamudi et al., 2005; Lopez-Fanarraga et al., 2007). In fact, TBCB gene silencing, using RNA interference (RNAi), enhances MT density in amoeboid microglia and stimulates axonal growth in neuronal cells (Lopez-Fanarraga et al., 2007; Fanarraga et al., 2009). These results are consistent with the implication of mammalian TBCB in tubulin heterodimer dissociation but raise the question of whether TBCB is required for tubulin dimerization and MT network formation in a metazoan organism. It is also unclear whether this protein is involved in cell division and polarity during development. Moreover, some studies have reported an association of TBCB with MTs and centrosomes (Feierbach et al., 1999; Vadlamudi et al., 2005). However, this distribution remains a matter of debate (Radcliffe et al., 1999; Kortazar et al., 2007; Lopez-Fanarraga et al., 2007; Fanarraga et al., 2009). A functional study of TBCB in a metazoan organism would therefore help to clarify the role of this protein in vivo.
Drosophila has been extensively used to study MT-dependent processes during development. During oogenesis, cyst divisions, oocyte differentiation, and establishment of the two main body axes of the future embryo all depend on MTs and polarized transport (Cooley and Theurkauf, 1994; Huynh and St Johnston, 2004; Becalska and Gavis, 2009). MTs are also essential for the apico-basal polarity of follicle cells, the somatic epithelial cells surrounding the germ cells (St Johnston and Ahringer, 2010). However, molecules triggering MT network organization and remodeling during oogenesis remain largely unknown. TBCs, by modulating the concentration of tubulin dimers available for MT polymerization, are possible candidates for regulating specific cellular functions during oogenesis, as well as other developmental processes. Indeed, dTBCE, the only tubulin cofactor studied in flies, was shown to be required for the normal development of neuromuscular synapses (Jin et al., 2009).
The Drosophila genome contains a single TBCB orthologue, annotated as CG11242 (Tweedie et al., 2009). Using mutant flies, we show that dTBCB is essential for viability, MT network integrity, and cell polarity but, surprisingly, not for cell proliferation. During oogenesis, loss of dTBCB causes delocalization of axis-determining mRNAs at midoogenesis and of PAR complex components in follicle epithelial cells. Throughout development, cell division can still occur, although probably with a considerable delay, as revealed in dividing neuroblasts. We therefore propose that dTBCB enhances tubulin dimer assembly in vivo, thereby regulating cell polarity and development of multicellular organisms.
RESULTS
CG11242 encodes a Drosophila orthologue of human TBCB
The CG11242 gene encodes a protein that we named dTBCB on the basis of its high degree of sequence similarity to TBCB proteins from yeast, plants, and mammals (Tian et al., 1997; Feierbach et al., 1999; Radcliffe et al., 1999; Dhonukshe et al., 2006). CG11242 and human TBCB share 61% similar residues, with 43% identical (Figure 1, B–D). Similar to its human orthologue, dTBCB has three conserved structural motifs: a ubiquitin-like domain (Ubl), a coiled-coil, and a cytoskeleton-associated protein (CAP)-glycine rich (Gly) domain (Figure 1, C and D). As a first step in investigating the in vivo function of dTBCB, we took advantage of genome-wide transgenic RNAi libraries in Drosophila combined with the Gal4/UAS system (Dietzl et al., 2007) to inactivate dTBCB. Ubiquitous overexpression of UAS-RNAi-dTBCB with daughterless-Gal4 (da-Gal4) strongly reduced viability, especially at the pupal stage (43% lethality, n = 539; Figure 2A). Western blot analysis with a polyclonal antibody that we raised against the full-length protein showed that this RNAi significantly reduces dTBCB levels at the larval stage (Figure 2B). Most of the flies that reached adulthood harbored altered wings (91% of the flies, n = 305; Figure 2, A and D). These particular effects of TBCB on pupal lethality and wing development were also obtained with engrailed-Gal4 (en-Gal4) and apterous-Gal4 (ap-Gal4) drivers (Figure 2, A and D).
FIGURE 2:
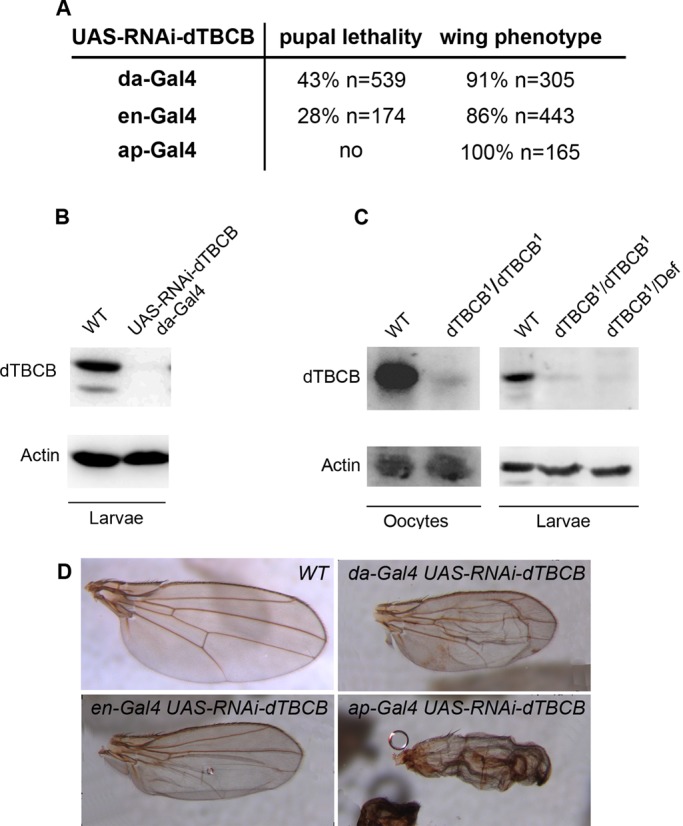
dTBCB is an essential gene. (A) Pupal and adult phenotypes obtained with UAS-RNAi-dTBCB flies combined with various transgenic Gal4 drivers: da-Gal4 is expressed ubiquitously in wing disks, en-Gal4 is expressed in the posterior compartment, and ap-Gal4 is expressed in the dorsal compartment. (B) Western blot of dTBCB RNAi knockdown transgenic flies. Third-instar larval stage extracts showing dTBCB protein levels. UAS-RNAi-dTBCB; da-Gal4: larvae carrying one copy of UAS-RNAi-dTBCB and one copy of da-Gal4. (C) Western blot of stage 14 oocyte and second-instar larval stage extracts showing dTBCB protein levels. dTBCB1/dTBCB1: homozygous mutant larvae or germ-line clones. dTBCB1/Def: larvae carrying one copy of dTBCB1 over a deficiency removing the dTBCB locus. (B and C) Actin is used as a loading control. (D) Wing phenotypes obtained with UAS-RNAi-dTBCB flies combined with various transgenic Gal4 drivers. Wings are orientated with the anterior compartment to the top.
To obtain a different knockdown of dTBCB, we took advantage of a chemical mutagenesis aimed at identifying new mutations affecting MT organization and polarized transport in the oocyte (Gervais et al., 2008). We identified a mutation that we named dTBCB1 (see Materials and Methods), which is homozygous lethal at the end of the second-instar larval stage. Clonal analysis of this allele reveals that dTBCB is also essential during oogenesis for egg chamber development (see below; Figures 4 and 6 later in the paper). In dTBCB1, the mutated Leu-87 is located in the Ubl domain and results in the insertion of a glutamine, a polar residue, in a hydrophobic environment. This mutation affects a very conserved residue predicted to be required for the tertiary structure stabilization of the domain (Lytle et al., 2004; Figure 1, B–D).
FIGURE 4:
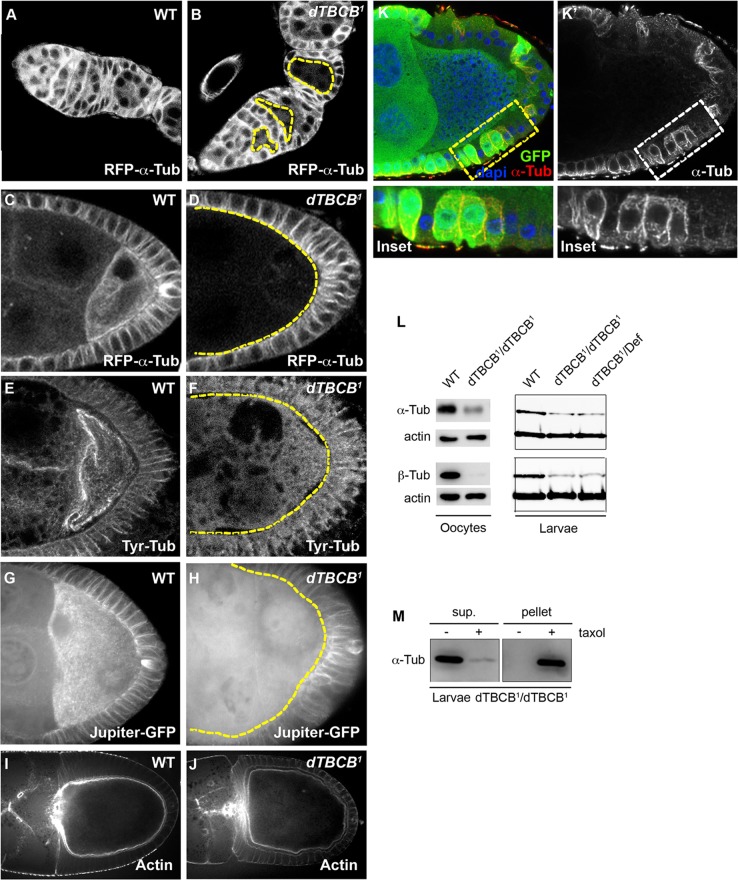
dTBCB is required for the integrity of the MT network. (A and B) MT distribution during the initial stages of oogenesis. dTBCB1 mutant cysts are delimited by yellow dashed lines. MTs were detected with a Ubi-α-RFP-tubulin transgene. dTBCB1 mutant cells highlighted by yellow dashed lines are marked by the absence of GFP (unpublished data). Germaria are oriented with anterior to the left. (C–H) MT distribution at stages 9–10 in wild-type (C, E, and G) and in dTBCB1 mutant (D, F, and H) oocytes. MTs were detected with Ubi-α-RFP-tubulin (C and D), an antibody specific for the tyrosinated form of α-tubulin (E and F) and the Jupiter MAP fused to GFP (G and H). All egg chambers were fixed, except (G and H), which are living egg chambers. Z-projections of stacks of 15 frames taken every 0.5 μm are shown (G and H). (I and J) Actin distribution at stages 9–10 in wild-type (J) and dTBCB1 mutant oocytes (K). F-actin is stained with phalloidin. (K and K′) MT distribution in follicle cells at a stage 9. dTBCB1 mutant cells are marked by the absence of GFP (green). MTs are stained with an anti–α-tubulin antibody (red in L and white in L′). Dashed boxes indicate the cells enlarged in the insets. DNA is shown in blue. (L) Western blot analysis to quantify α- and β-tubulin levels in stage 14 oocytes and second-instar larvae. dTBCB1/dTBCB1: homozygous larvae. dTBCB1/Def: larvae carrying one copy of dTBCB1 over a deficiency removing the dTBCB locus. Actin is used as a loading control. (M) MT pulldown experiments in the presence or absence of Taxol, in dTBCB1 larvae (sup, supernatant). The remaining tubulins present in dTBCB1 larvae are able to polymerize into MTs, indicating they are present as heterodimers.
FIGURE 6:
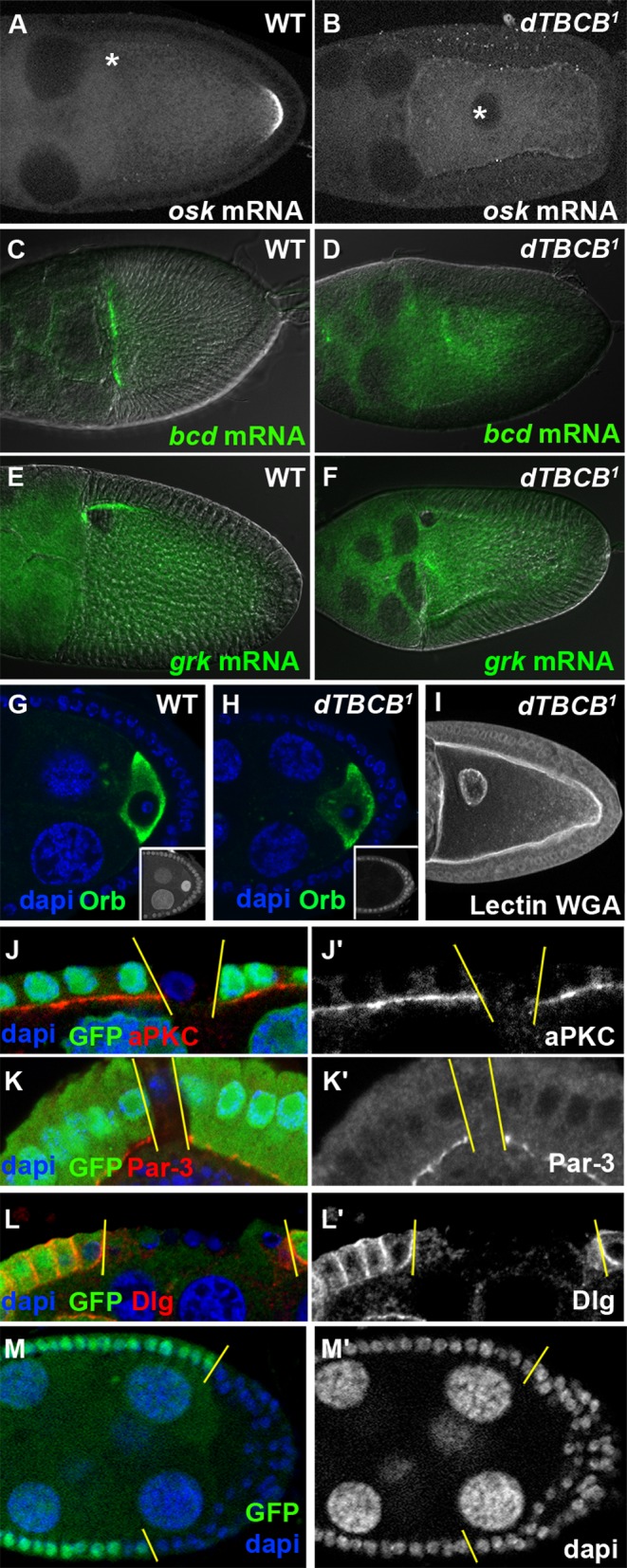
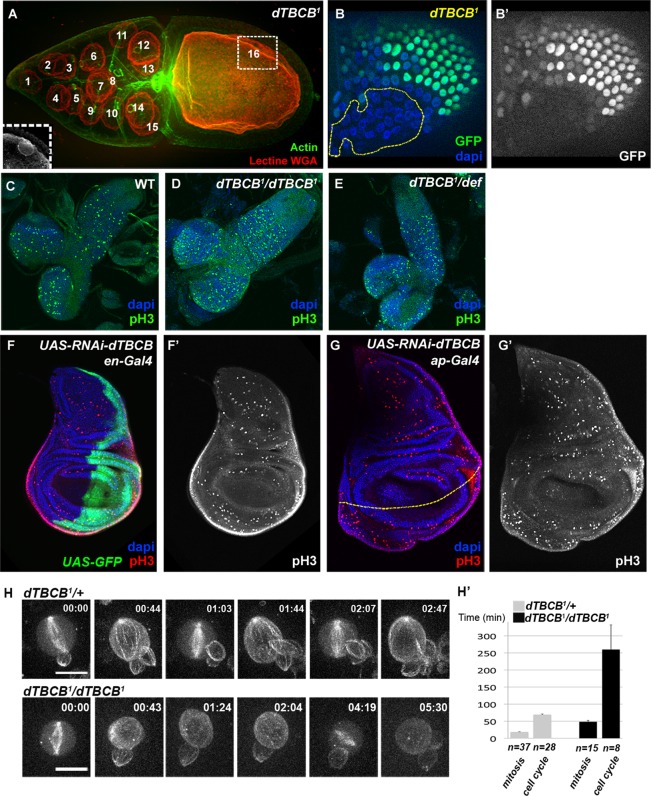
dTBCB is required for cell polarity. (A–F) Asymmetric transport in wild-type (A, C, and E) and in dTBCB1 mutant (B, D, and F) stage 10 oocytes. (A and B) osk mRNA, (C and D) bcd mRNA, and (E and F) grk mRNA. (C–F) DIC images were used to visualize chambers. Asterisk indicates oocyte nucleus (A and B). (G and H) Asymmetric accumulation of the Orb protein during stage 4 oocytes. Orb protein (green) specifically marks the oocyte. Insets show specific absence of nuclear GFP characterizing dTBCB1germ-line clone. DNA is shown in blue. (I) dTBCB1 mutant stage 10 oocyte with delocalized nucleus. Secreted proteins and the nucleus were detected with wheat germ agglutinin (WGA) lectin (white). (J–M′) Apico-basal polarity in follicle cells. dTBCB1 mutant cells are revealed by the absence of GFP (green in J, K, L, and M). The apical side of the epithelium is facing down (J–L′). Staining of the apical markers aPKC (J and J′, red and white) and Par3 (K and K’, red and white) and of the lateral marker Dlg (L and L′, red and white). (M and M′) dTBCB1 mutant follicle cells can form a disorganized multistratified epithelium. DNA is shown in blue or white. Chambers (A–I, M, and M′) are oriented with anterior to the left.
In dTBCB1 mutant larvae, dTBCB transcript detected by reverse transcriptase PCR is present at a normal level, indicating that transcription and stability of the mRNA are not affected (unpublished data). By contrast, Western blot analysis showed that dTBCB protein was almost undetectable in dTBCB1 ovarian and late second-instar larval extracts (Figure 2C). Furthermore, the larval lethality stage observed with homozygous dTBCB1 is similar to the one observed with dTBCB1 over a deficiency removing the complete dTBCB locus. Importantly, this lethality is fully rescued by a transgene expressing a genomic fragment encompassing dTBCB locus (see Materials and Methods). Taken together, these data led us to conclude that dTBCB1 mutation seems to abolish dTBCB function and is most likely amorphic. These results give the first striking evidence that TBCB is essential for the viability of a metazoan organism.
dTBCB is not restricted to the cytosolic compartment
As the intracellular localization of TBCB remains a matter of debate (Feierbach et al., 1999; Radcliffe et al., 1999; Vadlamudi et al., 2005; Kortazar et al., 2007; Lopez-Fanarraga et al., 2007; Fanarraga et al., 2009), we investigated its cellular localization. We generated transgenic flies ubiquitously expressing dTBCB fused to green fluorescent protein (GFP). By expressing this transgene in wild-type flies, we revealed that dTBCB-GFP is distributed mainly in the cytoplasm and in the nuclei during oogenesis (Figure 3, A–B′′). However, we found that a fraction of the protein is associated with the cell cortex, more particularly along the anterior margin of the oocyte, where MTs are particularly enriched (Figure 3, A–B′′). We found that dTBCB1 homozygous mutant flies expressing dTBCB-GFP are viable and fertile, indicating that the transgene is functional. Moreover, a similar expression pattern of dTBCB-GFP was obtained in the mutant background, suggesting that it reflects the endogenous dTBCB expression (Figure S1, B–B′′). To determine dTBCB distribution during mitosis, we live-imaged dTBCB-GFP in wild-type syncytial embryos in which a large set of nuclei dividing synchronously is particularly suitable to visualize mitotic MTs (Figure 3, C–D′′). In addition to being distributed in the cytoplasm, dTBCB-GFP can be observed in the nucleus, at the duplicated centrosomes during prophase (Figure 3, C–C′′), and along the mitotic spindle during metaphase/anaphase (Figure 3, D–D′′). We observed a similar distribution in fixed S2R+ Drosophila cells (Figure 3, E–F′′). Taken together, these results point to a complex subcellular dTBCB distribution. The cytosolic distribution of dTBCB is in accordance with previous reports (Feierbach et al., 1999; Radcliffe et al., 1999; Kortazar et al., 2007; Lopez-Fanarraga et al., 2007) and consistent with its requirement for the tubulin heterodimer assembly. The nuclear and the MT distributions may point to additional functions for TBCB.
FIGURE 3:
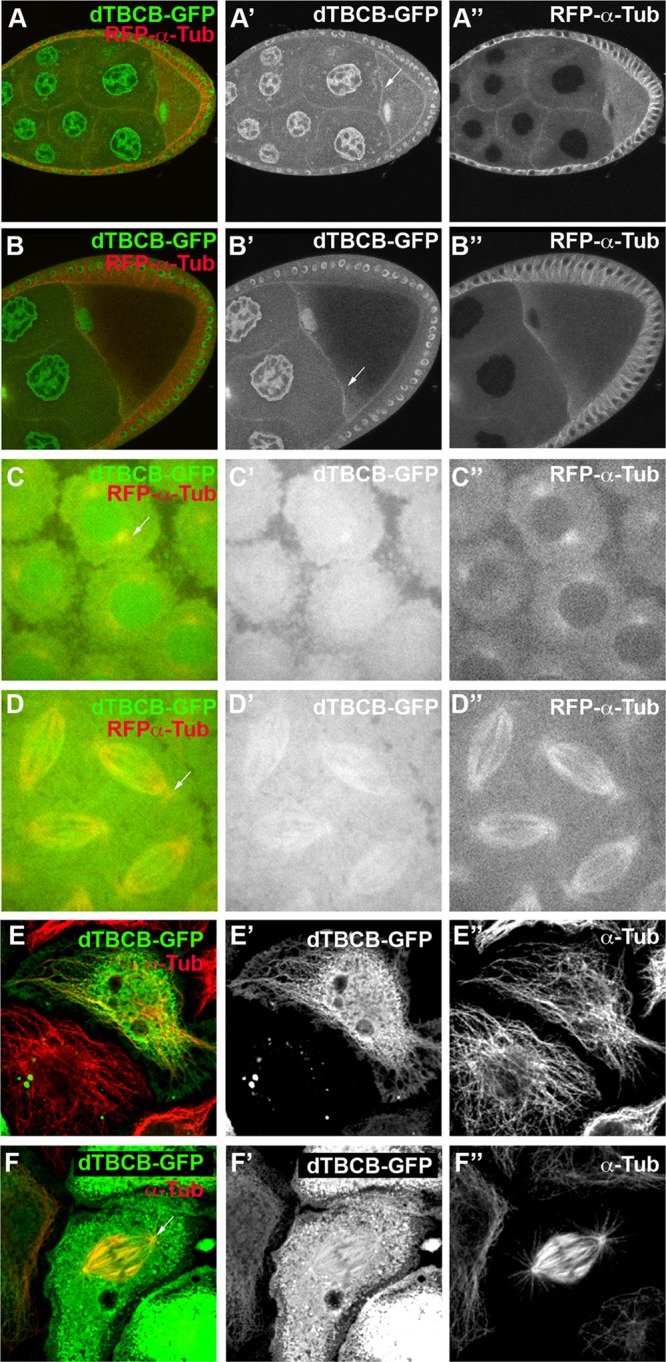
Cellular distribution of dTBCB. (A–A′′ and B–B′′) Immunofluorescence of stage 8 and 9 eggs expressing a Ubi-dTBCB-GFP transgene (green) and a Ubi-α-RFP-tubulin transgene (red). Chambers are oriented with anterior to the left. White arrows highlight the anterior margin of the oocyte. (C–D′′) Live image of a mitotic syncytial embryo coexpressing the Ubi-dTBCB-GFP (green) and Ubi-α-RFP-tubulin (red) transgenes. (C–C′′) Mitotic syncytial embryo during prophase. White arrow highlights a centrosome. (D–D′′) Mitotic syncytial embryo during metaphase/anaphase. White arrow highlights a centrosome. (E–F′′) Immunostaining of S2R+ cells expressing a Ubi-dTBCB-GFP transgene (green). MTs were detected with a α-tubulin antibody (red). (E–E′′) S2R+ cell during interphase. (F–F′′) S2R+ cell during mitosis. White arrow highlights a centrosome.
dTBCB is required for the integrity of the MT network
The involvement of TBCB from several species in α-β–tubulin dimerization (Tian et al., 1997; Radcliffe et al., 1999), raises the question of the role of dTBCB in MT regulation. Using dTBCB1 mutant germ-line clones and a constitutively expressed red fluorescent protein (RFP)–α-tubulin transgene for MT visualization, we observed that the MT network decreases significantly in dTBCB1 oocytes and nurse cells from early to late oogenesis, although a few MTs remain detectable (Figure 4, A–D). A fluorescence profile analysis for RFP−α-tubulin between dTBCB1 and wild-type germ-line cysts indicated that the diminution is ∼43% at any developmental stage (Supplemental Table S1). MT defects were also observed in vivo with a transgene encoding a MAP (Jupiter) fused to GFP (Karpova et al., 2006; Figure 4, G and H; Supplemental Movies S1 and S2) and in fixed tissues with an antibody specific for the tyrosinated form of α-tubulin (Figure 4, E and F). Moreover, actin cytoskeleton was not affected in dTBCB1 germ cells, indicating that the MT defects were not linked to actin disorganization (Figure 4, I and J). The clonal depletion of dTBCB also strongly affects the interphasic MT network in the epithelial follicle cells surrounding the oocyte, demonstrating that dTBCB is also required for MT integrity in somatic cells (Figure 4, K and K′). All these MT defects in dTBCB1 mutant clones are specific, since dTBCB1 mutant flies rescued by a dTBCB-GFP transgene harbor normal MT network (Supplemental Figure S1, B–B′′; 100%, n = 64).
As TBCB is predicted to stabilize α-tubulin by directing monomers to the heterodimerization pathway, we then investigated whether the observed MT defects were correlated with a decrease in cellular tubulin levels. In mutant larvae homozygotic for dTBCB1 or transheterozygous for dTBCB1 and a deficiency, levels of total α- and β-tubulin were 50% lower than in the wild type (Figure 4L). In dTBCB1 germ-line clones, levels of total α- and β-tubulin were also significantly reduced (from 30 to 60%; Figure 4L). Thus, although dTBCB is known to associate specifically with α-tubulin, it also indirectly maintains levels of β-tubulin.
We then investigated whether the remaining tubulins were monomers or successfully assembled heterodimers capable of polymerizing into MTs. In dTBCB1 mutant larval extracts, virtually all the residual α-tubulin can be pulled down following the addition of the MT-stabilizing agent Taxol (Figure 4M). This suggests that, in dTBCB1 mutant extracts, most of the residual α-tubulin is binding β-tubulin, forming functional heterodimers that can assemble into MTs. These results suggest that, although strongly affected, tubulin dimerization can still partially occur in dTBCB1 mutants. TBCB also participates, together with TBCE, in the dissociation of tubulin heterodimers (Kortazar et al., 2007). Accordingly, TBCB overexpression in yeast and mammals was shown to trigger MT network destabilization (Radcliffe et al., 1999; Wang et al., 2005; Kortazar et al., 2007; Fanarraga et al., 2009). Similarly, we observed that a strong overexpression of dTBCB-GFP in S2R+ Drosophila cells led to MT depolymerization (Figure S2).
Overall our results indicate that, in Drosophila, TBCB is essential for MT network integrity and for the maintenance of tubulin levels in various cell types and at various stages of development. These results could be explained by a reduced tubulin dimerization in dTBCB1 mutant cells.
dTBCB modulates cell cycle duration during development
The importance of dTBCB for MT integrity throughout oogenesis led us to assess more precisely the function of this protein during the different steps of egg chamber development, especially during cell proliferation. In the germ line, at the level of the germarium, a stem cell divides asymmetrically to self-renew and to produce a cystoblast that will undergo four synchronous divisions without cytokinesis to produce a 16-cell cyst. This cyst will differentiate into one oocyte and fifteen nurse cells. We found that clonal dTBCB1 cystoblasts always divide correctly into 16-cell germ-line cysts (Figure 5A). It is important to mention that this normal development of mutant germ-line cells seems unlikely to be due to persistence of the wild-type maternal TBCB or tubulin, because mutant clones were always studied from 13 to 28 d after their induction, and MT defects were always detectable, even in the germarium (Figure 4B). In the somatic follicle epithelium that surrounds the germ-line cyst, cell proliferation occurs from stage 1 to stage 6 of oogenesis. The recovery of large dTBCB1 follicle cell mutant clones (Figures 5, B and B′, and 6, M and M′) suggests dTBCB is also not essential for cell division in somatic cells.
FIGURE 5:
dTBCB is not essential for cell proliferation but modulates cell cycle duration during development. (A) dTBCB1 mutant stage 10 germ-line cyst. Nuclei were detected with WGA lectin (red). Actin labeling (green) reveals cellular outline. The 16 nuclei of the cyst are numbered. The oocyte nucleus (numbered 16) is in a different focal plane, which is shown in the inset. (B and B′) Cell proliferation is not impaired in dTBCB1 mutant follicle cells. A large dTBCB1 mutant follicle cell clone, GFP negative, is highlighted by a yellow dashed line. DNA is shown in blue and GFP in green and white. (C–E) Immunostainning of second-instar larval brains. Mitosis is detected with a pH3 antibody (green). DNA is shown in blue. (C) Wild type, (D) dTBCB1/dTBCB1: homozygous larvae, (E) dTBCB1/Def: larvae carrying one copy of dTBCB1 over a deficiency removing the dTBCB locus. (F–G′) Immunostainning of third-instar larval wing disks. Mitosis is detected with a pH3 antibody (red and white). DNA is shown in blue. (F–F′) UAS-RNAi-dTBCB flies combined with en-Gal4. (F) The expression domain of en-Gal4 at the posterior side is revealed by a UAS-GFP transgene (green). (G–G′) UAS-RNAi-dTBCB flies combined with ap-Gal4. (G) The dorso-ventral border of the wing disc is highlighted by a yellow dashed line. The expression domain of ap-Gal4 at the dorsal compartment stands at the upper side of the line. (H–H′) Selected time-lapse series of neuroblasts from cultured larval explant brains expressing the Ubi-α-RFP-tubulin transgene. Successive cell cycles are shown for the dTBCB1/+ control and the homozygous dTBCB1 mutant neuroblasts, with consecutive mitosis (frames 1, 3, and 5 for the control; frames 1 and 5 for the mutant). In the mutant conditions, the laser was increased for better visualization of MTs. (H′) Comparison of either cell cycle length (between two successive nuclear breakdowns) or mitosis length (nuclear breakdown to cytokinesis) in dTBCB1/+ control and homozygous dTBCB1mutant neuroblasts. n corresponds either to the number of mitoses or to the number of cell cycles. For dTBCB1/+, 10 independent cells were analyzed, and for homozygous dTBCB1, 20 independent cells were analyzed. Time stamp is hh:mm. Scale bar: 10 μm.
To independently corroborate these results, we tested whether dTBCB is dispensable for mitosis in other highly proliferative tissues, such as imaginal discs and larval brains. In third-instar larvae dTBCB1 homozygous mutant brains (n = 11) or transheterozygous for dTBCB1 and a deficiency (n = 18), we observed that the mitotic index, revealed by anti-phosphohistone H3 (anti-pH3), was similar to control brains (n = 18; Figure 5, C–E). Moreover, specific depletion of dTBCB in the posterior compartment of wing imaginal disc by expressing UAS-RNAi-dTBCB with engrailed-Gal4, which compromises adult wing development (Figure 2, A and D), does not prevent cells entering mitosis, as indicated by positive staining for pH3 in larval discs (Figure 5, F and F′). A similar result is also observed when dTBCB is specifically depleted in the dorsal compartment of the wing disc by expressing UAS-RNAi-dTBCB with apterous-Gal4 (Figures 2, A and D, and 5, G and G′). These results suggest dTBCB is not essential for cell proliferation during fly development.
To directly test the requirement for dTBCB in cell division, we then analyzed larval neuroblasts in culture by time-lapse microscopy. Both dTBCB1 homozygous and control dTBCB1/+ heterozygous larval neuroblasts were able to form mitotic spindles and to divide (Figure 5H; Supplemental Movies S3 and S4). However, while divisions in control heterozygous cells showed normal timing of mitosis and cell cycle, consistent with previous reports (Rebollo et al., 2007), homozygous mutant cells showed a delayed cell cycle progression. The majority of the mutant cells (13 out of 20; 65%) were delayed in interphase (4.2-fold increase), as well as in mitosis (2.6-fold increase; Figure 5H). The remaining homozygous mutant cells were either arrested in metaphase or in interphase (respectively 4 and 3 cells out of 20; unpublished data). Overall these data show that the majority of dTBCB1 mutant neuroblasts can divide, although with a delayed cell cycle progression.
dTBCB is required for cell polarity
In addition to mitosis, MTs are essential for cell polarity, a key feature for multicellular organism development. In the oocyte, from midoogenesis onward, MT-dependent polarized transport was strongly affected by the absence of dTBCB. At stages 9–10, osk, bcd, and grk mRNAs were not located at the posterior, anterior, and antero-dorsal poles of the oocyte, respectively (Figure 6, A–F; Becalska and Gavis, 2009). This mislocalization was fully penetrant for osk mRNA (76 of 76 oocytes examined), and highly penetrant for bcd and grk mRNAs, with residual localization occurring in only 18 of 36 and 13 of 37 oocytes, respectively. The antero-dorsal positioning of the nucleus was also impaired at this stage in 70% of the chambers (n = 64; Figure 6, B and I, asterisk in B). We then searched for polarity defects in earlier stages. During the first stages of oogenesis, accumulation of the Orb protein, which is a hallmark of differentiation of one of the 16 germ cells into an oocyte, is dependent on the MT cytoskeleton (Huynh and St Johnston, 2004). Interestingly, this process is not affected in dTBCB1 mutant cysts (Figure 6, G and H). This absence of phenotype was not due to protein persistence, since the mutant clones were analyzed up to 28 d after their induction, and MT defects were always detectable, even in the germarium (Figure 4B). Thus, dTBCB seems specifically required for oocyte cell polarity at midoogenesis.
We next investigated the role of dTBCB in the apico-basal polarity of the epithelial follicle cells (Muller, 2000; St Johnston and Ahringer, 2010). We observed that the distribution of the apical determinants PAR-3 and atypical protein kinase C (aPKC) and of the basolateral determinant DLG were completely disrupted in dTBCB1 mutant clones (Figure 6, J–L′). Consistent with cell polarity defects, we frequently observed large dTBCB1 mutant follicle cell clones forming multilayers (Figure 6, M and M′). We confirmed that these polarity defects were due to the absence of dTBCB by expressing a dTBCB-GFP transgene in dTBCB1 mutant cells. This construct rescued antero-posterior polarity in stage 9–10 oocytes (Figure S1, D–D′′; 100%, n = 62) and apico-basal polarity in the follicle cells (Figure S1, F–F′′; 100%, n = 66). Taken together, these results indicate that, in flies, an essential function of TBCB is to sustain some aspects of intracellular polarity.
To substantiate that the impaired polarity of dTBCB1 mutant cells is due to MT destabilization, we tested for a genetic interaction between dTBCB and the ubiquitously expressed α-tubulin 84B. Flies double heterozygous for dTBCB1 and α-tubulin 84B5 mutations were viable, but displayed a strongly reduced fertility (Figure S3A) and an attenuated aPKC localization in follicle cells compared with single heterozygote mutants (Figure S3, B–F). Overall these results indicate that, in Drosophila, dTBCB is required to generate a functional MT network that can sustain cell polarity and development.
DISCUSSION
The TBCB protein is part of an evolutionarily conserved tubulin-folding pathway crucial for the formation of the tubulin heterodimers. Indeed, TBCs are thought to regulate MT dynamics by modulating the concentration of dimers available for polymerization (Lopez-Fanarraga et al., 2001). Using Drosophila as a model system, we looked at the requirement for TBCB during development. We show that dTBCB is required for MT integrity and is essential for viability, MT-associated transport, and cell polarity.
TBCs are mostly cytoplasmic proteins, in accordance with their tubulin-chaperoning function (Lopez-Fanarraga et al., 2001). However, TBCE also accumulates at the Golgi apparatus of motor neurons, where it is essential for axonal tubulin routing (Schaefer et al., 2007). TBCD is concentrated at the centrosome, midbody, and cell junctions, where it participates in centriologenesis, spindle organization, cell abscission, and epithelial cell structure (Cunningham and Kahn, 2008; Shultz et al., 2008; Fanarraga et al., 2010). Our results indicate that dTBCB is largely cytoplasmic but also partly overlaps with MTs, corroborating some previous studies in different species (Feierbach et al., 1999; Vadlamudi et al., 2005; Kortazar et al., 2007). This partial colocalization is particularly clear in developing embryos and in S2 cells. Such distribution is not observed with dTBCE, the α-TBC interacting with TBCB (Jin et al., 2009). This possibly indicates that dTBCB fulfills additional functions independent of the other TBCs. Further experiments will be required to address these potential functions.
We show that dTBCB is required for the integrity of the MT network in different tissues. This is consistent with data obtained in yeast knockout, but not in mammalian knockdown cells (Radcliffe et al., 1999; Lopez-Fanarraga et al., 2007; Fanarraga et al., 2009). TBCB has been shown, together with TBCE, to stimulate tubulin heterodimer dissociation in vitro (Kortazar et al., 2007). In accordance with this, we found that strong dTBCB overexpression in S2 cells triggers MT depletion. The increased MT density observed in mammalian microglia, after TBCB depletion by RNAi, might therefore be due to a reduced tubulin heterodimer dissociation. The different phenotypes reported in previous studies might be due to species specificity or to the efficiency of the technical approaches, with the mutational approach probably being more efficient than RNAi. We propose that severely impaired tubulin dimerization is the cause of the strong MT defects in yeast and Drosophila. In mammalian knockdown cells, however, a sufficient pool of tubulin heterodimers is produced, and only the dissociation of the tubulin heterodimer seems affected. Altogether these results suggest that TBCB is required for tubulin dimerization and for tubulin heterodimer dissociation in vivo. The latter function might be more sensitive to variations in TBCB concentration and could be used to fine-tune MT polymerization in a spatiotemporal manner.
The α-TBC TBCE, a direct partner of TBCB, is required for viability and MT formation in Drosophila but does not affect α-tubulin levels (Jin et al., 2009). In dTBCB1 mutant egg chambers and larvae, however, we observed a strong reduction of α-tubulin levels. This suggests that the monomeric α-tubulins, which failed to bind dTBCB and to dimerize, are unstable. This is consistent with the fact that monomeric tubulins have been observed to be very unstable molecules in vitro (Tian et al., 1996, 1997). We also observed that the β-tubulin levels were reduced in dTBCB1, although TBCB is known to associate specifically with α-tubulin. This suggests that the monomeric form of β-tubulin in Drosophila is unstable, even in the presence of its putative dedicated chaperone, dTBCA. It is interesting to note that knocking down mammalian TBCA also induces a decrease in both α- and β-tubulin levels (Nolasco et al., 2005). In dTBCB1 mutant tissues, the observed tubulin levels decrease most likely causes the MT destabilization. However, dTBCB does not seem absolutely required to form tubulin heterodimers in vivo, since we find that a significant pool of dimers is still present in mutant extracts. Overall our results suggest that dTBCB enhances tubulin dimer formation in vivo to promote MT assembly.
In plants and yeasts, TBCB is essential for cell division (Radcliffe et al., 1999; Du et al., 2010). In flies, dTBCB is expressed throughout cell cycle, and during mitosis it colocalizes to some extend with the MT spindle. Surprisingly, dTBCB knockout in diverse tissues does not prevent cell division, suggesting that dTBCB is not essential for mitosis. We observed that mitotic spindles do form in mutant neuroblasts and allow mitosis, even if the overall cell cycle is slower than normal. Therefore the remaining levels of α- and β-tubulin are sufficient to fulfill cell division. Similarly, depletion of Drosophila TBCE decreases MT levels without precluding cell proliferation proceeding in embryos (Jin et al., 2009). Likewise, even though a null allele of α-tubulin 84B is cell-lethal, a hypomorphic mutation can affect larval viability without preventing cell proliferation (Matthews and Kaufman, 1987). This suggests that mitosis in flies is a robust process, resistant to a significant decrease in α- and β-tubulin amounts. It is possible that, at least in Drosophila, TBCs are not essentials to sustain the MT polymerization level necessary for mitosis.
We have found that dTBCB is required for cell polarity in the ovary. In the oocyte and in the overlying epithelial follicle cells, the MT-based transport is particularly important for the asymmetric distribution of determinants (St Johnston and Ahringer, 2010). We found that the drop in MT level observed in dTBCB mutant cells correlates with defects in localization of apico-lateral polarity proteins in epithelial cells, defects of mRNAs, and nucleus-polarized transport in oocyte at midoogenesis. However, low MT density does not trigger obvious polarity defects in early oogenesis polarization steps, since Orb accumulation occurs normally in mutant oocytes. It therefore seems that there is an interesting differential requirement for dTBCB between early and late stages of oogenesis that is not due to the persistence of the protein. This may point to a higher robustness of the MT-dependent processes in early stages of oogenesis: a partial depolymerization of the MT network may affect late but not early events, as suggested previously (Bolivar et al., 2001). Early polarity maintenance could be due to the smaller size of the early cysts that probably need a lower MT-nucleation activity to produce a functional MT network.
We have found that the simultaneous decreases of the expression levels of dTBCB and the ubiquitously expressed α-tubulin 84B prevent egg hatching and cause cell polarity defects in ovarian epithelial cells, suggesting that dTBCB regulates polarity most likely by controlling tubulin/MT integrity (Figure S3). Interestingly, this heteroallelic combination does not impair the formation/division of the follicle cell monolayer but affects its apico-basal polarity. We believe that these results confirm that mitotic cells are less sensitive to MT destabilization than interphasic cells, probably because of the much higher MT nucleation activity during mitosis. In accordance with this, it is interesting to note that nocodazol treatment depolymerizes interphasic MT networks much more efficiently than mitotic spindles (Cassimeris et al., 1990).
In humans, the presence of a mutated α-tubulin defective for TBCB binding has been correlated with the brain malformation lissencephaly (Tian et al., 2008), while high dTBCB levels have been reported in breast tumors (Vadlamudi et al., 2005). The results of this study may be of significant importance in understanding the molecular basis of the development of these pathologies.
MATERIALS AND METHODS
Identification of the dTBCB mutation and fly stocks
We carried out a genetic screen in germ-line clones for mutations disrupting the posterior distribution of Staufen (Stau) and the antero-dorsal positioning of the nucleus (Gervais et al., 2008). A lethal homozygous mutant line, 3.112, was identified in the screen. The mutated locus was mapped with the 2R Exelixis deficiencies collection (Parks et al., 2004). The 3.112 flies failed to complement overlapping Df(2R)ED3728 and Df(2R)BSC22 (Bloomington Drosophila Stock Center, Bloomington, IN) deficiencies, but did complement Df(2R)BSC26 (Bloomington) and Par-1w3 mutations (Shulman et al., 2000). Using these deficiencies, we were able to map the mutation to a 264-kb interval. By concentrating on genes predicted to regulate MTs and sequencing putative candidates, we identified a point mutation in the second exon of the CG11242 gene. We performed 3.112 rescue experiments, using a genomic CG11242 transgene or a CG11242 cDNA fused to GFP under the control of the ubiquitin promoter (Ubi-dTBCB-GFP). Homozygous mutant dTBCB larvae were selected with a CyO-GFP balancer chromosome. dTBCB1 germ-line clones were generated with the FLP/FRT system, with either the ovoD1 or GFP selector/marker genes (Chou and Perrimon, 1996; Blair, 2003). In both cases, clones were induced by heat shock for 2 h at 37°C during the second-instar larval stage, and females were dissected at least 5–20 d after emergence. Follicle cell clones were induced by heat shock for 1 h at 37°C during adulthood or at a larval stage, with the FLP/FRT system and the GFP selector gene (Luschnig et al., 2000). UASt-RNAi-dTBCB flies correspond to v103943 stock from the Vienna Drosophila RNAi center. da-Gal4, ap-Gal4, en-Gal4, and UAS-GFP were kindly provided by J. Silber (IJM, Paris, France). Knockdown of dTBCB was achieved by combining a single copy of UAS-RNAi transgene with a single copy of the dedicated driver. The Jupiter-GFP/TM6Ser (Karpova et al., 2006) fly line was also used. pUbi-RFP-α-Tub was kindly provided by J. Raff (Oxford University, UK). α-tubulin 84B5 flies were obtained from the Bloomington Drosophila Stock Center.
Molecular biology and biochemistry
The dTBCB1 mutation was sequenced in PCR-amplified genomic DNA extracted from homozygous mutant larvae. For transgenesis, we obtained dTBCB cDNA and genomic DNA respectively from w1118 larval total RNA and embryonic genomic DNA. The genomic dTBCB locus, containing introns, untranslated regions (UTRs), and the putative 5′ promoter region, was inserted into a pCasper vector. The dTBCB cDNA was inserted into the pUWG vector (Gateway, Invitrogen, Carlsbad, CA), to generate Ubi-dTBCB-GFP, and the pGex-6P1 vector (GE Healthcare, Waukesha, WI), to generate the recombinant GST-dTBCB protein. Recombinant glutathione S-transferase (GST)-dTBCB was produced in Escherichia coli BL-21 and affinity-purified on glutathione-Sepharose 4B (GE Healthcare). The GST tag was removed with the Prescission protease (GE Healthcare). Proteins were quantified using the Bradford assay (Bio-Rad, Hercules, CA).
Antibody production and immunostaining
Two polyclonal antisera against dTBCB were produced by immunizing rabbits with purified, untagged, full-length recombinant dTBCB (Eurogentec, Osaka, Japan). For Western blot analysis, we used the 2278 antibody at a dilution of 1/5000. Both affinity-purified and unpurified antibodies recognize a single band at the expected size of 27 kDa that was not revealed by the preimmune serum. Crude larval mutant extracts were prepared by squashing 6- to 7-d-old dTBCB1/Df(2R)ED3728 or dTBCB1 homozygous animal and third-instar larvae expressing the UASt-RNAi-dTBCB transgene under the daughterless driver. Germ-line mutant extracts were prepared by dissecting dTBCB1 clonal oocytes at stage 14. We used anti–α-tubulin DM1A antibody (Sigma-Aldrich, St. Louis, MO) at a dilution of 1:2500; anti-actin C4 antibody (MP Biomedicals, Solon, OH) at a dilution of 1:5000, and anti–β-tubulin E7 antibody (Developmental Studies Hybridoma Bank [DSHB], University of Iowa) at a dilution of 1:50.
In oocytes, RNAs were detected in situ as previously described (Gervais et al., 2008). For immunostaining, various antibodies and conjugated molecules were used in ovaries: anti–α-tubulin DM1A antibody (Sigma-Aldrich) at a dilution of 1:250; anti–actin C4 antibody (MP Biomedicals) at a dilution of 1:5000; anti-phalloidin Alexa Fluor 488–labeled antibody (Molecular Probes, Paisley, UK) at a dilution of 1:250; anti-lectin WGA Alexa Fluor 594–labeled antibody (Molecular Probes) at a dilution of 1:30; anti-aPKC antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:1000; anti–Par-3 antibody (Wodarz et al., 1999) at a dilution of 1:2000; anti-Dlg antibody (DSHB) at a dilution of 1:20; anti–Orb 6H4 antibody (DSHB) at a dilution of 1:30; 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) at a dilution of 1:200; anti–Tyr-tubulin YL1/2 antibody (Abcam, Cambridge, MA) at a dilution of 1:1000; and anti–Staufen dN16 antibody (Santa Cruz Biotechnology) at a dilution of 1:200. Ovaries were fixed by incubation for 20 min with 4% paraformaldehyde, except for tyrosine-tubulin staining, for which samples were pretreated with triton before cold methanol fixation (Januschke et al., 2006).
Larval brains and disks were fixed 30 min in 4% paraformaldehyde and immunostained with an anti-pH3 antibody (Millipore Upstate, Billerica, MA) at a dilution of 1:500.
S2R+ cells were transfected with Effectene (Qiagen, Valencia, CA) and fixed, after 3 day, by incubation in cold methanol or Cytoskelfix (Cytoskeleton, Denver, CO) at −20°C for 10 min. Images were acquired with a Leica (Wetzlar, Germany) TCS-SP5 AOBS inverted scanning microscope, analyzed with ImageJ software (National Institute of Health), and assembled with Photoshop (Adobe, San Jose, CA).
Fly wings were dissected in 70% ethanol and mounted in Hoyer's medium. Pictures were taken with a Zeis Lumar stereomicroscope and the AxioVision software.
Image processing and MT quantification
Images of RFP–α-tubulin in wild-type and dTBCB1 mutant cysts were acquired using the same confocal settings through different stages. Quantification of the RFP signal intensity was performed on a single confocal section using ImageJ software. Regions of interests (ROIs) corresponding to the entire germ-line cyst area for previtellogenic egg chambers and to the oocyte area for vitellogenic egg chambers were defined and signal intensity, reflecting the amount of RFP–α-tubulin, was determined. For the quantification, the RFP intensity signals measured were normalized across the corresponding ROIs.
Live imaging
Live imaging was carried out with oocytes and syncytial embryos on a Leica CSU10 inverted spinning-disk confocal microscope. Hand-dechorionated embryos and dissected ovarian chambers were covered with oil (Voltalef 10S). Images were acquired every 10 s for embryos and every minute for Jupiter-expressing oocytes (15 stacks of 0.5 μm for each time point). Images were acquired with MetaMorph 7 (Universal Imaging, Sunnyvale, CA), analyzed with ImageJ, and assembled with Photoshop.
Live imaging of individual neuroblasts in culture was performed according to Januschke et al. (2011) using a spinning-disk confocal system (Andor, Belfast, UK) mounted to an Olympus IX71 (Tokyo, Japan) microscope using a UPlanSApo 100×/1.4 numerical aperture objective. Z-stacks were acquired every 90 s. Owing to a lower MT intensity signal in dTBCB1 mutant cells compared with wild-type, a longer exposure time was used (15 vs. 75 ms). Movie files were processed using Fiji and assembled using After Effects 7.0 (Adobe).
MT pulldown assay
Experiments with second-instar larval extracts were carried out on 30 individuals homogenized in 50 μl BRB80 supplemented with 0.1% NP40, 1X protease inhibitors (Roche, Indianapolis, IN), 1 mM GTP, and 1 mM dithiothreitol at 4°C. Extracts were precleared by centrifugation for 5 min at 10,000 × g and 30 min at 100,000 × g at 4°C, and then treated with 10 μM Taxol (Sigma-Aldrich) for 30 min at room temperature. Centrifugation through 40% glycerol cushions (20 min 100,000 × g) was used to separate polymerized and free tubulins. This protocol was adapted from http://mitchison.med.harvard.edu/protocols.html.
Supplementary Material
Acknowledgments
We thank the IJM ImagoSeine bioimaging core facility. We acknowledge C. Gonzalez for providing the resources allowing us to live-image larval neuroblasts. We thank J. Raff and J. Silber for reagents and D. Chrétien, R. Karess, F. Perez, J. A. Lepesant, S. Claret, and V. Brodu for discussion and critical comments on the manuscript. Drosophila embryo injections were carried out by BestGene (Chino Hills, CA). A.B. and B.B. were supported by the Association pour la Recherche sur le Cancer. This work was supported by ANR (Cymenpol, program Blan06-3-139786) and by ARC (SL220100601358).
Abbreviations used:
- anti-pH3
anti-phosphohistone H3
- aPKC
atypical protein kinase C
- DSHB
Developmental Studies Hybridoma Bank
- dTBCB
Drosophila tubulin-binding cofactor B
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- MAP
microtubule-associated protein
- MT
microtubule
- RFP
red fluorescent protein
- RNAi
RNA interference
- ROI
regions of interest
- TBC
tubulin-binding cofactor
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0633) on August 1, 2012.
*These authors contributed equally to this work.
Present addresses: †Department of Pathology and Cell Biology, College of Physicians and Surgeons, Columbia University, New York, NY 10032
‡Faculté de Pharmacie, Université Paris Sud, EA4530 IFR 141 IPSIT, 92296 Châtenay-Malabry, France
§Host-Virus Interactions Group, Institut Cochin, 74014 Paris, France.
REFERENCES
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. Microtubule +TIPs at a glance. J Cell Sci. 2010;123:3415–3419. doi: 10.1242/jcs.062414. [DOI] [PubMed] [Google Scholar]
- Amos LA, Schlieper D. Microtubules and maps. Adv Protein Chem. 2005;71:257–298. doi: 10.1016/S0065-3233(04)71007-4. [DOI] [PubMed] [Google Scholar]
- Becalska A, Gavis E. Lighting up mRNA localization in Drosophila oogenesis. Development. 2009;135:2493–2503. doi: 10.1242/dev.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair SS. Genetic mosaic techniques for studying Drosophila development. Development. 2003;130:5065–5072. doi: 10.1242/dev.00774. [DOI] [PubMed] [Google Scholar]
- Bolivar J, Huynh JR, Lopez-Schier H, Gonzalez C, St Johnston D, Gonzalez-Reyes A. Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organisation of the microtubule cytoskeleton. Development. 2001;128:1889–1897. doi: 10.1242/dev.128.10.1889. [DOI] [PubMed] [Google Scholar]
- Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Rieder CL, Rupp G, Salmon ED. Stability of microtubule attachment to metaphase kinetochores in PtK1 cells. J Cell Sci. 1990;96:9–15. doi: 10.1242/jcs.96.1.9. [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Theurkauf WE. Cytoskeletal functions during Drosophila oogenesis. Science. 1994;266:590–596. doi: 10.1126/science.7939713. [DOI] [PubMed] [Google Scholar]
- Cunningham LA, Kahn RA. Cofactor D functions as a centrosomal protein and is required for the recruitment of the γ-tubulin ring complex at centrosomes and organization of the mitotic spindle. J Biol Chem. 2008;283:7155–7165. doi: 10.1074/jbc.M706753200. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Bargmann BO, Gadella TW., Jr. Arabidopsis tubulin folding cofactor B interacts with α-tubulin in vivo. Plant Cell Physiol. 2006;47:1406–1411. doi: 10.1093/pcp/pcl001. [DOI] [PubMed] [Google Scholar]
- Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Du Y, Cui M, Qian D, Zhu L, Wei C, Yuan M, Zhang Z, Li Y. AtTFC B is involved in control of cell division. Front Biosci (Elite Ed) 2010;2:752–763. doi: 10.2741/e135. [DOI] [PubMed] [Google Scholar]
- Fanarraga ML, Bellido J, Jaen C, Villegas JC, Zabala JC. TBCD links centriologenesis, spindle microtubule dynamics, and midbody abscission in human cells. PLoS One. 2010;5:e8846. doi: 10.1371/journal.pone.0008846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanarraga ML, Villegas JC, Carranza G, Castano R, Zabala JC. Tubulin cofactor B regulates microtubule densities during microglia transition to the reactive states. Exp Cell Res. 2009;315:535–541. doi: 10.1016/j.yexcr.2008.10.045. [DOI] [PubMed] [Google Scholar]
- Feierbach B, Nogales E, Downing KH, Stearns T. Alf1p, a CLIP-170 domain-containing protein, is functionally and physically associated with α-tubulin. J Cell Biol. 1999;144:113–124. doi: 10.1083/jcb.144.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais L, Claret S, Januschke J, Roth S, Guichet A. PIP5K-dependent production of PIP2 sustains microtubule organization to establish polarized transport in the Drosophila oocyte. Development. 2008;135:3829–3838. doi: 10.1242/dev.029009. [DOI] [PubMed] [Google Scholar]
- Holmfeldt P, Sellin ME, Gullberg M. Predominant regulators of tubulin monomer-polymer partitioning and their implication for cell polarization. Cell Mol Life Sci. 2009;66:3263–3276. doi: 10.1007/s00018-009-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh JR, St Johnston D. The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Curr Biol. 2004;14:R438–449. doi: 10.1016/j.cub.2004.05.040. [DOI] [PubMed] [Google Scholar]
- Januschke J, Gervais L, Gillet L, Keryer G, Bornens M, Guichet A. The centrosome-nucleus complex and microtubule organization in the Drosophila oocyte. Development. 2006;133:129–139. doi: 10.1242/dev.02179. [DOI] [PubMed] [Google Scholar]
- Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Pan L, Liu Z, Wang Q, Xu Z, Zhang YQ. Drosophila tubulin-specific chaperone E functions at neuromuscular synapses and is required for microtubule network formation. Development. 2009;136:1571–1581. doi: 10.1242/dev.029983. [DOI] [PubMed] [Google Scholar]
- Karpova N, Bobinnec Y, Fouix S, Huitorel P, Debec A. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil Cytoskeleton. 2006;63:301–312. doi: 10.1002/cm.20124. [DOI] [PubMed] [Google Scholar]
- Kortazar D, Fanarraga M, Carranza G, Bellido J, Villegas J, Avila J, Zabala J. Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp Cell Res. 2007;313:425–436. doi: 10.1016/j.yexcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Lopez-Fanarraga M, Avila J, Guasch A, Coll M, Zabala JC. Review: postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J Struct Biol. 2001;135:219–229. doi: 10.1006/jsbi.2001.4386. [DOI] [PubMed] [Google Scholar]
- Lopez-Fanarraga M, Carranza G, Bellido J, Kortazar D, Villegas JC, Zabala JC. Tubulin cofactor B plays a role in the neuronal growth cone. J Neurochem. 2007;100:1680–1687. doi: 10.1111/j.1471-4159.2006.04328.x. [DOI] [PubMed] [Google Scholar]
- Luschnig S, Krauss J, Bohmann K, Desjeux I, Nusslein-Volhard C. The Drosophila SHC adaptor protein is required for signaling by a subset of receptor tyrosine kinases. Mol Cell. 2000;5:231–241. doi: 10.1016/s1097-2765(00)80419-0. [DOI] [PubMed] [Google Scholar]
- Lytle BL, Peterson FC, Qiu SH, Luo M, Zhao Q, Markley JL, Volkman BF. Solution structure of a ubiquitin-like domain from tubulin-binding cofactor B. J Biol Chem. 2004;279:46787–46793. doi: 10.1074/jbc.M409422200. [DOI] [PubMed] [Google Scholar]
- Matthews KA, Kaufman TC. Developmental consequences of mutations in the 84B α-tubulin gene of Drosophila melanogaster. Dev Biol. 1987;119:100–114. doi: 10.1016/0012-1606(87)90211-9. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Muller HA. Genetic control of epithelial cell polarity: lessons from Drosophila. Dev Dyn. 2000;218:52–67. doi: 10.1002/(SICI)1097-0177(200005)218:1<52::AID-DVDY5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Nolasco S, Bellido J, Goncalves J, Zabala JC, Soares H. Tubulin cofactor A gene silencing in mammalian cells induces changes in microtubule cytoskeleton, cell cycle arrest and cell death. FEBS Lett. 2005;579:3515–3524. doi: 10.1016/j.febslet.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Parks AL, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Radcliffe PA, Hirata D, Vardy L, Toda T. Functional dissection and hierarchy of tubulin-folding cofactor homologues in fission yeast. Mol Biol Cell. 1999;10:2987–3001. doi: 10.1091/mbc.10.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Toda T. Characterisation of fission yeast alp11 mutants defines three functional domains within tubulin-folding cofactor B. Mol Gen Genet. 2000;263:752–760. doi: 10.1007/s004380000252. [DOI] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Schaefer MK, Schmalbruch H, Buhler E, Lopez C, Martin N, Guenet JL, Haase G. Progressive motor neuronopathy: a critical role of the tubulin chaperone TBCE in axonal tubulin routing from the Golgi apparatus. J Neurosci. 2007;27:8779–8789. doi: 10.1523/JNEUROSCI.1599-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, Benton R, St Johnston D. The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell. 2000;101:377–388. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Shultz T, Shmuel M, Hyman T, Altschuler Y. β-Tubulin cofactor D and ARL2 take part in apical junctional complex disassembly and abrogate epithelial structure. FASEB J. 2008;22:168–182. doi: 10.1096/fj.06-7786com. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded β-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Tian G, Kong X, Jaglin X, Chelly J, Keays D, Cowan N. A pachygyria-causing α-tubulin mutation results in inefficient cycling with CCT and a deficient interaction with TBCB. Mol Biol Cell. 2008;19:1152–1163. doi: 10.1091/mbc.E07-09-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Barnes CJ, Rayala S, Li F, Balasenthil S, Marcus S, Goodson HV, Sahin AA, Kumar R. p21-activated kinase 1 regulates microtubule dynamics by phosphorylating tubulin cofactor B. Mol Cell Biol. 2005;25:3726–3736. doi: 10.1128/MCB.25.9.3726-3736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R. On and around microtubules: an overview. Mol Biotechnol. 2009;43:177–191. doi: 10.1007/s12033-009-9193-5. [DOI] [PubMed] [Google Scholar]
- Wang W, Ding J, Allen E, Zhu P, Zhang L, Vogel H, Yang Y. Gigaxonin interacts with tubulin folding cofactor B and controls its degradation through the ubiquitin-proteasome pathway. Curr Biol. 2005;15:2050–2055. doi: 10.1016/j.cub.2005.10.052. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.