Oxysterol-binding protein (OSBP) is involved in endoplasmic reticulum (ER)-Golgi sterol transport, but how its activity is regulated is unknown. OSBP is phosphorylated at multiple sites, two of which regulate sterol binding and interaction with vesicle-associated, membrane protein–associated protein A in the ER. Phosphorylation does not affect phosphatidylinositol 4-phosphate binding, which could serve as a counter-ligand to facilitate sterol release in the Golgi.
Abstract
The endoplasmic reticulum (ER)-Golgi sterol transfer activity of oxysterol-binding protein (OSBP) regulates sphingomyelin (SM) synthesis, as well as post-Golgi cholesterol efflux pathways. The phosphorylation and ER-Golgi localization of OSBP are correlated, suggesting this modification regulates the directionality and/or specificity of transfer activity. In this paper, we report that phosphorylation on two serine-rich motifs, S381-S391 (site 1) and S192, S195, S200 (site 2), specifically controls OSBP activity at the ER. A phosphomimetic of the SM/cholesterol-sensitive phosphorylation site 1 (OSBP-S5E) had increased in vitro cholesterol and 25-hydroxycholesterol–binding capacity, and cholesterol extraction from liposomes, but reduced transfer activity. Phosphatidylinositol 4-phosphate (PI(4)P) and cholesterol competed for a common binding site on OSBP; however, direct binding of PI(4)P was not affected by site 1 phosphorylation. Individual site 1 and site 2 phosphomutants supported oxysterol activation of SM synthesis in OSBP-deficient CHO cells. However, a double site1/2 mutant (OSBP-S381A/S3D) was deficient in this activity and was constitutively colocalized with vesicle-associated membrane protein–associated protein A (VAP-A) in a collapsed ER network. This study identifies phosphorylation regulation of sterol and VAP-A binding by OSBP in the ER, and PI(4)P as an alternate ligand that could be exchanged for sterol in the Golgi apparatus.
INTRODUCTION
Mammalian oxysterol-binding protein (OSBP) and OSBP-related proteins (ORPs) constitute a large eukaryotic gene family characterized by a conserved C-terminal sterol-binding domain (Lehto and Olkkonen, 2003). Full-length OSBP/ORPs also have N-terminal pleckstrin homology (PH) domains that interact with phosphatidylinositol (PI) phosphates (Levine and Munro, 1998, 2002) and a two phenylalanines (FF) in an acidic tract (FFAT) motif that bind vesicle-associated membrane protein–associated protein A (VAP-A) on the cytoplasmic surface of the endoplasmic reticulum (ER; Wyles et al., 2002; Loewen et al., 2003). This domain organization suggests that a primary function of OSBP/ORPs is to transfer cholesterol, ergosterol, or oxysterols between target membranes and/or to transduce sterol-dependent signals at these points of contact (Prinz, 2007; Ngo et al., 2010).
Oxysterol-binding protein homologues (OSH) in Saccharomyces cerevisiae catalyze phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2)-dependent ergosterol transfer in vitro and regulate cholesterol delivery from the ER to the plasma membrane (PM; Raychaudhuri et al., 2006). However, Osh4p and other Osh proteins are also implicated in the regulation of PM phosphatidylinositol 4-phosphate (PI(4)P) levels (Stefan et al., 2011), PM sterol distribution (Georgiev et al., 2011), and polarized endocytosis by mechanisms that evoke sterol-sensing activity (Alfaro et al., 2011). The integration of PI(4)P and ergosterol distribution and function by Osh4p could be due to competitive, high-affinity binding and transfer of these two ligands (de Saint-Jean et al., 2011). OSBP family members bind cholesterol and oxysterols, and some have been implicated in sterol transfer between organelles (Suchanek et al., 2007; Ngo and Ridgway, 2009; Banerji et al., 2010). ORP9L and OSBP extract and transfer cholesterol between liposomes in vitro (Ngo and Ridgway, 2009). Both proteins localize to the ER and Golgi apparatus and influence the post-Golgi distribution of cholesterol in cultured cells, suggesting they directly or indirectly transfer cholesterol within the early secretory pathway. In the case of OSBP, sterol transfer between the ER and Golgi apparatus is coupled to activation of ceramide transport protein (CERT) and sphingomyelin (SM) synthesis through increased activity of PI4KIIα, a cholesterol-sensitive PI4-kinase (Waugh et al., 2006; Banerji et al., 2010). On the other hand, OSBP and ORPs have been implicated in regulation of diverse signaling pathways that could be the result of sterol-sensing or sterol-transduction activity (Wang et al., 2005; Johansson et al., 2007; Lessmann et al., 2007; Lehto et al., 2008; Ngo et al., 2010).
The ER is cholesterol-poor compared with later secretory compartments, indicating that transport proteins must utilize a mechanism to deliver ligands against a concentration gradient. In this regard, OSBP is subject to a rapid phosphorylation cycle that is linked to Golgi localization and the cholesterol and SM content of cells (Ridgway et al., 1998b; Storey et al., 1998). Rapid depletion of SM in the PM results in dephosphorylation of OSBP and localization to the Golgi apparatus. Similarly, depletion of cholesterol by prolonged inhibition of synthesis causes OSBP dephosphorylation and Golgi localization. Because SM and cholesterol are essential components of PM rafts, OSBP phosphorylation appears to be dependent on the integrity of these membrane domains. A serine-rich motif situated between the FFAT and sterol-binding domain of OSBP (S381, S384, and S387; Figure 1) is responsive to changes in sterol and SM; S381 is required for subsequent phosphorylation of S384 and S387, indicating these later two sites are phosphorylated by casein kinase 1 (CK1; Mohammadi et al., 2001). A similar cholesterol and SM-sensitive phosphorylation site in CERT negatively regulates PH domain interaction with PI(4)P in the Golgi apparatus (Kumagai et al., 2007). This serine-rich motif consists of a protein kinase D (PKD) phosphorylation site (S132; Fugmann et al., 2007) that regulates phosphorylation of nine adjacent serine and threonine residues by CK1γ2 (Tomishige et al., 2009). Dephosphorylation in the ER by protein phosphatase 2Cε relieves inhibition of the PH domain (Saito et al., 2008). This identified two functional states for CERT; a dephosphorylated form that associates with the Golgi and mediates ceramide transfer from the ER, and an inactive PKD/CK1 phosphorylated form that binds ceramide but does not interact with the Golgi apparatus. The regulation of CERT and OSBP at the level of sterol- and SM-mediated phosphorylation and the sterol-dependent activation of CERT by OSBP (Perry and Ridgway, 2006; Banerji et al., 2010), demonstrate that this functional pair have an important role in regulating membrane homeostasis in the secretory pathway.
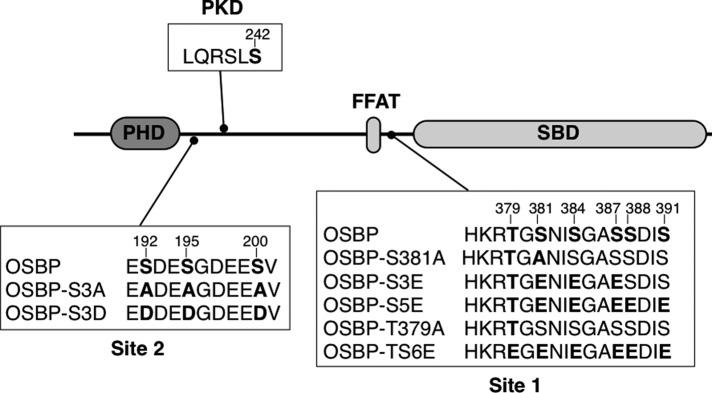
FIGURE 1:
OSBP phosphorylation sites and mutations. Site 1, site 2, and PKD phosphorylation sites and mutations reported in this study are indicated in bold. PHD, PH domain; SBD, sterol-binding domain.
To understand how phosphorylation regulates OSBP, we undertook a functional analysis of phosphorylation sites adjacent to the sterol-binding (site 1) and PH (site 2) domains. Results with phosphomimetic mutants indicate that the VAP-A–binding and sterol-binding phase of OSBP at the ER is regulated by phosphorylation at these two sites and is functionally linked to sterol-dependent activation of SM synthesis in the Golgi apparatus. We also demonstrate that the PI(4)P-binding activity of OSBP is competitive with cholesterol but insensitive to phosphorylation of site 1, indicating involvement in a Golgi-specific phase of the transfer cycle.
RESULTS
OSBP site 1 phosphorylation specifically increases sterol-binding activity
OSBP is phosphorylated between the sterol-binding and FFAT domains (site 1), as well as a PKD site at S242 (Mohammadi et al., 2001; Nhek et al., 2010). Recent phosphoproteome studies have extended site 1 to include phosphorylation at T379, S388, and S391, constituting a large CK1 motif that is activated by phosphorylation of S381 (Figure 1; Dephoure et al., 2008; Zahedi et al., 2008; Gauci et al., 2009). These studies also identified a cluster of three phosphoserine residues (positions 192, 195, and 200, designated site 2) in a potential casein kinase 2 motif next to the PH domain. To determine how phosphorylation at site 1 and site 2 regulates OSBP activity, we mutated serine and threonine residues to mimic fully dephosphorylated or phosphorylated species (summarized in Figure 1). In the case of site 1, OSBP-S381A prevents phosphorylation of downstream serine residues and mimics a dephosphorylated state with increased mobility on SDS–PAGE, while introduction of glutamine residues simulates a phosphorylated state with reduced mobility on SDS–PAGE (Figure 2 and Figure 5 later in the paper; Mohammadi et al., 2001).
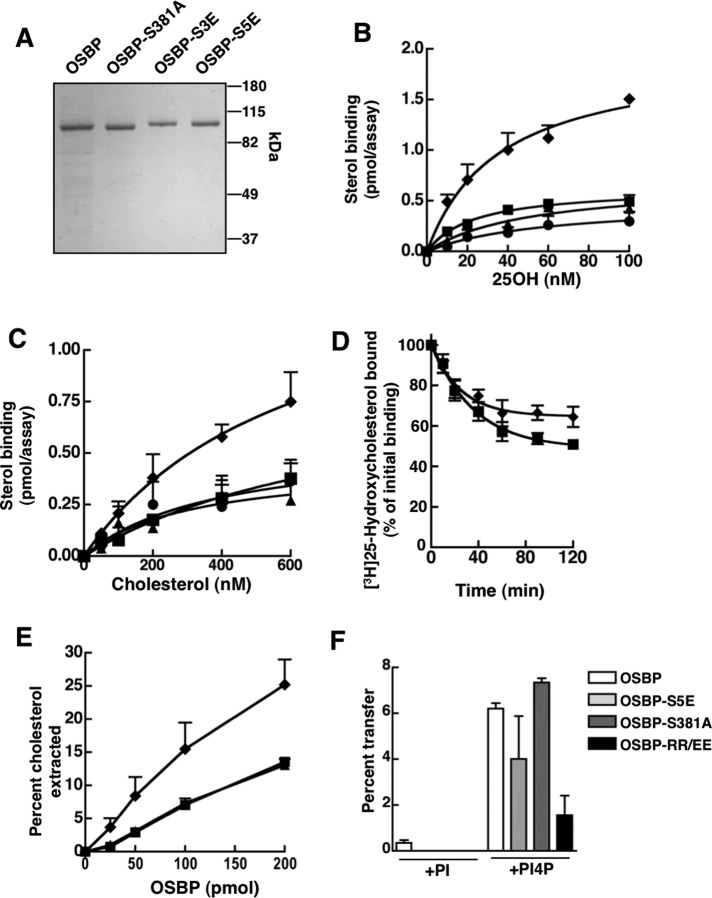
FIGURE 2:
Identification of an OSBP site 1 phosphomutant with increased sterol-binding activity. (A) Purified OSBP, OSBP-S381A, OSBP-S3E, and OSBP-S5E (2 μg each) were resolved by SDS–6%PAGE and stained with Coomassie Blue. (B and C) Specific binding of [3H]25OH and [3H]cholesterol by 12 pmol of OSBP (■), OSBP-S381A (▲), OSBP-S3E (●), or OSBP-S5E (♦) was assayed as described in Materials and Methods. (D) Following binding of 100 nM [3H]25OH, OSBP and OSBP-S5E were isolated on Talon resin and resuspended in binding buffer containing 100 nM 25OH at 20°C. At the indicated times, OSBP-bound 25OH was removed by centrifugation, and radioactivity in the supernatant was quantified. (E) Increasing amounts of OSBPs (see B and C for symbols) were incubated with liposomes containing 2 mol% [3H]cholesterol, and the extraction of radiolabel into the supernatant was measured. (F) Transfer of [3H]cholesterol between liposomes by OSBP, OSBP-S5E, OSBP-S381A, or OSBP-RR/EE was determined using a modified assay that involved preextraction of sterols from donor liposomes prior to addition of acceptor liposomes containing 2 mol% PI or PI(4)P. Results in all panels are the mean and SEM of three or more experiments using two to three different protein preparations.
FIGURE 5:
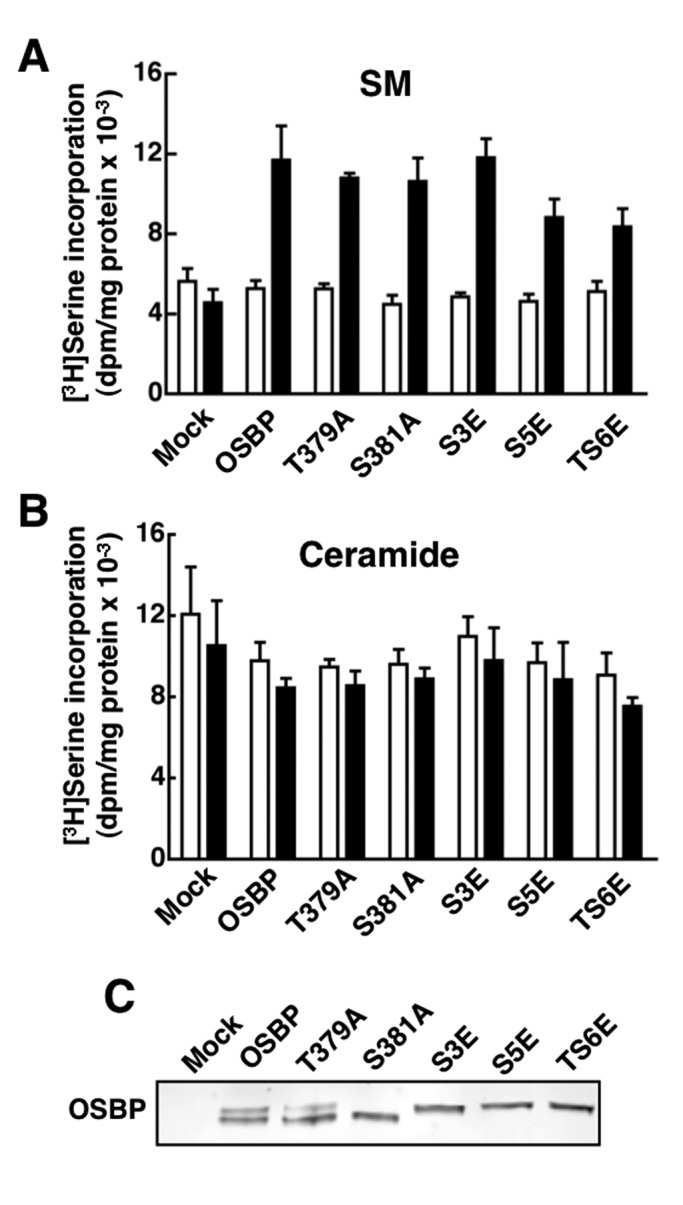
OSBP site 1 phosphomutants restore 25OH-activated SM synthesis in OSBP-depleted CHO cells. (A and B) CHO cells deficient in endogenous OSBP were transiently transfected for 48 h with wild-type OSBP and site 1 phosphomutants. SM (A) and ceramide (B) synthesis was measured by [3H]serine incorporation after treatment with 25OH (6 μM, black bars) or solvent control (empty bars) for 6 h. Results are the mean and SEM of three experiments. (C) Total cell lysates from mock and OSBP-transfected cells were resolved by SDS–6%PAGE and immunoblotted with an OSBP polyclonal antibody.
To establish whether phosphorylation of site 1 affects OSBP activity in vitro, we expressed and purified OSBP and OSBP-S381A, -S3E, and -S5E from Sf21 cells and assayed them for sterol binding, extraction, and transfer activities (Figure 2). The migration of each protein on SDS–PAGE was consistent with its proposed phosphorylation state; S381A migrated as a dephosphorylated species, while OSBP-S3E and -S5E mimicked a phosphorylated state with reduced mobility (Figure 2A). Wild-type OSBP expressed in Sf21 cells is dephosphorylated at site 1, as indicated by comigration with OSBP-S381A. OSBP displayed saturable binding of [3H]25-hydroxycholesterol (25OH; Kd 28 ± 5 nM; Bmax 0.63 ± 0.05) that was similar to OSBP-S381A (Kd 51 ± 12 nM; Bmax 0.68 ± 0.11) and OSBP-S3E (Kd 58 ± 14; Bmax 0.49 ± 0.15; Figure 2B). The Kd for oxysterol binding to OSBP-S5E (35 ± 9 nM) was similar to wild-type and the other mutants, but binding capacity (Bmax 2.0 ± 0.26) was increased threefold. Specific binding of [3H]cholesterol by OSBPs was saturable, but calculated binding constants were inconsistent due to nonlinear Scatchard plots (Figure 2C). However, it is evident that maximal cholesterol binding by OSBP-S5E was also increased greater than twofold compared with OSBP and the other phosphomutants. The rate of sterol dissociation was determined by incubating [3H]25OH-loaded OSBP or OSBP-S5E in buffer containing unlabeled 25OH at 20°C and measuring the appearance of radiolabeled oxysterol in the supernatant (Figure 2D). Under these conditions, [3H]25OH exchanged more slowly from OSBP-S5E compared with wild-type OSBP.
We next determined whether OSBP-S5E had altered cholesterol extraction and transfer activity using liposomal substrates. Extraction was measured by incubating increasing amount of OSBPs with liposomes containing 1% [3H]cholesterol and measuring the appearance of the radiolabel in the supernatant after donor liposome removal (Figure 2E). Similar to results with detergent dispersions of cholesterol (Figure 2C), OSBP-S5E extracted twofold more [3H]cholesterol from liposomes compared with OSBP and OSBP-S381A. The effect of site 1 phosphorylation on cholesterol transfer was determined used a modified assay in which OSBPs were first incubated with donor liposomes to initiate [3H]cholesterol extraction; this was followed by addition of acceptor liposomes (Figure 2F). The percentage of [3H]cholesterol transferred by all OSBPs to acceptor liposomes containing PI was negligible but was stimulated by the presence of 2 mol% PI(4)P. Although OSBP-S5E extracted twice as much cholesterol as OSBP and OSBP-S381A (Figure 2E), the percentage transferred to acceptor liposomes with PI(4)P was similar, indicating less efficient delivery of bound cholesterol. Transfer by a OSBP PH domain mutant (RR/EE) that does not interact with PI(4)P was reduced by 75%, confirming previous results indicating the PH domain has a stimulatory role in transfer (Ngo and Ridgway, 2009). Like Osh4p (de Saint-Jean et al., 2011), cholesterol transfer by OSBP is stimulated by PI(4)P in acceptor liposomes, but the N-terminal PH domain also has a positive role.
PI(4)P occupies the Osh4p sterol-binding pocket with its 4-phosphate residue associated with a phylogenetically conserved histidine pair, raising the possibility that other OSBP/ORPs bind both sterols and PI(4)P (de Saint-Jean et al., 2011). We therefore used competition and direct-binding assays to determine whether OSBP binds PI(4)P, and whether site 1 mutations affect this binding activity. First, OSBP extraction of [3H]cholesterol was assayed using liposomes that contained increasing amounts of unlabeled PI, PI(4)P, or PI(4,5)P2 (Figure 3A). Inclusion of PI or PI(4,5)P2 in donor liposomes did not inhibit [3H]cholesterol extraction compared with liposomes without PIPs. In contrast, inclusion of PI(4)P caused a 75% reduction in [3H]cholesterol extraction. We needed to exclude the possibility that this resulted from OSBP sequestration by PI(4)P-containing liposomes, so the distribution of OSBP between the supernatant and pellet fraction of the assay shown in Figure 3A was subjected to SDS–PAGE and protein staining (Figure 3B). It is evident that concentrations of PI(4)P that inhibited [3H]cholesterol to OSBP did not increase OSBP association with donor liposomes in the pellet fraction.
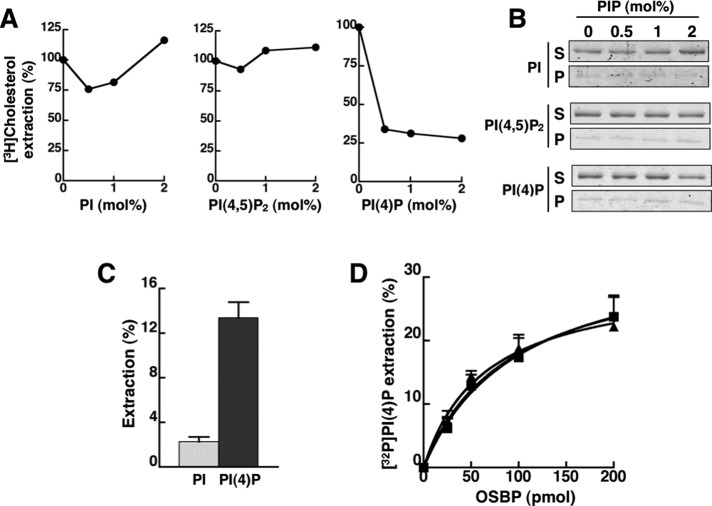
FIGURE 3:
PI(4)P binding by OSBP is competitive with cholesterol but unaffected by site 1 phosphorylation. (A) OSBP (50 pmol) was incubated with liposomes containing [3H]cholesterol (1 mol%) and increasing amounts PI, PI(4,5)P2, or PI(4)P (0–2 mol%). The extraction of [3H]cholesterol from liposomes by OSBP into the supernatant is expressed relative to activity in the absence of PIPs. Results are from a representative experiment. (B) The association of OSBP with liposomes during the extraction assay shown in (A) was determined by SDS–PAGE of supernatant (S) and pellet (P) fractions. (C) OSBP (50 pmol) was incubated with liposomes containing 0.5 mol% of [3H]PI or [32P]PI(4)P, and extraction of radioactivity into the supernatant was measured after precipitation of liposomes. Results are the mean and SEM of three experiments. (D) Increasing amounts of OSBP (■), OSBP-S381A (▲), or OSBP-S5E (●) were incubated with liposomes containing 0.5 mol% [32P]PI(4)P, and extraction of radioactivity into the supernatant was measured. The results are the mean and SEM of three experiments.
To verify the results from competition assays shown in Figure 3A, we measured direct binding and extraction of radiolabeled PI and PI(4)P by OSBP (Figure 3, C and D). OSBP extracted significantly more [32P]-labeled PI(4)P from liposomes compared with [3H]PI (Figure 3C). [32P]PI(4)P extraction by OSBP was dose-dependent but became nonlinear after 50 pmol (Figure 3D). Interestingly, OSBP-S5E extracted similar amounts of [32P]PI(4)P compared with wild type and OSBP-S381A, indicating that phosphorylation of site 1 specifically stimulates sterol-binding activity. Thus, in addition to binding PI(4)P via the PH domain, OSBP has an additional PI(4)P-binding site that is competitive with cholesterol.
OSBP site 1 phosphorylation does not affect oxysterol-activation of SM synthesis
Because OSBP-S5E has increased sterol-binding activity, and oxysterol and cholesterol binding is known to enhance OSBP association with the Golgi apparatus (Lagace et al., 1999), we tested whether OSBP-S5E and other site 1 phosphomutants have altered membrane-binding activity and Golgi localization (Figure 4). PH domain–dependent interactions of OSBPs were assessed by binding to phosphorylated PIs on nitrocellulose and in liposomes, although the later assay is complicated by the potential for PI(4)P to be bound and extracted by the sterol-binding domain (Figure 3). Wild-type OSBP and the site 1 phosphomutants had similar, preferential binding to PI(4)P that was immobilized on nitrocellulose filters (Figure 4A). Similarly, OSBP binding to liposomes containing 10 mol% PI(4)P or PI(4,5)P2 was unaffected by site 1 mutations, although this assay indicated a binding preference for PI(4,5)P2 (Figure 4B). For assessment of interaction of OSBP site 1 mutants with the Golgi apparatus, N-terminal mCherry fusions were transiently expressed in CHO cells, and cellular localization was determined by immunofluorescence microscopy in the presence and absence of 25OH (Figure 4C). OSBP and OSBP-S5E were primarily in a diffuse cytoplasmic/ER compartment in the absence of 25OH but became strongly localized to the perinuclear Golgi compartment in cells exposed to 25OH. OSBP-S381A had a slightly more pronounced perinuclear distribution in the absence of 25OH but translocated to the Golgi apparatus in oxysterol-treated cells. Untagged OSBP mutants showed similar behavior in response to 25OH treatment (unpublished data). Although the site 1 phosphomutants did not have altered ER-Golgi localization, the proximity of site 1 to the FFAT motif suggests that phosphorylation could increase negative charge in this region and influence VAP-A interaction. For assessing this in vitro, increasing amounts of recombinant OSBP site 1 mutants were incubated with GST-VAP-A and interaction was determined by pulldown and SDS–PAGE (Figure 4D). Binding of OSBPs by GST-VAP-A was relatively linear over the chosen concentration range, and there was no significant difference in the amount of OSBP-S381A or -S5E associated with GST-VAP-A compared with wild-type OSBP. These in vitro experiments show that site 1 phosphorylation specifically enhances sterol-binding capacity without affecting the FFAT and PH domain interactions of OSBP.
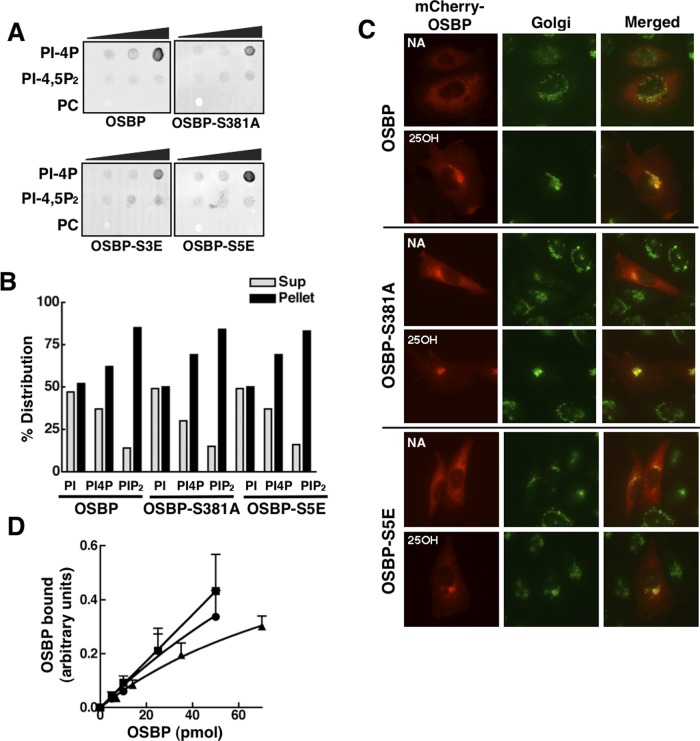
FIGURE 4:
PH-domain activity and Golgi localization is not affected by OSBP site 1 mutations. (A) Purified wild-type and OSBP mutants (200 pmol) were incubated with 0, 100, 200, and 500 pmol of PI(4)P, PI(4,5)P2, and PC that was immobilized on nitrocellulose filters. Filters were probed with an OSBP polyclonal and goat anti-rabbit IRDye 800–conjugated secondary antibodies. (B) The association of OSBP and phosphomutants with liposomes containing 10 mol% PI, PI(4)P, or PI(4,5)P2 was determined by quantification of distribution in the supernatant (light bars) and pellet (black bars) fractions after centrifugation. Results are from a representative experiment. (C) The indicated mCherry-OSBP fusions were transiently expressed for 24 h in OSBP-deficient CHO cells; this was followed by incubation with 25OH (6 μM) or solvent control (NA, no addition) for 2 h and immunostaining with a giantin antibody and goat anti-rabbit Alex Fluor 488. (D) The indicated amounts of recombinant OSBP (■), OSBP-S381A (▲), or OSBP-S5E (●) were incubated with GST-VAP-A (50 pmol), complexes were isolated by binding to glutathione-Sepharose, and bound OSBP was quantified by SDS–PAGE and Coomassie staining. Results are the mean and SEM of three experiments.
Sterol-mediated translocation of OSBP to the Golgi apparatus activates CERT-dependent ceramide delivery to the Golgi apparatus for conversion to SM (Perry and Ridgway, 2006). Reconstitution of SM synthesis (measured by [3H]serine incorporation) in OSBP-deficient CHO cells by cDNA transfection can be used as a functional readout to assess the influence of OSBP mutations on this pathway. Using this approach, OSBP site 1 mutants were tested for their ability to restore 25OH-activated SM synthesis in CHO cells depleted of endogenous OSBP (Figure 5). Compared with mock-transfected cells, OSBP expression in deficient CHO cells did not affect synthesis under unstimulated conditions but restored a 2.5-fold activation of SM synthesis by 25OH (Figure 5A). Expression of OSBP-S381A or the phosphomimics OSBP-S3E or -S5E also restored SM synthesis to a level that was not significantly different from wild type. T379 phosphorylation was also evaluated as a potential regulatory site of SM synthesis. OSBP-T379A migrated as a doublet on SDS–PAGE (Figure 5C), indicating that site 1 phosphorylation is intact and phosphorylation of S381 alone triggers phosphorylation of the adjacent four serine residues. Expression of OSBP-T379A or a mutant with all six threonine and serine residues changed to glutamate (OSBP-TS6E) restored SM synthesis (Figure 5A). [3H]serine incorporation into ceramide in mock- and OSBP-transfected CHO cells was similar under control and 25OH-treated conditions (Figure 4B). Collectively, this suggests that phosphorylation at site 1 is sufficient to enhance sterol binding but not subsequent steps in OSBP regulation of SM synthesis at the Golgi apparatus.
OSBP is phosphorylated at site 2 adjacent to the PH domain
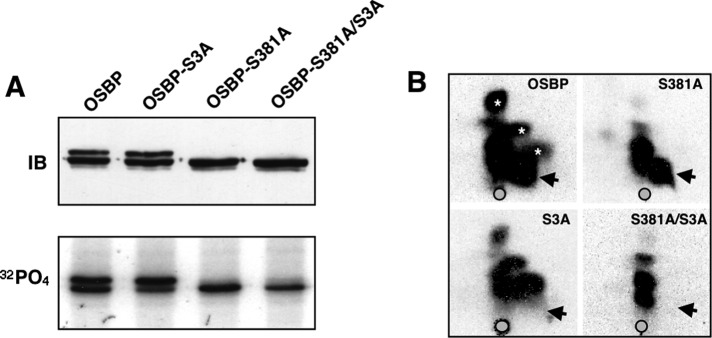
Phosphoproteome studies identified additional phosphoserines at positions 192, 195, and 200 that are embedded in a negatively charged region adjacent to the PH domain (Figure 1). For confirmation of phosphorylation at site 2, OSBP and single or double site 1/site 2 mutants that prevent phosphorylation (S3A and S381A) were transiently transfected into CHO cells, and phosphorylation status was monitored by immunoblotting and phosphopeptide analysis (Figure 6). OSBP-S3A had similar mobility as OSBP on immunoblots and reduced 32PO4 incorporation into the lower band but not the upper band, indicating that site 1 was unaffected (Figure 6A). OSBP-S381A lacked the upper phosphorylated species corresponding to site 1 phosphorylation but had similar incorporation of 32PO4 into the lower band, suggesting that site 2 phosphorylation was unaffected. The double mutant lacked the upper band and had reduced incorporation into the lower species. Tryptic digestion of immunoprecipitated OSBP and two-dimensional mapping revealed a single phosphopeptide (Figure 6B, arrows) containing site 2 phosphoserine residues that was absent in both S3A and S381A/S3A mutants (Figure 6B). The S3A mutation did not affect 32PO4 incorporation into the three phosphopeptides that constitute this site (Figure 6B, asterisks). The phosphopeptides remaining in OSBP-S381A/S3A could be due to phosphorylation of S242 by PKD and/or S353 by an unknown kinase (unpublished data). This confirms that site 2 is phosphorylated in CHO cells but it does not appear to be required for site 1 phosphorylation and vice versa.
FIGURE 6:
A site 2 phosphorylation motif is adjacent to the PH domain. (A) OSBP and the indicated site1 and site 2 mutants were transiently transfected into CHO cells for 48 h. Cells were then harvested for immunoblotting or labeled with 32PO4 (0.3 mCi/ml) for 4 h. 32PO4-labeled OSBPs were immunoprecipitated with monoclonal 11H9, resolved by SDS–PAGE, and subject to autoradiography for 12 h at −80°C. (B) 32PO4-labeled OSBPs were digested with trypsin and resolved by two-dimensional thin-layer electrophoresis and chromatography on cellulose-coated plates (Mohammadi et al., 2001). Arrows indicate the position of the site 2 phosphopeptide. Asterisks indicate the positions of site 1 phosphopeptides.
A site 1/site 2 double mutant is nonfunctional and aggregates in the ER
Restoration of 25OH-activated SM synthesis in OSBP-depleted CHO cells was used to assess the role of site 2 in OSBP activity (Figure 7). Expression of the site 2 phosphorylation mutants S3A and S3D restored 25OH-activated [3H]serine incorporation into SM and ceramide to the level of cells expressing OSBP (Figure 7A). Immunoblots of total cell lysates further established that site 2 mutations did not affect the slower migrating site 1 phosphospecies (Figure 7C). Immunofluorescence localization experiments confirmed that OSBP-S3A and -S3D localization was similar to wild type in control and 25OH-treated cells (Supplemental Figure S1).
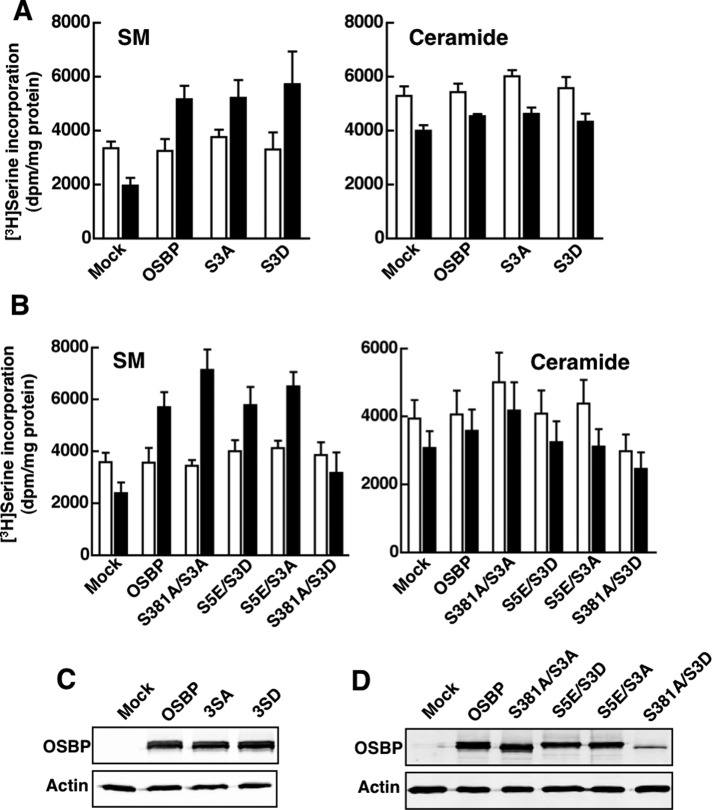
FIGURE 7:
Identification of a site 1/site 2 double mutant that does not restore sterol activation of SM synthesis. (A) OSBP site 2 mutants (S3A and S3D) were transiently expressed in OSBP-depleted CHO cells, and SM and ceramide synthesis was measured by [3H]serine incorporation as described in the legend to Figure 5. (B) A series of four OSBP site1/site2 double phosphomutants were transiently expressed in OSBP-depleted CHO cells, and SM and ceramide synthesis was measured as described above. The results shown in (A) and (B) are the means and SEM of three experiments. (C and D) Immunoblot analysis of site 2 (C) and site 1/site 2 double phosphomutants (D) expressed in OSBP-depleted CHO cells.
Because individual phosphorylation site mutations did not affect OSBP function, we decided to test a series of four site 1/site 2 double mutants (Figure 7, B and D). Three of these mutants (S381A/S3A, S5E/S3D, and S5E/S3A) restored a 2.5-fold activation of SM synthesis by 25OH without affecting basal synthesis. In contrast, expression of OSBP-S381A/S3D failed to restore oxysterol activation of SM synthesis without affecting basal SM or ceramide synthesis. OSBP-S381A/S3D was highly expressed compared with mock cells, in which RNA interference silencing is >95% effective, but was only 20–25% compared with the other double mutants (Figure 7D).
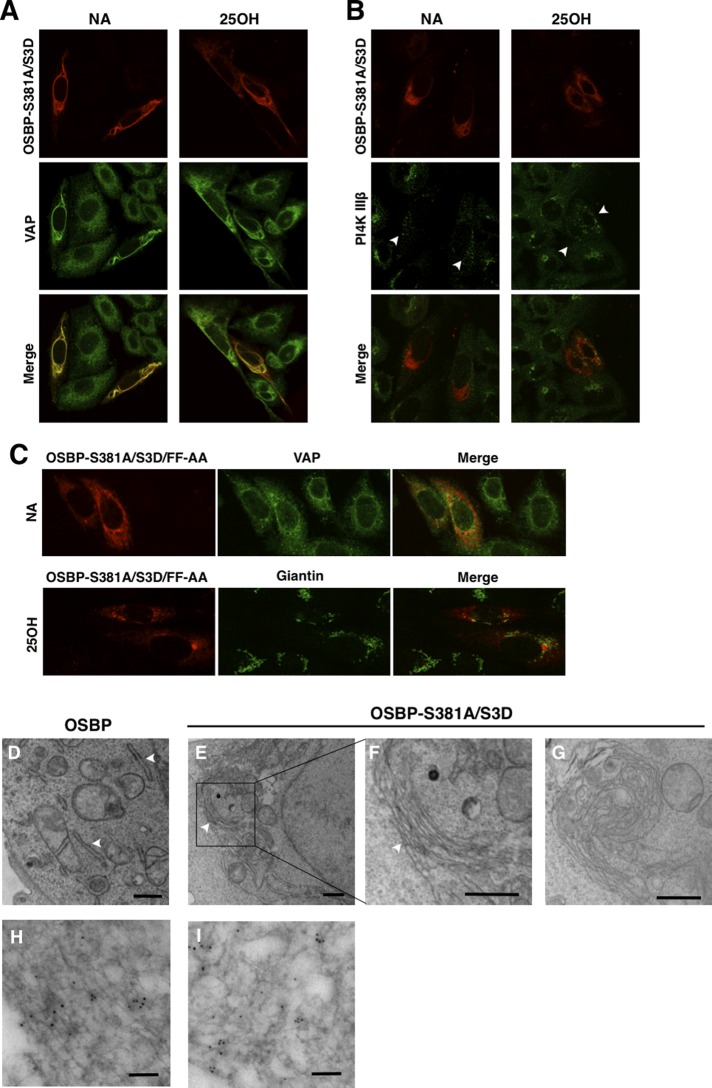
To determine why OSBP-S381A/S3D was defective with respect to sterol activation of SM synthesis, we examined its intracellular localization using immunofluorescence and transmission electron microscopy (TEM) in OSBP-depleted CHO cells (Figure 8). Site 1/site 2 double mutants that restored SM synthesis in CHO cells (Figure 7) showed the expected translocation from an ER/cytoplasmic compartment to the Golgi apparatus in response to oxysterol treatment (Figure S1). Interestingly, OSBP-S381A/S3D was exclusively localized to unusual filamentous structures around the nucleus of CHO cells (Figure 8A). These structures were absent in adjacent nonexpressing cells and unaffected by 25OH treatment and colocalized with VAP-A, which is normally present in an extended reticular network. These aggregated structures did not localize with the trans-Golgi enzyme PI4KIIIβ, but cells expressing OSBP-S381A/S3D had a dispersed PI4KIIIβ staining pattern (Figure 8B). A similar pattern of ER aggregation and colocalization of OSBP-A381A/S3D with VAP-A was observed in HeLa cells (unpublished data). To determine whether aggregation of the ER by OSBP-S381A/S3D was due to interaction with VAP-A, we mutated the FFAT motif (FF-AA), expressed the triple mutant in CHO cells, and determined its location relative to endogenous VAP-A (Figure 8C). Unlike the double phosphomutant, OSBP-S381A/S3D/FF-AA was dispersed in the cytoplasm or in small punctate structures and did not colocalize with VAP-A or cause its aggregation. Interestingly, OSBP-S381A/S3D/FF-AA did not localize to the Golgi apparatus when cell were treated with 25OH, suggesting a nonfunctional PH domain.
FIGURE 8:
OSBP S381A/S3D collapses and aggregates the ER. (A) OSBP-depleted CHO cells transiently expressing OSBP-S381A/S3D for 24 h were treated with 25OH (6 μg/ml) or solvent control (no addition, NA) for 2 h. Cells were immunostained with OSBP monoclonal 11H9 and Alexa Fluor 594–conjugated antibodies, which was followed by incubation with VAP-A polyclonal and Alexa Fluor 488–conjugated secondary antibodies. (B) Following 25OH treatment, CHO cells were immunostained with an OSBP polyclonal and Alexa Fluor 594–conjugated antibodies, which was followed by incubation with PI4K IIIβ monoclonal and Alexa Fluor 488–conjugated secondary antibodies. Arrowheads indicate dispersed Golgi staining in expressing cells. (C) CHO cells expressing OSBP-S381A/S3D/FF-AA, treated with or without 25OH, were coimmunostained for VAP-A or giantin, using Alexa Fluor 488–conjugated secondary antibodies. CHO cells expressing OSBP (D) or OSBP-S381A/S3D (E, F, and G) were processed for TEM, as described in Materials and Methods. The ER is indicated by arrowheads. Scale bar: 500 nm. (F) High magnification of the boxed area in (E). (H and I) OSBP-S381A/S3D was visualized in CHO cells with 11H9 and a goat anti-mouse secondary conjugated to 5-nm colloidal gold particles. Scale bar: 100 nm.
To better visualize ultrastructural changes to the ER induced by OSBP-S381A/S3D, we used TEM to analyze CHO cells expressing the mutant (Figure 8, D–I). CHO cells expressing OSBP displayed normal organelle morphology with many ribosome-studded ER tubules (Figure 8D, arrowheads). In contrast, CHO cells expressing OSBP-S381A/S3D contained extended aggregates of membrane tubules that were close to the nucleus and devoid of ribosomes (Figure 8, E–G). Cells containing these ER aggregates were also devoid of normal ER tubules elsewhere in the cell. Immunostaining for OSBP-S381A/S3D showed the presence of 5-nm colloidal gold particles in areas enriched in tubule aggregates (Figure 8, H and I). Thus OSBP-S381A/S3D caused the ER to collapse into filamentous bundles and dispersed the Golgi apparatus, perhaps contributing to its poor expression compared with other mutants (Figure 7D).
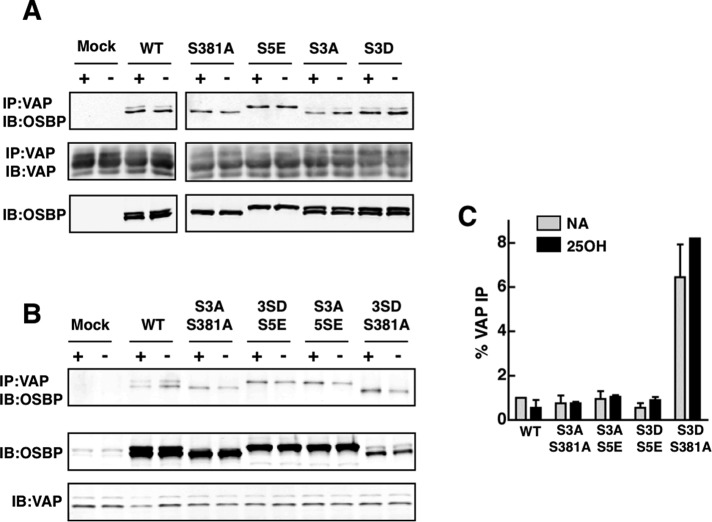
To assess whether the colocalization of OSBP-S381A/S3D and VAP-A observed by immunofluorescence was the result of enhanced physical interaction, and whether other phosphomutants might display altered binding, we assessed interaction with VAP-A using coimmunoprecipitation from OSBP-transfected CHO cells (Figure 9). Detergent extracts from cells expressing OSBP site 1 and site 2 phosphomutants were immunoprecipitated using a VAP-A antibody and probed for OSBP. There were no reproducible differences in the amount of single OSBP mutants coimmunoprecipitated with VAP-A under control or 25OH-treated conditions (Figure 9A). Compared with wild-type, OSBP-S381A/S3A, -S5E/S3D, and -S5E/S3A double mutants had similar levels of expression and interaction with VAP-A (Figure 9B). Despite reduced expression, OSBP-S381A/S3D showed similar levels of coimmunoprecipitation with VAP-A compared with the other double mutants. When quantified relative to input, approximately sixfold more OSBP-S381A/S3D was coimmunoprecipitated with VAP-A compared with the other three double mutants (Figure 9C). This further shows that OSBP-S381A/S3D contributes to ER aggregation due to enhanced interaction with VAP-A multimers.
FIGURE 9:
Enhanced interaction between VAP-A and OSBP-S381A/S3D. (A) Triton X-100 extracts were prepared from OSBP-depleted CHO cells transiently expressing the indicated single OSBP and site 1 or site 2 mutants and treated without (−) or with (+) 25OH (6 μM for 2 h). VAP-A was immunoprecipitated from extracts with a polyclonal antibody and immunoblotted for OSBP and VAP-A. (B) OSBP was coimmunoprecipitated with VAP-A from Triton X-100 extracts of OSBP-depleted CHO cells expressing site 1/site 2 double phosphomutants. (C) The amount of OSBP coimmunoprecipitated with VAP-A in (B) was quantified as a percentage of total OSBP input (average and range of two experiments).
DISCUSSION
The differential localization of OSBP between the ER and Golgi apparatus in response to exogenous and endogenous sterol ligands and the resultant effects on sterol-responsive activities in the Golgi apparatus suggest that OSBP transfers cholesterol and/or oxysterols between these organelles. This activity can be partially reconstituted in vitro if cholesterol extraction and transport occurs along a concentration gradient. This is unlikely to occur in vivo, since the ER is cholesterol-poor relative to the Golgi and other organelles (Ikonen, 2008). In this study, we identify two factors that could facilitate sterol loading and transport to the Golgi apparatus by OSBP; phosphorylation on two serine-rich motifs (sites 1 and 2) that control sterol binding and interaction with VAP-A in the ER and competition for sterol binding by PI(4)P. OSBP phosphorylation regulates the ER-specific sterol/VAP-A binding but not the binding of PI(4)P, a ligand highly enriched in the Golgi apparatus, where it could exchange with cholesterol.
Site 1 phosphorylation is initiated at S381, which triggers phosphorylation of four adjacent serine residues by a CKI-like activity (Mohammadi et al., 2001). The organization of site 1 is reminiscent of the serine-rich motif in CERT, which is initiated by PKD phosphorylation and sensitive to cellular cholesterol and SM content (Kumagai et al., 2007). However, OSBP-S381 is not phosphorylated by PKD, nor is PKD phosphorylation of S242 involved in site 1 phosphorylation (unpublished data). While the PH- and FFAT-domain activity was unaffected by site 1 mutations, introducing a constitutive negative charge in OSBP-S5E, which is proximal to the α-helical lid of the sterol-binding domain, caused a significant increase in the maximal binding of aqueous dispersions of 25OH without significantly affecting the apparent Kd. The partially phosphorylated mimetic OSBP-S3E had sterol-binding properties similar to OSBP and OSBP-S381A, indicating that maximal phosphorylation of site 1 is necessary to increase sterol binding. The cholesterol-binding capacity of OSBP-S5E for cholesterol dispersions was similarly increased relative to dephosphomimics, but dissociation constants could not be reproducibly measured due to irregular binding curves, possibly related to the poor ligand solubility and nonequilibrium conditions that were used to avoid protein inactivation. OSBP-S5E also extracted more cholesterol from liposomes compared with dephosphomimics, but the extracted cholesterol was not efficiently transferred to acceptor liposomes. This implies that phosphorylation at site 1 increases the initial capacity of OSBP to bind sterols at a donor membrane, and subsequent dephosphorylation facilitates the release of the sterol ligand to an acceptor membrane. In intact cells, 32PO4 pulse-chase experiments showed that phosphorylation of site 1 has a half-life of ∼20 min (Ridgway et al., 1998a). This is consistent with a dynamic phosphorylation cycle at this site that, in conjunction with other protein/lipid interactions and phosphorylation sites, regulates sterol binding and release.
Site 1 phosphorylation is correlated with changes in OSBP localization, particularly in response to SM and cholesterol metabolism and content (Ridgway et al., 1998b; Mohammadi et al., 2001). However, individual site 1 mutations did not affect in vitro interactions with VAP-A or PI(4)P or the distribution of OSBP between the ER and Golgi apparatus in CHO cells, indicating other variables are involved. Site 2 was of interest in this regard, since it encompasses three serine residues in a glutamate/asparate-rich region located next to the PH domain. 32PO4 labeling and peptide mapping of site1 and site 2 mutants indicated no interdependence between the sites and, similar to site 1, individual site 2 phosphomutants were normal with respect to cellular localization and restoration of 25OH-activated SM synthesis. However, expression of a mutant mimicking dephosphorylation of site 1 and phosphorylation of site 2 (OSBP-S381A/S3D) collapsed the peripheral ER into membrane bundles around the nucleus and strongly colocalized and associated with VAP-A. A similar ER morphology was observed in cells expressing an OSBP PH domain mutant (W174A; Wyles and Ridgway, 2004) and Nir proteins, PI-binding/transfer proteins that interact with VAP-A via FFAT domains (Amarilio et al., 2005; Peretti et al., 2008). In these instances, enhanced interaction with VAP-A causes cross-linking of ER tubules and collapse of the reticular network into membrane bundles and whorls. The similarity between OSBP W174A and OSBP-S381A/S3D with respect to ER perturbation and VAP-A interaction suggests that the latter mutant also has a nonfunctional PH domain. In support of this conclusion, mutation of the FFAT domain in OSBP-S381A/S3D prevented interaction with VAP-A and collapse of the ER, but the mutant did not regain the ability to localize to the Golgi in the presence of 25OH. This suggests a phosphorylation state that causes reduced cholesterol-binding activity, increased VAP-A binding, and reduced affinity for PH domain ligands in the Golgi apparatus. Based on the normal activity of other single and double site 1/site 2 mutants, subsequent changes in the phosphorylation status of either site 1 or 2, by as-yet-unknown kinases and/or phosphatases, would disengage OSBP from the ER and promote interaction with the Golgi apparatus.
OSBP homologues in diverse phyla are distinguished by two conserved histidine residues (H522 and H523 in human OSBP) situated at the entrance of the sterol-binding pocket (Lehto and Olkkonen, 2003; Im et al., 2005). This histidine pair is essential for Osh4p function but dispensable for in vitro sterol binding and transfer activity (Im et al., 2005; Raychaudhuri et al., 2006). Instead, these histidine residues form an essential contact with the 4-phosphate residue of PI(4)P that mediates competitive binding with dehydroergosterol (de Saint-Jean et al., 2011). This suggested that Osh4p transport of ergosterol to PI(4)P-enriched compartments is coupled to exchange and transport of PI(4)P in the opposite direction. This model is particularly suited to OSBP, which could transport cholesterol against a gradient to the PI(4)P-enriched trans-Golgi network in conjunction with PI(4)P transport in the opposite direction. Initial in vitro experiments lend support to this model; cholesterol extraction from liposomes by OSBP was specifically competed by PI(4)P, and radiolabeled PI(4)P was extracted by OSBP from cholesterol-free liposomes. Based on the percent of extraction of cholesterol and PI(4)P from liposomes by increasing amounts of OSBP (compare Figure 2E with Figure 3D), the relative affinity of OSBP for PI(4)P and cholesterol is comparable. Interestingly, the increased sterol-binding activity of OSBP-S5E toward cholesterol was not evident with PI(4)P. This indicates that phosphorylation events that affect the unique electrostatic and hydrogen-bonding contacts for sterols and PI(4)P could regulate an organelle-specific interaction with these ligands.
To test whether PI(4)P stimulated the transfer of cholesterol to acceptor liposomes, we used a modified assay that involved preextraction of cholesterol from donor liposomes. In this context, PI(4)P stimulated the transfer of cholesterol by OSBPs, suggesting an exchanged-based mechanism similar to Osh4 (de Saint-Jean et al., 2011). However, this is not the only explanation, since efficient cholesterol transfer by OSBP also required its N-terminal PH domain (Figure 2F; Ngo and Ridgway, 2009). The PH domains could facilitate exchange by targeting OSBP to PI(4)P-enriched membranes or to regulate ligand transfer by inter/intramolecular interactions.
This study provides a new perspective on OSBP activity in the ER-Golgi secretory pathway through identification of serine phosphorylation motifs that regulate the ER-specific phase of the OSBP transfer cycle involving VAP-A and sterol binding. The subsequent release of sterols in the Golgi apparatus could involve ligand exchange with PI(4)P. This exchange could be driven by OSBP-dependent activation of PI(4)P synthesis by the sterol-regulated PI4KIIα (Banerji et al., 2010). Alternatively, PKD could also regulate OSBP at the Golgi apparatus via direct phosphorylation (Nhek et al., 2010) or activation of PI(4)P synthesis by PI4KIIIβ (Hausser et al., 2005).
MATERIALS AND METHODS
Materials
Cholesterol and 25OH were purchased from Steraloids (Newport, RI). [3H]cholesterol, [3H]25OH, 32PO4, [3H]PI, and [3H]serine were from Perkin Elmer-Cetus (Waltham, MA). PI(4)P (dipalmitoyl and bovine brain) and PI(4,5)P2 (dipalmitoyl) were from Echelon (Salt Lake City, UT). Phosphatidylcholine (PC) was purchased from Avanti Polar Lipids (Alabaster, AL). OSBP polyclonal and monoclonal antibodies were as previously described (Ridgway et al., 1992; Mohammadi et al., 2001). A VAP-A–specific antibody was as previously described (Wyles et al., 2002). A colloidal, gold-conjugated secondary antibody was purchased from Sigma-Aldrich (St. Louis, MO). Alexa Fluor 488– and 594–conjugated secondary antibodies and Baculo-Direct expression system were from Life Technologies (Burlington, ON). IRDye 800– and 680–conjugated secondary antibodies were purchased from LI-COR Biosciences (Lincoln, NE). Site 1 phosphomutants of rabbit pOSBP (Mohammadi et al., 2001) were further mutagenized to produce pOSBP-S5E (S381,384,387,388,391E) and pOSBP-TS6E (T379E, S381,387,387,388,391E). Site 2 pOSBP-S192,195,200D (S3D) and pOSBP-S192,195,200A (S3A) were prepared by mutagenesis of pOSBP. Site 1 and site 2 double mutants were prepared by restriction digestion and ligation of DNA fragments containing mutated sites. All constructs were verified by sequencing and contained three silent mutations that prevented silencing in CHO cells expressing shOSBP.
Cell culture and transfection
CHO cells were cultured in DMEM with 5% fetal calf serum (Medium A). Stable silencing of OSBP in CHO cells was achieved by stable expression of a lentiviral shOSBP (Banerji et al., 2010). Plasmids encoding OSBPs were transfected into CHO cells using Lipofectamine 2000 according to the manufacturer's instructions. Sf21 insect cells were cultured in SF900-II medium containing 5% (vol/vol) fetal bovine serum, 10 μg/ml G418 and 0.25 μg/ml fungizone in monolayer or suspension at 27°C.
Baculovirus expression and purification of OSBP
The cDNAs for OSBP, OSBP-S381A, OSBP-S3E, OSBP-RR109,110EE (RR/EE), and OSBP-S5E were cloned into pENTR/d-Topo, verified by sequencing, inserted into linearized BaculoDirect (C-terminal V5-His–tagged) by recombination, and transduced into Sf21 cells until a titer of 1 × 108 pfu/ml was achieved. Sf21 cells were infected at an multiplicity of infection of 0.1 for 1 h at 20°C and resuspended in Sf-900 II media. After being cultured in suspension for 72 h, cells were lysed, and His-tagged OSBPs were purified by a two-step protocol (Ngo and Ridgway, 2009). Following the last step, OSBPs were concentrated to 1–1.5 mg protein/ml in Tris-HCl (pH 7.4) and 250 mM NaCl using a 30-kDa cutoff centrifugal filter concentrator (Millipore, Billerica, MA) and stored at −80°C.
Sterol binding, extraction, and transfer assays
The specific binding of 25OH by purified OSBP and OSBP mutants (12 pmol protein) was carried out in 75 μl of 10 mM HEPES (pH 7.4), 100 mM KCl, 2% (wt/vol) polyvinyl alcohol, and [3H]25OH at 4°C for 16 h. A charcoal-dextran slurry (45 μl) was added for 30 min, after which the mixture was sedimented by centrifugation at 10,000 × g for 10 min, and protein-bound sterol in the supernatant was measured by liquid scintillation counting. Specific binding is defined as total [3H]25OH binding minus binding in the presence of a 40-fold excess of unlabeled 25OH.
[3H]cholesterol-binding assays were similar to those described above, except 0.05% Triton X-100 was included. After incubation at 20°C for 2 h, OSBP-bound [3H]cholesterol was isolated by incubation with 12 μl of Talon affinity resin for 30 min with constant mixing. The resin was washed four times with 500 μl of 10 mM HEPES/100 mM KCl and collected by centrifugation at 5000 × g for 1 min. OSBP-bound [3H]cholesterol was released from the resin with 150 mM imidazole and quantified by liquid scintillation counting.
The liposomal [3H]cholesterol extraction assay was previously described (Ngo and Ridgway, 2009). The cholesterol transfer activity of OSBP and phosphomutants was assayed by a modified method (Ngo and Ridgway, 2009). Donor and acceptor liposomes composed of [14C]PC:PE:PS:[3H]cholesterol (69:20:10:1, mol/mol) and PC:PE:PS:PIPs:lactosyl-PE (58:20:10:2:10, mol/mol), respectively, were prepared by extrusion and sedimented at 15,000 × g for 5 min prior to use. [14C]PC was included in donor liposomes to correct for contamination of acceptor liposomes after sedimentation. The assay was modified to include a preextraction step prior to addition of acceptor liposomes. Donor liposomes (10 nmol) were incubated with 100 pmol of recombinant OSBPs and 3 μg fatty acid–free bovine serum albumin (BSA) in 80 μl of liposome buffer (25 mM HEPES, 150 mM NaCl, and 1 mM EDTA, pH 7.4) at 25°C. After 20 min, assays received 1) 20 μl of liposome buffer and were placed on ice (to measure extraction) or 2) 20 μl of acceptor liposomes (10 nmol) containing 2 mol% PI or PI(4)P for 10 min at 25°C before stopping the reaction on ice (to measure transfer). Acceptor liposomes were precipitated with 10 μg Ricinus communis agglutinin at 15,000 × g for 5 min, and radioactivity in the supernatant and pellet was measured by liquid scintillation counting. Extraction and transfer values were expressed as a percentage of total [3H]cholesterol input corrected for donor liposome contamination and transfer in the absence of OSBP.
PI(4)P competition and extraction assays
[32P]PI(4)P was isolated from HeLa cells incubated for 24 h in phosphate-free media containing 0.5 mCi/ml of 32PO4. Cells were harvested in 1 ml of phosphate-buffered saline (PBS) and 4 ml of CHCl3/MeOH/12N HCl (2:4:0.1, vol/vol), 1.2 ml of CHCl3, and 1.2 ml water were added. The organic phase was dried under nitrogen and resolved by TLC in CHCl3/MeOH/4M NH4OH (90:70:20, vol/vol), and [32P]PI(4)P was identified by autoradiography and comigration with an authentic standard (purification was repeated once more). Competitive extraction assays were performed using liposomes containing 1 mol% [3H]cholesterol and increasing mol% of PI, PI(4)P, or PI(4,5)P2. Direct extraction of 0.5% [3H]PI or [32P]PI(4)P by OSBPs from cholesterol-free liposomes was assayed as described for [3H]cholesterol.
Immunoblotting and immunoprecipitation
Total cell lysates were prepared in SDS–PAGE sample buffer (12.5% SDS, 30 mM Tris-HCl, 12.5% glycerol, and 0.01% bromophenol blue, pH 6.8), heated at 90°C for 5 min, resolved on SDS–PAGE, and transferred to nitrocellulose. Proteins were visualized by incubation with primary antibodies for 1–2 h at 20°C, which was followed by incubation with secondary IRDye 800– or 680–conjugated antibodies for infrared detection using an Odyssey Infrared Imaging System and quantification using Odyssey Application Software version 3.0 (LI-COR Biosciences, Lincoln, NE).
Coimmunoprecipitation of VAP-A and OSBP was carried out as previously described (Wyles et al., 2002). Briefly, CHO cells expressing OSBP phosphorylation mutants were lysed in PBS containing 5 mM KCl, 2 mM EDTA, 2 mM EGTA, and 0.5% Triton X-100, and were precleared by centrifugation at 14,000 × g for 15 min at 4°C. VAP-A was immunoprecipitated from supernatants using a polyclonal antibody and protein A-Sepharose, washed three times with PBS/0.1% Triton X-100, and resuspended in SDS–PAGE sample buffer. Immunoprecipitates were resolved by SDS–8%PAGE and immunoblotted for OSBP using monoclonal 11H9. The degree of VAP-A-OSBP coimmunoprecipitation was quantified relative to OSBP input.
Immunofluorescence and transmission electron microscopy
CHO cells cultured on glass coverslips were fixed with 4% (wt/vol) paraformaldehyde in PBS for 15 min and permeabilized with 0.5% (wt/vol) Triton X-100 for 20 min at 4°C. Coverslips were incubated sequentially with a primary antibody in PBS containing 1% (wt/vol) BSA at 20°C for 1 h, which was incubation with followed by an Alexa Fluor–conjugated secondary antibody for an additional 1 h at 20°C. Coverslips were mounted on glass slides with Mowiol 4–88, and images were captured using a Zeiss LSM510/AxioVert 200M inverted microscope with Plan-Apochromat 100×/1.40 numerical aperture (NA) oil-immersion objective or a Zeiss LSM510 laser-scanning confocal upright microscope with a Plan-Apochromat 63×/1.40 NA oil immersion objective (Jena, Germany).
Thin-section (80–100 nm) TEM of CHO cells was performed as described previously (Lagace and Ridgway, 2005). For immunoelectron microscopy, CHO-K1 cells were fixed with 4% (wt/vol) paraformaldehyde/0.5% (vol/vol) glutaraldehyde in 0.1M sodium cacodylate buffer for 30 min. Thin sections mounted on nickel grids were incubated with OSBP monoclonal 11H9 overnight at 4°C, which was followed by an 1-h incubation with 5-nm colloidal, gold-conjugated goat anti-mouse secondary antibody at 20°C. Grids were fixed with 2.5% glutaraldehyde and stained with 2% (wt/vol) uranyl acetate and lead citrate. Images were captured using a JEOL (Tokyo, Japan) JEM 1230 transmission electron microscope at 80 kV and Hamamatsu (Hamamatsu, Japan) ORCA-HR digital camera.
Analysis of PIP binding by OSBP phosphomutants
Liposomes composed of PC and PE and containing either PI, PI(4)P, or PI(4,5)P2 (70:20:10, mol/mol) were incubated with 100 pmol of purified protein for 25 min at 25°C. Samples were centrifuged at 100,000 × g for 30 min, and the supernatant and pellet fractions were analyzed by SDS–PAGE and stained with Coomassie Blue, and OSBP distribution was quantified using a LI-COR Odyssey IR imaging system.
Lipids (100–300 pmol) and solvent control were spotted onto a Hybond-C nitrocellulose membrane, which was incubated in blocking buffer (20 mM Tris HCl, pH 7.4, 150 mM NaCl, 3% [wt/vol] fatty acid–free BSA and 0.1% [vol/vol] Tween-20) at 4°C with 50 nM OSBPs for 1 h. Bound OSBPs were visualized with 11H9 monoclonal and IRDye 800–conjugated antibodies for infrared detection using an Odyssey Infrared Imaging System.
Supplementary Material
Acknowledgments
We thank Robert Zwicker and Cheong-Min Baek for excellent technical assistance. This work was supported by an operating grant from the Canadian Institutes of Health Research (MOP-15284) and an Issac Walton Killiam Graduate Studentship (C.-A.R.).
Abbreviations used:
- 25OH
25-hydroxycholesterol
- BSA
bovine serum albumin
- CERT
ceramide transport protein
- CK1
casein kinase 1
- ER
endoplasmic reticulum
- FFAT
two phenylalanines in an acidic tract
- NA
numerical aperture
- ORP
OSBP-related protein
- OSBP
oxysterol-binding protein
- OSH
oxysterol-binding protein homologues
- PBS
phosphate-buffered saline
- PH
pleckstrin homology
- PI
phosphatidylinositol
- PI(4)P
phosphatidylinositol 4-phosphate
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PKD
protein kinase D
- PM
plasma membrane
- SM
sphingomyelin
- TEM
transmission electron microscopy
- TGN
trans-Golgi network
- VAP-A
vesicle-associated, membrane protein–associated protein A
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0283) on August 8, 2012.
REFERENCES
- Alfaro G, Johansen J, Dighe SA, Duamel G, Kozminski KG, Beh CT. The sterol-binding protein Kes1/Osh4p is a regulator of polarized exocytosis. Traffic. 2011;11:1521–1536. doi: 10.1111/j.1600-0854.2011.01265.x. [DOI] [PubMed] [Google Scholar]
- Amarilio R, Ramachandran S, Sabanay H, Lev S. Differential regulation of endoplasmic reticulum structure through VAP-A-Nir protein interaction. J Biol Chem. 2005;280:5934–5944. doi: 10.1074/jbc.M409566200. [DOI] [PubMed] [Google Scholar]
- Banerji S, Ngo M, Lane CF, Robinson CA, Minogue S, Ridgway ND. Oxysterol binding protein (OSBP)-dependent activation of sphingomyelin synthesis in the Golgi apparatus requires PtdIns 4-kinase IIα. Mol Biol Cell. 2010;23:4141–4150. doi: 10.1091/mbc.E10-05-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann T, Hausser A, Schoffler P, Schmid S, Pfizenmaier K, Olayioye MA. Regulation of secretory transport by protein kinase D-mediated phosphorylation of the ceramide transfer protein. J Cell Biol. 2007;178:15–22. doi: 10.1083/jcb.200612017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81:4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- Georgiev AG, Sullivan DP, Kersting MC, Dittman JS, Beh CT, Menon AK. Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic. 2011;12:1341–1355. doi: 10.1111/j.1600-0854.2011.01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser A, Storz P, Martens S, Link G, Toker A, Pfizenmaier K. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIβ at the Golgi complex. Nat Cell Biol. 2005;7:880–886. doi: 10.1038/ncb1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor βIII spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai K, Kawano M, Shinkai-Ouchi F, Nishijima M, Hanada K. Interorganelle trafficking of ceramide is regulated by phosphorylation-dependent cooperativity between the PH and START domains of CERT. J Biol Chem. 2007;282:17758–17766. doi: 10.1074/jbc.M702291200. [DOI] [PubMed] [Google Scholar]
- Lagace TA, Byers DM, Cook HW, Ridgway ND. Chinese hamster ovary cells overexpressing the oxysterol binding protein (OSBP) display enhanced synthesis of sphingomyelin in response to 25-hydroxycholesterol. J Lipid Res. 1999;40:109–116. [PubMed] [Google Scholar]
- Lagace TA, Ridgway ND. The rate-limiting enzyme in phosphatidylcholine synthesis regulates proliferation of the nucleoplasmic reticulum. Mol Biol Cell. 2005;16:1120–1130. doi: 10.1091/mbc.E04-10-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto M, Mayranpaa MI, Pellinen T, Ihalmo P, Lehtonen S, Kovanen PT, Groop PH, Ivaska J, Olkkonen VM. The R-Ras interaction partner ORP3 regulates cell adhesion. J Cell Sci. 2008;121:695–705. doi: 10.1242/jcs.016964. [DOI] [PubMed] [Google Scholar]
- Lehto M, Olkkonen VM. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim Biophys Acta. 2003;1631:1–11. doi: 10.1016/s1388-1981(02)00364-5. [DOI] [PubMed] [Google Scholar]
- Lessmann E, Ngo M, Leitges M, Minguet S, Ridgway ND, Huber M. Oxysterol-binding protein-related protein (ORP) 9 is a PDK-2 substrate and regulates Akt phosphorylation. Cell Signal. 2007;19:384–392. doi: 10.1016/j.cellsig.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP-A. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi A, Perry RJ, Storey MK, Cook HW, Byers DM, Ridgway ND. Golgi localization and phosphorylation of oxysterol binding protein in Niemann-Pick C and U18666A-treated cells. J Lipid Res. 2001;42:1062–1071. [PubMed] [Google Scholar]
- Ngo M, Ridgway ND. Oxysterol binding protein-related protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol Biol Cell. 2009;20:1388–1399. doi: 10.1091/mbc.E08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo MH, Colbourne TR, Ridgway ND. Functional implications of sterol transport by the oxysterol-binding protein gene family. Biochem J. 2010;429:13–24. doi: 10.1042/BJ20100263. [DOI] [PubMed] [Google Scholar]
- Nhek S, Ngo M, Yang X, Ng MM, Field SJ, Asara JM, Ridgway ND, Toker A. Regulation of oxysterol-binding protein Golgi localization through protein kinase D-mediated phosphorylation. Mol Biol Cell. 2010;21:2327–2337. doi: 10.1091/mbc.E10-02-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP-A proteins and is essential for Golgi-mediated transport. Mol Biol Cell. 2008;19:3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RJ, Ridgway ND. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell. 2006;17:2604–2616. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA. Non-vesicular sterol transport in cells. Prog Lipid Res. 2007;46:297–314. doi: 10.1016/j.plipres.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Im YJ, Hurley JH, Prinz WA. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J Cell Biol. 2006;173:107–119. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway ND, Badiani K, Byers DM, Cook HW. Inhibition of phosphorylation of the oxysterol binding protein by brefeldin A. Biochim Biophys Acta. 1998a;1390:37–51. doi: 10.1016/s0005-2760(97)00167-7. [DOI] [PubMed] [Google Scholar]
- Ridgway ND, Dawson PA, Ho YK, Brown MS, Goldstein JL. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J Cell Biol. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway ND, Lagace TA, Cook HW, Byers DM. Differential effects of sphingomyelin hydrolysis and cholesterol transport on oxysterol-binding protein phosphorylation and Golgi localization. J Biol Chem. 1998b;273:31621–31628. doi: 10.1074/jbc.273.47.31621. [DOI] [PubMed] [Google Scholar]
- Saito S, Matsui H, Kawano M, Kumagai K, Tomishige N, Hanada K, Echigo S, Tamura S, Kobayashi T. Protein phosphatase 2Cε is an endoplasmic reticulum integral membrane protein that dephosphorylates the ceramide transport protein CERT to enhance its association with organelle membranes. J Biol Chem. 2008;283:6584–6593. doi: 10.1074/jbc.M707691200. [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Storey MK, Byers DM, Cook HW, Ridgway ND. Cholesterol regulates oxysterol binding protein (OSBP) phosphorylation and Golgi localization in Chinese hamster ovary cells: correlation with stimulation of sphingomyelin synthesis by 25-hydroxycholesterol. Biochem J. 1998;336:247–256. doi: 10.1042/bj3360247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek M, Hynynen R, Wohlfahrt G, Lehto M, Johansson M, Saarinen H, Radzikowska A, Thiele C, Olkkonen VM. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige N, Kumagai K, Kusuda J, Nishijima M, Hanada K. Casein kinase Iγ2 down-regulates trafficking of ceramide in the synthesis of sphingomyelin. Mol Biol Cell. 2009;20:348–357. doi: 10.1091/mbc.E08-07-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PY, Weng J, Anderson RG. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 2005;307:1472–1476. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- Waugh MG, Minogue S, Chotai D, Berditchevski F, Hsuan JJ. Lipid and peptide control of phosphatidylinositol 4-kinase IIα activity on Golgi-endosomal rafts. J Biol Chem. 2006;281:3757–3763. doi: 10.1074/jbc.M506527200. [DOI] [PubMed] [Google Scholar]
- Wyles JP, McMaster CR, Ridgway ND. Vesicle-associated membrane protein-associated protein-A (VAP-A-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J Biol Chem. 2002;277:29908–29918. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- Wyles JP, Ridgway ND. VAMP-associated protein-A regulates partitioning of oxysterol-binding protein-related protein-9 between the endoplasmic reticulum and Golgi apparatus. Exp Cell Res. 2004;297:533–547. doi: 10.1016/j.yexcr.2004.03.052. [DOI] [PubMed] [Google Scholar]
- Zahedi RP, Lewandrowski U, Wiesner J, Wortelkamp S, Moebius J, Schutz C, Walter U, Gambaryan S, Sickmann A. Phosphoproteome of resting human platelets. J Proteome Res. 2008;7:526–534. doi: 10.1021/pr0704130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.