Cyclin-dependent kinase-5 and the kinesin-3 family motor KLP-4 function in the same pathway to promote anterograde trafficking of glutamate receptors in the ventral nerve cord of Caenorhabditis elegans. This study reveals a cellular control mechanism by which receptor cargo is targeted for degradation in the absence of its motor.
Abstract
The transport of glutamate receptors from the cell body to synapses is essential during neuronal development and may contribute to the regulation of synaptic strength in the mature nervous system. We previously showed that cyclin-dependent kinase-5 (CDK-5) positively regulates the abundance of GLR-1 glutamate receptors at synapses in the ventral nerve cord (VNC) of Caenorhabditis elegans. Here we identify a kinesin-3 family motor klp-4/KIF13 in a cdk-5 suppressor screen for genes that regulate GLR-1 trafficking. klp-4 mutants have decreased abundance of GLR-1 in the VNC. Genetic analysis of klp-4 and the clathrin adaptin unc-11/AP180 suggests that klp-4 functions before endocytosis in the ventral cord. Time-lapse microscopy indicates that klp-4 mutants exhibit decreased anterograde flux of GLR-1. Genetic analysis of cdk-5 and klp-4 suggests that they function in the same pathway to regulate GLR-1 in the VNC. Interestingly, GLR-1 accumulates in cell bodies of cdk-5 but not klp-4 mutants. However, GLR-1 does accumulate in klp-4–mutant cell bodies if receptor degradation in the multivesicular body/lysosome pathway is blocked. This study identifies kinesin KLP-4 as a novel regulator of anterograde glutamate receptor trafficking and reveals a cellular control mechanism by which receptor cargo is targeted for degradation in the absence of its motor.
INTRODUCTION
The neurotransmitter glutamate is responsible for the majority of fast excitatory synaptic transmission in the mammalian CNS. Activity-induced changes in synaptic strength are believed to be the cellular mechanism underlying learning and memory (Bredt and Nicoll, 2003; Shepherd and Huganir, 2007; Kessels and Malinow, 2009). One mechanism by which neurons regulate synaptic strength is through altering the number of glutamate receptors (GluRs) in the postsynaptic membrane. Many studies have focused on how activity-dependent exocytosis and endocytosis of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)–type GluRs regulate synaptic GluR abundance; however, much less is known about the mechanisms involved in the anterograde trafficking of GluRs from the cell body to synapses (Bredt and Nicoll, 2003; Shepherd and Huganir, 2007; Kessels and Malinow, 2009; van der Sluijs and Hoogenraad, 2011).
Kinesin and dynein superfamily proteins are ATP-dependent molecular motors that transport diverse cellular cargoes along microtubules to various destinations in the cell (Vale, 2003; Goldstein et al., 2008; Hirokawa et al., 2010). Microtubule-dependent motors are essential for synapse development and synaptic transmission. Loss of motor function in Caenorhabditis elegans, Drosophila, and mammalian neurons can result in the accumulation of cargo early in the secretory pathway and the mistrafficking of cargo to inappropriate subcellular compartments. For example, loss of function of kinesin UNC-104/KIF1A or kinesin UNC-116/KIF5 results in the accumulation of presynaptic cargoes in neuronal cell bodies and axons and, in some cases, in the inappropriate trafficking of cargo into dendrites (Hall and Hedgecock, 1991; Ferreira et al., 1992; Okada et al., 1995; Yonekawa et al., 1998; Byrd et al., 2001; Sakamoto et al., 2005; Sieburth et al., 2005; Pack-Chung et al., 2007; Barkus et al., 2008; Ou et al., 2010; Goodwin et al. 2012). Aberrant accumulation of cargo in neuronal processes is observed in several neurological diseases and can result in synaptic transmission defects, paralysis, and neurodegeneration (Yonekawa et al., 1998; Martin et al., 1999; Hafezparast et al., 2003; Koushika et al., 2004; Duncan and Goldstein, 2006; De Vos et al., 2008). These studies illustrate the important role that motor proteins play in the function and viability of neurons and suggest a link between inappropriate motor activity and neurodegenerative disorders.
Several microtubule-based motors are implicated in transporting AMPA receptors to synapses in mammalian cultured neurons. The microtubule minus end–directed motor cytoplasmic dynein transports AMPA receptors into proximal dendrites, which contain microtubules of mixed polarity (Kim and Lisman, 2001; Kapitein et al., 2010). The scaffolding protein GRIP1 recruits AMPA receptors to the microtubule plus end–directed motor kinesin-1/KIF5 and steers the complex into dendrites (Kim and Lisman, 2001; Setou et al., 2002). Finally, the kinesin-3 family motor UNC-104/KIF1A, which transports presynaptic cargo to synapses (Hall and Hedgecock, 1991; Okada et al., 1995), has been implicated in trafficking AMPA receptors into dendrites via the scaffolding protein SYD-2/liprin-α (Wyszynski et al., 2002; Shin et al., 2003). These results illustrate a common theme in motor-dependent transport in which one cargo uses multiple motors to reach its ultimate destination in the cell (Vale, 2003; Guzik and Goldstein, 2004; Hirokawa et al., 2010).
The genetic model organism C. elegans has been used to identify many genes that are involved in GluR trafficking; however, the molecular motors involved in transporting GluRs from the cell body to synapses in C. elegans are not known. The C. elegans genome contains 10 glutamate receptor subunits: 2 NMDA type and 8 non-NMDA type (Brockie et al., 2001). GLR-1 is a non-NMDA receptor that is most homologous to AMPA-type glutamate receptors (Hart et al., 1995; Maricq et al., 1995; Brockie et al., 2001). GLR-1 is expressed in a population of ventral cord interneurons, where it is localized to sensory–interneuron and interneuron–interneuron synapses (Hart et al., 1995; Maricq et al., 1995; Rongo et al., 1998; Brockie et al., 2001; Burbea et al., 2002) and is required for several glutamate-dependent behaviors (Hart et al., 1995; Maricq et al., 1995; Zheng et al., 1999; Mellem et al., 2002; Chao et al., 2004). In this study, we identify the kinesin KLP-4 as a novel regulator of glutamate receptor trafficking in C. elegans.
KLP-4 is a kinesin-3 family member that is distinct from UNC-104/KIF1A and most homologous to Drosophila kinesin Khc-73 and mammalian kinesins KIF13A and KIF13B. In Drosophila, Khc-73 regulates neuroblast polarity and mitotic spindle orientation and has recently been shown to localize to endosomes (Siegrist and Doe, 2005; Huckaba et al., 2011). In mammals, KIF13A transports the mannose-6-phosphate receptor from the trans-Golgi network to the plasma membrane (Nakagawa et al., 2000), mediates endosomal sorting of cargo during melanosome biogenesis (Delevoye et al., 2009), and regulates cytokinesis by recruiting phosphatidylinositol 3-phosphate (PI3P) and scission machinery to the midbody (Sagona et al., 2010). KIF13B (also known as GAKIN; Hanada et al., 2000) mediates axonal polarity in hippocampal neurons in culture through its ability to transport PIP3-containing vesicles into axons (Venkateswarlu et al., 2005; Horiguchi et al., 2006).
We show here that KLP-4 promotes the abundance of the glutamate receptor GLR-1 in the ventral nerve cord (VNC). Genetic analysis and time-lapse data suggest that KLP-4 regulates the anterograde trafficking of GLR-1. We previously showed that cyclin-dependent kinase-5 (CDK-5) positively regulates the abundance of GLR-1 in the VNC (Juo et al., 2007). Our genetic analyses indicate that KLP-4 functions in the same pathway as CDK-5 to regulate GLR-1 trafficking. Furthermore, we find that in the absence of functional KLP-4 motors, GLR-1 does not accumulate in the cell body but is instead targeted for degradation in the multivesicular body (MVB)/lysosome pathway. Our results suggest that regulating the amount or availability of a motor can influence the cellular fate of its cargo.
RESULTS
Identification of KLP-4
We study GLR-1 trafficking in C. elegans by analyzing the distribution of GLR-1 tagged with green fluorescent protein (GFP) at an internal site in its cytoplasmic tail (GLR-1::GFP; Rongo et al., 1998). GLR-1::GFP, expressed under the glr-1 promoter, localizes to puncta in the VNC (Figure 1B). Greater than 80% of these puncta are closely apposed to presynaptic markers, indicating that the majority of GLR-1::GFP puncta in the VNC represent postsynaptic sites (Rongo et al., 1998; Burbea et al., 2002). Expression of GLR-1::GFP under its own promoter rescues the behavioral defects of glr-1–null mutant animals, indicating that the GFP-tagged receptor is functional (Rongo et al., 1998).
FIGURE 1:
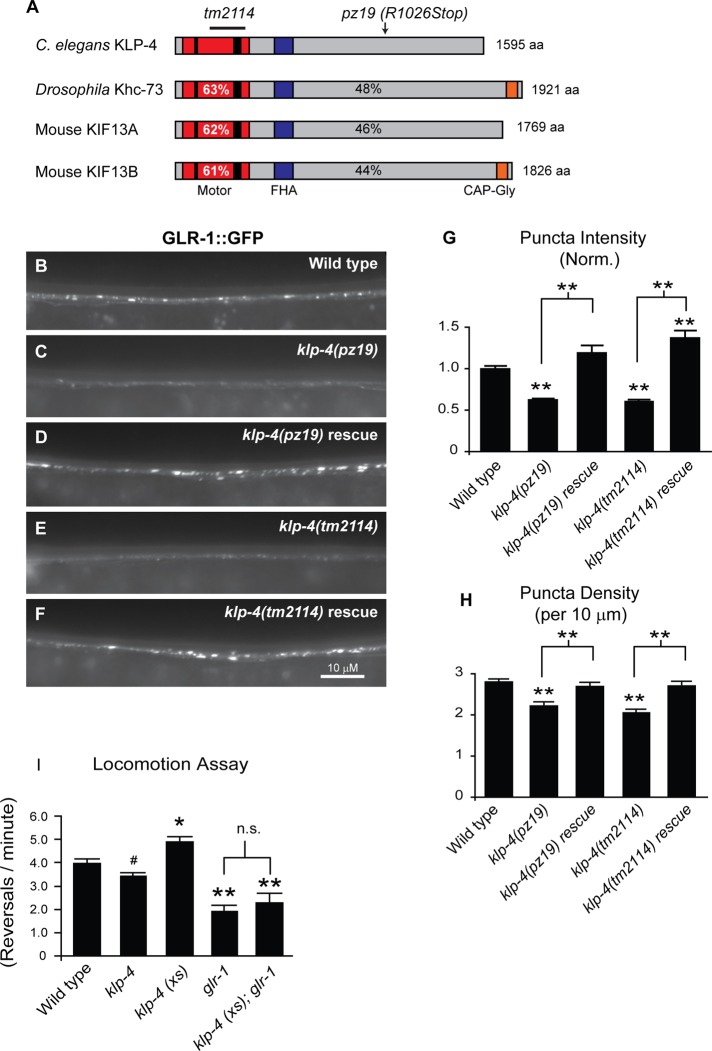
The kinesin KLP-4 functions in the VNC to regulate the abundance of GLR-1 and GLR-1–dependent behavior. (A) Protein domain organization of C. elegans KLP-4, Drosophila Khc-73, and mouse KIF13A and KIF13B. The N-terminal motor domain (red box) containing the signature ATP-binding motif (thin black bar) and microtubule-binding regions (thick black bar), the cargo-binding domain (gray box), the Forkhead-associated domain (FHA; purple box), and the microtubule plus end–binding domain (CAP-Gly; orange box) are shown. The percentage identity of the motor domain of C. elegans KLP-4 (percentages in white) or full-length KLP-4 (percentages in black) vs. Drosophila Khc73 and mouse KIF13A and KIF13B are indicated. (B–F) Representative images of the anterior VNC of larval stage 4 (L4) wild-type (B), klp-4(pz19) (C), rescued klp-4(pz19) (D), klp-4(tm2114) (E), and rescued klp-4(tm2114) (F) animals expressing an integrated GLR-1::GFP transgene (nuIs25). In these and all subsequent images anterior is to the left and ventral is up. (G, H) Quantification of GLR-1::GFP puncta intensities (normalized; G) and densities (H) for the strains pictured in B–F. Means and SEM are shown for n = 38 wild-type, n = 23 klp-4(pz19), n = 21 rescued klp-4(pz19), n = 20 klp-4(tm2114), and n = 21 rescued klp-4(tm2114) animals. (I) Quantification of number of spontaneous reversals per minute for n = 20 wild-type, n = 20 klp-4(tm2114), and n = 23 animals expressing an integrated klp-4 transgene under the control of the glr-1 promoter (klp-4(xs)) (pzIs20), n = 14 glr-1(n2461), and n = 13 glr-1(n2461) animals overexpressing klp-4 (klp-4(xs)). Values that differ significantly from wild type are indicated by asterisks above each bar, whereas other comparisons are marked by brackets (**p ≤ 0.001, *p ≤ 0.01, #p ≤ 0.05, Tukey–Kramer test).
To identify novel genes involved in GLR-1 trafficking, we took advantage of a previously identified gene, cyclin-dependent kinase-5 (cdk-5), which regulates the abundance of GLR-1 in the VNC of C. elegans (Juo et al., 2007). cdk-5 loss-of-function mutants have decreased GLR-1 in the VNC and a corresponding accumulation of GLR-1 in the cell body. In contrast, overexpression of cdk-5 results in increased levels of GLR-1 in the VNC (Juo et al., 2007). We performed a forward genetic suppressor screen to identify mutants that blocked the effects of cdk-5 overexpression on GLR-1 in the VNC. From this screen we isolated pz19 as a strong suppressor mutant. Genetic mapping and sequencing analysis indicate that the pz19 allele contains a mutation in the kinesin-3 family motor klp-4 (see Materials and Methods). The pz19 mutation corresponds to a single C-to-T point mutation (C3076T) in the cargo-binding tail of klp-4 that results in a premature stop codon (R1026Stop; Figure 1A). The C. elegans genome encodes three kinesin-3 family members, UNC-104/KIF1A, KLP-4/KIF13, and KLP-6/KIF28 (alignment shown in Supplemental Figure S1A; Peden and Barr, 2005). KLP-4 is homologous to Drosophila kinesin-73 (Khc-73; 48% identity) and mammalian KIF13A (46% identity) and KIF13B/GAKIN (44% identity; Figure 1A and Supplemental Figure S1B). KLP-4 is likely a microtubule plus end–directed motor because its N-terminal motor domain is >60% identical to the motor domains of plus end–directed kinesins Khc-73, KIF13A, and KIF13B (Figure 1A and Supplemental Figure S1B). In addition, KLP-4 contains the signature amino acids found in plus-end motors (Case et al., 1997; Supplemental Figure S1). Of the two mammalian homologues, KLP-4 appears to be more closely related to KIF13A based on sequence identity and the lack of a microtubule plus end–binding CAP-Gly domain, which is found at the C-terminus of KLP13B (Figure 1A).
To investigate the expression pattern of klp-4, we generated a transcriptional reporter strain consisting of the klp-4 promoter driving expression of GFP (Pklp-4::GFP). We found that Pklp-4::GFP is expressed in the pharynx, in the intestine, and throughout the nervous system, including the nerve ring and the lateral, dorsal, and ventral nerve cords (Supplemental Figure S2A-F and data not shown). This klp-4 expression pattern is consistent with previous expression studies (McKay et al., 2003; Hunt-Newbury et al., 2007). In addition, Pklp-4::GFP is expressed in many head, tail, and ventral cord neurons, including glr-1–expressing neurons, based on the overlapping expression pattern of Pklp-4::GFP and a glr-1 transcriptional reporter (Pglr-1::dsRED; Supplemental Figure S2, G–I).
Kinesin KLP-4 functions in GLR-1–expressing interneurons to positively regulate GLR-1 abundance
We initially identified klp-4 mutants by their ability to suppress the effects of cdk-5 overexpression on GLR-1. To determine whether mutation of klp-4 alone affects the abundance of GLR-1 in the VNC in the absence of cdk-5 overexpression, we examined the distribution of GLR-1::GFP in the anterior region of the VNC of klp-4(pz19) mutant animals. We found that klp-4(pz19) mutant animals expressing an integrated GLR-1::GFP transgene (nuIs25) exhibit a 37% decrease in GLR-1::GFP puncta intensity (p < 0.001) and a 21% decrease in puncta density (p < 0.001) compared with wild-type controls (Figure 1, B, C, G, and H; see Materials and Methods for quantification). We confirmed this effect by analyzing the distribution of GLR-1::GFP in a second, independent klp-4 mutant, klp-4(tm2114). The tm2114 mutant allele corresponds to a 747–base pair deletion in klp-4, resulting in the deletion of most of the motor domain, followed by a frame shift and a premature stop codon near the end of the motor domain (Figure 1A and Supplemental Figure S1). Thus tm2114 likely represents a functional null mutant of klp-4. We found that klp-4(tm2114) mutants exhibit a 39% decrease in GLR-1::GFP puncta intensity (p < 0.001) and a 27% decrease in puncta density (p < 0.001) compared with wild-type controls (Figure 1, B, E, G, and H). Expression of wild-type klp-4 cDNA under the control of the glr-1 promoter corrects the defects in GLR-1::GFP puncta intensities and densities in both klp-4(pz19) and klp-4(tm2114) mutants (Figure 1, D and F–H). In addition, GLR-1::GFP abundance decreases in klp-4 mutants in the posterior VNC and in the nerve ring, suggesting that KLP-4 promotes GLR-1 levels at synapses throughout the neurons (Supplemental Figure S3). Conversely, we found that increasing KLP-4 activity in wild-type animals by overexpressing klp-4 under control of the glr-1 promoter results in increased levels of GLR-1 in the VNC (Supplemental Figure S4). Taken together, these data indicate that KLP-4 functions in glr-1–expressing interneurons to positively regulate GLR-1 abundance in the VNC.
KLP-4 regulates GLR-1–dependent locomotion behavior
If KLP-4 regulates the synaptic levels of GLR-1, we might expect klp-4 loss-of-function mutants and animals overexpressing klp-4 to affect GLR-1 signaling and consequently glutamate-dependent behaviors, such as locomotion. C. elegans locomotion is characterized by periods of forward movement interrupted by brief periods of backward movement. The frequency of these spontaneous reversals is regulated by the level of glutamatergic signaling (Zheng et al., 1999). For example, animals with decreased glutamatergic signaling, such as mutants lacking glr-1 or the vesicular glutamate transporter eat-4/VGLUT, exhibit decreased reversal frequencies (Zheng et al., 1999; Burbea et al., 2002), and conversely, animals with increased glutamatergic signaling have increased reversal frequencies (Zheng et al., 1999; Burbea et al., 2002; Juo and Kaplan, 2004; Juo et al., 2007; Schaefer and Rongo, 2006). We found that klp-4(tm2114) mutants exhibit a small but significant (p < 0.05) decrease in reversal frequency, consistent with a decrease in glutamatergic signaling (Figure 1I). Conversely, we found that animals overexpressing klp-4 (under control of the glr-1 promoter) exhibit an increase in reversal frequency (p < 0.01) consistent with increased glutamatergic signaling (Figure 1I). Importantly, this increase in reversal frequency was completely blocked by mutations in glr-1, suggesting that the effect of KLP-4 on locomotion is dependent on endogenous GLR-1 (Figure 1I). Thus loss and gain of function of KLP-4 regulates GLR-1::GFP abundance in the VNC and has corresponding effects on GLR-1-dependent locomotion behavior.
KLP-4 does not affect the distribution of several other synaptic markers
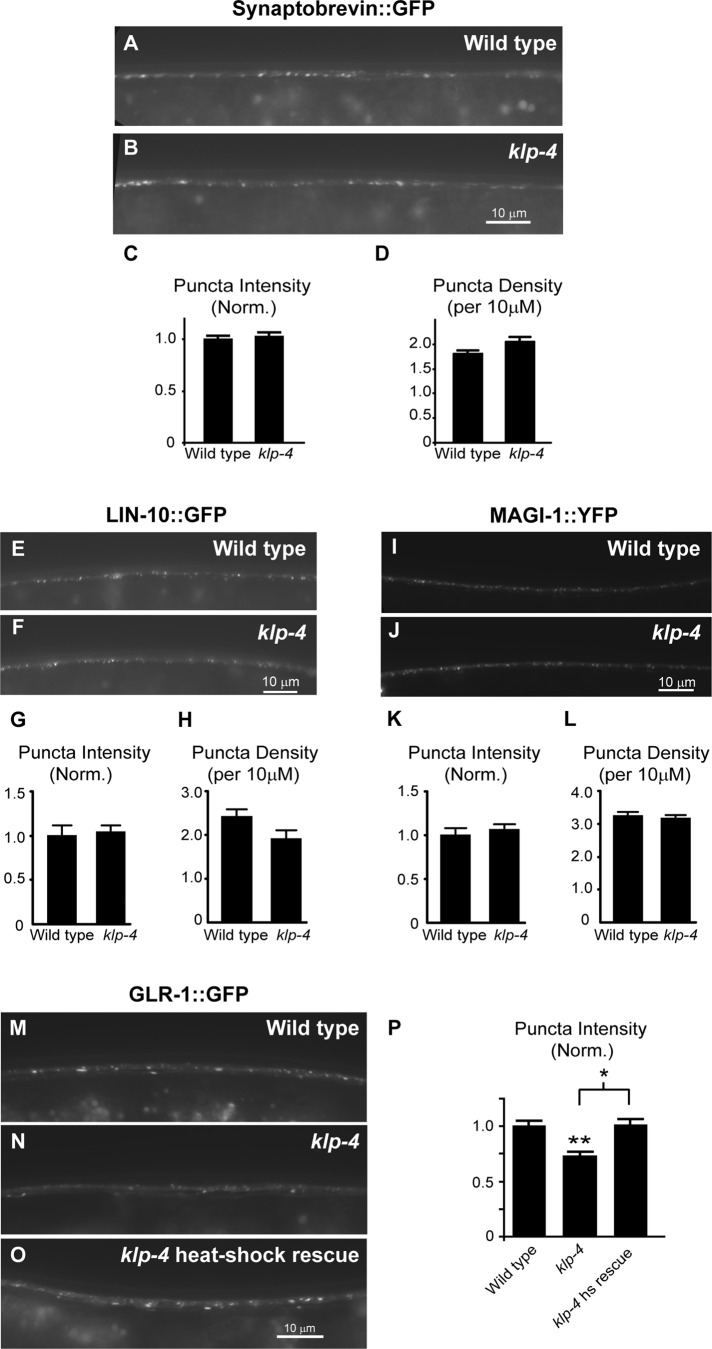
The decrease in GLR-1::GFP in the VNC of klp-4 mutants could be due to a decrease in synaptic connections. We tested this possibility by examining the distribution of the synaptic vesicle protein synaptobrevin tagged with GFP (SNB-1::GFP) in the VNC of wild-type and klp-4 mutant animals. We observed no difference in the intensity or density of SNB-1::GFP puncta in klp-4 mutants compared with wild-type controls (Figure 2, A–D). We also analyzed the distribution of two other synaptic proteins that have been shown to colocalize with GLR-1 in the VNC—the PDZ protein LIN-10/Mint1 and the MAGUK protein MAGI-1/S-SCAM (Rongo et al., 1998; Emtage et al., 2009). We found that the distributions of LIN-10::GFP (Figure 2, E–H) and MAGI-1::YFP (Figure 2, I–L) were unaltered in klp-4 mutants when compared with wild-type controls. These results suggest that klp-4 mutations do not result in a general defect in trafficking of synaptic proteins, and glr-1–expressing interneurons likely have normal synaptic inputs in klp-4 mutants. These data are consistent with a specific role for KLP-4 in regulating GLR-1 in the VNC.
FIGURE 2:
KLP-4 does not affect the distribution of multiple synaptic markers and is not required during early development to regulate GLR-1. (A, B, E, F, I, J) Representative images of the anterior VNC of L4 wild-type (A, E, I) and klp-4(tm2114) (B, F, J) animals expressing either synaptobrevin::GFP (nuIs125) (A, B), LIN-10::GFP (nuEx993) (E, F), or MAGI-1::YFP (nuEx1004) (I, J) under the control of the glr-1 promoter. (C, D, G, H, K, L) Quantification of puncta intensities (normalized; C, G, K) and densities (D, H, L) for the strains pictured. Mean and SEM are shown for at least n = 20 animals per genotype. No significant difference was found in puncta fluorescence intensities or densities between wild-type and klp-4 mutant animals for any of the synaptic markers (p > 0.05, Student's t test). (M–O) Representative images of the anterior VNC of adult wild-type (M), klp-4(tm2114) (N), and heat-shock–rescued klp-4(tm2114) (O) animals expressing GLR-1::GFP (nuIs25) after 16 h of heat shock. Heat-shock–rescued klp-4–mutant animals express wild-type klp-4::mCherry under control of the heat-shock–inducible promoter (pzEx237). (P) Quantification of GLR-1::GFP puncta intensities (normalized) for the strains pictured in M–O. Means and SEM are shown for n = 18 wild-type, n = 35 klp-4(tm2114), and n = 23 heat-shock–rescued klp-4(tm2114) animals. Values that differ significantly from wild type are indicated by asterisks above each bar, whereas other comparisons are marked by brackets (**p ≤ 0.001, *p ≤ 0.01, Tukey–Kramer test).
KLP-4 is not required during early development to regulate GLR-1 in the VNC
To test more directly whether the decreased levels of GLR-1 observed in klp-4 mutants could be due to defects in early development, we expressed klp-4 cDNA in klp-4 mutants using a heat-shock–inducible promoter (hsp16.2). We raised animals at 20°C until the L4 stage of development, when all our static imaging was performed. We then induced expression of klp-4 by shifting the animals to 30°C and imaged GLR-1::GFP 16h later in adult animals. Similar to non–heat-shocked animals, we found that heat-shocked klp-4(tm2114) mutant animals exhibited a 27% decrease (p < 0.001) in GLR-1::GFP puncta intensity in the VNC compared with heat-shocked wild-type controls (Figure 2, M–P). In contrast, heat-shocked klp-4 mutants expressing klp-4 cDNA under control of a heat-shock–inducible promoter (Phsp16.2::klp-4) completely restored GLR-1::GFP puncta intensities to wild-type levels (Figure 2, M–P). These results suggest that KLP-4 is not required during early development to regulate GLR-1 levels in the VNC and that KLP-4 activity may be continuously required to maintain GLR-1 levels in the mature nervous system.
KLP-4 functions before endocytosis of GLR-1 at synapses in the VNC
One possible mechanism by which KLP-4 could regulate GLR-1::GFP abundance in the VNC is through regulation of glr-1 transcription. If this were the case, we would expect decreased levels of glr-1 transcript in klp-4 mutants. We determined the relative amounts of glr-1 to act-1 (actin) mRNA in wild-type, klp-4(tm2114), and klp-4(pz19) mutant animals using real-time PCR. Rather than a decrease in transcription, we found that both klp-4 mutants had an increase in glr-1 transcript compared with wild-type controls (Supplemental Figure S5). This result indicates that the decrease in GLR-1::GFP in the VNC of klp-4 mutants is not due to a decrease in glr-1 transcription. The increased glr-1 transcripts observed in klp-4 mutants might be due to a compensatory feedback mechanism triggered by the decreased GLR-1 in the VNC.
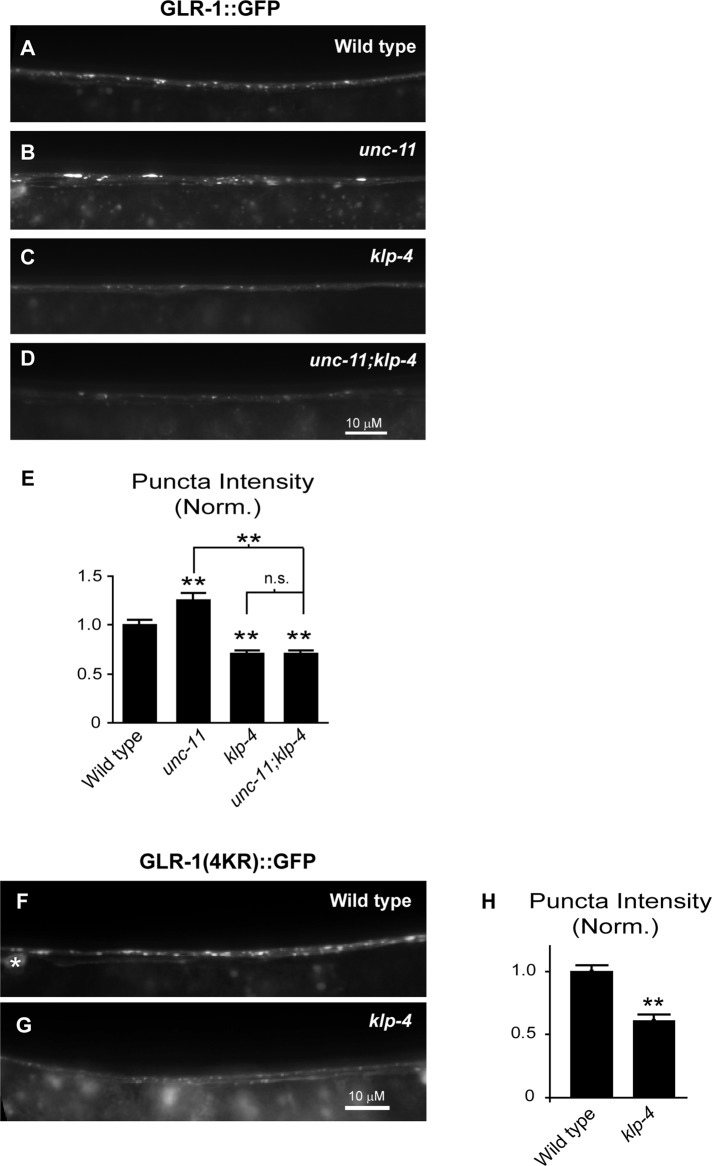
We next tested whether KLP-4 was involved in one of several GLR-1–trafficking steps. GluRs are transported from the cell body to synapses, inserted into the postsynaptic membrane via exocytosis, internalized via endocytosis, and subsequently either degraded in lysosomes or recycled back to the plasma membrane (Ehlers, 2000; Lin et al., 2000; Shepherd and Huganir, 2007; van der Sluijs and Hoogenraad, 2011). Decreased GLR-1 levels observed in the VNC of klp-4 mutants could be due to a function of KLP-4 in anterograde transport of GLR-1 from the cell body to the VNC or at one of the postendocytic trafficking steps (i.e., by inhibiting trafficking to the lysosome or by promoting reinsertion into the membrane). We tested whether klp-4 functions before clathrin-mediated endocytosis at the plasma membrane by analyzing GLR-1 in the VNC of klp-4;unc-11 double-mutant animals. Mutations in the clathrin adaptin unc-11/AP180 result in defects in clathrin-mediated endocytosis (Zhang et al., 1998; Nonet et al., 1999) and the accumulation of GLR-1::GFP in the VNC (Burbea et al., 2002). If KLP-4 acts at a step before GLR-1 endocytosis in the VNC (i.e., to promote the anterograde trafficking of GLR-1 from the cell body), we would expect klp-4;unc-11 double mutants to have decreased GLR-1::GFP in the VNC as observed in klp-4 single mutants. However, if KLP-4 functions at a postendocytic trafficking step, we would expect GLR-1::GFP to accumulate in the VNC of klp-4;unc-11 double mutants as observed in unc-11 single mutants. We found that klp-4;unc-11 double mutants had decreased GLR-1::GFP fluorescence intensities in the VNC, and this effect was indistinguishable from that for klp-4 single mutants (p = 0.99; Figure 3, A–E). Thus mutations in klp-4 prevent GLR-1::GFP accumulation in the VNC of unc-11 mutants. This result suggests that KLP-4 acts before GLR-1 endocytosis at the synapse and is consistent with a role for this motor in the anterograde trafficking of GLR-1.
FIGURE 3:
KLP-4 functions before GLR-1 endocytosis in the VNC. (A–D) Representative images of the anterior VNC of wild-type (A), unc-11(e47) (B), klp-4 (tm2114) (C), and unc-11(e47);klp-4(tm2114) (D) L4 animals expressing GLR-1::GFP (nuIs25). (E) Quantification of GLR-1::GFP puncta intensities (normalized) for the strains pictured in A–D. Mean and SEM are shown for n = 24 wild-type, n = 25 unc-11, n = 22 klp-4, and n = 22 unc-11;klp-4 animals. (F, G) Representative images of the anterior VNC of wild-type (F) and klp-4(tm2114) (G) L4 animals expressing GLR-1(4KR)::GFP under the control of the glr-1 promoter (nuIs108). The white asterisk marks a neuronal cell body. (H) Quantification of GLR-1(4KR)::GFP puncta intensities (normalized) for the strains pictured in F and G. Mean and SEM are shown for n = 25 wild-type and n = 19 klp-4 animals. (E, H) Values that differ significantly from wild type are indicated by asterisks above each bar, whereas other comparisons are marked by brackets (**p ≤ 0.001, Tukey–Kramer test). n.s., no significant difference (p > 0.05).
To investigate further whether KLP-4 acts before clathrin-mediated endocytosis and degradation of GLR-1 at the synapse, we tested whether klp-4 mutations affect the abundance of a nonubiquitinatable version of GLR-1::GFP. Clathrin-mediated endocytosis and degradation of GLR-1 in the lysosome is dependent on the ubiquitination of GLR-1 (Burbea et al., 2002; Chun et al., 2008; Kowalski et al., 2011). Ubiquitination is a process in which the 76–amino acid protein ubiquitin is conjugated to lysine residues on target proteins. A mutant version of GLR-1::GFP that has all four intracellular lysine residues mutated to arginines (GLR-1(4KR)::GFP) cannot be ubiquitinated and therefore accumulates in the VNC (Burbea et al., 2002). We analyzed the levels of GLR-1(4KR)::GFP in the VNC of klp-4 mutants to test whether KLP-4 functions before degradation of GLR-1 at the synapse. We found that the fluorescence intensity of GLR-1(4KR)::GFP in the VNC decreased by 39% in the VNC of klp-4 mutants compared with wild-type controls (p < 0.001; Figure 3, F–H). The magnitude of this decrease was similar to that observed with wild-type GLR-1::GFP in klp-4 mutants (Figure 1). These data suggest that KLP-4 functions before clathrin-mediated endocytosis and degradation of GLR-1 at synapses in the VNC, providing support for a role of KLP-4 in the anterograde trafficking of GLR-1.
KLP-4 promotes the anterograde transport of GLR-1
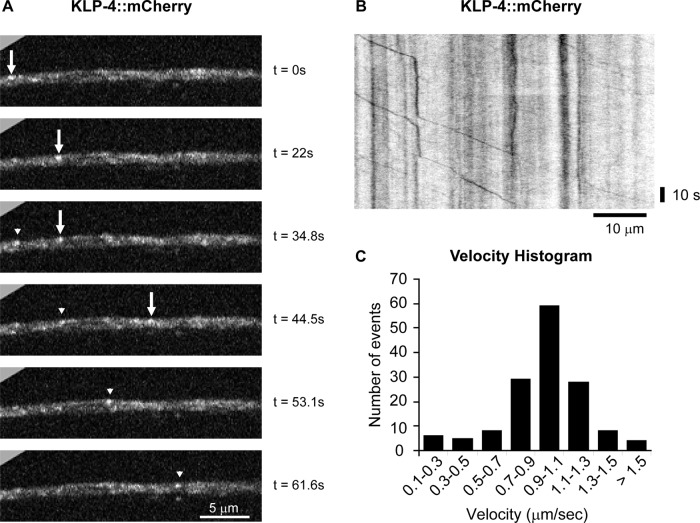
If KLP-4 functions as a motor to regulate the anterograde trafficking of GLR-1, we would expect KLP-4 to exhibit transport characteristics similar to those of other fast neuronal motors. We tagged KLP-4 with mCherry on its C-terminus and expressed the tagged motor in interneurons using the glr-1 promoter in order to investigate the subcellular localization and transport characteristics of KLP-4. We found that KLP-4::mCherry was localized to puncta in the cell body and throughout the VNC (Figure 4 and data not shown). Time-lapse microscopy and kymograph analysis revealed that KLP-4::mCherry puncta were mobile in cell bodies and the VNC and moved with an average velocity of 1.0 μm/s and an average run length of 9.2 μm (Figure 4 and Supplemental Videos S1 and S2). This velocity of KLP-4 is comparable to the average in vitro velocities observed for other kinesin-3 family motors, such as Khc-73, KIF13A, and KIF13B (Okada et al., 1995; Horiguchi et al., 2006; Huckaba et al., 2011), and the in vivo velocity of GFP-tagged UNC-104 (Zhou et al., 2001) and indicates that KLP-4 is a fast neuronal motor that is mobile in interneuron cell bodies and VNC processes.
FIGURE 4:
KLP-4 is a fast neuronal motor that moves in the VNC. (A) Individual frames from a time-lapse movie (Supplemental Video S1) show KLP-4::mCherry puncta in the anterior VNC of young adult wild-type animals expressing KLP-4::mCherry under the control of the glr-1 promoter (pzEx234). The arrow and arrowhead mark two different KLP-4::mCherry puncta moving from the head toward the tail in the interneuron processes of the VNC. Images are oriented with anterior to the left. Time in seconds is marked on the right. (B) A representative kymograph showing mobile and stationary KLP-4::mCherry puncta was generated from the same time-lapse movie (Supplemental Video S1) of KLP-4::mCherry in the anterior VNC. The kymograph is oriented with anterior to the left. (C) Velocity histogram of KLP-4::mCherry puncta moving in the VNC from the head toward the tail (n = 147 mobility events). The maximum KLP-4::mCherry velocity was 2.0 μm/s from the head toward the tail and 2.6 μm/s from the tail toward the head.
To test directly whether KLP-4 regulates GLR-1 transport, we first analyzed the movement of GLR-1::GFP puncta in the VNC of wild-type and klp-4 mutant animals using time-lapse microscopy and kymograph analysis (Supplemental Figure S6; see Materials and Methods). The vast majority of GLR-1::GFP–expressing ventral cord neurons possess cell bodies in the head of the animal and send processes toward the tail; however, two strongly expressing GLR-1::GFP neurons that contribute processes to the VNC possess cell bodies in the tail (White et al., 1976; Hart et al., 1995; Maricq et al., 1995; Brockie et al., 2001). In wild-type animals, we observed that GLR-1::GFP puncta moved with a maximum velocity of 2.2 μm/s from the head toward the tail and 2.7 μm/s from the tail toward the head. For GLR-1::GFP puncta moving from the head toward the tail, the average velocity was 1.0 μm/s and the average run length was 9.1 μm. Time-lapse analysis of klp-4 mutants revealed several differences in the movement of GLR-1::GFP puncta from the head toward the tail in the VNC compared with wild-type animals. First, there was a 31% decrease in the flux of GLR-1::GFP puncta in klp-4 mutants compared with controls (p < 0.005; Supplemental Figure S6). Second, the average run length of mobile GLR-1::GFP puncta was decreased by 45% (p < 0.005; Supplemental Figure S6), and the average pause frequency was increased by 56% in klp-4 mutants compared with wild type (average number of pauses/μm: wild type, 0.09 ± 0.008; klp-4, 0.14 ± 0.007; p < 0.001). This reduced run length combined with the increased pause frequency of GLR-1::GFP puncta in klp-4 mutants is consistent with an overall reduction in the processivity of cargo motion. Third, the average velocity of GLR-1::GFP puncta was decreased by 32% in klp-4 mutants compared with controls (average head-to-tail velocity: wild type, 1.0 ± 0.02 μm/s [n = 158]; klp-4, 0.67±0.05 μm/s [n = 196]; p < 0.001). We found no significant differences in any of the parameters for GLR-1::GFP puncta moving in the VNC from the tail toward the head (Supplemental Figure S6; average tail to head velocity: wild type, 1.29 ± 0.06 μm/s [n = 79]; klp-4, 1.29 ± 0.06 μm/s [n = 68]; p > 0.05). Taken together with our previous data, these results suggest that KLP-4 positively regulates trafficking of GLR-1::GFP from the head toward the tail in the VNC. Because the majority of glr-1–expressing cell bodies are in the head, these data implicate a role for KLP-4 in the anterograde trafficking of GLR-1.
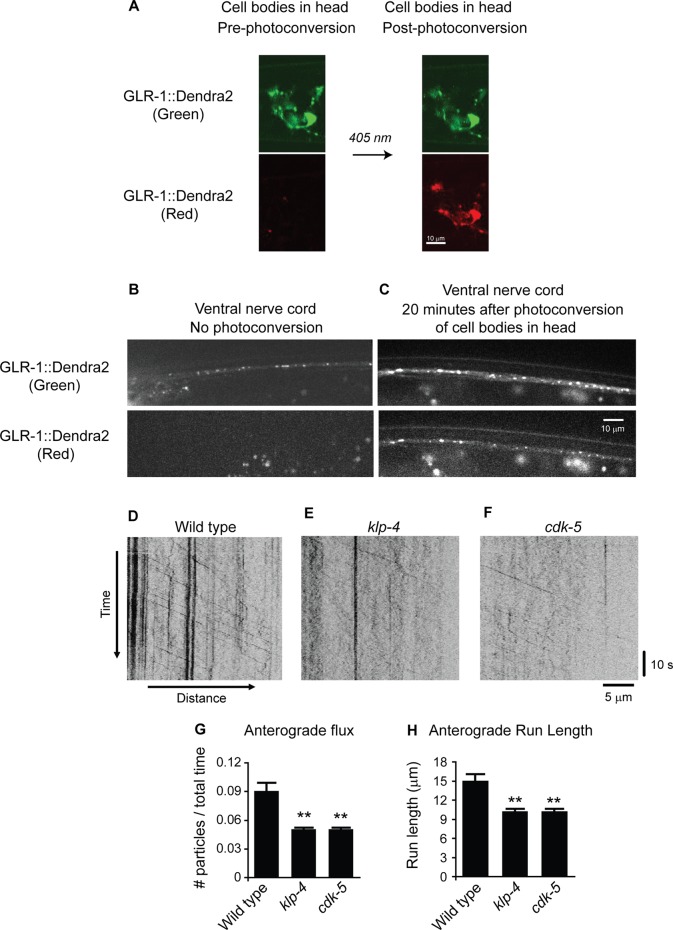
To measure anterograde trafficking of GLR-1 more accurately, we repeated our time-lapse analysis on transgenic animals (pzIs18) expressing GLR-1 tagged with the photoconvertible fluorescent protein Dendra2. Dendra2 fluorescence is irreversibly converted from green to red by ultraviolet (UV) light, providing an optical method to photolabel and track a defined population of proteins within cells (Gurskaya et al., 2006; Chudakov et al., 2007). We used a confocal microscope to locally photoconvert GLR-1::Dendra2 from green to red in cell bodies of glr-1–expressing head neurons (Figure 5A) of wild-type and klp-4 mutants and performed time-lapse microscopy of the red mobile GLR-1::Dendra2 puncta in the VNC ∼20 min postconversion (Figure 5, D–H). Control experiments show that before photoconversion or immediately after local photoconversion of neuron cell bodies in the head, there is no red GLR-1::Dendra2 signal in the VNC (Figure 5B and data not shown), whereas 20 min postconversion, we observe red GLR-1::Dendra2 puncta in the VNC (Figure 5C). Time-lapse analysis of red GLR-1::Dendra2 puncta moving in the anterograde direction revealed an average anterograde velocity of 1.0 μm/s in wild-type animals. Similar to our analysis of mobile GLR-1::GFP, we found that klp-4 mutants had a 44% decrease in anterograde flux (p < 0.005; Figure 5, D–E and G), a 32% decrease in anterograde run length (p < 0.005; Figure 5, D, E, and H) and a 26% decrease in anterograde velocity (average anterograde velocity: wild type, 1.0 ± 0.02 μm/s [n = 165]; klp-4, 0.73 ± 0.01 μm/s [n = 120]; p < 0.001) of red GLR-1::Dendra2 puncta compared with wild-type controls. The low number of retrogradely moving puncta at this time point precluded accurate quantitative analysis of retrograde movement. Taken together, these data suggest that KLP-4 promotes the anterograde trafficking of GLR-1 from neuronal cell bodies in the head to the VNC.
FIGURE 5:
KLP-4 and CDK-5 promote the anterograde trafficking of GLR-1 in the VNC. (A) Green (488 nm) and red (543 nm) images of cell bodies in the heads of young adult animals expressing integrated GLR-1::Dendra2 under the control of the glr-1 promoter (pzIs18) before and immediately after photoconversion using 405-nm light. (B, C) Images of the anterior VNC of young adult animals expressing GLR-1::Dendra2 without photoconversion (B) and 20 min after local photoconversion (C) of GLR-1::Dendra2 in cell bodies in the head. (D–F) Representative kymographs made from time-lapse imaging using a 543-nm laser showing mobile and stationary red GLR-1::Dendra2 puncta in the anterior VNC of young adult wild-type (D), klp-4(tm2114) (E), and cdk-5(gm336) (F) animals. For all kymographs, anterior is to the left (proximal) and posterior is to the right (distal). (G) Quantification of average anterograde flux for red GLR-1::Dendra2 puncta for wild-type (n = 86 mobility events), klp-4 (n = 42 mobility events), and cdk-5 (n = 45 mobility events) animals. (H) Quantification of average anterograde run length for red GLR-1::Dendra2 puncta for wild-type (n = 101 mobility events), klp-4 (n = 125 mobility events), and cdk-5 (n = 104 mobility events) animals. Mean and SEM are shown. Values that differ significantly from wild type are indicated by asterisks above each bar (**p < 0.001, Student's t test).
CDK-5 and KLP-4 act in the same genetic pathway to regulate GLR-1 in the VNC
Because we initially identified klp-4 in a cdk-5 genetic suppressor screen and previously implicated CDK-5 in regulating the anterograde trafficking of GLR-1 (Juo et al., 2007), we repeated our time-lapse analysis of mobile, red GLR-1::Dendra2 puncta in the VNC of cdk-5–mutant animals. As described earlier, GLR-1::Dendra2 was locally photoconverted in head neuron cell bodies, and red GLR-1::Dendra2 puncta movements in the VNC were measured in wild-type and cdk-5 mutant animals. Similar to our klp-4 mutant data, we found that cdk-5 mutants had a 42% decrease in anterograde GLR-1::Dendra2 flux (p < 0.005; Figure 5, D, F, and G) and a 32% decrease in anterograde run length (p < 0.005; Figure 5, D, F, and H). However, we observed no change in the anterograde velocity of GLR-1 in cdk-5–mutant animals (average anterograde velocity: wild type, 1.0 ± 0.02 μm/s [n = 165]; cdk-5, 1.0 ± 0.02 μm/s [n = 151]; p > 0.05). Time-lapse analysis of GLR-1::GFP in the VNC of cdk-5 mutants revealed similar decreases in GLR-1 transport (Supplemental Figure S6). These data indicate that, similar to KLP-4, CDK-5 promotes the anterograde trafficking of GLR-1 in the VNC.
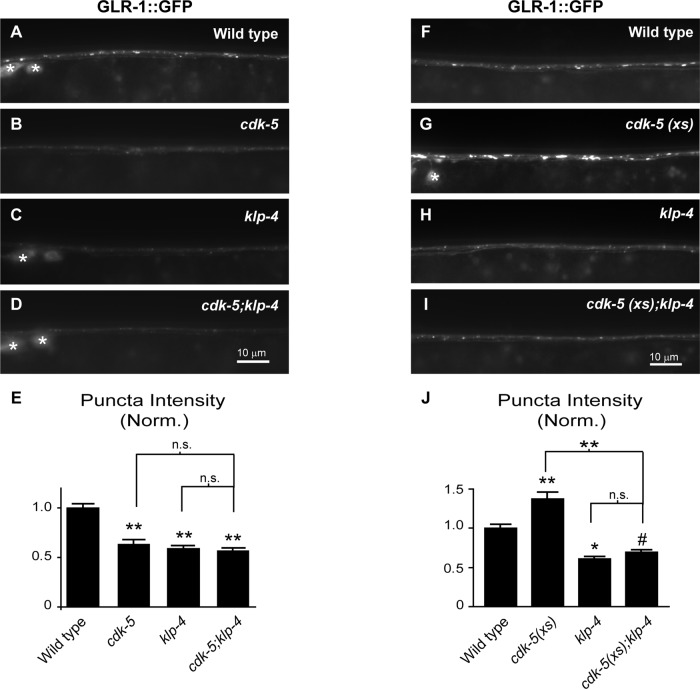
Because cdk-5 and klp-4 mutants have very similar defects in GLR-1 trafficking, we performed two complementary experiments to formally test whether cdk-5 and klp-4 function in the same genetic pathway. First, we analyzed the abundance of GLR-1::GFP in the VNC of animals with mutations in both cdk-5(gm336) and klp-4(tm2114) (Figure 6, A–E). The gm336 allele is a predicted molecular null (Juo et al., 2007). If cdk-5 and klp-4 act in separate genetic pathways, we would expect that the combination of the two mutations would cause an additive decrease in GLR-1::GFP levels in the VNC compared with either single mutant. Conversely, if cdk-5 and klp-4 act in the same genetic pathway, then cdk-5;klp-4 double mutants would have nonadditive effects on GLR-1::GFP levels compared with the single mutants. We found that the puncta intensity of GLR-1::GFP in the VNC of cdk-5;klp-4 double mutants decreased by 44% compared with wild-type controls (p < 0.001) and was not significantly different from either single mutant (p = 0.1 vs. klp-4; p = 0.5 vs. cdk-5; Figure 6, A–E). Similarly, GLR-1::GFP puncta densities in cdk-5;klp-4 double mutants were not significantly different from either single mutant (average puncta density [per 10 μm] ± SEM: wild type, 2.88 ± 0.08; cdk-5, 2.56 ± 0.14; klp-4, 2.55 ± 0.08; cdk-5; klp-4, 2.25 ± 0.08, p < 0.001 vs. wild type, p = 0.11 vs. cdk-5, p = 0.09 vs. klp-4), suggesting that cdk-5 and klp-4 function in a common pathway. Second, we tested whether klp-4 mutations could prevent the ability of overexpressed cdk-5 to increase GLR-1::GFP abundance in the VNC. We previously showed that overexpression of cdk-5 in glr-1–expressing neurons (cdk-5(xs)) causes an increase in GLR-1::GFP puncta intensities and widths in the VNC (Juo et al., 2007). If klp-4 acts in the same genetic pathway as cdk-5, we would expect mutations in klp-4 to completely occlude the effects of cdk-5 overexpression on GLR-1::GFP abundance. We found that GLR-1::GFP puncta intensity and width in cdk-5(xs);klp-4(tm2114) animals are identical to those for klp-4(tm2114) single-mutant animals (Figure 6, F–J) (average puncta width [μm] ± SEM: wild type, 0.84 ± 0.02; cdk-5(xs), 1.1 ± 0.03, p < 0.001; klp-4(tm2114), 0.83 ± 0.03; cdk-5(xs);klp-4, 0.74 ± 0.02; p = 0.2 vs. klp-4 and p < 0.001 vs. cdk-5(xs)). Similarly, the effects of overexpressed cdk-5 on GLR-1::GFP puncta intensity and width were occluded by a second klp-4(pz19) loss-of-function mutant (Supplemental Figure S7; average puncta width [μm] ± SEM: wild type, 0.89 ± 0.03; cdk-5(xs), 1.3 ± 0.06, p < 0.001 vs. wild type; klp-4(pz19), 0.87 ± 0.03; cdk-5(xs);klp-4, 0.84 ± 0.03, p = 0.4 vs. klp-4 and p < 0.001 vs. cdk-5(xs)). These results suggest that cdk-5 and klp-4 act in the same genetic pathway to regulate the trafficking of GLR-1 in the VNC.
FIGURE 6:
KLP-4 and CDK-5 act in the same genetic pathway to regulate GLR-1 levels in the VNC. (A–D) Representative images of the anterior VNC of wild-type (A), cdk-5(gm336) (B), klp-4(tm2114) (C), and cdk-5(gm336);klp-4(tm2114) double-mutant (D) L4 animals expressing GLR-1::GFP (nuIs25). White asterisks mark neuronal cell bodies. (E) Quantification of GLR-1::GFP puncta intensities (normalized) for the strains pictured in A–D. Mean and SEM are shown for n = 35 wild-type, n = 22 cdk-5, n = 29 klp-4, and n = 32 cdk-5;klp-4 animals. (F–I) Representative images of the anterior VNC of L4-stage wild-type animals (F), animals overexpressing cdk-5 under the control of the glr-1 promoter (cdk-5(xs)(pzIs2)) (G), klp-4(tm2114) mutants (H), and klp-4(tm2114) mutants overexpressing cdk-5 (cdk-5(xs)) (I). The white asterisk marks a neuronal cell body. (J) Quantification of GLR-1::GFP puncta intensities (normalized) for the strains pictured in F–I. Mean and SEM are shown for n = 21 wild-type, n = 24 cdk-5(xs), n = 21 klp-4, and n = 21 cdk-5(xs);klp-4 animals. Values that differ significantly from wild type are indicated by asterisks above each bar, whereas other comparisons are marked by brackets (#p ≤ 0.05, *p ≤ 0.01, **p ≤ 0.001, Tukey–Kramer test). n.s., no significant difference between the indicated strains (p > 0.05).
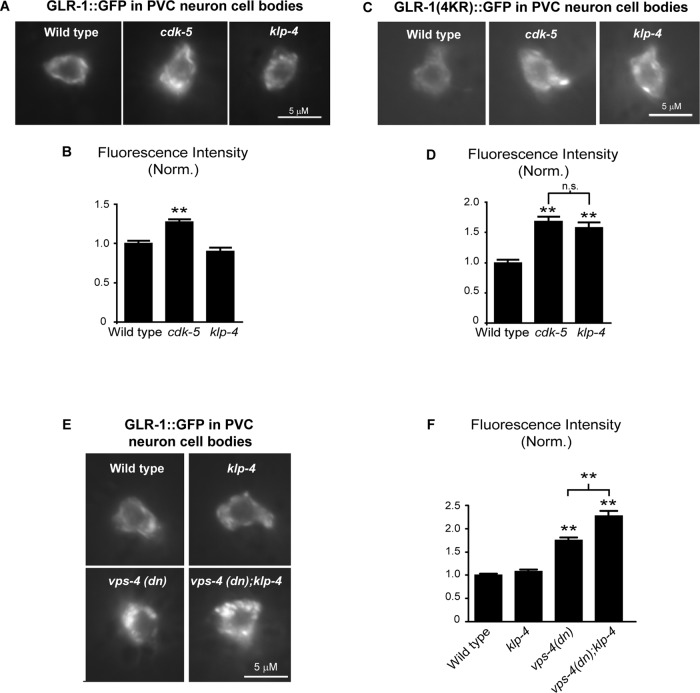
GLR-1 accumulates in klp-4 mutant cell bodies under conditions of decreased degradation
We previously showed that GLR-1::GFP decreases in the VNC and accumulates in cell bodies of cdk-5 mutant animals (Juo et al., 2007; Figure 6). Because our analysis of cdk-5 and klp-4 double mutants suggests that these genes function in the same genetic pathway, we tested whether klp-4 mutations also result in the accumulation of GLR-1::GFP in the cell body. Surprisingly, we observed no difference in the amount of GLR-1::GFP in klp-4 mutant cell bodies compared with wild-type controls (p = 0.08; Figure 7, A and B). One possible explanation for the lack of GLR-1::GFP accumulation in klp-4 mutant cell bodies is that in the absence of functional KLP-4 motors, GLR-1 is targeted for degradation in the cell body. We tested this hypothesis using two approaches to inhibit the degradation of GLR-1. GLR-1 is ubiquitinated and targeted for degradation in the MVB/lysosome pathway, and this process likely occurs in both the VNC and cell body (Burbea et al., 2002; Chun et al., 2008; Kowalski et al., 2011). As mentioned earlier, degradation of GLR-1 in this pathway can be prevented by using a nonubiquitinatable version of GLR-1 (GLR-1(4KR)::GFP; Burbea et al., 2002). In the first approach, we measured the levels of nonubiquitinatable GLR-1(4KR)::GFP in the cell bodies of wild-type and klp-4 mutant cells. We found that GLR-1(4KR)::GFP abundance in the cell body increases by 58% in klp-4 mutants compared with wild-type controls (Figure 7, C and D). Similarly, GLR-1(4KR)::GFP levels in the cell body also increases in cdk-5 mutants compared with controls (Figure 7, C and D). This result demonstrates that in contrast to wild-type GLR-1::GFP, which does not accumulate in klp-4–mutant cell bodies, GLR-1(4KR)::GFP does accumulate in klp-4–mutant cell bodies. In the second approach, we inhibited the degradation of GLR-1 in the MVB/lysosome pathway by expressing a dominant-negative version of vps-4 (vps-4(dn)). VPS-4 is an AAA-ATPase required for the formation of MVBs and degradation of ubiquitinated proteins in the MVB/lysosome pathway (Babst et al., 2002; Katzmann et al., 2002). Expression of vps-4(dn) in interneurons results in increased GLR-1::GFP abundance in the cell body and VNC (Figure 7; Chun et al., 2008; Kowalski et al., 2011). We measured the levels of GLR-1::GFP in cell bodies of wild-type and klp-4 mutant cells expressing vps-4(dn). We found that GLR-1::GFP abundance increased by 34% in klp-4 mutant cell bodies expressing vps-4(dn) compared with cell bodies expressing vps-4(dn) alone (Figure 7, E and F). Thus, inhibiting the degradation of GLR-1 in the MVB/lysosome pathway by expressing vps-4(dn) results in the accumulation of GLR-1::GFP in wild-type cell bodies, and this cell body accumulation is enhanced in klp-4 mutant animals. Taken together, these experiments suggest that in the absence of functional KLP-4 motors, GLR-1 is inefficiently trafficked to synapses in the VNC and, instead of accumulating in the cell body, is targeted for degradation in the MVB/lysosome pathway.
FIGURE 7:
GLR-1 accumulates in cell bodies of klp-4 mutants when receptor degradation is blocked. (A, C) Representative images of PVC neuron cell bodies of wild-type, cdk-5(gm336), and klp-4(tm2114) L4 animals expressing GLR::GFP (nuIs25) (A) or GLR-1(4KR)::GFP (nuIs108) (C). (B) Quantification of GLR-1::GFP mean fluorescence intensity (normalized) for the strains pictured in A. Mean and SEM are shown for n = 34 wild-type, n = 32 cdk-5, and n = 20 klp-4 animals. (D) Quantification of GLR-1(4KR)::GFP mean fluorescence intensity (normalized) for the strains pictured in C. Mean and SEM are shown for n = 24 wild-type, n = 25 cdk-5, and n = 23 klp-4 animals. (E) Representative images of GLR::GFP (nuIs24) in PVC neuron cell bodies of L4 stage wild-type animals, klp-4(tm2114) mutants, wild-type animals expressing vps-4(dn), and klp-4(tm2114) mutants expressing vps-4(dn). (F) Quantification of GLR-1::GFP mean fluorescence intensity (normalized) for the strains pictured in E. Mean and SEM are shown for n = 24 wild-type, n = 20 klp-4, n = 22 vps-4(dn), and n = 22 vps-4(dn);klp-4 animals. Values that differ significantly from wild type are indicated by asterisks above each bar (**p ≤ 0.001). n.s., no significant difference between the indicated strains (p > 0.05).
DISCUSSION
The trafficking of GluRs to synapses is important during synapse formation in development and during synaptic plasticity in the mature nervous system (Shepherd and Huganir, 2007; Kessels and Malinow, 2009; van der Sluijs and Hoogenraad, 2011). Although much research has focused on the insertion and removal of glutamate receptors at synapses, less is known about the mechanisms involved in transporting receptors from the cell body to synapses. Here we describe a function for the kinesin-3 family motor, KLP-4, for the first time and identify KLP-4 as a novel regulator of glutamate receptor trafficking in C. elegans.
KLP-4 regulates anterograde trafficking of GLR-1
Several pieces of evidence suggest that KLP-4 functions in interneurons to promote the anterograde trafficking of GLR-1. First, the abundance of GLR-1::GFP decreases in the VNC of two independent klp-4 loss-of-function mutants and is rescued by expression of wild-type klp-4 under the control of the glr-1 promoter (Figure 1). Second, KLP-4 functions before GLR-1 endocytosis and degradation at the synapse because klp-4 mutations prevent the accumulation of GLR-1 at synapses in the VNC when endocytosis is blocked with unc-11/AP180 mutations or when receptor ubiquitination and degradation is prevented (Figure 3). Third, time-lapse imaging revealed that several aspects of the anterograde trafficking of GLR-1, including anterograde flux, velocity, and run length, are decreased in klp-4 mutants (Figure 5 and Supplemental Figure S6). The role of KLP-4 in promoting the anterograde trafficking of GLR-1 receptors is consistent with the function of a mammalian homologue of KLP-4, KIF13A, which has been shown to transport mannose-6-phosphate receptors from the trans-Golgi network (TGN) to the plasma membrane in nonneuronal cells (Nakagawa et al., 2000).
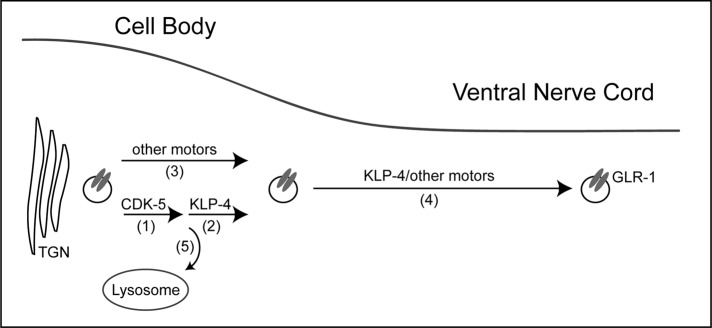
Previous work in mammalian neurons demonstrated that multiple motors are required for efficient GluR trafficking (Setou et al., 2000, 2002; Wyszynski et al., 2002; Shin et al., 2003; Kapitein et al., 2010). Our data are consistent with the idea that KLP-4 functions together with other motors to transport GLR-1 from the TGN in the cell body to synapses in the VNC. In klp-4 mutants, there is a 40% decrease in the abundance of GLR-1 (Figure 1) and a 38% decrease in the anterograde flux of GLR-1 (Figure 5 and Supplemental Figure S6) in the VNC. Thus, although GLR-1 trafficking is greatly reduced in klp-4 mutants, it is not eliminated, suggesting that other mechanisms exist to traffic GLR-1 to synapses in the VNC. We propose that KLP-4 functions together with other motors to regulate the anterograde trafficking of GLR-1 from the TGN to synapses in the VNC (see model in Figure 8).
FIGURE 8:
Model illustrating KLP-4 regulation of GLR-1 trafficking. (1, 2) In the cell body, CDK-5 and KLP-4 function in the same pathway to regulate the anterograde trafficking of GLR-1 receptors. (3) Other motors may function together with KLP-4 in the cell body to traffic GLR-1. (4) In the VNC processes, GLR-1 is transported to synapses by KLP-4 and/or other motors. (5) In the absence of KLP-4 motors, GLR-1 is targeted for degradation in the MVB/lysosome pathway.
One possible model is that KLP-4 functions in the cell body and VNC together with other motors to transport GLR-1 to synapses (Figure 8). In support of this model, our data show that KLP-4 can move processively in the VNC at a velocity that is comparable to that of GLR-1::GFP (Figure 4 and Supplemental Video S1). Alternatively, KLP-4 could function in the cell body to transport GLR-1–containing endosomes to the cell periphery, where GLR-1 is then loaded onto other motors for trafficking in the VNC (Figure 8). Consistent with this model, we were able to observe comigration of KLP-4::mCherry and GLR-1::GFP in neuronal cell bodies (Supplemental Video S2). Interestingly, KLP-4 homologues in other organisms have been shown to associate with and traffic RAB-5 and PIP3-containing early endosomes to the cell periphery (Hoepfner et al., 2005; Horiguchi et al., 2006; Delevoye et al., 2009; Huckaba et al., 2011). Although our initial attempts to observe comigration of KLP-4 and GLR-1::GFP in the VNC were unsuccessful, we cannot rule out a direct role of KLP-4 in transporting GLR-1 in the VNC because it remains possible that our imaging conditions are not sensitive enough to detect cargo being moved by one or a few motors. Regardless of whether KLP-4 functions with other motors in the cell body and/or the VNC, our data show that KLP-4 is required for the efficient trafficking and accumulation of GLR-1 at synapses. In the future, it will be interesting to identify other motors that function together with KLP-4 and to understand how multiple motors coordinate trafficking of GLR-1 cargo.
CDK-5 functions in the same pathway as KLP-4 to regulate GLR-1
Genetic analysis of the unc-11/AP180 clathrin adaptin mutant together with cdk-5 (Juo et al., 2007) or klp-4 mutants (Figure 3) suggests that both CDK-5 and KLP-4 act before endocytosis of GLR-1 in the VNC. In addition, genetic analysis of cdk-5;klp-4 double mutants and klp-4 mutants overexpressing cdk-5 suggests that CDK-5 and KLP-4 function in the same pathway to regulate GLR-1 in the VNC (Figure 6). Furthermore, our time-lapse analyses revealed that anterograde trafficking of GLR-1 is reduced to a similar extent in both cdk-5 and klp-4 mutants (Figure 5 and Supplemental Figure S6), suggesting that CDK-5 may directly regulate KLP-4–dependent trafficking of GLR-1. However, cdk-5 and klp-4 have different effects on GLR-1 in the cell body. Our data show that GLR-1 accumulates in cdk-5, but not klp-4 mutant cell bodies (Figure 7), suggesting that CDK-5 may also function before KLP-4 to make receptors competent for KLP-4–dependent anterograde trafficking. Further studies are necessary to investigate the precise mechanism by which CDK-5 regulates KLP-4–dependent GLR-1 trafficking. However, we anticipate that the effect of CDK-5 on trafficking will be specific to the particular cargo and neuronal cell type being investigated, given the many different effects of CDK-5 on axonal and dendritic trafficking (Ratner et al., 1998; Morfini et al., 2004; Juo et al., 2007; Ou et al., 2010; Park et al., 2011; Goodwin et al., 2012). Intriguingly, although GLR-1 does not accumulate in klp-4 mutant cell bodies, the receptor does accumulate if receptor degradation in the MVB/lysosome pathway is blocked by expression of vps-4(dn) or nonubiquitinatable GLR-1(4KR) (Figure 7). These data suggest that in the absence of the kinesin klp-4, GLR-1 is targeted, likely in a ubiquitin-dependent manner, for degradation in the MVB/lysosome pathway (Figure 8). Interestingly, studies in yeast indicate that membrane proteins can be targeted for degradation early in the secretory pathway. For example, ubiquitination of the amino acid permease Gap1p in the Golgi can target the protein to a pre–multivesicular endosome, where it can either be sent to the vacuole for degradation or recycled back to the Golgi for transport to the plasma membrane (Helliwell et al., 2001; Risinger and Kaiser, 2008).
Recent studies suggest that vesicular cargo may be targeted for degradation in the absence of motors or cargo adaptors. In mammalian neurons lacking the adaptor protein AP-4, AMPA receptors are not trafficked to dendrites but instead accumulate in autophagosomes, which often results in protein degradation when they fuse with lysosomes (Matsuda et al., 2008). In C. elegans, amyloid precursor protein APL-1 is normally trafficked from the cell body to the plasma membrane of neuronal processes by kinesins UNC-104/KIF1A and UNC-116/KIF5 (Wiese et al., 2010). Loss of function of either of these motors results in decreased levels of APL-1 in neuronal processes. However, instead of accumulating in neuronal cell bodies of these motor mutants, APL-1 protein levels decrease, implying that APL-1 may be degraded in the absence of its normal motors (Wiese et al., 2010). Another study demonstrated that antisense oligonucleotide knockdown of kinesin KIF17 expression results in decreased protein levels of its cargo, the N-methyl-d-aspartate (NMDA) receptor NR2B subunit (Guillaud et al., 2003). These studies are consistent with our data showing that GLR-1 is targeted for degradation in the MVB/lysosome pathway in the absence of KLP-4 motors. Together, these data suggest an interesting potential cellular regulatory mechanism that targets cargo for degradation in the absence of its appropriate anterograde motor or adaptor. Such a mechanism could prevent excess membrane proteins from accumulating early in the secretory pathway, which may be detrimental to the neuron. These results imply that the fate of GLR-1 receptors early in the secretory pathway, and consequently the abundance of GLR-1 at synapses, could be controlled by regulating the expression level or availability of KLP-4 motors.
Implications for synaptic plasticity
Regulation of the number of glutamate receptors in the postsynaptic membrane contributes to activity-dependent synaptic plasticity. In particular, many studies have focused on the activity-dependent regulation of exocytosis and endocytosis of AMPA receptors at the synapse (Shepherd and Huganir, 2007; van der Sluijs and Hoogenraad, 2011). However, other studies suggest that activity-dependent regulation of GluRs can also occur earlier in the secretory pathway. For example, activity blockade regulates alternative splicing of NMDA receptor subunit NR1, resulting in increased ER exit and synaptic levels of the receptor (Mu et al., 2003). In C. elegans, glr-1 splicing and the abundance of GLR-1 at synapses can be regulated by the RNA-binding protein GRLD-1 (Wang et al., 2010). Interestingly, several studies suggest that anterograde motors are limiting in neurons and that regulation of motor expression influences synaptic plasticity. For example, overexpression of KIF17 in the mouse forebrain increases expression and trafficking of its cargo NMDA receptors to synapses, resulting in increased spatial and working memory (Wong et al., 2002). Activity can influence motor/cargo expression because activity blockade of NMDA receptors results in the coordinated up-regulation of the NMDA receptor and its KIF17 motor (Guillaud et al., 2003). In addition, raising mice in an enriched-environment results in BDNF-dependent up-regulation of KIF1A and transport of its cargoes in the hippocampus (Kondo et al., 2012). Similarly, in Aplysia, neuronal activity results in increased expression of kinesin heavy chain motors and its cargoes, and up-regulation of the motor is required and sufficient for long-term synaptic plasticity (Puthanveettil et al., 2008). These studies show that regulation of motor expression can affect synaptic plasticity and that activity-dependent expression of motors is coordinated with expression of their respective cargoes. Our work identifies KLP-4 as a novel motor that regulates anterograde trafficking of GluRs and reveals an interesting regulatory mechanism by which expression or availability of the motor influences receptor degradation and ultimately the abundance of GLR-1 in the VNC. Future studies will be necessary to determine whether GLR-1 and KLP-4 expression are coordinated in vivo and whether this concerted regulation influences synaptic plasticity.
MATERIALS AND METHODS
Strains
The following strains were used in this study: nuIs25 (Pglr-1::glr-1::gfp), nuIs24 (Pglr-1::glr-1::gfp), nuIs108 (Pglr-1::glr-1(4kr)::gfp), nuIs125 (Pglr-1::snb-1::gfp), nuIs145 [Pglr-1::vps-4 (dn)], pzIs2(Pglr-1::cdk-5), pzIs15 (Pglr-1::dsRED), pzIs18 (Pglr-1::Dendra2), pzIs20 (Pglr-1::klp-4), nuEx993 (Pglr-1::lin-10::gfp), nuEx1004 (Pglr-1::magi1::yfp), pzEx167 (Pglr-1::klp-4), pzEx169 (Pglr-1::klp-4), pzEx188 (Pglr-1::klp-4::gfp), pzEx234 (Pglr-1::klp-4::mCherry), pzEx237 (Phsp-16.2::klp-4), pzEx244 (Pklp4::NLS::GFP::LacZ), pzEx256 (Pklp-4::GFP), klp-4(tm2114), klp-4(pz19), unc-11(e47), cdk-5(gm336), and glr-1(n2461). All strains were maintained at 20°C as described previously (Brenner, 1974).
Transgenes and germline transformation
Standard techniques were used to isolate transgenic strains by microinjection of various plasmids. nuIs24, nuIs25, nuIs125, nuEx993, nuEx1004, nuIs108, nuIs145, and pzIs2 have been described previously (Rongo et al., 1998; Burbea et al., 2002; Juo and Kaplan, 2004; Juo et al., 2007; Chun et al., 2008; Kowalski et al., 2011). The klp-4 (F56E3.3) open reading frame (ORF) was obtained by reverse transcription-PCR from cDNA isolated from wild-type animals. The klp-4 cDNA was put under control of the glr-1 promoter by subcloning klp-4 into pV6 using NheI/KpnI restriction sites to create Pglr-1::klp-4 (FJ#83). pzEx169 and pzEx167 were created by injecting FJ#83 plasmid at 25 ng/μl along with coinjection marker Pmyo-2::NLS::mCherry (10 ng/μl). pzIs20 (Pglr-1::klp-4) was created by UV integrating pzEx167. pzEx188 (Pglr-1::klp-4::gfp) and pzEx234 (Pglr-1::klp-4::mcherry) were constructed by subcloning GFP or mCherry flanked by NotI restriction sites into a plasmid containing (Pglr-1::klp-4) with a NotI restriction site engineered immediately before the klp-4 stop codon to create Pglr-1::klp-4::gfp (FJ#84) and Pglr-1::klp-4::mcherry (FJ#85), respectively, and injected at 25 ng/μl (pzEx188) or 100 ng/μl (pzEx234) along with injection marker Pmyo2::NLS-mCherry (5 ng/μl). pzEx237 (Phsp-16.2::klp-4::mCherry) was made by subcloning the klp-4::mCherry coding sequence from FJ#85 into the NheI/KpnI restriction sites of the heat-shock promoter plasmid pPD49.78 (gift from Andrew Fire, Stanford University, Palo Alto, CA) to create Phsp-16.2::klp-4::mCherry (FJ#86) and injected at 25 ng/μl along with injection marker Pmyo2::NLS-mCherry (5 ng/μl). pzEx244 (Pklp-4::NLS::GFP::LacZ) was made by PCR amplifying the 3-kb segment of genomic DNA immediately upstream of the klp-4 coding region, flanking it with PstI/BamHI restriction sites, and subcloning it into the GFP expression plasmid pPD96.04 (gift from Andrew Fire) to create Pklp-4::nls::gfp::lacz (FJ#87) and injected at 25 ng/μl. pzEx256 (Pklp-4::gfp) was generated by subcloning the klp-4 promoter from FJ#87 into the PstI/BamHI restriction sites of the GFP expression plasmid pPD95.75 (gift from Andrew Fire) to create Pklp-4::gfp (FJ#88) and injected at 25 ng/μl along with injection marker Pmyo2::NLS-mCherry (5 ng/μl). The plasmid KP#889 (Pglr-1::dsRED) (gift from Lars Dreier, UCLA, Los Angeles, CA) was injected at 25 ng/μl to make pzEx96 and UV integrated to generate pzIs15 (Pglr-1::dsRED). Dendra2 (gift from HoYi Mak, Stowers Institute, Kansas City, MO) was subcloned into HindIII sites flanking the gfp coding sequence in Pglr-1::glr-1::gfp to make Pglr-1::glr-1::dendra2 (FJ#89). Plasmid FJ#89 was injected at 25 ng/μl along with coinjection marker Pttx3::GFP at 50 ng/μl to make pzEx160 and subsequently UV integrated to generate pzIs18 (Pglr-1::glr-1::dendra2). All constructs were confirmed by sequencing. Full details of plasmids and oligonucleotides are available upon request.
Fluorescence imaging
All static images of the VNC were taken of the anterior portion of the ventral nerve cord, posterior to the RIG and AVG neuronal cell bodies in larval stage 4 (L4) animals unless otherwise stated. Images of the posterior nerve cord in Supplemental Figure S3 were taken posterior to the vulva. Nerve ring images in Supplemental Figure S3 were taken immediately anterior to the terminal bulb of the pharynx. Imaging of heat-shocked animals shown in Figure 2 was performed on young adult animals. For all images of the VNC, animals were immobilized in 30 mg/ml 2,3-butanedione monoxamine (Sigma-Aldrich, St. Louis, MO) for 5–7 min before imaging. Imaging was performed using an Axioimager M1 microscope (Carl Zeiss, Jena, Germany) with a 100× Plan Apochromat (1.4 numerical aperture [NA]) objective with GFP filter and captured with an Orca-ER charge-coupled device camera (Hamamatsu, Hamamatsu, Japan) using MetaMorph, version 7.1, software (Molecular Devices, Sunnyvale, CA). To quantitate VNC fluorescence, maximum intensity projections from Z-series stacks of 1 μm total depth were used. For nerve ring images Z-series stacks of 3 μm total depth were used. Exposure settings and gain were set to fill the 12-bit dynamic range and to avoid saturation. Settings were identical for each image acquired within an experiment for a given fluorescent marker. MetaMorph, version 6.0, software was used to generate line scans of maximum-intensity projection images of the VNC. Line scans were analyzed with Igor Pro, version 5 or 6 (WaveMetrics, Portland, OR), using custom-written software (gift from J. Dittman, Weill Cornell Medical College, New York) as described previously (Burbea et al., 2002). The fluorescence intensity of 0.5-μm FluoSphere beads (Invitrogen, Carlsbad, CA) was measured for each day of imaging and used to normalize daily arc lamp intensity. Puncta intensities were normalized to the average bead intensity for the corresponding day. Puncta intensities are shown normalized to wild type. Puncta widths were calculated by measuring the width of each punctum at half the maximal peak fluorescence intensity. Puncta densities represent the average number of puncta per 10 μm of the ventral nerve cord. For all imaging quantification in the VNC, average ± SEM values are reported, and statistical significance was determined using Student's t test for experiments in which two genotypes were compared or the Tukey–Kramer test for experiments comparing more than two genotypes.
To quantify the amount of GLR-1::GFP in interneuron cell bodies, images were taken of PVC cell bodies, and maximum-intensity projections were generated from Z-series stacks (2 μm total depth) as described previously (Juo et al., 2007; Kowalski et al., 2011). Average pixel intensities of two to three separate regions of each cell body were measured using MetaMorph, version 7.1, software.
Heat shock was performed by shifting L4 stage animals from normal growth conditions at 20°C to 30°C for 16 h. After heat shock, animals were allowed to recover at 20°C for at least 1 h before imaging.
Time-lapse imaging and data analysis
For all in vivo time-lapse imaging of the VNC, 1-d adult hermaphrodites were immobilized with 3 mM levamisole in M9 and mounted on a 2% agarose pad. VNC imaging was performed in an anterior region (70–80 μm) of the VNC just posterior to the RIG and AVG neuronal cell bodies.
VNC time-lapse image acquisition
GLR-1::GFP time-lapse images were obtained with an Olympus IX81 microscope (Olympus, Tokyo, Japan) using a Plan Apochromat objective (100×, 1.4 NA) attached with a spinning disk confocal head (CSU22; Yokogawa, Tokyo, Japan) and equipped with an electron-multiplying charge-coupled device (EMCCD) camera (iXon 897; Andor Technology, South Windsor, CT). KLP-4::mCherry time-lapse images were obtained with an Andor Spinning Disk confocal microscope.
Photoconversion and time-lapse imaging of GLR-1::Dendra2
GLR-1::Dendra2 was locally photoconverted in neuronal cell bodies in the head in a 10 × 20 μm region of interest using a 405-nm laser on a Zeiss LSM 510 meta-confocal with a 60×/1.45 NA oil objective (Carl Zeiss, Jena, Germany). Before photoconversion, interneuron cell bodies in the head were identified using a 488-nm argon laser (30 mW) at low 2% power (60× objective, scan speed of 400 Hz, and 100-μm pinhole size) to avoid accidental photoconversion of Dendra2. Photoconversion was performed using a 405-nm diode laser (25 mW) at 20% power for 4–5 min (60× objective, scan speed of 200 Hz, pinhole size 100 μm). Twenty minutes after local photoconversion of the cell bodies in the head, time-lapse images of the VNC were obtained using an Andor Spinning Disk confocal microscope using a Plan Apochromat objective (100×, 1.4 NA) attached to a spinning disk confocal head (CSU22; Yokogawa) and equipped with an EMCCD camera (iXon-897). We focused on the VNC using the 488-nm laser (25 mW) at 10% power (100× objective, 150-ms exposure). There was no accidental photoconversion of Dendra2 using these conditions. Time-lapse imaging of photoconverted red GLR-1::Dendra2 in the VNC was performed using a 561-nm laser (25 mW) at 60–80% power (100× objective, exposure of 300 ms/image).
VNC time-lapse analysis
Moving particles were defined as puncta that were displaced by at least three pixels in successive time frames, and stationary particles were defined as puncta that were immobile for more than three consecutive frames. The flux of particles was calculated as the number of puncta moving in either direction in a 15- to 20-μm region (just posterior to the RIG and AVG cell bodies) divided by the total time. Run length was calculated as the length in microns of each distinct movement event in the x-coordinate plane. The start and end of each movement event was defined by pauses, reversals, or length of movie. Images (512 × 512 pixels) were acquired at a constant frame rate of 3–4 frames/s for a total of 300–400 frames. Image analysis was performed using ImageJ software, version 1.37 (National Institutes of Health, Bethesda, MD), and statistical significance was determined using the Student's t test.
Comigration time-lapse imaging
Comigration time-lapse imaging of GLR-1::GFP and KLP-4::mCherry was performed on animals expressing both GLR-1::GFP (nuIs25) and KLP-4::mCherry under control of the glr-1 promoter (pzEx234). Time-lapse imaging was performed using an UltraView spinning disk confocal microscope (PerkinElmer, Waltham, MA) and assembled into time-lapse movies using Volocity (PerkinElmer) and QuickTime (Apple, Cupertino, CA) software.
Real-Time PCR
Total RNA was isolated from mixed-stage wild-type (nuIs25) and klp-4 mutant (nuIs25;klp-4(tm2114) or nuIs25; klp-4(pz19)) animals using an RNeasy fibrous tissue kit (Qiagen, Valencia, CA) as previously described (Kowalski et al., 2011). First-strand cDNA was synthesized using Superscript III Reverse Transcriptase (Invitrogen), and real-time PCR was performed using the Brilliant SYBR Green Master Mix and SureStart Taq (Stratagene, Santa Clara, CA) on the MX3000P real-time PCR machine (Tufts Center for Neuroscience Research, Boston, MA). Standard curves were used to calculate the efficiency of primers for both glr-1 and act-1, and the relative amount of glr-1 mRNA compared with act-1 mRNA in each of six replicate samples was determined as previously described (Pfaffl, 2001). The ratio of glr-1:act-1 mRNA in klp-4(tm2114) and klp-4(pz19) mutant animals was normalized to that in wild-type animals. For all genotypes mean ± SEM of n = 3 replicates of two different starting cDNA concentrations was determined.
Locomotion behavior assay
The spontaneous reversal assay was performed as previously described (Kowalski et al., 2011). Fresh standard nematode growth medium (NGM) agar plates were poured and allowed to dry at room temperature. Young adult hermaphrodites were transferred using halocarbon oil from a plate with food to the center of a fresh NGM standard plate without food and allowed to acclimate to the plate for 2 min. The number of spontaneous reversals was recorded over the subsequent 5 min. The genotypes of the animals being studied in the behavioral assay were coded so that they were unknown to the experimenter during the observation period. The average number of reversals per minute ± SEM was determined for each genotype.
Ethyl methane sulfonate mutagenesis and mutations
The klp-4(pz19) allele was isolated in a forward genetic suppressor screen for mutants that suppressed the effects of overexpressed cdk-5 (pzIs2) on GLR-1::GFP (nuIs24). An F1 clonal screen was performed by mutagenizing nuIs24;pzIs2 animals with ethyl methane sulfonate (EMS) and screening F2 progeny on a compound microscope for mutants with decreased abundance of GLR-1::GFP in the VNC. Briefly, synchronized L4 animals were incubated with 50 mM EMS at room temperature for 4 h as previously described (Brenner, 1974). After removal of EMS, the treated animals were allowed to recover on NGM plates with food until movement resumed.
Mutagenized L4 animals were transferred to fresh NGM plates and incubated at 20°C to allow for growth of the F1 generation. One hundred young adult F1 animals were singled and incubated at 20°C. F2 animals were screened for GLR-1::GFP abundance in the VNC as detailed earlier. pz19 was one of the strong suppressor mutants isolated in this screen and was mapped to the X chromosome using standard genetic mapping techniques. A parallel, independent RNA interference screen for genes that decrease GLR-1::GFP in the VNC also identified klp-4 as a gene that regulates GLR-1 and aided our mapping of pz19. Sequencing of the exons and intron/exon junctions of klp-4 revealed that the pz19 allele contains a C-to-T point mutation at nucleotide 3076, resulting in a premature stop codon at amino acid 1026 (Arg1026Stop) in the ORF of KLP-4 (numbering based on F56E3.3a splice form). The klp-4 deletion allele tm2114 was isolated by Shohei Mitani (National BioResource Project, Tokyo, Japan) and consists of a 747–base pair deletion. The tm2114 allele deletes exons 4–6 of klp-4, eliminating more than half of the motor domain, including the microtubule-binding sites, and thus likely represents a functional null mutant. The tm2114 deletion also causes a frameshift, resulting in a predicted premature stop codon after the first 167 amino acids of KLP-4, eliminating the entire cargo-binding tail. pz19 fails to complement tm2114 in a noncomplementation test.
Supplementary Material
Acknowledgments
We thank Josh Kaplan for strains and reagents, Ho-Yi Mak for Dendra2, Jeremy Dittman for custom-written software, and Gian Garriga, Shohei Mitani, and the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN) for strains. We thank Victor Hatini for use of the Zeiss LSM510 confocal microscope and Stephen Straub for help with the PerkinElmer UltraView confocal microscope. We also thank Annette McGehee and Steven Garafalo for help in generating plasmids and strains and the Juo lab for useful discussions and critical reading of the manuscript. This research was supported by National Institutes of Health Grant NS059953 (P.J.), a Basil O'Connor March of Dimes Scholar Award (P.J.), and the Tufts Center for Neuroscience Research (National Institutes of Health Grant P30 NS 047243). M.I.M. was supported in part by a Genetics and Development Training Grant (National Institutes of Health Grant T32 HD 049341). J.R.K. was supported by the Training in Education and Critical Research Skills Postdoctoral Program (National Institutes of Health Grant 5K12GM074869). S.A. was supported by a grant from the Department of Science and Technology and the Council of Scientific and Industrial Research, Government of India, to S.P.K. Research in the laboratory of S.P.K. is supported by the Department of Biotechnology and the Council of Scientific and Industrial Research, Government of India.
Abbreviations used:
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- CDK-5
cyclin-dependent kinase 5
- GLR-1
glutamate receptor-1
- GluR
glutamate receptor
- KLP-4
kinesin-like protein-4
- MVB
multivesicular body
- NMDA
N-methyl-d-aspartate
- VNC
ventral nerve cord
- VPS
vacuolar protein sorting
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0334) on August 1, 2012.
*Present address: Department of Biological Sciences, Butler University, Indianapolis, IN 46208.
REFERENCES
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Barkus RV, Klyachko O, Horiuchi D, Dickson BJ, Saxton WM. Identification of an axonal kinesin-3 motor for fast anterograde vesicle transport that facilitates retrograde transport of neuropeptides. Mol Biol Cell. 2008;19:274–283. doi: 10.1091/mbc.E07-03-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt DS, Nicoll RA. AMPA receptor trafficking at excitatory synapses. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbea M, Dreier L, Dittman JS, Grunwald ME, Kaplan JM. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron. 2002;35:107–120. doi: 10.1016/s0896-6273(02)00749-3. [DOI] [PubMed] [Google Scholar]
- Byrd DT, Kawasaki M, Walcoff M, Hisamoto N, Matsumoto K, Jin Y. UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron. 2001;32:787–800. doi: 10.1016/s0896-6273(01)00532-3. [DOI] [PubMed] [Google Scholar]
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Chao MY, Komatsu H, Fukuto HS, Dionne HM, Hart AC. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc Natl Acad Sci USA. 2004;101:15512–15517. doi: 10.1073/pnas.0403369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- Chun DK, McEwen JM, Burbea M, Kaplan JM. UNC-108/Rab2 regulates postendocytic trafficking in Caenorhabditis elegans. Mol Biol Cell. 2008;19:2682–2695. doi: 10.1091/mbc.E07-11-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Grierson AJ, Ackerley S, Miller CC. Role of axonal transport in neurodegenerative diseases. Annu Rev Neurosci. 2008;31:151–173. doi: 10.1146/annurev.neuro.31.061307.090711. [DOI] [PubMed] [Google Scholar]
- Delevoye C, et al. AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J Cell Biol. 2009;187:247–264. doi: 10.1083/jcb.200907122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JE, Goldstein LS. The genetics of axonal transport and axonal transport disorders. PLoS Genet. 2006;2:e124. doi: 10.1371/journal.pgen.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- Emtage L, Chang H, Tiver R, Rongo C. MAGI-1 modulates AMPA receptor synaptic localization and behavioral plasticity in response to prior experience. PLoS One. 2009;4:e4613. doi: 10.1371/journal.pone.0004613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A, Niclas J, Vale RD, Banker G, Kosik KS. Suppression of kinesin expression in cultured hippocampal neurons using antisense oligonucleotides. J Cell Biol. 1992;117:595–606. doi: 10.1083/jcb.117.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AY, Wang X, Schwarz TL. Axonal transport and the delivery of pre-synaptic components. Curr Opin Neurobiol. 2008;18:495–503. doi: 10.1016/j.conb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin PR, Sasaki JM, Juo P. CDK-5 regulates the polarized trafficking of neuropeptide-containing dense-core vesicles in C. elegans motor neurons. J Neurosci. 2012;32:8158–8572. doi: 10.1523/JNEUROSCI.0251-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaud L, Setou M, Hirokawa N. KIF17 dynamics and regulation of NR2B trafficking in hippocampal neurons. J Neurosci. 2003;23:131–140. doi: 10.1523/JNEUROSCI.23-01-00131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- Guzik BW, Goldstein LS. Microtubule-dependent transport in neurons: steps towards an understanding of regulation, function and dysfunction. Curr Opin Cell Biol. 2004;16:443–450. doi: 10.1016/j.ceb.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Hafezparast M, et al. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300:808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- Hanada T, Lin L, Tibaldi EV, Reinherz EL, Chishti AH. GAKIN, a novel kinesin-like protein associates with the human homologue of the Drosophila discs large tumor suppressor in T lymphocytes. J Biol Chem. 2000;275:28774–28784. doi: 10.1074/jbc.M000715200. [DOI] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Losko S, Kaiser CA. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J Cell Biol. 2001;153:649–662. doi: 10.1083/jcb.153.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Horiguchi K, Hanada T, Fukui Y, Chishti AH. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J Cell Biol. 2006;174:425–436. doi: 10.1083/jcb.200604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba TM, Gennerich A, Wilhelm JE, Chishti AH, Vale RD. Kinesin-73 is a processive motor that localizes to Rab5-containing organelles. J Biol Chem. 2011;286:7457–7467. doi: 10.1074/jbc.M110.167023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo P, Harbaugh T, Garriga G, Kaplan JM. CDK-5 regulates the abundance of GLR-1 glutamate receptors in the ventral cord of Caenorhabditis elegans. Mol Biol Cell. 2007;18:3883–3893. doi: 10.1091/mbc.E06-09-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo P, Kaplan JM. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr Biol. 2004;14:2057–2062. doi: 10.1016/j.cub.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Schlager MA, Kuijpers M, Wulf PS, van Spronsen M, MacKintosh FC, Hoogenraad CC. Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr Biol. 2010;20:290–299. doi: 10.1016/j.cub.2009.12.052. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kessels HW, Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron. 2009;61:340–350. doi: 10.1016/j.neuron.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Lisman JE. A labile component of AMPA receptor-mediated synaptic transmission is dependent on microtubule motors, actin, and N-ethylmaleimide-sensitive factor. J Neurosci. 2001;21:4188–4194. doi: 10.1523/JNEUROSCI.21-12-04188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Takei Y, Hirokawa N. Motor protein KIF1A is essential for hippocampal synaptogenesis and learning enhancement in an enriched environment. Neuron. 2012;73:743–757. doi: 10.1016/j.neuron.2011.12.020. [DOI] [PubMed] [Google Scholar]
- Koushika SP, Schaefer AM, Vincent R, Willis JH, Bowerman B, Nonet ML. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J Neurosci. 2004;24:3907–3916. doi: 10.1523/JNEUROSCI.5039-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski JR, Dahlberg CL, Juo P. The deubiquitinating enzyme USP-46 negatively regulates the degradation of glutamate receptors to control their abundance in the ventral nerve cord of Caenorhabditis elegans. J Neurosci. 2011;31:1341–1354. doi: 10.1523/JNEUROSCI.4765-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Ju W, Foster K, Lee SH, Ahmadian G, Wyszynski M, Wang YT, Sheng M. Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptor internalization. Nat Neurosci. 2000;3:1282–1290. doi: 10.1038/81814. [DOI] [PubMed] [Google Scholar]
- Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- Martin M, Iyadurai SJ, Gassman A, Gindhart JG, Jr, Hays TS, Saxton WM. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol Biol Cell. 1999;10:3717–3728. doi: 10.1091/mbc.10.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Miura E, Matsuda K, Kakegawa W, Kohda K, Watanabe M, Yuzaki M. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron. 2008;57:730–745. doi: 10.1016/j.neuron.2008.02.012. [DOI] [PubMed] [Google Scholar]
- McKay SJ, et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Mellem JE, Brockie PJ, Zheng Y, Madsen DM, Maricq AV. Decoding of polymodal sensory stimuli by postsynaptic glutamate receptors in C. elegans. Neuron. 2002;36:933–944. doi: 10.1016/s0896-6273(02)01088-7. [DOI] [PubMed] [Google Scholar]
- Morfini G, Szebenyi G, Brown H, Pant HC, Pigino G, DeBoer S, Beffert U, Brady ST. A novel CDK5-dependent pathway for regulating GSK3 activity and kinesin-driven motility in neurons. EMBO J. 2004;23:2235–2245. doi: 10.1038/sj.emboj.7600237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD. Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron. 2003;40:581–594. doi: 10.1016/s0896-6273(03)00676-7. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Setou M, Seog D, Ogasawara K, Dohmae N, Takio K, Hirokawa N. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell. 2000;103:569–581. doi: 10.1016/s0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Holgado AM, Brewer F, Serpe CJ, Norbeck BA, Holleran J, Wei L, Hartwieg E, Jorgensen EM, Alfonso A. UNC-11, a Caenorhabditis elegans AP180 homologue, regulates the size and protein composition of synaptic vesicles. Mol Biol Cell. 1999;10:2343–2360. doi: 10.1091/mbc.10.7.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Ou CY, et al. Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell. 2010;141:846–858. doi: 10.1016/j.cell.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack-Chung E, Kurshan PT, Dickman DK, Schwarz TL. A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat Neurosci. 2007;10:980–989. doi: 10.1038/nn1936. [DOI] [PubMed] [Google Scholar]
- Park M, Watanabe S, Poon VY, Ou CY, Jorgensen EM, Shen K. CYY-1/cyclin Y and CDK-5 differentially regulate synapse elimination and formation for rewiring neural circuits. Neuron. 2011;70:742–757. doi: 10.1016/j.neuron.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden EM, Barr MM. The KLP-6 kinesin is required for male mating behaviors and polycystin localization in Caenorhabditis elegans. Curr Biol. 2005;15:394–404. doi: 10.1016/j.cub.2004.12.073. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthanveettil SV, Monje FJ, Miniaci MC, Choi YB, Karl KA, Khandros E, Gawinowicz MA, Sheetz MP, Kandel ER. A new component in synaptic plasticity: upregulation of kinesin in the neurons of the gill-withdrawal reflex. Cell. 2008;135:960–973. doi: 10.1016/j.cell.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner N, Bloom GS, Brady ST. A role for cyclin-dependent kinase(s) in the modulation of fast anterograde axonal transport: effects defined by olomoucine and the APC tumor suppressor protein. J Neurosci. 1998;18:7717–7726. doi: 10.1523/JNEUROSCI.18-19-07717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risinger AL, Kaiser CA. Different ubiquitin signals act at the Golgi and plasma membrane to direct GAP1 trafficking. Mol Biol Cell. 2008;19:2962–2972. doi: 10.1091/mbc.E07-06-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- Sagona AP, Nezis IP, Pedersen NM, Liestol K, Poulton J, Rusten TE, Skotheim RI, Raiborg C, Stenmark H. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- Sakamoto R, Byrd DT, Brown HM, Hisamoto N, Matsumoto K, Jin Y. The Caenorhabditis elegans UNC-14 RUN domain protein binds to the kinesin-1 and UNC-16 complex and regulates synaptic vesicle localization. Mol Biol Cell. 2005;16:483–496. doi: 10.1091/mbc.E04-07-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer H, Rongo C. KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol Biol Cell. 2006;17:1250–1260. doi: 10.1091/mbc.E05-08-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417:83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- Shin H, Wyszynski M, Huh KH, Valtschanoff JG, Lee JR, Ko J, Streuli M, Weinberg RJ, Sheng M, Kim E. Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. J Biol Chem. 2003;278:11393–11401. doi: 10.1074/jbc.M211874200. [DOI] [PubMed] [Google Scholar]
- Sieburth D, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- van der Sluijs P, Hoogenraad CC. New insights in endosomal dynamics and AMPA receptor trafficking. Semin Cell Dev Biol. 2011;22:499–505. doi: 10.1016/j.semcdb.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Hanada T, Chishti AH. Centaurin-alpha1 interacts directly with kinesin motor protein KIF13B. J Cell Sci. 2005;118:2471–2484. doi: 10.1242/jcs.02369. [DOI] [PubMed] [Google Scholar]
- Wang GJ, Kang L, Kim JE, Maro GS, Xu XZ, Shen K. GRLD-1 regulates cell-wide abundance of glutamate receptor through post-transcriptional regulation. Nat Neurosci. 2010;13:1489–1495. doi: 10.1038/nn.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- Wiese M, Antebi A, Zheng H. Intracellular trafficking and synaptic function of APL-1 in Caenorhabditis elegans. PLoS One. 2010;5:e12790. doi: 10.1371/journal.pone.0012790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RW, Setou M, Teng J, Takei Y, Hirokawa N. Overexpression of motor protein KIF17 enhances spatial and working memory in transgenic mice. Proc Natl Acad Sci USA. 2002;99:14500–14505. doi: 10.1073/pnas.222371099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski M, Kim E, Dunah AW, Passafaro M, Valtschanoff JG, Serra-Pages C, Streuli M, Weinberg RJ, Sheng M. Interaction between GRIP and liprin-alpha/SYD2 is required for AMPA receptor targeting. Neuron. 2002;34:39–52. doi: 10.1016/s0896-6273(02)00640-2. [DOI] [PubMed] [Google Scholar]
- Yonekawa Y, Harada A, Okada Y, Funakoshi T, Kanai Y, Takei Y, Terada S, Noda T, Hirokawa N. Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J Cell Biol. 1998;141:431–441. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Koh YH, Beckstead RB, Budnik V, Ganetzky B, Bellen HJ. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 1998;21:1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Maricq AV. Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron. 1999;24:347–361. doi: 10.1016/s0896-6273(00)80849-1. [DOI] [PubMed] [Google Scholar]
- Zhou HM, Brust-Mascher I, Scholey JM. Direct visualization of the movement of the monomeric axonal transport motor UNC-104 along neuronal processes in living Caenorhabditis elegans. J Neurosci. 2001;21:3749–3755. doi: 10.1523/JNEUROSCI.21-11-03749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.