There are 221 experimentally validated, leucine-rich nuclear export signal (NES)–containing CRM1 cargoes in a database named NESdb. Entries in NESdb are annotated with sequence and structural information on both NES and cargo proteins, as well as with experimental evidence on NES-mapping and CRM1-mediated nuclear export.
Abstract
The leucine-rich nuclear export signal (NES) is the only known class of targeting signal that directs macromolecules out of the cell nucleus. NESs are short stretches of 8–15 amino acids with regularly spaced hydrophobic residues that bind the export karyopherin CRM1. NES-containing proteins are involved in numerous cellular and disease processes. We compiled a database named NESdb that contains 221 NES-containing CRM1 cargoes that were manually curated from the published literature. Each NESdb entry is annotated with information about sequence and structure of both the NES and the cargo protein, as well as information about experimental evidence of NES-mapping and CRM1-mediated nuclear export. NESdb will be updated regularly and will serve as an important resource for nuclear export signals. NESdb is freely available to nonprofit organizations at http://prodata.swmed.edu/LRNes.
INTRODUCTION
Dynamic nuclear–cytoplasmic trafficking of macromolecules controls many eukaryotic cellular processes, such as gene expression, signal transduction, cell differentiation, and immune response. The karyopherin-β family of transport factors recognizes targeting signals within cargo proteins for transport in and out of the nucleus. Nuclear localization signals direct proteins into the nucleus, and nuclear export signals (NESs) direct proteins into the cytoplasm (reviewed in Görlich and Kutay, 1999; Chook and Blobel, 2001; Conti and Izaurralde, 2001; Weis, 2003; Kutay and Güttinger, 2005; Tran et al., 2007; Xu et al., 2010).
The leucine-rich or classic NES is the only class of nuclear export signal that has been characterized. An NES is 8–15 amino acids long and contains regularly spaced hydrophobic residues. The name leucine-rich NES was coined because the first signals identified in the HIV-1 Rev and PKIα proteins are enriched with leucine residues (Fischer et al., 1994; Meyer and Malim, 1994; Wen et al., 1995). Since then, many more NES-containing proteins have been identified, and mutagenesis and computational analyses have shown the NES sequences to be more diverse and conform to the loose consensus sequence φ-X2-3-φ-X2-3-φ-X-φ, where φ is L, V, I, F, or M and X is any amino acid (Bogerd et al., 1996; Henderson and Eleftheriou, 2000; Engelsma et al., 2004; la Cour et al., 2004; Kutay and Güttinger, 2005). The NES is recognized by the export karyopherin CRM1, which is also known as exportin 1 (Fornerod et al., 1997; Fukuda et al., 1997; Neville et al., 1997; Ossareh-Nazari et al., 1997; Richards et al., 1997; Stade et al., 1997). Recently published crystal structures of CRM1 bound to several NESs showed that the signals adopt either combined α-helix–loop or all-loop structures that bind in a hydrophobic groove on the convex surface of CRM1 (Dong et al., 2009a, b; Monecke et al., 2009; Güttler et al., 2010). Leptomycin B (LMB) inhibits nuclear export by forming a covalent bond with Cys528 of human CRM1, which is located in the NES-binding groove, thus blocking access of the NES to its binding site (Kudo et al., 1999; Dong et al., 2009b; Monecke et al., 2009).
NESs have been identified in >300 proteins with diverse functions, such as transcription factors, cell cycle regulators, ribonucleoprotein complexes, translation factors, and viral proteins (Fischer et al., 1994; Wen et al., 1995; Fridell et al., 1996; Ho et al., 2000; Murdoch et al., 2002; Vissinga et al., 2009). Nuclear export of viral proteins by CRM1 is important for replication of many viruses that cause human diseases. Aberrant mislocalization of cellular CRM1 cargoes also interrupts numerous cellular processes, often resulting in diseases. Therefore controlling CRM1–NES interactions might be a potential therapeutic target for many disease conditions such as cancer and viral infections (Bogerd et al., 1995; Yi et al., 2002; Faustino et al., 2007; Noske et al., 2008).
A database of 80 NESs named NESbase 1.0 was compiled in 2003 (la Cour et al., 2003). More recently, Fu et al. (2011) published a list of 70 NES-containing proteins. Here, we present NESdb, an up-to-date and substantially larger NES database with 221 experimentally identified entries. Each entry is annotated with many detailed features related to the sequence, structure, and nuclear export activity of the NESs and cargo proteins. NESdb is a valuable information resource for the biomedical research community to learn about nuclear export signals that have already been identified. Analysis of the sequences and three-dimensional structures of NESs in NESdb and false-positive NESs generated from NESdb revealed some distinguishing features that might be important for the future development of accurate NES prediction algorithms (Xu et al., 2012).
DATABASE CONTENT AND DEVELOPMENT
NESdb contains 221 entries as of December 2011. Each entry is a protein that contains one or more NESs. All NESs listed in NESdb were experimentally identified and reported in the published literature. Both the PubMed and UniProt databases were searched using keywords “nuclear export signal,” “NES,” and “CRM1” (Jain et al., 2009; The UniProt Consortium, 2011). The returned literature was examined with the following criteria to identify the existence of an experimentally tested NES: 1) evidence of CRM1-dependent nuclear export, such as binding to CRM1, inhibition by LMB, nuclear retention at nonpermissive temperature in CRM1 temperature-sensitive yeast strains, or competition with other CRM1 cargoes; 2) the presence of a protein segment that matches the traditional NES consensus sequence φ-X2-3-φ-X2-3-φ-X-φ, which can target a reporter protein for nuclear export; and 3) the presence of mutations within the tested NES segment that abolished nuclear export of the full-length protein. All proteins in NESdb meet the first criterion, and many meet all three criteria. The collected information is manually entered into the database. NESdb was implemented as a MySQL database. PHP5 was used to connect to the database and dynamically generate HTML pages. Apache Web server hosted on a Linux cluster was used to serve the database.
DATABASE ACCESS AND USER INTERFACE
The NESdb database is freely available for nonprofit organizations at http://prodata.swmed.edu/LRNes. At this time, NESdb contains 221 experimentally identified CRM1 cargoes reported in the literature. The published literature is searched on a bimonthly basis and NESdb is updated with every 20 new entries. However, many sequences in the genome, especially those in amphipathic helices, match the NES consensus, thus making accurate NES identification difficult. It is likely that some published studies contain mistakenly identified NESs. As a caution to the research community, we separated the 221 proteins in NESdb into two groups. The first group is named “NESs” and contains experimentally identified NESs with no contradicting experimental evidence. The second group is named “NESs in doubt” and contains proteins that were initially reported as NESs but with doubts on their validity cast by subsequent experiments. Clicking the corresponding link on the main page brings up a list of proteins that belongs to each group. The list can be sorted alphabetically by protein names or numerically by protein ID numbers in NESdb. Users are able to positively or negatively flag specific NES-containing proteins on their individual pages. A tally of flags for each protein is displayed next to its name on the list. An entry with many negative flags will be reevaluated and moved to the “NESs in doubt” category or vice versa. The database is also equipped with a search button, which searches the full name, alternative names, and organism of proteins for the keywords. Clicking on a particular protein will load the individual page for the protein.
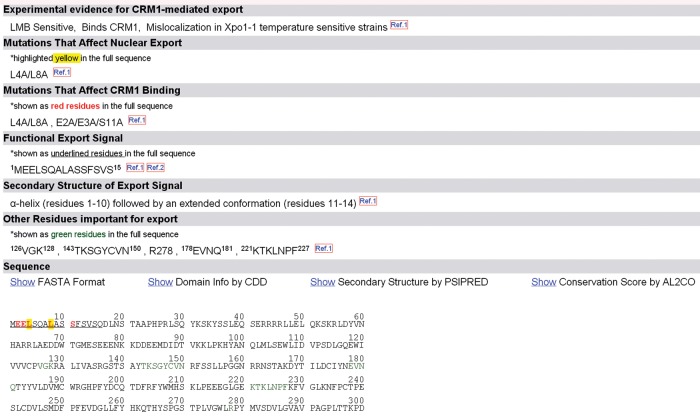
Each entry contains 14 features related to the sequence, structure, and nuclear export activity of the NESs and cargo proteins. A sample page for snurportin 1 (SNUPN) is shown in Figure 1. The NES features include the following:
Full name: the recommended name in UniProt database for the given protein, along with the link to its entry in UniProt (Jain et al., 2009; The UniProt Consortium, 2011).
Alternative names: other names of the protein that are commonly used in the literature.
Organism: the organism of the listed protein.
Experimental evidence for CRM1-mediated nuclear export: reported experimental evidence for CRM1-mediated nuclear export. Reports on whether 1) the protein binds CRM1, 2) the protein is retained in the nucleus by LMB, 3) nuclear export of the protein is affected by another CRM1 cargo such as the HIV-Rev protein, or 4) CRM1 is required for nuclear export in digitonin-permeabilization transport assays. All NESdb entries contain experimental evidence for CRM1-mediated nuclear export.
Mutations that affect nuclear export: mutations that have been shown to disrupt nuclear export in cells.
Mutations that affect CRM1 binding: mutations that have been shown to disrupt in vitro CRM1 binding.
Functional export signals: the protein segment that resembles an NES and when fused to a reporter protein can independently target the reporter for nuclear export. Such data define boundaries and sufficiency of the putative minimal NESs.
Secondary structure of the export signal: the explicitly denoted, experimentally determined secondary structure for the reported NES.
Other residues important for export: residues known to contact CRM1 or shown to affect CRM1 binding or nuclear export but are located outside of the NES segment. This information may be useful since cargoes may bind CRM1 in multipartite manner and contain additional binding epitopes.
Sequence: sequence of the full-length protein with the functional NES underlined. Mutations that disrupt nuclear export in cells are highlighted in yellow, mutations that disrupt in vitro CRM1 binding are in red, and other, non-NES residues reported to affect CRM1 binding or nuclear export are in green. There are four tabs associated with the sequence: 1) the sequence in FASTA format (Pearson and Lipman, 1988), 2) conserved domains of the protein obtained from the Conserved Domain Database (Marchler-Bauer et al., 2009), 3) predicted secondary structure of the protein by PSIPRED (McGuffin et al., 2000), and 4) conservation scores of the protein calculated by AL2CO (Pei and Grishin, 2001).
Three-dimensional structures: links to three-dimensional structures in the Protein Data Bank (PDB), if available.
Comments: a short summary of protein functions and experiments related to the identification of its transport signals.
References: the literature from which the features were extracted, with links to PubMed.
User input: because NESs are easily misidentified, a user input field at the bottom of each entry that allows users to positively or negatively flag the NES after submitting supporting comments/rationales.
FIGURE 1:
A sample page from NESdb that shows 7 of the 14 illustrated features of the NES from snurportin 1 (SNUPN). The features not shown include full name, alternative names, organism, three-dimensional structures, comments, references, and a user input form.
CONCLUSION
NESdb will contribute to the understanding of how protein function is controlled by intracellular localization and will serve as a useful resource for the development of inhibitors that target CRM1-mediated nuclear export. NESdb may be used to train and test new NES prediction algorithms to increase the reliability and accuracy of identifying vague and diverse NESs in the genome.
Acknowledgments
We thank Lisa Kinch for insightful suggestions for user interface, Ming Tang for technical assistance with Web server hosting, and Maarten Fornerod for discussion. This work is funded by the National Institutes of Health (F32GM093493 to D.X., R01-GM069909 to Y.M.C., and R01-GM094575 to N.V.G.), the Welch Foundation (I-1532 to Y.M.C. and I-1505 to N.V.G.), the Leukemia and Lymphoma Society Scholar Program (to Y.M.C.), the Cancer Prevention Research Institute of Texas (PR-101496 to Y.M.C.), and the UT Southwestern Endowed Scholars Program (to Y.M.C. and N.V.G.).
Abbreviations used:
- CRM1
chromosome region maintenance 1
- HIV
human immunodeficiency virus
- LMB
leptomycin B
- NES
nuclear export signal
- PKIα
cAMP-dependent protein kinase inhibitor alpha
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-01-0045) on July 25, 2012.
REFERENCES
- Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Madore S, Cullen BR. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol. 2001;13:310–319. doi: 10.1016/s0955-0674(00)00213-1. [DOI] [PubMed] [Google Scholar]
- Dong X, Biswas A, Chook YM. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol. 2009a;16:558–560. doi: 10.1038/nsmb.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Biswas A, Süel KE, Jackson LK, Martinez R, Gu H, Chook YM. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009b;458:1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsma D, Bernad R, Calafat J, Fornerod M. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 2004;23:3643–3652. doi: 10.1038/sj.emboj.7600370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino RS, Nelson TJ, Terzic A, Perez-Terzic C. Nuclear transport: target for therapy. Clin Pharmacol Ther. 2007;81:880–886. doi: 10.1038/sj.clpt.6100141. [DOI] [PubMed] [Google Scholar]
- Fischer U, Meyer S, Teufel M, Heckel C, Lührmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Fischer U, Lührmann R, Meyer BE, Meinkoth JL, Malim MH, Cullen BR. Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1 Rev. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu SC, Imai K, Horton P. Prediction of leucine-rich nuclear export signal containing proteins with NESsential. Nucleic Acids Res. 2011;39:e111. doi: 10.1093/nar/gkr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Güttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T, Ficner R, Sattler M, Görlich D. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol. 2010;17:1367–1376. doi: 10.1038/nsmb.1931. [DOI] [PubMed] [Google Scholar]
- Henderson BR, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Güttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Meyer BE, Malim MH. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- Monecke T, Güttler T, Neumann P, Dickmanns A, Görlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324:1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- Murdoch K, Loop S, Rudt F, Pieler T. Nuclear export of 5S rRNA-containing ribonucleoprotein complexes requires CRM1 and the RanGTPase cycle. Eur J Cell Biol. 2002;81:549–556. doi: 10.1078/0171-9335-00271. [DOI] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Noske A, Weichert W, Niesporek S, Röske A, Buckendahl AC, Koch I, Sehouli J, Dietel M, Denkert C. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer. 2008;112:1733–1743. doi: 10.1002/cncr.23354. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- Richards SA, Carey KL, Macara IG. Requirement of guanosine triphosphate-bound ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Bolger TA, Wente SR. SnapShot: nuclear transport. Cell. 2007;131:420. doi: 10.1016/j.cell.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Vissinga CS, Yeo TC, Warren S, Brawley JV, Phillips J, Cerosaletti K, Concannon P. Nuclear export of NBN is required for normal cellular responses to radiation. Mol Cell Biol. 2009;29:1000–1006. doi: 10.1128/MCB.01131-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr Opin Struct Biol. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Farmer A, Collett G, Grishin NV, Chook YM. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol Biol Cell. 2012;23:3677–3693. doi: 10.1091/mbc.E12-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Bogerd HP, Cullen BR. Recruitment of the Crm1 nuclear export factor is sufficient to induce cytoplasmic expression of incompletely spliced human immunodeficiency virus mRNAs. J Virol. 2002;76:2036–2042. doi: 10.1128/jvi.76.5.2036-2042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]