SRSF1 splicing factor and nuclear-localized MALAT1 RNA influence the assembly of nuclear speckles. Depletion of SRSF1 compromises the association of splicing factors to nuclear speckles and influences the levels of other SR proteins. SRSF1 regulates RNA polymerase II–mediated transcription.
Abstract
The mammalian cell nucleus is compartmentalized into nonmembranous subnuclear domains that regulate key nuclear functions. Nuclear speckles are subnuclear domains that contain pre-mRNA processing factors and noncoding RNAs. Many of the nuclear speckle constituents work in concert to coordinate multiple steps of gene expression, including transcription, pre-mRNA processing and mRNA transport. The mechanism that regulates the formation and maintenance of nuclear speckles in the interphase nucleus is poorly understood. In the present study, we provide evidence for the involvement of nuclear speckle resident proteins and RNA components in the organization of nuclear speckles. SR-family splicing factors and their binding partner, long noncoding metastasis-associated lung adenocarcinoma transcript 1 RNA, can nucleate the assembly of nuclear speckles in the interphase nucleus. Depletion of SRSF1 in human cells compromises the association of splicing factors to nuclear speckles and influences the levels and activity of other SR proteins. Furthermore, on a stably integrated reporter gene locus, we demonstrate the role of SRSF1 in RNA polymerase II–mediated transcription. Our results suggest that SR proteins mediate the assembly of nuclear speckles and regulate gene expression by influencing both transcriptional and posttranscriptional activities within the cell nucleus.
INTRODUCTION
The mammalian cell nucleus is organized into specialized nuclear domains or nuclear bodies that are generally characterized by the presence of a unique group of proteins and RNAs within them (Matera et al., 2009; Dundr and Misteli, 2010; Mao et al., 2011b). Nuclear domains coordinate specific functions, including the synthesis and processing of pre-rRNA and the initial assembly of ribosome subunits in the nucleolus (Matera et al., 2009; Dundr and Misteli, 2010; Dundr, 2012; Mao et al., 2011b). Nuclear bodies could either host-specific functions such as transcription and RNA processing or help to provide increased local concentrations of factors involved in related functions. Such an organization of molecules will significantly accelerate intermolecular interactions within a spatially restricted area and will also reduce nonspecific interactions with other nuclear factors, which could compete against or prevent a specific nuclear function (Matera et al., 2009; Dundr and Misteli, 2010; Dundr, 2012; Mao et al., 2011b). Unlike cytoplasmic structures or organelles, nuclear domains are not enclosed within a lipid membrane. It has been demonstrated that the molecular constituents within a nuclear domain display continuous and rapid exchange with the surrounding nucleoplasm, and this continuous flux of components is required for the efficient functioning of these nuclear compartments (Dundr and Misteli, 2010). The presence of discrete nuclear bodies even in the absence of a surrounding membrane suggests that the nuclear compartments are formed due to localized accumulations of proteins and RNAs through protein–protein or protein–RNA interactions. This supports a model of stochastic self-organization in which high-affinity interactions among molecules within a domain help to establish a steady-state residency time within these domains (Kaiser et al., 2008; Dundr and Misteli, 2010).
Nuclear speckles (also called SC35 domains or splicing speckles) are dynamic, irregularly shaped nuclear domains that are enriched with factors involved in pre-mRNA processing and RNA transport (Hall et al., 2006; Spector and Lamond, 2011). An interphase nucleus contains 20–50 nuclear speckles. Under the electron microscope (EM), nuclear speckles correspond to interchromatin granule clusters, which range from one to several micrometers in diameter and consist of 20- to 25-nm granules that are connected in places by thin fibrils (Thiry, 1995). Several studies found that transcriptionally active genes preferentially localize adjacent to nuclear speckles (Hall et al., 2006; Hu et al., 2009; Zhao et al., 2009). Furthermore, coordinately active genes tend to associate with the same nuclear speckle (Brown et al., 2008; Hu et al., 2008; Takizawa et al., 2008; Zhao et al., 2009). All these findings suggest that nuclear speckles spatially coordinate transcription and pre-mRNA processing by acting as functional centers to bring transcriptionally active genes near their periphery (perichromatin fibrils) so as to organize euchromatic neighborhoods (Zhao et al., 2009; Spector and Lamond, 2011). Nuclear speckles are also suggested to act as storage/assembly sites of various pre-mRNA processing factors, from where these factors are actively recruited to nearby transcription sites (Spector and Lamond, 2011).
Many of the pre-mRNA splicing factors, including spliceosomal small nuclear ribonucleoproteins (snRNPs) and SR-family factors, localize to nuclear speckles (Spector and Lamond, 2011). SR proteins consist of a group of essential splicing factors that are involved in both constitutive and alternative splicing (Bourgeois et al., 2004; Long and Caceres, 2009; Zhong et al., 2009). SR proteins contain RNA-recognition motif(s) (RRMs), as well as serine-arginine–rich dipeptide RS domains (Lin and Fu, 2007; Long and Caceres, 2009). SR proteins are phosphorylated at multiple serine residues in the RS domain, and the continuous phosphorylation/dephosphorylation cycle of SR proteins is required for efficient pre-mRNA splicing (Lin and Fu, 2007; Long and Caceres, 2009). Phosphorylation of SR proteins also influences their intranuclear distribution (Caceres et al., 1997; Stamm, 2008). The hyperphosphorylation of SR proteins by exogenous overexpression of the SR-protein kinases results in nuclear speckle disruption, further indicating the involvement of RS-domain phosphorylation in nuclear speckle assembly (Gui et al., 1994; Sacco-Bubulya and Spector, 2002). Based on these results, it was inferred that different domains within SR proteins (RRMs and RS) facilitate dynamic intermolecular interactions with other proteins and RNA(s) and that such associations could influence the assembly and maintenance of nuclear speckles at specific nuclear regions.
In addition to localizing a subset of proteins involved in pre-mRNA metabolism, nuclear speckles also harbor RNA molecules, including several noncoding RNAs (ncRNAs). Nuclear speckle–associated RNAs include uridine-rich small nuclear RNAs (UsnRNAs), 7SK RNA, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) long ncRNA (lncRNA), and a population of uncharacterized poly(A)+ RNAs (Spector and Lamond, 2011). MALAT1, one of the most abundant lncRNAs, interacts with several splicing factors, including SR proteins (Hutchinson et al., 2007; Bernard et al., 2010; Tripathi et al., 2010; Zong et al., 2011). Recent studies indicated that in human cells, MALAT1 regulates alternative splicing of pre-mRNAs by modulating the cellular distribution, phosphorylation, and splicing activity of SR splicing factors (Tripathi et al., 2010; Lin et al., 2011).
It is not clear how nuclear speckles are initially formed and maintained within the cell. It is speculated that they form through self-assembly of speckle-resident proteins and RNAs and are maintained by transient protein–protein or protein–RNA interactions (Dundr and Misteli, 2010; Mao et al., 2011b). SR proteins, with their ability to interact with RNA, as well as with other RS domain-containing proteins, are attractive candidates for initiating the stochastic assembly of nuclear speckles. A recent study also indicated that exogenously expressed intron-containing pre-mRNA could act as an initial nucleation site for speckle assembly (Shevtsov and Dundr, 2011). This is in conjunction with several recent studies demonstrating the role of RNAs, especially ncRNAs, as nucleation sites for the initial assembly of nuclear domains (Mao et al., 2011a; Shevtsov and Dundr, 2011).
In the present study, we examine the role of SR splicing factors and MALAT1 lncRNA in the organization of nuclear speckles. We demonstrate that SR proteins and MALAT1 facilitate the association of several of the pre-mRNA processing factors on a chromatin locus. Depletion of splicing factor SRSF1 results in the mislocalization of nuclear speckle proteins and snRNAs but not MALAT1 from nuclear speckles, indicating that SRSF1 along with MALAT1 could influence the recruitment of splicing factors to nuclear speckles. We also present evidence for differential expression and activity of members of the SR proteins upon depletion of SRSF1, implicating the compensatory mechanism used by the cell to coordinate pre-mRNA processing. Finally, SR protein-depleted cells show decreased RNA pol II–mediated transcription and aberrant recruitment of transcription factors.
RESULTS
SRSF1 depletion results in the mislocalization of splicing factors from nuclear speckles
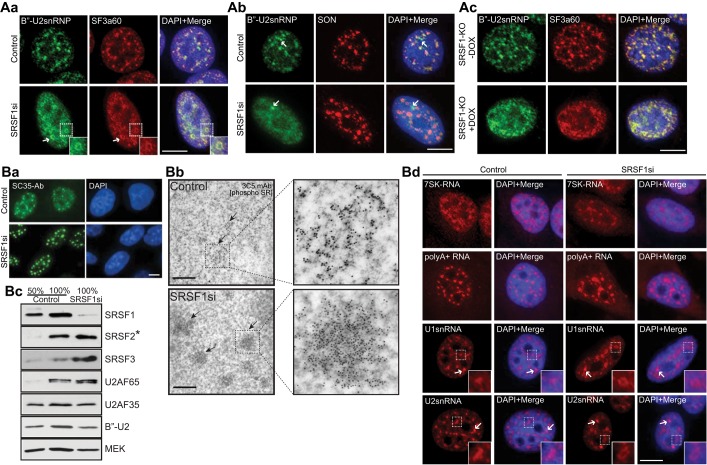
We previously demonstrated that SR proteins interact with nuclear speckle–localized MALAT1 lncRNA and that depletion of MALAT1 results in the mislocalization of several splicing factors, including SRSF1 (Tripathi et al., 2010). To test whether the SR proteins are involved in the nuclear speckle organization, we examined the distribution of splicing factors to nuclear speckles upon depletion of SR proteins. In comparison to control small interfering RNA (siRNA)–treated HeLa cells, SRSF1-depleted cells (Figure 1B, c, and Supplemental Figure S1, A, a, and C, a) showed mislocalization of several of the nuclear speckle components (Figure 1, A, a, b, and d, and B, a, and Supplemental Figure S1A, b and c). In control cells, both B′′-U2snRNP and SF3a60, components of U2snRNP, localized to nuclear speckles, whereas SRSF1-depleted cells displayed a more homogeneous distribution of these proteins (Figure 1A, a and b). In a fraction of SRSF1-depleted cells (∼20%, n = 150), B′′-U2snRNP and SF3a60 localized in the form of ring-like structures within the nucleus (Figure 1A, a, see arrows). A similar doughnut-shaped localization of splicing factors was previously observed upon depletion of nuclear speckle–localized SON pre-mRNA splicing factor (Sharma et al., 2010). Immunolocalization of snRNP-specific proteins in SRSF1 knockout (KO) mouse embryonic fibroblasts (MEF; S1Ad) further confirmed the involvement of SRSF1 in the localization of these proteins to nuclear speckles (Figure 1A, c, and Supplemental Figure S1A, e). In addition to snRNP components, SRSF1-depleted cells also displayed altered localization of other nuclear speckle–associated pre-mRNA processing factors, including SF1, UAP56, and U170K (Supplemental Figure S1A, b and c). Immunolocalization analysis using the Y12 antibody that preferentially recognizes B′/B and D polypeptides of the Sm complex further confirmed the mislocalization of snRNPs from nuclear speckles in SRSF1-depleted cells (Supplemental Figure S1A, c). However, SRSF1 knockdown did not result in the physical disruption of nuclear speckles, as other speckle proteins, such as SON, continued to decorate nuclear speckles even in absence of SRSF1 (Figure 1A, b). We also determined the intranuclear distribution of SR proteins upon SRSF1 depletion. Other SR proteins continue to localize to nuclear speckles in SRSF1-depleted cells as enlarged nuclear speckles. This was observed either by immunolocalization analyses using antibodies, which preferentially detect phospho-SRSF2 (Figure 1B, a [SC35 antibody]) and phosphorylated pan-SR proteins (Supplemental Figure S1A, c [phospho SR]) or by cyan fluorescent protein (CFP)/yellow fluorescent protein (YFP)/red fluorescent protein (RFP)–tagged SRSF2 and SRSF3 localization (Supplemental Figures S1A, b [CFP-SRSF3], and S2A; also see Figure 6B, o–o″, later in the paper). Such “rounded-up” speckles in SRSF1-depleted cells were also confirmed by immuno-EM analyses (Figure 1B, b).
FIGURE 1:
Depletion of SRSF1 results in the disorganization of nuclear speckle components. (A, a and b) Immunofluorescence staining using antibodies against B′′-U2snRNP, SF3a60, and SON in control and SRSF1 siRNA-treated HeLa cells. The arrows (A, b) designate Cajal bodies. (A, c) Immunofluorescence staining using antibodies against B′′-U2snRNP and SF3a60 in control and SRSF1-knockout MEFs. Immunofluorescence (B, a; SC35 antibody) and immune-EM (B, b; 3C5 antibody) analyses using antibodies that preferentially detect phosphorylated SR proteins in control and SRSF1-siRNA–treated HeLa cells. (B, c) Immunoblot analyses using antibodies against various pre-mRNA processing factors in control and SRSF1-depleted total cellular extracts. Asterisk, SRSF2 antibody preferentially recognizes the unphosphorylated or hypophosphorylated forms of SRSF2 (Bubulya et al., 2004). (B, d) RNA-FISH (red) of 7SK RNA, poly(A)+ RNA, and U1 and U2 snRNA in control and SRSF1-depleted HeLa cells. The DNA is counterstained with DAPI. Scale bars, fluorescent and EM images, 5 and 1 μm, respectively.
FIGURE 6:
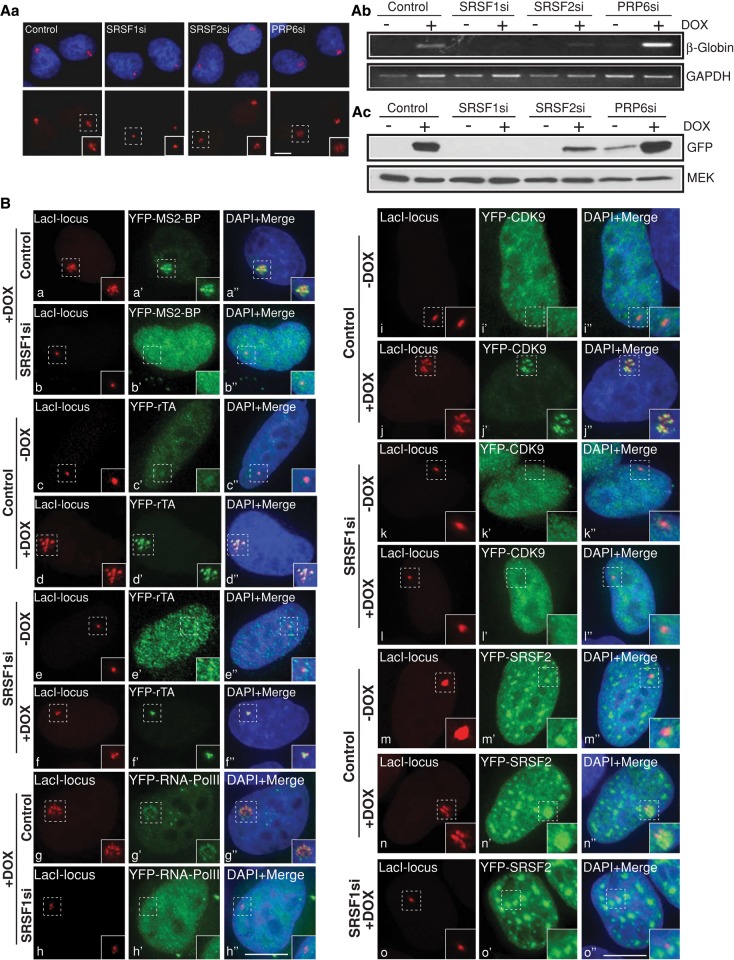
SRSF1-depleted cells show reduced cellular transcription. (A, a) LacI-mCherry–localized gene locus in the DOX-treated control and SRSF1, SRSF2, and PRP6 siRNA–transfected CLTon cells. (A, b and c) RT-PCR and immunoblot analyses show reduced reporter RNA (β-globin) and protein (GFP) levels in the SR protein–depleted (SRSF1 and SRSF2) cells. GAPDH RNA and MEK protein are used as loading controls. (B) Localization of MS2BP–YFP as an indicator of active transcription at the gene locus (+DOX) in control (a–a′′) and SRSF1-depleted (b–b′′) CLTon cells. (c). Localization of YFP-rTa (c′, d′, e′, f′), YFP-RNA pol II (g′, h′), and YFP-CdK9 (i′, j′, k′, l′) at the gene locus (+DOX) in control and SRSF1-depleted CLTon cells. Note the absence of MS2-BP-YFP (b–b′′), RNA pol II (h–h′′Cdk9 (l–l′′), and YFP-SRSF2 (o–o′′), and the presence of YFP-rTa (f–f′′) at the DOX-induced gene locus of SRSF1-depleted cells. The YFP-SRSF2 in the SRSF1-depleted cells (o′, o′′) are imaged with less exposure time compared with control siRNA–treated cells for the better clarity of nuclear speckles. DNA is counterstained with DAPI. Scale bars, 5 μm.
Finally, we examined the distribution of nuclear-speckle localized ncRNAs in SRSF1-depleted cells. SRSF1 depletion did not affect the nuclear speckle distribution of 7SK and poly(A)+ RNAs (Figure 1B, d). However, SRSF1-depleted HeLa cells showed defects in the distribution of UsnRNAs to nuclear speckles (Figure 1B, d). Similar to B′′-U2snRNP, SF3a60, and U170K, SRSF1-depleted cells displayed doughnut-shaped localization of U1 and U2snRNAs (Figure 1Bd). The nuclear speckle–resident MALAT1 lncRNA continued to show similar distribution in control and SRSF1-depleted mouse (Supplemental Figure S1A, f) and human cells (Tripathi et al., 2010). Of interest, the localization of splicing factors and RNAs to Cajal bodies— distinct subnuclear bodies where snRNPs localize during their biogenesis for late maturation steps and recycling after splicing—remains unaltered upon SRSF1 depletion, indicating that depletion of SRSF1 compromised only the distribution of splicing factors in nuclear speckles and not in other nuclear domains (arrows in Figure 1, A, b, and B, d [U1 and U2snRNAs] and Supplemental Figure S1A, c [U170K, Y12]).
Next we determined whether the altered distribution of nuclear speckle components observed in SRSF1-depleted cells was sensitive to the total cellular transcription. We incubated HeLa cells with or without SRSF1 with DRB to inhibit RNA pol II transcription and compared the distribution of nuclear speckle proteins. Intriguingly, RNA pol II transcription–inhibited control- and SRSF1 siRNA–treated cells showed similar localization of splicing factors to nuclear speckles (Supplemental Figure S1A, g). However, upon transcription reactivation after DRB washout, the SRSF1-depleted cells displayed doughnut-shaped localization of splicing factors. From these data we conclude that SRSF1 influences the nuclear speckle association of pre-mRNA processing factors in transcriptionally active cells.
To examine the specificity of the SRSF1 siRNA (siRNA was designed from the 3′ untranslated region [UTR] of the human Srsf1 gene) used in the present study, we conducted a rescue experiment in which HeLa cells stably expressing YFP-SRSF1 cDNA (lacking the 3′UTR targeted by the SRSF1 siRNA) was transfected with SRSF1 siRNA and the intranuclear distribution of splicing factors was examined (Supplemental Figure S1B; Bubulya et al., 2004; Prasanth et al., 2010). SRSF1 siRNA–treated enhanced YFP (eYFP)-SRSF1–expressing cells showed reduced levels of endogenous SRSF1 but did not show changes in the localization of nuclear speckle proteins, further confirming that the SRSF1 influences the cellular levels and distribution of splicing factors (Supplemental Figure S1B, a and c). We also examined the involvement of other SR proteins in the cellular levels and distribution of splicing factors (Supplemental Figure S1C). SRSF2-depleted HeLa cells or KO-MEFs showed unaltered cellular levels and normal speckle localization of RNA and splicing factors (Supplemental Figure S1, A, f, and C, a–d). Our results suggest that SRSF1 and not SRSF2 specifically influences the distribution of several nuclear speckle–localized pre-mRNA processing factors and ncRNAs.
SRSF1 depletion results in the stabilization of SRSF2
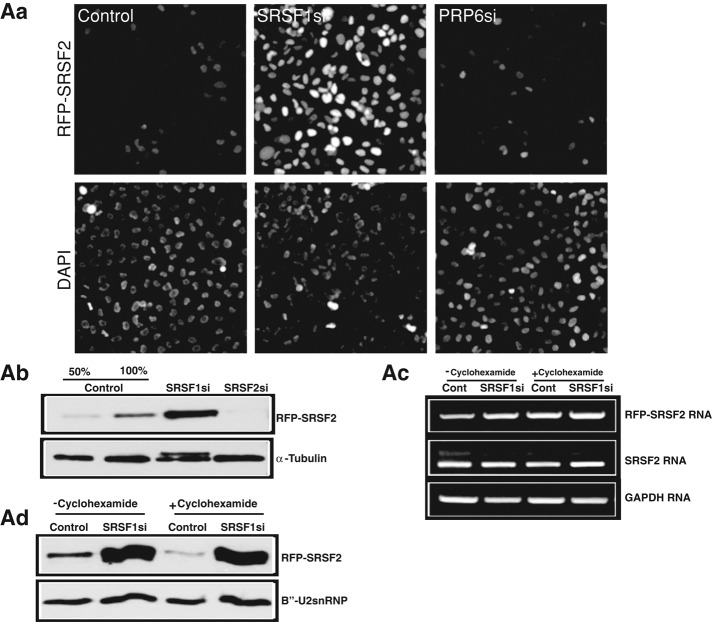
Immunoblot analysis from whole-cell extracts of SRSF1-depleted cells displayed increased levels of several endogenous SR proteins, including SRSF2 (∼1.6-fold increase) and SRSF3 (∼4.5-fold increase), but not the levels of other speckle-localized pre-mRNA processing factors (Figure 1B, c). To understand how SRSF1 influences other SR protein levels, we examined the changes in the cellular levels and stability of stably expressing monomeric RFP (mRFP)- or YFP-SRSF2 upon SRSF1 depletion. SRSF1-depleted cells showed a pronounced increase in the levels of fluorescently tagged SRSF2 (Figure 2A, a, and Supplemental Figure S2A). However, depletion of human PRP6, an essential, non-SR splicing factor that is involved in U4/U6.U5 tri-snRNP assembly (Figure 2A, a), or B′′-U2snRNP (unpublished data) did not alter the levels of SRSF2, indicating that the changes in the abundance of SR proteins observed in SRSF1-depleted cells are not merely due to pre-mRNA splicing defects. Immunoblot analyses further confirmed the increase in SRSF2 levels upon SRSF1 depletion (Figure 2A, b, and Supplemental Figure S2, b). Reverse transcription (RT)-PCR results reveal that the SRSF1-depleted cells did not show any significant change in the endogenous or exogenous (RFP-SRSF2) mRNA levels (Supplemental Figure S2, c), indicating that SRSF2 is being regulated at the translational level. To test this possibility, we compared SRSF2 mRNA and protein levels from control and SRSF1-depleted cells that were incubated with cycloheximide, a translation inhibitor. SRSF1-depleted cells grown in presence or absence of cycloheximide did not show any change in the SRSF2 mRNA levels (Figure 2A, c) but continued to show increased levels of SRSF2 protein (Figure 2A, d). On the basis of these results, we conclude that SRSF1 depletion results in the stabilization of the already existing pool of SRSF2 and/or other SR proteins.
FIGURE 2:
SRSF1-depletion results in the stabilization of the cellular pool of SRSF2. (A, a) SRSF1-depleted HeLa cells show increased cellular levels of RFP-SRSF2. (A, b) Immunoblot analysis using RFP antibody from total-cell extracts of control or SRSF1- or SRSF2-depleted HeLa cells (RFP-SRSF2 stable cell line) display increased expression of RFP-SRSF2 upon SRSF1 depletion. α-Tubulin is used as a loading control. (A, c) RT-PCR using indicated primers from control or SRSF1 siRNA–treated HeLa cells stably expressing RFP-SRSF2. (A, d) Immunoblot analysis using RFP antibody from total cell extracts of control and SRSF1-depleted HeLa cells in presence or absence of cycloheximide. B′′-U2snRNP is used as a loading control. GAPDH is used as loading control (A, c).
MALAT1 and SR proteins facilitate the association of splicing factors to a chromatin locus and/or nucleate the de novo assembly of nuclear speckles
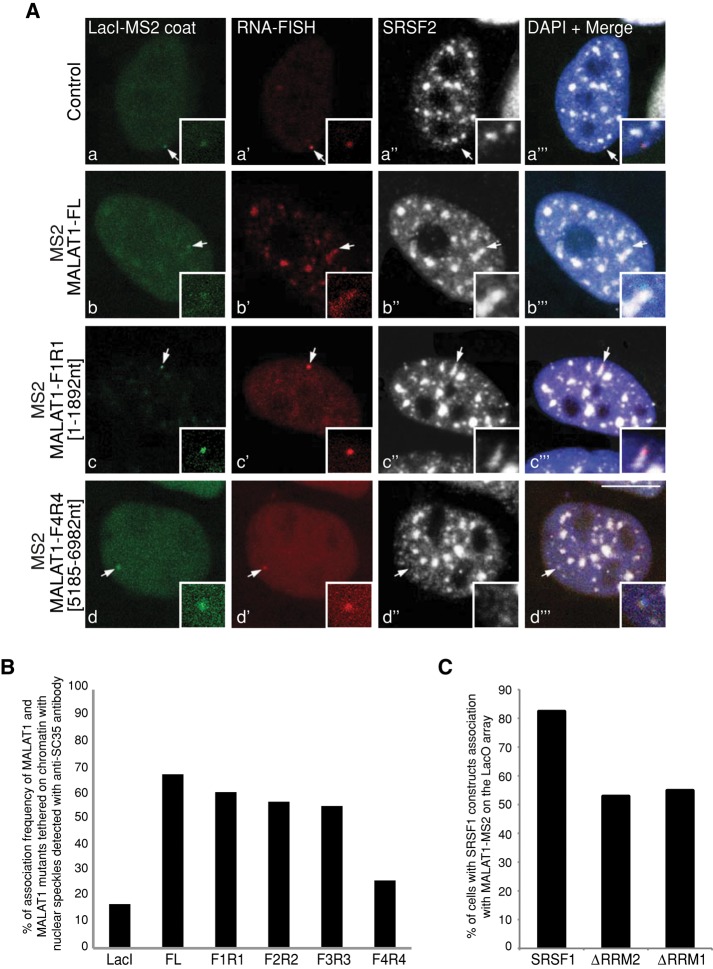
Most of the splicing factors that showed aberrant nuclear speckle distribution upon SRSF1 depletion also displayed similar changes in their localization after depletion of MALAT1 (Tripathi et al., 2010). In addition, MALAT1-depleted human cells also showed mislocalization of SRSF1 from nuclear speckles but not vice versa (Supplemental Figure S1A, f; Tripathi et al., 2010). This indicates that MALAT1 could act upstream of SR proteins in the nuclear speckle assembly pathway. To examine the involvement of MALAT1 in the association of splicing factors to nuclear speckles, we used an experimental cell system in which MALAT1 RNA tagged with a bacteriophage MS2 stem loop was targeted and immobilized on a Lac operon (LacO)-repeat array (256 repeats of LacO sequence stably integrated at a single integration site on chromosome 7) in the chromatin of modified HeLa cells by Lac repressor (LacI)-NLS-GFP-MS2 coat protein, which selectively binds the MS2 loop (Shevtsov and Dundr, 2011). Using this approach, we examined whether the immobilized MALAT1-MS2 RNA could specifically recruit splicing factors to chromatin sites (Figure 3). The tethered LacI-MS2 coat protein alone did not recruit endogenous SRSF2 (Figure 3A, a–a′′′) and SRSF1 or B′′-U2snRNP (unpublished data) to the locus, whereas the locus containing immobilized full-length MALAT1-MS2 RNA colocalized with SRSF2-containing (Figure 3, A, b–b′′′, and B) or SRSF1- or B′′-U2snRNP-containing (unpublished data) nuclear speckles. RNA-immunoprecipitation (IP) and CLIP-seq results revealed that MALAT1 interacts with several members of the SR protein family, and each of the SR proteins has several potential binding sites on MALAT1 (Sanford et al., 2009; Tripathi et al., 2010; Anko et al., 2012). For example, each MALAT1 RNA contains ∼50 potential SRSF1-binding sites (Sanford et al., 2009; Tripathi et al., 2010). Most of these sites are located in the 5′ half (first 5 kb of ∼7-kb RNA) of the MALAT1 sequence, whereas the 3′ end contains few SRSF1-binding sites. We therefore immobilized several deletion mutants of MALAT1 RNA on chromatin and examined their involvement in the recruitment of SR proteins to the genomic locus. The F1-R1 (nucleotides [nts] 1–1892 of MALAT1), F2-R2 (nts 1677–3600 of MALAT1), and F3-R3 (nts 3476–5331 of MALAT1) mutants recruited SRSF2 with similar efficiency to full-length MALAT1 (Figure 3, A, c–c′′′, and B). However, the F4-R4 (nts 5185–6982 of MALAT1) mutant failed to recruit SR proteins to the locus (Figure 3, A, d–d′′′, and B). This is consistent with our previous RNA-IP results showing reduced interaction of SRSF1 with the F4-R4 region of MALAT1 (Tripathi et al., 2010). To identify the domain(s) within SR proteins that facilitate the recruitment of SR proteins to the MALAT1-tethered nuclear speckle, we examined the recruitment of CFP-SRSF1 mutants to the de novo–formed speckle (Figure 3C). In ∼80% of the cells analyzed, full-length CFP-SRSF1 was recruited to the MALAT1-tethered locus (n = 50). However, in comparison to the full-length SRSF1, we observed a reduction in the recruitment of SRSF1-ΔRRM1 and SRSF1-ΔRRM2 mutants to the locus (∼50%; n = 50). This result indicates that deletion of any of the two RRMs somewhat compromises the association of SRSF1 to the MALAT1-tethered locus. This result corroborates our previous RNA-IP studies in which both the RRM domains of SRSF1 are required for the efficient interaction of SRSF1 to MALAT1 (Tripathi et al., 2010). Taken together, our results imply that MALAT1 RNA can efficiently initiate the assembly of splicing factors, including SR proteins at a specific chromatin site.
FIGURE 3:
Immobilization of MALAT1-MS2 RNA on chromatin leads to association with nuclear speckle or de novo formation of nuclear speckle. (A) RNA-FISH using a probe against vector (a′) or mouse MALAT1 (b′, c′, d′; red) combined with immunofluorescence staining using SRSF2 antibody (white) on the LacO-containing HeLa cells transiently cotransfected with vector or full-length or mutant MS2-MALAT1 and GFP-lacI-NLS-MS2 coat protein (green). Note that the immobilized wild-type and F1-R1 mutant MALAT1 RNA associate with SRSF2-containing nuclear speckle, whereas F4-R4 MALAT1 mutant RNA does not localize to nuclear speckles. (B) Quantitative analysis of association of tethered wild-type and mutant MALAT1 RNA with existing speckle or de novo speckle formation. (C) Quantitative analysis of association of full-length and RRM mutants of CFP-SRSF1 to the MALAT1-MS2–tethered locus. Values represent averages (n = 50–60) from two independent experiments. DNA is counterstained with DAPI. Scale bar, 5 μm.
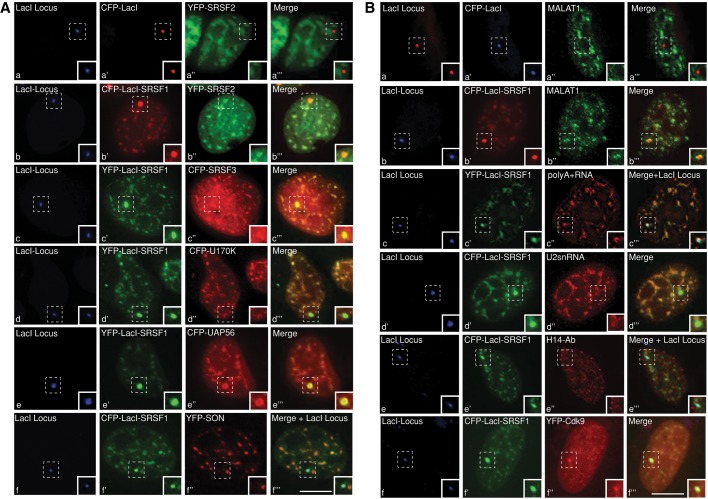
Next we analyzed the role of SR proteins in de novo speckle assembly. For this assay, we used a modified version of the original U2OS 2-6-3 in vivo cell system that was developed by David Spector's group (Janicki et al., 2004). We stably integrated Cherry-LacI and rTa (tet-activator) into the original U2OS 2-6-3 cells (U2OS 2-6-3 CLTon). This approach enabled us to readily visualize the single Lac operator locus in the CLTon cells by the presence of Cherry-LacI (Shen et al., 2010; Sathyan et al., 2011). A triple-fusion protein expressing YFP- or CFP-LacI-SRSF1 and -SRSF2 was generated so that the SR proteins could be immobilized at the stably integrated chromatin locus in 2-6-3 CLTon via a LacO-LacI interaction. By immobilizing YFP- or CFP-LacI-SRSF1 and -SRSF2 to the locus, we were able to examine the recruitment of various pre-mRNA processing factors and nuclear speckle–resident ncRNAs to the locus (Figures 4 and 5A and Supplemental Figure S3, A–C). As a negative control, we tethered the YFP- or CFP-LacI fusion proteins and analyzed the recruitment of the specific proteins to the YFP- or CFP-LacI–immobilized locus (Figure 4A, a–a′′′, and Supplemental Figure S3, D and E). Targeting of LacI-SRSF1 to the locus resulted in specific association/recruitment of several pre-mRNA splicing factors to the chromatin locus. These proteins include SRSF2 (Figure 4A, b–b′′′), SRSF3 (Figure 4A, c–c′′′), U170K (Figure 4A, d–d′′′), and UAP56 (Figure 4A, e–e′′′). SON splicing factor, although colocalized with SRSF1 in the nuclear speckles, was not recruited to the LacI-SRSF1–containing chromatin locus (Figure 4A, f–f′′′, and Supplemental Figure S3A, a–a′′′). In addition to the pre-mRNA processing factors, LacI-SRSF1 also recruited several of the speckle-associated RNAs to the chromatin locus, including MALAT1 (Figure 4B, b–b′′′), poly(A)+ RNA (Figure 4B, c–c′′′), and U2snRNA (Figure 4B, d–d′′′). Nuclear speckles also contain a population of RNA pol II and several other transcription factors, including pTEFb kinase complex, and SR proteins are known to interact with both the RNA pol II and pTEFb complex (Spector and Lamond, 2011). We therefore examined whether SR proteins could recruit RNA pol II and pTEFb complex to the locus. LacI-SRSF1–immobilized locus did not recruit RNA pol II. Both the initiation (as detected by H14 antibody staining; Figure 4B, e–e′′′) and elongation competent forms (as detected by H5 antibody staining; Supplemental Figure S3A, b–b′′′) of RNA pol II were absent from the locus. Of interest, LacI-SRSF1 successfully recruited Cdk9, a component of the pTEFb complex, to the locus (Figure 4B, f–f′′′).
FIGURE 4:
Immobilization of SRSF1 on chromatin leads to association with nuclear speckle or de novo formation of nuclear speckle. (A) CLTon cells are cotransfected with CFP-LacI vector (a′), CFP-LacI-SRSF1 (b′, f′), or YFP-LacI-SRSF1 (c′, d′, e′) and YFP-SRSF2 (a′′, b′′), CFP-SRSF3 (c′′), CFP-U170K (d′′), CFP-UAP56 (e′′), and YFP-SON (f′′). (B, a–d) RNA-FISH shows the nuclear distribution of endogenous MALAT1 (a′′, b′′), poly(A)+ RNA (c′′), and U2snRNA (d′′) in CLTon cells that are transfected with CFP-LacI vector (a′), CFP-LacI-SRSF1 (b′, d′), or YFP-LacI-SRSF1 (c′). Immunolocalization of RNA pol II (H14 antibody; e′′) and YFP-Cdk9 localization (f′′) in CLTon cells, which are transfected with CFP-LacI-SRSF1 (e′, f′). Note that CFP-LacI-SRSF1 fails to recruit YFP-SON and RNA pol II to the de novo–formed speckles. A and B, a–f, represent the stably integrated LacI locus (blue) in the CLTon cells. Scale bars, 5 μm.
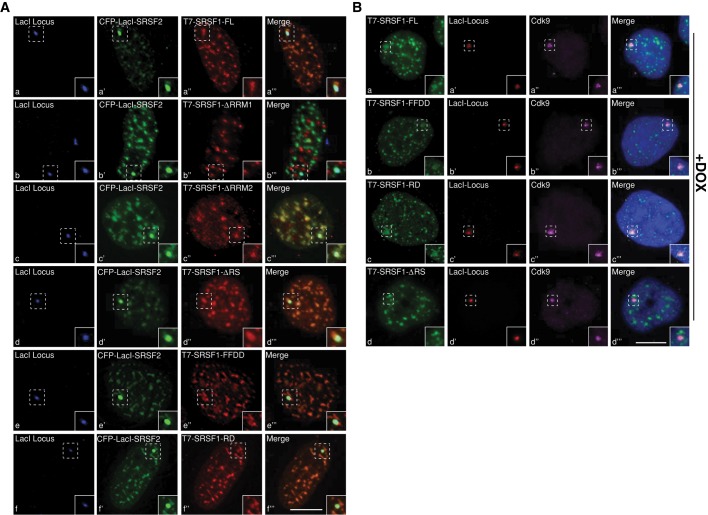
FIGURE 5:
RRM1 and RS domains are required for the recruitment of SR proteins to de novo–formed nuclear speckles and to transcription sites, respectively. (A) CLTon cells are cotransfected with CFP-LacI-SRSF2 (a′, b′, c′, d′, e′, f′) and T7-tagged, full-length or mutant SRSF1 constructs. Note that the T7-SRSF1-ΔRRM1 (b–b′′′) fail to localize to the de novo–formed nuclear speckle. (B) Immunofluorescence localization of transiently expressed, T7-tagged, full-length or mutant SRSF1 and endogenous Cdk9 in DOX-induced CLTon cells. Note that T7-SRSF1-ΔRS (d–d′′′) does not localize to the transcriptionally active gene locus. The DNA is counterstained with DAPI. Scale bars, 5 μm.
Next we determined the involvement of specific modular domains of SRSF1 in the recruitment of splicing factors to the de novo–formed speckles. We examined the role of the RS domain, a region that is primarily involved in protein–protein interactions, in the recruitment of other SR proteins to de novo–formed speckles. Various SRSF1 mutants were tethered to the locus, and the recruitment of SRSF2 was analyzed (Supplemental Figure S3A, c–e). The CFPlacI-SRSF1ΔRS (82%; n = 50; Supplemental Figure S3A, d) and CFPlacI-SRSF1ΔRRM1 (48%; n = 50; Supplemental Figure S3A, e) mutants efficiently recruited SRSF2 to the locus. These results indicate that the RS domain of SRSF1 is dispensable for the recruitment of SRSF2 to the locus. Similar to full-length SRSF1, SRSF2 also facilitated the recruitment of a similar set of splicing factors and RNA molecules to the locus (Supplemental Figure S3, B and C). SR proteins specifically mediate the association of only the nuclear speckle–resident proteins and RNAs to the chromatin locus. In contrast, factors that are localized to other nuclear bodies did not associate with SR protein-immobilized genomic locus (coilin and promyelocytic leukemia [PML] protein, structural components of Cajal and PML nuclear bodies, respectively; unpublished data).
Different modular domains of SRSF1 dictate its association to the de novo–formed nuclear speckles and to gene transcription sites
The RRM domains of an SR protein specify its RNA-binding properties, whereas the RS domain acts as a protein–protein interaction module and recruits components of the core splicing machinery to promote splice-site selection (Sanford et al., 2005). The RS domain of SR proteins has also been demonstrated to interact directly with the branch point and 5′ splice site of pre-mRNA (Shen and Green, 2004) and, furthermore, to dictate several cellular functions, including determining the nucleocytoplasmic shuttling of SR proteins and influencing the localization of SR proteins to nuclear speckles and sites of transcription (Caceres et al., 1997; Long and Caceres, 2009; Zhong et al., 2009). We examined the involvement of various SR protein domains in the localization of SR proteins to the SR protein–immobilized nuclear speckles. We coexpressed CFP-LacI-SRSF2 along with T7-tagged full-length (FL) and various mutant forms of SRSF1 (Caceres et al., 1997) in the CLTon cells and analyzed their recruitment to the locus-immobilized CFP-LacI-SRSF2 (Figures 5A). The SRSF1-FL, SRSF1ΔRRM2 (contains intact RRM1 and RS domains), and SRSF1ΔRS (contains intact RRM1 and 2) proteins colocalized with CFP-LacI-SRSF2 at the chromatin locus (Figures 5A, a–a′′′, c–c′′′, and d–d′′′). Although SRSF1ΔRRM1 showed a speckle-like distribution, it failed to associate with the SRSF2-immobilized chromatin locus (Figure 5A, b–b′′′). To understand the involvement of RRM1 and RS-domains of SR proteins in their assembly onto the de novo formed speckles, we next examined the recruitment of SRSF1 RRM1 (FF-DD) and RS domain phospho-mimetic (SRSF1-RD) mutants to the SRSF2-immobilized nuclear speckle (Cazalla et al., 2002). The RRM1 FF-DD (the Phe-56 and -58 in RRM1 were replaced with Asp residues) did not support constitutive splicing and displayed weak RNA-binding activity (Caceres and Krainer, 1993). Of interest, both mutants showed similar association to the locus containing CFP-LacI-SRSF2 (Figure 5A, e–e′′′ and f–f′′′). On the basis of these results, we conclude that SRSF1 requires the RRM1 domain for its association with the de novo–formed nuclear speckles. Furthermore, an RS domain and its phosphorylation do not influence the association of SRSF1 to an SR protein–immobilized nuclear speckle–like structure.
In addition to nuclear speckles, a specific combination of SR proteins associates with pre-mRNAs at the transcription sites (Bjork et al., 2009). To assess RS domain involvement in the recruitment of SR proteins to a transcriptionally active gene locus, we examined the association of T7-SRSF1 RRM1 and RS mutants at the doxycycline (DOX)-induced transcriptionally active gene locus in CLTon cells (Figure 5B; Janicki et al., 2004). Treatment with DOX activated transcription from the gene locus, as observed by the association of Cdk9 (Figure 5B, a′′, b′′, c′′, and d′′) and RNA pol II (unpublished data) at the locus (Sathyan et al., 2011). SRSF1 FL, FF-DD, and RD mutants (Figure 5B, a–c) were recruited to the transcriptionally active gene locus, whereas the ΔRS mutant failed to associate with the gene locus (Figure 5B, d–d′′′). This indicates that the RS domain is essential for the recruitment of SRSF1 to a transcription site but not necessary for its association to the de novo–formed nuclear speckles. These findings are in agreement with a previous study demonstrating the role of RS domains in the recruitment of SR proteins to the transcription sites (Misteli et al., 1998).
Our experiments using locus-immobilized SR proteins or MALAT1 demonstrate that SR proteins and their interacting partner, MALAT1, can either successfully nucleate the assembly of nuclear speckle proteins and RNAs to a de novo–formed nuclear speckle at a chromatin locus or target the chromatin locus to an already existing nuclear speckle. In specific instances, immobilized-MALAT1 and -SR proteins on the genomic array protruded toward an existing nuclear speckle and formed a part of the speckle (∼38% [n = 100]; Figures 3A, b–b′′′and c–c′′′, and 5A, a–a′′′ and d–f). In other instances (∼62% [n = 100]), the locus completely overlapped with an independent nuclear speckle (Figure 4A, b–b′′′, and Supplemental Figure S3A, b–b′′′ and j–j′′′). Furthermore, the SR protein–immobilized locus did not contain all of the bona fide nuclear speckle components (examples include SON and phosphorylated RNA pol II), supporting the argument that the tethered SR proteins at the locus initiate the assembly of a new nuclear speckle or nuclear speckle–like structure.
SR proteins modulate RNA polymerase II–mediated transcription
Besides pre-mRNA processing and mRNA export, SR proteins are also implicated in other functions, including translation, nonsense-mediated mRNA decay (NMD), and genome stability (Zhong et al., 2009). A study demonstrated the involvement of SR proteins in transcription elongation (Lin et al., 2008). SR protein–depleted (SRSF1 and SRSF2) HeLa cells showed reduced overall RNA pol II–mediated transcription, as observed by in vitro Br-UTP pulse incorporation assays (unpublished data). We therefore decided to examine the role of SR proteins in the transcriptional induction at a single-cell level and the involvement of SR proteins in the recruitment of various transcription and pre-mRNA processing factors to the transcription site. On DOX addition, the tet-inducible reporter gene in the CLTon cells activated transcription and displayed dramatic decompaction of the chromatin locus (Janicki et al., 2004; Prasanth et al., 2010; Tripathi et al., 2010; Sathyan et al., 2011; Figure 6A, a, and Supplemental Figure S4). In contrast, the gene locus in the SR protein–depleted cells (especially SRSF1-depleted cells) continued to stay at a highly condensed state (Figure 6A, a, and Supplemental Figure S4). Similarly, RT-PCR and immunoblot analyses using probes that detect reporter RNA (rabbit β-globin) and reporter protein (GFP) in the CLTon locus (Janicki et al., 2004) showed a dramatic reduction in reporter gene activation upon SR protein depletion (Figure 6A, b and c). However, knockdown of the splicing factor PRP6 did not affect chromatin decondensation, transcription, and translation from the reporter gene locus (Figure 6A, a–c). Surprisingly, PRP6-depleted cells showed increased levels of reporter RNA and protein compared with control siRNA–treated cells, indicating that PRP6 negatively influences transcription from the reporter gene locus. In this context it is interesting to note that human PRP6 factor was initially identified as a transcription factor, and it was shown to interact with androgen receptor and also regulate RNA pol II–mediated transcription (Zhao et al., 2002)
To determine how SR proteins regulate transcription, we examined the recruitment of various transcription and pre-mRNA processing factors to the reporter gene locus in SRSF1-depleted CLTon cells. In control siRNA–treated cells, DOX treatment activated transcription from the reporter gene, as observed by accumulation of YFP-MS2-binding protein (BP; YFP-MS2-BP specifically recognizes the bacteriophage MS2 repeat present in the reporter RNA; Janicki et al., 2004) at the gene locus in ∼50% of the cells (n = 80; Figure 6Ba). In contrast, none of the SRSF1-depleted, DOX-induced cells show accumulation of YFP-MS2-BP at the gene locus and instead showed homogeneous nuclear distribution of YFP-MS2-BP, which is indicative of transcription repression (Figure 6B,b). Next we examined the recruitment of transcription activator (rTa) to the gene locus in presence or absence of SRSF1. Both control and SRSF1-depleted cells showed robust accumulation of rTa to the locus, indicating that SR protein depletion does not affect the association of transcription activator to the gene locus (44% in control siRNA–treated cells vs. 42% in SRSF1-depleted cells; n = 50; Figure 6B, c–f). However, SRSF1-depleted cells showed reduced association of RNA pol II (38% in control siRNA-treated cells vs. 4% in SRSF1-depleted cells; n = 50; Figure 6B, g and h), pTEFb kinase complex (as observed by Cdk9 localization; 52% in control siRNA–treated cells vs. 8% in SRSF1-depleted cells; n = 50; Figure 6B, i–l), and pre-mRNA processing factors (30% in control siRNA–treated cells vs. 8% in SRSF1-depleted cells; n = 50; SRSF2; Figure 6B, m–o) to the DOX-treated gene locus. These results indicate that SR proteins influence RNA pol II–mediated transcription.
DISCUSSION
In the interphase nucleus, nonmembranous nuclear bodies are speculated to form due to high- or low-affinity dynamic interactions between individual components present in each of these nuclear compartments (Matera et al., 2009; Dundr and Misteli, 2010; Dundr, 2012; Mao et al., 2011b). In the present study, we examined the role of nuclear speckle–associated SR splicing factors and MALAT1 lncRNA in nuclear speckle assembly. We demonstrate that both SR proteins and MALAT1 RNA assemble multiple nuclear speckle components to a chromatin site. Loss-of-function studies in human cancer cells revealed that depletion of MALAT1 or SRSF1 results in dissociation of several pre-mRNA processing factors from nuclear speckles. In particular, MALAT1 influenced the speckle association of SRSF1 in a phosphorylation-independent manner (Tripathi et al., 2010). However, SR protein depletion did not affect the distribution of MALAT1 to nuclear speckles. These results indicate that MALAT1 acts upstream of SR proteins in coordinating the assembly of splicing factors to nuclear speckles. Recent studies strongly argue in favor of the roles played by resident RNA molecules in the initial assembly of specific nuclear domains (Dundr, 2011; Mao et al., 2011a; Shevtsov and Dundr, 2011). In general, nuclear speckles are not sites of active gene transcription, and most of the pre-mRNA splicing initiates at transcription sites that are preferentially located at the periphery of nuclear speckles (Spector and Lamond, 2011). We propose that long ncRNAs such as MALAT1 recruit SR proteins and act as nucleation sites for the assembly of nuclear speckles. The SR proteins further interact with other pre-mRNA processing factors and recruit them to the nuclear speckles. Specific domains within SR proteins (RRMs and RS domains) modulate the intermolecular interactions among SR proteins and other splicing factors and RNAs that localize to nuclear speckles. Our results indicate that the SR protein–mediated, de novo–formed nuclear speckles do not contain all the nuclear speckle constituents. This suggests that nuclear speckles contain many subcomplexes and that member(s) of a complex interact with constituents of another complex to influence their association into nuclear speckles. Several studies indicated that specific gene networks could be organized around individual nuclear speckles (Zhong et al., 2009; Spector and Lamond, 2011). However, it is not clear how such a specific organization is achieved. MALAT1 RNA has recently been shown to be involved in organizing the transcriptionally active genes at the nuclear speckle periphery (Yang et al., 2011). SR proteins also interact with the transcriptionally active genes through their direct interaction with the histone 3 tail of chromatin (Loomis et al., 2009; Sapra et al., 2009). We hypothesize that SR proteins would be ideal candidates to coordinate the organization of nuclear speckles and gene networks in close proximity, which in turn will facilitate cellular processes, including transcription, pre-mRNA splicing, and mRNA export (Zhong et al., 2009).
In the present study, we showed that depletion of SRSF1 but not SRSF2 alters the nuclear speckle distribution of several of the pre-mRNA processing factors. Mammalian cells contain several SR and SR-like splicing factors (Shepard and Hertel, 2009). Initial in vitro studies suggest that SR proteins show functional redundancy, as individual SR proteins could often substitute for one another in biochemical splicing assays with cell-free extracts (Bourgeois et al., 2004; Long and Caceres, 2009; Zhong et al., 2009). Similarly, such functional redundancy was also observed in vivo in Caenorhabditis elegans (Longman et al., 2000). However, a large number of studies in other organisms, including mammalian cells, have clearly established the distinct nonoverlapping roles played by individual SR proteins (Wang et al., 1996, 2001; Li and Manley, 2005; Xiao et al., 2007). Besides their role as a general regulator of pre-mRNA splicing, individual members of the SR protein family are also known to control the alternative splicing of several nonoverlapping sets of pre-mRNAs (Zhong et al., 2009). SRSF1 is a shuttling protein and is involved in transcription, mRNA export, NMD, translation, and maintenance of genome stability, whereas SRSF2 is primarily a nuclear protein and confines its function within the nuclear compartment (Long and Caceres, 2009; Zhong et al., 2009). Genetic inactivation of SRSF1 results in cell lethality, which could not be rescued by other members of the SR protein family (Zhong et al., 2009). Our results indicate that SRSF1 plays a separate, nonoverlapping role in organizing nuclear speckle components. Future studies will address whether additional members of the SR and SR-like protein families play key roles in the organization of nuclear speckle components.
We also demonstrate that the SRSF1-depleted cells showed increased cellular levels of SR proteins, including SRSF2 and SRSF3. This is achieved preferentially by the stabilization of the existing pool of SR proteins, although the mechanism for how this occurs is unknown. A previous study reported the up-regulation of SRSF2 in SRSF1-depleted chicken DT-40 cells (Liu et al., 2003). In SRSF1 knockout B cells, activation resulted in elevated expression of SRSF2 and rescued RNA splicing at the proximal 3′ splice sites of a reporter bovine papilloma virus type 1 late pre-mRNA (Liu et al., 2003). The authors suggested the involvement of phosphatidylinositol 3-kinase/Akt pathways in the increased expression of SR proteins in the SRSF1-depleted cells (Liu et al., 2003). Cellular levels of SR proteins are tightly regulated by several mechanisms, including the existence of unproductive splicing in the ultraconserved DNA elements located within the SR genes (Sureau et al., 2001; Lareau et al., 2007; Sun et al., 2010). SRSF2 is autoregulated by transcribing unstable alternatively spliced mRNA isoforms that undergo NMD (Sureau et al., 2001). A recent study reported that the expression of SRSF1 is also autoregulated at multiple levels, including alternative splicing and translation regulation (Sun et al., 2010). SRSF3 is also known to regulate the expression of its own mRNA (Jumaa and Nielsen, 1997). Overexpression of SRSF3 promotes the synthesis of an SRSF3 mRNA isoform that produces a truncated protein. Of interest, SRSF1 negatively regulates the synthesis of this truncated SRSF3 mRNA isoform and thereby facilitates the production of full-length SRSF3 mRNA. Specific cellular signals and viral infections are also known to alter the expression of specific SR genes. For instance, the human papilloma virus (HPV) E2 transcription factor induces the expression of Srsf1 and Srsf3 in HPV-infected cells, preferentially through chromosomal translocation of the SR genes (Mole et al., 2009).
Our data suggest a role for SR proteins in RNA pol II–mediated transcription. SR proteins are known to function in coordinating transcription, as well as in pre-mRNA processing (Zhong et al., 2009). For example, pre-mRNA splicing efficiency is strongly enhanced if SR proteins are recruited to the gene before transcription activation but not if they are brought in after transcription (Das et al., 2007). This indicates that the initial recruitment of SR proteins to the nascent pre-mRNA is required for the efficient splicing of the nascent transcript. Such a functional coupling is primarily mediated by the carboxy-terminal domain (CTD) of RNA pol II, which interacts with SR proteins and facilitates cotranscriptional recruitment of SR proteins to nascent pre-mRNA transcripts (Misteli and Spector, 1999; Das et al., 2007). SR proteins are also known to interact with several transcription factors, and such an association could also influence the recruitment of SR proteins to specific gene promoters (Bourgeois et al., 2004; Zhong et al., 2009). In the present study, SRSF1-depleted cells showed defects in chromatin decondensation and decreased RNA pol II–mediated transcription at a gene locus. The SR-like protein Npl3p in budding yeast is also involved in transcription elongation (Dermody et al., 2008). A similar role for mammalian SRSF2 in transcription elongation was recently reported, in which in vivo depletion of SRSF2 affected the chromatin loading of RNA pol II and nascent RNA synthesis (Lin et al., 2008). In murine cells, SRSF2 coimmunoprecipitated with Cdk9 and SRSF2-depleted cells showed reduced association of Cdk9 with RNA pol II and CTD phosphorylation (Lin et al., 2008). Furthermore, chromatin immunoprecipitation studies revealed that SRSF2-depleted cells showed reduced association of Cdk9 to the body of specific genes. This indicates that the SR proteins might modulate transcription elongation either by recruiting key factors to transcription sites or by stabilizing the association of specific transcription factors with core transcription machinery (Fong and Zhou, 2001; Pandit et al., 2008; Zhong et al., 2009). We observed decreased association of Cdk9 to the transcription site in the SRSF1-depleted cells. Our in vivo localization studies further confirm the role of SR proteins in the recruitment of Cdk9 to the transcription sites. Our results support a model in which SR proteins initially associate at the transcription sites through their interaction with RNA pol II and transcription factors. This enables efficient recruitment of pTEFb components to the nascent pre-mRNA and positively regulates the phosphorylation of RNA pol II CTD by pTEFb complex to facilitate transcription elongation.
MATERIALS AND METHODS
Cell culture and treatment
U2OS and HeLa cells were grown in DMEM containing high glucose (Invitrogen, Carlsbad, CA) supplemented with penicillin–streptomycin and 10% fetal bovine serum (FBS; Hyclone, Logan, UT). The U2OS 2-6-3 CLTon cells were grown in DMEM high-glucose media containing 10% Tet-free FBS (Clontech, Mountain View, CA) and G418 and hygromycin. SRSF1 and SRSF2 KO-MEFs were grown in DMEM high-glucose media containing 10% Tet-free FBS. The SRSF1 and SRSF2 KO MEFs stably express a Tet-responsive, hemagglutinin (HA)-tagged SRSF1 and SRSF2, respectively (Lin et al., 2005; Xiao et al., 2007). To deplete the exogenously expressed SR proteins in the SR KO MEFs, we added DOX (10 μg/ml) in the media for 3–4 d.
To inhibit translation, cells were incubated with cycloheximide (50 μg/ml; Sigma-Aldrich, St. Louis, MO) for 2–4 h. Nascent transcription sites were detected using Br-UTP incorporation assays using a previously published protocol (Prasanth et al., 2003).
cDNA constructs and transfection
The T7-tagged plasmids used in the present study include pCGT vector, pCGT-SRSF1, pCGT-SRSF1ΔRRM1, pCGT-SRSF1ΔRRM2, pCGT-SRSF1ΔRS (a kind gift from Adrian Krainer, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), pCGT-SRSF1-FFDD, and pCGT-SRSF1-RD. Fluorescent protein–tagged plasmid DNAs used in the present study include pCMV-eGFP [GFP], eYFP [YFP], or eCFP [CFP]–tagged SF1, U2AF65, U2AF35, and UAP56 (gift from M. Carmo-Fonseca, Lisbon, Portugal); SRSF3 and U170K (gift from J. Caceres, MRC, University of Edinburgh, UK); SON (gift from P. Bubulya, Wright State University, Dayton, OH); and SRSF1 and SRSF2, MS2-BP, rTa, and Cdk9. The coding region of human SRSF1 FL and mutant cDNAs and SRSF2 full-length cDNA was PCR amplified and cloned into eYFP or eCFP-LacI (Kaiser et al., 2008) vectors. The pSV2-MS2-mMALAT1 FL and mutant constructs were generated by cloning the 24X MS2 repeats at the 5′ end of the mMALAT1 FL and mutant cDNAs.
For transient transfection experiments, plasmid DNA (1–2 μg) was transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After transfection, cells were processed for siRNA treatment, followed by immunofluorescence (IF) localization or RNA–fluorescence in situ hybridization (FISH; Prasanth et al., 2005). For the tethering assays in U2OS 2-6-3 CLTon cells, 500 ng of YFP- or CFP-LacI constructs were used along with 1 μg of the desired protein cDNA constructs. Cells were fixed in 2% formaldehyde 16–18 h posttransfection and processed for RNA-FISH or immunostaining. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted in p-phenylenediamine (PPD). DOX, 1 μg/ml, was added for 3 h to the cells in order to induce transcription from the reporter gene locus in CLTon cells.
HeLa cells stably expressing the integrated LacO array (Shevtsov and Dundr, 2011) growing on glass coverslips were cotransfected with mMALAT FL and mutant constructs along with LacI-GFP-MS2 coat protein and mCherry-LacI by Lipofectamine 2000. Cells were further processed for immunostaining or RNA-FISH after 16–24 h posttransfection.
siRNA knockdown
Depletion of human SRSF1, SRSF2, and PRP6 was performed by using double-stranded siRNAs (Tripathi et al., 2010). The siRNAs targeting human SRSF1 (SRSF1 3′UTR-siRNA sense, UUGGCAGUAUUGACCUUAdTdT; SRSF1 3′UTR-siRNA antisense, UAAGGUCAAUACUGCCAAdTdT), SRSF2 (SRSF2-siRNA sense, AAAUCCAGGUCGCGAUCGAdTdT; SRSF2-siRNA antisense, UCGAUCGCGACCUGGAUUUdTdT), PRP6 (PRP6 siRNA sense, GCUACAAGUAGCUCGGAACCUdTdT; PRP6 siRNA antisense, AGGUUCCGAGCUACUUGUAGCdTdT), and control luciferase were synthesized by Dharmacon (Lafayette, CO). siRNAs were delivered into cells at a final concentration of 50–100 nM, mediated by Lipofectamine RNAiMAX (Invitrogen), and were delivered twice at a gap of 24 h.
Antibodies
Antibodies used in the present study include the following: B′′-U2-snRNP (mouse immunoglobulin G [mIgG]; IF, 1:100; Western blot [WB], 1:250), SF3a60 (rabbit IgG [rIgG]; IF, 1:200; WB, 1:2000; Kramer et al., 1994), SRSF1 (mIgG, mAb103; IF, 1:300; Hanamura et al., 1998), SRSF1 (mIgG, mAb96; WB, 1:1000; Hanamura et al., 1998), SRSF2 (mIgG; WB, 1:20; Cavaloc et al., 1999), SRSF2 (SC35, mIgG; IF, 1:300; Fu and Maniatis, 1990), SRSF3 (rIgG; WB, 1:1000; Invitrogen), U170K (mIgG; IF, 1:150; S. Hoch, Agouron Institute, La Jolla, CA), 3C5 (mIgM; IF, 1:100; Turner and Franchi, 1987), U2AF-65 (rIgG; WB, 1:1000), U2AF-35 (rIgG; WB, 1:1000), SON (rIgG; IF, 1:5000; Sharma et al., 2010), Y12 (anti-Sm, mIgG; IF, 1:20; Abcam, Cambridge, MA), RNA polymerase II (H5 antibody, mIgM; IF, 1:100), RNA pol II (H14 antibody, mIgM; IF, 1:100), Cdk9 (rIgG; IF, 1:100; Santa Cruz Biotechnology, Santa Cruz, CA), MEK (rIgG; WB, 1:2000), T7 (mIgG; IF, 1:1000, WB, 1:5000; Novus Biologicals, Littleton, CO), α-tubulin (mIgG; WB, 1:5000; Sigma-Aldrich), GFP (mIgG; WB, 1:500; Covance, Berkeley, CA), mRFP (WB, 1:500; Chemicon International, Temecula, CA), and HA tag (clone 12CA5, mIgG; WB, 1:500).
RNA-FISH and immunofluorescence staining
RNA-FISH to detect MALAT1, U1 and U2 snRNAs, poly(A)+ RNA, and β-tropomyosin reporter RNA was conducted as described previously (Huang et al., 1994; Sacco-Bubulya and Spector, 2002; Prasanth et al., 2005; Tripathi et al., 2010). Cells were fixed in 4% formaldehyde in phosphate-buffered saline (PBS; pH 7.4) for 15 min at room temperature. Hybridization was performed using either nick-translated cDNA probe (for MALAT1 and β-tropomyosin reporter; Abbott Molecular, Des Plaines, IL) or fluorescently labeled oligonucleotide probes (for U1 and U2 snRNAs and oligo dT probes for poly(A+) RNA) in a moist chamber at 37°C for 12–16 h as described earlier (Prasanth et al., 2005).
Immunofluorescence was performed as described previously (Prasanth et al., 2003). Briefly, cells were fixed in 2% formaldehyde for 15 min at room temperature and permeabilized in 0.5% Triton X-100 in PBS for 10 min on ice. One percent normal goat serum in PBS was used as blocking agent. Cells were incubated with primary antibodies in a humidified chamber for 1–2 h and further with secondary antibody for 45 min and finally stained with DAPI and mounted in PPD. For 3C5 antibody staining, cells were fixed and permeabilized in chilled methanol for 5 min.
Image acquisition and processing
Fluorescence images were acquired using an Axio Imager Z1 (63×/1.4 numerical aperture [NA] oil objective, Carl Zeiss, Jena, Germany), DeltaVision RT (Olympus, 60×/1.42 NA oil objective; Applied Precision, Issaquah, WA), or Zeiss LSM510 confocal microscope (100×/1.4 NA Plan Apochromat oil objective). Images were collected as vertical z-stacks spanning the entire nuclei and were processed using either AxioVision (Axio Imager), SoftWoRx (DeltaVision), or Zeiss software.
Immuno-EM analysis
Control and SRSF1 siRNA–treated cells were grown on Thermanox coverslips (Electron Microscopy Sciences, Fort Washington, PA), fixed with 2% formaldehyde and 0.2% glutaraldehyde in PBS, and then dehydrated with ethanol by the progressive lowering of temperature, followed by embedding and polymerization in Lowicryl K4M (Electron Microscopy Sciences) at –35°C. Thin sections collected on nickel grids were first incubated with anti–phospho-SR antibody (3C5) for 1 h at room temperature, washed in PBS, and incubated for 15 min at 37°C in secondary antibody conjugated to 10-nm colloidal gold (Amersham Biosciences Piscataway, NJ). The grids were further washed, air dried, and then counterstained with aqueous uranyl acetate.
RNA Isolation and RT-PCR
Total cellular RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions, DNase I treated (amplification grade; Invitrogen), and further reverse transcribed into cDNA using Multiscribe Reverse Transcriptase and Random Hexamers (Applied Biosystems, Foster City, CA). Semiquantitative RT-PCR was performed using gene-specific primers in the Eppendorf Thermal Cycler (Eppendorf, Hauppauge, NY). Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control for all the experiments.
PCR Primers
GAPDH: forward primer (FP), TCACCAGGGCTGCTTTTAAC; GAPDH reverse primer (RP), TTCTAGACGGCAGGTCAGGT. RFP: FP, CGAGGACGTCATCAAGGAGT; RP, GGTACAGCTTCTCGGTGGAG. SRSF2: FP, TTAAAGCTGCGGTCTCCTGT; RP, TTCCTGGCCAAATAACCAAG. KLF6: FP, TGCTCCCCATGTGCAGCATC; RP, TTCAGTTCGGATTCCTCC. Rabbit β-globin: FP, GTTCATTAGATCCTGAGAACTTCAG; RP, AAAGATCTCAGTGGTATTTGTGAGC.
Supplementary Material
Acknowledgments
We thank P. Bubulya (SON antibody and plasmid constructs, YFP-SRSF1, HeLa cells), M. Carmo-Fonseca (SF1, U2AF65, 35, and UAP56 plasmids), J. Caceres (U170K and SRSF3 constructs), J. Caceres, J. Sanford, and A. Krainer (SRSF1 WT and mutant constructs), M. Hübner (YFP-CdK9), and J. Stevenin (SRSF2 monoclonal antibody) for their kind gift of reagents. We thank lab members for the help in subcloning the splicing factors into LacI vectors. We thank D. L. Spector (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for valuable discussions. We also thank P. Bubulya, M. Bellini, S. Ceman, A. Lal, P. Newmark, S. G. Prasanth, and the Prasanth lab members for critical reading of the manuscript. Research in K.V.P.’s lab is supported by grants from the American Cancer Society (RSG-11-174-01-RMC) and the National Institutes of Health/National Institute of General Medical Sciences (GM088252). Research in X.D.F.’s lab is funded by Grant GM052872 and that in D.M.’s lab by Grant GM090156 from the National Institutes of Health/National Institute of General Medical Sciences.
Abbreviations used:
- CLIP-Seq
cross-linking and immunoprecipitation-sequencing
- CLTon
Cherry-LacI-Tet-on
- DOX
doxycycline
- DRB
5,6-dichloro-1-d-ribofuranosylbenzimidazole
- GFP/YFP/CFP/RFP
green/yellow/cyan/red fluorescent protein
- IGCs
interchromatin granule clusters
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- MEFs
mouse embryonic fibroblasts
- NMD
nonsense-mediated decay
- RNA-IP
RNA-immunoprecipitation
- RRM
RNA recognition motif
- RS
arginine-serine
- snRNPs
small nuclear ribonucleoproteins
- SRSF1
serine/arginine-rich splicing factor 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-03-0206) on August 1, 2012.
*These authors equally contributed in the work.
REFERENCES
- Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork P, Jin S, Zhao J, Singh OP, Persson JO, Hellman U, Wieslander L. Specific combinations of SR proteins associate with single pre-messenger RNAs in vivo and contribute different functions. J Cell Biol. 2009;184:555–568. doi: 10.1083/jcb.200806156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubulya PA, Prasanth KV, Deerinck TJ, Gerlich D, Beaudouin J, Ellisman MH, Ellenberg J, Spector DL. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J Cell Biol. 2004;167:51–63. doi: 10.1083/jcb.200404120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Caceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M. Seed and grow: a two-step model for nuclear body biogenesis. J Cell Biol. 2011;193:605–606. doi: 10.1083/jcb.201104087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol. 2012;24:415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kramer A, Legrain P, Mulhauser F, Groning K, Brosi R, Bilbe G. Splicing factor SF3a60 is the mammalian homologue of PRP9 of S. cerevisiae: the conserved zinc finger-like motif is functionally exchangeable in vivo. Nucleic Acids Res. 1994;22:5223–5228. doi: 10.1093/nar/22.24.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Liu X, Mayeda A, Tao M, Zheng ZM. Exonic splicing enhancer-dependent selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site can be rescued in a cell lacking splicing factor ASF/SF2 through activation of the phosphatidylinositol 3-kinase/Akt pathway. J Virol. 2003;77:2105–2115. doi: 10.1128/JVI.77.3.2105-2115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Longman D, Johnstone IL, Caceres JF. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 2000;19:1625–1637. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011a;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011b;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- Mole S, Milligan SG, Graham SV. Human papillomavirus type 16 E2 protein transcriptionally activates the promoter of a key cellular splicing factor, SF2/ASF. J Virol. 2009;83:357–367. doi: 10.1128/JVI.01414-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Camiolo M, Chan G, Tripathi V, Denis L, Nakamura T, Hubner M, Spector DL. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell. 2010;21:4184–4196. doi: 10.1091/mbc.E10-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol Biol Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco-Bubulya P, Spector DL. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J Cell Biol. 2002;156:425–436. doi: 10.1083/jcb.200107017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Ellis J, Caceres JF. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem Soc Trans. 2005;33:443–446. doi: 10.1042/BST0330443. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34:179–190. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- Sathyan KM, Shen Z, Tripathi V, Prasanth KV, Prasanth SG. A BEN-domain-containing protein associates with heterochromatin and represses transcription. J Cell Sci. 2011;124:3149–3163. doi: 10.1242/jcs.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell. 2010;21:650–663. doi: 10.1091/mbc.E09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, Wang F, Prasanth KV, Prasanth SG. A WD-repeat protein stabilizes ORC binding to chromatin. Mol Cell. 2010;40:99–111. doi: 10.1016/j.molcel.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harbor Perspect Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008;22:489–498. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M. The interchromatin granules. Histol Histopathol. 1995;10:1035–1045. [PubMed] [Google Scholar]
- Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM, Franchi L. Identification of protein antigens associated with the nuclear matrix and with clusters of interchromatin granules in both interphase and mitotic cells. J Cell Sci. 1987;87:269–282. doi: 10.1242/jcs.87.2.269. [DOI] [PubMed] [Google Scholar]
- Wang HY, Xu X, Ding JH, Bermingham JR, Jr, Fu XD. SC35 plays a role in T cell development and alternative splicing of CD45. Mol Cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- Wang J, Takagaki Y, Manley JL. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Goto K, Saitoh M, Yanase T, Nomura M, Okabe T, Takayanagi R, Nawata H. Activation function-1 domain of androgen receptor contributes to the interaction between subnuclear splicing factor compartment and nuclear receptor compartment. Identification of the p102 U5 small nuclear ribonucleoprotein particle-binding protein as a coactivator for the receptor. J Biol Chem. 2002;277:30031–30039. doi: 10.1074/jbc.M203811200. [DOI] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong X, Tripathi V, Prasanth KV. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 2011;8:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.