The polarity regulator Bazooka (Par-3) is shown to directly bind the signaling lipid phosphatidic acid via its PDZ domains in vitro. In vivo, the interaction appears to support Bazooka functions important for Drosophila embryonic epithelial structure. Thus Bazooka has a closer connection to membrane lipids than previously recognized.
Abstract
Bazooka (Par-3) is a conserved polarity regulator that organizes molecular networks in a wide range of cell types. In epithelia, it functions as a plasma membrane landmark to organize the apical domain. Bazooka is a scaffold protein that interacts with proteins through its three PDZ (postsynaptic density 95, discs large, zonula occludens-1) domains and other regions. In addition, Bazooka has been shown to interact with phosphoinositides. Here we show that the Bazooka PDZ domains interact with the negatively charged phospholipid phosphatidic acid immobilized on solid substrates or in liposomes. The interaction requires multiple PDZ domains, and conserved patches of positively charged amino acid residues appear to mediate the interaction. Increasing or decreasing levels of diacylglycerol kinase or phospholipase D—enzymes that produce phosphatidic acid—reveal a role for phosphatidic acid in Bazooka embryonic epithelial activity but not its localization. Mutating residues implicated in phosphatidic acid binding revealed a possible role in Bazooka localization and function. These data implicate a closer connection between Bazooka and membrane lipids than previously recognized. Bazooka polarity landmarks may be conglomerates of proteins and plasma membrane lipids that modify each other's activities for an integrated effect on cell polarity.
INTRODUCTION
Bazooka (Baz; Drosophila Par-3) and its homologues play essential roles in the polarization of epithelial and single cells. Through dynamic interactions with PAR-6 and atypical protein kinase C (aPKC), Baz (Par-3) helps to define molecular associations with the plasma membrane at one pole of a cell. This polarization can guide the assembly of cellular junctions, orient microtubule networks, regulate the actin cytoskeleton, and/or direct membrane trafficking. In epithelia, Baz (Par-3) functions as a landmark to organize the apical domain (reviewed in Nelson, 2003; Suzuki and Ohno, 2006; Goldstein and Macara, 2007; St Johnston and Ahringer, 2010).
Baz (Par-3) is a scaffold protein composed of three conserved regions: conserved region 1 contains an N-terminal oligomerization domain (OD), conserved region 2 contains three PDZ (postsynaptic density 95, discs large, zonula occludens-1) domains, and conserved region 3 contains an aPKC-binding site (reviewed in Laprise and Tepass, 2011; Figure 1A). The PDZ domain is a versatile molecular interaction module that can bind proteins using its peptide-binding pocket or other sequences (reviewed in Nourry et al., 2003). Indeed, such interactions are integral to the assembly of functional Baz polarity landmarks (reviewed in Laprise and Tepass, 2011). In addition, PDZ domains can bind lipids (Wu et al., 2007; reviewed in Nourry et al., 2003). For example, rat Par-3 PDZ2 can bind phosphoinositide species (Wu et al., 2007). Although key sequences involved are not conserved in Baz (Wu et al., 2007), a close relationship with phosphoinositide signaling is common to Baz and vertebrate Par-3. Specifically, Baz contains a distinct phosphoinositide-binding site in its C-terminal region downstream of the PDZ domains (Krahn et al., 2010), and both Baz and Par-3 have been shown to interact with the lipid phosphatase PTEN through their PDZ domains (von Stein et al., 2005; Feng et al., 2008).
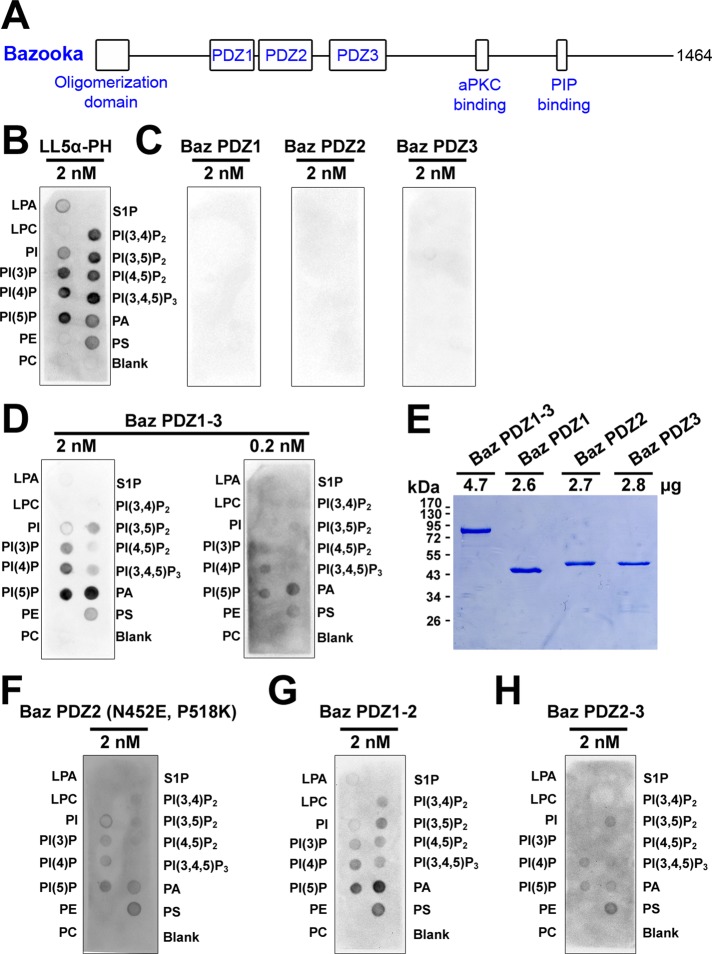
FIGURE 1:
Preferential binding of tandem Bazooka PDZ domains to immobilized phosphatidic acid. (A) A schematic of Baz. (B–D, F–H) GST antibody detection of GST-fusion proteins bound to immobilized lipids: lysophosphatidic acid (LPA), lysophosphocholine (LPC), phosphatidylinositol (PI), phosphatidylinositol (3) phosphate (PI(3)P), phosphatidylinositol (4) phosphate (PI(4)P), phosphatidylinositol (5) phosphate (PI(5)P), phosphatidylethanolamine (PE), phosphatidylcholine (PC), sphingosine 1-phosphate (S1P), phosphatidylinositol (3,4) bisphosphate (PI(3,4)P2), phosphatidylinositol (3,5) bisphosphate (PI(3,5)P2), phosphatidylinositol (4,5) bisphosphate (PI(4,5)P2), phosphatidylinositol (3,4,5) trisphosphate (PI(3,4,5)P3), phosphatidic acid (PA), phosphatidylserine (PS). (B) The LL5α-PH domain (MultiPIP Grip) preferentially binds phosphoinositides. (C) Individual Baz PDZ domains show no detectable lipid binding. (D) Baz PDZ1-3 preferentially binds PA over a 10-fold concentration range. (E) 10% SDS–PAGE and staining with Coomassie brilliant blue shows that the PDZ proteins used in C and D had similar concentrations and stabilities. (F) Converting amino acid residues in Baz PDZ2 to those responsible to phosphoinositide binding in Rat PDZ2 (Wu et al., 2007) conveys lipid binding activity. (G) Baz PDZ1-2 preferentially binds PA. (H) Baz PDZ2-3 also binds lipids but with a reduced preference for PA.
Phosphoinositides also play a central role in cell polarization. In single migratory cells, leading and trailing edges are organized by the polarization of different phosphoinositide species in response to extracellular cues (reviewed in Janetopoulos and Firtel, 2008). Similarly, the polarization of phosphoinositide species helps to organize the apical and basal domains of epithelial cells (reviewed in Shewan et al., 2011). Phosphoinositide species are regulated by lipid kinases and phosphatases and engage downstream effectors through a number of domains, including pleckstrin-homology (PH) and PDZ domains (reviewed in Shewan et al., 2011). Of note, the effects of phosphoinositides are also influenced by other lipids in the local membrane environment (reviewed in Stace and Ktistakis, 2006).
Phosphatidic acid (PA) is a negatively charged phospholipid with two fatty acid chains that makes up 1–4% of total lipids in mammalian cells. It can be produced from diacylglycerol by diacylglycerol kinase (DGK) or from phosphatidylcholine (PC) by phospholipase D (PLD). Although there is no specific binding module for PA, proteins typically bind to PA through clusters of surface-exposed, positively charged amino acid residues. PA can recruit proteins to the plasma membrane and/or activate enzyme activity. PA can also affect membrane curvature due to its conical shape. In addition, PA works in conjunction with other signaling lipids, such as phosphoinositides (reviewed in Stace and Ktistakis, 2006). For example, in mammalian neutrophils, plasma membrane recruitment of the Rac guanine nucleotide exchange factor DOCK2 is initiated by phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and stabilized by PA (Nishikimi et al., 2009). In Drosophila, PA has been implicated in apical membrane transport during rhabdomere biogenesis (Raghu et al., 2009) and the speed of embryo cellularization (LaLonde et al., 2006).
We find that the PDZ domains of Baz directly bind PA in vitro. Mapping experiments implicate multiple positively charged surface patches in the interaction. By manipulating PA levels and mutating the positive residues in vivo, we find that PA supports Baz activity in the embryonic ectoderm. We propose that Baz–PA interactions contribute to a specific lipid environment important for the function of Baz polarity landmarks.
RESULTS
Bazooka PDZ domains bind immobilized phosphatidic acid when present in tandem
To test lipid-binding properties of the Baz PDZ domains, we first probed lipids immobilized on solid supports with various purified glutathione S-transferase (GST) fusion proteins. The commercially available LL5α-PH domain (MultiPIP Grip) was used a positive control and bound phosphoinositide species most strongly (Figure 1B). In contrast, equimolar amounts of Baz PDZ1, Baz PDZ2, or Baz PDZ 3 displayed no detectable binding to any lipid species (Figure 1C). However, all three Baz PDZ domains in tandem (Baz PDZ1-3) bound lipids to a similar degree as equimolar amounts of the LL5α-PH domain, although Baz PDZ1-3 preferentially bound PA (Figure 1D). Reducing the concentration of Baz PDZ1-3 10-fold still revealed a preferential binding to PA (Figure 1D). Of importance, the Baz PDZ1, PDZ2, and PDZ3 proteins were just as stable as Baz PDZ1-3 (Figure 1E shows equimolar amounts analyzed by SDS–PAGE) and appeared more stable than PDZ1-3 in other purification conditions tested (unpublished data). Moreover, Baz PDZ2 gained lipid-binding properties when nonconserved residues shown to be important for rat Par-3 PDZ2-phosphoinositide binding were mutated to the rat residues (N452E, P518K; Wu et al., 2007; Figure 1F). Overall, these results indicate that the Baz PDZ domains preferentially bind PA but only when present in tandem. Both Baz PDZ1-2 and Baz PDZ2-3 displayed lipid binding, with Baz PDZ1-2 showing greater preference for PA in this assay (Figure 1, G and H), indicating that multiple binding sites are involved.
Tandem Bazooka PDZ domains bind phosphatidic acid in liposomes
To test interactions between Baz PDZ domains and PA in a more natural context, we pursued liposome sedimentation assays. To test whether PDZ1-3-PA binding was due to charge alone, we incubated PDZ1-3 with either PA or phosphatidylserine (PS) mixed with phosphatidylcholine (PC) in liposomes. After liposome sedimentation, protein amounts in the pellet and the supernatant were assessed by SDS–PAGE and Coomassie brilliant blue staining. Staining and quantification showed that greater than one-half of PDZ1-3 sedimented with the PA-PC liposomes but that <10% of PDZ1-3 sedimented with equal amounts of PS-PC liposomes (Figure 2A). Thus PDZ1-3–PA binding does not appear to be due to nonspecific ionic interactions.
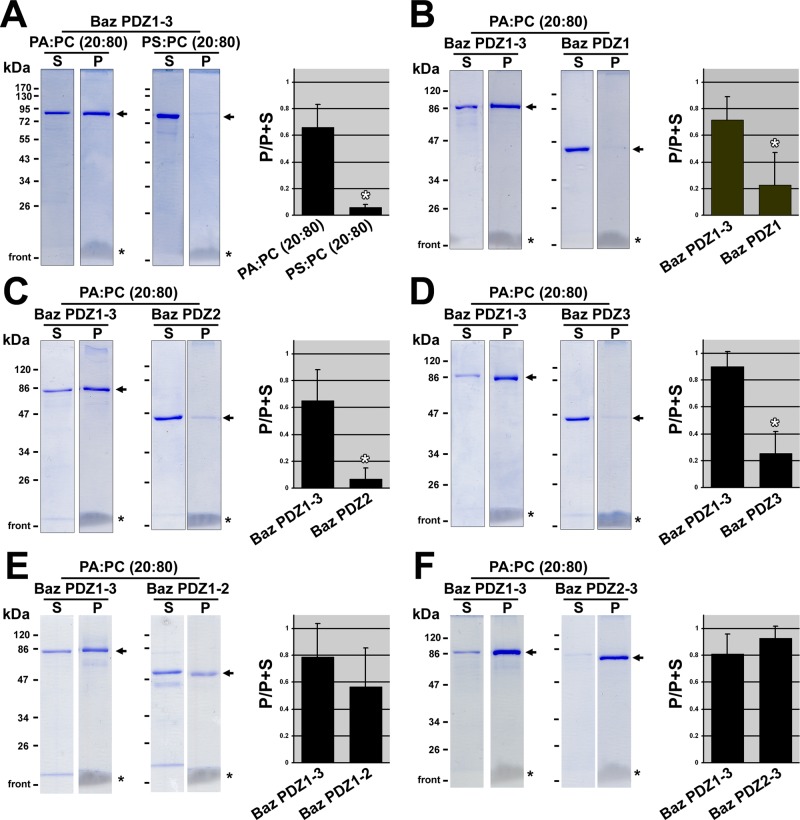
FIGURE 2:
Preferential binding of tandem Bazooka PDZ domains to phosphatidic acid in liposomes. Use of 10% SDS–PAGE and staining with Coomassie brilliant blue shows the distribution of GST-PDZ proteins (arrows) between the supernatant (S) and the liposome pellet (P) after liposome-binding assays. Equal proportions of the supernatants and pellets were loaded. Lanes with the same markers were cropped from the same gels. Note the migrating lipids at the base of the pellet lanes (asterisks). Quantifications of three separate binding assays are shown to the right. The Coomassie-stained bands were quantified in ImageJ, and the proportion of the total protein in the pellet is shown. Means shown with standard deviations. White asterisks indicate statistically significant differences (p < 0.01; t tests). (A) Baz PDZ1-3 binds PA more strongly than PS. (B) PA binds Baz PDZ1-3 more strongly than Baz PDZ1. (C) PA binds Baz PDZ1-3 more strongly than Baz PDZ2. (D) PA binds Baz PDZ1-3 more strongly than Baz PDZ3. (E) PA binds Baz PDZ1-3 and Baz PDZ1-2 similarly. (F) PA binds Baz PDZ1-3 and Baz PDZ2-3 similarly.
Next we confirmed the PA binding of Baz PDZ1, PDZ2, PDZ3, PDZ1-2, and PDZ2-3 in the liposome assay. PDZ1, PDZ2, and PDZ3 each showed significantly lower PA-PC liposome binding versus PDZ1-3, which was tested as a positive control in each assay (Figure 2, B–D). In contrast, PDZ1-2 and PDZ2-3 had indistinguishable binding to PA-PC liposomes versus PDZ1-3 (Figure 2, E and F). Thus multiple PDZ domains are also required for PA binding in liposome assays. Considering the fact that GST dimerizes (Wilce and Parker, 1994), it appears that two PA-binding sites on a dimeric GST-Baz protein are not sufficient for PA binding. On the assumption that binding is mediated by multiple separate binding sites, four (or possibly three) of these sites may be the minimum needed for a Baz protein dimer to bind PA. Alternately, having a minimum of two PDZ domains in tandem in the same Baz protein monomer may be the specific determinant for PA binding.
Genetic interaction studies indicate that phosphatidic acid supports Bazooka activity in embryos
The protective cuticle secreted from the embryonic epidermis provides a readout of the integrity and development of the ectodermal epithelium. Zygotic bazXi106 mutants contain a maternal supply of baz gene product that can mediate early epithelial development. However, this maternal supply becomes undetectable by stage 12 (Tanentzapf and Tepass, 2003; our observations), and, as a result of this depletion, the mutants display a range of cuticle phenotypes (Figure 3A). The severity of this phenotypic range can be modified by mutations affecting proteins that function with Baz (Harris and Peifer, 2005; Shao et al., 2010; Sawyer et al., 2011). We examined two genes responsible for PA synthesis—rdgA (encoding DGK) and Pld—which were previously analyzed to assess PA activities in the Drosophila eye (Raghu et al., 2009) and mammalian cell culture (Antonescu et al., 2010).
FIGURE 3:
Enzymes that produce phosphatidic acid promote Bazooka activity but not localization in vivo. (A) Categories of cuticle phenotypes scored in experiments. (B–D) Quantification of phenotypic ranges from specific crosses shown. Each data set is an average of two separate experiments (N = 98–406 embryonic cuticles per experiment). (B) Overexpression of DGK partially rescues the baz mutant cuticle phenotype, but expression of PLD has minimal effect. bazXi106, gal4 is short form for bazXi106, maternal-α4-tubulin-GAL4-VP16. (C) Reducing the levels of DGK (rdgA encodes DGK) or PLD enhances the bazXi106 mutant cuticle phenotype. rdgAKS60 and PldNull alleles were used. (D) Combined reduction of DGK and PLD enhances the bazXi106 mutant cuticle phenotype further. Combined reduction of DGK and PLD also enhances the bazGD21 mutant cuticle phenotype vs. an outcrossed control. rdgAKS60 and PldNull alleles were used in each case. (E) Staining for Baz in baz zygotic mutants vs. their heterozygous siblings with or without DGK overexpression shows that the partial rescue of the baz mutant cuticle phenotype with DGK overexpression is not due to a detectable stabilization of the maternal supply of Baz protein around the apical cortex. Each image was collected and adjusted with the same settings. Histograms below show the numbers of pixels for each grayscale value for images collected with the same settings. The pixel numbers are averages for the sample sizes indicated for each genotype. Note the higher-intensity values forming shoulders in the heterozygous sibling distributions and the similar shapes of the hemizygous mutant distributions with or without DGK overexpression.
To test whether overexpression of the enzymes could rescue the baz mutant cuticle phenotype, we expressed them zygotically in bazXi106 mutants using UAS-transgenes and the maternal-α4tubulin-GAL4-VP16 driver. In this mating scheme, populations without transgene expression displayed cuticles mainly composed of small residual sheets (Figure 3B). With DGK expression, populations shifted to less severe cuticles that were largely intact, with one to three holes (Figure 3B). PLD expression had no detectable effect (Figure 3B), which could be due to a difference in expression or activity level. Expression of either enzyme alone had no effect on embryonic lethality. In addition, there was no modification of the bazXi106 mutant phenotype when crosses were performed with the UAS-DGK line but in the absence of GAL4.
To test whether loss of the enzymes could enhance the baz mutant cuticle phenotype, we first generated mothers heterozygous for bazXi106 and rdgAKS60 or PldNull and crossed them to wild-type males to assess the hemizygous bazXi106 mutant progeny (baz is X linked). The bazXi106 allele was outcrossed from its balancer stock to produce mothers with the bazXi106 allele in-trans to a wild-type X chromosome. These mothers were crossed to wild-type males, and the hemizygous bazXi106 mutant progeny displayed mild cuticle phenotypes that mainly appeared normal or had one hole (Figure 3C; the phenotypic distribution of the bazXi106 mutant was more severe in Figure 3B because of genetic interactions with the balancer chromosome needed in the mothers; Shao et al., 2010). Additional maternal heterozygosity for rdgAKS60 or PldNull substantially enhanced the zygotic baz mutant cuticle phenotype (Figure 3C). Crossing mothers heterozygous for bazXi106 and rdgAKS60 to males homozygous for PldNull enhanced the phenotype further (Figure 3D). To confirm the specificity of the genetic interaction with baz, we also tested genetic interactions with the bazGD21 allele. Crossing mothers heterozygous for bazGD21 and rdgAKS60 to males homozygous for PldNull also enhanced the bazGD21 mutant cuticle phenotype versus an out-crossed control (Figure 3D). We crossed mothers heterozygous for bazXi106 and Pld to males homozygous for rdgAKS60 but saw no effect, as expected, as only bazXi106/Y progeny would be lethal and would lack the rdgA allele since it is X linked. Both rdgAKS60 and PldNull homozygotes are adult viable and fertile on their own (Masai et al., 1993; LaLonde et al., 2006). Taken together, these results suggest that PA contributes to Baz activity in the embryonic ectoderm.
Phosphatidic acid does not promote Bazooka localization
Because Baz functions as a polarity landmark, PA could contribute to Baz activity by promoting its localization. To test this idea, we probed the maternal contribution of Baz at stage 11 in bazXi106 mutants with or without overexpression of DGK (using the same experimental setup that partially rescued the bazXi106 mutant phenotype). The detection of maternal Baz in the mutants was similar with or without DGK expression, as was its intensity relative to the detection of maternal and zygotic Baz protein in heterozygous siblings on the same slides (Figure 3E). Thus the partial rescue ability of DGK is apparently not due to an effect on Baz localization. Instead it may affect activities downstream of Baz or parallel pathways. As one candidate, we probed for aPKC at stage 11 in bazXi106 mutants with or without overexpression of DGK, but aPKC levels appeared similarly reduced in each case compared with the respective heterozygous siblings (unpublished data).
Phosphatidic acid binding requires positively charged patches on the Baz PDZ domains
To further dissect the role of Baz-PA binding, we first pursued the binding sites required for the interaction. Because PA binding typically involves surface patches of positively charged amino acid residues (Stace and Ktistakis, 2006), we considered a patch of positive amino acid residues previously revealed on the crystal structure of rat Par-3 PDZ2 and implicated in phosphoinositide binding (Wu et al., 2007; Figure 4A). It is intriguing that the amino acid residues of this patch are conserved in Drosophila PDZ2 (Figure 4A). Because our data indicate that each Baz PDZ domain contributes to PA binding, similar patches would be expected on the other domains. The crystal structure of rat Par-3 PDZ3 has also been determined (Feng et al., 2008) and displays a patch of positive amino acid residues in a similar local configuration and at a similar overall position on the PDZ domain relative to the peptide-binding pocket (Figure 4B). The amino acid residues of this patch are also conserved in Drosophila PDZ3 (Figure 4B). Crystallographic information is not available for PDZ1, but positively charged amino acid residues are present after α-helix-A and β-strand-F (Figure 4C), as in Baz PDZ2 and PDZ3 (Figure 4, A and B), but these are not conserved in Rat PDZ1 (Figure 4C). Thus each Baz PDZ domain appears to have a positive patch of surface amino acid residues at a similar position.
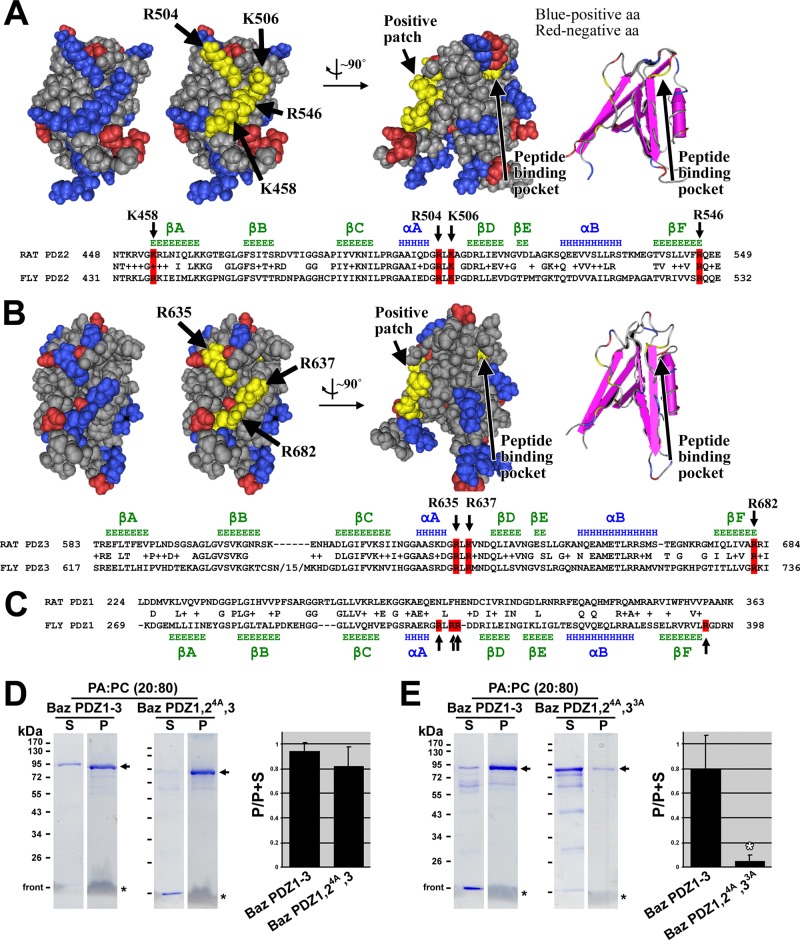
FIGURE 4:
Mapping residues on Bazooka PDZ domains responsible for phosphatidic acid binding. (A) A Cn3D space-filling representation of the solution structure of rat Par-3 PDZ2 (Molecular Modeling Database [MMDB] ID: 61497; Protein Data Bank [PDB] ID: 2OGP) shows negatively charged residues in red and positively charged residues in blue. A positively charged patch is highlighted in yellow, and the residues involved are indicated with arrows. Rotation of the structure by ∼90° shows the relative position of the peptide-binding pocket (the residue at the base of the pocket is highlighted in yellow, and the pocket is also shown with secondary structures, far right). A sequence alignment between rat Par-3 PDZ2 and Baz PDZ2 is shown below, with conserved residues of the positive patch highlighted in red. Secondary structure data above the alignment are based on the structure of rat Par-3 PDZ2 (β-strands in green and α-helices in blue; Wu et al., 2007). (B) A Cn3D space-filling representation of the solution structure of rat Par-3 PDZ3 (MMDB ID: 64814; PDB ID: 2K1Z) with negatively charged residues in red and positively charged residues in blue. A positively charged patch is highlighted in yellow, and the residues involved are indicated with arrows. Rotation of the structure by ∼90° shows the relative position of the peptide-binding pocket (the residue at the base of the pocket is highlighted in yellow, and the pocket is also shown with secondary structures, far right). A sequence alignment between rat Par-3 PDZ3 and Baz PDZ3 is shown below, with conserved residues of the positive patch highlighted in red. Secondary structure data above the alignment are based on the structure of rat Par-3 PDZ3 (β-strands in green and α-helices in blue; Feng et al., 2008). (C) A sequence alignment between rat Par-3 PDZ1 and Baz PDZ1. Secondary structure data below the alignment are based on secondary structure predictions of Baz PDZ1 using Jpred3, HNN, Prof, SOOPMA, and PORTER secondary structure prediction tools (β-strands in green and α-helices in blue). Positively charged residues found at similar positions as those in Baz PDZ2 and 3 are indicated in red. These are not conserved in rat Par-3 PDZ1. (D) Mutation of the four positive amino acid residues in PDZ2 to alanine in Baz PDZ1-3 has no effect on PA liposome binding. (E) Mutation of the four positive amino acid residues in PDZ2 plus the three positive amino acid residues in PDZ3 to alanine in Baz PDZ1-3 abrogates PA liposome binding. The experiments in D and E were conducted in the same way as those in Figure 2.
To test the role of the positive patches in PA binding, we mutated their residues to alanine in Baz PDZ1-3, which was isolated as a GST fusion protein, and tested for PA binding in the liposome assay. Mutating the four positive amino acid residues in PDZ2 had no effect on PA binding versus wild-type PDZ1-3 tested alongside (Figure 4D). This result is consistent with two PA-binding sites remaining in the molecule. Thus the three positive amino acid residues of PDZ3 were additionally mutated to alanine. Mutation of both the PDZ2 and PDZ3 patches significantly reduced PA binding versus the nonmutated PDZ1-3 tested alongside (Figure 4E). The binding site in PDZ1 appears to have limited PA-binding activity with the other PDZ domains mutated, consistent with the limited PA-binding activity of PDZ1 alone. These results implicate the amino acids residues responsible for Baz PDZ domain binding to PA.
The positively charged patches promote Bazooka activity and have context-dependent effects on Baz localization
To test the impact of the PA-binding regions, we first tested their effects on Baz cortical localization. The cortical localization of Baz involves multiple redundant sequences along the protein (McKinley et al., 2012). We performed three experiments to probe the PA-binding regions in Baz constructs with other major localization activities removed. For the first two experiments, we followed up on findings that sequences in PDZ1 and PDZ3 outside of their peptide-binding pockets promote apical circumferential and surface recruitment (McKinley et al., 2012). Specifically, a green fluorescent protein (GFP)–tagged construct with the OD and all three PDZ domains deleted (BazΔOD,ΔPDZ1-3-GFP) fails to localize cortically, but adding PDZ1 with its peptide-binding pocket mutated into the construct (BazΔOD,PDZ15A,ΔPDZ2-3-GFP) or adding PDZ3 with its peptide-binding pocket mutated into the construct (BazΔOD,ΔPDZ1-2,PDZ35A-GFP) enables strong cortical localization (the alanine mutations in PDZ1 and PDZ3 are predicted to disrupt their peptide-binding pockets; McKinley et al., 2012). Because these cortical recruitment activities reside outside of the peptide-binding pockets of PDZ1 and PDZ3 and thus cover the PA-binding positive patches, we tested whether PA could enhance them by coexpressing DGK with BazΔOD,PDZ15A,ΔPDZ2-3-GFP or BazΔOD,ΔPDZ1-2,PDZ35A-GFP. However, we observed no effect on the intensity levels or distributions of the proteins (Figure 5A). In the third experiment, we tested whether the positively charged patch of PDZ3 is responsible for the cortical localization of BazΔOD,ΔPDZ1-2,PDZ35A-GFP by additionally mutating residues of the positive patch to alanine (as done in vitro in Figure 4E). However, the levels and distribution of this protein were indistinguishable from those of the parent molecule (Figure 5B). The results of these three experiments, together with our analysis of residual maternal Baz protein with DGK expression (Figure 3E), all suggest that PA has minimal effect on Baz localization.
FIGURE 5:
Context-dependent effects of phosphatidic acid–binding residues on Bazooka activity and localization in vivo. (A) DGK overexpression has no effect on the localization of BazΔOD,PDZ15A,ΔPDZ2-3-GFP or BazΔOD,ΔPDZ1-2,PDZ35A-GFP. The cortical localization of these proteins relies on sequences in PDZ1 and PDZ3, respectively, outside of their peptide-binding pockets (McKinley et al., 2012). For each Baz construct, each image shown was collected and adjusted with the same settings. Signal intensities were quantified by determining average grayscale values across single xy-planes (200 by 200 pixels). Means and standard deviations are shown for these quantifications from the numbers of embryos indicated. (B) Mutation of the three positive amino acid residues responsible for PA binding to alanine in PDZ3 of BazΔOD,ΔPDZ1-2,PDZ35A-GFP has no effect on the localization of the protein. Each image shown was collected and adjusted with the same settings. Signal intensities were quantified as in A. (C) BazΔPDZ1-2 partially rescues the baz mutant cuticle phenotype, but mutation of the three positive amino acid residues responsible for PA binding in PDZ3 to alanine abrogates this activity. Each data set is an average of two separate experiments (N = 122–393 embryonic cuticles per experiment). (D) Baz∆PDZ1-2-GFP and Baz∆PDZ1-2,PDZ33A-GFP both localize around the apical cortex of baz mutant epithelial cells, although Baz∆PDZ1-2,PDZ33A-GFP does so at lower levels. Each image shown was collected and adjusted with the same settings. Signal intensities were quantified as in A. In C and D, baz, gal4 is short form for baz, maternal-α4-tubulin-GAL4-VP16. (E) Expression of Baz∆PDZ1-2-GFP and Baz∆PDZ1-2,PDZ33A-GFP with a stronger GAL4 driver in a wild-type background led to a greater difference in their cortical levels. Each image shown was collected and adjusted with the same settings. Signal intensities were quantified as in A.
Finally, we tested whether the PA-binding site of PDZ3 has an effect on Baz function by mutating it and performing rescue experiments. Constructs with the oligomerization domain absent have minimal rescue activity (McKinley et al., 2012). Thus we mutated the positive patch of PDZ3 in BazΔPDZ1-2-GFP, which was previously shown to partially rescue the bazXi106 zygotic mutant phenotype (McKinley et al., 2012). Indeed, Baz∆PDZ1-2-GFP partially rescued the embryonic lethality of baz mutants from 27.2 ± 2.2% to 17.7 ± 3.2% (n = 4 experiments; 300 embryos each; p < 0.01) and shifted the cuticle distribution to less severe phenotypes (Figure 5C). Mutation of the positive patch reversed both effects, with 26.7 ± 1.7% lethality (n = 4 experiments, 300 embryos each, p < 0.01, vs. Baz∆PDZ1-2) and a cuticle distribution indistinguishable from that of the nontransgene control. Analysis of the baz mutant embryos revealed that both constructs were expressed and localized to the cortex—Baz∆PDZ1-2-GFP at low cortical levels and Baz∆PDZ1-2,PDZ33A-GFP at somewhat lower cortical levels (Figure 5D). Expression of Baz∆PDZ1-2-GFP and Baz∆PDZ1-2,PDZ33A-GFP with a stronger GAL4 driver in a wild-type background led to an even greater difference in their cortical levels (Figure 5E). We confirmed that both constructs were inserted at the same chromosomal site using PCR, as done previously (McKinley et al., 2012). In addition, the lower levels of Baz∆PDZ1-2,PDZ33A-GFP versus Baz∆PDZ1-2-GFP may not be due to a general protein-folding issue since the same three alanines were mutated in BazΔOD,ΔPDZ1-2,PDZ35A-GFP without effect. Although it is unclear why mutating the positive patch affects Baz∆PDZ1-2-GFP but not BazΔOD,ΔPDZ1-2,PDZ35A-GFP, the data implicate the PA-binding site of PDZ3 in the localization and function of the Baz∆PDZ1-2-GFP construct.
DISCUSSION
Our results identified an in vitro interaction between Baz PDZ domains and PA. Each PDZ domain appears to have a positive surface patch capable of binding PA, but multiple sites must be present in Baz for PA binding. By manipulating the enzymes responsible for generating PA and mutating the PA-binding sites in Baz, Baz-PA interactions appear to have minimal, or context-specific, effects on Baz recruitment to the plasma membrane and may influence downstream effects of Baz important for epithelial structure (Figure 6).
FIGURE 6:
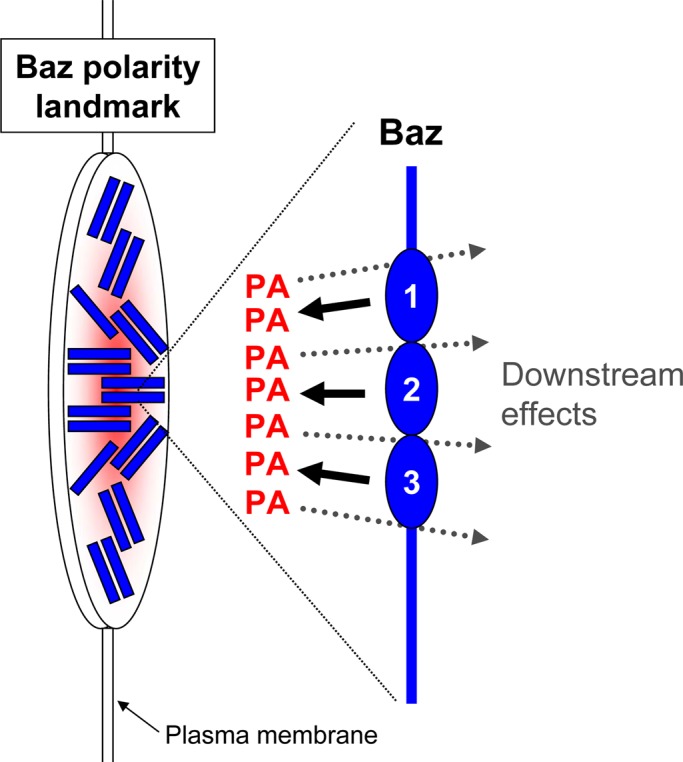
A model for the role of phosphatidic acid at Bazooka polarity landmarks. Individual Baz proteins bind PA through positive patches on each their three PDZ domains (labeled 1–3). These interactions can affect Baz recruitment to the plasma membrane and promote the downstream effects of Baz important for epithelial tissue structure. The oligomerization of Baz at Baz polarity landmarks leads to local stabilization and enrichment of PA to form a specialized plasma membrane domain.
The abilities of GST-tagged Baz PDZ1-3, PDZ1-2, and PDZ2-3, but not single PDZ domains, to bind PA indicate that each PDZ domain can contribute to PA binding but that GST dimers of single PDZ domains are insufficient for binding. These data suggest that the binding interaction involves multiple low-affinity sites that require tandem organization and/or oligomerization for an effective interaction with PA. Consistent with this requirement, the full-length Baz protein contains an oligomerization domain (Benton and St Johnston, 2003) that is expected to form extended multimers through front-to-back interactions between positively and negatively charged faces on the domain, based on studies of the homologous mammalian Par-3 domain (Feng et al., 2007). Such extended oligomerization may allow Baz to form platforms for engaging and stabilizing local pools of PA (Figure 6). It may also explain the ability of PDZ3 to confer PA binding site–dependent localization and activity in the Baz∆PDZ1-2 construct (Figure 5C). Indeed, with the oligomerization domain deleted, mutation of the PA binding site of PDZ3 had no detectable effect on the localization of a construct missing PDZ1 and PDZ2.
The oligomerization of Baz is expected to draw multiple binding partners together to form Baz polarity landmarks. In addition, multiple redundant binding sites have been shown to recruit Baz to the apical circumference of Drosophila embryonic ectodermal cells (McKinley et al., 2012). However, by stripping these recruitment sites from the protein or by analyzing sensitive pools of the protein, it was difficult to find evidence for PA recruiting Baz to the plasma membrane (it was suggested in the results of one experiment— Figure 5, D and E—but not from four other experiments— Figures 3E and 5, A and B). Alternatively, PA may influence effects downstream of Baz. Some of these effects could involve parallel pathways, but the ability of Baz to bind PA in vitro and the effects of mutating the PA binding site in vivo suggest that PA functions at Baz polarity landmarks. Unfortunately, the lack of probes for PA prevents the visualization of PA in developing epithelial cells.
Of the known binding partners of Baz, two have been shown to be influenced by PA in other systems—phosphoinositides and aPKC. Baz can directly bind the lipid phosphatase PTEN (von Stein et al., 2005) and its substrate PIP3 (Krahn et al., 2010), suggesting that it may facilitate the production of phosphatidylinositol 4,5-biphosphate (von Stein et al., 2005; Krahn et al., 2010), which has been shown to direct apical membrane identity in other systems (Shewan et al., 2011). The Baz phosphoinositide-binding site also contributes to its cortical localization through a mechanism involving multiple low-affinity interactions (Krahn et al., 2010). PA can influence phosphoinositide production (Stace and Ktistakis, 2006) and thus could potentially affect the cortical localization of Baz via phosphoinositides, but the apparent role of PA in mediating effects downstream of Baz suggests that any role in phosphoinositide signaling might be downstream. As discussed, the plasma membrane recruitment of DOCK2 is initiated by PIP3 and stabilized by PA (Nishikimi et al., 2009). Such cooperation may be prevalent, as a number of other proteins appear to have binding sites for both PA and phosphoinositides (Stace and Ktistakis, 2006). Thus an accumulation of PA at Baz polarity landmarks could influence the effects of phosphoinositides in the local membrane.
Baz also binds aPKC to dynamically form the PAR complex (Wodarz et al., 2000; Morais-de-Sá et al., 2010; Walther and Pichaud, 2010). It is intriguing that PA has been shown to bind mammalian aPKC (PKCζ) in vitro and addition of PA to cell extracts activates the kinase (Limatola et al., 1994). Similarly, exogenous PLD can increase PKCζ activity in cell culture (Limatola et al., 1997). More recently, DGK activity has been linked to increased PKCζ/ι activity as part of a signaling pathway linking hepatocyte growth factor to Rac activation and membrane ruffling in mammalian cell culture (Chianale et al., 2010). Thus local recruitment of PA to Baz polarity landmarks has the potential to affect local aPKC activity.
PDZ domains of many proteins have been shown to bind phosphoinositides (Nourry et al., 2003; Wu et al., 2007). Our results expand the lipid repertoire that PDZ domains can bind. For Baz specifically, our data implicate a closer and more complex connection to membrane lipids than previously recognized. Thus Baz polarity landmarks can be viewed as conglomerates of proteins and plasma membrane lipids that likely modify each other's activities for an integrated effect on cellular organization.
MATERIALS AND METHODS
GST-fusion proteins
A pENTR vector containing the Baz cDNA (McKinley et al., 2012) was used as the template for PCR. PDZ domain positions were predicted using the InterProScan Sequence Search (www.ebi.ac.uk/Tools/InterProScan/), and PCR primers were designed to add 10 amino acid residues to the predicted ends of each domain, as done by others (Tonikian et al., 2008). The PCR primers also generated restriction sites for cloning into pGEX 6P vectors. The vectors and PCR products were cut with EcoRI and XhoI and ligated using T4 ligase. The insertions were confirmed by sequencing. Primers and vectors are listed in Supplemental Table 1. All proteins were tagged with GST at their N-termini.
The GST-fusion protein constructs were transformed into DL21 Escherichia coli for protein expression. Bacteria were grown in 2 ml of LB/ampicillin (Amp) culture medium overnight at 37°C, and this sample was used to inoculate a 50-ml LB/Amp culture, which was shaken for 2 h at 28°C, induced with 0.1 mM isopropyl-β-d-thiogalactoside, and then shaken for 3 h at 28°C. Bacteria were pelleted at 3500 × g for 20 min at 4°C. The pellets were frozen overnight and then resuspended in 20 ml of PBS-T buffer (phosphate-buffered saline [PBS] with 1% Triton X-100, 1 mg/ml lysozyme, and a protease inhibitor cocktail (Complete Mini, EDTA free [Roche, Indianapolis, IN]; one tablet per 7 ml, pH 7.5). The suspensions were kept at room temperature for 40 min to lyse the bacteria. After centrifugation at 12,000 × g for 20 min, the supernatants were passed through 0.2-μm filters. The filtrates were applied to glutathione-agarose columns (∼0.4 ml of packed beads; Sigma-Aldrich, St. Louis, MO). The flowthrough was reapplied to the columns three times. The columns were then washed once with PBS-T, and the GST proteins were eluted with 1 ml of elution buffer (PBS with 10 mM glutathione, 1 mM dithiothreitol and 15% glycerol). Purity was determined by SDS–PAGE. Concentration was determined by Bradford protein assay (Sigma-Aldrich).
Probing lipids on solid supports
PIP strips were probed with purified GST proteins using supplier instructions (Echelon, Salt Lake City, UT; www.echelon-inc.com/content/EBI/product/files/PROTOCOL_Strip_Array.v9.pdf). The LL5α-PH domain (MultiPIP Grip; Echelon) was used as a positive control. Rabbit antibodies against GST (generated in our lab), horseradish peroxidase (HRP)–conjugated secondary antibodies (Thermo Fisher Scientific, Waltham, MA), HRP detection reagents (Thermo Fisher Scientific), and a FluorChem 8900 imaging system (Alpha Innotech, Santa Clara, CA) were used to detect the GST proteins.
Liposome-binding assays
To prepare liposomes, 8 mg of dipalmitoylphosphatidylcholine and 2 mg of dipalmitoylphosphatidic acid or dipalmitoylphosphatidylserine (each from Avanti Polar Lipids, Alabaster, AL) were dissolved in 1 ml of chloroform:methanol (1:0.4). The solvent was evaporated with nitrogen gas. The lipid mixtures were resuspended in 1 ml HBS buffer (0.01 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% Sulfactant P20 [Biacore, GE Healthcare Biosciences, Piscataway, NJ]) by stirring for 1 h at 25°C. Then the suspension was sonicated at 80°C for 15 min to make unilamellar vesicles and frozen at −20°C for later use. This approach was based on published assays (Fang et al., 2001; Zhao et al., 2007; Nishikimi et al., 2009). Initially, 5, 10, and 20% PA concentrations were tested in PA:PC liposomes. Baz PDZ1-3 showed no detectable binding to 5% PA liposomes, weak binding to 10% PA liposomes, and relatively strong binding to 20% PA liposomes, which were used in the assays in Figure 2.
To test liposome binding, we mixed 40 μl of the thawed liposome suspension with 5 μg of each GST protein in Tris-buffered saline (TBS; pH 7.5) containing protease inhibitor cocktail (Complete Mini, EDTA free; one tablet per 7 ml). TBS containing the protease inhibitor cocktail was added to the suspension for a final volume 500 μl, and the mixture was rocked at room temperature for 1.5 h. The suspension was then centrifuged for 30 min at 14,000 × g at room temperature. After collecting the supernatant, we washed the pellet four times with 200 μl of TBS containing protease inhibitor cocktail. The supernatant was mixed with 500 μl of methanol by vortexing, and then 125 μl chloroform was added, and the mixture was vortexed. The mixture was centrifuged for 1 min at 14,000 × g at 25°C, and the top aqueous layer was removed. Then 500 μl of methanol was added, and the mixture was vortexed. After centrifugation for 2 min at 14,000 × g at 25°C, the supernatant was removed, and the pellet was dried with a SpeedVac. Both the liposome pellet and precipitated protein from the supernatant were boiled for 5 min in SDS–PAGE sample buffer, and equal proportions of each sample were separated by 10% SDS–PAGE and stained with Coomassie brilliant blue.
Site-directed mutagenesis
Nucleotides were changed by PCR-mediated site-directed mutagenesis, as described previously (McKinley et al., 2012). Templates were from E. coli C7510 cells, and parental strands were destroyed with DpnI. The templates were the GST-PDZ1-3 construct described earlier and BazΔOD,ΔPDZ1-2,PDZ35A and Baz∆PDZ1-2, which were generated previously in the pENTR vector (McKinley et al., 2012). See Supplemental Table 2 for the list of primers and mutations made.
Mutated BazΔOD,ΔPDZ1-2,PDZ35A and Baz∆PDZ1-2 were recombined by Gateway cloning (Invitrogen, Carlsbad, CA) into pPWG for C-terminal EGFP tagging and placement downstream of the UASp promoter. An attb recombination site in the pPWG Nsi1 site allowed vector targeting to the attp2 recombination site on chromosome 3 in transgenic flies (Genetic Services, Cambridge, MA).
Fly stocks and genetics
In addition to those already cited, stocks included UAS-DGK and UAS-PLD flies (gifts of R. Padinjat, Tata Institute for Fundamental Research, Bangalore, India), bazXi106 mutants (gift of A. Wodarz, University of Göttingen, Göttingen, Germany), bazGD21 mutants (gift of J. Zallen, Sloan-Kettering Institute, New York, NY), rdgAKS60 mutants (#31426; Bloomington Drosophila Stock Center, Bloomington, IN,), PldNull mutants (gift of M. Frohman, Stony Brook University, Stony Brook, NY), and daughterless (da)-Gal4 flies (gift of U. Tepass, University of Toronto, Toronto, Canada). WT was yellow white.
A recombinant chromosome containing maternal-α4-tubulin-GAL4-VP16 (gift of M. Peifer, University of North Carolina at Chapel Hill, Chapel Hill, NC) and the bazXi106 allele allowed construct expression in baz zygotic mutants, as done previously (McKinley et al., 2012). For genotyping, the chromosome was balanced over FM7[twi-GAL4, UAS-GFP] (#6873; Bloomington Drosophila Stock Center), and females were crossed to males with Red-Stinger (#6873; Bloomington Drosophila Stock Center) on the X chromosome and UAS constructs on the autosomes.
Embryonic lethality rates and cuticle preparations
For embryonic lethality rates, flies laid eggs for up to 24 h at 25°C. Three hundred eggs were collected and incubated for 48 h. The percentage of unhatched embryos was determined versus total embryo number (unfertilized eggs excluded). For cuticle preparations, all unhatched eggs were collected, washed, dechorionated with 50% bleach, mounted on slides with Hoyer's mountant:lactic acid (1:1), and baked at 60°C overnight.
Embryo staining and imaging
Embryos were washed with 0.1% Triton solution; dechorionated with 50% bleach; washed; fixed for 20 min in 1:1 3.7% formaldehyde/PBS:heptane; subjected to methanol devitellinization; blocked and stained with rabbit anti-Baz antibodies (1:2000; raised in our lab against GST-Baz 1-311) and Alexa 647 secondary antibodies (1:1000; Invitrogen) in PBS/1% goat serum/0.1% Triton X-100/1% sodium azide.
Dechorionated live embryos were mounted in halocarbon oil (series 700; Halocarbon Products, Beech Island, SC) on petriPERM dishes (Sigma-Aldrich). Fixed embryos were mounted in Aqua Polymount (Polysciences, Warrington, PA). Images were collected with a spinning-disk confocal system (Quorum Technologies, Guelph, Canada) at room temperature with a 63× Plan Apochromat numerical aperture 1.4 objective (Carl Zeiss, Jena, Germany), a piezo top plate, an electron-multiplying charge-coupled device camera (Hamamatsu Photonics, Hamamatsu, Japan), and Volocity software (PerkinElmer, Waltham, MA).
Supplementary Material
Acknowledgments
We thank Julie Brill for critiques and suggestions and R. Padinjat, A. Wodarz, M. Frohman, U. Tepass, J. Zallen, and M. Peifer for reagents. This work was supported by a Canadian Institutes of Health Research operating grant. T. Harris holds a Tier 2 Canada Research Chair.
Abbreviations:
- aPKC
atypical protein kinase C
- Baz
Bazooka
- DGK
diacylglycerol kinase
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- OD
oligomerization domain
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PDZ
postsynaptic density 95, discs large, zonula occludens-1
- PH
pleckstrin homology
- PLD
phospholipase D
- PS
phosphatidylserine
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-03-0196) on July 25, 2012.
REFERENCES
- Antonescu CN, Danuser G, Schmid SL. Phosphatidic acid plays a regulatory role in clathrin-mediated endocytosis. Mol Biol Cell. 2010;21:2944–2952. doi: 10.1091/mbc.E10-05-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, St Johnston D. A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol. 2003;13:1330–1334. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- Chianale F, et al. Diacylglycerol kinase alpha mediates HGF-induced Rac activation and membrane ruffling by regulating atypical PKC and RhoGDI. Proc Natl Acad Sci USA. 2010;107:4182–4187. doi: 10.1073/pnas.0908326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. EMBO J. 2007;26:2786–2796. doi: 10.1038/sj.emboj.7601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M. Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J Biol Chem. 2008;283:23440–23449. doi: 10.1074/jbc.M802482200. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol. 2005;170:813–823. doi: 10.1083/jcb.200505127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetopoulos C, Firtel RA. Directional sensing during chemotaxis. FEBS Lett. 2008;582:2075–2085. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn MP, Klopfenstein DR, Fischer N, Wodarz A. Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr Biol. 2010;20:636–642. doi: 10.1016/j.cub.2010.01.065. [DOI] [PubMed] [Google Scholar]
- LaLonde M, et al. A role for phospholipase D in Drosophila embryonic cellularization. BMC Dev Biol. 2006;6:60. doi: 10.1186/1471-213X-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprise P, Tepass U. Novel insights into epithelial polarity proteins in Drosophila. Trends Cell Biol. 2011;21:401–408. doi: 10.1016/j.tcb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Limatola C, Barabino B, Nista A, Santoni A. Interleukin 1-beta-induced protein kinase C-zeta activation is mimicked by exogenous phospholipase D. Biochem J. 1997;321:497–501. doi: 10.1042/bj3210497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limatola C, Schaap D, Moolenaar WH, van Blitterswijk WJ. Phosphatidic acid activation of protein kinase C-zeta overexpressed in COS cells: comparison with other protein kinase C isotypes and other acidic lipids. Biochem J. 1994;304:1001–1008. doi: 10.1042/bj3041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai I, Okazaki A, Hosoya T, Hotta Y. Drosophila retinal degeneration A gene encodes an eye-specific diacylglycerol kinase with cysteine-rich zinc-finger motifs and ankyrin repeats. Proc Natl Acad Sci USA. 1993;90:11157–11161. doi: 10.1073/pnas.90.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley RF, Yu CG, Harris TJ. Assembly of Bazooka polarity landmarks through a multifaceted membrane-association mechanism. J Cell Sci. 2012;125:1177–1190. doi: 10.1242/jcs.091884. [DOI] [PubMed] [Google Scholar]
- Morais-de-Sá E, Mirouse V, St Johnston D. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 2010;141:509–523. doi: 10.1016/j.cell.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikimi A, et al. Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemotaxis. Science. 2009;324:384–387. doi: 10.1126/science.1170179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP. PDZ domain proteins: plug and play! Sci STKE. 2003;2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- Raghu P, et al. Rhabdomere biogenesis in Drosophila photoreceptors is acutely sensitive to phosphatidic acid levels. J Cell Biol. 2009;185:129–145. doi: 10.1083/jcb.200807027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JK, Choi W, Jung KC, He L, Harris NJ, Peifer M. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol Biol Cell. 2011;22:2491–508. doi: 10.1091/mbc.E11-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Wu J, Chen J, Lee DM, Tishkina A, Harris TJC. A modifier screen for Bazooka/PAR-3 interacting genes in the Drosophila embryo epithelium. PLoS ONE. 2010;5:e9938. doi: 10.1371/journal.pone.0009938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewan A, Eastburn DJ, Mostov K. Phosphoinositides in cell architecture. Cold Spring Harb Perspect Biol. 2011;3:a004796. doi: 10.1101/cshperspect.a004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Stace CL, Ktistakis NT. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim Biophys Acta. 2006;1761:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- Tanentzapf G, Tepass U. Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat Cell Biol. 2003;5:46–52. doi: 10.1038/ncb896. [DOI] [PubMed] [Google Scholar]
- Tonikian R, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stein W, Ramrath A, Grimm A, Muller-Borg M, Wodarz A. Direct association of Bazooka/PAR-3 with the lipid phosphatase PTEN reveals a link between the PAR/aPKC complex and phosphoinositide signaling. Development. 2005;132:1675–1686. doi: 10.1242/dev.01720. [DOI] [PubMed] [Google Scholar]
- Walther RF, Pichaud F. Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr Biol. 2010;20:1065–1074. doi: 10.1016/j.cub.2010.04.049. [DOI] [PubMed] [Google Scholar]
- Wilce MC, Parker MW. Structure and function of glutathione S-transferases. Biochim Biophys Acta. 1994;1205:1–18. doi: 10.1016/0167-4838(94)90086-8. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Grimm A, Knust E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol. 2000;150:1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell. 2007;28:886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat Cell Biol. 2007;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.