Background: Few approaches exist to rationally engineer live, attenuated virus vaccines.
Results: Converting the conserved catalytic regulator lysine of the viral polymerase to arginine produces a slow, attenuated virus that elicits a protective immune response.
Conclusion: We have developed a polymerase mechanism-based strategy for viral attenuation and vaccine development.
Significance: This strategy may be used to create live, attenuated vaccines for other viruses.
Keywords: Enzyme catalysis, Enzyme mechanisms, Positive-strand RNA viruses, RNA viruses, Vaccine development, Viral polymerase, Viral replication
Abstract
Live, attenuated vaccines have prevented morbidity and mortality associated with myriad viral pathogens. Development of live, attenuated vaccines has traditionally relied on empirical methods, such as growth in nonhuman cells. These approaches require substantial time and expense to identify vaccine candidates and to determine their mechanisms of attenuation. With these constraints, at least a decade is required for approval of a live, attenuated vaccine for use in humans. We recently reported the discovery of an active site lysine residue that contributes to the catalytic efficiency of all nucleic acid polymerases (Castro, C., Smidansky, E. D., Arnold, J. J., Maksimchuk, K. R., Moustafa, I., Uchida, A., Götte, M., Konigsberg, W., and Cameron, C. E. (2009) Nat. Struct. Mol. Biol. 16, 212–218). Here we use a model RNA virus and its polymerase to show that mutation of this residue from lysine to arginine produces an attenuated virus that is genetically stable and elicits a protective immune response. Given the conservation of this residue in all viral polymerases, this study suggests that a universal, mechanism-based strategy may exist for viral attenuation and vaccine development.
Introduction
The current state of the art for vaccinology limits the development of a vaccine as a first response to an epidemic caused by the natural or intentional spread of a new virus. Live, attenuated virus vaccines are most efficacious because both humoral and cellular immunity are generally elicited. In addition, transmission of the vaccine strain can lead to herd immunity. Creation of a live, attenuated virus vaccine usually involves random approaches, for example adaptation of a virus to growth at low temperatures or in nonhuman cell cultures (2). The process of adaptation leads to numerous genetic changes, only a subset of which may be responsible for the attenuated phenotype in humans (3, 4). The regulatory process now requires an absolute determination of the genetic basis for the attenuated phenotype (5), allowing assessment of the “stability” of the attenuated phenotype. This regulatory requirement represents a high hurdle for vaccine development such that if the Sabin poliovirus (PV)3 vaccines needed to meet this criterion, they would not be approved today.
There has been a flurry of activity by many laboratories to develop rational approaches for vaccine development (2, 6–10). One of these approaches involved increasing the replication fidelity of the RNA-dependent RNA polymerase (RdRp) of a prototypical positive strand RNA virus, PV (10). Enhanced replication fidelity of the viral RdRp has two major outcomes. First, the reduced genetic variation is attenuating because variants in the population required to evade barriers of the host are not generated at the appropriate frequency (11). Second, the enhanced fidelity reduces the frequency of genetic reversion to a wild-type, pathogenic phenotype (12). There is now evidence in several systems that RdRp fidelity is a determinant of viral pathogenesis and virulence (11–18). Unfortunately, none of the mutations in the RdRp gene that lead to increased replication fidelity are at positions conserved across virus families, making it difficult to extrapolate attenuation by enhanced fidelity to other systems.
In this study, we show that mutating the conserved active site lysine residue that serves as a general acid catalyst during nucleotide incorporation to arginine produces a viral RdRp that replicates slower with higher fidelity. This virus variant is genetically stable, replicates well in cell culture, is attenuated (fails to cause disease), and elicits a protective immune response in vivo. Due to the conservation of this residue in viral polymerases, this study suggests that a universal, mechanism-based strategy may exist for viral attenuation and vaccine development.
EXPERIMENTAL PROCEDURES
Expression and Purification of PV RdRp
Construction of pET26Ub-PV-3D expression plasmids were described previously (1, 19). PV RdRp was expressed and purified as described previously (1, 19).
Kinetic Analysis of PV RdRp
Nucleotide incorporation experiments were performed essentially as described previously (1, 23).
Sequence Alignments
All sequences were obtained from the National Center for Biotechnology Information (NCBI) Database. Sequences were aligned using ClustalW2 and based upon alignments previously published (22).
Cells and Viruses
HeLa S3 cells were obtained from the American Type Culture Collection (ATCC) and grown in DMEM/F-12 medium plus 10% fetal bovine serum, 100 units/ml penicillin, and 100 units/ml streptomycin. PV type 1 Mahoney was used throughout this study.
Construction of Mutated Viral cDNA Clones and Replicons
The expression plasmid encoding the K359R mutation (1) was digested and inserted into an intermediate plasmid, pUC18-BglII-EcoRI-3CD (referred to as pUC-3CD in Ref. 20), and then the fragment between BglII and ApaI was cloned into either the subgenomic replicon, pRLucRA, or viral cDNA, pMovRA. Plasmids were linearized and used in RNA transcription reactions as described previously (21).
Infectious Center Assays
Infectious center assays were performed as described previously (21).
Virus Isolation, Titer, and One-step Growth Curves
Virus was harvested and titered by plaque assay as described previously (21). For TCID50, virus was serially diluted in medium, and each dilution was added to 10 wells of a 96-well plate previously seeded with 5 × 103 HeLa cells. Plates were incubated at 37 °C for 7 days before being manually scored for Cytopathic effect. The percentage of wells with CPE for each dilution was used to calculate viral titer (in TCID50/ml). When the same stock was titered by both methods, 1 pfu/ml was approximately equivalent to 20 TCID50/ml. To analyze one-step virus growth, cells in 12-well plates were infected with virus at a multiplicity of infection of 10. Virus was allowed to adsorb, cells were washed with PBS, medium was added to each well, and cells were incubated at 37 °C for various times after infection. Virus was harvested by three repeated freeze-thaw cycles, and virus titers were performed by TCID50 assays.
Northern Blot Analysis
Northern blots were performed as described previously (21).
Ribavirin Sensitivity Assay
HeLa cell monolayers in 6-well plates were pretreated for 1 h with various concentrations of ribavirin and then infected with 50 pfu of WT, G64S, or K359R PV, incubated for 20 min to allow for virus adsorption, and then washed and overlaid with 0.5% agarose medium containing the same concentration of ribavirin. Plates were incubated for 3–4 days at 37 °C before overlays were removed, and monolayers were stained with crystal violet. Plaques were counted and compared with the untreated (0 mm ribavirin) control dish. To determine the IC50 value, the percentage of plaques was plotted as a function of ribavirin concentration and fit to a sigmoidal dose response equation (Equation 1)
where Y is the percentage of plaques relative to untreated cells, A is the minimum percentage of plaques, and H is the Hill Slope.
Subgenomic Replicon Assays
Subgenomic replicon assays were performed as described previously using a luciferase-expressing, subgenomic replicon (21).
Mouse Infection and Protection
Mice were bred and housed in standard ventilated caging for all experiments. Viral stocks generated in serum-free medium, harvested and titered as above, were diluted to 2 × 109 TCID50 per 3 ml of serum-free medium. Six-to-eight-week-old outbred (ICR) mice transgenic for the PV receptor (cPVR) were infected with PV by intraperitoneal injection in 3 ml of serum-free medium (or mock-infected with 3 ml of serum-free medium alone). Mice were observed for 14 days for signs of disease and were euthanized upon showing dual limb paralysis or paralysis such that their ability to obtain food and water was compromised; this was in accordance with approval by the Institutional Animal Care and Use Committee (IACUC) at Penn State. Surviving mice were challenged using the same methods with a lethal dose (2 × 109 TCID50) of WT PV 1 month after infection and observed for 14 days as above.
Statistical Analysis
Survival curves were performed using the product limit method of Kaplan and Meier and survival curve comparisons using the log rank test as provided by GraphPad Prism 4. p values are indicated.
RESULTS AND DISCUSSION
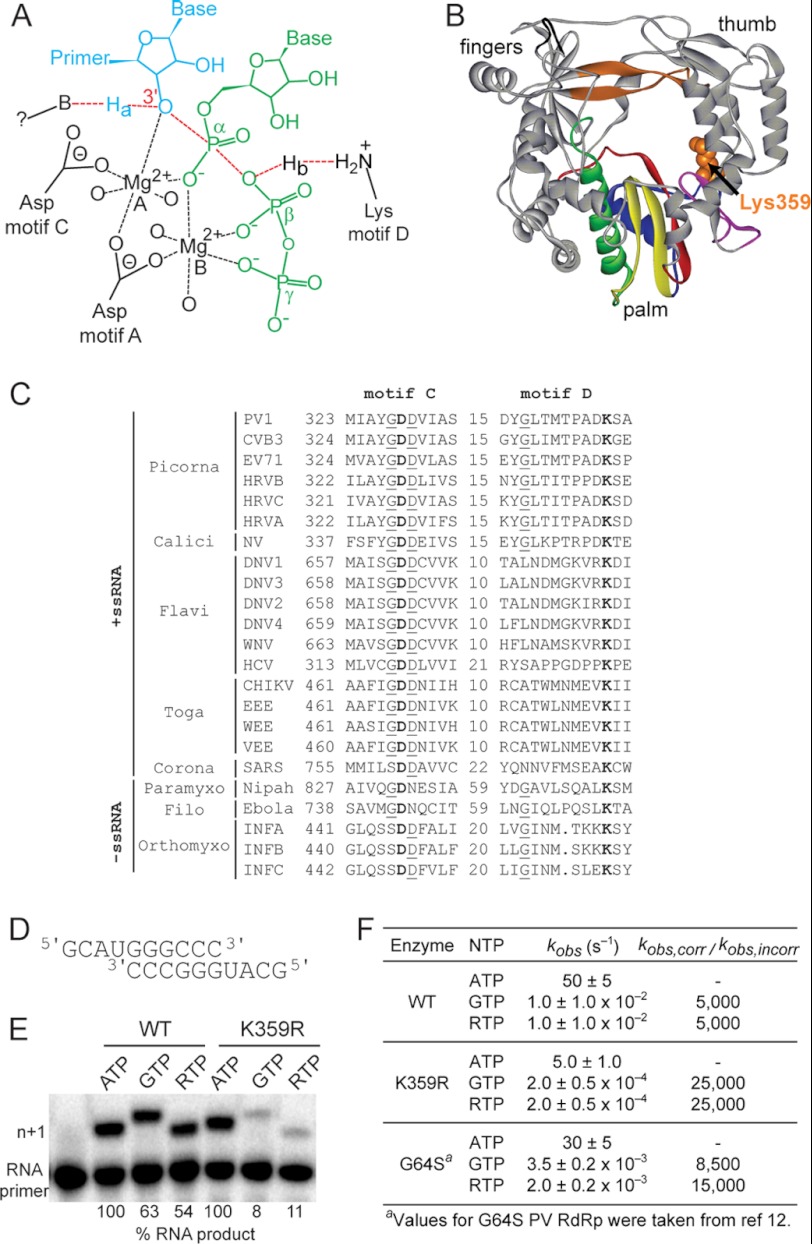
It is long established that nucleic acid polymerases use a two-metal-ion mechanism for nucleotidyl transfer (Fig. 1A)(24). In this mechanism, two magnesium ions are used to organize the reactants, 3′-OH of primer and α-phosphorous atom of the nucleoside triphosphate (NTP), and one magnesium ion lowers the pKa of the 3′-OH for deprotonation by a base that has yet to be identified. We recently expanded the chemical mechanism of nucleotidyl transfer to include a general acid, which protonates the pyrophosphate leaving group of the NTP substrate and contributes a 50–2000-fold rate enhancement to nucleotidyl transfer (Fig. 1A)(1, 23). In the case of PV RdRp, the general acid is Lys-359. This residue is located in a motif termed D that is conserved in the structures of all RdRps and reverse transcriptases (Fig. 1B). Importantly, an orthologous residue is predicted in RNA viruses for which vaccines could be of great benefit (Fig. 1C).
FIGURE 1.
PV RdRp Lys-359 is a determinant of catalytic efficiency and fidelity. A, Lys-359 functions as a general acid catalyst. As the transition state of nucleotidyl transfer is approached, primer 3′-OH proton, Ha, is transferred to an unidentified base (B), and pyrophosphate leaving group is protonated (Hb) by a conserved basic amino acid in the active site, in the case of PV RdRp, Lys-359 (1). B, structure of PV RdRp. Palm, fingers, and thumb subdomains are indicated. Conserved structural motifs are colored: A, red; B, green; C, yellow; D, blue; E, purple; F, orange; G, black. Lys-359 in motif D is indicated (Corey-Pauling-Koltun, orange). C, the motif D lysine that functions as a general acid is conserved in viral RdRps. Sequence alignments of motifs C and D from the indicated positive- and negative-strand viral RdRps are shown. Numbers indicate position from first amino acid of the RdRp domain and length separating motifs C and D. Conserved residues are shown in boldfaced type. Residues conserved within a virus group are underlined. D, primed template use is referred to as sym/sub-U and contains a uracil as the first templating base. E, reaction products from PV RdRp-catalyzed nucleotide incorporation using indicated nucleotide and sym/sub-U. K359R PV RdRp incorporated GMP and ribavirin monophosphate less efficiently than WT. The percentage of RNA product relative to the correct nucleotide (ATP) is shown. F, K359R PV RdRp is more faithful than the WT enzyme. Observed rate constants for correct and incorrect nucleotide incorporation catalyzed by WT, K359R, and G64S PV RdRp are shown. The relative fidelity or error frequency is shown. a, Values for G64S PV RdRp taken from a previous study (12).
The RdRp from PV is the only enzyme in this superfamily of enzymes for which a detailed, kinetic description of the mechanism and fidelity exists (1, 23, 25). PV RdRp will assemble stable elongation complexes on symmetrical RNA primer-templates, referred to as sym/sub (Fig. 1D)(26). Nucleotide addition can be evaluated by monitoring changes in extension of 32P end-labeled RNA (26). The PV RdRp with Lys-359 substituted by Arg (referred to as K359R RdRp) is known to catalyze nucleotidyl transfer at a rate 10-fold lower than wild type (1). However, the impact of this change on fidelity has not been studied. A slow polymerase with high incorporation fidelity could contribute to a stable, attenuated phenotype in the context of a virus. Fidelity was evaluated by two approaches using sym/sub-U, which has a uridine as the first templating nucleotide (Fig. 1D). The first approach monitored utilization of GTP (misincorporation); the second approach monitored utilization of ribavirin triphosphate (RTP). RTP was chosen because the first high fidelity variant of PV RdRp, G64S, was isolated by selection of mutants with reduced sensitivity to ribavirin, which was caused by reduced utilization of RTP (12, 16). Under conditions in which AMP incorporation went to completion for both WT and K359R RdRps (lanes 2 and 5, Fig. 1E), a substantial reduction in GMP misincorporation and ribavirin monophosphate incorporation was observed for K359R RdRp (Fig. 1E, lanes 6 and 7) relative to WT RdRp (Fig. 1E, lanes 3 and 4). To obtain a more quantitative perspective of the change in fidelity, we evaluated the kinetics of incorporation of the various nucleotides at saturating concentrations. K359R RdRp was on the order of 5-fold more faithful than WT RdRp (Fig. 1F). Under the same conditions, G64S RdRp exhibited only a 2–3-fold enhancement in fidelity (Fig. 1F)(12). Because GMP incorporation opposite uridine represents the most facile misincorporation event, this study only establishes a lower limit for the increase in fidelity caused by the K359R substitution.
To determine how the biochemical changes associated with the K359R RdRp impact virus multiplication in cell culture, we constructed a PV genome encoding the K359R RdRp. The AAA codon encoding lysine was changed to the CGT codon encoding arginine. Therefore, genetic reversion requires two transversion mutations, a very inefficient event that provides some barrier to reversion. Transcripts encoding the K359R RdRp were on the order of 10-fold less cytolytic to HeLa cells than those encoding WT RdRp (Fig. 2A). K359R PV recovered from this type of experiment exhibited a reduced rate of virus production, with a final yield 10-fold less than that of WT PV (Fig. 2B). Despite the reduced fitness of K359R PV, both the K359R change and the corresponding phenotype were stable over eight serial passages at a multiplicity of infection of infection from 0.1 to 1 (data not shown). Interestingly, the duration of neither the eclipse phase (lag) nor the exponential phase of infectious virus production was reduced by the magnitude of the nucleotidyl transfer rate (10-fold) measured in vitro (Fig. 2B). It is therefore possible that RNA synthesis does not contribute directly to the rate-limiting step for virus production. To address this possibility, we used Northern blotting to evaluate RNA synthesis as a function of time after infection. We found that the kinetics of RNA accumulation were clearly diminished (Fig. 2C). To increase the signal for replication, we engineered the K359R substitution into a PV subgenomic replicon that permits RNA synthesis to be evaluated indirectly by monitoring luciferase activity. In this experiment, it was clear that at least a 10-fold reduction occurred in all phases of genome replication (Fig. 2D). Collectively, these data suggest that the biochemical defect associated with the Arg-359 substitution is manifested in cells. We interpret the reduction in viral genomes, and consequentially reduced infectious virus, to mean that some innate process capable of squelching RNA synthesis is manifested when replication requires longer than 4 h for completion. A PV variant exhibiting increased nucleotide incorporation fidelity exhibits reduced sensitivity to the antiviral nucleoside, ribavirin (12, 16). Therefore, to assess K359R fidelity in cells, we evaluated the sensitivity of K359R PV to ribavirin. By plaque assay, there was a clear reduction in sensitivity of K359R PV to ribavirin when compared with WT PV (Fig. 2E). The ribavirin resistance phenotype was on par with that observed for G64S PV (Fig. 2E), consistent with biochemical data for the corresponding polymerases (Fig. 1F). More rigorous analysis revealed an intermediate resistance for K359R PV relative to WT and G64S PVs (Fig. 2F). It is possible that the reduced speed of genome replication is antagonistic with the increased fidelity of genome replication, leading to the observation that K359R PV is less fit than G64S PV in the presence of ribavirin.
FIGURE 2.
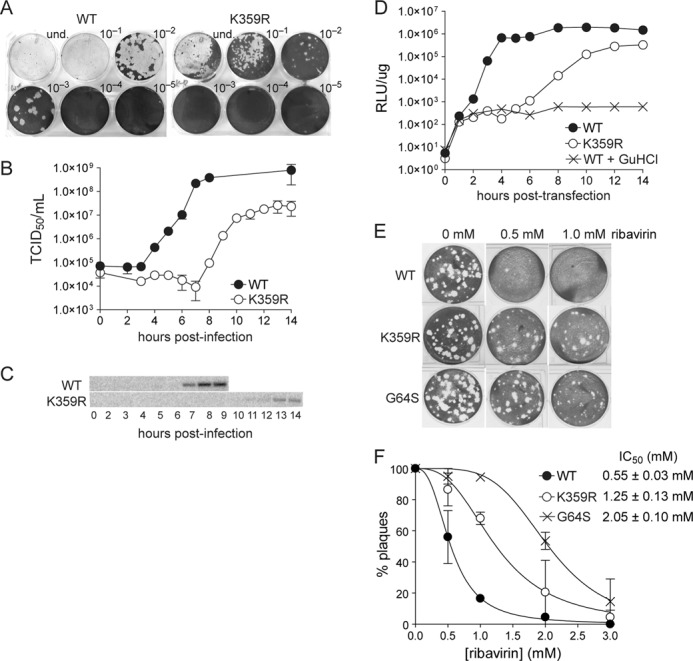
K359R PV is viable but attenuated in cell culture. A, infectious center assay. The specific infectivity of viral RNA was 2 × 104 and 2 × 103 for WT and K359R, respectively. und. indicates addition of undiluted sample. B and C, kinetics of virus growth for K359R PV. Viral titer (TCID50/ml) was plotted as a function of time after infection. Error bars indicate S.D. Duplicate infected samples were used for RNA isolation and Northern blot analysis (C). D, kinetics of RNA synthesis using a luciferase-expressing, subgenomic replicon. Luciferase-specific activity is reported in relative light units (RLU) per microgram of total protein in the extract. WT + guanidine hydrochloride (GuHCl) represents a control for translation of input RNA without replication. Shown is one representative data set. E and F, ribavirin sensitivity. Plaque numbers were plotted against ribavirin concentration normalized to untreated (0 mm) control (F). The solid line represents the fit of the data to a sigmoidal dose response equation (Equation 1), yielding IC50 values of 0.55 ± 0.03, 2.05 ± 0.10, and 1.25 ± 0.13 mm for WT, G64S, and K359R PV, respectively.
In cells, K359R PV is delayed in growth but replicates with high fidelity. These characteristics predict that K359R PV should be more attenuated in animals than G64S PV. We compared the virulence of K359R PV with G64S and WT PVs in a mouse transgenic for the PV receptor (27). In this system, infection by WT PV is generally lethal (27). At the highest dose of K359R PV readily attainable, none of the inoculated mice showed any symptoms of infection (Fig. 3A). At the same dose, both G64S and WT PVs caused two-limb paralysis (IACUC-approved end point for these experiments) (Fig. 3A). Previous studies of G64S PV used a different end point, thus permitting the diminished virulence of G64S PV relative to WT PV to be observed (11). To determine whether K359R PV actually replicates in the mice and has potential as a vaccine candidate, we performed a challenge experiment with WT PV. Mice were inoculated with K359R PV. Four weeks after inoculation, the mice were challenged with a lethal dose of WT PV. All of the K359R PV-inoculated mice survived this lethal challenge; none of the mock-infected (naive) mice survived (Fig. 3B). We conclude that K359R PV replicates in these animals as UV-inactivated, replication-incompetent PV is incapable of eliciting immunity sufficient to protect against a lethal challenge with WT PV (10, 11). To place the protection afforded by this mechanism-based approach into context, we compared the efficacy of K359R PV vaccination with that of Sabin-1 PV. Doses of K359R PV below 2 × 109 infectious units were less effective than the corresponding dose of Sabin-1 PV (Fig. 3C). Overall, the efficacy of K359R PV appeared on the order of one log lower than Sabin-1 PV in this model (Fig. 3C). Given that Sabin-1 PV has 57 nucleotide changes that cause 21 amino acid changes relative to the Mahoney strain of PV (3), the attenuation and stability afforded by this single change of lysine to arginine is remarkable.
FIGURE 3.
K359R PV is attenuated and protective in the cPVR mouse model. A, mouse infections with WT, K359R, and G64S PVs. cPVR mice were infected by intraperitoneal injection with serum-free medium containing 2 × 109 TCID50 of WT, K359R, or G64S PV. K359R PV-infected mice showed no signs of infection or disease; n = 10 for WT and K359R PV and n = 15 for G64S PV. Survival curves for mice infected with K359R and G64S PV are significantly higher than those for mice infected with WT PV (p < 0.0001, p value of 0.002, respectively, Kaplan-Meier test). B, mouse protection studies. Mice immunized with K359R PV as above or mock-immunized with serum-free medium alone (Naive) were infected 4 weeks after immunization with a lethal dose of WT PV (2 × 109 TCID50); n = 10. Survival curves for mice immunized with K359R PV are significantly higher than those for mice immunized with control serum-free medium (p < 0.0001, Kaplan-Meier test). C, comparison of protection between K359R PV and the Sabin type 1 PV vaccine strain. The percentage of protection is shown. Mice immunized with either K359R or Sabin-1 PV with the indicated dose were infected 4 weeks after immunization with a lethal dose of WT PV (2 × 109 TCID50); n = 5, errors indicate S.E. Two independent trials were performed with K359R PV.
We have used the PV system to show that the general acid of the viral RdRp can be changed to create a slower, more faithful RdRp derivative that produces an attenuated virus capable of protecting mice against a lethal challenge with wild-type virus. This is the first polymerase mechanism-based approach for viral attenuation. Importantly, this attenuation strategy can be easily and rapidly deployed in any positive-strand RNA virus, and perhaps other RNA and DNA viruses as well. As long as the polymerase belongs to one of the known superfamilies, sequence alone should be sufficient to identify the general acid (e.g. Fig. 1C). The polymerases of negative- and double-strand RNA viruses are all predicted to have this conserved lysine residue (22, 28). In addition, the replicative polymerases of herpes-pox and adenoviruses, all DNA viruses, are B-family polymerases predicted to have this conserved lysine (29). Evaluating the impact of the Lys-to-Arg substitution in other viruses will be necessary to determine how generalizable this attenuation strategy will be.
A great benefit associated with this attenuation mechanism is that it should combine in a predictable manner with other rational attenuation strategies, especially those that alter codon pair bias (7, 8) or incorporate microRNAs (6, 9). The increased fidelity of RNA synthesis may further diminish the rate of genetic reversion and restoration of the virulent phenotype.
An immediate application of this technology is in the production of seed stocks for creation of inactivated PV vaccine. Currently, wild-type strains are used, increasing production costs due to containment and limiting the number of sites globally capable of producing the vaccine, which leads to expenses for shipment and storage. An attenuating mutation in polymerase-coding sequence will not alter the stability or antigenicity of the capsid. It is also easy to envision using this polymerase mechanism-based strategy when the toxicity associated with virus is sufficiently high that virus yield is negatively impacted. This circumstance may have contributed to the shortage of vaccine for the swine flu outbreak in 2009.
Viruses encoding polymerases with a substitution of the general acid may also prove to be useful reagents to study virus molecular and cellular biology. The 10-fold reduction in polymerase activity appears to prolong the kinetics of virus multiplication. Therefore, processes that occur early during infection may now be slow enough to interrogate experimentally. For example, during the first 30 min to 2 h after infection of HeLa cells by PV, the site of replication changes from the Golgi apparatus to the endoplasmic reticulum (30). Expanding the duration of time required for this transition will facilitate further dissection of this process and identification of host factors participating in the transition.
This work was supported, in whole or in part, by National Institutes of Health Grant AI053531 (to C. E. C.).
- PV
- poliovirus
- RdRp
- RNA-dependent RNA polymerase
- TCID50
- median tissue culture infective dose
- RTP
- ribavirin triphosphate.
REFERENCES
- 1. Castro C., Smidansky E. D., Arnold J. J., Maksimchuk K. R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C. E. (2009) Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 16, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lauring A. S., Jones J. O., Andino R. (2010) Rationalizing the development of live attenuated virus vaccines. Nat. Biotechnol. 28, 573–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nomoto A., Omata T., Toyoda H., Kuge S., Horie H., Kataoka Y., Genba Y., Nakano Y., Imura N. (1982) Complete nucleotide sequence of the attenuated poliovirus Sabin-1 strain genome. Proc. Natl. Acad. Sci. U.S.A. 79, 5793–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Racaniello V. R. (2006) One hundred years of poliovirus pathogenesis. Virology 344, 9–16 [DOI] [PubMed] [Google Scholar]
- 5. Goetz K. B., Pfleiderer M., Schneider C. K. (2010) First-in-human clinical trials with vaccines — what regulators want. Nat. Biotechnol. 28, 910–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barnes D., Kunitomi M., Vignuzzi M., Saksela K., Andino R. (2008) Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe 4, 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coleman J. R., Papamichail D., Skiena S., Futcher B., Wimmer E., Mueller S. (2008) Virus attenuation by genome-scale changes in codon pair bias. Science 320, 1784–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mueller S., Coleman J. R., Papamichail D., Ward C. B., Nimnual A., Futcher B., Skiena S., Wimmer E. (2010) Live attenuated influenza virus vaccines by computer-aided rational design. Nat. Biotechnol. 28, 723–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perez J. T., Pham A. M., Lorini M. H., Chua M. A., Steel J., tenOever B. R. (2009) MicroRNA-mediated species-specific attenuation of influenza A virus. Nat. Biotechnol. 27, 572–576 [DOI] [PubMed] [Google Scholar]
- 10. Vignuzzi M., Wendt E., Andino R. (2008) Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 14, 154–161 [DOI] [PubMed] [Google Scholar]
- 11. Vignuzzi M., Stone J. K., Arnold J. J., Cameron C. E., Andino R. (2006) Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439, 344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arnold J. J., Vignuzzi M., Stone J. K., Andino R., Cameron C. E. (2005) Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 280, 25706–25716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arias A., Arnold J. J., Sierra M., Smidansky E. D., Domingo E., Cameron C. E. (2008) Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J. Virol. 82, 12346–12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coffey L. L., Beeharry Y., Bordería A. V., Blanc H., Vignuzzi M. (2011) Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc. Natl. Acad. Sci. U.S.A. 108, 16038–16043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crotty S., Maag D., Arnold J. J., Zhong W., Lau J. Y., Hong Z., Andino R., Cameron C. E. (2000) The broad spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6, 1375–1379 [DOI] [PubMed] [Google Scholar]
- 16. Pfeiffer J. K., Kirkegaard K. (2003) A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. U.S.A. 100, 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Severson W. E., Schmaljohn C. S., Javadian A., Jonsson C. B. (2003) Ribavirin causes error catastrophe during Hantaan virus replication. J. Virol. 77, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sierra M., Airaksinen A., González-López C., Agudo R., Arias A., Domingo E. (2007) Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J. Virol. 81, 2012–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gohara D. W., Ha C. S., Kumar S., Ghosh B., Arnold J. J., Wisniewski T. J., Cameron C. E. (1999) Production of “authentic” poliovirus RNA-dependent RNA polymerase (3Dpol) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17, 128–138 [DOI] [PubMed] [Google Scholar]
- 20. Gohara D. W., Crotty S., Arnold J. J., Yoder J. D., Andino R., Cameron C. E. (2000) Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 275, 25523–25532 [DOI] [PubMed] [Google Scholar]
- 21. Oh H. S., Pathak H. B., Goodfellow I. G., Arnold J. J., Cameron C. E. (2009) Insight into poliovirus genome replication and encapsidation obtained from studies of 3B-3C cleavage site mutants. J. Virol. 83, 9370–9387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poch O., Sauvaget I., Delarue M., Tordo N. (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8, 3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Castro C., Smidansky E., Maksimchuk K. R., Arnold J. J., Korneeva V. S., Götte M., Konigsberg W., Cameron C. E. (2007) Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 104, 4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brautigam C. A., Steitz T. A. (1998) Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 8, 54–63 [DOI] [PubMed] [Google Scholar]
- 25. Arnold J. J., Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43, 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arnold J. J., Cameron C. E. (2000) Poliovirus RNA-dependent RNA polymerase (3Dpol): assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275, 5329–5336 [DOI] [PubMed] [Google Scholar]
- 27. Crotty S., Hix L., Sigal L. J., Andino R. (2002) Poliovirus pathogenesis in a new poliovirus receptor transgenic mouse model: age-dependent paralysis and a mucosal route of infection. J. Gen. Virol. 83, 1707–1720 [DOI] [PubMed] [Google Scholar]
- 28. Poch O., Blumberg B. M., Bougueleret L., Tordo N. (1990) Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71, 1153–1162 [DOI] [PubMed] [Google Scholar]
- 29. Wang T. S., Wong S. W., Korn D. (1989) Human DNA polymerase α: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 3, 14–21 [DOI] [PubMed] [Google Scholar]
- 30. Hsu N. Y., Ilnytska O., Belov G., Santiana M., Chen Y. H., Takvorian P. M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T., Cameron C. E., Ehrenfeld E., van Kuppeveld F. J., Altan-Bonnet N. (2010) Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141, 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]