Abstract
Ryanodine receptors (RyRs) are huge ion channels that are responsible for the release of Ca2+ from the sarco/endoplasmic reticulum. RyRs form homotetramers with a mushroom-like shape, consisting of a large cytoplasmic head and transmembrane stalk. Ca2+ is a major physiological ligand that triggers opening of RyRs, but a plethora of modulatory proteins and small molecules in the cytoplasm and sarco/endoplasmic reticulum lumen have been recognized. Over 300 mutations in RyRs are associated with severe skeletal muscle disorders or triggered cardiac arrhythmias. With the advent of high-resolution structures of individual domains, many of these can be mapped onto the three-dimensional structure.
Keywords: Calcium Channels, Excitation-Contraction Coupling, Genetic Diseases, Malignant Hyperthermia, Ryanodine Receptor, Central Core Disease, Structural Biology
Introduction
The South American plant Ryania speciosa has long been recognized for its insecticidal properties (1). Its active compound, an alkaloid known as ryanodine, targets a eukaryotic membrane protein known as the ryanodine receptor (RyR).2 Long before their isolation, RyRs had already been visualized in thin section or negative stain electron microscopy studies of muscle ultrastructure. These images showed the presence of intracellular junctions between the sarcoplasmic reticulum (SR) and transverse tubular invaginations of the plasma membrane. Electron dense protrusions, named “feet,” were found to span these junctions (2). Although the identities of the feet structures were initially unknown, later purification and electron microscopy studies confirmed them to be RyRs (3, 4).
RyRs form homotetrameric assemblies and constitute the largest ion channels known to date, with molecular masses of ∼2.2 MDa and each monomer consisting of ∼5000 amino acid residues (3, 4). They are responsible for the release of Ca2+ from the endoplasmic reticulum (ER) and SR and thus control many Ca2+-dependent processes within the cell. Ryanodine binds RyRs preferentially in the open state. At nanomolar concentrations, it “locks” the channel in a subconductance state, but at concentrations >100 μm, it inhibits Ca2+ release (5).
In mammalian organisms, RyRs are found in a wide variety of cell types, including neurons, exocrine cells, epithelial cells, lymphocytes, and many more (6). They are known mostly for their involvement in excitation-contraction coupling, releasing Ca2+ from the SR and thus driving muscle contraction. Three different isoforms (RyR1–3) have been found to date. RyR1 is widely expressed in skeletal muscle and was the first one to be cloned (7, 8). RyR2 is found primarily in the heart (9, 10), and RyR3 was originally identified in the brain (11), although each isoform is found in many different cell types (6). They share ∼65% sequence identity, and the largest degree of difference is found in three “divergent regions” throughout the sequence, known as D1 (residues 4254–4631 in RyR1), D2 (residues 1342–1403), and D3 (residues 1872–1923). Lower organisms express fewer RyR isoforms. Non-mammalian vertebrates express two isoforms, RyRα and RyRβ, whereas a single isoform was found to be expressed in lower organisms, including nematodes, fruit flies, and lobster (6).
The primary trigger for RyR opening is Ca2+, the same ion it permeates. In cardiac myocytes, for example, depolarization of the plasma membrane leads to the opening of L-type voltage-gated calcium channels, resulting in an influx of Ca2+ from the extracellular space. Calcium sensors within RyR2 then bind Ca2+ and facilitate opening, resulting in release of Ca2+ from the SR. Under these circumstances, the RyR acts as a signal amplifier, and the process is known as Ca2+-induced Ca2+ release (12, 13). However, as Ca2+ levels in the cytoplasm rise, Ca2+ can trigger closing of the RyR. This shows that there are multiple Ca2+-binding sites with different affinities and binding kinetics. A plot of the open probability of the channel as a function of Ca2+ concentration therefore displays a bell-shaped curve (14). In addition, RyRs can sense the Ca2+ concentrations in the SR/ER lumen. For example, under conditions whereby the ER is overloaded with Ca2+, RyRs can also open spontaneously in a process known as store overload-induced calcium release (SOICR) (15, 16).
Although Ca2+ is a major triggering ligand, it is not absolutely required to elicit channel opening. In the case of RyR1, the skeletal muscle isoform, it is widely assumed that there exists a physical link with the voltage-gated calcium channel CaV1.1 (17–20). In this case, voltage changes across the plasma membrane can cause opening of RyR1 in the absence of extracellular Ca2+. An intracellular loop of CaV1.1 that connects transmembrane repeats II and III (“II-III linker”) appears to be the primary interaction site for RyR1. However, not every RyR1 is near a CaV1.1 channel, and such channels are likely activated by Ca2+ released from neighboring channels.
Electron Microscopy Imaging
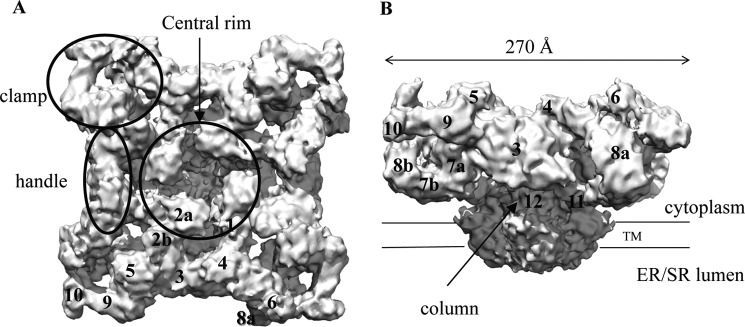
Since their purification from muscle tissues, the structures of RyRs have been studied extensively (4, 21). To date, crystal structures that describe the entire channel structure are not available, but several cryo-EM studies have reached resolutions near 10 Å (22–24). These studies agree very well on the overall structure of the receptor (Fig. 1). The basic architecture can be described as a mushroom, with a large cap representing ∼80% of the volume located in the cytoplasm and the stalk crossing the membrane into the SR/ER lumen. The transmembrane region measures 120 × 120 × 60 Å, whereas the cytoplasmic area measures ∼270 × 270 × 100 Å. These two major parts are connected via four thick columns. An interesting and important feature is that the RyR cytoplasmic head does not form a rigid block. Instead, there are many solvent-filled cavities and numerous globular masses that may correspond to individual or groups of folded domains. To aid with the structural description of RyRs, several portions have received names, including “clamps,” “handles,” and a “central rim” that surrounds a central cavity (Fig. 1). The globular portions have received identifying numbers and are often referred to as “subregions” (Fig. 1) (23, 25). Although most cryo-EM studies have been performed on RyR1, there are reconstructions for RyR2 and RyR3 as well, albeit at much lower resolutions. These show that the overall shape is very similar for all RyR isoforms (26, 27).
FIGURE 1.
Overall structure. Shown is a cryo-EM reconstruction of RyR1 at 9.6 Å (Electron Microscopy Data Bank entry 1275) (24). A, top view from the cytoplasm, looking toward the SR/ER. B, side view showing the large cytoplasmic head. Labels show the structural elements and the numbered subregions. TM, transmembrane domain.
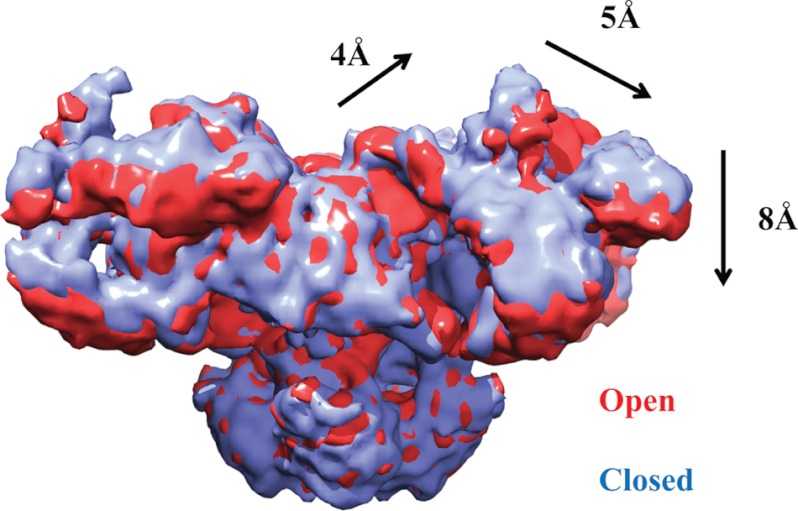
There has been much debate about the number of transmembrane helices, but the overall consensus now is that there are either six or eight segments per subunit (28). Five or six of these can be detected in the cryo-EM maps (23, 24). The inner helices create the pore-forming region, and sequence homology suggests an arrangement similar to various tetrameric ion channel structures. A major point of interest lies with the motions the channel undergoes during opening and closing. Based on different cryo-EM reconstructions of RyR1, there is still some debate about the movements the inner helices undergo during channel opening (22–24). A systematic investigation of the open and closed states at 10.2 Å seems to indicate that the inner helices kink, thus widening the pore during channel opening (22). A 9.6 Å cryo-EM structure of RyR1, reported to be in the closed state, shows that these helices are already kinked and thus suggests an alternative mechanism for channel opening (24). It is of course possible that the extraction and purification conditions in this latter study were not favorable for a closed state, but higher resolution studies will be needed to shed light on this matter. It is also clear from these studies that the RyR is a bona fide allosteric protein, as channel opening results in substantial structural rearrangements in the cytoplasmic portion. The most prominent motions occur near the central rim and the clamp regions (Fig. 2) (22). The movements in the transmembrane and cytoplasmic portions are most likely transmitted via the columns. Due to this allosteric coupling, binding of ligands or auxiliary proteins to any mobile portion of the cytoplasmic region can influence the ability of the pore region to open.
FIGURE 2.
Allosteric movements. Shown is the superposition of RyR1 cryo-EM maps in the closed (blue) and open (red) states (Electron Microscopy Data Bank entries 1606 and 1607) (22). The arrows and distances show the overall motions in the cytoplasmic region as the RyR opens.
In a physiological context, many RyRs will not be found in isolation but rather in a cluster with neighboring channels. An interesting feature of the RyR is that it can form regular arrays at the SR-plasma membrane junctions. Using purified protein, it has been shown that RyR1 can form planar crystalline arrangements, forming checkerboard patterns in the absence of any other protein (29). Two-dimensional crystallization experiments show that subregion 6 in the clamp region is responsible for the interprotein contacts (30). As the clamp region has been shown to undergo substantial motions during opening and closing (22), motions in one RyR channel can thus be transmitted to neighboring RyRs. This may underlie the phenomenon of coupled gating, whereby opening of one channel can induce opening in neighboring channels through physical interactions (31). Although the ability to form two-dimensional crystals holds promise for improved resolutions, so far these have not yet exceeded the single-particle cryo-EM studies.
Regulators
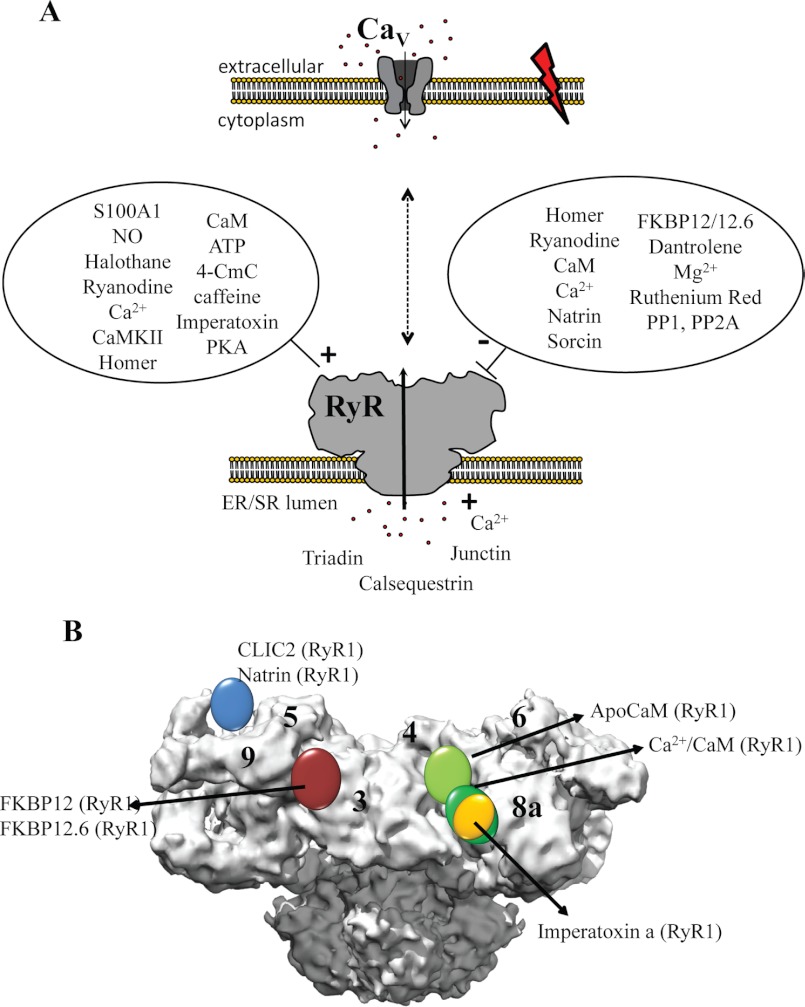
A Ca2+-selective pore could be made with just a few transmembrane α-helices. Why is it then that we have evolved >2-MDa giants to fulfill the same function? The answer undoubtedly lies with regulation: as Ca2+ is a very potent messenger, its entry into the cytoplasm through RyRs is tightly controlled by a myriad of proteins, small molecules, and post-translational modification events that affect opening or closing of the channels. By providing a large cytoplasmic mass and many solvent-filled cavities, each RyR yields ∼500,000 Å2 of surface area (transmembrane area included) onto which many regulators can dock. In addition to the Ca2+ and voltage-gated calcium channels mentioned above, these modulators include binding partners in both the cytoplasmic and SR/ER luminal portions that can provide either positive or negative input signals (Fig. 3A). The list is too extensive to cover every binding partner in detail, but several excellent reviews have provided comprehensive overviews (6, 32). Here, I describe a number of regulators that have received a lot of attention.
FIGURE 3.
Binding partners and ligands. A, schematic overview of the RyR and voltage-gated calcium channel (CaV), present in two different membranes, along with several binding partners in the cytoplasmic and luminal areas. 4-CmC, 4-chloro-m-cresol. B, locations of several protein-binding partners based on difference cryo-EM.
Well known binding partners of RyRs are FK506-binding proteins (FKBPs). Named according to their molecular mass, both FKBP12 and FKBP12.6 can associate with all three RyR isoforms (33). The affinity seems to be quite strong, as FKBP12 co-purifies with RyR1, and FKBP12.6 co-purifies with RyR2. They stabilize the closed state of the channels and prevent the formation of subconductance states in RyR1 (34). Using cryo-EM studies, FKBPs were found to bind in a site near subdomains 3, 5, and 9 (35, 36), and this was confirmed via FRET studies (37, 38). On the amino acid level, it was reported that a Val-Pro (RyR1 and RyR3) or Ile-Pro (RyR2) motif formed the binding site for FKBPs, but several other studies disagree (e.g. see Ref. 39).
Other well studied regulators are EF-hand-containing proteins that can bind Ca2+. Calmodulin (CaM) can associate with the RyR under both apo- and Ca2+-loaded conditions and fine-tune the effect of Ca2+. The functional effect of CaM depends on both the Ca2+ concentration and the RyR isoform. At high Ca2+ levels, CaM can inhibit both RyR1 and RyR2. At low Ca2+ levels, however, it activates RyR1 but inhibits RyR2 (40–43). Many studies have focused on identifying the sequences in RyRs that can bind CaM. Currently, there is one crystal structure available for Ca2+/CaM bound to a peptide from RyR1 (residues 3614–3643). In this case, both CaM lobes wrap around an amphipathic α-helix in an antiparallel arrangement, with the N-terminal lobe bound to the C-terminal half and the C-terminal lobe bound to the N-terminal half of the helix (44). However, several other RyR peptides are found to bind CaM (e.g. Ref. 45), and it is likely that the Ca2+/N-terminal lobe binds a different segment in intact RyRs (44, 46). Interestingly, the same RyR1 peptide can bind the EF-hand protein S100A1, which can enhance opening of both RyR1 and RyR2 (47). An NMR structure shows that its binding site overlaps with the Ca2+/CaM-binding site (48), and it is therefore likely that S100A1 acts by displacing CaM at high Ca2+ levels, thus abolishing the latter's inhibitory effects. Sorcin is yet another EF-hand-containing protein that can associate with RyR2 at elevated Ca2+ concentrations and mediate inhibition (49).
Using cryo-EM reconstructions, the binding of both apo-CaM and Ca2+/CaM has been visualized on intact RyRs (50). These show that the location of CaM changes upon binding Ca2+, in accordance with the differential functional effects of CaM at various Ca2+ levels (Fig. 3B). FRET-based measurements between CaM and FKBP suggest that the N-terminal lobe is closer to FKBP than the C-terminal lobe (37). As both CaM and FKBP are located in a region of the channel that undergoes substantial motions upon opening and closing (22), these allosteric modulators likely act by stabilizing or destabilizing individual states.
Calsequestrin is a major Ca2+-buffering protein in the SR lumen. It can form oligomers and interact with the membrane-associated proteins junctin and triadin. Together, this complex is thought to either increase or decrease the RyR activity depending on the calsequestrin isoform, although little is known about the exact molecular mechanisms (51).
Homer 1c is an adaptor protein that can activate RyR1 and inhibit RyR2 (49). In addition to various protein-binding partners, RyRs are also modulated by small molecules like Mg2+ (inhibition) and ATP (activation) (14), toxins like natrin (inhibition) and imperatoxin (activation), and several non-physiological ligands (caffeine, 4-chloro-m-cresol, ruthenium red, volatile anesthetics, and dantrolene). Importantly, RyRs are sensitive to redox conditions and can be activated, for example, by NO (Fig. 3A).
Phosphorylation
RyRs are the target for several kinases (PKA, PKG, and Ca2+/CaM-dependent protein kinase II (CaMKII)) and phosphatases (PP1, PP2A, and PDE4D3). Some of these enzymes are anchored to RyRs via scaffolding proteins, allowing for specific and compartmentalized regulation (32). Phosphorylation by PKA has received a lot of attention, as it forms the link between physiological stress and RyRs via activation of β-adrenergic receptors. PKA targets several cytoplasmic proteins, and at least two RyR residues (Ser-2843 in human RyR1 and Ser-2030 and Ser-2808 in RyR2) can be phosphorylated by PKA. However, a great deal of controversy exists about which phosphorylation event predominates and the role of this in heart failure. On the one hand, Ser-2808 was found to be hyperphosphorylated by PKA in heart failure, leading to the dissociation of FKBP12.6. As the latter stabilizes the closed state, the dissociation would then lead to enhanced activity of the RyR (52). However, several groups have failed to detect PKA hyperphosphorylation in heart failure and did not observe a dissociation of FKBPs by RyR phosphorylation (e.g. Refs. 53 and 54). For example, one report found that Ser-2808 is already highly phosphorylated in the basal state and that instead Ser-2030 is the major PKA target residue in RyR2 (55). Most likely, much of the controversy has to do with details in the preparations. For example, RyRs are sensitive to redox conditions, and a different amount of oxidation in the various studies could therefore influence the affinity for FKBPs (56).
CaMKII is regulated by intracellular Ca2+ concentrations through CaM. Like PKA, it can phosphorylate Ser-2843 in RyR1 and Ser-2808 in RyR2 but also seems to have a unique phosphorylation site in RyR2 (Ser-2814). CaMKII was found to increase the open probability and Ca2+ sensitivity of the channel (57) and has also been shown to contribute to cardiac arrhythmia and contractile dysfunction (58).
Disease Mutations
Although the roles and mechanisms of RyRs in heart failure are still under scrutiny, it has become clear that mishandling of Ca2+ in the cytoplasm due to mutations in the ryr genes can give rise to severe conditions. No disease phenotype has been associated with RyR3 mutations, but both RyR1 and RyR2 have been linked to a number of genetic diseases that are due mostly to their prominent role in muscle contraction (59–61). Malignant hyperthermia (MH) is a pharmacogenetic disorder, characterized by muscle rigidity and fatal rises in body temperature (62, 63). It is typically triggered by the combination of a RyR1 mutation and an external compound such as a volatile anesthetic or succinylcholine, a muscle relaxant. In some cases, stress may serve as an alternative external trigger (64). In pigs, the RyR1 mutation R615C was found to cause the related porcine stress syndrome (65, 66), and the corresponding mutation in humans was soon found to underlie MH (67). In a MH event, an excessive leak of Ca2+ from the SR results in a hypermetabolic state, depleting the ATP pool and leading to acidosis. Dantrolene is a clinically approved drug to treat MH and acts by decreasing the intracellular Ca2+ concentration (68). Several studies suggest a direct interaction between dantrolene and RyR1 (69, 70). Dantrolene inhibits RyR1 even when expressed in a heterologous system such as HEK293 cells and appears to inhibit SOICR (71). However, the single-channel behavior of RyR1 incorporated in planar lipid bilayers appears to be unaffected, and the precise mechanism of action is therefore still unknown.
In addition to MH, RyR1 mutations can cause congenital myopathies such as central core disease (CCD) (72, 73). The latter is characterized by cores of metabolically inactive tissue in the center of muscle fibers, which can lead to muscle weakness. In contrast to MH, CCD does not require an external trigger. Although RyR1 is abundant in skeletal muscle, it is also widely expressed in the brain, and a CCD mutation was recently shown to have a neuronal phenotype (74). Other defects associated with RyR1 mutations include heat/exercise-induced exertional rhabdomyolysis (75), multiminicore disease (76), and atypical periodic paralysis (77).
Because RyR2 plays a major role in cardiac excitation-contraction coupling, mutations in this isoform can give rise to cardiac arrhythmias. Catecholaminergic polymorphic ventricular tachycardia (CPVT) is a condition often triggered by emotional or physical stress, leading to bidirectional ventricular tachycardia that may result in sudden cardiac death (78). A popular model assumes that the leakage of Ca2+ through RyR2 increases the activity of the Na+/Ca2+ exchanger, which allows for the electrogenic influx of three Na+ ions in exchange for efflux of one Ca2+ ion, resulting in delayed afterdepolarizations (59). In addition to CPVT, RyR2 mutations associate with arrhythmogenic right ventricular dysplasia, in which the right ventricular muscle is gradually replaced by fibrofatty deposits (79). As RyR2 is also widely expressed in the brain, there may be neuronal effects of the mutations as well, and a mouse model has shown that RyR2 mutations can also give rise to seizures (80).
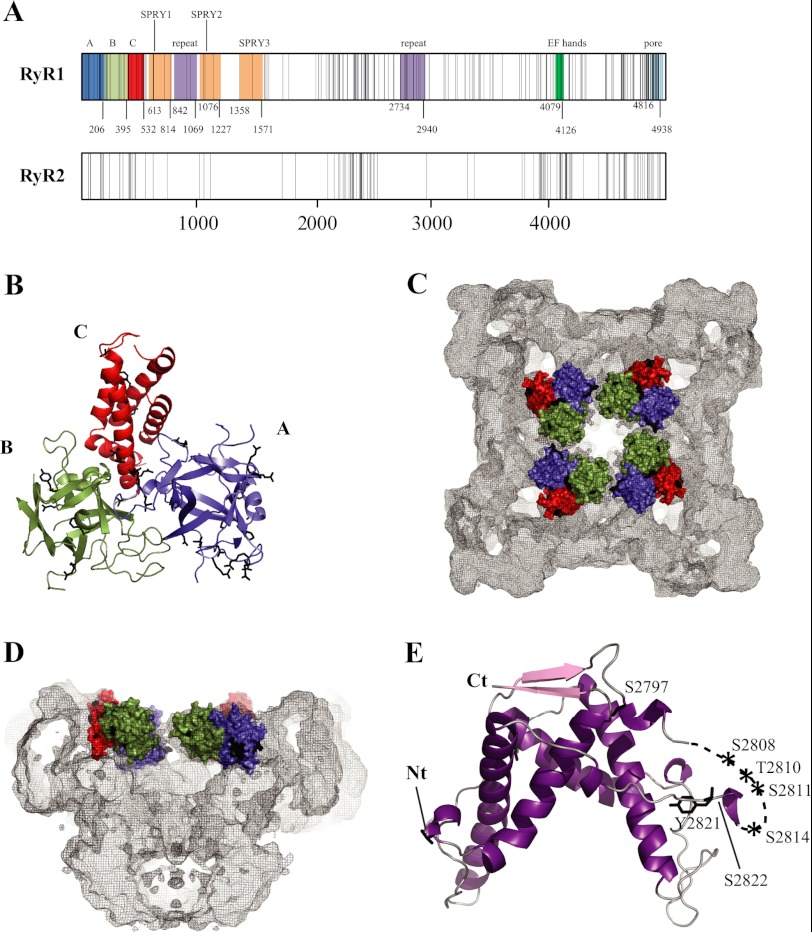
Since the initial links between RyR mutations and disease were established, >300 disease mutations have been identified. In both RyR1 and RyR2, most of these cluster in three or four different “hot spots,” located in the N-terminal region (first ∼600 amino acids), a central region (amino acids ∼2100–2500), and the C-terminal area (amino acid ∼3900–end) (Fig. 4A) (60, 81). However, the appearance of clusters may be due, in part, to sequencing bias, and mutations are increasingly found outside of these three segments. The C-terminal portion encodes the transmembrane area, and a subset of mutations could thus interfere directly with the passage of Ca2+. However, this third hot spot also encodes a substantial amount of cytoplasmic components. Interestingly, many CCD mutations are found in the C-terminal region.
FIGURE 4.
Disease hot spots. A, linear view of the RyR sequences, with each vertical line representing a disease mutation. The shaded areas indicate experimentally verified (domains A–C) and some key predicted domains. Another truncated repeat may be present between SPRY2 and SPRY3 (not shown). The domain boundaries are numbered according to the RyR1 sequence. Except for domains A–C, the numbering is only a prediction and may vary (Ref. 97 and the Phyre2 server, Structural Informatics Group, Imperial College London). The predicted pore-forming area is based on homology to the pore-forming region of a bacterial voltage-gated sodium channel (Protein Data Bank code 3RVZ). B, crystal structure of the RyR1 ABC region (Protein Data Bank code 2XOA) (93). Disease mutations are shown as black sticks. Mutations in flexible loops have been omitted for clarity. C and D, location of the RyR1 ABC domains in the RyR1 cryo-EM map. The domains are shown in surface representation. Black patches correspond to the locations of disease mutations. Shown are a view from the cytoplasm (C) and a side view with one-half omitted (D). E, crystal structure of the RyR2 phosphorylation domain. The labels indicate phosphorylation target sites within the phosphorylation loop.
A general observation is that most disease mutations cause a gain of function, although there are exceptions (82). These mutations then lead to premature or prolonged release of Ca2+ in the cytoplasm. It has been shown that CPVT mutations in RyR2 lower the threshold for activation by luminal Ca2+, thus affecting SOICR (16, 83). In addition, many studies have shown that the mutations increase the sensitivity of the channels to activating agents (e.g. Refs. 84 and 85). One hypothesis suggests that there is a direct interaction between the N-terminal and central hot spots and that this interaction is “unzipped” during channel opening through allosteric coupling (86, 87). Disease mutations clustered at this interface would thus weaken the interaction and facilitate channel opening. This theory is based on spectroscopic measurements and the ability of peptides to interfere with channel function, but a direct interaction between individually folded domains of the N-terminal and central hot spots has not yet been shown. Another theory, similar to the effect of PKA phosphorylation, proposes that the disease mutations weaken the interaction with FKBPs, and the resulting dissociation of FKBP then leads to increased single-channel activity under conditions that simulate stress (88). Despite the differences, the combined data converge on a gain of function for most disease mutations, with a concomitant increased sensitivity toward cytoplasmic or luminal activation mechanisms and decreased binding of modulators that preferentially bind to the closed state.
Crystal Structures and Domain Architecture
Despite the availability of high-quality cryo-EM maps, the current resolution is too small to pinpoint the location of individual amino acids. In trying to match primary with tertiary structure, many studies have benefited from raising antibodies against particular stretches of the RyR sequence or from inserting GFP at various locations in the sequence. Cryo-EM reconstructions were then made for the RyR-antibody complexes and fusion proteins, which created restraints on the position of the sequence in the three-dimensional structure (for a review, see Ref. 21). Using this approach, it was shown, for example, that insertions in the N-terminal and central disease hot spots both yield difference density in the clamp region, suggesting that these segments may be in close proximity (89), in agreement with the zipper hypothesis.
Although no well diffracting crystals have been reported for the intact RyR, four reports have provided high-resolution information on individually folded RyR domains, corresponding to the N-terminal domains of RyR1 (residues 1–205) (90, 91) and RyR2 (91, 92) and a larger N-terminal region of RyR1 (residues 1–559) (93). The latter study represents the bulk of the N-terminal disease hot spot and shows that it is built up by three individually folded domains (A–C) that form a compact arrangement (Fig. 4B). Domains A and B form β-trefoils, containing 12 β-strands each, whereas domain C consists of a five-helix bundle. Docking of this hot spot in several RyR1 cryo-EM maps shows that it is located in the cytoplasmic portion, forming a vestibule around the 4-fold symmetry axis (Fig. 4, C and D). Although this position may seem at odds with a GFP insertion study (89), the data can be reconciled when the length of the linkers for the fusion protein is taken into consideration (93).
Over 55 disease mutations (RyR1 and RyR2 combined) can be located in the crystal structure and pseudo-atomic model. Most of these are found at domain-domain boundaries, either in between the three domains or at interfaces with neighboring RyR domains. This suggests that some domain interactions may be disrupted during channel opening and that mutations at such functional interfaces facilitate the process by weakening the contacts. Six mutations were found to be buried within individual domains, and these could alter the local folding of the domains, thereby having a less specific effect. Most of the mutations are at interfaces with other N-terminal hot spot domains, either within or across subunits. The zipper hypothesis, involving interactions with the central hot spot region, can therefore apply to only less than one-third of the N-terminal mutations.
Most disease mutations consist of point substitutions, but a very severe form of CPVT is caused by deletion of the entire third exon of RyR2, consisting of 35 amino acids (94). This exon encodes a β-strand and α-helix in RyR2 domain A, but rather than causing misfolding, the deletion increases the thermal stability of the domain (91, 92). A crystal structure of the mutant domain shows that this is accomplished by insertion of a flexible loop, encoded by the fourth exon, into the β-trefoil core (92). In non-diseased individuals, this may represent a case of alternative splicing, whereby two short stretches “compete” for a β-strand position. This allows for the formation of an alternative N-terminal domain, which further fine-tunes the RyR activity.
RyRs also encode tandem repeats that are present at least two times in the sequence (Fig. 4A). Crystal structures of this domain in the central region of RyR1, RyR2, and RyR3 show a mostly α helical structure with 2-fold symmetry (Fig. 4E) (95). The repeats are separated by a flexible loop that contains up to 7 different phosphorylation target sites, including the well-studied Ser-2808 and Ser-2814 sites in RyR2. In addition to the crystallized portions, the RyR genes encode a number of predicted cytoplasmic domains (Fig. 4A). This includes three SPRY domains encoded in the N-terminal one-third of the sequence. SPRY domains were first detected in the SplA kinase and in RyRs. The core of the SPRY structure consists of a sandwich formed by two four-stranded antiparallel β-sheets. They generally form protein-protein interaction domains and thus may serve as docking sites for auxiliary proteins in RyRs. The second SPRY domain in RyR1 has been suggested to form a docking site for the CaV1.1 II-III loop (96). Tandem EF-hands, forming likely Ca2+-binding sites, are encoded closer to the C terminus (97).
RyR and the Inositol 1,4,5-Trisphosphate Receptor
The ER membrane is home to another Ca2+ release channel, modulated by inositol 1,4,5-trisphosphate (IP3). Although much smaller (∼1 MDa), this IP3 receptor (IP3R) also forms a tetrameric assembly and shares many structural features with RyRs, predominantly in the transmembrane and N-terminal regions. A recent cryo-EM structure at ∼9.5 Å of IP3R isoform 1 displays an overall mushroom shape, although with a smaller cap than for the RyR (98). Several crystallographic studies on the N-terminal region of IP3Rs show a striking similarity to domains A–C in RyRs (99, 100). IP3R domain A (also known as the “suppressor domain”) and domains B and C (together known as the IP3-binding core) display a similar overall arrangement and domain interactions as in the RyR1 ABC structure. Docking in the 9.5 Å IP3R cryo-EM map yields a similar position, whereby four N-terminal ABC domains form a continuous ring around the 4-fold symmetry axis (99). Binding of IP3 between domains B and C causes a rearrangement of the three N-terminal domains, with domain A moving ∼3–4 Å toward domains B and C (99, 100). An overall theme thus seems to emerge in both Ca2+ release channels. Affecting the interfaces between the ABC domains, either by ligand binding in IP3Rs or by disease mutations in RyRs, seems to facilitate channel opening in both receptors. The overall similarity in structure and mechanisms between both channels is further corroborated by the fact that both domain A and the transmembrane area can be functionally swapped (99).
Conclusion
Much remains unknown about the molecular mechanisms that underlie opening and closing of the RyR. In the absence of full-length crystal structures, the construction of pseudo-atomic models holds much promise. Smaller domains may prove more cumbersome to dock reliably, but together with restraints provided by difference cryo-EM and FRET measurements, there is great promise to solve this three-dimensional puzzle in the years to come.
This article is part of the Thematic Minireview Series on Ins and Outs of Calcium Transport.
- RyR
- ryanodine receptor
- SR
- sarcoplasmic reticulum
- ER
- endoplasmic reticulum
- SOICR
- store overload-induced calcium release
- FKBP
- FK506-binding protein
- CaM
- calmodulin
- CaMKII
- Ca2+/CaM-dependent protein kinase II
- MH
- malignant hyperthermia
- CCD
- central core disease
- CPVT
- catecholaminergic polymorphic ventricular tachycardia
- IP3
- inositol 1,4,5-trisphosphate
- IP3R
- IP3 receptor.
REFERENCES
- 1. Rogers E. F., Koniuszy F. R., Shavel J., Jr., Folkersand K. (1948) Plant insecticides. I. Ryanodine, a new alkaloid from Ryania speciosa Vahl. J. Am. Chem. Soc. 70, 3086–3088 [DOI] [PubMed] [Google Scholar]
- 2. Franzini-Armstrong C. (1970) Studies of the triad. I. Structure of the junction in frog twitch fibers. J. Cell Biol. 47, 488–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lai F. A., Erickson H. P., Rousseau E., Liu Q. Y., Meissner G. (1988) Purification and reconstitution of the calcium release channel from skeletal muscle. Nature 331, 315–319 [DOI] [PubMed] [Google Scholar]
- 4. Inui M., Saito A., Fleischer S. (1987) Purification of the ryanodine receptor and identity with feet structures of junctional terminal cisternae of sarcoplasmic reticulum from fast skeletal muscle. J. Biol. Chem. 262, 1740–1747 [PubMed] [Google Scholar]
- 5. Meissner G. (1986) Ryanodine activation and inhibition of the Ca2+ release channel of sarcoplasmic reticulum. J. Biol. Chem. 261, 6300–6306 [PubMed] [Google Scholar]
- 6. Lanner J. T., Georgiou D. K., Joshi A. D., Hamilton S. L. (2010) Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harb. Perspect. Biol. 2, a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takeshima H., Nishimura S., Matsumoto T., Ishida H., Kangawa K., Minamino N., Matsuo H., Ueda M., Hanaoka M., Hirose T. (1989) Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature 339, 439–445 [DOI] [PubMed] [Google Scholar]
- 8. Zorzato F., Fujii J., Otsu K., Phillips M., Green N. M., Lai F. A., Meissner G., MacLennan D. H. (1990) Molecular cloning of cDNA encoding human and rabbit forms of the Ca2+ release channel (ryanodine receptor) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 265, 2244–2256 [PubMed] [Google Scholar]
- 9. Otsu K., Willard H. F., Khanna V. K., Zorzato F., Green N. M., MacLennan D. H. (1990) Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J. Biol. Chem. 265, 13472–13483 [PubMed] [Google Scholar]
- 10. Nakai J., Imagawa T., Hakamat Y., Shigekawa M., Takeshima H., Numa S. (1990) Primary structure and functional expression from cDNA of the cardiac ryanodine receptor/calcium release channel. FEBS Lett. 271, 169–177 [DOI] [PubMed] [Google Scholar]
- 11. Hakamata Y., Nakai J., Takeshima H., Imoto K. (1992) Primary structure and distribution of a novel ryanodine receptor/calcium release channel from rabbit brain. FEBS Lett. 312, 229–235 [DOI] [PubMed] [Google Scholar]
- 12. Endo M., Tanaka M., Ogawa Y. (1970) Calcium-induced release of calcium from the sarcoplasmic reticulum of skinned skeletal muscle fibers. Nature 228, 34–36 [DOI] [PubMed] [Google Scholar]
- 13. Fabiato A. (1983) Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am. J. Physiol. 245, C1–C14 [DOI] [PubMed] [Google Scholar]
- 14. Meissner G., Darling E., Eveleth J. (1986) Kinetics of rapid Ca2+ release by sarcoplasmic reticulum. Effects of Ca2+, Mg2+, and adenine nucleotides. Biochemistry 25, 236–244 [DOI] [PubMed] [Google Scholar]
- 15. Palade P., Mitchell R. D., Fleischer S. (1983) Spontaneous calcium release from sarcoplasmic reticulum. General description and effects of calcium. J. Biol. Chem. 258, 8098–8107 [PubMed] [Google Scholar]
- 16. Jiang D., Xiao B., Yang D., Wang R., Choi P., Zhang L., Cheng H., Chen S. R. (2004) RyR2 mutations linked to ventricular tachycardia and sudden death reduce the threshold for store overload-induced Ca2+ release (SOICR). Proc. Natl. Acad. Sci. U.S.A. 101, 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Block B. A., Imagawa T., Campbell K. P., Franzini-Armstrong C. (1988) Structural evidence for direct interaction between the molecular components of the transverse tubule/sarcoplasmic reticulum junction in skeletal muscle. J. Cell Biol. 107, 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanabe T., Beam K. G., Adams B. A., Niidome T., Numa S. (1990) Regions of the skeletal muscle dihydropyridine receptor critical for excitation-contraction coupling. Nature 346, 567–569 [DOI] [PubMed] [Google Scholar]
- 19. Tanabe T., Beam K. G., Powell J. A., Numa S. (1988) Restoration of excitation-contraction coupling and slow calcium current in dysgenic muscle by dihydropyridine receptor complementary DNA. Nature 336, 134–139 [DOI] [PubMed] [Google Scholar]
- 20. Rios E., Brum G. (1987) Involvement of dihydropyridine receptors in excitation-contraction coupling in skeletal muscle. Nature 325, 717–720 [DOI] [PubMed] [Google Scholar]
- 21. Kimlicka L., Van Petegem F. (2011) The structural biology of ryanodine receptors. Sci. China Life Sci. 54, 712–724 [DOI] [PubMed] [Google Scholar]
- 22. Samsó M., Feng W., Pessah I. N., Allen P. D. (2009) Coordinated movement of cytoplasmic and transmembrane domains of RyR1 upon gating. PLoS Biol. 7, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Samsó M., Wagenknecht T., Allen P. D. (2005) Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat. Struct. Mol. Biol. 12, 539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ludtke S. J., Serysheva I. I., Hamilton S. L., Chiu W. (2005) The pore structure of the closed RyR1 channel. Structure 13, 1203–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Serysheva I. I., Ludtke S. J., Baker M. L., Cong Y., Topf M., Eramian D., Sali A., Hamilton S. L., Chiu W. (2008) Subnanometer resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc. Natl. Acad. Sci. U.S.A. 105, 9610–9615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharma M. R., Penczek P., Grassucci R., Xin H. B., Fleischer S., Wagenknecht T. (1998) Cryo-electron microscopy and image analysis of the cardiac ryanodine receptor. J. Biol. Chem. 273, 18429–18434 [DOI] [PubMed] [Google Scholar]
- 27. Sharma M. R., Jeyakumar L. H., Fleischer S., Wagenknecht T. (2000) Three-dimensional structure of ryanodine receptor isoform three in two conformational states as visualized by cryo-electron microscopy. J. Biol. Chem. 275, 9485–9491 [DOI] [PubMed] [Google Scholar]
- 28. Du G. G., Sandhu B., Khanna V. K., Guo X. H., MacLennan D. H. (2002) Topology of the Ca2+ release channel of skeletal muscle sarcoplasmic reticulum (RyR1). Proc. Natl. Acad. Sci. U.S.A. 99, 16725–16730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yin C. C., Lai F. A. (2000) Intrinsic lattice formation by the ryanodine receptor calcium release channel. Nat. Cell Biol. 2, 669–671 [DOI] [PubMed] [Google Scholar]
- 30. Yin C. C., Blayney L. M., Lai F. A. (2005) Physical coupling between ryanodine receptor calcium release channels. J. Mol. Biol. 349, 538–546 [DOI] [PubMed] [Google Scholar]
- 31. Marx S. O., Ondrias K., Marks A. R. (1998) Coupled gating between individual skeletal muscle Ca2+ release channels (ryanodine receptors) Science 281, 818–821 [DOI] [PubMed] [Google Scholar]
- 32. Zalk R., Lehnart S. E., Marks A. R. (2007) Modulation of the ryanodine receptor and intracellular calcium. Annu. Rev. Biochem. 76, 367–385 [DOI] [PubMed] [Google Scholar]
- 33. Chelu M. G., Danila C. I., Gilman C. P., Hamilton S. L. (2004) Regulation of ryanodine receptors by FK506-binding proteins. Trends Cardiovasc. Med. 14, 227–234 [DOI] [PubMed] [Google Scholar]
- 34. Ahern G. P., Junankar P. R., Dulhunty A. F. (1997) Subconductance states in single-channel activity of skeletal muscle ryanodine receptors after removal of FKBP12. Biophys. J. 72, 146–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Samsó M., Shen X., Allen P. D. (2006) Structural characterization of the RyR1-FKBP12 interaction. J. Mol. Biol. 356, 917–927 [DOI] [PubMed] [Google Scholar]
- 36. Benacquista B. L., Sharma M. R., Samsó M., Zorzato F., Treves S., Wagenknecht T. (2000) Amino acid residues 4425–4621 localized on the three-dimensional structure of the skeletal muscle ryanodine receptor. Biophys. J. 78, 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cornea R. L., Nitu F., Gruber S., Kohler K., Satzer M., Thomas D. D., Fruen B. R. (2009) FRET-based mapping of calmodulin bound to the RyR1 Ca2+ release channel. Proc. Natl. Acad. Sci. U.S.A. 106, 6128–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cornea R. L., Nitu F. R., Samsó M., Thomas D. D., Fruen B. R. (2010) Mapping the ryanodine receptor FK506-binding protein subunit using fluorescence resonance energy transfer. J. Biol. Chem. 285, 19219–19226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Masumiya H., Wang R., Zhang J., Xiao B., Chen S. R. (2003) Localization of the 12.6-kDa FK506-binding protein (FKBP12.6)-binding site to the NH2-terminal domain of the cardiac Ca2+ release channel (ryanodine receptor). J. Biol. Chem. 278, 3786–3792 [DOI] [PubMed] [Google Scholar]
- 40. Ikemoto T., Iino M., Endo M. (1995) Enhancing effect of calmodulin on Ca2+-induced Ca2+ release in the sarcoplasmic reticulum of rabbit skeletal muscle fibers. J. Physiol. 487, 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tripathy A., Xu L., Mann G., Meissner G. (1995) Calmodulin activation and inhibition of skeletal muscle Ca2+ release channel (ryanodine receptor). Biophys. J. 69, 106–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Buratti R., Prestipino G., Menegazzi P., Treves S., Zorzato F. (1995) Calcium-dependent activation of skeletal muscle Ca2+ release channel (ryanodine receptor) by calmodulin. Biochem. Biophys. Res. Commun. 213, 1082–1090 [DOI] [PubMed] [Google Scholar]
- 43. Fuentes O., Valdivia C., Vaughan D., Coronado R., Valdivia H. H. (1994) Calcium-dependent block of ryanodine receptor channel of swine skeletal muscle by direct binding of calmodulin. Cell Calcium 15, 305–316 [DOI] [PubMed] [Google Scholar]
- 44. Maximciuc A. A., Putkey J. A., Shamoo Y., Mackenzie K. R. (2006) Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode. Structure 14, 1547–1556 [DOI] [PubMed] [Google Scholar]
- 45. Chen S. R., MacLennan D. H. (1994) Identification of calmodulin-, Ca2+-, and ruthenium red-binding domains in the Ca2+ release channel (ryanodine receptor) of rabbit skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 269, 22698–22704 [PubMed] [Google Scholar]
- 46. Xiong L. W., Newman R. A., Rodney G. G., Thomas O., Zhang J. Z., Persechini A., Shea M. A., Hamilton S. L. (2002) Lobe-dependent regulation of ryanodine receptor type 1 by calmodulin. J. Biol. Chem. 277, 40862–40870 [DOI] [PubMed] [Google Scholar]
- 47. Prosser B. L., Hernández-Ochoa E. O., Schneider M. F. (2011) S100A1 and calmodulin regulation of ryanodine receptor in striated muscle. Cell Calcium 50, 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wright N. T., Prosser B. L., Varney K. M., Zimmer D. B., Schneider M. F., Weber D. J. (2008) S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J. Biol. Chem. 283, 26676–26683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bers D. M. (2004) Macromolecular complexes regulating cardiac ryanodine receptor function. J. Mol. Cell. Cardiol. 37, 417–429 [DOI] [PubMed] [Google Scholar]
- 50. Samsó M., Wagenknecht T. (2002) Apocalmodulin and Ca2+/calmodulin bind to neighboring locations on the ryanodine receptor. J. Biol. Chem. 277, 1349–1353 [DOI] [PubMed] [Google Scholar]
- 51. Beard N. A., Wei L., Dulhunty A. F. (2009) Ca2+ signaling in striated muscle: the elusive roles of triadin, junctin, and calsequestrin. Eur. Biophys. J. 39, 27–36 [DOI] [PubMed] [Google Scholar]
- 52. Marx S. O., Reiken S., Hisamatsu Y., Jayaraman T., Burkhoff D., Rosemblit N., Marks A. R. (2000) PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell 101, 365–376 [DOI] [PubMed] [Google Scholar]
- 53. George C. H., Higgs G. V., Lai F. A. (2003) Ryanodine receptor mutations associated with stress-induced ventricular tachycardia mediate increased calcium release in stimulated cardiomyocytes. Circ. Res. 93, 531–540 [DOI] [PubMed] [Google Scholar]
- 54. Jiang M. T., Lokuta A. J., Farrell E. F., Wolff M. R., Haworth R. A., Valdivia H. H. (2002) Abnormal Ca2+ release, but normal ryanodine receptors, in canine and human heart failure. Circ. Res. 91, 1015–1022 [DOI] [PubMed] [Google Scholar]
- 55. Xiao B., Zhong G., Obayashi M., Yang D., Chen K., Walsh M. P., Shimoni Y., Cheng H., Ter Keurs H., Chen S. R. (2006) Ser-2030, but not Ser-2808, is the major phosphorylation site in cardiac ryanodine receptors responding to protein kinase A activation upon β-adrenergic stimulation in normal and failing hearts. Biochem. J. 396, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Andersson D. C., Betzenhauser M. J., Reiken S., Meli A. C., Umanskaya A., Xie W., Shiomi T., Zalk R., Lacampagne A., Marks A. R. (2011) Ryanodine receptor oxidation causes intracellular calcium leak and muscle weakness in aging. Cell Metab. 14, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wehrens X. H., Lehnart S. E., Reiken S. R., Marks A. R. (2004) Ca2+/calmodulin-dependent protein kinase II phosphorylation regulates the cardiac ryanodine receptor. Circ. Res. 94, e61–e70 [DOI] [PubMed] [Google Scholar]
- 58. Ai X., Curran J. W., Shannon T. R., Bers D. M., Pogwizd S. M. (2005) Ca2+/calmodulin-dependent protein kinase modulates cardiac ryanodine receptor phosphorylation and sarcoplasmic reticulum Ca2+ leak in heart failure. Circ. Res. 97, 1314–1322 [DOI] [PubMed] [Google Scholar]
- 59. Betzenhauser M. J., Marks A. R. (2010) Ryanodine receptor channelopathies. Pflugers Arch. 460, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Priori S. G., Chen S. R. (2011) Inherited dysfunction of sarcoplasmic reticulum Ca2+ handling and arrhythmogenesis. Circ. Res. 108, 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maclennan D. H., Zvaritch E. (2011) Mechanistic models for muscle diseases and disorders originating in the sarcoplasmic reticulum. Biochim. Biophys. Acta 1813, 948–964 [DOI] [PubMed] [Google Scholar]
- 62. Denborough M. A., Lovell R. R. H. (1960) Anesthetic deaths in a family. Lancet 276, 45 [Google Scholar]
- 63. Rosenberg H., Davis M., James D., Pollock N., Stowell K. (2007) Malignant hyperthermia. Orphanet J. Rare Dis. 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Capacchione J. F., Muldoon S. M. (2009) The relationship between exertional heat illness, exertional rhabdomyolysis, and malignant hyperthermia. Anesth. Analg. 109, 1065–1069 [DOI] [PubMed] [Google Scholar]
- 65. Fujii J., Otsu K., Zorzato F., de Leon S., Khanna V. K., Weiler J. E., O'Brien P. J., MacLennan D. H. (1991) Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science 253, 448–451 [DOI] [PubMed] [Google Scholar]
- 66. MacLennan D. H., Phillips M. S. (1992) Malignant hyperthermia. Science 256, 789–794 [DOI] [PubMed] [Google Scholar]
- 67. Gillard E. F., Otsu K., Fujii J., Khanna V. K., de Leon S., Derdemezi J., Britt B. A., Duff C. L., Worton R. G., MacLennan D. H. (1991) A substitution of cysteine for arginine 614 in the ryanodine receptor is potentially causative of human malignant hyperthermia. Genomics 11, 751–755 [DOI] [PubMed] [Google Scholar]
- 68. Krause T., Gerbershagen M. U., Fiege M., Weisshorn R., Wappler F. (2004) Dantrolene–a review of its pharmacology, therapeutic use, and new developments. Anaesthesia 59, 364–373 [DOI] [PubMed] [Google Scholar]
- 69. Paul-Pletzer K., Yamamoto T., Bhat M. B., Ma J., Ikemoto N., Jimenez L. S., Morimoto H., Williams P. G., Parness J. (2002) Identification of a dantrolene-binding sequence on the skeletal muscle ryanodine receptor. J. Biol. Chem. 277, 34918–34923 [DOI] [PubMed] [Google Scholar]
- 70. Zhao F., Li P., Chen S. R., Louis C. F., Fruen B. R. (2001) Dantrolene inhibition of ryanodine receptor Ca2+ release channels. Molecular mechanism and isoform selectivity. J. Biol. Chem. 276, 13810–13816 [DOI] [PubMed] [Google Scholar]
- 71. Jiang D., Chen W., Xiao J., Wang R., Kong H., Jones P. P., Zhang L., Fruen B., Chen S. R. (2008) Reduced threshold for luminal Ca2+ activation of RyR1 underlies a causal mechanism of porcine malignant hyperthermia. J. Biol. Chem. 283, 20813–20820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Quane K. A., Healy J. M., Keating K. E., Manning B. M., Couch F. J., Palmucci L. M., Doriguzzi C., Fagerlund T. H., Berg K., Ording H. (1993) Mutations in the ryanodine receptor gene in central core disease and malignant hyperthermia. Nat. Genet. 5, 51–55 [DOI] [PubMed] [Google Scholar]
- 73. Zhang Y., Chen H. S., Khanna V. K., De Leon S., Phillips M. S., Schappert K., Britt B. A., Browell A. K., MacLennan D. H. (1993) A mutation in the human ryanodine receptor gene associated with central core disease. Nat. Genet. 5, 46–50 [DOI] [PubMed] [Google Scholar]
- 74. De Crescenzo V., Fogarty K. E., Lefkowitz J. J., Bellve K. D., Zvaritch E., MacLennan D. H., Walsh J. V., Jr. (2012) Type 1 ryanodine receptor knock-in mutation causing central core disease of skeletal muscle also displays a neuronal phenotype. Proc. Natl. Acad. Sci. U.S.A. 109, 610–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Capacchione J. F., Sambuughin N., Bina S., Mulligan L. P., Lawson T. D., Muldoon S. M. (2010) Exertional rhabdomyolysis and malignant hyperthermia in a patient with ryanodine receptor type 1 gene, L-type calcium channel α-1 subunit gene, and calsequestrin-1 gene polymorphisms. Anesthesiology 112, 239–244 [DOI] [PubMed] [Google Scholar]
- 76. Ferreiro A., Monnier N., Romero N. B., Leroy J. P., Bönnemann C., Haenggeli C. A., Straub V., Voss W. D., Nivoche Y., Jungbluth H., Lemainque A., Voit T., Lunardi J., Fardeau M., Guicheney P. (2002) A recessive form of central core disease, transiently presenting as multiminicore disease, is associated with a homozygous mutation in the ryanodine receptor type 1 gene. Ann. Neurol. 51, 750–759 [DOI] [PubMed] [Google Scholar]
- 77. Zhou H., Lillis S., Loy R. E., Ghassemi F., Rose M. R., Norwood F., Mills K., Al-Sarraj S., Lane R. J., Feng L., Matthews E., Sewry C. A., Abbs S., Buk S., Hanna M., Treves S., Dirksen R. T., Meissner G., Muntoni F., Jungbluth H. (2010) Multiminicore disease and atypical periodic paralysis associated with novel mutations in the skeletal muscle ryanodine receptor (RYR1) gene. Neuromuscul. Disord. 20, 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Priori S. G., Napolitano C., Tiso N., Memmi M., Vignati G., Bloise R., Sorrentino V., Danieli G. A. (2001) Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 103, 196–200 [DOI] [PubMed] [Google Scholar]
- 79. Muthappan P., Calkins H. (2008) Arrhythmogenic right ventricular dysplasia. Prog. Cardiovasc. Dis. 51, 31–43 [DOI] [PubMed] [Google Scholar]
- 80. Lehnart S. E., Mongillo M., Bellinger A., Lindegger N., Chen B. X., Hsueh W., Reiken S., Wronska A., Drew L. J., Ward C. W., Lederer W. J., Kass R. S., Morley G., Marks A. R. (2008) Leaky Ca2+ release channel/ryanodine receptor 2 causes seizures and sudden cardiac death in mice. J. Clin. Invest. 118, 2230–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Robinson R., Carpenter D., Shaw M. A., Halsall J., Hopkins P. (2006) Mutations in RYR1 in malignant hyperthermia and central core disease. Hum. Mutat. 27, 977–989 [DOI] [PubMed] [Google Scholar]
- 82. Jiang D., Chen W., Wang R., Zhang L., Chen S. R. (2007) Loss of luminal Ca2+ activation in the cardiac ryanodine receptor is associated with ventricular fibrillation and sudden death. Proc. Natl. Acad. Sci. U.S.A. 104, 18309–18314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jiang D., Wang R., Xiao B., Kong H., Hunt D. J., Choi P., Zhang L., Chen S. R. (2005) Enhanced store overload-induced Ca2+ release and channel sensitivity to luminal Ca2+ activation are common defects of RyR2 mutations linked to ventricular tachycardia and sudden death. Circ. Res. 97, 1173–1181 [DOI] [PubMed] [Google Scholar]
- 84. Tong J., Oyamada H., Demaurex N., Grinstein S., McCarthy T. V., MacLennan D. H. (1997) Caffeine and halothane sensitivity of intracellular Ca2+ release is altered by 15 calcium release channel (ryanodine receptor) mutations associated with malignant hyperthermia and/or central core disease. J. Biol. Chem. 272, 26332–26339 [DOI] [PubMed] [Google Scholar]
- 85. Jiang D., Xiao B., Zhang L., Chen S. R. (2002) Enhanced basal activity of a cardiac Ca2+ release channel (ryanodine receptor) mutant associated with ventricular tachycardia and sudden death. Circ. Res. 91, 218–225 [DOI] [PubMed] [Google Scholar]
- 86. Oda T., Yano M., Yamamoto T., Tokuhisa T., Okuda S., Doi M., Ohkusa T., Ikeda Y., Kobayashi S., Ikemoto N., Matsuzaki M. (2005) Defective regulation of interdomain interactions within the ryanodine receptor plays a key role in the pathogenesis of heart failure. Circulation 111, 3400–3410 [DOI] [PubMed] [Google Scholar]
- 87. Liu Z., Wang R., Tian X., Zhong X., Gangopadhyay J., Cole R., Ikemoto N., Chen S. R., Wagenknecht T. (2010) Dynamic, inter-subunit interactions between the N-terminal and central mutation regions of cardiac ryanodine receptor. J. Cell Sci. 123, 1775–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wehrens X. H., Lehnart S. E., Huang F., Vest J. A., Reiken S. R., Mohler P. J., Sun J., Guatimosim S., Song L. S., Rosemblit N., D'Armiento J. M., Napolitano C., Memmi M., Priori S. G., Lederer W. J., Marks A. R. (2003) FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell 113, 829–840 [DOI] [PubMed] [Google Scholar]
- 89. Wang R., Chen W., Cai S., Zhang J., Bolstad J., Wagenknecht T., Liu Z., Chen S. R. (2007) Localization of an NH2-terminal disease-causing mutation hot spot to the “clamp” region in the three-dimensional structure of the cardiac ryanodine receptor. J. Biol. Chem. 282, 17785–17793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Amador F. J., Liu S., Ishiyama N., Plevin M. J., Wilson A., MacLennan D. H., Ikura M. (2009) Crystal structure of type I ryanodine receptor amino-terminal β-trefoil domain reveals a disease-associated mutation “hot spot” loop. Proc. Natl. Acad. Sci. U.S.A. 106, 11040–11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lobo P. A., Van Petegem F. (2009) Crystal structures of the N-terminal domains of cardiac and skeletal muscle ryanodine receptors: insights into disease mutations. Structure 17, 1505–1514 [DOI] [PubMed] [Google Scholar]
- 92. Lobo P. A., Kimlicka L., Tung C. C., Van Petegem F. (2011) The deletion of exon 3 in the cardiac ryanodine receptor is rescued by β-strand switching. Structure 19, 790–798 [DOI] [PubMed] [Google Scholar]
- 93. Tung C. C., Lobo P. A., Kimlicka L., Van Petegem F. (2010) The amino-terminal disease hot spot of ryanodine receptors forms a cytoplasmic vestibule. Nature 468, 585–588 [DOI] [PubMed] [Google Scholar]
- 94. Bhuiyan Z. A., van den Berg M. P., van Tintelen J. P., Bink-Boelkens M. T., Wiesfeld A. C., Alders M., Postma A. V., van Langen I., Mannens M. M., Wilde A. A. (2007) Expanding spectrum of human RYR2-related disease: new electrocardiographic, structural, and genetic features. Circulation 116, 1569–1576 [DOI] [PubMed] [Google Scholar]
- 95. Yuchi Z., Lau K., Van Petegem F. (2012) Disease mutations in the ryanodine receptor central region: crystal structures of a phosphorylation hot spot domain. Structure 20, 1201–1211 [DOI] [PubMed] [Google Scholar]
- 96. Tae H. S., Cui Y., Karunasekara Y., Board P. G., Dulhunty A. F., Casarotto M. G. (2011) Cyclization of the intrinsically disordered α1S dihydropyridine receptor II-III loop enhances secondary structure and in vitro function. J. Biol. Chem. 286, 22589–22599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hamilton S. L., Serysheva I. I. (2009) Ryanodine receptor structure: progress and challenges. J. Biol. Chem. 284, 4047–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ludtke S. J., Tran T. P., Ngo Q. T., Moiseenkova-Bell V. Y., Chiu W., Serysheva I. I. (2011) Flexible architecture of IP3R1 by cryo-EM. Structure 19, 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Seo M. D., Velamakanni S., Ishiyama N., Stathopulos P. B., Rossi A. M., Khan S. A., Dale P., Li C., Ames J. B., Ikura M., Taylor C. W. (2012) Structural and functional conservation of key domains in InsP3 and ryanodine receptors. Nature 483, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lin C. C., Baek K., Lu Z. (2011) Apo and InsP3-bound crystal structures of the ligand-binding domain of an InsP3 receptor. Nat. Struct. Mol. Biol. 18, 1172–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]