Background: α-Synuclein accumulation alters adult neurogenesis by down-regulation of Notch1.
Results: p53 is a negative regulator of Notch1 transcription, and interaction with α-synuclein further represses Notch1 expression.
Conclusion: p53 mediates the α synuclein-induced alterations of adult neurogenesis.
Significance: This might be the first link between p53 effects and aberrant neurogenesis in synucleopathies.
Keywords: Alpha-Synuclein, Notch, p53, Parkinson Disease, Transcription Regulation, Adult Neurogenesis
Abstract
Parkinson disease is characterized by the loss of dopaminergic neurons mainly in the substantia nigra. Accumulation of α-synuclein and cell loss has been also reported in many other brain regions including the hippocampus, where it might impair adult neurogenesis, contributing to nonmotor symptoms. However, the molecular mechanisms of these alterations are still unknown. In this report we show that α-synuclein-accumulating adult rat hippocampus neural progenitors present aberrant neuronal differentiation, with reduction of Notch1 expression and downstream signaling targets. We characterized a Notch1 proximal promoter that contains p53 canonical response elements. In vivo binding of p53 represses the transcription of Notch1 in neurons. Moreover, we demonstrated that α-synuclein directly binds to the DNA at Notch1 promoter vicinity and also interacts with p53 protein, facilitating or increasing Notch1 signaling repression, which interferes with maturation and survival of neural progenitors cells. This study provides a molecular basis for α-synuclein-mediated disruption of adult neurogenesis in Parkinson disease.
Introduction
Parkinson disease (PD)2 is the second most common neurodegenerative disorder in the elderly, affecting 2% of the population over 60 years old (1). The classic form of PD is manifested clinically as a slowly progressive movement disorder with resting tremor and postural instability. Neuropathologically, PD is characterized by the loss of dopaminergic neurons mainly in the substantia nigra pars compacta accompanied by the formation of intracytoplasmic inclusions known as Lewy bodies, containing α-synuclein (α-syn) (2, 3). Whereas neurodegeneration in PD affects primarily the striatonigral system (4), more widespread degeneration is observed in cases with cognitive impairment, affecting neuronal populations in the striatum, hippocampus, and neocortex (5, 6). Accumulation of α-syn in these structures affects nonmotor functions such as neurogenesis and olfaction (7), contributing to PD pathology.
Neurogenesis in the adult brain occurs in the subventricular zone/olfactory bulb region and in the hippocampus (8), where neural precursor cells (NPCs) of the subgranular zone generate new neuronal populations in the dentate gyrus contributing to its role in learning and memory (8–11). We previously demonstrated that accumulation of α-syn results in reduced neurogenesis in the olfactory bulb and in the hippocampus of adult transgenic mice expressing human α-syn, mainly because of diminished survival of NPCs in neurogenic regions (12, 13). More recently, we showed aberrant neurogenesis in α-syn-overexpressing mouse embryonic stem cells and in the dentate gyrus of α-syn transgenic mice, an effect that was accompanied by down-regulation of Notch1 expression (14).
Notch1 signaling has profound effects on the development of the nervous system, including maintenance of stem cell self-renewal, proliferation, neuronal differentiation, and glial determination (15). Notch1 inhibits the differentiation of neurons and astrocytes from their precursors, coordinating the temporal maturation during brain development (16). Modulation of Notch1 levels is implicated in the regulation of proliferation in the adult dentate gyrus (17), in neurogenic responses in epilepsy (18), and as a compensatory mechanism in neurodegeneration (19, 20). Several components of Notch1 signaling are highly expressed in the adult hippocampus, suggesting the role of this pathway in adult neurogenesis. These discoveries, in addition to our previous findings of reduced neurogenesis in PD models, prompted us to investigate the effects of abnormal α-syn accumulation in a model of mammalian adult neurogenesis.
In the present work we report abnormal neuronal differentiation of adult rat hippocampus NPCs (ARH-NPCs) that overexpress α-syn, mediated by a reduction in the expression of Notch1 and its downstream targets Hes1 and Hes5. While investigating the molecular mechanisms responsible for Notch1 down-regulation in ARH-NPCs, we characterized a proximal promoter region of Notch1 that contains conserved elements for p53 binding. We show that p53 binds to these response elements in vivo to repress the transcription driven by the Notch1 promoter. Moreover, we present evidence for the direct binding of α-syn to the DNA and for the interaction of α-syn and p53 proteins, events that contribute to repress Notch1 transcription. The identification of the interactions between p53 and α-syn might shed light on the molecular mechanisms by which α-syn aggregation contributes to the disruption of adult neurogenesis in PD and related synucleopathies.
EXPERIMENTAL PROCEDURES
Cell Culture
Adult rat hippocampal NPCs (generously provided by Dr. F. Gage, Salk Institute, La Jolla, CA) were cultured routinely for expansion essentially as described previously (21) with some modifications (22). Briefly, cells were grown for expansion in DMEM/F-12 medium (Mediatech, Manassas, VA) containing B27 supplement, 1× l-glutamine, and 1× penicillin/streptomycin (all from Invitrogen). For induction of neuronal differentiation, the cells were plated onto polyornithine/laminin-coated (Sigma-Aldrich) plates or coverslips and transferred the next day to differentiation media containing N2 supplement (Invitrogen), 1 μm all-trans-retinoic acid (Sigma-Aldrich), 5 μm forskolin (Sigma-Aldrich), and 1% FBS. Except when otherwise stated, the cells were differentiated for 4 days, and fresh differentiation medium was added at day 2. The cells were grown at 37 °C in 5% CO2, 95% air.
Production of Lentiviral (LV) Constructs
Vector plasmids were constructed for the production of third generation LV vectors that expressed wild type human α-syn (LV-α-syn), Myc-tagged human α-syn (LV-myc-α-syn), rat siRNA for p53 (LV-si p53), rat siRNA for α-syn (LV-shRNA-α-synuclein), siRNA for luciferase (LV-si Luc), or the empty plasmid backbone as a control (LV-CT/LV-control). The human CMV promoter was used to drive expression of the transgenes. Lentiviral vectors were generated by transient transfection with the vector and packaging plasmids in 293T cells as described previously (23).
Lentivirus-mediated Infection of Cells
For delivery of α-syn or the desired siRNAs, the LV system was utilized. The day before infection, 3 × 106 cells were plated on coated dishes in exponential medium. After 1 day, the medium was changed, and the corresponding lentiviral constructs were added to the dishes at a multiplicity of infection (MOI) of 30 (unless otherwise indicated). The cells were incubated for 24–72 h in a 5% CO2, 95% air at 37 °C.
Immunohistochemistry
Briefly, as described previously (12, 13), cells grown on coated glass coverslips were fixed in 4% paraformaldehyde and treated with 0.6% H2O2 in TBS (0.15 m NaCl, 0.1 m Tris-HCl, pH 7.5) for 30 min and incubated with antibodies against doublecortin (1:100; Santa Cruz Biotechnologies, Santa Cruz, CA), tubulin III (Tuj1 clone, 1:500; Chemicon, Temecula, CA), GFP (1:250; Millipore, Temecula, CA), α-syn (1:500; Chemicon), Notch1 (1:350; Santa Cruz), or p53 (1:500; Santa Cruz) overnight at 4 °C. The cells were incubated for 1 h with biotinylated secondary antibodies directed against mouse, goat, or rabbit. Following rinses in TBS, avidin-biotin-peroxidase complex was applied for 1 h, and then peroxidase detection was performed for 10 min (25 mg/ml diaminobenzidine, 0.01% H2O2, 0.04% NiCl in TBS).
Double Immunocytochemical Analysis and Confocal Microscopy
Neuronal differentiated ARH-NPCs grown on coverslips were double labeled with polyclonal antibodies against α-syn and Notch1 (1:1000) and detected with the Tyramide Signal AmplificationTM-Direct (Red) system (1:100; PerkinElmer Life Sciences), and the mouse monoclonal antibodies against tubulin III (Tuj1 clone; 1:100) or p53 (1:500), detected with FITC-conjugated secondary antibodies (1:75; Vector Laboratories, Burlingame, CA) (24). All sections were processed simultaneously under the same conditions, and experiments were performed twice to assess reproducibility. Sections were imaged with a Zeiss 63× (N.A. 1.4) objective on an Axiovert 35 microscope (Zeiss) with an attached MRC1024 LSCM system (Bio-Rad) (24). To confirm the specificity of primary antibodies, control sections were incubated overnight in the absence of primary antibody (deleted) or preimmune serum and primary antibody alone.
Real Time PCR Analysis
Real time PCR experiments were performed using the iCycler iQ5 real time PCR detection system (Bio-Rad) as described previously (25). Total RNA was extracted from cultured cells using the RNeasy mini kit (Qiagen) and was reverse transcribed using iScript cDNA synthesis kit (Bio-Rad) with 1 μg of total RNA. Amplification was performed on a cDNA amount equivalent to 100 ng of total RNA with the iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions. PCRs were performed on four independent sets of templates. Specific primers for each studied sequence and for endogenous controls were designed using Oligo perfect software (Invitrogen), and their specificity for binding to the desired sequences was searched against NCBI database (supplemental Table S1). Standard curves were generated for each gene of interest using serial dilutions of cDNAs. Experimental samples and no-template controls were all run in duplicate. The PCR cycling parameters were: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 94 °C for 15 s, and 60 °C for 1 min. Finally, a dissociation protocol was also performed at the end of each run to verify the presence of a single product with the appropriate melting point temperature for each amplicon. The amount of studied cDNA in each sample was calculated using Bio-Rad software by the comparative threshold cycle (Ct) method and expressed as 2exp(Ct) using β-actin as an internal control. For calculations applying the Ct method, the cells infected with empty lentivirus (LV-CT) were used as calibrator samples.
Western Blots
After washing with PBS, the cells were harvested and disrupted in cell lysis buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 50 mm NaF, 1 mm Na3VO4, 1% Triton X-100) supplemented with protease inhibitor mixture (Calbiochem, San Diego, CA). The lysed samples were transferred to microcentrifuge tubes, incubated on ice for 20 min, and then cleared by centrifugation (13,000 × g, 2 min) at 4 °C. Lysate protein concentration was measured by the bicinchoninic acid assay (Pierce). For electrophoretic analysis, 4× SDS sample buffer was added to cell lysates. The samples were loaded on SDS-polyacrylamide gels for electrophoresis and subsequently transferred onto polyvinylidene fluoride membranes (Invitrogen). After blocking with 3% BSA, 0.01% Tween 20, PBS, the membranes were incubated with gentle agitation overnight at 4 °C with the specific primary antibodies against β-III tubulin (tubulin III, Tuj1 clone, 1:1000; Chemicon), α-syn (1:500; Chemicon), full-length Notch1 (1:1000; Santa Cruz), p53 (1:500; Active Motif, Carlsbad, CA), doublecortin (1:100; Santa Cruz), GFAP (1:500; Chemicon), S100 (1:250; Sigma), or Nestin, PCNA, Hes1, Hes5, anti-Myc, or β-actin (all 1:1000; Chemicon). After washing, the membranes were incubated with HRP-conjugated secondary antibody for 1 h at room temperature. Detection was performed using the VersaDoc imaging system (Bio-Rad).
Electrophoretic Mobility Shift Assay
RE900 and RE500 DNA fragments were generated by PCR amplification from gDNA extracted from B103 rat neuroblastoma cells using a DNAeasy blood and tissue kit (Qiagen) and using the following primers: RE900F, 5′-ggtgcagtatgaggtcaggg-3′ and RE900R 5′-gaaggggctattaatcaagc-3′; RE500F, 5′-cgagaatagggtggaattcc-3′ and RE500R 5′-caacctccacccttgcttca-3′, under standard PCR conditions and using Faststart Taq DNA polymerase (Roche Applied Science). The EMSA kit (Invitrogen) was used according to the manufacturer's instructions. 100 ng of each DNA template were incubated with 2 or 5 μg of human recombinant α-syn (EMD, La Jolla, CA) in 1× binding buffer for 30 min at room temperature prior to electrophoresis through a 6% agarose gel run in 0.5× TBE. DNA was visualized by SYBR® green staining, and detection was performed using the VersaDoc imaging system (Bio-Rad). To control for specific binding of α-syn to the DNA, a similar EMSA was run using 100 ng of a double-stranded randomized oligonucleotide with or without preincubation with human α-syn.
Chromatin Immunoprecipitation Assay
ChIP assays were performed using the ChIP-Enzymatic Express kit as recommended by the manufacturer (Active Motif) with slight modifications. Briefly, 4.5 × 107 rat neuroblastoma B103 cells were infected with LV-α-syn or LV-CT at MOI 30. After 48 h of incubation, the cells were cross-linked with 1% formaldehyde at room temperature for 5 min, repeatedly washed with ice-cold PBS, and lysed using a Dounce homogenizer followed by centrifugation. The nuclear pellet was resuspended in enzymatic shearing mixture and digested at 37 °C for 10 min to shear DNA. Ten percent of the mixture of protein-DNA complex was taken for “input DNA” analysis. An equal amount of the protein-DNA complex (7 μg) was then incubated with 2 μg of rat p53 antibody (Active Motif) or 2 μg of human α-syn antibody (Chemicon) and protein G magnetic beads at 4 °C overnight in a rotor. Immunoprecipitated DNA was eluted from protein G beads, cross-linking was reversed, and the DNA was purified. The rat Notch1 promoter region corresponding to bases −1002 and −890, comprising the putative p53 RE900, or the region containing residues −580 to −432 covering RE500 (GenBankTM accession number NM_001105721) were PCR-amplified using the following primers: RE900F, 5′-ggtgcagtatgaggtcaggg-3′; RE900R, 5′-ggcgcaggatttccaaactt-3′; RE500F, 5′-atgcctgcgaacaggtattg-3′; and RE500R, 5′-cgtgggaaaaagcgacagct-3′. In addition, 7 μg of chromatin were immunoprecipitated with 2 μg of rat IgG antibody (negative control) or 2 μg of rat RNA polymerase II antibody (positive control), and the eluted DNA was amplified using the Notch1 promoter primers (negative) or β-actin control primers (positive) from a Chip-IT Control kit (Active Motif) (data not shown). The PCR amplification was performed for 35 cycles under standard conditions using the Faststart Taq DNA polymerase (Roche Applied Science). Detection of DNA was performed using the VersaDoc imaging system (Bio-Rad).
Luciferase Assay
Rat neuroblastoma B103 cells were plated at a density of 4 × 105 cells/well in 6-well tissue culture plates. The following day the cells were infected with LV-α-syn or LV-CT at MOI 30. 24 h post-infection, the cells were transfected with p53 expression vector (full-length cDNA cloned into pExpress plasmid (Open Biosciences)) and/or reporter plasmids (2 μg of total DNA) containing rat Notch1 promoter fragments NP1002 comprising bases −1002 to +81 (relative to TSS) or NP580 comprising bases −580 to +81, subcloned into pGLuc Basic reporter vector (New England Biolabs, Ipswich, MA), which contains the “humanized” coding sequence for the secreted Gaussia luciferase (GLuc) without a promoter. Empty pGLuc vector was used as control. Transfections were carried out using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions. 48 h after transfection, the cells were analyzed for luciferase activity. For luciferase assays, 25 μl of cell medium containing exported luciferase was incubated with 50 μl of luciferase assay substrate and buffer (New England Biolabs) in 96-well plates. Luciferase was measured at 530 nm in a DTX 880 Multimode Detector (Beckman Coulter, Fullerton, CA). Assays were performed in triplicate, and luciferase expression was normalized to the number of viable cells (determined in 15,000 cells at the end of the assay by PrestoblueTM cell viability reagent (Invitrogen) as per the manufacturer's instructions).
Co-immunoprecipitation Experiments
For the co-immunoprecipitation of interacting proteins, NPC-derived neuronal progeny were infected with LV-Myc-α-syn lentivirus at MOI 30 and treated with 0.5 μg/ml of doxorubicin (G Biosciences, St. Louis, MO) for 24 h to induce p53 expression (26). 48 h post-infection the cells were washed with PBS, and the nuclear protein fraction was extracted using the Epiquik nuclear extraction kit (Epigentek, Farmingdale, NY) as per the manufacturer's instructions. Aliquots containing 650 μg of protein were brought to a final volume of 500 μl with NET-BSA buffer (50 mm Tris/HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.1% (v/v) Nonidet P-40, 0.25% w/v BSA, and 0.02% sodium azide). Homogenates were incubated for 3 h with either 1 μg of rat p53 antibody (Active Motif), 1 μg of human α-syn antibody (Chemicon), or 1 μg of a normal rabbit IgG (Santa Cruz Biotechnology) as control, on a rocker at 4 °C. 40 μl of protein A-Sepharose with PBS 50:50 (v/v) was added to the homogenates and incubated overnight on a rocker at 4 °C. Collection of the conjugates was done by centrifuging the samples at 12,000 × g for 30 s and washing pellets for 10 min in a rocker at 4 °C using 1 ml of NET-BSA buffer and finally with 1 ml of 10 mm Tris/HCl, pH 7.5, 0.1% Nonidet P-40. Protein-Sepharose A beads were resuspended in 50 mm Tris/HCl, pH 6.5, 1× NuPAGE loading buffer (Invitrogen) and 1× reducing reagent (Invitrogen) and heated at 100 °C for 5 min before separation by SDS-PAGE and subsequent Western blotting.
Knockdown of p53 Expression
For specific knockdown of p53 expression, we generated a lentiviral construct containing the shRNA directed against rat p53 with the following sequence: 5′-GTACATGTGCAACAGCTCC-3′, subcloned in the p156RRL vector backbone and under the H1 promoter (LV-si p53). Rat neuroblastoma B103 cells and NPC-derived neuronal progeny were infected with LV-si p53 at MOI 30. p53 expression was monitored at 24-h intervals for 4 days post-infection to determine the optimal time point for protein knockdown. At 24 h p53 was reduced to 48% of control cells, and at 48 h the expression was ∼36% of the untreated cells. Because longer incubation did not yield significant reductions on p53 beyond the levels reached at 48 h, that was the time point used in Notch1 expression assays.
Knockdown of Rat Endogenous α-Syn Expression
For specific knockdown of α-syn expression, we generated a lentiviral construct containing the shRNA directed against rat α-syn with the sequence: 5′-GAAGGACCAGATGGGCAAG-3′, subcloned in the pSIH1-copGFP vector backbone under the H1 promoter and containing GFP as reporter (LV-sh-α-synuclein). ARH-NPCs were infected with LV-sh-α-synuclein MOI 30, and α-syn expression as monitored at 24-h intervals for 4 days post-infection by Western blot to determine the optimal time point for protein knockdown (not shown). At 24 h α-syn levels were 70% of control cells, and at 48 h the expression was ∼54% of the untreated cells. Longer incubation times did not yield significant increase in α-syn knockdown; therefore shRNA experiments were conducted 48 h post-infection.
Statistics
Student's t test was used to determine differences in expression of studied genes between control and α-syn-expressing cells. One-way ANOVA followed by Tukey's post-tests was used for multiple comparisons, and Student's t test was used for determination of exact p values. All statistical tests were performed using GraphPad software (GraphPad Prism, San Diego, CA).
RESULTS
Accumulation of α-Synuclein Impairs Neuronal Differentiation and Notch1 Signaling in Adult Rat Hippocampal Neural Progenitor Cells
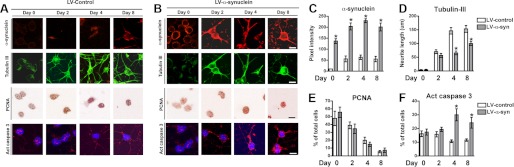
We have previously reported that accumulation of α-syn results in aberrant neurogenesis in both mouse embryonic stem cells and adult α-syn transgenic mice (14). To gain further insight into the mechanisms underlying adult neurogenesis, we investigated the effects of α-syn overexpression in the neuronal differentiation using an in vitro cell culture model. Cultured ARH-NPCs were infected with lentiviral constructs containing either human α-syn (LV-α-syn) or empty vector (LV-control) and were differentiated into neuronal cells during 8 days (Fig. 1). Significant overexpression of α-syn was evident in the cell body and neurites of LV-α-syn-infected cells, which was accompanied by reduced tubulin expression, shorter neurite length and slight increase of proliferating cell nuclear antigen (PCNA) expression in the NPC-derived neuronal progeny (Fig. 1, A–E). In addition, α-syn-overexpressing cells showed higher levels of active caspase 3, indicating ongoing apoptosis (Fig. 1, A, B, and F). Taken together, these alterations are suggestive of reduced neuronal differentiation and survival. Detailed analysis of proliferation and maturation markers, as well as markers for neuronal and glial cell types in the NPC-derived progeny at different time points during the neuronal differentiation protocol indicated that at day 4 our in vitro system contains mainly neuronal phenotypes; therefore, we continued the present work with an abbreviated neuronal differentiation protocol of 4 days (supplemental Fig. S1, A–C).
FIGURE 1.
α-Synuclein impairs neuronal maturation and survival in adult rat hippocampus neural progenitor cells. A and B, adult rat hippocampal NPCs were infected at day 0 with a lentiviral vector expressing human α-syn (LV-α-synuclein) or an empty vector as control (LV-Control) at a MOI of 30. Immunohistochemical analysis of the expression of α-syn, tubulin III (as a marker of neuronal maturation), PCNA (as a marker of proliferation), and active caspase 3 (apoptosis marker) was performed at days 0, 2, 4, and 8 of neuronal differentiation. Compared with LV-control, the NPC-derived neural progeny from LV-α-syn-infected cells showed shorter β-tubulin-immunoreactive neuritis, a slight increase of PCNA by day 8, and increased apoptosis. Scale bar, 10 μm. C–F, quantitative image analysis showing levels of α-syn, tubulin III, PCNA, and active caspase 3. *, p < 0.05 compared with LV-control-infected cells by one-way ANOVA with post hoc Tukey's test.
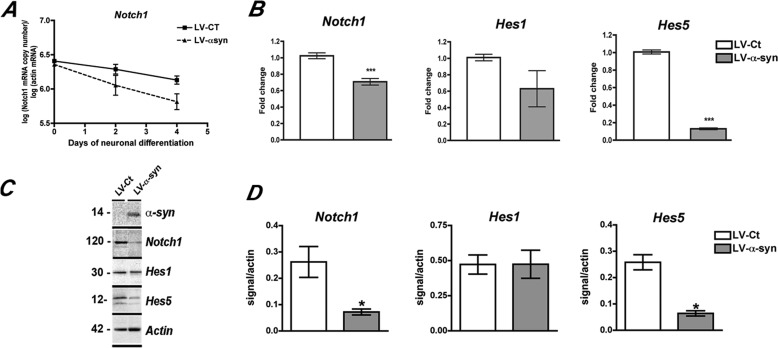
Notch1 signaling is known to be involved in fate determination and survival of progenitor cells. Because we previously reported that α-syn overexpression decreases Notch1 levels in mouse embryonic stem cells (14), we investigated whether Notch1 might also be altered by α-syn accumulation in the context of adult neurogenesis. As expected, real time quantitative PCR analysis showed a decrease in Notch1 as neuronal differentiation proceeds, but at day 4 of differentiation Notch1 mRNA levels were significantly lower in LV-α-syn-infected rat hippocampal NPCs that in control-infected cells (Fig. 2, A and B). Moreover, and evidencing the extent of α-syn effects on Notch1 signaling pathway, mRNA levels of two Notch1 downstream targets Hes1 and Hes5 were also altered, with Hes5 being significantly reduced in LV-α-syn-infected cells at day 4 of neuronal differentiation (Fig. 2B). Decreased mRNA expression of Notch1 signaling pathway components resulted in concomitant reductions of Notch1 and Hes5 proteins in cells accumulating α-syn (Fig. 2, C and D). These results show that overexpression of α-syn results in down-regulation of Notch1 and its downstream targets in the rat hippocampal NPCs, which might account for the abnormal neuronal differentiation/survival observed.
FIGURE 2.
Alterations in the expression of Notch1 signaling components in NPC-derived neural progeny expressing α-syn. A, absolute quantification of Notch1 mRNA levels throughout neuronal differentiation, showing decay of signal as neuronal differentiation progress. Notch1 levels were significantly lower in α-syn-overexpressing cells by day 4. B, quantitative real time PCR analysis of the mRNA levels of Notch1, Hes1, and Hes5 in NPC-derived neuronal progeny of cells infected with LV-α-syn in comparison with LV-control-infected cells at day 4. C, detection of α-syn, Notch1, Hes1, and Hes5 levels by Western blotting at day 4 of neuronal differentiation. D, image analysis showing integrated pixel intensity of the immunoreactivity of Notch1 signaling components. ***, p < 0.001 by paired Student's t test. *, p < 0.05 compared with LV-control-infected cells by one-way ANOVA with post hoc Tukey's test.
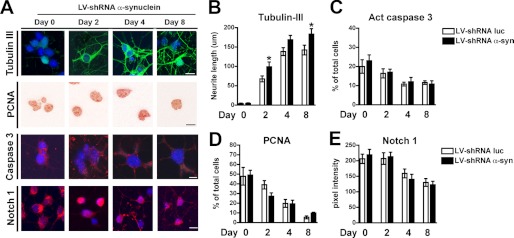
To further evaluate the role of endogenous α-syn in adult neurogenesis, we partially knocked down endogenous α-syn expression in ARH-NPCs by lentiviral-mediated delivery of a siRNA construct previously reported to effectively reduce in vivo the expression of α-syn in rat brains (27). Cells were infected 48 h prior to the starting of the neuronal differentiation protocol to allow for the decay of endogenous α-syn. Reduction of α-syn levels of up to 70% were obtained by day 4 of neuronal differentiation (supplemental Fig. S2). Knockdown of α-syn resulted in earlier appearance of neuronal phenotypes as evidenced by increased tubulin III expression (Fig. 3, A and B), whereas no significant changes in proliferation on caspase 3 activation were observed (Fig. 3, A, C, and D). Decreased α-syn slightly affected Notch1 signaling, with no significant changes detected at the protein level (Fig. 3, A and E) and with less than 10% reduction in Notch1 and 20% decrease in Hes5 mRNA levels (supplemental Fig. S3). These results suggest that α-syn expression is required by neuronal progenitors for the proper regulation of neurogenesis.
FIGURE 3.
Decreased endogenous α-synuclein levels alter neuronal maturation in adult rat hippocampus neural progenitor cells. A, ARH-NPCs were infected with a lentiviral vector expressing a shRNA directed to the rat α-syn (LV-shRNA-α-syn::GFP) or an shRNA specific for luciferase as control (LV-shRNA-Luc::GFP) at a MOI of 30. 48 h post-infection, when endogenous expression of α-syn reached the lowest level, neuronal differentiation was started (day 0). Immunohistochemical analysis of the expression of tubulin III (as a marker of neuronal maturation), PCNA (as a marker of proliferation), active caspase 3 (apoptosis marker), and Notch1 was performed at days 0, 2, 4, and 8 of neuronal differentiation. Compared with LV-shRNA-Luc, the NPC-derived neural progeny from LV-shRNA-α-syn-infected cells showed an earlier appearance of more mature neuronal phenotypes as evidenced by longer β-tubulin-immunoreactive neurites, whereas no significant changes were detected in proliferation, apoptosis, or Notch1 expression. Scale bar, 10 μm. B–E, quantitative image analysis showing levels of tubulin III, PCNA, active caspase 3, and Notch1. *, p < 0.05 compared with LV-shRNA-Luc-infected cells by one-way ANOVA with post hoc Tukey's test.
p53 Is a Negative Regulator of Notch1 Transcription in Adult Rat NPCs
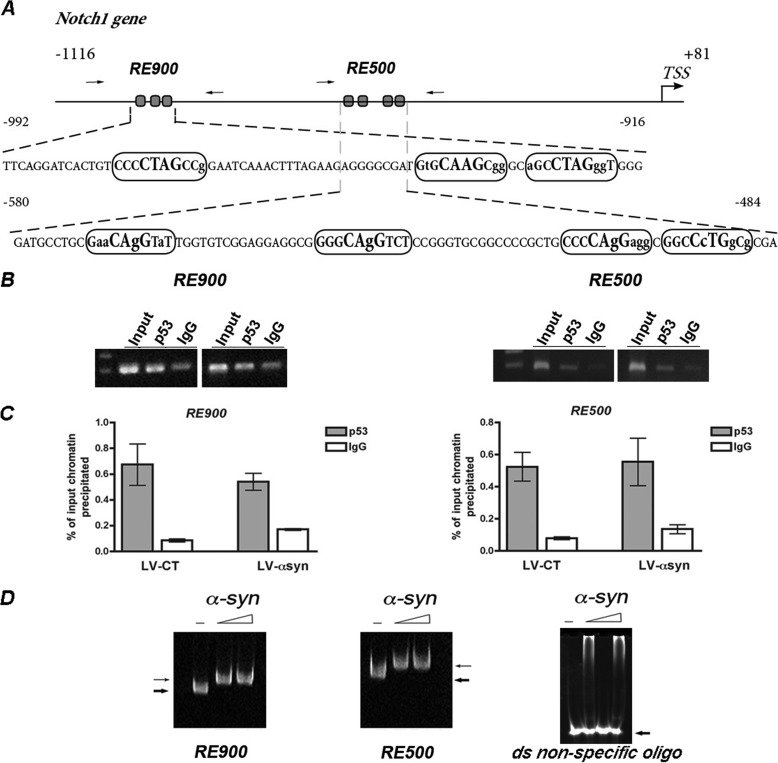
We next explored the mechanisms by which α-syn might disrupt Notch1 transcription. As a first step in the characterization of the Notch1 promoter in rat hippocampal NPCs, we retrieved a putative regulatory region upstream of the Notch1 coding sequence, containing nucleotides −1116 to + 81 relative to the translation start site (Fig. 4A). In silico analysis of this promoter region using SiteSeer software (28) revealed the presence of consensus sites for binding of multiple transcription factors. We focused our analysis on the most promising candidate: p53, which has been reported to directly regulate Notch1 transcription in epithelial cells (29). p53 binds to the DNA at p53 response elements (REs) composed of two decamer motifs (RRRCWWGYYY, where W is A/T, R is C/G, and Y is C/T) spaced by a stretch of 0–12 nucleotides (30). We identified at least two p53 REs in the Notch1 promoter with high homology to canonical sites (Fig. 4A). To further analyze the regulation of Notch1 transcription in vivo, we performed chromatin immunoprecipitation assays using an anti-p53 antibody followed by PCR amplification of the regions comprising −1002 to −890 (RE900) and −580 to −432 (RE500) of the rat Notch1 promoter. The B103 rat neuroblastoma cell line was chosen for these experiments because it shares many typical neuronal properties with other commonly used neuronal cell lines, including outgrowth of neurites upon differentiation, synthesis of neurotransmitters, possession of neurotransmitter receptors, and electrical excitability of surface membranes (31). ChIP assays showed binding of p53 to both RE900 and RE500, suggesting their functionality in vivo. Moreover, overexpression of α-syn seems to have no effect on p53 binding (Fig. 4, B and C). α-Synuclein has been reported to bind directly to the DNA at CG-rich regions (32). Although ChIP experiments using anti-α-syn antibody did not pull down the chromatin fragment containing the Notch REs, in vitro incubation of increasing amounts of human α-syn with either RE900 or RE500 was able to retard the migration of both DNA fragments in EMSA. Preincubation with α-syn did not alter the electrophoretic mobility of a double-stranded randomized oligonucleotide, indicating specific binding of α-syn to the Notch1 promoter vicinity in vitro (Fig. 4D).
FIGURE 4.
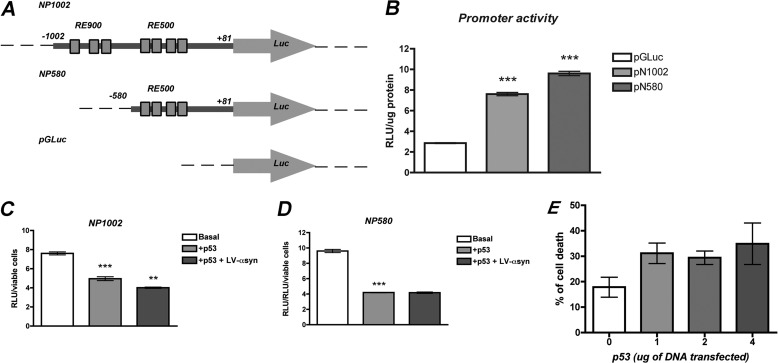
p53 and α-syn binding to Notch1 promoter. A, schematic representation of a proximal regulatory region in the rat Notch1 promoter depicting two potential p53 response elements (RE900 and RE500). Boxes show the sequence of the RE. The bold bigger font represents the core element, and bold smaller fonts show regulatory flanking sequences. Capital letters indicate conserved residues from the consensus site, and lowercase letters show deviation from consensus. Arrows indicate the approximate position of the primers used in PCR amplification. B, binding of endogenous p53 to the Notch1 promoter was assessed in vivo by ChIP assays in B103 cells infected with LV-control (LV-CT) or LV-α-syn. ChIP was performed using specific antibodies against p53 or control IgG. Input chromatin represents the portion of the enzymatic-sheared chromatin prior to immunoprecipitation. The immunoprecipitated chromatin was analyzed by PCR amplification with primers flanking each RE tested. C, quantification of the immunoprecipitated DNA was analyzed by quantitative real time PCR and is shown as a percentage of the total DNA input. D, EMSAs show shifts in mobility when increasing concentrations of human recombinant α-syn (2 and 5 μg) were incubated with both RE900 and RE500, suggesting direct binding to DNA. α-Synuclein did not alter the mobility of a control double-stranded randomized oligonucleotide.
As a transcriptional regulator, p53 can function either as activator or as repressor. Therefore, we explored how p53 modulates Notch1 transcription by generating Notch1 promoter-driven luciferase-reporter vectors. Two promoter fragments, NP1002 (extending from −1002 to + 81 and containing RE900 and RE500) and NP580 (containing the fragment −580 to +81 and only RE500), were subcloned upstream of the luciferase reporter gene in the pGLuc Basic vector (Fig. 5A). Transfection of B103 cells with NP1002 and NP580 resulted in more than 4-fold increase of luciferase expression over the promoterless construct (pGluc Basic) (Fig. 5B). To study the effects of p53, we measured luciferase expression in cells transfected with each promoter construct and with or without co-transfection of an expression plasmid harboring the full-length rat p53. Overexpression of p53 resulted in a significant decrease of luciferase from both NP1002 and NP580 promoters (Fig. 5, C and D). At the levels assayed, transfection with p53 expression vector moderately increased cell death by ∼10% over basal levels, most probably by induction of apoptosis. Luciferase expression was therefore normalized to viable cell number (Fig. 5E). These results showed a role of p53 as a negative regulator of Notch1 expression in neuronal progenitor cells.
FIGURE 5.
p53 is a negative regulator of Notch1 promoter-driven transcription and α-syn further represses its expression. A, schematic representation of Notch1 promoter fragments fused to luciferase-reporter vectors to test the effects of p53 on the transcription driven by Notch1 promoter. NP1002 expands from −1002 to + 81 and contains both RE900 and RE500. NP580 contains the fragment −580 to +81 and only RE500. Promoter-less vector pGLuc Basic was used as control of basal luciferase expression. B, transfection of 293T cells with NP1002 or NP580 resulted in a more than 4-fold increase of luciferase expression in comparison with pGluc control plasmid. ***, p < 0.001 by paired Student's t test. C and D, p53 represses the transcription from NP1002 (C) and NP580 (D) as observed by a decrease in luciferase expression in cells transfected with 2 μg of a plasmid expressing rat p53. Overexpression of α-syn by infection of the cells with LV-α-syn aggravates the repression of transcription mediated by p53 only for NP1002. ***, p < 0.001; **, p < 0.01 compared with control cells by one-way ANOVA with post hoc Tukey's test. E, transfection of 293T cells with increasing amounts of the p53-expression plasmid resulted in a 10% increase of cell death relative to mock-transfected cells.
We next evaluated a possible effect of α-syn on p53-mediated transcriptional repression of Notch1. Cells co-transfected with each promoter construct and p53 were also infected with LV-α-syn or LV-control (to account for possible fluctuations in expression caused by transcription factors recruitment by the lentivirus). Although α-syn binds to both p53 REs in vitro, overexpression of α-syn only enhanced p53-mediated repression of Notch1 from NP1002 promoter fragment in B103 cells (Fig. 5, C and D). This result highlights the functional relevance of RE900 for the interaction of α-syn and p53 regulatory effects and suggest that RE900 and RE500 might play different roles in the regulation of Notch1 transcription in vivo, where their activities might be affected by the binding of additional regulatory proteins.
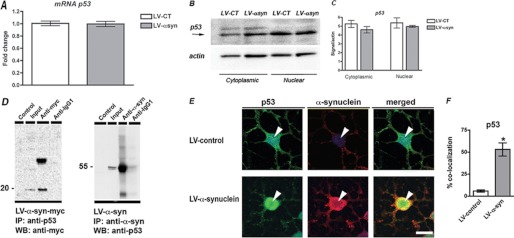
Overexpression of α-syn did not result in up-regulation of p53 expression, because levels of p53 mRNA remained unchanged in ARH-NPCs infected with LV-α-syn in comparison with control cells (Fig. 6A) and p53 protein levels in cytoplasmic and nuclear compartments were also indistinguishable between control and α-syn-overexpressing cells (Fig. 6, B and C). In a similar fashion, knockdown of endogenous α-syn did not affect the expression of p53 (supplemental Fig. S4).
FIGURE 6.
α-Synuclein physically interacts with p53. A, overexpression of α-syn in the neuronal progeny derived from ARH-NPCs does not alter p53 expression, as shown by quantitative PCR detection. B, Western blot analysis showing similar levels of p53 protein in the cytoplasmic as well as in the nuclear compartments of both ARH-NPCs infected with LV-α-syn and LV-control lentivirus. C, image analysis showing integrated pixel intensity of the immunoreactivity of p53. D, immunoprecipitation (IP) assays showed association of p53 with α-syn in ARH-NPCs infected with LV-α-syn-myc or LV-α-syn. The cells were treated with doxorubicin to increase p53 expression 24 h prior to lysis and nuclear protein extraction. E, p53 co-localizes with α-syn in ARH-NPCs infected with LV-α-syn. The bar represents 10 μm. F, image analysis showing percentages of p53 immunoreactive nuclei in which co-localization of α-syn was also detected. *, p < 0.05 by paired Student's t test. WB, Western blot.
p53 Is Associated with α-syn in Adult Rat NPCs
Because we observed that α-syn binds to Notch1 promoter near p53 REs in ARH-NPCs overexpressing α-syn contributing to Notch1 transcriptional repression without altering p53 levels, we hypothesized that p53 and α-syn proteins might directly interact. To investigate this hypothesis, we performed co-immunoprecipitation assays in rat adult hippocampal NPCs. The NPC-derived neuronal progeny was infected with a lentiviral construct expressing myc-tagged human α-syn (LV-α-syn-myc) and p53 expression was induced by doxorubicin treatment (26). Nuclear protein complexes immunoprecipitated with anti-p53 antibody revealed the presence of myc-α-syn, whereas the reverse immunoprecipitation with anti-α-syn antibody showed the presence of p53 in Western blot analysis (Fig. 6D). To determine whether this interaction holds true in vivo, we investigated their intracellular localization by immunohistochemistry. Co-localization of p53 and α-syn was evident in the nucleus of rat adult hippocampal NPCs (Fig. 6, E and F), supporting the in vivo association of these proteins.
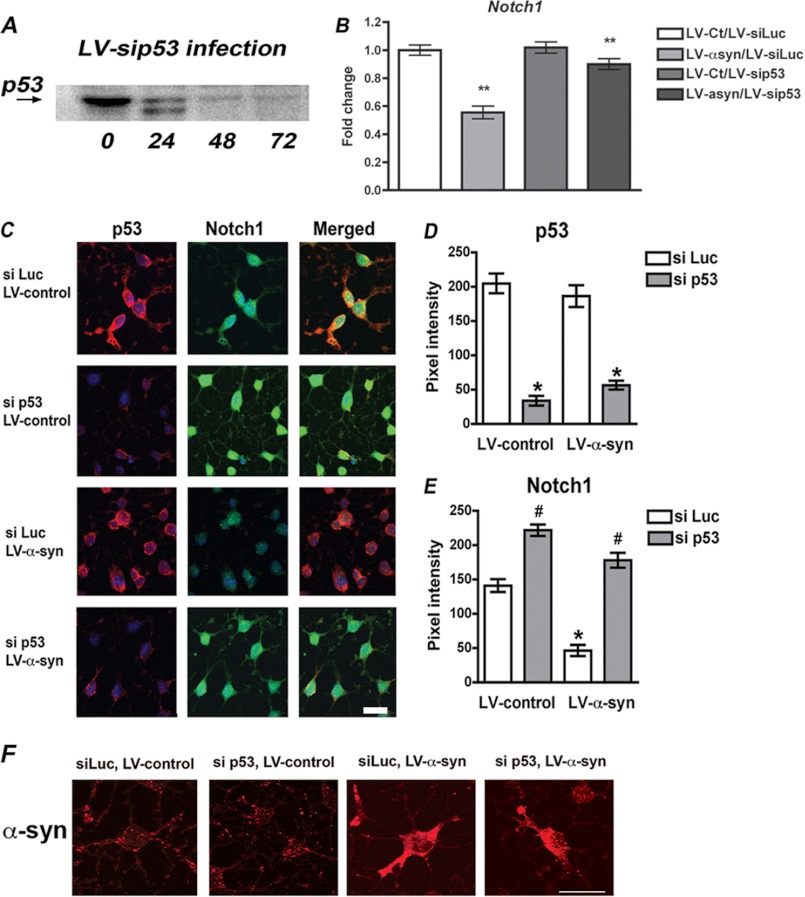
Notch1 Down-regulation Mediated by α-syn Can Be Rescued by Knockdown of p53
Finally, to confirm that down-regulation of Notch1 signaling as a result of α-syn accumulation in adult rat NPCs is mediated by p53 transcriptional repression, we monitored Notch1 expression in cells where p53 was knocked down by lentiviral delivery of rat-specific siRNA (Fig. 7A). Notch1 mRNA was quantified by real time PCR in cells infected with LV-α-syn with or without LV-si p53 or their corresponding control viruses (Fig. 7B). As described above, LV-α-syn infection resulted in significant decline of Notch1 mRNA levels. When p53 expression was knocked down by co-infection with LV-si p53, this reduction in Notch1 was almost completely restored to normal levels, despite α-syn accumulation. Immunohistochemical detection of Notch1 and p53 proteins in adult rat NPCs showed robust expression of both proteins in control conditions, whereas reduction of Notch1 levels was evident in LV-α-syn-infected cells (Fig. 7, C–E). Knockdown of endogenous p53 expression relieved the repression of Notch1 in α-syn accumulating cells, strongly suggesting that p53 mediates the effects of α-syn in the transcriptional repression of Notch1 in ARH-NPCs.
FIGURE 7.
Knockdown of p53 expression partially rescues the repression of Notch1 transcription induced by α-syn. A, delivery of specific siRNA directed toward rat p53 by lentiviral vectors (LV-si p53) to ARH-NPCs resulted in ∼65% reduction of p53 protein levels 48 h post-infection. B, quantitative real time PCR analysis of the mRNA levels of Notch1 in NPC-derived neuronal progeny of cells infected with LV-α-syn in comparison with LV-control-infected cells and co-infected with LV-si p53 to knock down p53 expression, or LV-si Luc as control for double viral infection. **, p < 0.01 by paired Student's t test. C, immunohistochemical analysis of p53 and Notch1 expression in the NPC-derived neuronal cells infected with LV-α-syn or control and with or without knocking down of p53 expression (si p53 or si Luc). Compared with LV-α-syn/si Luc cells, the cells that received si p53 show higher expression of Notch1, suggesting release of transcriptional repression. Scale bar, 20 μm. D and E, quantitative image analysis showing levels of p53 and Notch1. F, expression of human α-syn in cells infected with the different lentiviral constructs assayed, showing robust expression also in cells co-infected with si p53 and si Luc. * and #, p < 0.05 compared with control cells by one-way ANOVA with post hoc Tukey's test.
DISCUSSION
In the present work, we describe the involvement of accumulated α-syn in the aberrant regulation of Notch signaling during adult neurogenesis. Overexpression of α-syn in adult rat hippocampal neuronal progenitor cells facilitates p53-mediated repression of Notch1, a mechanism that might underlie the alterations in adult neurogenesis that we reported previously in different PD models (12–14).
Adult hippocampal neurogenesis is a multistep process initiated with the proliferation of precursor cells and ending with the existence of new granule cells in the dentate gyrus. During the initial expansion of the precursor cells, the proliferation is regulated by many nonspecific stimuli until reaching a postmitotic maturation phase, during which only a subset of the new cells survive (33). Notch1 is a key player on developmental neurogenesis that participates in the specification of cell fate and that is also involved in the survival of more mature phenotypes by regulation of apoptosis. Notch1 first regulates the number of cells acquiring neuronal potential and later determines whether the progeny will adopt a neuronal or a glial fate (34).
Although neurogenesis in the adult brain recapitulates many features from development, the microenvironment of the adult neurogenic niche is far more heterogeneous and complex, where proliferation, fate choice, migration, and maturation of new neurons overlap in time and space and proceed more slowly (35, 36). The role of Notch1 signaling in the adult brain is still not fully elucidated because of the multiple roles that it plays during neurogenesis. The fact that Notch pathway components are expressed throughout the brain suggests that Notch is also involved in other functions beyond regulation of stem cell maintenance and differentiation, including structural and synaptic plasticity (37).
We previously reported that accumulation of α-syn not only affects mouse embryonic stem cells but also impairs neurogenesis in transgenic mice models (12). Here we investigated the effects of α-syn on adult neurogenesis in a cellular model of adult rat hippocampal NPCs that overexpress human α-syn. These cells presented alterations in neuronal differentiation and increased apoptosis, changes that were accompanied by decreased levels of Notch1 and Hes5 transcripts and proteins.
To gain further insight into the transcriptional regulation of Notch1, we characterized a putative promoter region expanding ∼1 kb upstream of Notch1 TSS. We identified two clusters of p53-responsive elements that share high homology with the consensus binding sites.
p53 is a tumor suppressor/proapoptotic protein that regulates the transcription of a wide range of target genes involved in apoptosis, growth control, and senescence by acting either as an activator or repressor (38). p53 can recruit transcription factors and chromatin modifier proteins (39, 40) or can directly bind to the REs, as reported for the activation of Notch1 transcription in epithelial cells (29).
It has been proposed recently that nucleotide polymorphisms in the RE can affect the transcriptional behavior of p53, determining its fate as activator or repressor (41). Detailed sequence analysis of the REs that we identified at the Notch1 promoter revealed a partial enrichment in C/G residues and higher variability of nucleotides from the consensus, characteristics more likely found in repressor p53 REs. We provide evidence for the in vivo binding of p53 to these predicted REs, and in agreement with the in silico analysis, we report that p53 is a repressor of Notch1 transcription in rat adult hippocampal neural progenitor cells.
The cross-talk between Notch and p53 is a multidirectional and complex interaction. Notch signaling can either suppress or increase p53 activity in a context-dependent manner. Notch reduces p53 levels in T cell acute lymphoblastic leukemias (42), but when acting as a proapoptotic factor, Notch can activate p53 in hepatocellular cells (43) and during early embryonic development (44). On the other hand, p53 can also regulate Notch, and positive and negative feedback loops have been reported from p53 to Notch (45). p53 induces Notch expression in keratinocytes, stimulating differentiation and preventing tumor formation (46). In addition, p53 suppression of Notch was reported at the level of Notch ligand expression and γ-secretase activation, associated with increased apoptosis and neuronal death in the brains of p53-deficient mice (47). The data we present here support the role of p53 as a negative regulator of Notch transcription in adult rat hippocampal neuronal progenitors. Although p53 acts as a positive regulator of Notch transcription in non-neuronal cell models, the regulatory relationship between p53 and Notch appears to be different in neurogenic niches, where both proteins have predominant roles. p53 has been implicated in fine-tuning the balance between cell proliferation and cell death in adult neurogenesis (48) and negatively regulates proliferation, survival, and self-renewal of adult neural stem cells (49), processes that are under the control of Notch signaling pathway.
We also show here that α-syn directly binds in vitro to DNA fragments containing p53 REs and that this interaction does not seem to disrupt the binding of p53 to its consensus sites. So far, only one group has reported the binding of α-syn to the DNA (50), a process that might induce conformational changes in both α-syn and DNA (32, 51). It is interesting to note that α-syn has been reported to bind to CG-rich DNA regions, residues abundant in the REs that we identified at the rat Notch1 promoter. Moreover, those regions are common CpG islands highly susceptible to DNA methylation, opening the intriguing possibility that epigenetic mechanisms, like the ones we reported recently to be dysregulated in PD (52), could also be at play in the regulation of Notch1 transcription.
Aberrant protein-DNA interactions mediated by misfolded proteins have been proposed as a novel pathological mechanism common to a myriad of neurological disorders (53). The fact that we did not observed increased levels of p53 in cells accumulating α-syn led us to hypothesize that aggravated repression of Notch1 might be a result of the interaction of α-syn and p53, which we further demonstrated to occur in adult hippocampal neural progenitors. In addition, direct binding of α-syn to the DNA could trigger local changes in chromatin conformation, facilitating the interaction of p53 with its REs. Another interesting possibility is that the interaction of α-syn with p53 could recruit additional repressor factors, including the histone deacetylase Sin3A, as recently reported to occur in other systems (40, 54). We present additional evidence for p53-mediated repression of Notch transcription, because partial knockdown of endogenous p53 in ARH-NPCs restored Notch expression to basal levels in cells overexpressing α-syn.
The association between α-syn and p53 was previously investigated by Alves da Costa et al. (55, 56), who reported that α- and β-synucleins lowered p53 expression and transcriptional activity, thus reducing p53-dependent caspase activation in response to apoptotic stimuli in TSM1 neuronal cells. Aggregation of α-syn induced by 6-hydroxidopamine abolished this protective effect, whereas β-synuclein, which lacks the NAC domain necessary for aggregation, was not affected by 6-hydroxidopamine and was also able to rescue the antiapoptotic effects of α-syn in the presence of the toxin (56). In adult rat hippocampal NPCs, the overexpression of human α-syn did not altered the levels of p53, although it clearly affected neuronal differentiation, and Notch signaling also increasing caspase 3 activation. On the other hand, partial knockdown of endogenous rat α-syn also altered neurogenesis inducing early neuronal maturation, without affecting p53 expression. These results strongly suggest that our cellular model, which recapitulates the complex adult neurogenic niche where differentiation and apoptotic cues co-exist, is more sensitive to α-syn dosage than mature cortical TSM1 neurons. In that regard, ARH-NPCs are more similar to 6-hydroxidopamine-treated TSM1 cells, in which aggregated α-syn is not able to decrease p53 (55).
In summary, we propose a model in which nuclear α-syn accumulation results in the direct binding of α-syn to the Notch1 promoter vicinity, inducing changes in chromatin conformation that facilitate the binding of p53 to its core elements. Alternatively, direct interaction of α-syn and p53 might stabilize p53 on its REs, sustaining the repression of Notch1. Either of these models of molecular regulation, which are not mutually exclusive, results in down-regulation of Notch1 signaling components, interfering with the neuronal maturation and survival of NPCs.
In the adult neurogenic niche cell fate coordination among neighbors is necessary to sustain a pool of undifferentiated cells that ensures neurogenesis. When this finely coordinated process, in which Notch1 is a main player, is disturbed, the organism suffers from either premature exhaustion of progenitors or insufficient generation of functional neurons (57). Dysregulation of Notch1 signaling resulting from the interaction of p53 and accumulated α-syn might therefore underlie the impairments in adult neurogenesis that we previously reported as an important contributor to PD pathology.
This work was supported, in whole or in part, by National Institutes of Health Grants AG5131, AG18440, AG22074, AG3197, AG10435, and NS057096 (to E. M.).

This article contains supplemental Figs. S1–S4.
- PD
- Parkinson disease
- α-syn
- α-synuclein
- NPC
- neural precursor cell
- ARH
- adult rat hippocampus
- LV
- lentiviral
- MOI
- multiplicity of infection
- LV-CT
- B103 cells infected with LV-control
- Ct
- comparative threshold cycle
- ANOVA
- analysis of variance
- PCNA
- proliferating cell nuclear antigen
- RE
- response element.
REFERENCES
- 1. Dauer W., Przedborski S. (2003) Parkinson's disease. Mechanisms and models. Neuron 39, 889–909 [DOI] [PubMed] [Google Scholar]
- 2. Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M. (1997) α-Synuclein in Lewy bodies. Nature 388, 839–840 [DOI] [PubMed] [Google Scholar]
- 3. Takeda A., Mallory M., Sundsmo M., Honer W., Hansen L., Masliah E. (1998) Abnormal accumulation of NACP/α-synuclein in neurodegenerative disorders. Am. J. Pathol. 152, 367–372 [PMC free article] [PubMed] [Google Scholar]
- 4. Hirsch E. C., Hunot S., Faucheux B., Agid Y., Mizuno Y., Mochizuki H., Tatton W. G., Tatton N., Olanow W. C. (1999) Dopaminergic neurons degenerate by apoptosis in Parkinson's disease. Mov. Disord. 14, 383–385 [DOI] [PubMed] [Google Scholar]
- 5. Harding A. J., Lakay B., Halliday G. M. (2002) Selective hippocampal neuron loss in dementia with Lewy bodies. Ann. Neurol. 51, 125–128 [DOI] [PubMed] [Google Scholar]
- 6. McKeith I., Mintzer J., Aarsland D., Burn D., Chiu H., Cohen-Mansfield J., Dickson D., Dubois B., Duda J. E., Feldman H., Gauthier S., Halliday G., Lawlor B., Lippa C., Lopez O. L., Carlos Machado J., O'Brien J., Playfer J., Reid W. (2004) Dementia with Lewy bodies. Lancet Neurol. 3, 19–28 [DOI] [PubMed] [Google Scholar]
- 7. Fleming S. M., Tetreault N. A., Mulligan C. K., Hutson C. B., Masliah E., Chesselet M. F. (2008) Olfactory deficits in mice overexpressing human wildtype α-synuclein. Eur. J. Neurosci. 28, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gage F. H., Kempermann G., Palmer T. D., Peterson D. A., Ray J. (1998) Multipotent progenitor cells in the adult dentate gyrus. J. Neurobiol. 36, 249–266 [DOI] [PubMed] [Google Scholar]
- 9. Brown J., Cooper-Kuhn C. M., Kempermann G., Van Praag H., Winkler J., Gage F. H., Kuhn H. G. (2003) Enriched environment and physical activity stimulate hippocampal but not olfactory bulb neurogenesis. Eur. J. Neurosci. 17, 2042–2046 [DOI] [PubMed] [Google Scholar]
- 10. van Praag H., Christie B. R., Sejnowski T. J., Gage F. H. (1999) Running enhances neurogenesis, learning, and long-term potentiation in mice. Proc. Natl. Acad. Sci. U.S.A. 96, 13427–13431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Praag H., Schinder A. F., Christie B. R., Toni N., Palmer T. D., Gage F. H. (2002) Functional neurogenesis in the adult hippocampus. Nature 415, 1030–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Winner B., Lie D. C., Rockenstein E., Aigner R., Aigner L., Masliah E., Kuhn H. G., Winkler J. (2004) Human wild-type α-synuclein impairs neurogenesis. J. Neuropathol. Exp. Neurol. 63, 1155–1166 [DOI] [PubMed] [Google Scholar]
- 13. Winner B., Rockenstein E., Lie D. C., Aigner R., Mante M., Bogdahn U., Couillard-Despres S., Masliah E., Winkler J. (2008) Mutant α-synuclein exacerbates age-related decrease of neurogenesis. Neurobiol. Aging 29, 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crews L., Mizuno H., Desplats P., Rockenstein E., Adame A., Patrick C., Winner B., Winkler J., Masliah E. (2008) α-Synuclein alters Notch-1 expression and neurogenesis in mouse embryonic stem cells and in the hippocampus of transgenic mice. J. Neurosci. 28, 4250–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Louvi A., Artavanis-Tsakonas S. (2006) Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93–102 [DOI] [PubMed] [Google Scholar]
- 16. Greenberg D. A., Jin K. (2006) Turning neurogenesis up a Notch. Nat. Med. 12, 884–885 [DOI] [PubMed] [Google Scholar]
- 17. Breunig J. J., Silbereis J., Vaccarino F. M., Sestan N., Rakic P. (2007) Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc. Natl. Acad. Sci. U.S.A. 104, 20558–20563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elliott R. C., Khademi S., Pleasure S. J., Parent J. M., Lowenstein D. H. (2001) Differential regulation of basic helix-loop-helix mRNAs in the dentate gyrus following status epilepticus. Neuroscience 106, 79–88 [DOI] [PubMed] [Google Scholar]
- 19. Kawai T., Takagi N., Nakahara M., Takeo S. (2005) Changes in the expression of Hes5 and Mash1 mRNA in the adult rat dentate gyrus after transient forebrain ischemia. Neurosci. Lett. 380, 17–20 [DOI] [PubMed] [Google Scholar]
- 20. Nagarsheth M. H., Viehman A., Lippa S. M., Lippa C. F. (2006) Notch-1 immunoexpression is increased in Alzheimer's and Pick's disease. J. Neurol. Sci. 244, 111–116 [DOI] [PubMed] [Google Scholar]
- 21. Ray J., Gage F. H. (2006) Differential properties of adult rat and mouse brain-derived neural stem/progenitor cells. Mol. Cell Neurosci. 31, 560–573 [DOI] [PubMed] [Google Scholar]
- 22. Crews L., Adame A., Patrick C., Delaney A., Pham E., Rockenstein E., Hansen L., Masliah E. (2010) Increased BMP6 levels in the brains of Alzheimer's disease patients and APP transgenic mice are accompanied by impaired neurogenesis. J. Neurosci. 30, 12252–12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tiscornia G., Singer O., Verma I. M. (2006) Production and purification of lentiviral vectors. Nat. Protoc. 1, 241–245 [DOI] [PubMed] [Google Scholar]
- 24. Masliah E., Rockenstein E., Veinbergs I., Mallory M., Hashimoto M., Takeda A., Sagara Y., Sisk A., Mucke L. (2000) Dopaminergic loss and inclusion body formation in α-synuclein mice. Implications for neurodegenerative disorders. Science 287, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 25. Desplats P. A., Lambert J. R., Thomas E. A. (2008) Functional roles for the striatal-enriched transcription factor, Bcl11b, in the control of striatal gene expression and transcriptional dysregulation in Huntington's disease. Neurobiol. Dis. 31, 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Waldman T., Kinzler K. W., Vogelstein B. (1995) p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 55, 5187–5190 [PubMed] [Google Scholar]
- 27. Gorbatyuk O. S., Li S., Nash K., Gorbatyuk M., Lewin A. S., Sullivan L. F., Mandel R. J., Chen W., Meyers C., Manfredsson F. P., Muzyczka N. (2010) In vivo RNAi-mediated α-synuclein silencing induces nigrostriatal degeneration. Mol. Ther. 18, 1450–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boardman P. E., Oliver S. G., Hubbard S. J. (2003) SiteSeer. Visualisation and analysis of transcription factor binding sites in nucleotide sequences. Nucleic Acids Res. 31, 3572–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yugawa T., Handa K., Narisawa-Saito M., Ohno S., Fujita M., Kiyono T. (2007) Regulation of Notch1 gene expression by p53 in epithelial cells. Mol. Cell Biol. 27, 3732–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. el-Deiry W. S., Kern S. E., Pietenpol J. A., Kinzler K. W., Vogelstein B. (1992) Definition of a consensus binding site for p53. Nat. Genet. 1, 45–49 [DOI] [PubMed] [Google Scholar]
- 31. Schubert D., Heinemann S., Carlisle W., Tarikas H., Kimes B., Patrick J., Steinbach J. H., Culp W., Brandt B. L. (1974) Clonal cell lines from the rat central nervous system. Nature 249, 224–227 [DOI] [PubMed] [Google Scholar]
- 32. Hegde M. L., Vasudevaraju P., Rao K. J. (2010) DNA induced folding/fibrillation of α-synuclein. New insights in Parkinson's disease. Front. Biosci. 15, 418–436 [DOI] [PubMed] [Google Scholar]
- 33. Kempermann G., Song H., Gage F. (2008) in Adult Neurogenesis (Gage F., Kempermann G., Song H., eds) pp. 159–174, Cold Spring Harbor Laboratories, Cold Spring Harbor, NY [Google Scholar]
- 34. Bray S. J. (2006) Notch signalling. A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 35. Duan X., Kang E., Liu C. Y., Ming G. L., Song H. (2008) Development of neural stem cell in the adult brain. Curr. Opin. Neurobiol. 18, 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson M. A., Ables J. L., Eisch A. J. (2009) Cell-intrinsic signals that regulate adult neurogenesis in vivo. Insights from inducible approaches. BMB Rep. 42, 245–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ables J. L., Breunig J. J., Eisch A. J., Rakic P. (2011) Not(ch) just development. Notch signalling in the adult brain. Nat. Rev. Neurosci. 12, 269–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 39. Murphy M., Ahn J., Walker K. K., Hoffman W. H., Evans R. M., Levine A. J., George D. L. (1999) Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13, 2490–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Solá S., Xavier J. M., Santos D. M., Aranha M. M., Morgado A. L., Jepsen K., Rodrigues C. M. (2011) p53 interaction with JMJD3 results in its nuclear distribution during mouse neural stem cell differentiation. PLoS One 6, e18421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang B., Xiao Z., Ren E. C. (2009) Redefining the p53 response element. Proc. Natl. Acad. Sci. U.S.A. 106, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Demarest R. M., Ratti F., Capobianco A. J. (2008) It's T-ALL about Notch. Oncogene 27, 5082–5091 [DOI] [PubMed] [Google Scholar]
- 43. Qi R., An H., Yu Y., Zhang M., Liu S., Xu H., Guo Z., Cheng T., Cao X. (2003) Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 63, 8323–8329 [PubMed] [Google Scholar]
- 44. Yang X., Klein R., Tian X., Cheng H. T., Kopan R., Shen J. (2004) Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 269, 81–94 [DOI] [PubMed] [Google Scholar]
- 45. Dotto G. P. (2009) Crosstalk of Notch with p53 and p63 in cancer growth control. Nat. Rev. Cancer 9, 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rangarajan A., Talora C., Okuyama R., Nicolas M., Mammucari C., Oh H., Aster J. C., Krishna S., Metzger D., Chambon P., Miele L., Aguet M., Radtke F., Dotto G. P. (2001) Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20, 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Amson R., Lassalle J. M., Halley H., Prieur S., Lethrosne F., Roperch J. P., Israeli D., Gendron M. C., Duyckaerts C., Checler F., Dausset J., Cohen D., Oren M., Telerman A. (2000) Behavioral alterations associated with apoptosis and down-regulation of presenilin 1 in the brains of p53-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 97, 5346–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Medrano S., Scrable H. (2005) Maintaining appearances. The role of p53 in adult neurogenesis. Biochem. Biophys. Res. Commun. 331, 828–833 [DOI] [PubMed] [Google Scholar]
- 49. Meletis K., Wirta V., Hede S. M., Nistér M., Lundeberg J., Frisén J. (2006) p53 suppresses the self-renewal of adult neural stem cells. Development 133, 363–369 [DOI] [PubMed] [Google Scholar]
- 50. Hegde M. L., Jagannatha Rao K. S. (2003) Challenges and complexities of α-synuclein toxicity. New postulates in unfolding the mystery associated with Parkinson's disease. Arch. Biochem. Biophys. 418, 169–178 [DOI] [PubMed] [Google Scholar]
- 51. Hegde M. L., Rao K. S. (2007) DNA induces folding in α-synuclein. Understanding the mechanism using chaperone property of osmolytes. Arch. Biochem. Biophys. 464, 57–69 [DOI] [PubMed] [Google Scholar]
- 52. Desplats P., Spencer B., Coffee E., Patel P., Michael S., Patrick C., Adame A., Rockenstein E., Masliah E. (2011) α-Synuclein sequesters Dnmt1 from the nucleus. A novel mechanism for epigenetic alterations in Lewy body diseases. J. Biol. Chem. 286, 9031–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiménez J. S. (2010) Protein-DNA interaction at the origin of neurological diseases. A hypothesis. J. Alzheimers Dis. 22, 375–391 [DOI] [PubMed] [Google Scholar]
- 54. Kontopoulos E., Parvin J. D., Feany M. B. (2006) α-Synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum. Mol. Genet. 15, 3012–3023 [DOI] [PubMed] [Google Scholar]
- 55. Alves Da Costa C., Paitel E., Vincent B., Checler F. (2002) α-Synuclein lowers p53-dependent apoptotic response of neuronal cells. Abolishment by 6-hydroxydopamine and implication for Parkinson's disease. J. Biol. Chem. 277, 50980–50984 [DOI] [PubMed] [Google Scholar]
- 56. da Costa C. A., Masliah E., Checler F. (2003) β-Synuclein displays an antiapoptotic p53-dependent phenotype and protects neurons from 6-hydroxydopamine-induced caspase 3 activation. Cross-talk with α-synuclein and implication for Parkinson's disease. J. Biol. Chem. 278, 37330–37335 [DOI] [PubMed] [Google Scholar]
- 57. Buchen E., Pleasure S. J. (2008) in Adult Neurogenesis (Gage F., Kempermann G., Song H., eds) pp. 267–282, Cold Spring Harbor Laboratories, Cold Spring Harbor, NY [Google Scholar]