Background: Huntington disease (HD) involves neurological and metabolic pathologies.
Results: Euglycemic therapeutics that affect hypothalamic transcription ameliorate multiple aspects of HD pathology.
Conclusion: Hypothalamic targeting in HD may yield enhanced HD drug efficacy.
Significance: Wider consideration of the metabolic aspects of neurological disorders may be important for drug design.
Keywords: Bioinformatics, Diabetes, Huntington Disease, Hypothalamus, Transcriptomics, Exendin-4, GLP-1, GLP-1-Tf, Insulin
Abstract
Our aim was to employ novel analytical methods to investigate the therapeutic treatment of the energy regulation dysfunction occurring in a Huntington disease (HD) mouse model. HD is a neurodegenerative disorder that is characterized by progressive motor impairment and cognitive alterations. Changes in neuroendocrine function, body weight, energy metabolism, euglycemia, appetite function, and gut function can also occur. It is likely that the locus of these alterations is the hypothalamus. We determined the effects of three different euglycemic agents on HD progression using standard physiological and transcriptomic signature analyses. N171–82Q HD mice were treated with insulin, Exendin-4, and the newly developed GLP-1-Tf to determine whether these agents could improve energy regulation and delay disease progression. Blood glucose, insulin, metabolic hormone levels, and pancreatic morphology were assessed. Hypothalamic gene transcription, motor coordination, and life span were also determined. The N171–82Q mice exhibited significant alterations in hypothalamic gene transcription signatures and energy metabolism that were ameliorated, to varying degrees, by the different euglycemic agents. Exendin-4 or GLP-1-Tf (but not insulin) treatment also improved pancreatic morphology, motor coordination, and increased life span. Using hypothalamic transcription signature analyses, we found that the physiological efficacy variation of the drugs was evident in the degree of reversal of the hypothalamic HD pathological signature. Euglycemic agents targeting hypothalamic and energy regulation dysfunction in HD could potentially alter disease progression and improve quality of life in HD.
Introduction
Huntington disease (HD)2 is one of the most prevalent inherited neurodegenerative disorders and is caused by an expansion of the trinucleotide repeat (CAG) in the gene encoding the huntingtin protein (htt) (1). HD is characterized by chorea, poor balance, cognitive impairments, weight loss, metabolic abnormalities, and eventual death (2). Conventionally, HD is considered to be primarily a neurological disorder. However, recent evidence indicates the importance of neuroendocrine alterations and peripheral dysglycemia in HD. Many HD patients, as well as HD mouse models, suffer from progressive weight loss, alterations in appetite and metabolic function, and dysglycemia (3). The cause of this is currently unclear (4), but a recent study has suggested that weight loss could be due to increased involuntary motor activity (5) or increased metabolic rate (6), as severe weight loss can occur despite adequate caloric intake (7). Various studies have demonstrated that HD patients and animal models exhibit increased hunger and appetite, possess elevated circulating levels of orexigenic hormones, and reduced circulating levels of anorexigenic hormones (7–9). Further reinforcing the relevance of neuroendocrine alterations in HD, sleep disturbances and disruptions in circadian rhythm often occur in HD patients (10, 11). Although htt disruption can affect central neurons, htt is ubiquitously expressed and is involved in multiple functions, e.g. protein scaffolding, neurotrophin transport, and cell metabolism (12). Multiple HD mouse models display dysglycemia involving alterations in pancreatic β-cell mass and reduced insulin secretion, presumably caused by mutant htt (mhtt) accumulation in the islets of Langerhans (9, 13). In HD mouse models, mhtt expression has been demonstrated in gonads, and HD patients often exhibit significantly reduced circulating testosterone and luteinizing hormone levels (14, 15). These effects could be due to pathology of gonadal and hypothalamic gonadotropin-releasing hormone-containing neurons (16).
A crucial link between peripheral endocrine system activity and central nervous system activity is the hypothalamus, as this organ facilitates hunger, thirst, reproductive function, and circadian rhythms (12). The potential hypothalamic alterations that occur in N171–82Q mice are currently poorly characterized, yet a deeper understanding of the changes that occur may assist the development of therapeutics that target hypothalamic dysfunction in HD. We have previously shown that the glucagon-like peptide 1 (GLP-1) analog, Exendin-4 (Ex-4), demonstrates effective therapeutic activity in the N171–82Q mouse model of HD (9). As the hypothalamus may represent an important therapeutic locus in HD, we have investigated hypothalamic transcriptional changes in the N171–82Q HD mouse model induced by three functionally related euglycemic agents. These agents are structurally distinct pharmacological compounds that possess convergent euglycemic mechanisms as follows: insulin (Lantus), Ex-4, and a novel long acting GLP-1 receptor agonist GLP-1-Tf (17). Ex-4, but not GLP-1-Tf, can enter the brain (17). As recent research has demonstrated HD to be a more multidimensional disorder than previously thought, encompassing neurodegeneration, protein aggregation, and metabolic disruption, we decided to develop a methodology that could appreciate, at a multifactorial level, the likely multitude of therapeutic mechanisms that could combine to ameliorate the complex HD pathophysiology. Therefore, using biochemical, behavioral, immunohistochemical, and multiple bioinformatic analyses, we have investigated the pathological phenotype of the N171–82Q mouse hypothalamus as well as the multidimensional functional efficacy of these three euglycemic agents for improving energy regulation, pancreatic morphology, hypothalamic function, motor coordination, and life span.
EXPERIMENTAL PROCEDURES
Animals, Drug Administration, and Hormone Measurements
All animal procedures were approved by the Animal Care and Use Committee of the NIA, National Institutes of Health. Male B6C3-Tg(HD82Gln)81Dbo/J (N171–82Q) mice and age-matched WT mice were used. Ex-4 (n = 10, 300 μl of 0.1 μmol/liter solution, Bachem, Torrance, CA), the GLP-1-Tf (n = 10, 1 mg/kg), insulin (n = 10, 5 units, Lantus), or control treatment (n = 10, phosphate-buffered saline (PBS); Sigma) was administered by a once-daily subcutaneous injection. Injections were started when animals were pre-symptomatic (2 months of age) and continued for 5 weeks. Body weight and glucose levels (using a Glucometer Elite XL) were measured weekly. At the end of the study, mice were euthanized, and hypothalami were isolated. Blood samples and pancreata were collected as described previously (9). Plasma levels of insulin, leptin, total ghrelin, and adiponectin were measured using standard ELISA and radioimmunoassay methods, as described previously (9).
Pancreatic Insulin and Glucagon Immunohistochemistry
Pancreatic sections were immunostained as described previously (9), using guinea pig anti-swine insulin, 1:300 (DakoCytomation, Carpinteria, CA), or guinea pig anti-glucagon, 1:500 (Millipore, Billerica, MA).
Motor Performance Assessment
Motor coordination was assessed using an accelerating RotaRod (Med Associates, Georgia, VT) as described previously (9).
RNA Extraction and Oligonucleotide Microarray Hybridization
RNA isolation from three individual animals in each experimental group was carried out using a Qiagen RNeasy mini kit (Qiagen, Inc., Valencia, CA), as described previously (18). RNA conversion to cDNA and subsequent hybridization with Sentrix MouseRef-8 Expression BeadChips (Illumina, San Diego) was performed as described previously (18). Microarray data were analyzed using DIANE 6.0, a spreadsheet-based microarray analysis program based on the SAS JMP7.0 system. Raw microarray data were subjected to filtering and z normalization and tested for significant changes as described previously (18). Initial filtering identified genes with a z ratio of ≥1.50, with the z ratio derived from the difference between the averages of the observed gene z scores, divided by the standard deviation of all of the differences for that particular comparison. Genes were then refined by calculating the false discovery rate, which controls for the expected proportion of falsely rejected hypotheses, and including only those genes with false discovery rate <0.05. These data were further analyzed using analysis of variance with significance set at p < 0.05. This allowed us to identify transcripts that differed in their intensity across all of the animal replicates and the various experimental conditions of the mice employed in this study. We have deposited the raw data at GEO/ArrayExpress under accession number GSE39586, and we can confirm all details are MIAME-compliant.
Protein Expression Analysis
Microarray results were validated by Western blots. Briefly, tissues were homogenized using sonication followed by fractionation using a Qiagen Q-proteome kit according to the manufacturer's instructions (Qiagen, Inc., Valencia, CA). For all experiments, cytoplasmic fractions were used. Each tissue homogenate was loaded onto a BisTris 4–12% polyacrylamide gel (Invitrogen) before electrotransfer to a PVDF membrane (Thermo Scientific, Rockford, IL). Proteins were identified using primary antisera at 1:1000 dilutions, followed by species-specific alkaline phosphatase-conjugated secondary antibodies (Sigma) at a 1:7000 dilution. Primary antisera specific for signal transducer and activator of transcription-3 (STAT3), transferrin receptor (TFR), Glypican 3 (GPC3), growth-associated protein 43 (GAP43), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta (YWHAH, 14-3-3-η), NADH dehydrogenase (ubiquinone) 1α subcomplex 4 (NDUFA4), and actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Glycogen synthase kinase β (GSK3-β), protein-tyrosine kinase-2 (FAK), neurotrophic receptor tyrosine kinase 2 (TRKB), Regulator of G protein signaling 4 (RGS4), and ribosomal protein S3 (RPS3) antisera were obtained from Cell Signaling Technology (Danvers, MA). The anti-Discs large homology 2 (DLG2) sera were obtained from GenWay Biotech (San Diego). Specific anti-DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (DDX1), receptor accessory protein 5 (REEP5), and histone deacetylase 3 (HDAC3) sera were obtained from Abcam (Cambridge, MA). PVDF-bound immune complexes were identified using enzyme-linked chemifluorescence and quantified using a Typhoon 9410 PhosphorImager (GE Healthcare). All antibodies used have been previously validated.
Bioinformatic Analysis
Array-derived gene lists were analyzed using multiple forms of functional annotational clustering, i.e. Gene Ontology (GO), Parametric Analysis of Geneset Enrichment (PAGE), and Latent Semantic Indexing (LSI). We employed a cutoff of at least two genes (from the original filtered/analysis of variance Geneset) needing to be present to fully populate a GO term group or an MSigDB gene collection, with a probability (p) of enrichment value of ≤0.05. Parametric Geneset enrichment analyses such as GO or MSigDB-PAGE are primarily performed using raw datasets to test significance of enrichment at a group/collection level rather than at the individual gene significance level (19). Ingenuity Pathway Analysis was employed for BioFunction Diseases and Disorders analysis. For Diseases and Disorders analysis, a hybrid score process was employed to generate a single index for each disorder significantly (p < 0.05) associated with input gene datasets. Ingenuity Pathway Analysis BioFunction Diseases and Disorders hybrid scores were generated by multiplication of the negative log10 of enrichment probability (p) with the number of significantly enriched genes in each Diseases and Disorders group. Latent Semantic Indexing (LSI, Computable Genomix, Memphis, TN) analysis was performed as described previously (18). Gene Indexer correlates the strength of association, using LSI, between specific genes in a dataset with user-defined interrogation terms. Gene Indexer employs a comprehensive murine database of over 2 × 106 scientific abstracts to perform this text-gene correlation analysis. LSI facilitates the specific textual interrogation of an input dataset with a specific interrogation term, e.g. diabetes, to ascertain which of the input dataset genes are associated with the interrogation term. The possible LSI correlation scores for a gene to be associated with an input interrogation term range from 0 to 1, with the stronger correlation scores approaching 1. A correlation score of ≥0.1 indicates at least an implicit correlation, between the specific gene and the user-defined input interrogation term.
Statistical Analyses
In each histogram, data represent the means ± S.E. Statistical analyses (Student's t test) were performed using GraphPad Prism (GraphPad Software, San Diego). p ≤ 0.05 was considered statistically significant.
RESULTS
Hypothalamic Gene Alterations in the N171–82Q Model of HD
Hypothalamic gene expression in symptomatic HD mice was compared with age-matched wild types (WT). 133 gene transcripts were significantly up-regulated and 194 were significantly down-regulated in HD hypothalamus compared with WT (Fig. 1A and supplemental Table S1). Various significantly altered genes were validated by Western blot as follows: Tfr, Dlg2, Ptk2, Ddx1, Reep5, and Hdac3 (up-regulated), and Stat3, Gsk3-β, TrkB, Rgs4, Gpc3, and Gap43 (down-regulated) (Fig. 1, B–N). These specific factors were chosen from the transcriptomic data as they demonstrate a variety of expression change polarities (up- and down-regulation), possess important neurophysiological roles, and represent a diverse set of functional entities (transmembrane receptors, kinases, transcription factor regulators, and neurosynaptic factors). To investigate whether the hypothalamic transcriptomic signatures could be considered an appropriate microcosm of the Huntington disease pathophysiology, we functionally annotated, using un-biased Ingenuity Pathway Analysis BioFunction-Diseases and Disorders analysis, the transcripts significantly and differentially regulated (supplemental Table S1) in HD compared with wild-type mice (Fig. 1O and supplemental Table S2). Applying a class-based statistical appraisal of the significantly populated (p < 0.05) BioFunction-Diseases and Disorders groups, we found that six disorder groups possessed a statistically significant greater hybrid score compared with the other BioFunction-Diseases and Disorders groups (supplemental Table S2). The functional nature of these six disorder groups (Fig. 1O) are all strongly associated with classical HD-related pathology; this result therefore suggests that our hypothalamic transcriptomic signatures functionally encrypt a microcosm of the overall genomic disorder.
FIGURE 1.
Transcriptomic alterations in the hypothalamus of N171–82Q mice. A, z ratio distribution for gene transcripts significantly and differentially expressed in N171–82Q HD mice compared with age/gender-matched wild-type (WT) controls. B, Western blot validation of multiple protein products from significantly regulated transcripts in N171–82Q mouse hypothalami. The protein levels were assessed in three WT mice (lanes 1–3) and three HD mice (lanes 4–6). Quantification of the representative Western blots is indicated for the WT (black bars) and HD (shaded bars) for the respective proteins: Tfr (C); Dlg2 (D); Ptk2 (E); Ddx1 (F); Reep5 (G); Hdac3 (H); STAT3 (I); GSK3-β (J); TrkB (K); Rgs4 (L); Gpc3 (M); and Gap43 (N). Expression variation was normalized to actin expression. O, BioFunction Disease and Disorder annotation of statistically significant gene transcript differentials between HD and wild-type mice. Box and Whisker plot in the upper panel demonstrates mean (blue line) and 95% confidence limits. Six Disease and Disorder groups (red circles) possess a calculated group hybrid score (number of genes in group multiplied by the negative log10 of the enrichment probability) significantly different from the other identified annotated Disease and Disorder groups. The lower panel describes the phenotypic nature of the six significantly different Disease and Disorder groups. Assessment of statistical significance was performed with GraphPad Prism (Version 5) using a Student's t test. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
In addition to the appreciation of the HD hypothalamic “signature” at the transcriptomic level, we also employed Gene Ontology (GO) and Parametric Analysis of Geneset Enrichment (PAGE) investigation of the hypothalamic HD molecular signature. Using unbiased GO term enrichment analysis (supplemental Table S3), we found a considerable enrichment in the HD mice of GO term groups related to cellular stress (Apoptosis, GO0006915; Response to Lipopolysaccharide, GO0032496), inflammatory signaling (NFκB cascade, GO0077249), and DNA damage (Response to UV, GO0009411; Response to DNA damage stimulus, GO0006974) by multiple transcripts up-regulated in the HD mice compared with WT (supplemental Table S3). In contrast, considerable enrichment of GO term groups associated with mitochondrial function (Organelle inner membrane, GO0019866), neuronal architecture (Neuron differentiation, GO0030182), protein translation (Ribonucleoprotein complex, GO0030529), neuronal receptor signaling (Neuropeptide hormone activity, GO0005184), and energy regulation (insulin-like growth factor binding, GO0005520) was observed by transcripts down-regulated in the HD mice compared with WT (supplemental Table S3). The transcriptomic collections at the Molecular Signatures Database (MSigDB) facilitate a capacity for experimental analogy to be drawn between different datasets. Using PAGE to interrogate the MSigDB, we were able to uncover multiple important molecular signatures within the hypothalamic transcriptomic differences in the HD mice compared with WT (supplemental Table S4). Using PAGE analysis, we found considerable MSigDB enrichment of transcript collections associated with DNA damage/cell cycle control (CELL_CYCLE_CHECKPOINT), stem-cell activation (STEMCELL_COMMON_UP), apoptosis/inflammatory signaling (STRESSPATHWAY), metabolic stress (BETAOXIDATIONPATHWAY), and neurodegenerative disease (PARKINPATHWAY) by multiple transcripts up-regulated in the HD mice compared with WT (supplemental Table S4). Similarly to what we observed using GO term enrichment analysis, we again found considerable enrichment of MSigDB collections associated with energy regulatory mechanisms (ELECTRON_TRANSPORT_CHAIN), protein translation (RIBOSOMAL_PROTEINS), neuronal architecture (CORTEX_ENRICHMENT_EARLY_UP), and cell-cell communication (CELL_ADHESION_RECEPTOR_ACTIVITY) by transcripts down-regulated in the HD mice compared with WT (supplemental Table S4).
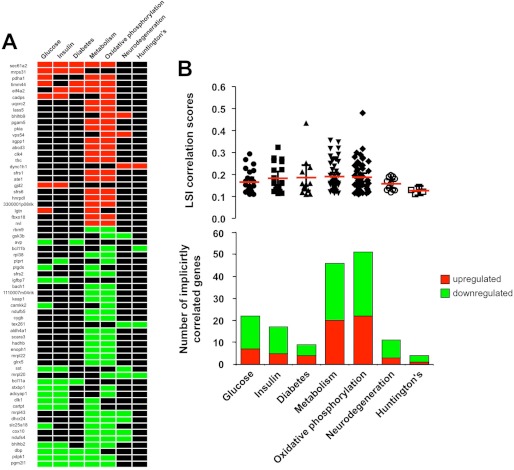
To further investigate the potential pathophysiological and metabolic ramifications of the hypothalamic HD transcriptomic signature, in an unbiased manner, we performed LSI upon the significantly altered genes (supplemental Table S5). LSI analysis facilitates a targeted association approach between user-defined input terms and transcriptomic activity. Using various “metabolic” and “neurodegenerative” input terms, we created a genomic heat map (>2 genes per input LSI term) that demonstrated the relative correlation strength of 63 statistically correlated genes (Fig. 2A). The input terms that demonstrated the most number of correlating terms from the HD hypothalamic transcriptomic signature were “metabolism” and “oxidative phosphorylation.” With subsequent analysis of mean transcript correlation values (Fig. 2B, upper panel) as well as the total numbers of implicitly correlating genes for each input LSI term (Fig. 2B, lower panel), again it was clear of the strong role metabolic tissues play in HD hypothalamic pathophysiology. The multiple disease- and pathophysiology-related functions, significantly populated by HD-regulated hypothalamic genes, suggest that unbiased bioinformatic analyses of this tissue again reveal a potential functional microcosm of the whole-animal pathophysiological phenotype. Hypothalamic transcriptomic alterations may therefore be highly correlated to the development of HD pathophysiology. HD pathophysiology in our mice and the amelioration of these pathophysiologies by our experimental compounds were next investigated.
FIGURE 2.
Genomic correlations in the hypothalamic N171–82Q HD mouse transcriptomic signature. A, genomic heat map representation of the significantly regulated transcripts that possess an implicit semantic correlation (LSI score >0.1) with at least two of the user-defined input interrogator terms (glucose, insulin, diabetes, metabolism, oxidative phosphorylation, neurodegeneration, and Huntington disease). Red blocks indicate the correlation of a correlated gene up-regulated in HD compared with WT controls, and green blocks indicate the correlation of a down-regulated HD-associated gene. Black blocks indicate no significant gene-LSI interrogator term correlation. B, upper panel indicates the mean (red bar) ± S.E. LSI correlation score from all the specifically correlating HD-associated genes with the input LSI interrogator terms. The lower panel displays the numbers and direction of transcript regulation of HD-associated genes correlating with the specific LSI interrogator terms.
Glycemic Hormones Are Differentially Altered by the Euglycemic Agents
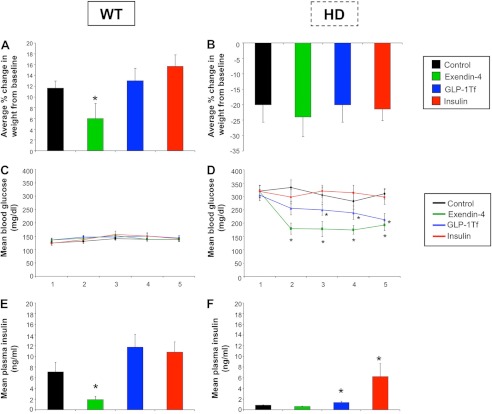
Presymptomatic male HD mice and age-matched controls were treated daily with insulin, Ex-4, GLP-1-Tf, or saline (control). Body weight and circulating levels of glucose and insulin were measured (Fig. 3). WT control mice demonstrated progressive weight gain during the study, and this was not significantly affected by daily injection of GLP-1-Tf or insulin (Fig. 3A). Ex-4, as expected, caused significant weight loss in WT animals (Fig. 3A). Control HD mice, however, demonstrated progressive weight loss during the study, and Ex-4, GLP-1-Tf, and insulin all failed to attenuate this effect (Fig. 3B). The HD mice demonstrated a profound dysglycemic phenotype (9). In WT animals, the three treatments did not significantly alter blood glucose levels (Fig. 3C), but Ex-4 treatment significantly lowered plasma insulin levels as expected (Fig. 3E). These findings corroborate previous findings on the effects of these compounds on glycemic hormone levels in WT animals (9, 17). In contrast, both Ex-4 and GLP-1-Tf treatments significantly lowered blood glucose levels in HD mice (Fig. 3D), with Ex-4 being the most effective. Interestingly, insulin treatment had no effect on blood glucose levels in HD mice, illustrating severe insulin resistance (Fig. 3D); however, both insulin and GLP-1-Tf treatments significantly increased plasma insulin levels in HD mice (Fig. 3F).
FIGURE 3.
Body weight and glycemic hormone levels are altered by Ex-4, GLP-1-Tf, or insulin treatments. Average % change in body weight from base line for WT (A) and HD (B) mice throughout the study. Mean blood glucose concentrations for WT (C) and HD (D) mice throughout the study. Mean plasma insulin concentrations in WT (E) and HD (F) mice. Values are mean ± S.E. *, p ≤ 0.05; n = 10 per group.
Euglycemic Agents Differentially Improve Pancreatic Structure in HD Mice
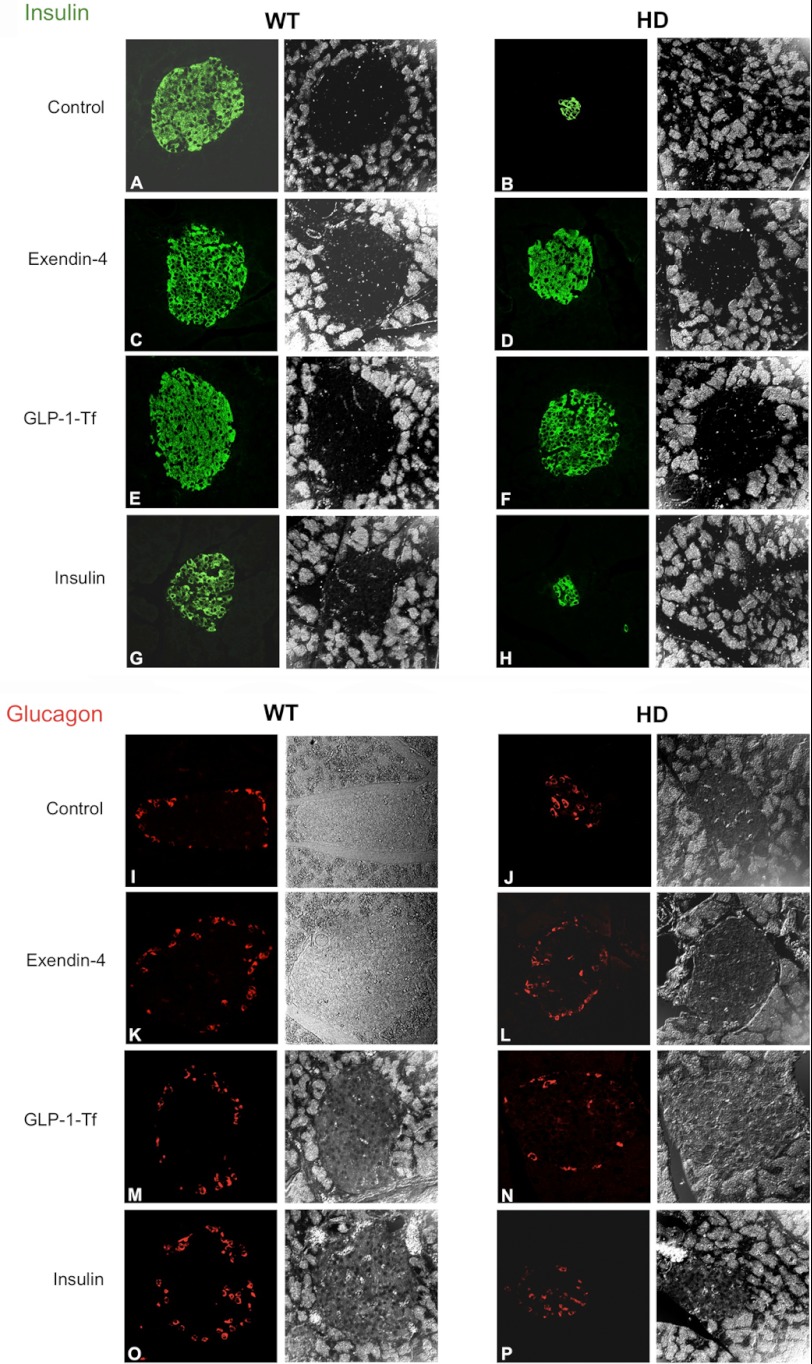
Previous studies have shown that HD mouse models demonstrate a buildup of mhtt aggregates within pancreatic islets, which may contribute to their diabetic-like condition (9). We assessed the effects of Ex-4, GLP-1-Tf, and insulin on islet beta-cell and alpha-cell organization. As shown previously, HD mice demonstrated significantly smaller islets and a smaller area of beta-cells, compared with WT mice (Fig. 4, A and B) (9). Both the Ex-4 and GLP-1-Tf treatments restored islet size and numbers of beta-cells in HD mice close to those of WT mice (Fig. 4, A–F). Ex-4 and GLP-1-Tf are effective treatments for type 2 diabetes, as they not only protect pancreatic beta-cell function but also significantly improve insulin sensitivity (9, 17, 20). Insulin treatment did not alter islet size and beta-cell numbers in HD mice (Fig. 4, G and H). In HD mice, some of the alpha-cells were present in the center of the islets (Fig. 4, I–P), causing abnormal islet structure. Ex-4 and GLP-1-Tf treatments restored the alpha-cell topography of islets in these HD mice (Fig. 4, K–N). Insulin treatment, however, did not (Fig. 4P). Global energy metabolism is not solely controlled by insulin but also by additional plasma hormones such as leptin, ghrelin, and adiponectin. We therefore assessed the effects of Ex-4, GLP-1-Tf, and insulin on these hormones.
FIGURE 4.
Drug-mediated modulation of beta- and alpha-cell distribution in islets. Pancreatic beta-cells were identified in WT and HD mice via insulin immunostaining (A–H), and pancreatic alpha-cells were identified in WT and HD mice via glucagon immunostaining (I–P). Effects of vehicle (A and B), Ex-4 (C and D), GLP-1-Tf (E and F), and insulin (G and H) treatment upon beta-cell population for WT and HD mice, respectively, are shown. Effects of vehicle (I and J), Ex-4 (K-L), GLP-1-Tf (M and N), and insulin (O and P) treatment upon alpha-cell population for WT and HD mice, respectively, are shown. n = 10 per group.
Euglycemic Agents Affect Energy-regulating Plasma Hormones in HD Mice
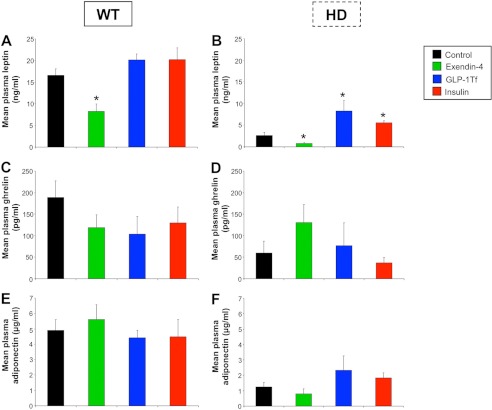
An alteration in energy homeostasis has been reported in HD patients and in HD mouse models (8, 9). As expected from their lower body mass, control HD animals demonstrated significantly lower leptin levels compared with WT mice (p ≤ 0.001; Fig. 5, A and B). In both HD and WT mice, Ex-4 treatment caused a significant decrease in plasma leptin levels (p ≤ 0.05). GLP-1-Tf and insulin treatments, however, caused a significant increase in plasma leptin levels in HD mice (p ≤ 0.05) but did not significantly affect plasma leptin levels in WT mice (Fig. 5, A and B). Plasma ghrelin levels were reduced in Ex-4-, GLP-1-Tf-, and insulin-treated WT mice compared with controls (Fig. 5C), and similar effects have been reported in humans with regard to Ex-4 (21). Although control HD mice had significantly lower ghrelin levels compared with WT mice, Ex-4 treatment restored ghrelin levels in HD mice to that of their WT counterparts (Fig. 5, C and D). GLP-1-Tf and insulin did not significantly affect ghrelin levels in HD mice (Fig. 5D). Ex-4, GLP-1-Tf, and insulin treatments did not alter plasma adiponectin levels in HD and WT mice (Fig. 5, E and F), although plasma adiponectin levels were significantly lower in HD mice compared with the WT mice (p ≤ 0.001). Decreased adiponectin levels are often present in insulin-resistant or diabetic states (22).
FIGURE 5.
Modification of plasma levels of energy-regulating hormones by Ex-4, GLP-1-Tf, and insulin treatments. Mean plasma leptin concentrations for WT (A) and HD (B) mice are shown. Mean plasma ghrelin concentrations in WT (C) and HD (D) mice are shown. Mean plasma adiponectin concentrations in WT (E) and HD (F) mice are shown. Values are the mean ± S.E., *, p ≤ 0.05; n = 10 per group.
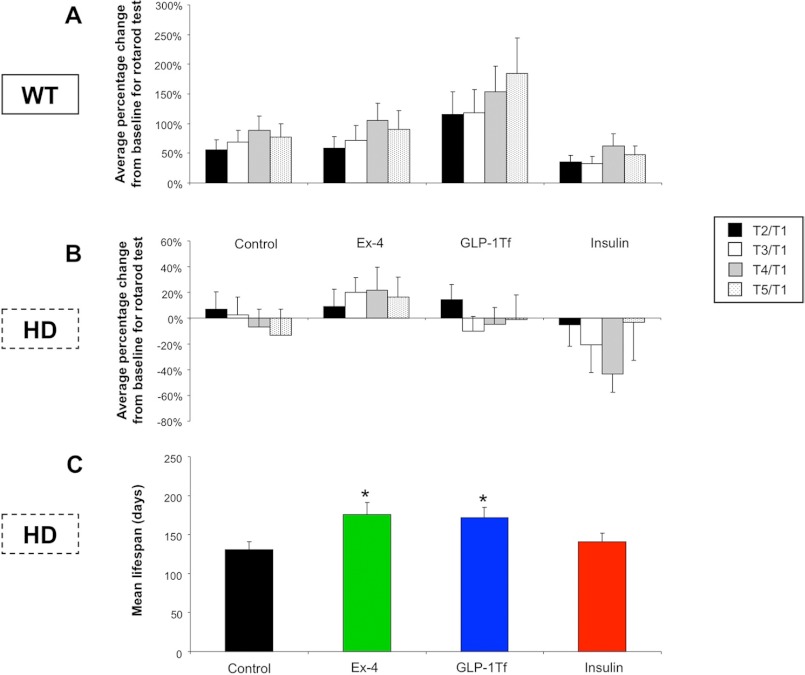
Euglycemic Agents Differentially Improve Motor Control in HD Mice
The N171–82Q model demonstrates a progressive decline of motor performance (9). We assessed RotaRod performance (time on RotaRod) as a percentage change of performance compared with performance in week 1 for 5 consecutive weeks in WT (Fig. 6A) and HD (Fig. 6B) mice. HD control mice spent significantly less time on the RotaRod than WT mice, which indicates that motor function was significantly impaired in the HD control mice (Fig. 6, A and B). For WT mice, Ex-4 treatment enhanced their ability to stay on the RotaRod (p ≤ 0.05). Additionally, GLP-1-Tf treatment also caused an increase in time spent on the RotaRod, which may be connected to increases in physical activity that is often observed with GLP-1 dosing. Insulin treatment had no effect on RotaRod performance in WT mice (Fig. 6A). Control HD mice demonstrated a progressive reduction in their ability to remain on the RotaRod during successive testing periods (Fig. 6B), indicated by the negative percentage change of performance compared with their initial base-line (T1) test. Ex-4-treated HD mice showed a reversal of this motor performance decline and actually demonstrated successive increases in performance throughout the study. GLP-1-Tf treatment partially attenuated performance loss in HD mice but, unlike Ex-4, failed to enhance performance score. Insulin treatment of HD mice exacerbated the degree of progressive decline in motor performance compared with the control HD group (Fig. 6B).
FIGURE 6.
RotaRod performance and mean life span are altered by Ex-4, GLP-1-Tf, and insulin treatments. Average percentage change from base line on the RotaRod test for WT (A) or HD mice (B) treated with either saline (control), Ex-4, GLP-1-Tf, or insulin during the study period. T1, RotaRod test 1; T2, RotaRod test 2; T3, RotaRod test 3; T4, RotaRod test 4. C, percent survival for HD animals throughout the study. Values are the mean ± S.E., *, p ≤ 0.05; n = 10 per group.
Euglycemic Agents Differentially Affect Life Span of HD Mice
HD mice demonstrated a significantly reduced life span (∼140 days) compared with WT mice (>730 days) (9). No premature deaths occurred in the WT animal cohort in this study. Ex-4 and GLP-1-Tf treatment, but not insulin treatment, significantly increased the mean life span of HD mice (Fig. 6C).
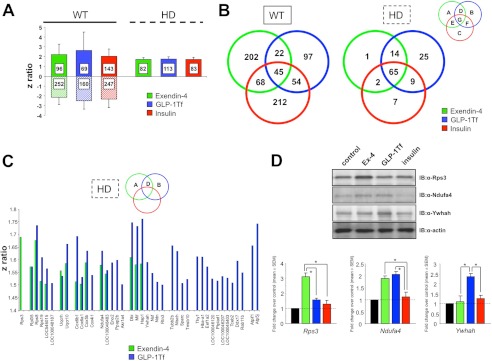
Euglycemic Agent-mediated Transcriptomic Alterations in Hypothalamus Are Related to Their Pharmacological Activity
We next used genomic analyses to investigate the effects of the euglycemic agents on the hypothalamic transcriptomic signatures of WT (Ex-4, GLP-1-Tf, and insulin: supplemental Tables S6–S8, respectively) and HD mice (Ex-4, GLP-1-Tf, and insulin: supplemental Tables S9–S11, respectively) (Fig. 7A). All three compounds significantly regulated multiple gene transcripts in both genotypes, and a numerically greater effect was observed in WT mice. In contrast to WT mice, only hypothalamic transcriptional up-regulation was observed in HD mice in response to Ex-4, GLP-1-Tf, or insulin. Specific transcriptional drug effects in WT mice were more unique to the individual drug compound than in HD mice (Fig. 7B). As our data suggested an HD therapeutic order of potency of Ex-4 > GLP-1-Tf > insulin (Figs. 2, 3, and 5), we assessed whether this signature could be detected in the hypothalamic transcriptome data (9). A greater similarity of function was seen between Ex-4 and GLP-1-Tf (the two most effective agents) compared with either compound or insulin (Venn intersection D, Fig. 7B). The ribosomal protein S3 (Rps3) was the only Ex-4-uniquely controlled hypothalamic gene. 25 transcripts were uniquely controlled by GLP-1-Tf, and 14 genes were co-regulated by Ex-4 and GLP-1-Tf. As only Ex-4 and GLP-1-Tf exerted therapeutic effects in HD mice, the z ratios for the genes that may elicit these actions are shown in Fig. 7C (Venn intersections A, B, and D). We validated the specific expression profiles of multiple protein products from this potentially therapeutic gene subset using Western blotting (Rps3, Ndufa4, and Ywhah, Fig. 7D). Despite the demonstrated effective physiological effects of the euglycemic agents (Figs. 3–6), a relatively small number of individual associated genomic factors were strongly and significantly modulated (Fig. 7). It is therefore possible that a better appreciation of the evident palliative effects of these drug compounds could instead be generated at a functional signature level through the coherent but small regulation of multiple transcripts and transcript groups (19). Therefore, rather than only assuming that drug actions occur via modulation of singular linear signaling pathways, they may instead exist at a systemic network level and thus a “higher order” level of appreciation may be required to understand and quantify their efficacy (23).
FIGURE 7.
Euglycemic agent-mediated transcriptomic alterations in hypothalamus are related to their pharmacological activity. A, hypothalamic z ratio ranges (mean ± S.D.), compared with control, for Ex-4-, GLP-1-Tf-, or insulin-treated WT or HD mice. B, Venn diagram separation of genes significantly regulated by Ex-4, GLP-1-Tf, or insulin treatment of WT or HD mice (green circles, Ex4; blue circles, GLP-1-Tf; red circles, insulin). C, z ratios of genes specifically regulated by only Ex-4 and/or GLP-1-Tf in HD mice. D, representative Western blot validation of Ex-4- and/or GLP-1-Tf-regulated genes in HD mice (controlled by actin expression). Associated histograms represent mean ± S.E. for the fold changes in expression of the specific proteins compared with control animal levels after Ex-4, GLP-1-Tf, or insulin treatment.
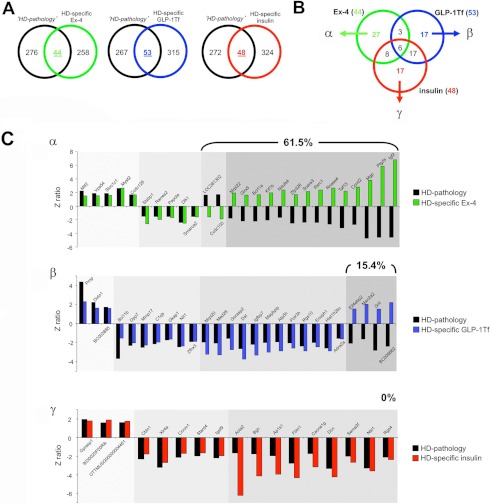
Euglycemic Agent Reversal of Hypothalamic HD Transcriptomic Signature Pathology Is Related to Eventual Functional Efficacy
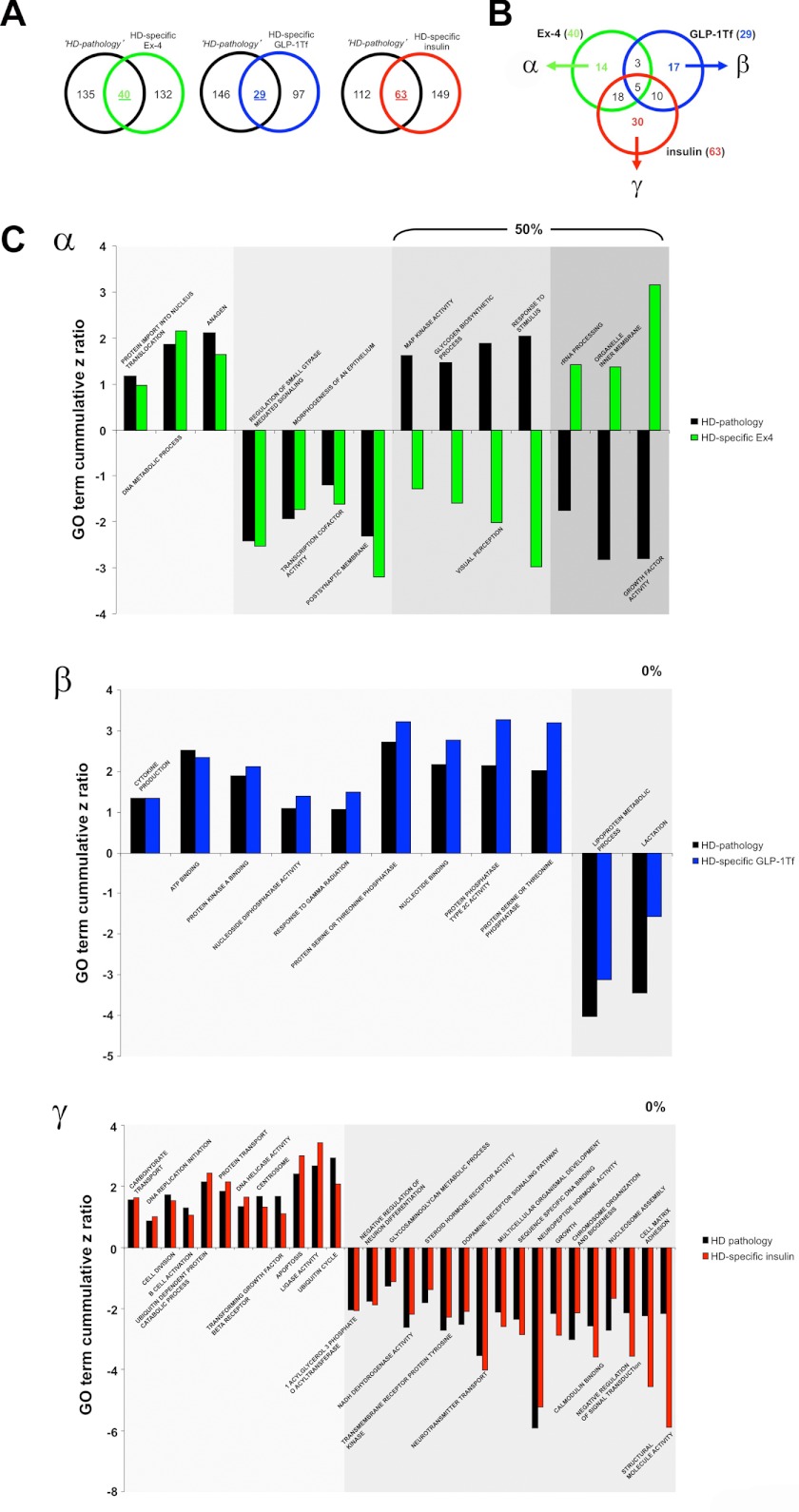
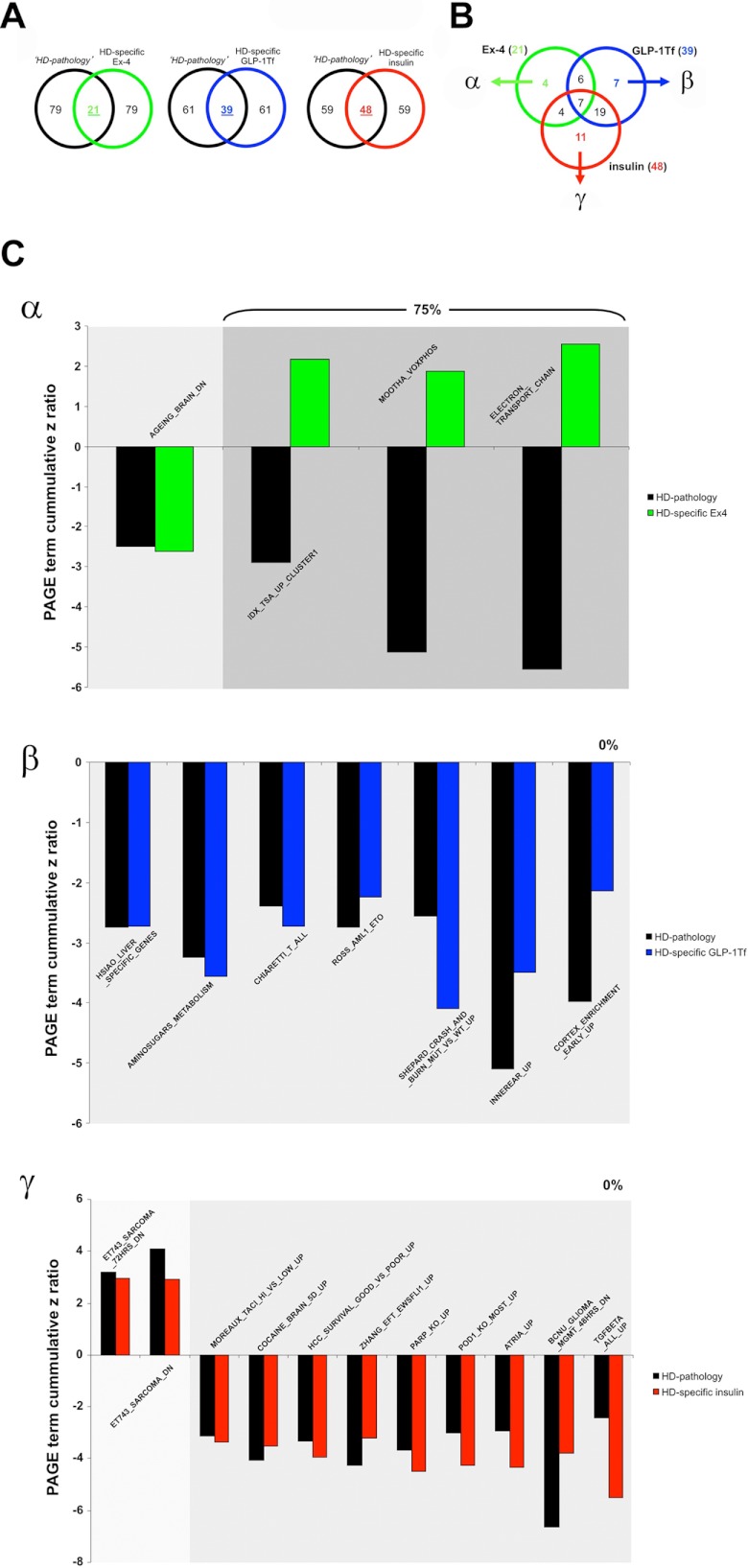
Using transcriptomic analysis, we identified a distinct set of genes associated with hypothalamic pathophysiology in HD mice (Fig. 1 and supplemental Table S1). To objectively compare the genotropic effects that the three euglycemic agents exert upon this “pathological” Geneset, we investigated the intersections between this pathological Geneset (control HD compared with control WT, supplemental Table S1) and the HD-specific actions of the three euglycemic agents (Ex-4/GLP-Tf/insulin-treated HD compared with Ex-4/GLP-1-Tf/insulin-treated WT, supplemental Tables S12–S14, respectively). Venn analysis of the pathological Geneset intersection with the experimental compound-controlled genesets is indicated in Fig. 8A. We found 44, 53, and 48 genes that were significantly regulated in both the pathological Geneset and the Ex-4/GLP-1-Tf/insulin-treated HD-specific genesets. Of these three gene groups, 27 transcripts were unique to Ex-4 activity alone (group α); 17 were unique to GLP-1-Tf activity alone (group β), and 17 were unique to insulin activity alone (group γ) (Fig. 8B). We found that 61.5% of the genes common to Ex-4 HD-specific activity and the pathological set showed a reversal in expression polarity; furthermore, we found that a considerably lower degree of similar transcript expression reversal was seen with GLP-1-Tf (15.4%) and insulin (0%) (Fig. 7C) activity compared with the HD pathological Geneset. Therefore, Ex-4, which of the three compounds demonstrated the most beneficial actions in HD mice, also seems to possess the greatest ability to directly reverse the direction of expression of hypothalamic genes associated with the HD genotype. We have shown that the extent of drug-induced reversal of the transcriptomic pathological signature of HD is strongly correlated to the agent's eventual efficacy. We next investigated the potential functional aspects, using GO terms and MSigDB pathways, of these beneficial drug signature reversal effects. Using the same Venn diagram-mediated approach employed in Fig. 8, A and B, we generated the α-, β-, and γ-specific subsets of significantly populated GO term group annotation (supplemental Tables S3 and S15–S17) from the drug treatment paradigms indicated in Fig. 9, A and B. Of these three GO term clusters, 14 GO terms were unique to Ex-4 activity alone (group α), 17 were unique to GLP-1-Tf activity alone (group β), and 30 were unique to insulin activity alone (group γ) (Fig. 9B). We found that 50% of the GO terms common to Ex-4 HD-specific activity and the pathological set (α) showed a reversal in GO term z score polarity. In contrast we found a complete absence of pathological GO term z score polarity reversal with both GLP-1-Tf (0%, β) and insulin (0%, γ) treatment (Fig. 9C). Therefore, as with the actual transcriptomic pathological signature (Fig. 8C), the most efficacious agent, Ex-4, demonstrated the greatest ability to reverse the functional GO term pathological HD signature as well. Reinforcing the concept that Ex-4 may engender a systemic therapeutic action via a multidimensional support of metabolic function, we found that many of the Ex-4-reversed pathological GO terms were associated with energy regulation, e.g. glycogen biosynthetic process, organelle inner membrane, and growth factor activity (Fig. 9C). GO term annotation is an excellent mechanism to identify broad functional aspects of a specific complex dataset; however, a stronger phenotypic and signaling prediction can be gained with MSigDB PAGE collection analysis. We therefore repeated the HD pathological signature analysis (Figs. 8 and 9) using the statistically significant drug-related MSigDB PAGE collection annotations generated (supplemental Tables S4 and S18–S20). Using the same Venn diagram-mediated approach employed in Figs. 8, A and B, and 9, A and B, we generated the α-, β-, and γ-specific subsets of significantly populated MSigDB PAGE collection annotation (supplemental Tables S4 and S18–S20) from the drug treatment paradigms indicated in Fig. 10, A and B. Four PAGE collections were unique to Ex-4 activity alone (group α); seven were unique to GLP-1-Tf activity alone (group β), and 11 were unique to insulin activity alone (group γ) (Fig. 10B). We found that 75% of the PAGE collections common to Ex-4 HD-specific activity and the pathological set (α) showed a reversal in GO term z score polarity. As with the GO term analysis, we found that no z score reversals of pathological collections were observed with both GLP-1-Tf (0%, β) or insulin (0%, γ) treatment (Fig. 10C). With specific respect to the functional MSigDB PAGE collection pathological signature reversed by Ex-4, a clear emphasis upon energy regulation again was evident, i.e. MSigDB PAGE collections reversed included ELECTRON_TRANSPORT_CHAIN and MOOTHA_VOXPHOS, both of which are critically involved in global metabolism (Fig. 10C).
FIGURE 8.
Euglycemic agents can reverse hypothalamic HD transcriptomic pathology to different degrees. Isolation of significantly regulated genes common between the HD mouse hypothalamic pathology set (black circles, HD pathology) and the HD-specific, Ex4- (green circle), GLP-1-Tf- (blue circle), or insulin (red circle)-regulated transcriptomic sets (A). Venn diagram isolation of Ex-4/GLP-1-Tf/insulin-unique HD pathology-related genes. Ex-4-unique HD pathology-related Geneset is designated α, GLP-1-Tf-unique HD pathology-related Geneset is designated β, and insulin-unique HD pathology-related Geneset is designated γ (B). C, z ratio distributions of α, β, and γ Geneset constituents. HD pathology-regulated genes are denoted in black, and the drug-dependent gene regulatory behavior is color-coded as follows: green bars, Ex-4; blue bars, GLP-1-Tf; red bars, insulin. The numerical percentage of these genes in the sets α, β, or γ that display a reversed polarity of z ratio regulation between the HD pathology data and the drug-controlled data is indicated in each panel.
FIGURE 9.
Euglycemic agents can reverse hypothalamic HD GO term pathology to different degrees. Isolation of significantly regulated GO terms are common between the HD mouse hypothalamic pathology set (black circles, HD pathology) and the HD-specific Ex4- (green circle), GLP-1-Tf- (blue circle), or insulin (red circle)-regulated GO term annotated datasets (A). Venn diagram isolation of Ex-4/GLP-1-Tf/insulin-unique HD pathology-related GO term groups is shown. Ex-4-unique HD pathology-related GO term set is designated α, GLP-1-Tf-unique HD pathology-related GO term set is designated β, and insulin-unique HD pathology-related GO term set is designated γ (B). C, z score distributions of α, β, and γ GO term set constituents. HD pathology-regulated GO term groups are denoted in black, and the drug-dependent GO term regulatory behavior is color-coded as follows: green bars, Ex-4; blue bars, GLP-1-Tf; red bars, insulin. The numerical percentage of these GO terms in the sets α, β, or γ that display a reversed polarity of z score regulation between the HD pathology data and the drug-controlled data is indicated in each panel.
FIGURE 10.
Euglycemic agents can reverse hypothalamic HD MSigDB PAGE collection pathology to different degrees. Isolation of significantly regulated PAGE collections common between the HD mouse hypothalamic pathology set (black circles, HD pathology) and the HD-specific Ex4- (green circle), GLP-1-Tf- (blue circle), or insulin (red circle)-regulated MSigDB PAGE annotated datasets is shown (A). Venn diagram isolation of Ex-4/GLP-1-Tf/insulin-unique HD pathology-related PAGE collections is shown. Ex-4-unique HD pathology-related PAGE collection set is designated α; GLP-1-Tf-unique HD pathology-related PAGE collection set is designated β, and insulin-unique HD pathology-related PAGE collection set is designated γ (B). C, z score distributions of α, β, and γ PAGE collection set constituents. HD pathology-regulated MSigDB PAGE collections are denoted in black, and the drug-dependent PAGE collection regulatory behavior is color-coded as follows: green bars, Ex-4; blue bars, GLP-1-Tf; red bars, insulin. The numerical percentage of these PAGE collections in the sets α, β, or γ that display a reversed polarity of z score regulation between the HD pathology data and the drug-controlled data is indicated in each panel.
Quantitative Bioinformatic Therapeutic Signature Analysis
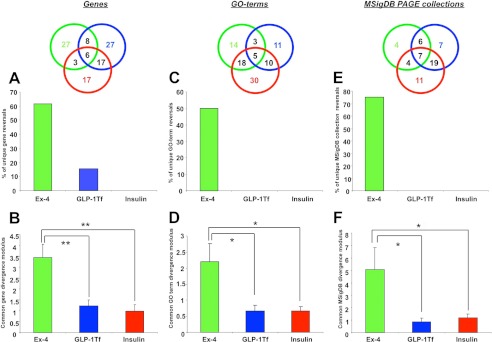
We have shown that Ex-4 has the greatest ability to reverse the regulatory direction of the largest number of HD pathology-related hypothalamic transcripts. Simply reversing the direction of gene transcription regulation (up versus down), however, may not be the sole therapeutic mechanism, i.e. small quantitative changes without direction reversal may also contribute to observed therapeutic effects. To investigate this, we measured the mean gene transcript divergence modulus (modulus of z ratio difference between pathological gene regulations and the drug-mediated gene regulations in sets α, β, and γ) using the three previously described gene groups α, β, and γ (Fig. 8C). Calculating the mean transcript divergence moduli for Ex-4, GLP-1-Tf, and insulin (from genes in groups α, β, or γ) revealed that the greatest mean divergence from the HD pathological genes was generated by Ex-4 (Fig. 11, A and B). This HD pathology-related gene divergence induced by Ex-4 was significantly greater than that for GLP-1-Tf (p ≤0.001) and insulin (p ≤0.001). In addition to analyzing drug activity at the gene level, we also conducted genomic analyses for the analogous GO terms and the MSigDB PAGE gene collection annotations of the same genesets. These two forms of bioinformatic annotation provide a prediction of the physiological actions of the significantly regulated genesets (24). We used our pathology versus drug-related Geneset analysis (as used in Fig. 8) with GO term (pathology set, supplemental Table S3; GO terms Ex-4, GLP-1-Tf and insulin sets, supplemental Tables S15–S17, respectively; Fig. 9) and MSigDB PAGE collections (pathology set, supplemental Table S4; MSigDB PAGE-Ex-4, GLP-1-Tf, and insulin sets, supplemental Tables S18–S20, respectively; Fig. 10). For GO term analysis, Ex-4 reversed the greatest number of HD pathology-related GO term groups (Fig. 11C) and, compared with GLP-1-Tf and insulin, generated a significantly greater GO term z score divergence modulus (p ≤0.05; Fig. 11D). For MSigDB PAGE collection analysis, we found that compared with GLP-1-Tf and insulin, Ex-4 reversed the greatest number of HD pathology-related MSigDB PAGE gene collections (Fig. 11E) and generated a significantly greater MSigDB PAGE collection z score divergence modulus (p ≤ 0.05; Fig. 11F). Our mixture of classical empirical physiological data with advanced informatic analyses provide evidence that it could be feasible to investigate the relative efficacy of therapeutic compounds using focused tissue-based bioinformatic approaches to illuminate drug-mediated reversals of pathological signatures.
FIGURE 11.
Quantitative bioinformatic therapeutic analysis. A, percentage of Ex-4/GLP-1-TF or insulin-unique HD-related genes with a reversed polarity of regulation compared with those in the HD pathology set. B, mean ± S.E. gene z ratio divergence modulus between the HD pathology Geneset and the drug-unique controlled HD-related genesets. C, percentage of Ex-4/GLP-1-TF or insulin-unique HD-related GO term groups with a reversed polarity of regulation compared with those in the HD pathology set. D, mean ± S.E. gene GO term group z score divergence modulus between the HD pathology GO terms groups and the drug-unique-controlled HD-related GO term groups. E, percentage of Ex-4/GLP-1-TF or insulin-unique HD-related MSigDB PAGE collections with a reversed polarity of regulation compared with those in the HD pathology MSigDB PAGE collections. F, mean ± S.E. MSigDB PAGE collection z score divergence modulus between the HD pathology MSigDB PAGE collections and the drug-unique controlled HD-related MSigDB PAGE collections.
DISCUSSION
We have previously shown that HD is a neurological disorder with significant metabolic/dysglycemic co-morbidities (2, 9). In this study, we demonstrated significant transcriptomic alterations in the hypothalamus of the N171–82Q HD mice. It is interesting to note that although the initiation of the disorder is monogenic, there is a considerable transcriptomic effect, or signature, in the hypothalamus and probably other organs as well, e.g. the pancreas. The hypothalamus provides a vital link between the central nervous system and the periphery and could play an important role in HD pathophysiology. Unbiased LSI analysis of the relationships between the hypothalamic genes significantly altered in HD mice indicated that a strong neurodegenerative and metabolic signature was present (Figs. 1 and 2). Many transcripts altered in the HD hypothalamus were associated with neurodegenerative disorders. Down-regulated genes in HD mice compared with WT animals included arginine vasopressin (Avp), prostaglandin D2 synthase (Ptgds), insulin-like growth factor 2 (Igf2), cocaine- and amphetamine-regulated transcript (Cart), mitogen-activated protein kinase 8 interacting protein (Mapk8ip, also known as JNK-interacting protein 1), and somatostatin (Sst). Avp is reduced in brain tissue and cerebrospinal fluid of patients with Alzheimer disease and Down syndrome (25, 26). Igf2 can exert neuroprotective activity in Alzheimer disease (27), whereas genetic disruption of Igf2 has been shown to exacerbate Parkinson disease (28). Mapk8ip expression is strongly associated with neuroprotective activity in Parkinson disease patients (29). Cart is involved in hypothalamic dysfunction in neurodegenerative disorders such as Dementia with Lewy Bodies (30). Reduced Sst levels have also been demonstrated in HD patient hypothalami (31). Neuropathology-related transcripts elevated in HD hypothalami include prion protein (Prnp), transferrin receptor (Tfr), sarcoglycan β (Sgcb), and Abelson helper integration site (Ahi1). Prnp has been shown to be involved in the etiology of numerous neurodegenerative disorders such as HD (32), Parkinson disease (33), and Hereditary Sensory and Motor Neuropathy (34), whereas Tfr expression has also been implicated in multiple neurodegenerative disorders as tissue iron levels are strongly correlated with neuronal oxidative damage (35). Mutations in Sgcb, a vital component of the dystrophin-associated glycoprotein scaffolding complex, causes significant movement disorders (36). Altered Ahi1 expression has also been linked to multiple neurological disorders, e.g. cerebellar ataxia (37).
As the hypothalamic signature in HD mice demonstrated a strong pathophysiological imprint (supplemental Tables S2–S4), we investigated how treatment with compounds that possess multiple activities, including euglycemic and neuroprotective roles, are associated with changes in hypothalamic gene transcription. In a previous study, we found that Ex-4 was able to ameliorate some of the pathophysiologies in N171–82Q mice (9). Here, we compared the HD therapeutic activity of Ex-4 to our novel non-brain penetrant GLP-1 analog (GLP-1-Tf (17)) and long lasting insulin (Lantus). For multiple indices of HD pathology, e.g. elevated blood glucose (Fig. 3), altered pancreatic islet morphology (Fig. 4), motor dysfunction (Fig. 6), and life span (Fig. 6), we found Ex-4 to be the most effective, whereas GLP-1-Tf was comparatively less effective, and insulin was completely ineffective. Our observation of a small but significant overall life span extension with Ex-4/GLP-1-Tf, may be associated with even more impressive effects in potential human studies, as this and other murine models possess considerably greater CAG repeat numbers (82–150) than human HD patients (36–50). This pathophysiological divergence, between animal models and humans, has been considered to be a profound hindrance for the demonstration of effective HD therapeutics in murine models (38).
If the neurological-endocrinological characteristics of HD are indeed associated with hypothalamic disruption, then it may be possible that the differential therapeutic transcriptional signatures of these three compounds are evident in the hypothalamus. As even in the cases of monogenic disorders such as HD, it is clear that multiple physiological alterations occur in the disease process, then perhaps appreciating and eventually treating such conditions is best approached in a systemic network manner (23). Therefore, in this study we strove to investigate the therapeutic interface between the disease and drug effects in a rational tissue locus, i.e. the hypothalamus. When we analyzed the effects of Ex4, GLP-1-Tf, and insulin on HD hypothalamic transcriptomes, we found that these functionally related compounds controlled many common genes (Fig. 7B). As many of these 65 common genes were shared with the ineffective insulin treatment, it is likely that the genes regulated only by Ex-4 or GLP-1-Tf were associated with their therapeutic activity in the hypothalamus. Only one gene was uniquely controlled in the hypothalamus by the most efficacious compound Ex-4, ribosomal protein S3 (Rps3) (Fig. 6C). Of all the treatment paradigms, hypothalamic Rps3 up-regulation was only observed with Ex-4 treatment of HD mice (Fig. 7D). Ex-4-mediated therapeutic activity may be strongly regulated by elevation of Rps3, as this protein is associated with both Akt-1-dependent neuroprotection and DNA-damage repair response processes (39) that together may combine to ameliorate hypothalamic damage. Among the potentially therapeutic transcripts co-regulated by Ex-4 and GLP-1-Tf, several strongly represented functional groups were present as follows: metabolic controllers (Uqcrh (40) and Uqcr10), cyclooxygenases (Cox4–6 isoforms) (37), and proteins associated with neuro- and diabetic pathophysiologies (Hap1, Mif, and Ndufa4) (41–43). Potentially therapeutic transcripts solely stimulated by GLP-1-Tf included Atpif1 (44), Ywhah/14-3-3-eta (45), and Tubb2b (46). Reinforcing our hypothesis that hypothalamic functionality could represent a keystone aspect of HD pathology, many of the transcripts controlled by Ex4/GLP1-Tf are simultaneously associated with neuroprotective and anti-diabetic activity as follows: Rps3 (37, 47); Mif (42, 48); Hap1 (41, 49); Ywhah/14-3-3-eta (45, 50); Nisch (51); and Sparc (52–53). In addition to the coherent nature of transcription patterns altered by the effective compounds, we were also able to demonstrate that drug-induced quantitative transcriptomic signature patterns, at multiple functional levels, in the hypothalamus strongly correlated to their physiological efficacy (Figs. 8–10). We found that Ex-4 treatment reversed the expression polarity of the greatest number of hypothalamic HD-specific pathological genes, GO term groups, and MSigDB PAGE collections compared with the other compounds (Figs. 8–10). Indeed, the overall gene transcript expression differences (divergence modulus from HD-related genes) for HD-specific transcripts, GO term groups, and MSigDB PAGE collections were significantly greater for Ex-4 compared with GLP-1-Tf or insulin (Fig. 11). Through our use of combined transcriptomic signature analyses, we were able to appreciate the drug-disease interactions at both the gene level and a higher functional organizational level. From this form of analysis, we were able to demonstrate that perhaps the most important mechanism by which Ex-4 was exerting its beneficial effects was via a broad systemic mechanism of metabolic support (Fig. 10). Our informatic demonstration of efficacy based upon a network style of drug effect is in line with previous data concerning the HD benefits of normalizing energy balance (54, 55). Therefore, rather than focusing upon individual response pathways, an additional complementary approach for drug discovery and efficacy measurement may be needed to assess the interactions between the pathological and drug-induced transcriptomic signatures. This may be especially important for therapeutic situations in which a broad, multitissue level systemic efficacy, e.g. somatic energy metabolism, may prove the best solution. Clearly at the present time standard drug investigation paradigms of singular drug-target analysis constitute the primary effective discovery mechanism. However, considering our modern ability to appreciate disease complexity, with quantitative transcriptomics and proteomics, at a functional signature level then perhaps the study of therapeutic drug effects should also be performed at this same intensive and thorough level of investigation. In this study, we found that our multiple bioinformatic analyses strongly recapitulated our standard HD therapeutic physiological findings and therefore may pave the way for the generation of complementary techniques for analysis of drug activity in the context of highly complex disease processes.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program, NIA.

This article contains supplemental Tables 1–20.
- HD
- Huntington disease
- Tfr
- transferrin receptor
- LSI
- latent semantic indexing
- GO
- Gene Ontology
- MSigDB PAGE
- Molecular Signatures Database Parametric Analysis of Geneset Enrichment
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Landles C., Bates G. P. (2004) Huntingtin and the molecular pathogenesis of Huntington disease. Fourth in molecular medicine review series. EMBO Rep. 5, 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martin B., Golden E., Keselman A., Stone M., Mattson M. P., Egan J. M., Maudsley S. (2008) Therapeutic perspectives for the treatment of Huntington disease. Treating the whole body. Histol. Histopathol. 23, 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Petersén A., Björkqvist M. (2006) Hypothalamic-endocrine aspects in Huntington disease. Eur. J. Neurosci. 24, 961–967 [DOI] [PubMed] [Google Scholar]
- 4. Aziz N. A., Swaab D. F., Pijl H., Roos R. A. (2007) Hypothalamic dysfunction and neuroendocrine and metabolic alterations in Huntington disease. Clinical consequences and therapeutic implications. Rev. Neurosci. 18, 223–251 [DOI] [PubMed] [Google Scholar]
- 5. Gaba A. M., Zhang K., Marder K., Moskowitz C. B., Werner P., Boozer C. N. (2005) Energy balance in early-stage Huntington disease. Am. J. Clin. Nutr. 81, 1335–1341 [DOI] [PubMed] [Google Scholar]
- 6. Van der Burg J. M., Bacos K., Wood N. I., Lindqvist A., Wierup N., Woodman B., Wamsteeker J. I., Smith R., Deierborg T., Kuhar M. J., Bates G. P., Mulder H., Erlanson-Albertsson C., Morton A. J., Brundin P., Petersen A., Bjorkqvist M. (2007) Increased metabolism in the R6/2 mouse model of Huntington disease. Neurobiol. Dis. 29, 41–51 [DOI] [PubMed] [Google Scholar]
- 7. Trejo A., Tarrats R. M., Alonso M. E., Boll M. C., Ochoa A., Velásquez L. (2004) Assessment of the nutrition status of patients with Huntington disease. Nutrition 20, 192–196 [DOI] [PubMed] [Google Scholar]
- 8. Popovic V., Svetel M., Djurovic M., Petrovic S., Doknic M., Pekic S., Miljic D., Milic N., Glodic J., Dieguez C., Casanueva F. F., Kostic V. (2004) Circulating and cerebrospinal fluid ghrelin and leptin. Potential role in altered body weight in Huntington disease. Eur. J. Endocrinol. 151, 451–455 [DOI] [PubMed] [Google Scholar]
- 9. Martin B., Golden E., Carlson O. D., Pistell P., Zhou J., Kim W., Frank B. P., Thomas S., Chadwick W. A., Greig N. H., Bates G. P., Sathasivam K., Bernier M., Maudsley S., Mattson M. P., Egan J. M. (2009) Exendin-4 improves glycemic control, ameliorates brain and pancreatic pathologies, and extends survival in a mouse model of Huntington disease. Diabetes 58, 318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morton A. J., Wood N. I., Hastings M. H., Hurelbrink C., Barker R. A., Maywood E. S. (2005) Disintegration of the sleep-wake cycle and circadian timing in Huntington disease. J. Neurosci. 25, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Petersén A., Gil J., Maat-Schieman M. L., Björkqvist M., Tanila H., Araújo I. M., Smith R., Popovic N., Wierup N., Norlén P., Li J. Y., Roos R. A., Sundler F., Mulder H., Brundin P. (2005) Orexin loss in Huntington disease. Hum. Mol. Genet. 14, 39–47 [DOI] [PubMed] [Google Scholar]
- 12. Hult S., Schultz K., Soylu R., Petersén A. (2010) Hypothalamic and neuroendocrine changes in Huntington disease. Curr. Drug Targets 11, 1237–1249 [DOI] [PubMed] [Google Scholar]
- 13. Björkqvist M., Fex M., Renström E., Wierup N., Petersén A., Gil J., Bacos K., Popovic N., Li J. Y., Sundler F., Brundin P., Mulder H. (2005) The R6/2 transgenic mouse model of Huntington disease develops diabetes due to deficient beta-cell mass and exocytosis. Hum. Mol. Genet. 14, 565–574 [DOI] [PubMed] [Google Scholar]
- 14. Sathasivam K., Hobbs C., Turmaine M., Mangiarini L., Mahal A., Bertaux F., Wanker E. E., Doherty P., Davies S. W., Bates G. P. (1999) Formation of polyglutamine inclusions in non-CNS tissue. Hum. Mol. Genet. 8, 813–822 [DOI] [PubMed] [Google Scholar]
- 15. Markianos M., Panas M., Kalfakis N., Vassilopoulos D. (2005) Plasma testosterone in male patients with Huntington disease. Relations to severity of illness and dementia. Ann. Neurol. 57, 520–525 [DOI] [PubMed] [Google Scholar]
- 16. Papalexi E., Persson A., Björkqvist M., Petersén A., Woodman B., Bates G. P., Sundler F., Mulder H., Brundin P., Popovic N. (2005) Reduction of GnRH and infertility in the R6/2 mouse model of Huntington disease. Eur. J. Neurosci. 22, 1541–1546 [DOI] [PubMed] [Google Scholar]
- 17. Kim B. J., Zhou J., Martin B., Carlson O. D., Maudsley S., Greig N. H., Mattson M. P., Ladenheim E. E., Wustner J., Turner A., Sadeghi H., Egan J. M. (2010) Transferrin fusion technology. A novel approach to prolonging biological half-life of insulinotropic peptides. J. Pharmacol. Exp. Ther. 334, 682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chadwick W., Zhou Y., Park S. S., Wang L., Mitchell N., Stone M. D., Becker K. G., Martin B., Maudsley S. (2010) Minimal peroxide exposure of neuronal cells induces multifaceted adaptive responses. PLoS One 5, e14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately down-regulated in human diabetes. Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 20. Doyle M. E., Egan J. M. (2007) Mechanisms of action of glucagon-like peptide 1 in the pancreas. Pharmacol. Ther. 113, 546–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pérez-Tilve D., González-Matías L., Alvarez-Crespo M., Leiras R., Tovar S., Diéguez C., Mallo F. (2007) Exendin-4 potently decreases ghrelin levels in fasting rats. Diabetes 56, 143–151 [DOI] [PubMed] [Google Scholar]
- 22. Altinova A. E., Toruner F., Bukan N., Yasar D. G., Akturk M., Cakir N., Arslan M. (2007) Decreased plasma adiponectin is associated with insulin resistance and HDL cholesterol in overweight subjects. Endocr. J. 54, 221–226 [DOI] [PubMed] [Google Scholar]
- 23. Schadt E. E., Friend S. H., Shaywitz D. A. (2009) A network view of disease and compound screening. Nat. Rev. Drug. Discov. 8, 286–295 [DOI] [PubMed] [Google Scholar]
- 24. Martin B., Brenneman R., Golden E., Walent T., Becker K. G., Prabhu V. V., Wood W., 3rd, Ladenheim B., Cadet J. L., Maudsley S. (2009) Growth factor signals in neural cells. Coherent patterns of interaction control multiple levels of molecular and phenotypic responses. J. Biol. Chem. 284, 2493–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Labudova O., Fang-Kircher S., Cairns N., Moenkemann H., Yeghiazaryan K., Lubec G. (1998) Brain vasopressin levels in Down syndrome and Alzheimer disease. Brain Res. 806, 55–59 [DOI] [PubMed] [Google Scholar]
- 26. Mazurek M. F., Growdon J. H., Beal M. F., Martin J. B. (1986) CSF vasopressin concentration is reduced in Alzheimer disease. Neurology 36, 1133–1137 [DOI] [PubMed] [Google Scholar]
- 27. Jarvis K., Assis-Nascimento P., Mudd L. M., Montague J. R. (2007) β-Amyloid toxicity and reversal in embryonic rat septal neurons. Neurosci. Lett. 423, 184–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sutherland G., Mellick G., Newman J., Double K. L., Stevens J., Lee L., Rowe D., Silburn P., Halliday G. M. (2008) Haplotype analysis of the IGF1-INS-TH gene cluster in Parkinson disease. Am. J. Med. Genet. B 147, 495–499 [DOI] [PubMed] [Google Scholar]
- 29. Xia X. G., Harding T., Weller M., Bieneman A., Uney J. B., Schulz J. B. (2001) Gene transfer of the JNK interacting protein-1 protects dopaminergic neurons in the MPTP model of Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 98, 10433–10438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schultz K., Wiehager S., Nilsson K., Nielsen J. E., Lindquist S. G., Hjermind L. E., Andersen B. B., Wallin A., Nilsson C., Petersén A. (2009) Reduced CSF CART in dementia with Lewy bodies. Neurosci. Lett. 453, 104–106 [DOI] [PubMed] [Google Scholar]
- 31. Timmers H. J., Swaab D. F., van de Nes J. A., Kremer H. P. (1996) Somatostatin 1–12 immunoreactivity is decreased in the hypothalamic lateral tuberal nucleus of Huntington disease patients. Brain Res. 728, 141–148 [DOI] [PubMed] [Google Scholar]
- 32. Jankoviç N., Kecmanoviç M., Dimitrijeviç R., Keckareviç-Markoviç M., Dobriciç V., Keckareviç D., Saviç Paviceviç D., Savić Paviceviç D., Romac S. (2008) HD phenocopies. Possible role of Saitohin gene. Int. J. Neurosci. 118, 391–397 [DOI] [PubMed] [Google Scholar]
- 33. Gossrau G., Herting B., Möckel S., Kempe A., Koch R., Reichmann H., Lampe J. B. (2006) Analysis of the polymorphic prion protein gene codon 129 in idiopathic Parkinson disease. J. Neural. Transm. 113, 331–337 [DOI] [PubMed] [Google Scholar]
- 34. Koop O., Timmerman V., de Jonghe P., Ringelstein B., Young P., Kuhlenbäumer G. (2005) Absence of mutations in the prion-protein gene in a large cohort of HMSN patients. Neuromuscul. Disord. 15, 549–551 [DOI] [PubMed] [Google Scholar]
- 35. LaVaute T., Smith S., Cooperman S., Iwai K., Land W., Meyron-Holtz E., Drake S. K., Miller G., Abu-Asab M., Tsokos M., Switzer R., 3rd, Grinberg A., Love P., Tresser N., Rouault T. A. (2001) Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 27, 209–214 [DOI] [PubMed] [Google Scholar]
- 36. Zimprich A., Grabowski M., Asmus F., Naumann M., Berg D., Bertram M., Scheidtmann K., Kern P., Winkelmann J., Müller-Myhsok B., Riedel L., Bauer M., Müller T., Castro M., Meitinger T., Strom T. M., Gasser T. (2001) Mutations in the gene encoding ϵ-sarcoglycan cause myoclonus-dystonia syndrome. Nat. Genet. 29, 66–69 [DOI] [PubMed] [Google Scholar]
- 37. Tory K., Lacoste T., Burglen L., Morinière V., Boddaert N., Macher M. A., Llanas B., Nivet H., Bensman A., Niaudet P., Antignac C., Salomon R., Saunier S. (2007) High NPHP1 and NPHP6 mutation rate in patients with Joubert syndrome and nephronophthisis. Potential epistatic effect of NPHP6 and AHI1 mutations in patients with NPHP1 mutations. J. Am. Soc. Nephrol. 18, 1566–1575 [DOI] [PubMed] [Google Scholar]
- 38. Kim J., Bordiuk O. L., Ferrante R. J. (2011) Experimental models of HD and reflection on therapeutic strategies. Int. Rev. Neurobiol. 98, 419–481 [DOI] [PubMed] [Google Scholar]
- 39. Lee S. B., Kwon I. S., Park J., Lee K. H., Ahn Y., Lee C., Kim J., Choi S. Y., Cho S. W., Ahn J. Y. (2010) Ribosomal protein S3, a new substrate of Akt, serves as a signal mediator between neuronal apoptosis and DNA repair. J. Biol. Chem. 285, 29457–29468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu H., Yang Y., Allister E. M., Wijesekara N., Wheeler M. B. (2008) The identification of potential factors associated with the development of type 2 diabetes. A quantitative proteomics approach. Mol. Cell. Proteomics 7, 1434–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu L. L., Fan Y., Li S., Li X. J., Zhou X. F. (2010) Huntingtin-associated protein-1 interacts with pro-brain-derived neurotrophic factor and mediates its transport and release. J. Biol. Chem. 285, 5614–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pan W., Kastin A. J. (2007) From MIF-1 to endomorphin: the Tyr-MIF-1 family of peptides. Peptides 28, 2411–2434 [DOI] [PubMed] [Google Scholar]
- 43. Barkalifa R., Yagil Y., Yagil C. (2010) Sex-specific genetic dissection of diabetes in a rodent model identifies Ica1 and Ndufa4 as major candidate genes. Physiol. Genomics 42, 445–455 [DOI] [PubMed] [Google Scholar]
- 44. Hong Y., Piao F., Zhao Y., Li S., Wang Y., Liu P. (2009) Subchronic exposure to arsenic decreased Sdha expression in the brain of mice. Neurotoxicology 30, 538–543 [DOI] [PubMed] [Google Scholar]
- 45. Chen J., Lee C. T., Errico S. L., Becker K. G., Freed W. J. (2007) Increases in expression of 14-3-3η and 14-3-3ζ transcripts during neuroprotection induced by Δ9-tetrahydrocannabinol in AF5 cells. J. Neurosci. Res. 85, 1724–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jaglin X. H., Poirier K., Saillour Y., Buhler E., Tian G., Bahi-Buisson N., Fallet-Bianco C., Phan-Dinh-Tuy F., Kong X. P., Bomont P., Castelnau-Ptakhine L., Odent S., Loget P., Kossorotoff M., Snoeck I., Plessis G., Parent P., Beldjord C., Cardoso C., Represa A., Flint J., Keays D. A., Cowan N. J., Chelly J. (2009) Mutations in the β-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat. Genet. 41, 746–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mokhtari D., Barbu A., Mehmeti I., Vercamer C., Welsh N. (2009) Overexpression of the nuclear factor-κB subunit c-Rel protects against human islet cell death in vitro. Am. J. Physiol. Endocrinol. Metab. 297, E1067–E1077 [DOI] [PubMed] [Google Scholar]
- 48. Khan R. S., Yu C., Kastin A. J., He Y., Ehrensing R. H., Hsuchou H., Stone K. P., Pan W. (2010) Brain activation by peptide Pro-Leu-Gly-NH2 (MIF-1). Int. J. Pept. 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Niu S. N., Huang Z. B., Wang H., Rao X. R., Kong H., Xu J., Li X. J., Yang C., Sheng G. Q. (2011) Brainstem Hap1-Ahi1 is involved in insulin-mediated feeding control. FEBS Lett. 585, 85–91 [DOI] [PubMed] [Google Scholar]
- 50. Pers T. H., Hansen N. T., Lage K., Koefoed P., Dworzynski P., Miller M. L., Flint T. J., Mellerup E., Dam H., Andreassen O. A., Djurovic S., Melle I., Børglum A. D., Werge T., Purcell S., Ferreira M. A., Kouskoumvekaki I., Workman C. T., Hansen T., Mors O., Brunak S. (2011) Meta-analysis of heterogeneous data sources for genome-scale identification of risk genes in complex phenotypes. Genet. Epidemiol. 35, 318–322 [DOI] [PubMed] [Google Scholar]
- 51. Head G. A., Mayorov D. N. (2006) Imidazoline receptors, novel agents, and therapeutic potential. Cardiovasc. Hematol. Agents Med. Chem. 4, 17–32 [DOI] [PubMed] [Google Scholar]
- 52. Scuteri A., Ravasi M., Pasini S., Bossi M., Tredici G. (2011) Mesenchymal stem cells support dorsal root ganglion neurons survival by inhibiting the metalloproteinase pathway. Neuroscience 172, 12–19 [DOI] [PubMed] [Google Scholar]
- 53. Kos K., Wilding J. P. H. (2010) SPARC. A key player in the pathologies associated with obesity and diabetes. Rev. Endocrinol. 6, 225–235 [DOI] [PubMed] [Google Scholar]
- 54. Schilling G., Coonfield M. L., Ross C. A., Borchelt D. R. (2001) Coenzyme Q10 and remacemide hydrochloride ameliorate motor deficits in a Huntington disease transgenic mouse model. Neurosci. Lett. 315, 149–153 [DOI] [PubMed] [Google Scholar]
- 55. Hickey M. A., Zhu C., Medvedeva V., Franich N. R., Levine M. S., Chesselet M. F. (2012) Evidence for behavioral benefits of early dietary supplementation with CoEnzymeQ10 in a slowly progressing mouse model of Huntington disease. Mol. Cell. Neurosci. 49, 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]