Background: Crystal structures of an inward-facing CaiT conformation with four substrate sites are reported.
Results: This conformation is substrate-releasing. Arg262 facilitates release from the primary site.
Conclusion: Unbinding pathway from the primary site is determined by a broken helical portion of a transmembrane domain.
Significance: Mechanism of substrate release may be relevant to structural homologues such as neurotransmitter transporters.
Keywords: Biophysics, Carnitine, Membrane Proteins, Molecular Dynamics, Transport
Abstract
CaiT is a homotrimeric antiporter that exchanges l-carnitine (CRN) with γ-butyrobetaine (GBB) across the bacterial membrane. Three structures have been resolved to date for CaiT, all in the inward-facing state: CRN-bound (with four CRNs per subunit), GBB-bound (two GBBs per subunit), and apo. One of the reported binding sites is the counterpart of the primary site observed in structurally similar transporters. However, the mechanism and pathway(s) of CRN/GBB unbinding and translocation, or even the ability of the substrates to dislodge from the reported binding sites, are yet to be determined. To shed light on these issues, we performed a total of 1.3 μs of molecular dynamics simulations and examined the dynamics of substrate-bound CaiT structures under different conditions. We find that both CRN and GBB are able to dissociate completely from their primary site into the cytoplasm. Substrate molecules initially located at the secondary sites dissociate even faster (within tens of nanoseconds) into the extra- or intracellular regions. Interestingly, the unbinding pathway from the primary site appears to be dictated by the geometry of the unwound part of the transmembrane (TM) helix 3, mostly around Thr100 therein. Arg262 on TM7, which apparently mimics the role of Na+ in CaiT structural homologues, plays a key role in triggering the dissociation of the substrate away from the primary site and guiding its release to the cytoplasm provided that the unwound part of TM3 switches from a shielding to a yielding pose.
Introduction
In eukaryotes, l-carnitine (CRN)2 is a ubiquitous polar compound that is essential for the transport of activated fatty acids across the inner mitochondrial membrane. CRN deficiency causes critical metabolic disorders such as hypoglycemia, skeletal muscle myopathy, and cardiomyopathy (1–3). In bacteria, it plays important roles in various metabolic pathways (4, 5). It serves as an electron acceptor under certain anaerobic conditions, upon conversion into γ-butyrobetaine (GBB), the excreted end product (6, 7).
The antiporter CaiT exchanges extracellular (EC) CRN for intracellular (IC) GBB. CaiT has been crystallized in three forms: (i) CRN-bound, (ii) GBB-bound, and (iii) substrate-free (8, 9). In all three resolved structures, CaiT has an inward-facing conformation, i.e. its central region is exposed to the cytoplasm. The substrate-bound structures were derived from Escherichia coli (EcCaiT), and the substrate-free structures were derived from Proteus mirabilis (PmCaiT) (8, 9). The antiporters from these two species share a sequence identity of 87%, and the three structures exhibit a backbone root mean square deviation of 1.2 Å only between their monomeric units (8). The backbone conformation is also similar to that of the crystallized sodium/betaine transporter BetP, another member of the family of betaine/choline/carnitine transporters (10), although the primary site for betaine binding is occluded in BetP (11).
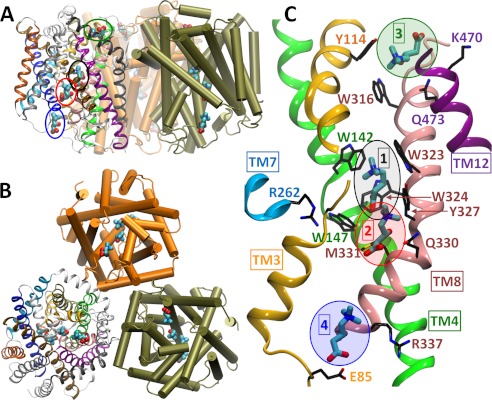
The crystal structures show that CaiT forms a homotrimer (Fig. 1, A and B), a finding supported by biochemical data (12). Each monomer is composed of 12 transmembrane (TM) helices, with the transport core formed by TM3–12. The CRN-bound structure contains four CRN molecules in each of the three subunits. The corresponding binding sites are labeled 1–4 in Fig. 1C, and we will refer to the carnitines originally bound to those sites as CRN1–CRN4. Site 1 is the so-called primary binding site, equivalent to that of leucine or galactose in the structurally homologous leucine transporter (LeuT) (13) and sodium-coupled galactose symporter (vSGLT) (14), respectively. As in the related BetP (11), the primary site is a “tryptophan box,” lined by Trp142 and Trp147 at the unwound region of TM4 (green) and Trp323, Trp324, and Tyr327 on the highly strained central part of TM8 (pink) (Fig. 1C and supplemental Fig. S1). Sites 2–4 are secondary binding sites: site 2 is at the base of the IC vestibule, ∼6 Å from the primary site; site 3 is in a shallow cavity on the EC surface ∼12 Å away from the primary site (9); and site 4 is at the entrance to the IC vestibule, ∼15 Å away from the primary site (Fig. 1 and supplemental Fig. S1). The GBB-bound structure contains two substrates only, at sites 1 and 3. The CRN and GBB binding poses at the equivalent sites overlap substantially in space and orientation (supplemental Fig. S1).
FIGURE 1.
Structure of CaiT trimer with four CRN binding sites on each monomer. Shown here is the crystallographically resolved trimeric structure of CaiT (PDB 3HFX). Each monomer contains four CRN molecules (enclosed by ellipses), as viewed through the membrane (A) or from the extracellular side (B). A close-up on the bound substrates at sites 1–4 is shown together with the adjacent helices and interacting residues as viewed from the side (C). Note the confinement of the substrate bound to site 1 (primary binding site) to a cage-like region surrounded by Trp142, Trp147, Trp323, Trp324, and Tyr327. The structure of the GBB-bound CaiT (PDB 2WSX; not shown) is very similar (backbone root mean square deviation = 1.2 Å). Therein, substrates are observed at the equivalent sites 1 and 3 only as illustrated in supplemental Fig. S1.
The cytoplasmic sides of CaiT monomers show a funnel-like cavity ∼25 Å deep and 15 Å wide that narrows down to roughly 7 Å in the center. In BetP, a sodium ion located at the so-called Na1 site coordinates the substrate betaine (11, 15), in a manner similar to the coordination of the carboxyl group of leucine by Na+ in LeuT (13, 16). In contrast to LeuT, vSGLT, and other structural homologues where transport activity is sodium-dependent, CaiT is sodium-independent, and so neither CaiT structure shows a cation at the Na1 site. Instead, in the GBB-bound structure, the sulfur of a methionine (Met331) is seen in the close proximity of the substrate carboxyl group (Fig. 1C and supplemental Fig. S1). This methionine is conserved in the bacterial CaiT and in the mammalian organic cation/CRN transporters. In the CRN-bound CaiT structure, Met331 sulfur is ∼5 Å away from the carboxyl group of substrates 1 and 2 (Fig. 1 and supplemental Fig. S1A).
In addition to the Na1 site, a more general cation binding site, referred to as Na2, is shared by several members of LeuT fold family. In LeuT, the Na2 site lies between TM1 and TM8 (13), the counterparts of TM3 and TM10, respectively, in CaiT. This sodium ion is known to provide structural stability and/or increased substrate affinity to Na+-coupled symporters (17–19). In CaiT, the same location, also modeled as the Na2 site in vSGLT and BetP (11, 13, 14), is occupied by Arg262 on TM7 (Fig. 1C and supplemental Fig. S1). Arg262 interacts with residues Cys99 and Thr100 on the unwound portion of TM3 in CRN-bound CaiT (Fig. 1 and supplemental Fig. S1) (9). In the GBB-bound structure, it interacts with Ala97, Cys99, and Thr100 on TM3, as well as Thr421 on TM10 (8).
As important as these crystal structures are, they represent a few out of many conformations visited by the transporter during the substrate transport/antiport cycle. They are not informative with regard to the mobility of the substrates, the mechanism and pathway(s) of substrate translocation, or the role of specific interactions with neighboring residues in mediating the movement of the substrates into or out of the antiporter. To shed light on such questions, we have used these crystallographic data to construct and simulate the dynamics of the two substrate-bound structures over relatively extensive time periods (up to 400 ns per run) under different conditions.
To our knowledge, these are the most extensive simulations reported to date for such a trimeric membrane protein of ∼170 kDa, with explicit water molecules and lipid bilayer (a system of ∼1.7 × 105 atoms). These simulations were made possible upon using the high performance (Terascale) computing resources provided by the Extreme Science and Engineering Discovery Environment (XSEDE). Our study provides insights into the mechanism of substrate release to the EC or IC regions by CaiT and highlights the key role of Arg262 near the unwound region of TM3 in facilitating substrate dislocation from the primary binding site. The study thus improves our understanding of the mechanism of action of a structural homologue to the important mammalian neuronal transporters belonging to the neurotransmitter:sodium symporter family, the targets of many natural and synthetic psychoactive drugs.
MATERIALS AND METHODS
System Preparation and Equilibration
Crystal structures of EcCait in the presence of CRN or GBB (Protein Data Bank (PDB) codes 3HFX and 2WSX, respectively) were placed in a 1-palmitoyl-2-oleoyl-phosphatidylethanolamine bilayer and solvated in water with 150 mm NaCl. To ensure optimal comparison between the two structures, we opted to construct very similar initial systems and used identical protocols and parameters for equilibration and productive runs. A total of six runs have been performed, the properties of which are listed in supplemental Table S1. The equilibration and simulation protocols are described in detail in the supplemental material. In summary, two sets of three runs were performed, labeled 1a–c and 2a–c, where the respective PDB structures 3HFX and 2WSX were used for the initial coordinates of CaiT atoms, hereafter referred to as conformations (conf) I and II, respectively. Runs a and b were performed with the crystal structure substrates, and run c was performed after swapping the substrates to assess the dependence of the trajectories on the specific protein conformer and substrate.
Simulation Properties
All runs, equilibration and productive, were performed using NAMD (version 2.8) (20) under NPT conditions at 1 atm and 310 K in a flexible cell, with periodic boundary conditions, 2-fs time steps, and rigid bonds for all hydrogen atoms. Particle mesh Ewald summations were adopted for full electrostatic interactions (spacing of 1 Å), along with a 10 Å cutoff distance (switching at 8 Å) for van der Waals and short-range electrostatic interactions. CHARMM36 force-field parameters were used for lipids, and CHARMM27 parameters were used for the protein and water molecules. VMD (version 1.9) (21) was used for visualization and construction of the systems and for trajectory analysis. The productive runs 1a, 1c, 2a, and 2c (supplemental Table S1) were unbiased molecular dynamics simulations, whereas in 1b and 2b, the backbone atoms of the residues that make close interatomic contacts (within 5 Å distance, based on conf I) with the substrates (a total of 34 residues) were constrained using spring forces of 10 kcal/(mol Å).
RESULTS
CaiT Substrates at Secondary Binding Sites Exhibit Rapid Dissociation/Unbinding Behavior
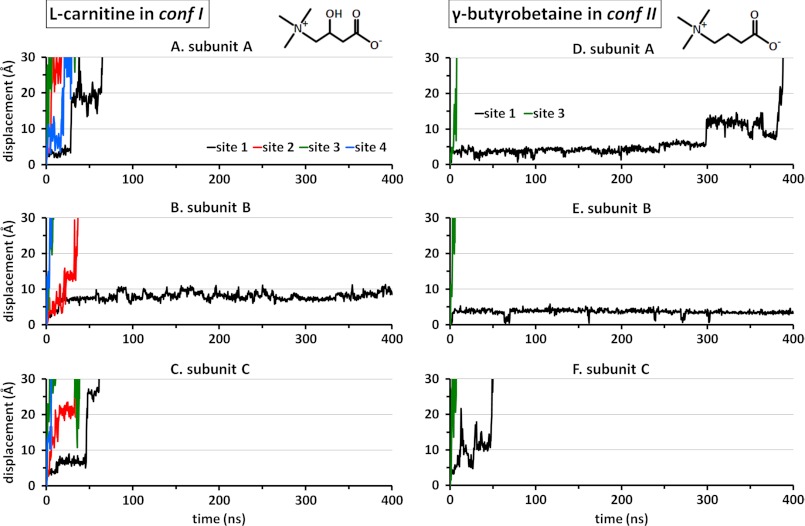
The two antiporter systems were highly stable over the entire 400-ns runs in the presence of CRN and GBB (runs 1a and 2a, respectively, supplemental Table S1). The backbone root mean square deviation of the α-helical domains remained smaller than 3 Å for CRN-bound CaiT and smaller than 2 Å for the GBB-bound structure. The most prominent feature was the spontaneous dissociation of the substrates located at the secondary binding sites (CRN2–CRN4) once the constraints imposed during equilibration were released. The rapid displacements of the substrates away from their original (secondary) binding sites are depicted in Fig. 2 by the colored curves. In particular, the diffusion of CRN3 to the EC environment was almost instantaneous (within the first few nanoseconds) for all subunits as was that of GBB3 (Fig. 2, A–C and D–F, for the respective confs I and II; green curves). CRN2 and CRN4 also exhibited fast (within tens of nanoseconds) unbinding and release to the cytoplasm (red and blue curves, respectively). On the other hand, the substrates bound to the primary site 1 displayed a different behavior in both CRN-bound and GBB-bound structures and will be the focus of the present study.
FIGURE 2.
Displacement in CRN and GBB away from their original binding sites. Results are displayed for the three respective subunits of each structure, based on the geometric center of each substrate molecule. The curves are color-coded according to the substrate binding sites, as indicated. A–C show the release of the CRNs bound to secondary sites 2–4 in all subunits and that of the CRNs bound to the primary site (site 1) on subunits A and C. The CRN bound to site 1 on subunit B (B; black curve) remains within the confines of site 1 during the course of (400-ns) simulations. D–F display the behavior of the two GBB molecules originally bound onto conf II, again confirming the rapid release of the substrate at site 3 and the resistance to translocation exhibited by GBB at the primary site, although the substrate is eventually released in two (A and C) of the three subunits.
Substrates at Secondary Binding Sites Unbind Even in the Absence of CaiT Backbone Rearrangements
As a further validation of the rapid unbinding and release properties of CRN2–CRN4, we generated two additional trajectories of 100 ns each (runs 1b and 2b, supplemental Table S1). The goal in this new set of simulations was to assess whether the unbinding and release within 1–10 nanoseconds from the secondary sites required accommodating motions from the part of the protein. To this aim, the backbone atoms of substrate-coordinating residues were spatially constrained, whereas their side chains were allowed to move. In parallel with previous observations, all substrates at sites 2–4 left the antiporter completely (supplemental Fig. S2). The main difference was a time delay in the release of CRN2 and CRN3 (whereas CRN4 remained unaffected). In particular, the CRN2 bound to subunit B (supplemental Fig. S2B, red curve) took more than 80 ns to be released, presumably due to the spatial constraints imposed on on-pathway residues. However, the eventual release of all substrates in sites 2–4 within 100 ns, even in the absence of accommodating rearrangements by the CaiT backbone, suggests that the present conformers are predisposed to substrate release, and the unbinding from the secondary sites involves only low energy barriers, if any.
Unbinding, Translocation, and Release of Substrate from the Primary Binding Site Takes Up to Hundreds of Nanoseconds and Requires Accommodating Rearrangements by the CaiT Backbone
We next examined the behavior of the substrates bound to the primary sites. The corresponding trajectories (Fig. 2, A–C for CRN1 and D–F for GBB1; black curves) show that four out of six substrates (those on subunits A–C in both structures) find their way out to the cytoplasm, but this process takes at least an order of magnitude longer time when compared with that of substrates bound to secondary sites. The subunit B substrates in both confs I and II remained bound for 400 ns. Notably, GBB1 in subunit A was released at 380 ns only, suggesting that the timescale reached by current terascale computations is just on the verge of possibly detecting translocation events from the primary sites.
We further observed whether backbone motions played a role in enabling the dissociation of the substrates from the primary binding site and in facilitating their translocation across the protein. In the presence of constraints, only one (CRN1 in subunit C) out of six substrates was observed to unbind and reached the cytoplasm (supplemental Fig. S2C, black curve). All others remained tightly bound at/near their primary site, revealing the functional significance of protein backbone accommodating motions.
Based on our simulations, it is difficult to assess whether CaiT has different affinities for the two substrates. This is due to the small number of unbinding events and the fact that dissociation from only two out of three subunits was observed for each substrate in the unbiased simulations. However, it is worth noting that all substrates (except for CRN3) move toward the cytoplasm, not to the EC region, suggesting an intrinsic property of the inward-facing conformation of CaiT.
In contrast to other transporters where water influx plays a role in dislocating the substrate, the solvation of the primary binding site does not appear to precipitate substrate unbinding in CaiT. The present simulations suggest that there was practically no energetic barrier to water influx into the primary site. Water molecules could reach the site even before the constraints adopted for energy minimization were lifted, and the substrates interacted with ∼8–9 water molecules most of the time. This is true for all subunits regardless of whether the substrate was able to leave or not and regardless of the constraints on the backbone atoms. In supplemental Fig. S2, A and D, the inset panels illustrate the hydration of the primary binding site early on during the simulations, which, contrary to expectation, does not trigger the dislocation of the substrate. In harmony with this is the observation that at the primary site/pocket, there is enough space for the substrate to move 4–5 Å away from its original pose, an observation made for all substrates in the confines of site 1.
The Pathway of CRN Translocation from the Primary Binding Site to the IC Region Involves Salt-bridge Formations with Arg170 and Arg337 prior to Release to the Cytoplasm
To determine whether a preferred pathway of substrate translocation from the primary binding site to the IC region exists, we evaluated the closest distances of approach and pairwise interaction energies between the substrates and select on-pathway residues observed during the course of simulations. We will focus on the substrates that are released to the cytoplasm, i.e. CRN1, CRN2, CRN4, and GBB1.
The first two residues that are encountered by CRN1 and GBB1 upon direct downward translation into the IC region are Gln330 and Met331 on TM8, directly below Tyr327 that lines the binding pocket (Fig. 1). Both residues are close to CRN2 in conf I, suggesting that they might act as attractors for substrate channeling. To examine whether they play a role in dislodging the substrate from site 1 to site 2, we calculated the interaction energy between each substrate and either of these two residues during each unbinding event from site 1. Only weak interactions were observed, in accord with the relatively fast unbinding of CRN2 in the original structure (supplemental Fig. S3). These observations suggest that these residues may promote the movement of the substrates toward the IC region, but do not hold back their translocation toward the IC cavity.
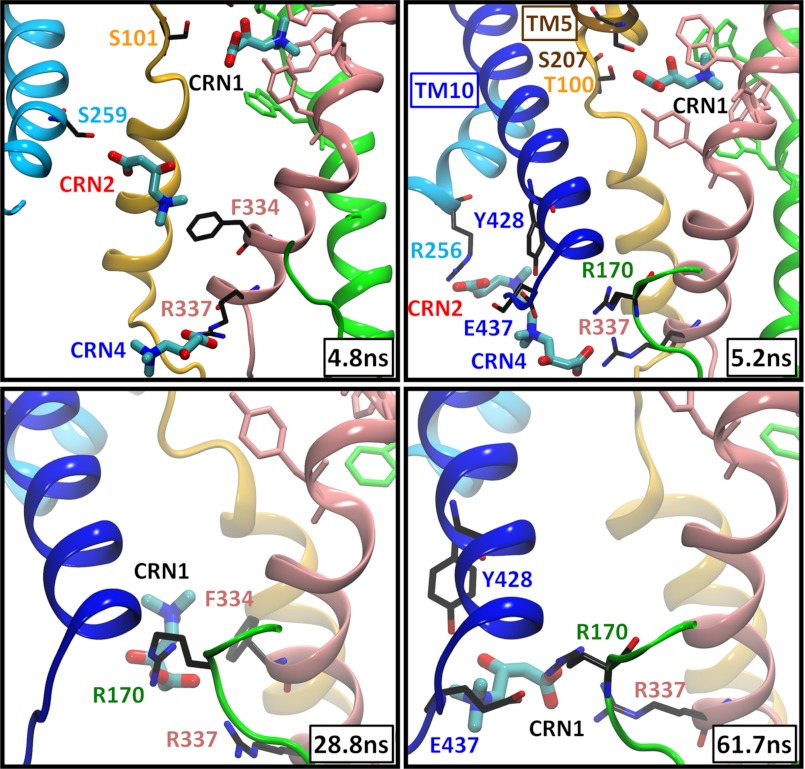
In the absence of any constraints (run 1a), CRN1 spend ∼30–50 ns in the confines of site 1. During this period, CRN2 and CRN4 are dislocated from their binding sites and released to the cell interior. Strikingly, the translocation of CRN1 does not occur by a smooth diffusion, but by almost hopping from one site to another. For example, CRN1 in subunit A is dislodged around t = 28 ns from site 1, to settle shortly in the IC cavity in the vicinity of site 4, where it resides for ∼30 ns, prior to its complete release to the cytoplasm (Fig. 3 and supplemental Movie S1). Likewise, CRN1 in subunit C follows approximately the same trajectory, with residence times in the respective sites being about 50 and 10 ns, respectively. In both cases, the sites 4 (and 2) are vacated by CRN4 (and CRN2) prior to the translocation of CRN1 from site 1. In the newly “settled” region, CRN1 is mainly coordinated by Arg170 on the loop connecting TM4 and TM5 and by Arg337 at the IC end of TM8. This behavior is consistently reproduced in both constraint-free (Fig. 4A and supplemental Fig. S4A) and constrained (Fig. 4B) runs.
FIGURE 3.
CRN translocation and release to the IC region and key role of Arg170 and Arg337. Snapshots (taken from subunit A in run 1a) display the instantaneous poses of the substrates initially bound at the sites 1, 2 and 4. TM helices are colored as in Fig. 1, and residue labels are colored after the corresponding TM helices. Aromatic residues at the primary site (Trp142, Trp147, Trp323, Trp324, and Tyr327) are colored according to the TM helix they are located on. In this case, CRN1 practically hops from site 1 to site 4 around t = 28 ns (after the dislocation of CRN2 and CRN4), where it stays coordinated by Arg170 and Arg337, and later by Tyr428 and Glu437, until it is released to the cytoplasm at t = 62 ns (see also Fig. 2A). Residues that make close (≤3 Å) interactions with the substrates are displayed.
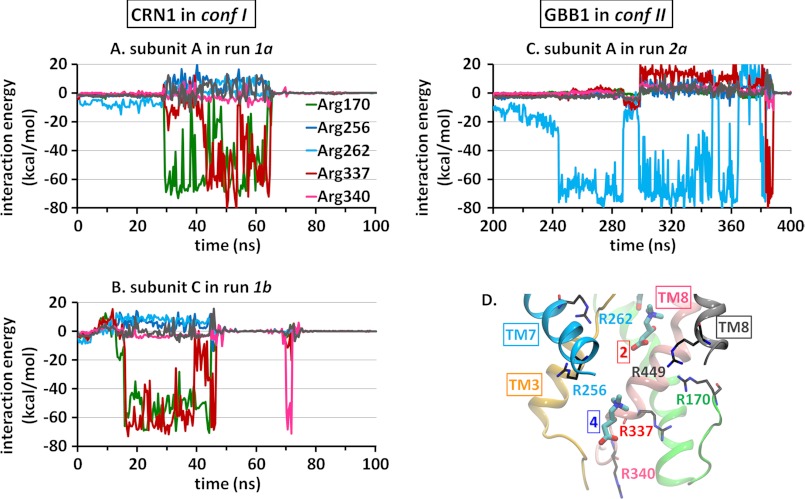
FIGURE 4.
Time evolution of the interactions with basic residues during the unbinding and release of CRN1 and GBB1. Plots indicate the interaction energy (kcal/mol) of the leaving CRN1 (A and B) or GBB1 (C) with each of six arginines located in the cytoplasmic cavity of CaiT. The relevant portions of the trajectories are displayed in each case. D shows the position of these residues in the CRN-bound structure.
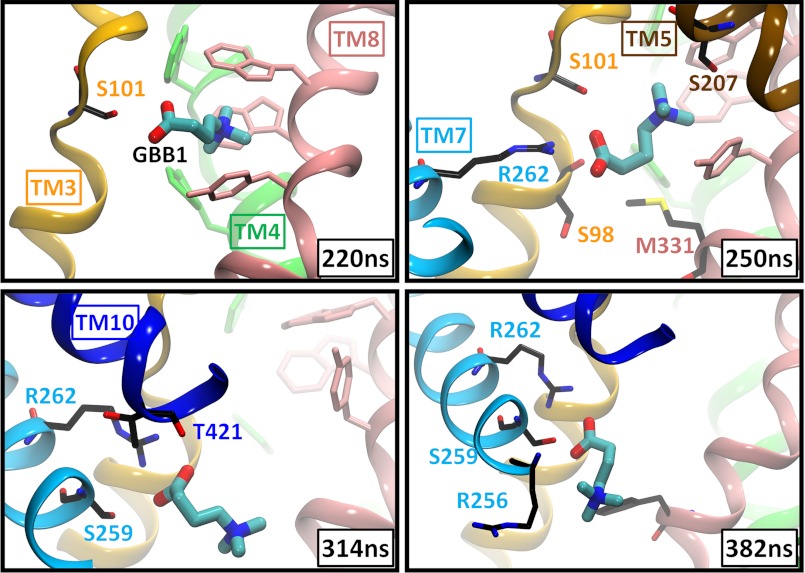
GBB1 Unbinding and Release Occur via a Different Pathway Controlled by Arg262, a Residue Equivalent to Na2 in the LeuT Fold of Sodium-coupled Transporters
Despite the close similarity in the binding poses of CRN1 and GBB1, GBB1 consistently chose a path different from CRN1 in exiting the primary site. This path is characterized by major involvement of Arg262. Arg262 is located on TM7 just across the unwound part of TM3 and in the vicinity of TM10 (similar to the position of Na2 between the equivalent helices TM1 and TM8 of LeuT). Its side chain is positioned ∼9 Å away from the carboxyl group of CRN2 and ∼13 and 11 Å away from CRN1 and GBB1, respectively. Notably, a strong attraction (salt bridge) between GBB1 carboxylate and Arg262 guanidinium group, mediated by 3 serines (Ser101 and Ser98 on TM3 unwound region, and Ser259 on TM7), essentially determines GBB1 translocation path (Fig. 5 and supplemental Movie S2). This behavior was consistently repeated in both events of unbinding in the absence of constraints (Fig. 4C and supplemental Fig. S4B).
FIGURE 5.
Release of GBB1 into the IC solvent and key role of Arg262. Snapshots (taken from subunit A in run 2a) are shown here for GBB1 viewed from the side. TM helices and residues are colored as in Fig. 3. Residues within 3 Å from the substrate are displayed in stick representation. Arg262 plays a key role in guiding the translocation of GBB1 across the protein, assisted by polar residues (Ser98 and Ser101 on TM3; Ser259 on TM7 and Thr421 on TM10) in the close neighborhood.
The key role of Arg262 in driving substrate permeation was specific to GBB in conf II as little or no such interaction was observed with five other basic residues located within the IC cavity (arginines 170, 256, 337, 340, and 449) (Fig. 4 and supplemental Fig. S4) or with CRN1 in conf I as shown previously. We also examined the potential role of acidic residues Glu85, Glu167, Asp260, Glu437, and Asp440 at the IC cavity (supplemental Fig. S5). Although Glu437 and Glu85 temporarily interacted with the CRNs near site 4, the total interaction energy of the substrate with these amino acids was significantly weaker than those with basic residues (supplemental Fig. S5), further supporting the distinctive function of Arg262 in conf II.
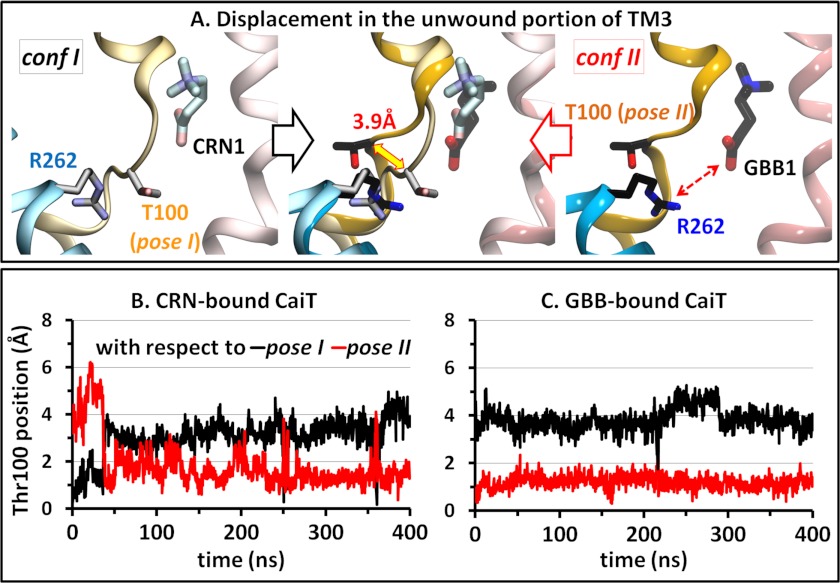
A Slight Geometric Difference between the Two Crystal Structures at the Unwound Part of TM3 Determines the Unbinding Pathway
Different unbinding pathways for different substrates from the same site can arise from differences in the substrates or in the protein at/close to the binding site, or in both. CRN differs from GBB by the presence of a hydroxyl group on the central carbon atom (Fig. 2). However, the presence/absence of a polar group seems unlikely to contribute to the differential unbinding pathway considering that the observed substrate-protein interactions are determined mainly by charged groups in each. In addition, the Arg262 side chain is located at the same distance from the binding sites in the two conformations and has a similar orientation toward the primary binding site. A closer look at the vicinity of the binding site between the two crystallized structures reveals, however, an interesting difference at the unwound helical part of TM3, which lies between the substrate at site 1 and Arg262. In conf I, this loop shields Arg262 from the substrate, whereas in conf II, it is shifted away from the space in between. This unwound part includes the segment Ser98-Cys99-Thr100-Ser101, with the most pronounced difference at the position of Thr100, which we will refer to as pose I or II (Fig. 6A). In addition to this structural difference, this portion of TM3 exhibits distinctive intrinsic dynamics in the two conformations; although the geometry (pose II) of the “yielding” structure (conf II) is maintained over the entire trajectories generated for GBB-bound structures, that (pose I) of the “shielding” one is not, and often switches to a pose similar to pose II for substantial durations of time (Fig. 6, B and C).
FIGURE 6.
Different geometries of the unwound portion of TM3 in the two resolved substrate-bound structures of CaiT. A, superposition of confs I and II shows a shift of ∼4 Å in the position of the Cα atom of Thr100 on the TM3 unwound region, which can either inhibit or favor the electrostatic interaction (dashed red arrow) between the substrate at site 1 and Arg262. The two respective conformers of Thr100 are called pose I and pose II. Although the Thr100 position is stable over the entire duration of simulations initiated with conf II (C), it can often switch from pose I to pose II when conf I is used as initial structure. Plots B and C display the time evolution of Thr100 Cα atom position, with respect to its pose I (black) or pose II (red), obtained upon alignment of the entire TM3 domain. The curves refer to subunit A in both runs.
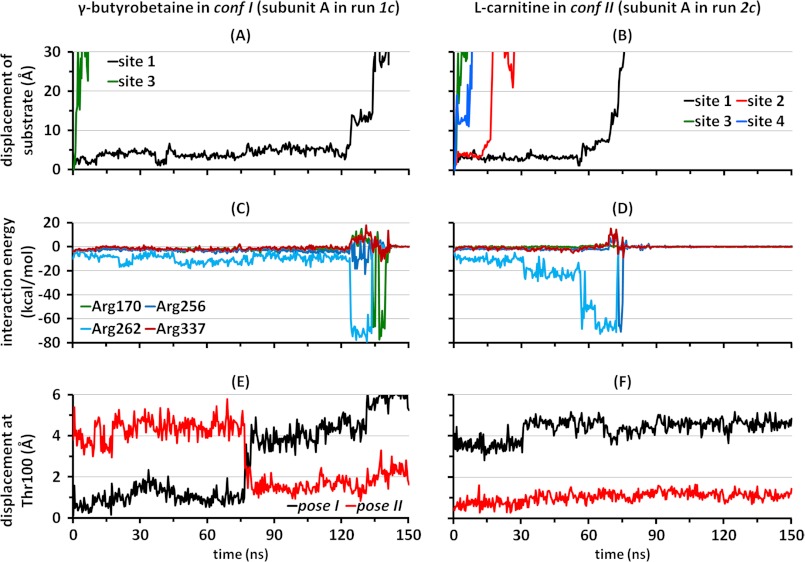
To definitively determine whether the structural dynamics of the unwound TM3 segment dictate the substrate release pathway, we switched the substrates between the two CaiT structures; the four CRN molecules were placed in conf II, and the two GBBs were placed in conf I. After short equilibration, with slight constraints on the protein backbone and on the substrates to facilitate the accommodation of the new substrates, constraint-free runs (1c and 2c) were performed over 150 ns in each case.
In harmony with the previous observation that the crystallized structure is a substrate-releasing one, in both runs, all substrates at secondary positions left the antiporter within up to 60 ns (Fig. 7, A and B, and supplemental Fig. S6, A and B). With regard to the substrate at the primary site, complete unbinding was seen in one subunit with GBB in conf I and in two subunits (A and B) with CRN in conf II (runs 1c and 2c, respectively). As expected from the hydroxyl group in CRN having little or no effect on the energetics of interaction with the protein, in both unbinding events, CRN left the primary binding site mainly upon interaction with Arg262, just as previously seen with GBB in the same conf II (see results for subunits A and B in Fig. 7D and supplemental Fig. S6D, respectively). A temporary salt bridge formed with Arg256, just before release to the IC region was observed in both cases (similar to that formed by CRN2 in the original CRN-bound structure (Fig. 3)). As in the previous run with conf II, the geometry of the unwound helix, and in particular the pose II of Thr100 that favors a strong attraction between Arg262 and the substrate, was maintained throughout the whole run (Fig. 7F and supplemental Fig. S6F).
FIGURE 7.
Interaction with Arg262 upon release from the primary site is dependent on the geometry of the unwound segment of TM3. Data here show characteristics of the dissociation of GBB bound to conf I (A, C, and E) or CRN bound to conf II (B, D, and F). Displacement of the geometric center of GBB at sites 1 and 3 (A) or CRN at sites 1–4 (B) away from the initial position (runs 1c and 2c, respectively) indicates that release from site 1 correlates primarily with interaction with Arg262 in both cases, shown here as a function of the interaction energy between the substrate and Arg262 (kcal/mol) (C and D). This release pattern is favored when Thr100 in the unwound TM3 segment assumes the pose II (E and F). The black and red curves refer to the distance of Thr100 α-carbon from its position in poses I and II, respectively (see also supplemental Fig. S6).
In the case of GBB bound to conf I (run 1c), no strong interaction with Arg262 took place until Thr100 in subunit A switched to pose II at t = 75 ns (Fig. 7, C and E). This conformational switch was succeeded, after a time delay of ∼50 ns, by a strong interaction of Arg262 with GBB1 that practically dislodged the substrate and prompted its release to the cytoplasm (after a temporary salt bridge formation with Arg170). A change in loop conformation from pose I to II also took place in subunit B in the same run, but no departure from GBB away from site 1 was seen within the simulated timescale. Overall, pose II is confirmed by all runs to be more stable; it undergoes no switches to pose I, and it persistently assists the translocation of the substrate (GBB or CRN) by favoring its interaction with Arg262. Even pose I tends to undergo a transition to pose II to facilitate substrate release, although in pose I, CRN (and possibly GBB) has access to an alternative path (via the Arg170-Arg337 pair near site 4) to the cytoplasm.
DISCUSSION
Despite the huge amount of data revealed by a crystal structure, fully atomic computational tools are often required to view the specifics of substrate dynamics and interaction patterns in the resolved structures. This approach is also required to assess the molecular mechanisms associated with substrate binding/unbinding and translocation events and has been so far applied to several transporters/antiporters that are structurally related to CaiT to draw the whole picture of how the transport/antiport cycle takes place.
For instance, in the structurally similar vSGLT, obtained in an inward-facing conformation as CaiT, molecular dynamics simulations have shown that no conformational changes were required for the release of galactose despite the apparent substantial occlusion of the substrate from the internal solvent by the full side chain of a tyrosine (22). In addition, the obtained structure was shown to be a sodium-releasing one in light of the spontaneous and repeated escape of Na+ modeled in the Na2 site (23, 24). This finding explained why no electron density of Na+ was seen in the crystal. In contrast, no such spontaneous events were seen in the outward-facing LeuT (13) where the substrate is occluded from the external solution mostly by the aromatic rings of a tyrosine and a phenylalanine and where an external force is required to pull the bound leucine out of the primary binding site (25). In the structurally related arginine/agmatine antiporter, AdiC, which is sodium-independent like CaiT, specific substrate extrusion from the primary site has been reported to be induced/accelerated by the physiologically relevant protonation of a key acidic residue, Glu208, near the binding site (26).
Our results include independent cases of spontaneous release of two different types of substrates from several sites reported in the crystallized inward-facing CRN/GBB antiporter CaiT. Overall, we have observed eight such events from the primary site, in addition to departure from all secondary binding sites in all runs (see supplemental Table S2 for a summary). The substrates in the primary site enjoy substantial water accessibility, and no gating residues are seen to clearly control the inward release thereof. This, together with the fact that most substrates bound to primary sites in each conformation in the constraint-free runs are able to spontaneously and completely dissociate into the IC region, strongly supports that the crystallized state of CaiT represents a substrate-releasing conformer. In light of the rapid dissociation of all reported substrates at the two secondary IC sites (2 and 4), it is unlikely that these represent sites that are stably occupied under physiological conditions. On the other hand, salt-bridge formations between the translocating substrates the basic residues in the vicinity of these sites (e.g. Arg262 and Arg256 near site 2, and Arg337 near site 4) suggest that these serve as temporary on-pathway stabilization sites that guide substrate translocation to the cytoplasm.
With regard to residues lining site 3 (that is, practically exposed to the EC region), the mutation W316L was previously reported to decrease transport of CRN by ∼70% in EcCaiT (9), whereas Y114L had a minor effect (see their location in Fig. 1C and supplemental Fig. S1, B and E). Furthermore, the W316A mutation was found to significantly impact the affinity of the substrate to PmCaiT, as well as the transport rate (8). In light of the fast dissociation observed in our simulations, it is unlikely that these amino acids present a barrier to substrate translocation in the particular crystallized conformations of CaiT. Neither substrate appears to be closely interacting with the aromatic groups of Tyr114 or Trp316. Also, the fact that none of the substrates at site 3 remained bound longer than a few nanoseconds supports the view that protein-substrate interactions at this location in the present conformations are weak at the most, and slight rearrangements in local geometry (to eliminate crystallographic artifacts, if any) are not likely to yield significant change in binding affinity.
Both CaiT crystals were grown using the hanging-drop vapor diffusion method. The structure with CRN molecules was grown by mixing the protein with the reservoir solution containing 5 mm CRN at 20 °C, whereas the GBB-bound structure was crystallized at 4 °C. Previous studies have shown that CRN has an external Km of 40–100 μm, and Kd values of 2–6 mm have been measured for CRN and GBB depending on the system used (6, 8, 9). Therefore, CaiT can bind more substrates than those being effectively transported, and current crystallization conditions may include occupied sites that are not necessarily functional. We also note that the resolution of 3.15 or 3.5 Å of these two respective crystals may render it difficult to determine with certainty the nature and pose of the molecules at the proposed binding sites. For instance, the only observed interaction between CRN3 and CaiT is between the hydroxyl group of CRN3 and the backbone carbonyl of Gly311. Such a single hydrogen bond is not likely to hold CRN at this site that is highly accessible to the aqueous environment, as confirmed by the water molecules that effectively destabilize this interaction at early stages of the simulations. Likewise, CRN4 is held in place by a hydrogen bond with Arg337, whereas experiencing an electrostatic repulsion with Glu85. Although the structure in the crystal environment represents an immobilized conformation, molecular dynamics simulations provide us with a dynamic view under physiological conditions.
Trajectories of hundreds of nanoseconds as those performed in this study are generally sufficient to view small motions, intrinsic or induced, that have the potential of affecting mechanisms such as substrate binding/unbinding or permeation processes. In this study, we have seen how a slight difference between the two crystallized structures can shape substrate dissociation pathway; a conformational switch in the unwound part of TM3 that leads to a 4 Å α-carbon displacement and side chain reorientation in Thr100 determines whether the substrate in the primary site is exposed to Arg262 on TM7, which can, in turn, facilitate its exit into the IC solvent. Such a difference stands out when the overall helical backbone root mean square deviation is only 0.6 Å. In the CRN-bound crystal structure (conf I of CaiT), the unwound TM3 acts as a barrier hindering the attraction between Arg262 and the substrate. However, this energetically “frustrated” region also possesses a high potential to undergo a conformational switch from a shielding to yielding state, allowing for Arg262 to attract/destabilize the bound substrate. This ability of Arg262 conferred by the conformational switch at the unwound portion of TM3 seems to dominate regardless of the pose and identity of the substrate in the primary site, being an intrinsic feature of the CaiT binding core itself. Upon replacement of GBB with CRN in the yielding structure (conf II), although the carboxyl group of the substrate was 13 rather than 11 Å away from the guanidinium group of Arg262, the long-range electrostatic interaction between the charged groups was still able to trigger the dislodging of the substrate and drive its translocation to the cytoplasm in both events of dissociation.
In the initial CRN-bound crystal structure (conf I of CaiT), mercury ions (Hg2+) were included, and one of them was coordinated by the side-chain sulfhydryl of Cys99 and hydroxyl of Ser101. This is clearly the cause of the difference at this segment between the two crystals. Our simulations show that although the local conformation is relatively unstable and tends to switch to the second yielding one, the second (conf II) is highly stable over the entire duration of all runs. Thus, it seems that substrate release from the primary binding site is more likely to be via attraction by Arg262 than by simple downward translation into the IC.
Our simulations also show that Arg170 and Arg337 play a key role in substrate release via an alternative pathway in the CRN-bound conformation. Whether Arg170 and Arg337 together constitute a secondary IC site is not evident from the resolved structures because their side-chain orientations differ in the two crystallized structures. In conf I, where they can attract the leaving substrate, their side chains point toward each other, whereas in conf II, that of Arg170 is maintained, whereas that of Arg337 points away from the former. However, Arg337 is located on an IC loop and may thus enjoy significant mobility to cooperate with Arg170 to capture the substrate and assist its release to the IC region, as observed in runs 1a and 1b, with multiple CRNs (including those originally bound to site 1 and 4). These observations suggest that these two residues may cooperatively define an alternative pathway for substrate release, especially when the TM3 unwound region cannot undergo a switch to the pose II.
This study thus sheds light on how small geometric changes around the binding site, and especially unwound regions of TM helices near the binding site, can affect the binding/unbinding pathway and suggest that attention should be given to the intrinsic dynamics of the protein when making conclusions about the mechanism of such processes based on a single crystal structure. In the structurally related transporter LeuT, the presence of a secondary binding located in the EC aqueous cavity, ∼10 Å “above” the primary site, has been experimentally and computationally supported (27, 28). This is a site where the substrate can bind prior to entry into the primary site from the EC solvent. However, no cytoplasmic secondary site has been experimentally proven to date in any member in this structural family of transporters/antiporters.
This research was supported in part by the National Science Foundation through resources provided by the Extreme Science and Engineering Discovery Environment (XSEDE) (Grants TG-MCB100144 and 3P41 RR006009 20S1) and by National Institutes of Health Grant R01 GM086238 from the NIGMS (to I. B.).

This article contains supplemental Figs. S1–S6, Tables S1 and S2, and Movies S1 and S2.
- CRN
- l-carnitine
- GBB
- γ-butyrobetaine
- TM
- transmembrane
- EC
- extracellular
- IC
- intracellular
- vSGLT
- sodium-coupled galactose symporter
- LeuT
- sodium-coupled leucine transporter.
REFERENCES
- 1. Fritz I. B., Yue K. T. (1964) Effects of carnitine on acetyl-CoA oxidation by heart muscle mitochondria. Am. J. Physiol. 206, 531–535 [DOI] [PubMed] [Google Scholar]
- 2. Treem W. R., Stanley C. A., Finegold D. N., Hale D. E., Coates P. M. (1988) Primary carnitine deficiency due to a failure of carnitine transport in kidney, muscle, and fibroblasts. N. Engl. J. Med. 319, 1331–1336 [DOI] [PubMed] [Google Scholar]
- 3. Nal̸ecz K. A., Miecz D., Berezowski V., Cecchelli R. (2004) Carnitine: transport and physiological functions in the brain. Mol. Aspects Med. 25, 551–567 [DOI] [PubMed] [Google Scholar]
- 4. Jung K., Jung H., Kleber H. P. (1987) Regulation of l-carnitine metabolism in Escherichia coli. J. Basic Microbiol. 27, 131–137 [DOI] [PubMed] [Google Scholar]
- 5. Kleber H. P. (1997) Bacterial carnitine metabolism. FEMS Microbiol. Lett. 147, 1–9 [DOI] [PubMed] [Google Scholar]
- 6. Jung H., Buchholz M., Clausen J., Nietschke M., Revermann A., Schmid R., Jung K. (2002) CaiT of Escherichia coli, a new transporter catalyzing l-carnitine/γ -butyrobetaine exchange. J. Biol. Chem. 277, 39251–39258 [DOI] [PubMed] [Google Scholar]
- 7. Lindstedt G. (1967) Hydroxylation of γ-butyrobetaine to carnitine in rat liver. Biochemistry 6, 1271–1282 [DOI] [PubMed] [Google Scholar]
- 8. Schulze S., Köster S., Geldmacher U., Terwisscha van Scheltinga A. C., Kühlbrandt W. (2010) Structural basis of Na+-independent and cooperative substrate/product antiport in CaiT. Nature 467, 233–236 [DOI] [PubMed] [Google Scholar]
- 9. Tang L., Bai L., Wang W. H., Jiang T. (2010) Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nat. Struct. Mol. Biol. 17, 492–496 [DOI] [PubMed] [Google Scholar]
- 10. Ziegler C., Bremer E., Krämer R. (2010) The BCCT family of carriers: from physiology to crystal structure. Mol. Microbiol. 78, 13–34 [DOI] [PubMed] [Google Scholar]
- 11. Ressl S., Terwisscha van Scheltinga A. C., Vonrhein C., Ott V., Ziegler C. (2009) Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature 458, 47–52 [DOI] [PubMed] [Google Scholar]
- 12. Vinothkumar K. R., Raunser S., Jung H., Kühlbrandt W. (2006) Oligomeric structure of the carnitine transporter CaiT from Escherichia coli. J. Biol. Chem. 281, 4795–4801 [DOI] [PubMed] [Google Scholar]
- 13. Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 14. Faham S., Watanabe A., Besserer G. M., Cascio D., Specht A., Hirayama B. A., Wright E. M., Abramson J. (2008) The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perez C., Koshy C., Ressl S., Nicklisch S., Krämer R., Ziegler C. (2011) Substrate specificity and ion coupling in the Na+/betaine symporter BetP. EMBO J. 30, 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singh S. K., Piscitelli C. L., Yamashita A., Gouaux E. (2008) A competitive inhibitor traps LeuT in an open-to-out conformation. Science 322, 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E. P., Rutherford N. G., Hadden J. M., O'Reilly J., Ma P., Saidijam M., Patching S. G., Hope R. J., Norbertczak H. T., Roach P. C., Iwata S., Henderson P. J., Cameron A. D. (2008) Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Farwick M., Siewe R. M., Krämer R. (1995) Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J. Bacteriol. 177, 4690–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanner B. I., Zomot E. (2008) Sodium-coupled neurotransmitter transporters. Chem. Rev. 108, 1654–1668 [DOI] [PubMed] [Google Scholar]
- 20. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 22. Li J., Tajkhorshid E. (2012) A gate-free pathway for substrate release from the inward-facing state of the Na+-galactose transporter. Biochim. Biophys. Acta 1818, 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li J., Tajkhorshid E. (2009) Ion-releasing state of a secondary membrane transporter. Biophys. J. 97, L29-L31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zomot E., Bahar I. (2010) The sodium/galactose symporter crystal structure is a dynamic, not so occluded state. Mol. Biosyst. 6, 1040–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Celik L., Schiøtt B., Tajkhorshid E. (2008) Substrate binding and formation of an occluded state in the leucine transporter. Biophys. J. 94, 1600–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zomot E., Bahar I. (2011) Protonation of glutamate 208 induces the release of agmatine in an outward-facing conformation of an arginine/agmatine antiporter. J. Biol. Chem. 286, 19693–19701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi L., Quick M., Zhao Y., Weinstein H., Javitch J. A. (2008) The mechanism of a neurotransmitter:sodium symporter–inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell 30, 667–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quick M., Shi L., Zehnpfennig B., Weinstein H., Javitch J. A. (2012) Experimental conditions can obscure the second high affinity site in LeuT. Nat. Struct. Mol. Biol. 19, 207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]