Background: Apicularen is a specific V-ATPase inhibitor that binds to the VO complex of the holoenzyme.
Results: Apicularen binds at the interface of the VO subunits a and c.
Conclusion: The binding site for apicularen is in the vicinity of those for bafilomycin and archazolid.
Significance: We propose the first model of binding site arrangement for these three classes of V-ATPase inhibitors.
Keywords: Antibiotics, Cancer, Membrane Proteins, Osteoporosis, Vacuolar ATPase, Apicularen, Archazolid, Bafilomycin, Manduca sexta, Saccharomyces cerevisiae
Abstract
The investigation of V-ATPases as potential therapeutic drug targets and hence of their specific inhibitors is a promising approach in osteoporosis and cancer treatment because the occurrence of these diseases is interrelated to the function of the V-ATPase. Apicularen belongs to the novel inhibitor family of the benzolactone enamides, which are highly potent but feature the unique characteristic of not inhibiting V-ATPases from fungal sources. In this study we specify, for the first time, the binding site of apicularen within the membrane spanning VO complex. By photoaffinity labeling using derivatives of apicularen and of the plecomacrolides bafilomycin and concanamycin, each coupled to 14C-labeled 4-(3-trifluoromethyldiazirin-3-yl)benzoic acid, we verified that apicularen binds at the interface of the VO subunits a and c. The binding site is in the vicinity to those of the plecomacrolides and of the archazolids, a third family of V-ATPase inhibitors. Expression of subunit c homologues from Homo sapiens and Manduca sexta, both species sensitive to benzolactone enamides, in a Saccharomyces cerevisiae strain lacking the corresponding intrinsic gene did not transfer this sensitivity to yeast. Therefore, the binding site of benzolactone enamides cannot be formed exclusively by subunit c. Apparently, subunit a substantially contributes to the binding of the benzolactone enamides.
Introduction
Vacuolar-type ATPases (V-ATPases)2 are found in the endomembrane system of all eukaryotic cells and in the plasma membranes of many animal cells. They energize multiple transport processes and regulate the pH in cells and organelles by coupling ATP hydrolysis to proton pumping (1, 2). Their heteromultimeric structure includes two main complexes, the catalytic V1 complex with a subunit composition of A3B3CDE3FG3H and the proton translocating VO complex with the subunits a, cn, c″ (in fungi additional subunit c′), d, and e (2, 3). ATP hydrolysis takes place at the hexameric headpiece built by three A catalytic subunits and three B regulatory subunits. The emerging free energy is converted into a rotation of the central stalk subunits D, F, and d and the ring formed by the proteolipid subunit c (2). For transmembrane transport, protons enter the proteolipid ring via the cytosolic hemichannel of subunit a and bind reversibly to the conserved essential glutamate in each c subunit. After rotation of the c-ring, protons leave via the luminal hemichannel of subunit a (2, 4).
By regulating the intracellular or intraorganellar pH, organellar V-ATPases operate in diverse processes such as receptor-mediated endocytosis, protein processing, and degradation as well as intracellular transport. Plasma membrane V-ATPases are involved in bone resorption, extracellular acidification, or energization of secondary active transport processes (2, 5). The connection of V-ATPase function to diseases such as osteoporosis or cancer and the verification of the V-ATPase as a suitable therapeutic target certainly require a comprehensive investigation of the V-ATPase, its inhibitors, and the intermolecular interactions. In the 1980s the plecomacrolide bafilomycin was identified as the first specific V-ATPase inhibitor, exhibiting nanomolar IC50 values (6). Until now, a reasonable number of other inhibitory compounds from different sources have been discovered (7). Among them are the macrolactone archazolid and the benzolactone enamide apicularen, both of which show a similar inhibitory efficacy as the popular plecomacrolides bafilomycin and concanamycin (8–10). However, the benzolactone enamides exhibit a unique feature as they do not, in contrast to the other inhibitors, affect V-ATPases from fungi, and therefore they are the first source specific V-ATPase inhibitors (7, 11). In consequence, this class of compounds may be one of the most promising groups of candidates for a therapeutic use of V-ATPase inhibitors.
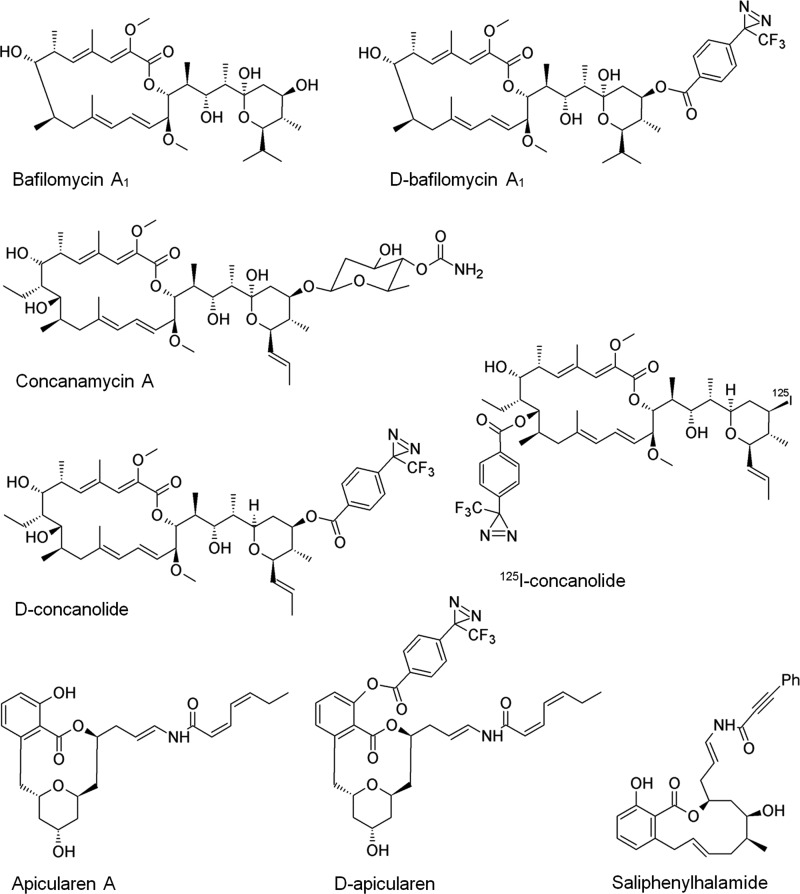
Concerning the inhibitor-binding sites and the mechanisms of V-ATPase inhibition, two discrete approaches using plecomacrolides have led to first insights as follows. Labeling experiments with the purified V1VO holoenzyme of Manduca sexta and a 125I-labeled derivative of concanamycin (Fig. 1) revealed the binding of plecomacrolides to the VO subunit c (12). Simultaneously, mutational analysis of the VO subunit c in Neurospora crassa disclosed that certain single amino acid exchanges in the sequence of this subunit altered the affinity of bafilomycin to the V-ATPase (13). Later on, additional single amino acid exchanges in subunit c of the V-ATPases from N. crassa and Saccharomyces cerevisiae resulted in a more precise localization of the plecomacrolide-binding site, which accordingly resides at the interface between helices 1 and 2 of one subunit c and helix 4 of an adjacent subunit c in the ring (14, 15). Interestingly, the c-ring did not appear to contain the whole plecomacrolide-binding site because mutations in subunit a of the yeast V-ATPase also conferred resistance to bafilomycin (16). In our previous photoaffinity labeling (PAL) studies using the concanamycin derivative mentioned above, the photoactivatable cross-linking diazirinyl group was bound to the macrocyclic ring of the inhibitor that led to an exclusive label at subunit c (12). Labeling of merely subunit c was surprising regarding the length (6.4 Å) and flexibility of the attached diazirinyl. However, with respect to the mutational analysis and modeling of the binding site within the c-ring, this was a strong indication that position C9 of concanamycin may be deeply buried between two adjacent c subunits. In this study, we used derivatives of bafilomycin and concanamycin modified with the newly developed 14C-labeled 4-(3-trifluoromethyl-diazirin-3-yl)benzoic acid (17). By repositioning the diazirinyl moiety to the opposite side of the plecomacrolide structures (Fig. 1), we anticipated labeling not only of subunit c but now also of subunit a. For the modification at C23, we did not expect strong influence on the inhibitory efficacy, as in previous studies it had already been shown that this position has only a negligible effect and that it does not seem to belong to the major pharmacophore (18–20).
FIGURE 1.
Structures of the PAL inhibitor derivatives D-bafilomycin, D-concanolide, 125I-concanolide, D-apicularen, saliphenylhalamide, and parent compounds.
The binding site of the archazolids originally had been presumed to overlap to a large extent with that of the plecomacrolides as archazolid prevented binding of a concanamycin derivative (10). However, the binding site for archazolids is relocated to the equatorial region of the c-ring and therefore overlaps with the plecomacrolide-binding site to a minor extent than previously thought (21). This revision has been derived from recent site-directed mutagenesis of the yeast V-ATPase subunit c and labeling of the M. sexta V-ATPase using a radioactive derivative of archazolid A as well as the fluorescent dicyclohexylcarbodiimide derivative NCD-4.
Up to now, information concerning the binding site of the benzolactone enamides is rare. For the benzolactone enamide salicylihalamide A, it has been reported that it binds to a different site than the plecomacrolides, although it inhibits proton translocation through the VO complex (12, 22). Recent labeling experiments in the presence of apicularen revealed no interference of plecomacrolide, archazolid, or NCD-4 binding to subunit c (10, 21). Yet it was not possible to elucidate where apicularen binds within the VO complex. The development of the 14C-labeled 4-(3-trifluoromethyldiazirin-3-yl)benzoic acid mentioned above now provided a convenient way to prepare an apicularen derivative that irreversibly cross-links to the protein upon UV exposure and therefore could be used to identify the interacting V-ATPase subunit(s) (17).
Furthermore, we used the radioactive derivatives of apicularen, bafilomycin, and concanamycin as well as nonradioactive compounds in competition assays to gain new insights into the interaction of the inhibitors. Considering the fact that the fungal V-ATPases are insensitive to benzolactone enamides, we used yeast deletion mutants deficient in subunit Vma3, Vph1, or Stv1 for the heterologous expression of their human or insect homologues to prove whether it is possible to transfer sensitivity against apicularen to the yeast-human or yeast-insect hybrid V-ATPase. Summing up all available information, we provide a model predicting the arrangement of the binding sites for plecomacrolides, archazolids, and benzolactone enamides within the VO complex.
EXPERIMENTAL PROCEDURES
Inhibitors
Bafilomycin A1, concanamycin A, 21-deoxyconcanolide A, apicularen A, archazolid A, and saliphenylhalamide were isolated or prepared as published (8, 9, 18, 23). 21-O-[4-(3-Trifluoromethyldiazirin-3-yl)benzoyl]bafilomycin A1 (D-bafilomycin), 23-O-[4-(3-trifluoromethyldiazirin-3-yl)benzoyl]-21-deoxyconcanolide A (D-concanolide), and their 1-14C-labeled derivatives were prepared according to Ref. 17. Based on previous evaluation of apicularen analogues, we favored position C3 to attach the diazirinyl moiety (7, 24). For synthesis of 3-O-[4-(3-trifluoromethyldiazirin-3-yl) benzoyl]apicularen (D-apicularen), to a solution of apicularen A (1.5 mg, 3.4 μmol) in CH2Cl2 (5 ml) was added 4-(dimethylamino)-pyridine (0.9 mg, 7 μmol), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (1.1 mg, 5.7 μmol), and 4-(3-trifluoromethyldiazirin-3-yl)benzoic acid (1.1 mg, 4.8 μmol), and the solution was stirred for 2.5 h in the dark at 25 °C. Column chromatography of the mixture on silica gel (CHCl3/MeOH 25:1) furnished 1.4 mg (63%) of D-apicularen (Rf = 0.17, CHCl3/MeOH 50:1). NMR data of D-apicularen is as follows: 1H NMR (600 MHz, [D6]-acetone): δ = 0.99 (t, J = 7.5 Hz, 3H, 25-H3), 1.46–1.56 (m, 3H, 14-Hb, 10-Hb, 12-Hb), 1.65 (ddd, J = 5.3, 7.8, 13.1 Hz, 1H, 12-Ha), 1.78 (dt, J = 10.9, 14.7 Hz, 1H, 14-Ha), 1.95 (dt, J = 4.6, 13.0 Hz, 1H, 10-Ha), 2.00–2.06 (m, 2H, 16-H2), 2.27 (dquint, J = 1.6, 7.5 Hz, 2H, 24-H2), 2.60 (dd, J = 1.5, 14.6 Hz, 1H, 8-Hb), 3.44 (dd, J = 10.4, 14.6 Hz, 1H, 8-Ha), 3.83–3.89 (m, 2H, 9-H, 11-OH), 3.98 (mc, 1H, 11-H), 4.23 (mc, 1H, 13-H), 4.97 (dt, J = 7.4, 14.5 Hz, 1H, 17-H), 5.39 (mc, 1H, 15-H), 5.69 (d, J = 11.5 Hz, 1H, 20-H), 5.79 (mc, 1H, 23-H), 6.71 (dd, J = 10.4, 14.5 Hz, 1H, 18-H), 6.85 (dt, J = 1.1, 11.5 Hz, 1H, 21-H), 7.23 (mc, 2H, 4-H, 6-H), 7.44 (dd, J = 7.7, 8.2 Hz, 1H, 5-H), 7.50 (d, J = 8.5 Hz, 2H, Ar-H), 7.51 (mc, 1H, 22-H), 8.23 (d, J = 8.7 Hz, 2 H, Ar-H), 8.92 (d, J = 10.4 Hz, 1 H, NH). 13C NMR (150.8 MHz, [D6]-acetone) is as follows: δ = 14.3 (C-25), 21.0 (C-24), 29.9 (CN2, hidden), 35.8 (C-16), 38.5 (C-14), 39.0 (C-10), 39.8 (C-8), 40.1 (C-12), 64.7 (C-11), 67.5 (C-13), 73.7 (C-9), 74.6 (C-15), 106.9 (C-17), 120.7 (C-20), 121.6 (C-6), 123.0 (q, JC-F = 275 Hz, CF3), 125.4 (C-22), 126.4 (C-18), 127.8 (C-Ar), 129.0 (C-4), 130.5 (C-5), 130.6 (C-7), 131.4 (C-Ar), 131.7 (C-2) 134.7 (C-Ar), 136.9 (C-21), 140.8 (C-Ar), 141.5 (C-23), 147.7 (C-3), 163.5 (C-19), 164.1 (C-1′), 167.9 (C-1).
Using [1-14C]4-(3-trifluoromethyldiazirin-3-yl)benzoic acid, the 14C-labeled apicularen derivative was prepared accordingly with a specific activity of 37.9 mCi/mmol as determined with a liquid scintillation counter. All inhibitors were dissolved in DMSO and stored in stock solutions (10 mm) at −80 °C.
Photoaffinity Labeling
Thirty μg of M. sexta V1VO holoenzyme, 20 μg of V1 complex, or 10 μg of VO complex, respectively, were incubated with 52 μm 21-O-[4-(3-trifluoromethyldiazirin-3-yl)-[1-14C]benzoyl]bafilomycin A1 (14C-D-bafilomycin), 52 μm 23-O-[4-(3-trifluoromethyldiazirin-3-yl)-[1-14C]benzoyl]-21-deoxyconcanolide A (14C-D-concanolide), or 100 μm 3-O-[4-(3-trifluoromethyldiazirin-3-yl)-[1-14C]benzoyl]apicularen (14C-D-apicularen) for 5 min at 25 °C. In initial experiments, the presence of ATP apparently led to a slightly better label efficacy (data not shown). This may be the consequence of an increased number of V-ATPase inhibitor complexes forced into these specific conformations in which the inhibitor is, as Bowman and co-workers suggested, bound “like a stone in the gears” (14). Therefore, 1 mm ATP in 1.5 mm MgCl2 was added to a final volume of 40 μl. For competition assays, 52 μm of the 14C-diazirinylbenzoyl-labeled inhibitors were used after a preincubation with a 10-fold excess of a nonradioactive inhibitor for 5 min at 25 °C. Cross-linking was induced by irradiating the samples for 1 min with UV light (366 nm) on ice. Subsequently, the samples were separated by SDS-PAGE (T 17%, C 0.4%) and stained with Coomassie. The gels were then dried on Whatman paper, exposed to a phosphor screen for 72 h, and analyzed with a phosphorimager (GE Healthcare).
Minimum Energy Calculation for D-Concanolide
For a theoretical insight from MM2-force field calculations on isolated systems in the gas phase the diazirinyl-labeled concanolides were investigated as example for nanomolar V-ATPase inhibitors. We calculated in respect to the possible positions which the flexible diazirinyl group may occupy in comparison to the pharmacophore using CS Chem3D Pro 12.0.2.1076, Gaussian 98, (CambridgeSoft, Cambridge, MA) and the energy minimization with the MM2-algorithm.
Yeast Strains and Growth Conditions
The yeast strains used in this study were BMA64-1B (MATα, ura3-52; trp1Δ2; leu2-3_112; his3-11; ade2-1; can1-100) (Euroscarf, Frankfurt, Germany) and BMA64-1BΔvma3 (MATα, ura3-52, trp1Δ2, leu2-3_112, his3-11, ade2-1, can1-100, vma3::HIS3) (21). The complemented strain BMA64-1BΔvma3+VMA3 was obtained by transformation of the plasmid carrying the VMA3 gene pRS415/VMA3 (21) into the vma3 deletion mutant by electroporation.
The mutant strain BY4741Δvph1Δstv1 was obtained by crossing the haploid strains BY4741Δvph1 and BY4742Δstv1 (both Euroscarf, Frankfurt, Germany). Each 500 μl of the appropriate overnight cultures were mixed and pelleted and afterward incubated overnight at 30 °C on agar plates containing YPDA, pH 5.5. Small amounts of the resulting zygotes were resuspended in 100 μl of H2O, applied on a new agar plate, separated by using a micromanipulator (Singer MSM), and again incubated at 30 °C. To obtain haploid double deletion mutants, the diploid strains were sporulated on agar plates containing 2% potassium acetate for 8 days at 30 °C. Again, a small amount of spores were resuspended in 100 μl of H2O, treated with 100 μg of zymolyase (100 units/mg, MP Biomedicals) for 10 min, and applied on an agar plate (YPDA, pH 5.5), and the tetrads were separated by using a micromanipulator (Singer MSM). By selection on plates containing 250 μg/μl G418, those tetrads of which two of the colonies grew on media with G418 and two colonies that did not grow on G418 were used for a PCR analysis of the genotype. By using two different primer combinations, (a) TACTCACGGCCGGCTAGTAATCCAGTTGCCGAGC and ATCGCGAGCCCATTTATACC and (b) GACATAGGCCCACGAAGGTG and ATCGCGAGCCCATTTATACC, as well as the isolated genomic DNA of the four tetrad colonies as templates, the deletions of the genes vph1 and stv1 were confirmed.
Cells were usually grown on YPD, pH 5.5, additionally containing 0.02% adenine (YPDA, pH 5.5). The medium was buffered to pH 5.5 with 50 mm MES, 50 mm MOPS. For YPDA, pH 7.5, the pH was adjusted using NaOH. YPDA, pH 5.5, medium with 0.1 m CaCl2 was obtained by adding 10% of a sterile 1 m CaCl2 stock solution after autoclaving the medium. For plates, 2% agar was added. Selection of cells was carried out by using plates with SD medium containing 0.67% yeast nitrogen base without amino acids, 2% glucose, 1.5% agar, and 10% drop out amino acid solution (21) lacking the respective amino acid. The medium was buffered with 50 mm MES, 50 mm MOPS and adjusted to pH 5.5 using HCl.
Plasmid Construction
The plasmid pXJ40-KKO-16k containing the coding sequence of the gene ATP6VOC of the human V-ATPase subunit c was kindly provided by M. A. Skinner (25). The ATP6VOC gene was amplified by PCR using the plasmid pXJ40-KKO-16k as template and the forward primer TACTCAAAGCTTATGTCCGAGTCCAAGAGC and the reverse primer TACTCAAAGCTTCTAAGCGTAATCTGGTACGTCGTATGGGTACTTTGTGGAGAGGATGAG. Notably, the second primer additionally contains the coding sequence for a single C-terminal HA tag. The PCR product was cloned into the yeast expression vector pGADT7 by using the HindIII restriction sites resulting in a plasmid called pGADT7/ATP6VOC. The gene of the M. sexta proteolipid subunit c was amplified by PCR using a pBluescript plasmid containing the accordant coding sequence (26) as template as well as the forward primer TACTCAAAGCTTATGGCCGAAAATCCAATC and the reverse primer TACTCAAAGCTTTTACTGTTTCGTGTACAG. The PCR product was cloned into the vector pGADT7 by using the HindIII restriction sites. The resulting plasmid pGADT7/Msc and the plasmid pGADT7/ATP6VOC were transformed into the vma3 deletion mutant BMA64-1BΔvma3 by electroporation, and the cells were selected on SD medium without leucine.
The construction of a plasmid carrying the coding sequence for a hybrid subunit a containing the N-terminal half of yeast VPH1 and the C-terminal half of the human a4 subunit was performed by homologous recombination in the yeast wild type strain BY4741 (Euroscarf, Frankfurt, Germany). For this purpose, the C-terminal half of the human a4 subunit encoded by the amino acids 396–840 and additional homologous regions were amplified by PCR using the primers CGGTATTGCTCAGTACAGAGAAATCAATGCTGGTTTACCCTACACCATCATCACCTTCCCCTTCC and GGTGGATTGGATTGCAAGTCTAACGTTTTCATGAGATAAGCTACTCCTCGGCTGTGCCATCCA and the plasmid pJV97ha4 (a gift of Fiona Karet) as template. For the plasmid pRS415/VPH1, the yeast VPH1 gene and in addition 400 bp upstream and downstream implying its native promoter and terminator was cloned into the yeast CEN vector pRS415. Afterward, the plasmid was linearized by the restriction enzyme BsgI. This open pRS415/VPH1 as well as the C-terminal half of the human a4 subunit were transformed into the yeast wild type strain BY4741. By homologous recombination, the C-terminal half of VPH1 encoded by the amino acids 411–840 was replaced by the C-terminal half of the human a4 subunit resulting in a yeast human hybrid subunit a. The plasmid was called pRS415/VPH1/a4.
Serial Drop Dilution Assay
To analyze the V-ATPase function, serial drop dilution assays were carried out. Yeast strains were grown overnight in 5 ml of YPDA, pH 5.5, and diluted to 105 cells/ml in distilled water. Serial 10-fold dilutions from 104 cells/ml to 100 cells/ml were prepared, and each 5 μl was dropped onto plates with different media. After 3 days of incubation at 30 °C, pictures were taken.
Other Methods
The V1VO-ATPase holoenzyme, the VO complex, and the V1 complex were isolated from M. sexta midgut as published previously (12, 27). Yeast vacuolar membranes were purified as described by Bockelmann et al. (21). V-ATPase assays using the M. sexta V1VO holoenzyme or V1 complex or yeast vacuolar membranes were carried out as already reported (21, 27, 28). Protein content was determined using Amido Black 10B (28). SDS-PAGE, Western blotting on nitrocellulose, and immunostaining were performed as described previously (12, 29). For immunodetection, the primary antibodies against the V-ATPase subunits A (30) and e (31) from M. sexta, Vma1 (A6422, Invitrogen), and Vma6 (gift from Christian Ungermann, University of Osnabrück) from yeast, the HA tag (Y-11) (sc-805, Santa Cruz Biotechnology), and the secondary antibodies against rabbit (A3687, Sigma) and mouse (A3562, Sigma) were used.
RESULTS AND DISCUSSION
Labeling of the V-ATPase with Radioactive PAL Inhibitors
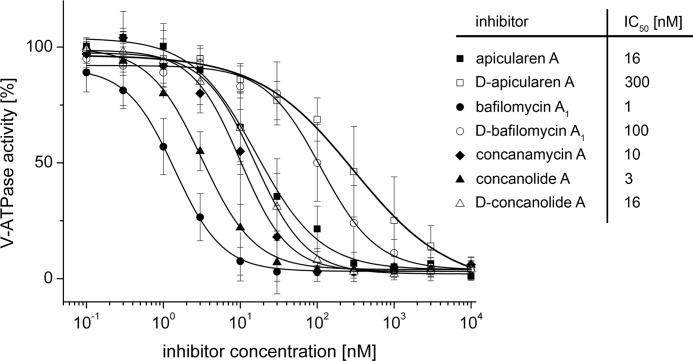
Before the diazirinyl-labeled derivatives of bafilomycin, concanolide, and apicularen (for structures see Fig. 1) were used for photoaffinity labeling, we tested in enzyme activity assays whether the modifications had an influence on V-ATPase inhibition. Compared with the natural products, the labeled derivatives exhibited reductions of their inhibitor potential by a factor of only 5–100 (Fig. 2) and therefore appeared to be highly suitable for the labeling assays.
FIGURE 2.
Inhibition of the ATPase activity of the M. sexta V1VO holoenzyme by bafilomycin, concanamycin, apicularen, and their derivatives. Values represent the means ± S.D. of experiments with three different preparations. The specific enzyme activity of the controls without inhibitors was 1.6 ± 0.4 μmol·mg−1·min−1.
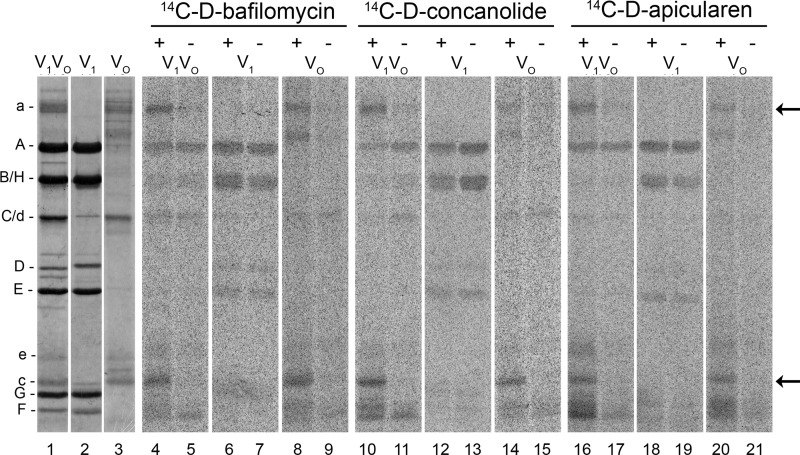
To determine which subunits of the V-ATPase interact with the inhibitors, the V1VO holoenzyme, the VO complex, or as a control, the V1 complex were incubated with the 14C-diazirinyl-labeled derivatives of the inhibitors, and the cross-link reactions were induced by the exposition to UV light. The results of autoradiography after SDS-PAGE are shown in Fig. 3. Generally speaking, labeling patterns appeared to be rather similar for all three inhibitors. The comparison of the exposed (lanes with even numbers, 4–20) and unexposed samples (lanes with odd numbers, 5–21) revealed a clear labeling of the VO subunits a and c in the V1VO holoenzyme as well as in the VO complex.
FIGURE 3.
Photoaffinity labeling of M. sexta V1VO holoenzyme, V1 complex, and VO complex with the 14C-labeled derivatives of D-bafilomycin, D-concanolide, and D-apicularen. For the labeling assays, 30 μg of V1VO holoenzyme, 20 μg of V1 complex, or 10 μg of VO complex were first incubated for 5 min at 25 °C with 52 μm 14C-D-bafilomycin, 52 μm 14C-D-concanolide, or 100 μm 14C-D-apicularen, respectively. Before the samples were exposed to UV light (366 nm) for 1 min (+) or kept in the dark (−), 1 mm Mg-ATP was added. Afterward, the protein subunits were separated by SDS-PAGE, stained with Coomassie Blue, and exposed to a phosphor screen. Lanes 1–3, typical staining with Coomassie Blue; lanes 4–21, readout of the phosphor screen.
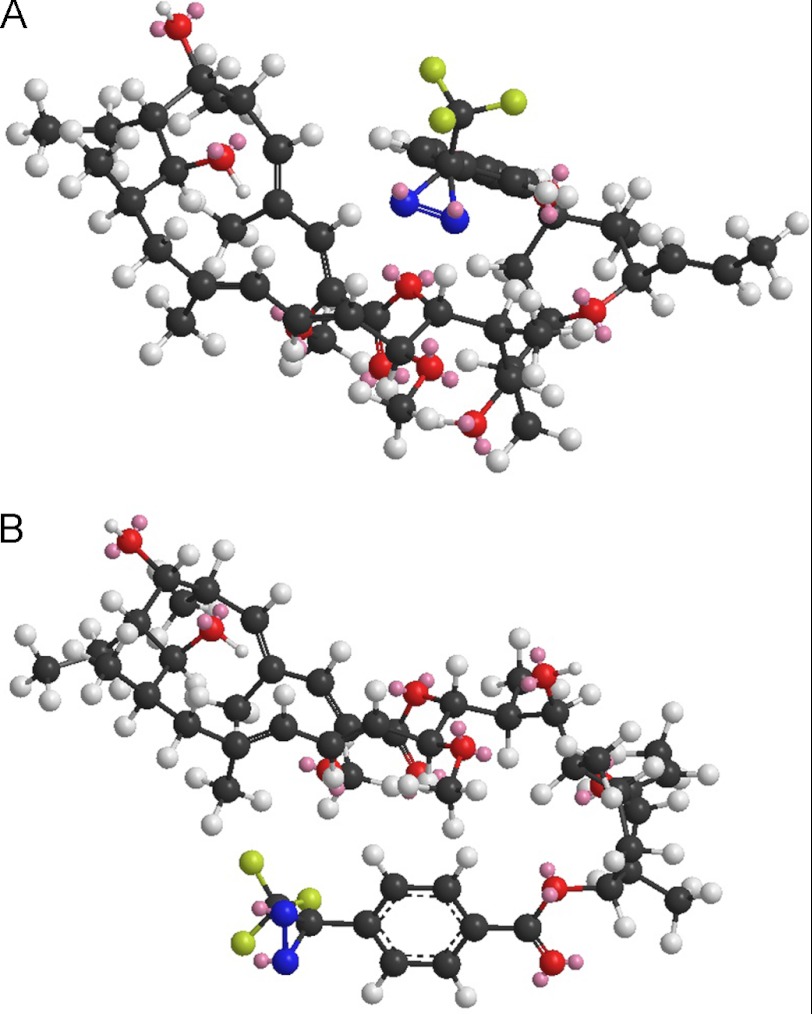
To date, subunit c was the only polypeptide of the VO complex that could be labeled successfully by inhibitors such as concanamycin, archazolid, or NCD-4 (12, 21). Yet the appearance of a label at subunit a was not unexpected as its participation in plecomacrolide binding had already been shown by site-directed mutagenesis studies in the yeast V-ATPase (16). Additionally, it was anticipated that placement of the diazirinyl in the concanamycin derivative at position C23 instead of position C9 as in the previous 125I-concanolide used in 2002 (12, 17) would lead to a different labeling pattern. This concept is based on the assumption that the diazirinyl attached to the C9 of the inhibitor is, as already mentioned above, buried in the binding site between two adjacent c subunits, whereas the diazirinyl at position C23 probably is localized at the interface between subunits a and c. For a rough estimation of the space-compassing manner of the diazirinyl derivatives and how they label two opposing subunits, we exemplarily performed a minimum energy calculation for D-concanolide. The model provides a view on the theoretical positions and spanned distances of the diazirinyl group, which is, with respect to the pharmacophore, flexible around its single bonds of the ester moiety. The results of the calculations pointed to two preferred conformations, one with the diazirinyl moiety above and one below the macrolactone ring close to the molecule (Fig. 4) and a small distance of the label. These theoretical data support the hypothesis derived from the functional assays that labeling of both subunits a and c is conclusive even though the inhibitor is bound in one certain orientation in the binding site.
FIGURE 4.
Minimum energy calculation for D-concanolide. For 21-deoxyconcanolide A, the space of the connected diazirinyl moiety is placed preferably at the east hemisphere. The length of the benzoyl-diazirinyl label can be calculated to 6.4 Å and its distance from the macrolactone ring to 5.4 Å (A). Minimum energy calculations suggest that the diazirinyl group is situated preferably above and below the macrolactone ring very close to the molecule (A and B). A, benzoyl diazirinyl system is on the other side of the carbonyl group of the macrolactone ester. B, benzoyl diazirinyl system is on the same side of the carbonyl group of the macrolactone ester.
The irradiated samples of the VO complex exhibited a further band below subunit a, which was most probably a degradation product of subunit a and also visible in the Coomassie staining of the VO complex. Labeling of the V1 subunits in the V1VO holoenzyme and in the V1 complex are considered unspecific as it occurred in both the irradiated and nonirradiated samples. In addition, we may exclude specific inhibitory interactions with the V1 complex as it was already shown previously that the ATPase activity of the V1 complex of M. sexta and yeast is not affected by the plecomacrolide concanamycin and that salicylihalamide, another benzolactone enamide, did not inhibit the V1 complex of bovine clathrin-coated vesicles (22, 27, 32). In this study, we confirmed these results for the V1 complex from M. sexta using either 30 μm of D-bafilomycin, D-concanolide, D-apicularen, or their parent compounds. None of these inhibitors had an effect, neither on the Ca2+-ATPase nor Mg2+-ATPase activity of the isolated V1 complex (data not shown).
Additional labeling of V1 subunits may thus be interpreted as a consequence of unspecific binding of the derivatives due to the high protein and inhibitor concentrations in the assays, as they appeared in both the exposed and unexposed samples and even in the purified V1 complex. A further label close to the V1 subunits F and G was found in the V1VO holoenzyme and in the VO complex but not in the V1 complex. To check the origin of this unexpected label, pieces containing subunits F and G as well as the areas above, in between, and below these subunits were excised from a gel for further analysis. Scintillation counting revealed no radioactivity in pieces containing subunits F and G, but in contrast, radioactivity was detected at the interspace between the subunits and below subunit F (data not shown). In addition, mass spectrometric analysis (ESI-MS) verified subunits F and G, but no protein was detected in the areas above, in between, and below the subunits. Therefore, we suppose that these labels result from cross-links of the diazirinyl-derivatives with lipid or detergent molecules that remain bound specifically to the VO complex during purification. This assumption is supported by the results of von Ballmoos et al. (33) who observed cross-links between lipids and diazirinyl-labeled carbodiimide inhibitors interacting with the FO subunit c of the ATP synthase.
Treatment of the V1VO holoenzyme as well as the VO complex with 14C-D-apicularen exhibited an additional radioactively labeled band with an approximate molecular mass of 20 kDa (Fig. 3, lanes 16 and 20). This size suggests that two polypeptides of the VO complex could be labeled subunit c″ and/or subunit e. Although a gene encoding a putative subunit c″ was identified in the M. sexta genome project with a calculated molecular mass of 27 kDa, we have no indication that a product of this gene is an integral part of the purified V-ATPase derived from the plasma membrane of the midgut from M. sexta. Extensive analysis of all proteins of the purified V1VO holoenzyme, the purified VO complex, and chloroform/methanol extracts of both by MALDI-MS and ESI-MS supplemented by N-terminal sequencing revealed so far no evidence for the presence of a subunit c″ (12, 34).3 Based on this state of knowledge, we so far do not see any evidence for the presence of subunit c″ in the plasma membrane V-ATPase of M. sexta. However, Western blot analysis of the 14C-D-apicularen-labeled V1VO holoenzyme using monoclonal antibodies against subunit e clearly identified this labeled band as subunit e (Fig. 5, lane 3). Being a part of the VO complex probably associated with subunit a (35), it is very likely that subunit e gets labeled due to its close proximity to subunit a, its position opposite to the c-ring, or even due to a direct contribution to the binding site. Anyhow, the result that D-apicularen is the first inhibitor also labeling subunit e indicates again that the binding site of the benzolactone enamides is indeed different from the binding sites for the plecomacrolides and the archazolids and that the mechanism by which the benzolactone enamides inhibit V-ATPases may also be a different one. Unfortunately, nearly nothing is known about the exact localization or the function of subunit e in the V-ATPase, except that it is an integral and essential part of the VO complex (12, 31, 36, 37). Although subunit e is positioned next to subunit a in some models of the VO complex, until now there are no experimental data supporting a direct interaction between subunit e and any other subunit of the VO complex (36, 38). Therefore, it appears difficult to integrate subunit e into a concise model of the localization of the inhibitor-binding sites for which reason we will draw our model without subunit e. Anyway, taken together, the cross-linking studies with all 14C-diazirinylbenzoyl-labeled inhibitors clearly identified both the VO subunits a and c to harbor parts of the binding sites.
FIGURE 5.
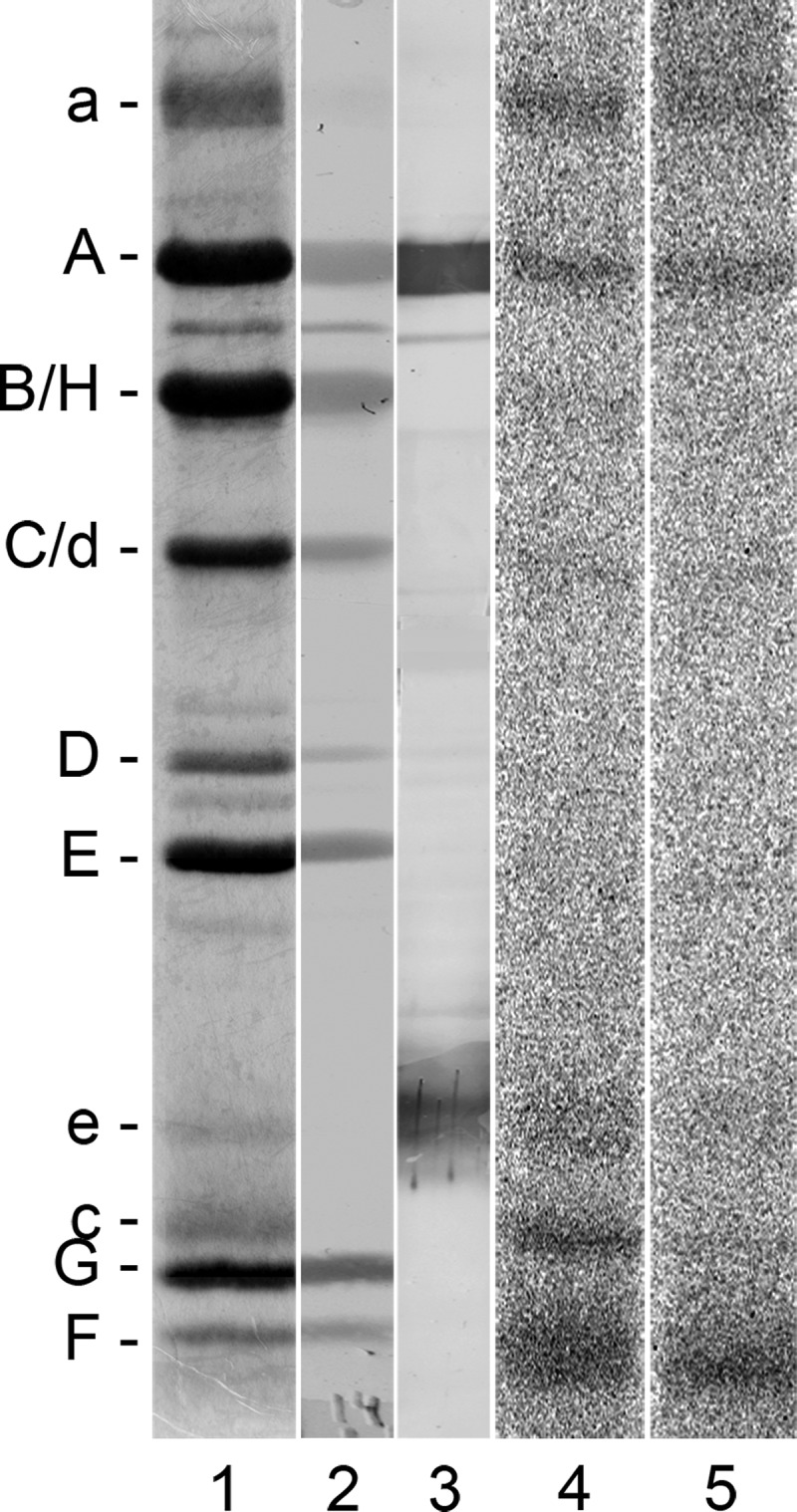
Western blot analysis of the V-ATPase with 14C-D-apicularen. For the identification of the 14C-D-apicularen-labeled band with the approximate molecular mass of subunit e 30 μg of the V1VO holoenzyme were incubated with 100 μm 14C-D-apicularen, and before the samples were exposed to UV light (366 nm) or kept in the dark, 1 mm Mg-ATP was added. Afterward, the protein subunits were separated by SDS-PAGE. Lane 1, staining of the SDS-gel with Coomassie Blue; lane 2, staining of the proteins with Ponceau S after electro-transfer onto a nitrocellulose membrane; lane 3, immunostaining with monoclonal anti-A (221-9) and anti-e (224-3) antibodies; lane 4, autoradiography of the irradiated sample; lane 5, autoradiography of the nonirradiated sample.
Specification of the Arrangement of the Binding Sites for Bafilomycin, Concanamycin, and Apicularen by Competitive Labeling
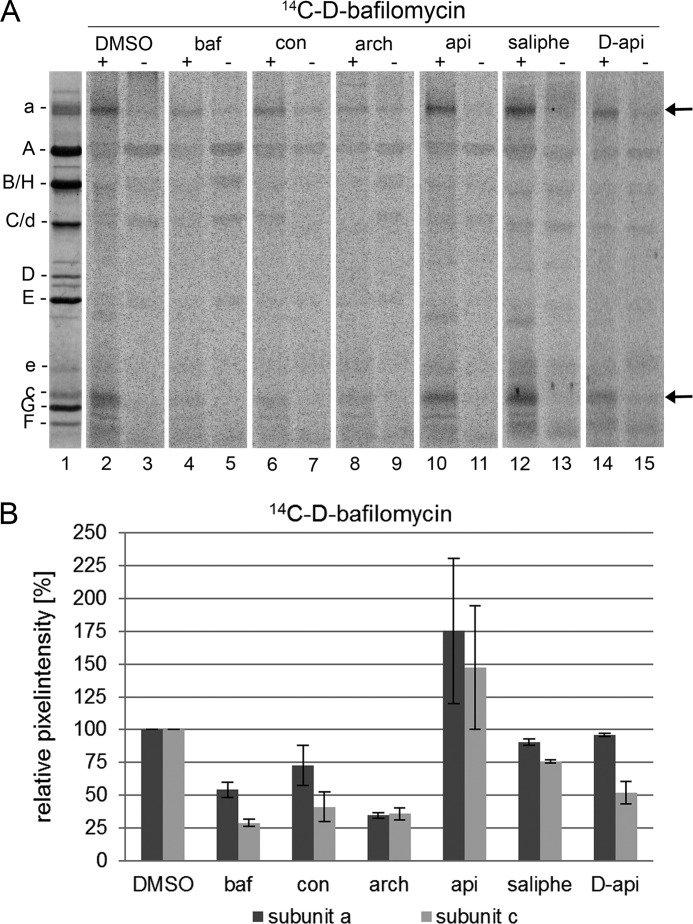
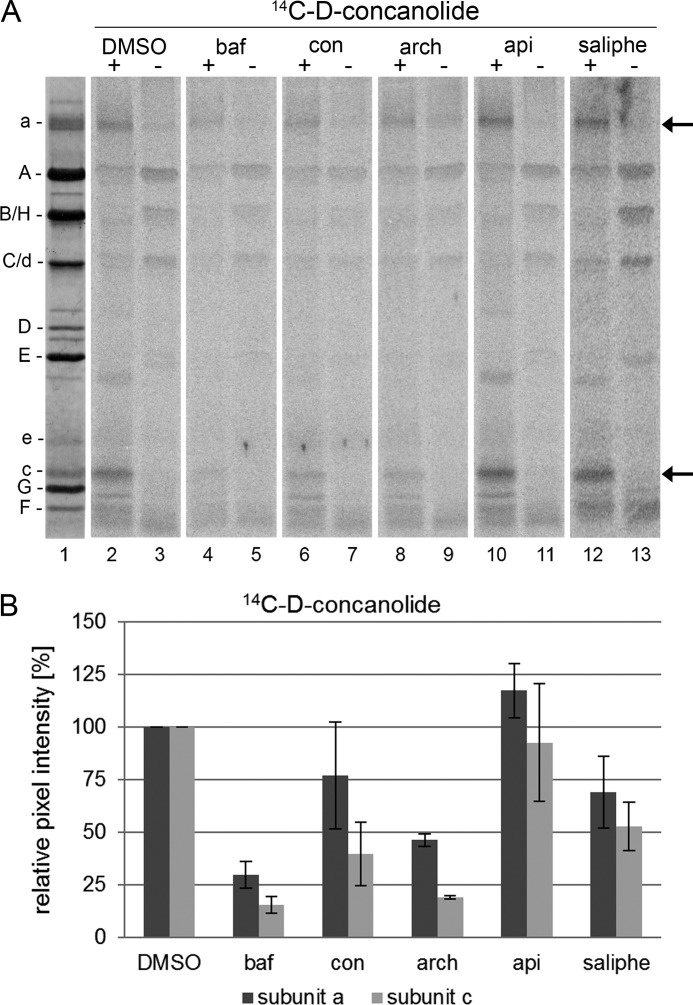
To investigate to what extent the binding sites for apicularen, concanamycin, and bafilomycin overlap within the VO complex, we performed photoaffinity labeling assays with the 14C-diazirinyl-containing derivatives in the presence of a 10-fold excess of the natural compounds, i.e. the V1VO holoenzyme was preincubated with DMSO, bafilomycin, concanamycin, archazolid, apicularen, or saliphenylhalamide, respectively. For 14C-D-bafilomycin, preincubation with the plecomacrolides bafilomycin and concanamycin as well as with the macrolide archazolid led to a significant reduction of the label in the VO subunit c (Fig. 6, lanes 4, 6, and 8). This outcome confirmed the results obtained in analogous experiments performed with bafilomycin, concanamycin, and archazolid where labeling of the VO subunit c by 125I-concanolide A or 14C-(bis-diazirinyl)-archazolid was suppressed in a similar manner (10, 12, 21). Evidently, the interaction with the VO subunit a also was significantly reduced by an excess of these compounds. The same effects can be seen in the 14C-D-concanolide series, where bafilomycin, concanamycin, and archazolid impeded labeling of both VO subunits a and c (Fig. 7, lanes 4, 6, and 8). Notably, in neither series did the benzolactone enamides apicularen and saliphenylhalamide prevent labeling by 14C-D-plecomacrolides (Figs. 6, lanes 10 and 12, and 7, lanes 10 and 12). This result supports the previous assumption that the binding site for the benzolactone enamides is significantly different from that for the plecomacrolides (10, 12, 22).
FIGURE 6.
Photoaffinity labeling of the V-ATPase with 14C-D-bafilomycin after preincubation with different inhibitors. Samples with 30 μg of V1VO holoenzyme were preincubated for 5 min at 25 °C with DMSO, 625 μm bafilomycin (baf), concanamycin (con), archazolid (arch), apicularen (api), or saliphenylhalamide (saliphe), respectively. Then the samples were incubated for 5 min at 25 °C with 52 μm 14C-D-bafilomycin. Before the samples were exposed to UV light (366 nm) for 1 min (+) or kept in the dark (−), 1 mm Mg-ATP was added. Then protein subunits were separated by SDS-PAGE, stained with Coomassie Blue, and exposed to a phosphor screen. A, lane 1, typical staining with Coomassie Blue; lanes 2–15, readout of the phosphor screen. B, analysis of the pixel intensity of the labeled bands with ImageQuant. Values show the means ± S.D. of two independent preparations, except for DMSO (n = 6) and apicularen (n = 5).
FIGURE 7.
Photoaffinity labeling of the V-ATPase with 14C-D-concanolide after preincubation with different inhibitors. Samples with 30 μg of V1VO holoenzyme were preincubated for 5 min at 25 °C with DMSO, 625 μm bafilomycin (baf), concanamycin (con), archazolid (arch), apicularen (api), or saliphenylhalamide (saliphe), respectively. Then samples were incubated for 5 min at 25 °C with 52 μm 14C-D-concanolide. Before the samples were exposed to UV light (366 nm) for 1 min (+) or kept in the dark (−), 1 mm Mg-ATP was added. Afterward protein subunits were separated by SDS-PAGE, stained with Coomassie Blue, and exposed to a phosphor screen. A, lane 1, typical staining with Coomassie Blue; lanes 2–13, readout of the phosphor screen. B, analysis of the pixel intensity of the labeled bands with ImageQuant. Values show the means ± S.D. of two independent preparations, except for DMSO (n = 8) and apicularen (n = 5).
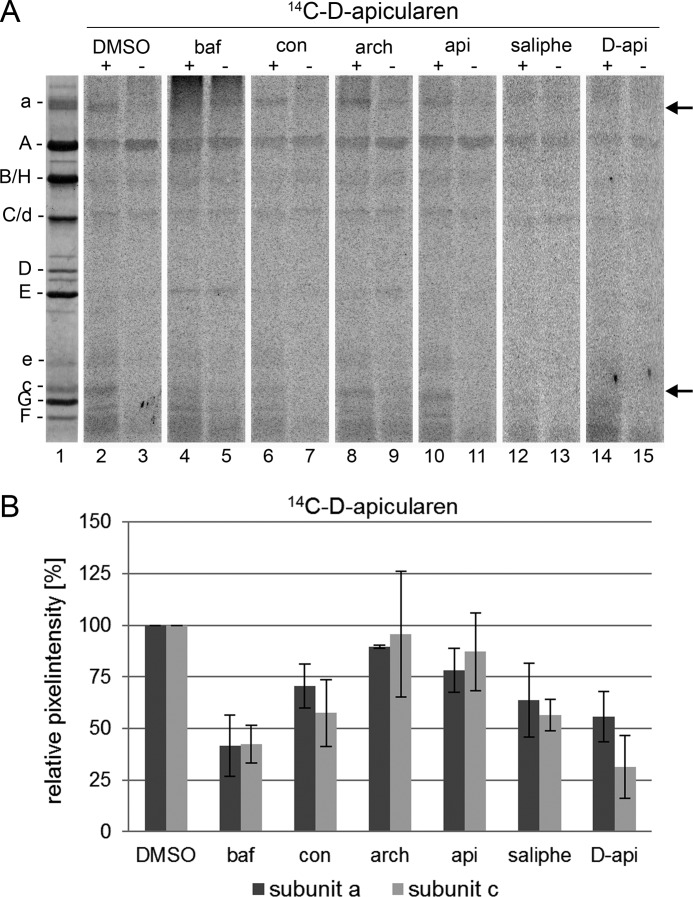
Competition assays using 14C-D-apicularen led to the unexpected result that the native apicularen itself did nearly not prevent the labeling (Fig. 8, lane 10). A more effective reduction of labeling was achieved by preincubation with the nonradioactive D-apicularen and with saliphenylhalamide, another benzolactone enamide (Fig. 8, lanes 12 and 14). Therefore, we suggest that the less bulky native apicularen might be too small to cover the complete binding site of 14C-D-apicularen. By contrast, the nonradioactive D-apicularen has the same size, and saliphenylhalamide has a comparable size (Fig. 1), and therefore both compounds are more suitable to displace 14C-D-apicularen. Furthermore, we may exclude binding of D-apicularen to an alternative binding site due to the sheer presence of the attached diazirinyl group, because a control compound modified with a diazirinyl was, in contrast to D-apicularen, not able to inhibit the V-ATPase (39). Surprisingly, an excess of the plecomacrolides bafilomycin or concanamycin also reduced labeling by 14C-D-apicularen (Fig. 8, lanes 4 and 6), whereas apicularen did not reduce labeling by 14C-D-bafilomycin as described above. Yet, in experiments using D-apicularen and 14C-D-bafilomycin, a partial reduction of the radioactive label was detectable (Fig. 6, lane 14). Based on these results, we suspect the diazirinyl group of D-apicularen to extend into the plecomacrolide-binding site leading to a displacement by the plecomacrolides or the other way around. Yet the binding site for the native apicularen is obviously different from that for the plecomacrolides as they were not displaced by apicularen in labeling experiments (see above).
FIGURE 8.
Photoaffinity labeling of the V-ATPase with 14C-D-apicularen after preincubation with different inhibitors. Samples with 30 μg of V1VO holoenzyme were preincubated for 5 min at 25 °C with DMSO, 625 μm bafilomycin (baf), concanamycin (con), archazolid (arch), apicularen (api), or saliphenylhalamide (saliphe). Then the samples were incubated for 5 min at 25 °C with 52 μm 14C-D-apicularen. Before the samples were exposed to UV light (366 nm) for 1 min (+) or kept in the dark (−), 1 mm Mg-ATP was added. Then protein subunits were separated by SDS-PAGE, stained with Coomassie Blue, and exposed to a phosphor screen. A, lane 1, typical staining with Coomassie Blue; lanes 2–15, readout of the phosphor screen. B, analysis of the pixel intensity of the labeled bands with ImageQuant. Values show the means ± S.D. of two independent preparations, except for DMSO (n = 7), apicularen (n = 5), and D-apicularen (n = 3).
The macrolactone archazolid did not prevent labeling of the V-ATPase by 14C-D-apicularen (Fig. 8, lane 8). This was expected from our previously published results with apicularen, NCD-4, and archazolid that had elucidated that archazolid binds to subunit c of the V1VO holoenzyme and has an overlapping but different binding site as bafilomycin (21). Taken together, our results now show that apicularen has a binding site within the subunit a or c of the VO complex, which differs from those for archazolid and for the plecomacrolides, but a slight overlap of the binding sites for the plecomacrolides and apicularen cannot be excluded.
Exploration of the Apicularen-binding Site by Genetic Manipulation of the Yeast V-ATPase
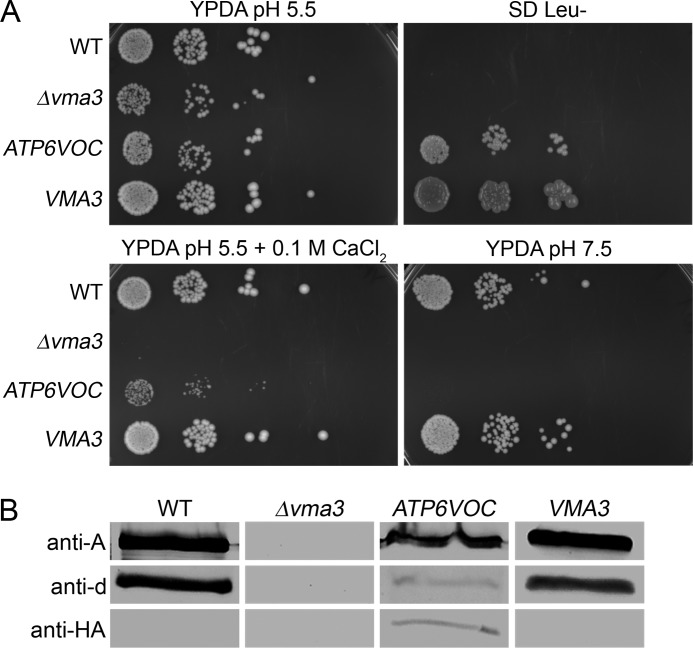
To find out whether subunit a or c harbors the apicularen-binding site, we took advantage of the facts that bakers' yeast can easily be genetically manipulated and furthermore that apicularen does not inhibit the fungal V-ATPases. The expression of the apicularen-sensitive homologues of either human V-ATPase subunit a or c in appropriate yeast deletion mutants should result in a hybrid V-ATPase, which is active and sensitive to apicularen. This would elegantly prove to which of these two subunits apicularen binds. The possibility to complement a yeast vma3 deletion mutant strain with c subunits of other organisms has already been reported, e.g. for the 16-kDa proteolipid from Nephrops norvegicus or Drosophila melanogaster (40, 41). In our approach, we expressed the human ATP6VOC in a yeast Δvma3 mutant strain that resulted in a partially restored vma− phenotype as this strain grew on media with an elevated CaCl2 concentration, whereas growth on media with alkaline pH failed (Fig. 9A) (42, 43). This moderately recovered V-ATPase activity was also confirmed by Western blot analysis of purified vacuolar membranes that revealed an appropriate amount of assembled V1VO holoenzyme by immunodetection of the V1 subunit A as well as the VO subunits d and human c (Fig. 9B). From ATPase activity assays with isolated vacuolar membranes, a specific enzyme activity of 0.064 ± 0.005 μmol·mg−1·min−1 was determined for the ATP6VOC-expressing strain, which is about 16% of the wild type activity and, according to Leng et al. (44), nearly enough activity to exhibit wild type growth. Even though this hybrid V-ATPase containing ATP6VOC was highly sensitive to bafilomycin A1, it still was not sensitive to apicularen (Fig. 10). In parallel, we also expressed the proteolipid subunit c of M. sexta in a yeast Δvma3 mutant strain. The analysis of the growth of the resulting strain on media with an elevated CaCl2 concentration or an alkaline pH as well as the Western blot analysis of the V-ATPase assembly in isolated vacuolar membranes and ATPase activity assays revealed the same results (data not shown) as for the expression of the human ATP6VOC. Similarly, this hybrid V-ATPase expressing the M. sexta VO subunit c was not sensitive to apicularen. Therefore, subunit c of the VO complex obviously does not harbor the binding site of apicularen.
FIGURE 9.
A, growth of the human Vma3 homologue ATP6VOC-expressing yeast strain. Serial 10-fold dilutions of the yeast wild type strain BMA64-1B, the deletion mutant BMA64-1BΔvma3, and the deletion mutant BMA64-1BΔvma3 complemented with pRS415/VMA3 were dropped on different media. After 3 days of incubation at 30 °C, pictures of the plates were taken. B, assembly of the V-ATPase in isolated vacuolar membranes of the ATP6VOC-expressing strain. Each 10 μg of isolated vacuolar membranes of the wild type, the deletion mutant, and the strains complemented with ATP6VOC or VMA3 were separated by SDS-PAGE, transferred to nitrocellulose, and analyzed by immunostaining with anti-A, anti-d, and anti-HA antibodies.
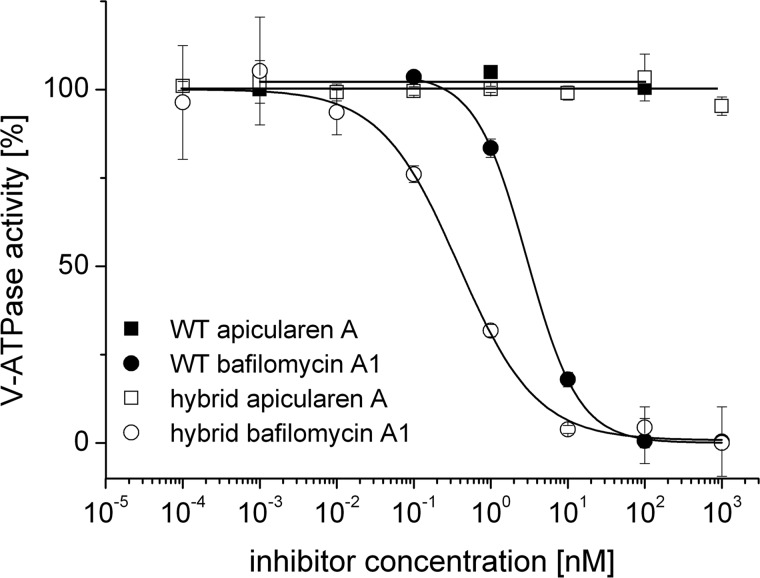
FIGURE 10.
Inhibition of the V-ATPase activity of vacuolar membranes from the wild type strain BMA64-1B (WT) and the ATP6VOC-expressing strain (hybrid) by bafilomycin A1 and apicularen A. Values represent the means of two independent preparations ± S.D. The specific enzyme activity of the controls was 0.064 ± 0.005 μmol·mg−1·min−1 (ATP6VOC-expressing strain) and 0.41 ± 0.1 μmol·mg−1·min−1 (wild type strain BMA64-1B).
The next step was to express the human isoform a4 of the V-ATPase in a Δvph1Δstv1 mutant strain to check whether this hybrid V-ATPase is sensitive to apicularen, and therefore subunit a may host its binding site. Unfortunately, this approach did not lead to an active V-ATPase (data not shown). Furthermore, the expression of a yeast-human hybrid subunit a containing the N-terminal half of yeast Vph1, which is important for targeting and regulation of the V-ATPase (45), connected with the C-terminal half of the human a4 subunit did not lead to the restoration of V-ATPase activity (data not shown). Nevertheless, we can conclude that subunit c does not exclusively form the binding site for apicularen and that subunit a contributes substantially to the binding of apicularen.
Modeling of the Binding Site for Apicularen in the Vicinity of Those for Plecomacrolides and Archazolid
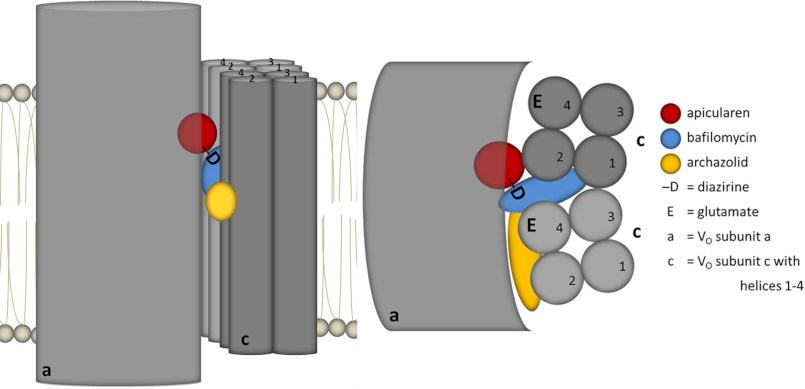
Taken together, the information from our results presented here and from previously published data on the binding sites for archazolid and for bafilomycin can be integrated in a concise model arranging the binding sites for three different classes of V-ATPase inhibitors within the VO complex (Fig. 11). Based on mutagenesis studies of the c subunit performed in N. crassa and S. cerevisiae, the binding site for the plecomacrolides is localized between two adjacent c subunits in the cytosolic half of the membrane bilayer (15, 21), indicated in blue in our model. The archazolid-binding site, displayed in yellow, has been shown to reside in the equatorial region of one single subunit c covering the essential glutamate (21). Now we suggest that the binding site for apicularen, indicated in red, is located at the interface of subunits a and c, above the equatorial region and in the cytosolic half of the membrane bilayer. Our assumption is based on the following results: (i) apicularen labels both subunits a and c; (ii) it does not interfere with archazolid binding, and (iii) it does not interfere with bafilomycin binding but may interact in the vicinity of bafilomycin, because D-apicularen, with its attached diazirinyl group, obviously interferes with the binding of the competing inhibitor (Fig. 6, lane 14). As already mentioned above, we decide to not include subunit e into our model as there is no reliable information on its localization available.
FIGURE 11.
Model of the inhibitor-binding site arrangement in the membrane. Left, side view; right, top view from cytosol. For simplicity, the N-terminal part of subunit a is drawn as a single sphere, and only two c subunits of the c-ring are shown. The four transmembrane helices of the c subunits are numbered (1–4). The suggested locations of the inhibitor-binding sites of apicularen, bafilomycin, and archazolid are colored in red, blue, and yellow, respectively. D, the position of the diazirinyl group attached to apicularen. E, the position of the essential glutamate in helix 4.
To gain further insights into the binding site of apicularen, it would be illuminative to construct a benzolactone enamide-sensitive subunit a in yeast by exchanging amino acids using the sequences of the human and insect subunits a as a guideline. This effort should be accompanied by elucidating the peptides cross-linked to D-apicularen by mass spectrometric analysis. In this context, modified derivatives with the diazirinyl group placed at another position or one of the improved diazirinylbenzoic acids with perfluorobutyl and perfluorooctyl chains (F-PAL) (39) could also be applied.
Acknowledgments
We thank Florenz Sasse (Braunschweig, Germany) for providing the inhibitor archazolid A; Xiao-Song Xie (Dallas, TX) for the inhibitor saliphenylhalamide, and Fiona E. Karet (Cambridge, UK), Mhairi A. Skinner (Guelph, Canada), and Julian A. S. Dow (Glasgow, UK) for supplying the plasmids carrying the coding sequences for subunit a of the human V-ATPase and subunit c from the V-ATPases of H. sapiens and M. sexta, respectively. We also thank Florian Zubeil (Eberhard Karls Universität Tübingen, Germany) for the minimum energy calculations and Martin Dransmann for excellent technical assistance.
M. Huss and H. Wieczorek, unpublished results.
- V-ATPase
- vacuolar-type ATPase
- PAL
- photoaffinity labeling
- NCD-4
- N-cyclohexyl-N′(4-(dimethylamino)naphthyl)-carbodiimide
- D-
- diazirinylbenzoyl-labeled inhibitor.
REFERENCES
- 1. Beyenbach K. W., Wieczorek H. (2006) The V-type H+ ATPase. Molecular structure and function, physiological roles, and regulation. J. Exp. Biol. 209, 577–589 [DOI] [PubMed] [Google Scholar]
- 2. Forgac M. (2007) Vacuolar ATPases. Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 3. Muench S. P., Huss M., Song C. F., Phillips C., Wieczorek H., Trinick J., Harrison M. A. (2009) Cryoelectron microscopy of the vacuolar ATPase motor reveals its mechanical and regulatory complexity. J. Mol. Biol. 386, 989–999 [DOI] [PubMed] [Google Scholar]
- 4. Toei M., Toei S., Forgac M. (2011) Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. J. Biol. Chem. 286, 35176–35186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wieczorek H., Beyenbach K. W., Huss M., Vitavska O. (2009) Vacuolar-type proton pumps in insect epithelia. J. Exp. Biol. 212, 1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bowman E. J., Siebers A., Altendorf K. (1988) Bafilomycins. A class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. U.S.A. 85, 7972–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huss M., Wieczorek H. (2009) Inhibitors of V-ATPases. Old and new players. J. Exp. Biol. 212, 341–346 [DOI] [PubMed] [Google Scholar]
- 8. Sasse F., Steinmetz H., Höfle G., Reichenbach H. (2003) Archazolids, new cytotoxic macrolactones from Archangium gephyra (myxobacteria). Production, isolation, physicochemical, and biological properties. J. Antibiot. 56, 520–525 [DOI] [PubMed] [Google Scholar]
- 9. Kunze B., Jansen R., Sasse F., Höfle G., Reichenbach H. (1998) Apicularens A and B, new cytostatic macrolides from Chondromyces species (Myxobacteria). Production, physicochemical, and biological properties. J. Antibiot. 51, 1075–1080 [DOI] [PubMed] [Google Scholar]
- 10. Huss M., Sasse F., Kunze B., Jansen R., Steinmetz H., Ingenhorst G., Zeeck A., Wieczorek H. (2005) Archazolid and apicularen. Novel specific V-ATPase inhibitors. BMC Biochem. 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boyd M. R., Farina C., Belfiore P., Gagliardi S., Kim J. W., Hayakawa Y., Beutler J. A., McKee T. C., Bowman B. J., Bowman E. J. (2001) Discovery of a novel antitumor benzolactone enamide class that selectively inhibits mammalian vacuolar-type (H+)-ATPases. J. Pharmacol. Exp. Ther. 297, 114–120 [PubMed] [Google Scholar]
- 12. Huss M., Ingenhorst G., König S., Gassel M., Dröse S., Zeeck A., Altendorf K., Wieczorek H. (2002) Concanamycin A, the specific inhibitor of V-ATPases, binds to the Vo subunit c. J. Biol. Chem. 277, 40544–40548 [DOI] [PubMed] [Google Scholar]
- 13. Bowman B. J., Bowman E. J. (2002) Mutations in subunit c of the vacuolar ATPase confer resistance to bafilomycin and identify a conserved antibiotic-binding site. J. Biol. Chem. 277, 3965–3972 [DOI] [PubMed] [Google Scholar]
- 14. Bowman E. J., Graham L. A., Stevens T. H., Bowman B. J. (2004) The bafilomycin/concanamycin-binding site in subunit c of the V-ATPases from Neurospora crassa and Saccharomyces cerevisiae. J. Biol. Chem. 279, 33131–33138 [DOI] [PubMed] [Google Scholar]
- 15. Bowman B. J., McCall M. E., Baertsch R., Bowman E. J. (2006) A model for the proteolipid ring and bafilomycin/concanamycin-binding site in the vacuolar ATPase of Neurospora crassa. J. Biol. Chem. 281, 31885–31893 [DOI] [PubMed] [Google Scholar]
- 16. Wang Y., Inoue T., Forgac M. (2005) Subunit a of the yeast V-ATPase participates in binding of bafilomycin. J. Biol. Chem. 280, 40481–40488 [DOI] [PubMed] [Google Scholar]
- 17. Bender T., Huss M., Wieczorek H., Grond S., von Zezschwitz P. (2007) Convenient synthesis of a [1-14C]diazirinylbenzoic acid as a photoaffinity label for binding studies of V-ATPase inhibitors. Eur. J. Org. Chem. 2007, 3870–3878 [Google Scholar]
- 18. Dröse S., Boddien C., Gassel M., Ingenhorst G., Zeeck A., Altendorf K. (2001) Semisynthetic derivatives of concanamycin A and C, as inhibitors of V- and P-type ATPases. Structure-activity investigations and developments of photoaffinity probes. Biochemistry 40, 2816–2825 [DOI] [PubMed] [Google Scholar]
- 19. Gagliardi S., Gatti P. A., Belfiore P., Zocchetti A., Clarke G. D., Farina C. (1998) Synthesis and structure-activity relationships of bafilomycin A1 derivatives as inhibitors of vacuolar H+-ATPase. J. Med. Chem. 41, 1883–1893 [DOI] [PubMed] [Google Scholar]
- 20. Ingenhorst G., Bindseil K. U., Boddien C., Dröse S., Gassel M., Altendorf K., Zeeck A. (2001) Synthesis of a doubly labeled concanamycin derivative for ATPase binding studies. Eur. J. Org. Chem. 2001, 4525–4532 [Google Scholar]
- 21. Bockelmann S., Menche D., Rudolph S., Bender T., Grond S., von Zezschwitz P., Muench S. P., Wieczorek H., Huss M. (2010) Archazolid A binds to the equatorial region of the c-ring of the vacuolar H+-ATPase. J. Biol. Chem. 285, 38304–38314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie X. S., Padron D., Liao X., Wang J., Roth M. G., De Brabander J. K. (2004) Salicylihalamide A inhibits the VO sector of the V-ATPase through a mechanism distinct from bafilomycin A1. J. Biol. Chem. 279, 19755–19763 [DOI] [PubMed] [Google Scholar]
- 23. Jansen R., Kunze B., Reichenbach H., Höfle G. (2000) Apicularen A and B, cytotoxic 10-membered lactones with a novel mechanism of action from Chondromyces species (Myxobacteria). Isolation, structure elucidation, and biosynthesis. Eur. J. Org. Chem. 2000, 913–919 [Google Scholar]
- 24. Petri A. F., Sasse F., Maier M. E. (2005) Synthesis and biological evaluation of apicularen A analogues. Eur. J. Org. Chem. 2005, 1865–1875 [Google Scholar]
- 25. Skinner M. A., Wildeman A. G. (2001) Suppression of tumor-related glycosylation of cell surface receptors by the 16-kDa membrane subunit of vacuolar H+-ATPase. J. Biol. Chem. 276, 48451–48457 [DOI] [PubMed] [Google Scholar]
- 26. Dow J. A., Goodwin S. F., Kaiser K. (1992) Analysis of the gene encoding a 16-kDa proteolipid subunit of the vacuolar H+-ATPase from Manduca sexta midgut and tubules. Gene 122, 355–360 [DOI] [PubMed] [Google Scholar]
- 27. Gräf R., Harvey W. R., Wieczorek H. (1996) Purification and properties of a cytosolic V1-ATPase. J. Biol. Chem. 271, 20908–20913 [DOI] [PubMed] [Google Scholar]
- 28. Wieczorek H., Cioffi M., Klein U., Harvey W. R., Schweikl H., Wolfersberger M. G. (1990) Isolation of goblet cell apical membrane from tobacco hornworm midgut and purification of its vacuolar-type ATPase. Methods Enzymol. 192, 608–616 [DOI] [PubMed] [Google Scholar]
- 29. Wieczorek H., Putzenlechner M., Zeiske W., Klein U. (1991) A vacuolar-type proton pump energizes K+/H+ antiport in an animal plasma membrane. J. Biol. Chem. 266, 15340–15347 [PubMed] [Google Scholar]
- 30. Sumner J. P., Dow J. A., Earley F. G., Klein U., Jäger D., Wieczorek H. (1995) Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J. Biol. Chem. 270, 5649–5653 [DOI] [PubMed] [Google Scholar]
- 31. Merzendorfer H., Huss M., Schmid R., Harvey W. R., Wieczorek H. (1999) A novel insect V-ATPase subunit M9.7 is glycosylated extensively. J. Biol. Chem. 274, 17372–17378 [DOI] [PubMed] [Google Scholar]
- 32. Parra K. J., Keenan K. L., Kane P. M. (2000) The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J. Biol. Chem. 275, 21761–21767 [DOI] [PubMed] [Google Scholar]
- 33. von Ballmoos C., Appoldt Y., Brunner J., Granier T., Vasella A., Dimroth P. (2002) Membrane topography of the coupling ion binding site in Na+-translocating F1F0-ATP synthase. J. Biol. Chem. 277, 3504–3510 [DOI] [PubMed] [Google Scholar]
- 34. Huss M., Wieczorek H. (2007) Influence of ATP and ADP on dissociation of the V-ATPase into its V1 and VO complexes. FEBS Lett. 581, 5566–5572 [DOI] [PubMed] [Google Scholar]
- 35. Muench S. P., Trinick J., Harrison M. A. (2011) Structural divergence of the rotary ATPases. Q. Rev. Biophys. 44, 311–356 [DOI] [PubMed] [Google Scholar]
- 36. Compton M. A., Graham L. A., Stevens T. H. (2006) Vma9p (subunit e) is an integral membrane VO subunit of the yeast V-ATPase. J. Biol. Chem. 281, 15312–15319 [DOI] [PubMed] [Google Scholar]
- 37. Sambade M., Kane P. M. (2004) The yeast vacuolar proton-translocating ATPase contains a subunit homologous to the Manduca sexta and Bovine e subunits that is essential for function. J. Biol. Chem. 279, 17361–17365 [DOI] [PubMed] [Google Scholar]
- 38. Toei M., Saum R., Forgac M. (2010) Regulation and isoform function of the V-ATPases. Biochemistry 49, 4715–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burkard N., Bender T., Westmeier J., Nardmann C., Huss M., Wieczorek H., Grond S., von Zezschwitz P. (2010) New fluorous photoaffinity labels (F-PAL) and their application in V-ATPase inhibition studies. Eur. J. Org. Chem. 2010, 2176–2181 [Google Scholar]
- 40. Finbow M. E., Goodwin S. F., Meagher L., Lane N. J., Keen J., Findlay J. B., Kaiser K. (1994) Evidence that the 16-kDa proteolipid (subunit c) of the vacuolar H+-ATPase and ductin from gap junctions are the same polypeptide in Drosophila and Manduca. Molecular cloning of the Vha16k gene from Drosophila. J. Cell Sci. 107, 1817–1824 [DOI] [PubMed] [Google Scholar]
- 41. Harrison M. A., Jones P. C., Kim Y. I., Finbow M. E., Findlay J. B. (1994) Functional properties of a hybrid vacuolar H+-ATPase in Saccharomyces cells expressing the Nephrops 16-kDa proteolipid. Eur. J. Biochem. 221, 111–120 [DOI] [PubMed] [Google Scholar]
- 42. Nelson H., Nelson N. (1990) Disruption of genes encoding subunits of yeast vacuolar H+-ATPase causes conditional lethality. Proc. Natl. Acad. Sci. U.S.A. 87, 3503–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ohya Y., Umemoto N., Tanida I., Ohta A., Iida H., Anraku Y. (1991) Calcium-sensitive cls mutants of Saccharomyces cerevisiae showing a pet− phenotype are ascribable to defects of vacuolar membrane H+-ATPase activity. J. Biol. Chem. 266, 13971–13977 [PubMed] [Google Scholar]
- 44. Leng X. H., Manolson M. F., Liu Q., Forgac M. (1996) Site-directed mutagenesis of the 100-kDa subunit (Vph1p) of the yeast vacuolar (H+)-ATPase. J. Biol. Chem. 271, 22487–22493 [DOI] [PubMed] [Google Scholar]
- 45. Kawasaki-Nishi S., Bowers K., Nishi T., Forgac M., Stevens T. H. (2001) The N-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the C-terminal domain affects coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 276, 47411–47420 [DOI] [PubMed] [Google Scholar]