Background: Chronic and low-dose arsenic exposure causes cancer in humans through an as yet unknown mechanism.
Results: PcG proteins, Bmi1 and Suz12, are required for arsenic-induced cell transformation through their inhibition of tumor suppressors.
Conclusion: PcG proteins play a critical role in cell transformation caused by low-dose arsenic exposure.
Significance: PcG protein function is enhanced by arsenic and is required for arsenic-induced carcinogenesis.
Keywords: Carcinogenesis, Polycomb, Signal Transduction, Transformation, Tumor Suppressor Gene, Arsenic
Abstract
Inorganic arsenic is a well-documented human carcinogen associated with cancers of the skin, lung, liver, and bladder. However, the underlying mechanisms explaining the tumorigenic role of arsenic are not well understood. The present study explored a potential mechanism of cell transformation induced by arsenic exposure. Exposure to a low dose (0.5 μm) of arsenic trioxide (As2O3) caused transformation of BALB/c 3T3 cells. In addition, in a xenograft mouse model, tumor growth of the arsenic-induced transformed cells was dramatically increased. In arsenic-induced transformed cells, polycomb group (PcG) proteins, including BMI1 and SUZ12, were activated resulting in enhanced histone H3K27 tri-methylation levels. On the other hand, tumor suppressor p16INK4a and p19ARF mRNA and protein expression were dramatically suppressed. Introduction of small hairpin (sh) RNA-BMI1 or -SUZ12 into BALB/c 3T3 cells resulted in suppression of arsenic-induced transformation. Histone H3K27 tri-methylation returned to normal in BMI1- or SUZ12-knockdown BALB/c 3T3 cells compared with BMI1- or SUZ12-wildtype cells after arsenic exposure. As a consequence, the expression of p16INK4a and p19ARF was recovered in arsenic-treated BMI1- or SUZ12-knockdown cells. Thus, arsenic-induced cell transformation was blocked by inhibition of PcG function. Taken together, these results strongly suggest that the polycomb proteins, BMI1 and SUZ12 are required for cell transformation induced by organic arsenic exposure.
Introduction
Arsenic is a well-documented human carcinogen and widespread environmental contaminant. Chronic low-dose exposure of arsenic caused skin discoloration, chronic indigestion, hypertension, peripheral vascular disease, ischemic heart disease, and internal cancers. Chronic exposure of arsenic contributes to an increased risk of skin, lung, bladder, liver, and kidney cancers (1). Current studies suggest that long-term and low-dose exposure to arsenic is involved in an increased rate of malignant transformation of human keratinocyte HaCaT cells (2–3), human small airway epithelial cells (4), human prostate epithelial cells (5–6), mouse epidermal JB6 Cl41 cells (7), and rat liver epithelial TRL1215 cells (8). Although arsenic causes carcinogenesis in humans, the mechanism explaining the effect of chronic arsenic exposure in tumor development is still undefined.
Polycomb group (PcG)2 complexes have important functions as global epigenetic repressors of transcription and key regulators of cell fate in proliferation, senescence and tumorigenesis (9). PcG proteins comprise two well-known polycomb repressive complexes 1 and 2 (PRC1 and PRC2). BMI1, a member of PRC1, was identified as a c-Myc-collaborating proto-oncogene in lymphomas (10) and is strongly linked to neoplastic development in several human cancer cell types. BMI1 also plays a crucial role in the maintenance of normal and cancer stem cells (11).
PRC2 proteins catalyze histone H3K27 di- or tri-methylations through enhancer of zeste homolog 2 (EZH2), a catalytic subunit of PRC2, to repress target genes such as Hox (12). A core of the PRC2 protein, suppressor of zeste 12 (SUZ12) is required for EZH2 activity with the embryonic ectoderm development (EED) protein and repression of gene transcription (13). SUZ12 is up-regulated in many human cancers including colon, breast and liver cancers (14). PRC2 recruitment of PcG target genes is critical to maintain the repression of genes mediated by PRC1 recognition (15). PRC1 and PRC2 proteins also bind to p16INK4a and p19ARF promoter regions and suppress their protein expression in multiple cell types (16–17).
Although PcG proteins, including BMI1 and SUZ12, are involved in cell transformation and human cancer development, the role of PcG proteins is not clear in arsenic exposure-induced cell transformation. Our results herein provide a mechanism showing that PcG proteins, including BMI1 and SUZ12, are required for the cell transformation caused by low-dose arsenic exposure through the repression of tumor suppressor expression.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Arsenic trioxide (As2O3) and Basal Medium Eagle (BME) were purchased from Sigma-Aldrich. Prestained protein marker and protease inhibitor cocktails were from GenDEPOT. Antibodies against BMI1 or tri-methylated histone H3 at Lys27 were purchased from Millipore and SUZ12 or total histone H3 was from Cell Signaling Technology, Inc. Antibodies to detect p16INK4a and p19ARF proteins were purchased from Santa Cruz Biotechnology, Inc. and Novus Biologicals, respectively.
Cell Culture and Construction of Arsenic-induced Transformed Cells
Wild-type and stable knockdown BMI1 or SUZ12 BALB/c 3T3 cells were grown in 10% CS/DMEM supplemented with penicillin/streptomycin (100 units/ml; Invitrogen) at 37 °C in a humidified 5% CO2 incubator. To construct arsenic-induced transformed BALB/c 3T3 cells, 0.5 mm As2O3 (final conc. 0.5 μm) in 0.1 m NaHCO3, or only 0.1 m NaHCO3 as a control, was included in culture medium to treat cells every 2 days over 2 or 4 weeks.
In Vivo Xenograft Mouse Model
Athymic nude mice (Cr:NIH(S), NIH Swiss nude, 5 weeks old) purchased from Jackson Laboratory were divided into two groups (n = 10) and injected intraperitoneally with 1 × 106 untreated control BALB/c 3T3 or 0.5 μm arsenic-treated BALB/c 3T3 cells. For this study, tumor volumes (following formula; mm3 = length × width × height × 0.52) of mice was calculated from measurements of the individual tumors for 25 days. Tumors were allowed to grow until most of mice had tumors measuring 1 cm3, which is the end point allowed by University of Minnesota Institutional Animal Care and Use Committee.
Establishing BMI1- or SUZ12-knockdown Stable Cells
To construct the knockdown of BMI1 or SUZ12 in BALB/c 3T3 cells, shRNA-BMI1 (sh-BMI1), shRNA-SUZ12 (sh-SUZ12), or shRNA-GFP (sh-GFP) control vector (RNAi Core Facility, BioMedical Genomic Center, University of Minnesota) based on the pKLO.1 lentiviral vector were infected into BALB/c 3T3 cells following the recommended protocols. Infected cells were selected in medium containing 2 μg/ml puromycin, and the expression level of the BMI1 or SUZ12 protein was confirmed by Western blot analysis.
MTS Assay
To estimate cell proliferation, arsenic-induced transformed BALB/c 3T3 cells (1 × 103) were seeded into 96-well plates in 100 μl of 10% CS/DMEM and incubated in a 37 °C, 5% CO2 incubator. After culturing for 12 h, 20 μl of the CellTiter 96® Aqueous One Solution (Promega) were added to each well, and cells were then incubated for 1 h at 37 °C in a 5% CO2 atmosphere. To stop the reaction, 25 μl of a 10% SDS solution were added, and absorbance was measured at 492 and 690 nm.
Anchorage-independent Cell Transformation Assay
In brief, cells (8 × 103/ml) were cultured in 1 ml of 0.3% Basal Medium Eagle (BME) agar containing 10% CS. The cultures were maintained in a 37 °C, 5% CO2 incubator for 7 days, and cell colonies were scored using a microscope and the Image-Pro PLUS (v.6) computer software program (Media Cybernetics).
Cell Cycle Analysis
Arsenic-induced transformed BALB/c 3T3 cells (1 × 105/ml) were seeded into 60-mm dishes and cultured for 48 h at 37 °C in a 5% CO2 incubator. The cells were harvested with trypsin, fixed with ice-cold methanol, stained with propidium iodide (PI), and then analyzed for cell cycle phase by flow cytometry.
RT-PCR
For RT-PCR, total RNA was extracted from each cell type using an RNA isolation solvent (TEL-TEST, INC.) following the manufacturer's instructions. First-strand cDNAs were synthesized using the AmfiRivert II cDNA Synthesis Master mix (GenDEPOT) and specific primers. PCR was performed for p16INK4a, p19ARF or β-actin transcription as follows: denaturation at 95 °C for 30 s, annealing at 55 °C for 1 min and extension at 72 °C for 1 min.
Isolation of Histone Proteins
Histone proteins were prepared as previously reported (18).
RESULTS
Low Dose Exposure to Arsenic Causes Cell Transformation
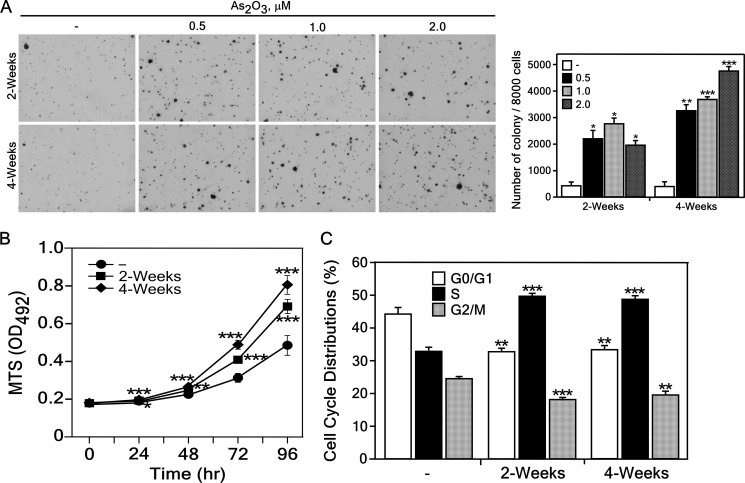
To determine whether low dose exposure to arsenic causes cell transformation, we tested the cell transforming activity of arsenic in BALB/c 3T3 cells. We found that exposure of BALB/c 3T3 cells to arsenic significantly increased anchorage-independent growth in a dose- and time-dependent manner (Fig. 1A). To characterize the arsenic-induced transformed BALB/c 3T3 cells, we measured proliferation using the MTS assay with untreated or 0.5 μm of As2O3-treated cells for 2 or 4 weeks. Results indicated that transformed cells showed a marked increase in the rate of proliferation compared with untreated control cells (Fig. 1B). Using these cell lines, we studied differences in cell cycle phase of arsenic-induced transformed BALB/c 3T3 cells results in that many arsenic-transformed BALB/c 3T3 cells occupied the S-phase and less accumulated in the G1/G0- or G2/M-phase of the cell cycle compared with untreated control cells (Fig. 1C). These results show that low-dose exposure arsenic caused cell transformation.
FIGURE 1.
Low dose exposure to arsenic induces transformation of BALB/c 3T3 cells. A, BALB/c 3T3 cells were assessed for transformation ability in an anchorage-independent soft agar assay after treatment with the different concentration of As2O3 (0, 0.5, 1, or 2 μm) in 0.1 m NaHCO3 or no treatment for 2 or 4 weeks. Cells (8 × 103) were seeded in 1 ml of 0.3% Basal Medium Eagle (BME) agar containing 10% calf serum (CS). The cultures were maintained at 37 °C in a 5% CO2 atmosphere for 7 days and then colonies were counted using a microscope and the Image-Pro PLUS (v.6) computer software program and representative plates are shown (left panels). The average colony number was calculated from three separate experiments and data are shown as means ± S.D. (right panel). B, arsenic-treated BALB/c 3T3 cells exhibit enhanced proliferation. Cell proliferation was estimated by reading absorbance at 492 nm every 24 h up to 96 h. Data are presented as means ± S.D. of triplicate experiments. C, arsenic-treated or untreated BALB/c 3T3 (1 × 105) cells were seeded into 60-mm dishes and cultured 48 h. The cells were harvested, fixed with methanol, stained with PI, and then analyzed for cell cycle phase. Data are expressed as the percentage of cells in G1/G0, S, or G2/M phase and are represented as means ± S.D. of values obtained from triplicate experiments. Statistical differences were evaluated using the Student's t test and the respective asterisks indicate a significant change in arsenic-treated cells compared with control cells (*, p < 0.001; **, p < 0.0005; ***, p < 0.0001).
Arsenic-induced Transformed Cells Form Tumors in Nude Mice
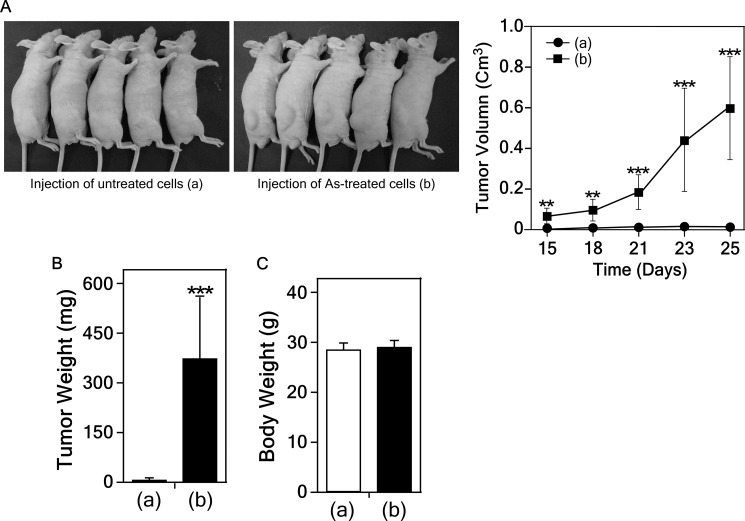
Next, we determined whether the arsenic-induced transformed cells could form tumors in vivo using mice injected with untreated control or treated cells (each 1 × 106 cells) with arsenic (0.5 μm) for 2 weeks. Tumors were allowed to grow in mice (n = 10 mice per group) and tumor and body weights of mice were measured weekly for 4 weeks. The results indicated that the arsenic-induced transformed cells, but not the untreated control cells, developed tumors (Fig. 2). This result clearly demonstrated that exposure to arsenic causes cell transformation.
FIGURE 2.
Arsenic-induced transformed cells develop tumor in xenograft mouse model. Cells transformed by arsenic exposure had tumorigenic activity in SKH-1 hairless mice (A, left panels). Average tumor volumes (A, right panel), weight (B) and body weight (C) are shown. Data are represented as means ± S.D. Statistical differences were evaluated using the Student's t test and the asterisks (** and ***) indicates a significant difference (p < 0.0005 and p < 0.0001, respectively) between the groups injected with arsenic-transformed cells compared with non-transformed cells.
Exposure of Arsenic Increases the Expression of PcG Complexes
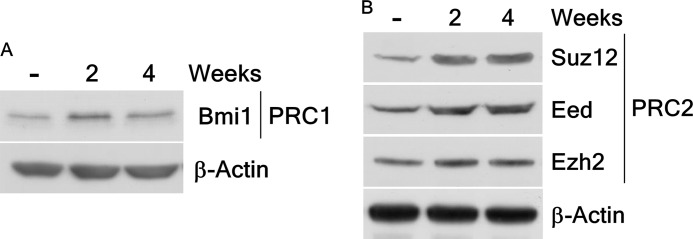
PcG complexes are key regulators in the transcription of many genes controlling cell-fate decisions including cancer development (19). To study this in arsenic-induced cell transformation, we detected the expression of PcG proteins in arsenic-induced transformed BALB/c 3T3 cells that were exposed to low-doses of arsenic. Results showed that the expression levels of the oncogene BMI1, a component of the PRC1 protein complex (Fig. 3A), and proteins of the PRC2 complex, including SUZ12, EED, and EZH2 (Fig. 3B), were increased in arsenic-induced transformed cells. This finding indicated that exposure to arsenic dramatically increases expression of several PcG proteins.
FIGURE 3.
Arsenic exposure increases the expression of PcG complexes in BALB/c 3T3 cells. A, the expression of PcG protein BMI1, a component of polycomb repressive complex 1 (PRC1), is enhanced in BALB/c 3T3 cells after arsenic treatment. B, arsenic markedly raises the polycomb repressive complex 2 (PRC2) PcG protein expression, including SUZ12, EED, and EZH2, in BALB/c 3T3 cells. Lysate proteins were resolved by 10% SDS-PAGE, and the protein bands were visualized by Western blotting with a specific BMI1, SUZ12, EED, or EZH2 primary antibody and an HRP-conjugated secondary antibody. Detection of total β-actin was used to verify equal protein loading.
Knockdown of BMI1 or SUZ12 Suppresses Cell Transformation Induced by Arsenic Exposure
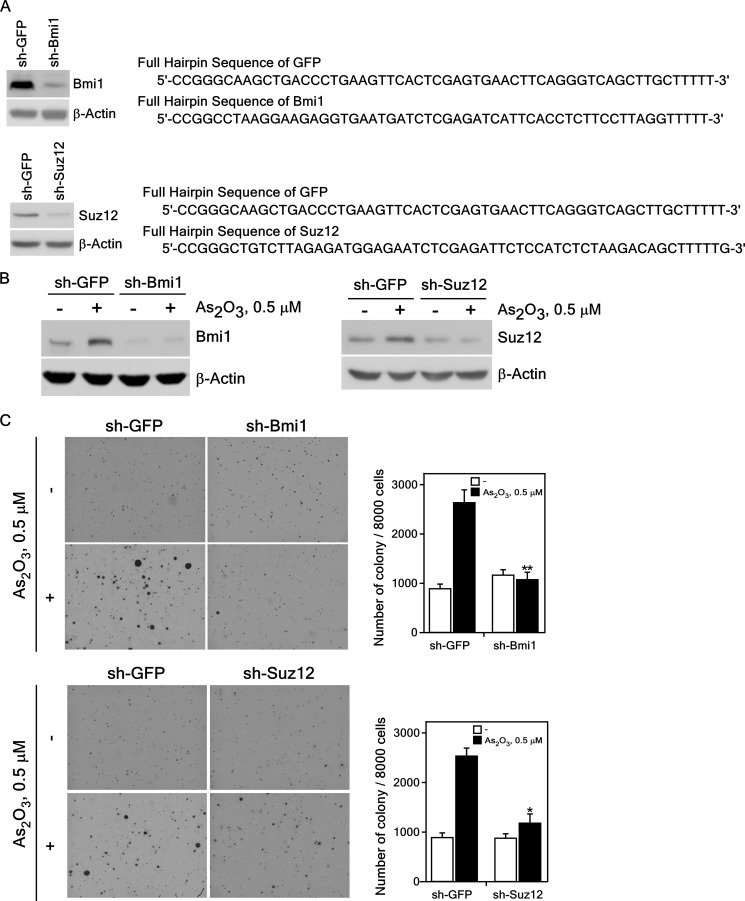
To determine whether PcG proteins are involved in cell transformation induced by low-dose exposure to arsenic, we established knockdown of BMI1 or SUZ12 in BALB/c 3T3 cells using sh-BMI1 or sh-SUZ12 or a control vector (sh-GFP). Immunoblot analysis of sh-BMI1-, sh-SUZ12-, and sh-GFP-infected BALB/c 3T3 cells revealed suppressed expression of endogenous BMI1 (Fig. 4A, upper left) and SUZ12 (Fig. 4A, lower left) of up to 90% compared with cells expressing sh-GFP.
FIGURE 4.
Knockdown of PcG proteins suppresses the anchorage-independent growth of BALB/c 3T3 cells exposed to a low dose of arsenic. A, knockdown of BMI1 or SUZ12 efficiently suppresses the endogenous BMI1 (upper left panel) or SUZ12 (bottom left panel) protein level. The DNA sequences of the sh-GFP, sh-BMI1 (upper right panel) and sh-SUZ12 (bottom right panel) are shown. Knockdown BMI1 or SUZ12 BALB/c 3T3 cells were constructed as described under “Experimental Procedures.” The expression of BMI1 (upper left panel) or SUZ12 (bottom left panel) was analyzed in the stable transfected BALB/c 3T3 cells. B, expression of BMI1 (left panel) or SUZ12 (right panel) proteins in BALB/c 3T3 cells treated with arsenic for 2 weeks. Lysate proteins were resolved by 10% SDS-PAGE, and the protein bands were visualized by Western blotting with a specific BMI1 or SUZ12 primary antibody and an HRP-conjugated secondary antibody. Detection of total β-actin was used to verify equal protein loading. C, cell transforming activity induced by arsenic is decreased in knockdown BMI1 (upper panel) or SUZ12 (bottom panel) stably infected BALB/c 3T3 cells. sh-GFP, sh-BMI1 or sh-SUZ12 stably infected cells were exposed to 0.5 μm of As2O3 in 0.1 m NaHCO3 for 2 weeks and these cells (8 × 103) were cultured in 1 ml of 0.3% BME agar containing 10% CS. The cultures were maintained at 37 °C in a 5% CO2 atmosphere for 7 days and then colonies were counted using a microscope and the Image-Pro PLUS (v.6) computer software program. The average colony number was calculated and photographed from three separate experiments, and representative plates are shown. Significant differences were evaluated using the Student's t test, and the respective asterisks indicate a significant decrease in arsenic-induced sh-Bmi1 or sh-SUZ12 cell transformation compared with sh-GFP cells (*, p < 0.01; **, p < 0.001).
We then examined the effect of suppressing BMI1 or SUZ12 expression on arsenic-induced cell transformation. These cell lines were stimulated with As2O3 (0.5 μm in 0.1 m NaHCO3) in culture medium and incubated at 37 °C in a 5% CO2 incubator for 2 weeks. We confirmed that the endogenous expression of the BMI1 or SUZ12 protein in these cells was still inhibited by knockdown of BMI1 or SUZ12 after arsenic treatment for 2 weeks (Fig. 4B). The colony-forming activity induced by arsenic in sh-BMI1 (Fig. 4C, upper panels) and sh-SUZ12 (Fig. 4C, lower panels) cells was dramatically decreased compared with sh-GFP control cells. Our results demonstrated that the knockdown of BMI1 or SUZ12, components of PcG complexes, suppressed colony formation induced by low doses of arsenic exposure. Overall, these data further demonstrated that PcG proteins play a key role in cell transformation induced by exposure to arsenic.
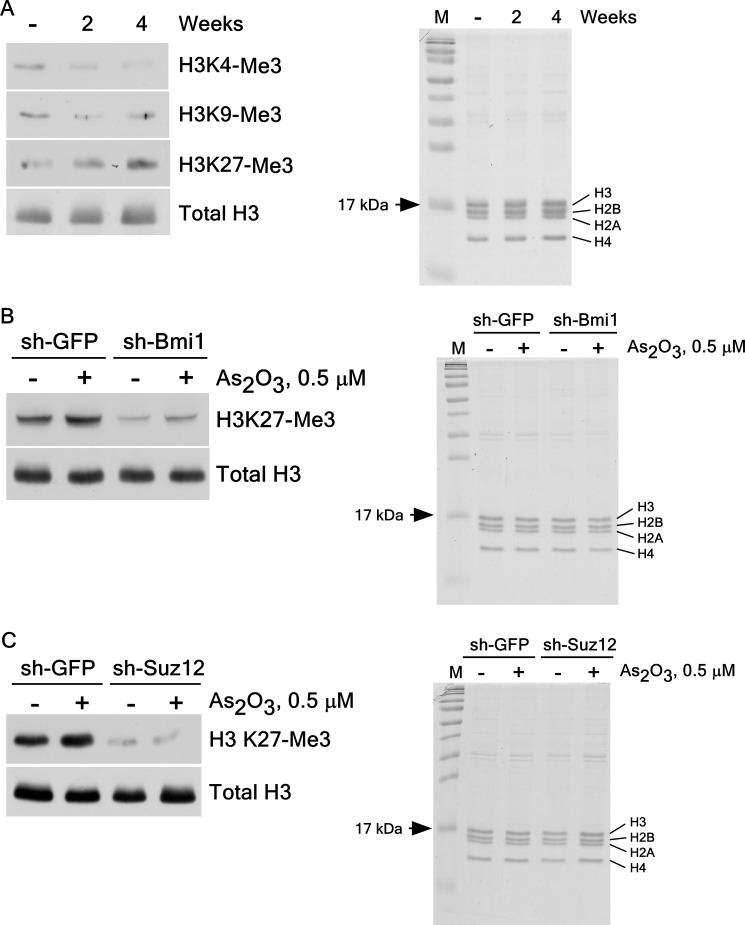
Histone H3K27 Tri-methylation Is Enhanced in PcG-mediated Cell Transformation Induced by Arsenic Exposure
PRC2 proteins increase histone H3K27 di- or tri-methylations through PRC1 mediation of PcG target genes (13, 15). To understand the role of histone H3 tri-methylation in arsenic-induced cell transformation, we measured the tri-methylation level of histone H3 in arsenic-induced transformed BALB/c 3T3 cells (Fig. 5A). We found increases in the tri-methylation of histone H3K27 in arsenic-induced transformed BALB/c 3T3 cell (Fig. 5A, left panel). We also examined the effect of BMI1 or SUZ12 knockdown on histone H3K27 tri-methylation in these cells. Results confirmed that arsenic-induced histone H3K27 tri-methylation is effectively suppressed in BMI1 (Fig. 5B, left panel) and SUZ12 (Fig. 5C, left panel) knockdown cells compared with sh-GFP control cells. This finding demonstrated that BMI1 and SUZ12 are essential for arsenic-induced histone H3K27 tri-methylation. Taken together, the results provide strong evidence showing that PcG-mediated histone H3K27 tri-methylation is involved in arsenic-induced cell transformation.
FIGURE 5.
Effect of arsenic exposure on histone H3K27 tri-methylation. A, histone H3K27 tri-methylation (H3K27-Me3) was increased in BALB/c 3T3 cells by arsenic treatment. B and C, histone H3K27 tri-methylation was inhibited by knockdown of BMI1 (B) or SUZ12 (C) in arsenic-induced transformed BALB/c 3T3 cells. Histone proteins (1 μg) were first resolved by SDS-15% polyacrylamide gel electrophoresis (A, B, or C, right panels) to show the same histone proteins, and then proteins were transferred to membranes for Western blot analysis. The tri-methylation of histone H3 expression was detected with specific antibodies. Total histone H3 was used to verify equal protein loading.
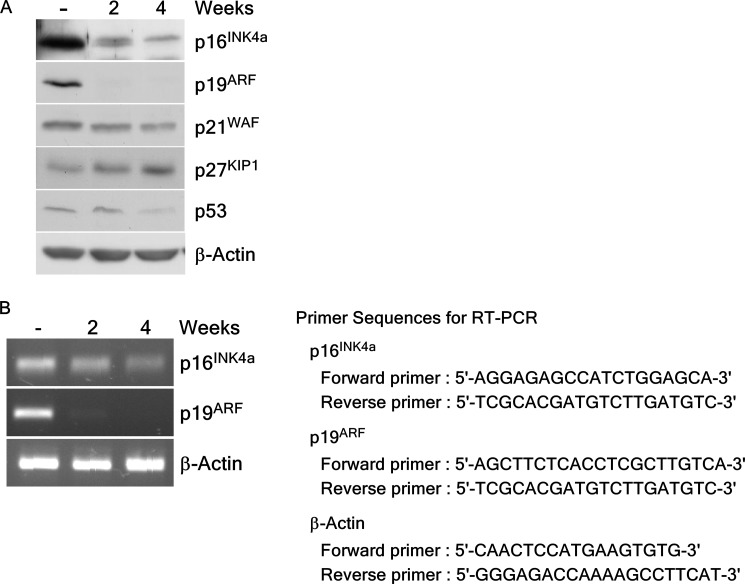
Exposure to Arsenic Reduces Tumor Suppressor p16INK4a and p19ARF Expression
We investigated whether arsenic exposure could affect the expression of several tumor suppressors in arsenic-induced transformed BALB/c 3T3 cells. Results showed that long-term exposure to arsenic led to reduced protein levels of p16INK4a and p19ARF (mouse homologue of p14ARF) in these cells (Fig. 6A). To examine the effect of arsenic on transcriptional levels of p16INK4a and p19ARF, we performed RT-PCR to determine the mRNA levels of p16INK4a and p19ARF in transformed cells. Long-term treatment with arsenic dramatically suppressed the mRNA levels of p16INK4a and p19ARF in these cells (Fig. 5B). This result suggested that arsenic exposure caused cell transformation through the inhibition of both protein and gene expression of the tumor suppressors p16INK4a and p19ARF.
FIGURE 6.
Expression of tumor suppressor proteins in BALB/c 3T3 cells after arsenic exposure. A, arsenic treatment reduces expression of tumor suppressor proteins such as p16INK4a or p19ARF in BALB/c 3T3 cells. Lysate proteins were resolved by 10% SDS-PAGE and the protein bands were visualized by Western blotting with specific primary antibodies and an HRP-conjugated secondary antibody. Detection of total β-actin was used to verify equal protein loading. B, exposure of arsenic inhibited mRNA expression of p16INK4a or p19ARF in BALB/c 3T3 cells. RT-PCR analysis was performed to detect gene transcription using specific primers for p16INK4a, p19ARF, or β-actin (B, right panel). Total RNA was isolated from BALB/c 3T3 cells treated or not treated with arsenic as described under “Experimental Procedures.”
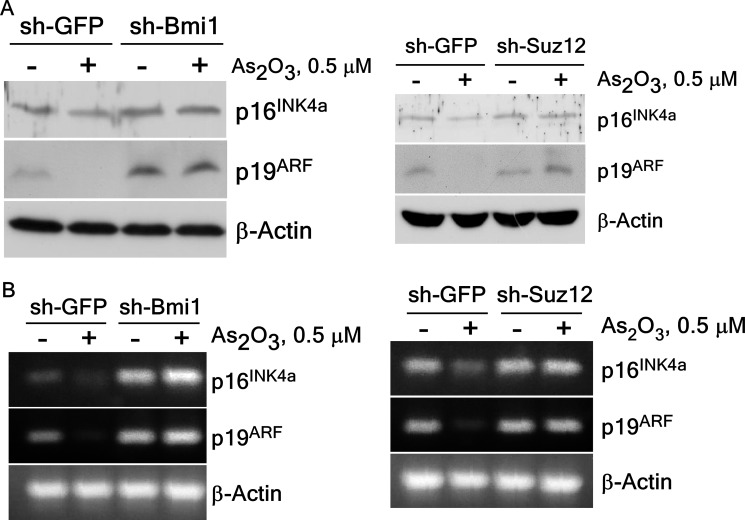
Knockdown of PcG Proteins Down-regulates Tumor Suppressor p16INK4a and p19ARF Expression
PcG proteins activate the INK4A-ARF locus, coinciding with an enhanced level of associated histone H3K27 tri-methylation (20). To verify the effect of knockdown of BMI1 or SUZ12 on the expression of p16INK4a and p19ARF in arsenic-induced cell transformation, we determined the p16INK4a and p19ARF proteins levels in sh-BMI1 or sh-SUZ12 BALB/c 3T3 cells exposed to low dose arsenic treatment. The protein levels of p16INK4a and p19ARF were restored in sh-BMI1 and sh-SUZ12 cells compared with the control sh-GFP cells (Fig. 7A). The mRNA levels of p16INK4a and p19ARF were also recovered in knockdown BMI1 and SUZ12 cells compared with arsenic treated control sh-GFP cells (Fig. 7B). This finding suggests that PcG proteins, BMI1 and SUZ12, are involved in regulating p16INK4a and p19ARF expression in arsenic-induced cell transformation.
FIGURE 7.
Expression of p16INK4a or p19ARF is recovered after exposure to arsenic in BALB/c 3T3 cells expressing knockdown PcG complexes. A, p16INK4a or p19ARF protein expression was recovered in knockdown BMI1 (left panel) or SUZ12 (right panel) stably infected BALB/c 3T3 cells after arsenic treatment. The proteins were prepared and subjected to Western blotting with specific primary antibodies and an HRP-conjugated secondary antibody. Detection of total β-actin was used to verify equal protein loading. B, the mRNA level of p16INK4a or p19ARF was also restored in knockdown BMI1 (left panel) or SUZ12 (right panel) stably infected BALB/c 3T3 cells. RT-PCR analysis was performed to detect the p16INK4a, p19ARF, or β-actin transcripts in BALB/c 3T3 cells as described under “Experimental Procedures.”
Finally, our findings provide a valuable model and mechanism regarding arsenic-induced cell transformation. Taken together, these results provide strong evidence showing that PcG proteins, BMI1 and SUZ12 are required for cell transformation induced by arsenic exposure, which might be regulated through repression of tumor suppressor p16INK4a and p19ARF expression.
DISCUSSION
Arsenic is one of the most well-known human carcinogens and also is a major environmental problem in many parts of the world (1, 21). Chronic exposure to low doses of arsenic leads to malignant transformation of HaCaT keratinocytes (2–3, 22) and several other epithelial cell lines (4–6, 8). A few reports focusing on the mechanism of arsenic-induced carcinogenesis revealed that a low level of arsenic affected the up-regulation of Hdm2 and subsequent p53 inactivation (23) and the mot2-mediated repressive effect of NF-κB on p53 (2) in the malignant transformation of human keratinocytes. Arsenic-induced transformation in human keratinocytes involves PI3-K/Akt-mediated cyclin D1 expression (22) and transcription factor Nrf-2 activation (3). Long-term treatment with arsenic also increased c-H-Ras, c-Myc, and c-Fos protein expression in transformed human small airway epithelial cells (4). To examine the mechanism of arsenic-induced carcinogenesis, we determined the transforming activity of low dose arsenic exposure in BALB/c 3T3 cells. This cell line is one of the cell lines useful for examining neoplastic transformation (24–25). In this study, we showed that treatment with arsenic trioxide (As2O3, 0.5 μm) for 2 weeks significantly increased anchorage-independent growth of BALB/c 3T3 cells (Fig. 1A) and the cells transformed by arsenic could form tumors in a xenograft mouse model assay (Fig. 2). Although arsenic induces cell transformation, the mechanism of arsenic-induced carcinogenesis is not clear. But our findings indicate that this model system appears to be a valuable tool for understanding chronic arsenic exposure-induced cell transformation.
Some investigators have proposed mechanisms of long-term exposure arsenic-induced malignant transformation. Arsenic-mediated DNA methylation is involved in the silencing of tumor suppressor genes or activation of oncogene expression in arsenic-induced carcinogenesis (5, 26–28). Arsenic exposure also altered histone H3 methylation in human lung carcinoma A549 cells (29). However, we hypothesized that arsenic affects epigenetic regulators, leading to histone H3 methylation in gene silencing and activation of PcG complexes during arsenic-associated carcinogenesis. We tested this hypothesis using arsenic-induced transformed BALB/c 3T3 cells. Our findings demonstrated that key regulators of PcG complexes, BMI1 and SUZ12, were substantially increased by arsenic exposure (Fig. 3); whereas the transforming activity induced by low-dose exposure to arsenic was totally suppressed in knockdown BMI1 and SUZ12 cells (Fig. 4). These results strongly suggested that PcG complexes have important roles in arsenic-induced cell transformation, but it is still unclear for the reason of PcG proteins' activation by arsenic.
PcG complexes comprise the machinery of transcriptional regulatory processing in homeotic (HOX) gene expression (12, 30), and several cellular responses, including cell cycle (31), senescence (20), cancer (11, 32–34), cell fate decisions (33) and stem cell differentiation or maintenance (11, 35). PcG consists of two distinct protein complexes, PRC1 and PRC2, which are best characterized in the repression of transcription. PRC2 maintains and establishes tumor suppressor gene silencing during carcinogenesis (36–37). Increases in SUZ12, a key component of PRC2, coincide with transformation and many cancers (14, 34, 36, 38). PRC1 recognizes the histone H3K27 methylation and recruits PRC2 to PcG target genes resulting in transcriptional repression (15). The PRC1 protein BMI1 is up-regulated and correlates with several human cancers and tumorigenesis (11, 34, 39–40). However PcG proteins including BMI1 and SUZ12 are involved in cancer development and maintenance. Our results support the function of the PcG complexes in carcinogenesis induced by arsenic exposure, but the regulation mechanism of PcG proteins by arsenic is still not clear. Recently, the relationship of p38 activation and BMI1 was reported (41–42). We found that p38 signaling was involved the low-dose arsenic induced-transformation, which could account for activation of PcG by low-dose arsenic exposure. Arsenic increased the expression of p38 and expression of endogenous BMI1 and SUZ12 was significantly reduced in stable p38 BALB/c 3T3 knockdown cells compared with wild type cells (supplemental Fig. S1). Taken together, PcG proteins play a key role in arsenic-induced transformation through p38 activation.
PcG complexes directly bind to tumor suppressor p16INK4a and p19ARF promoter regions and repress expression of these genes (16–17, 20). PRC2 proteins increased the tri-methylation level of H3K27 at this locus thus repressing p16INK4a and p19ARF expression in a pRB-dependent manner (43). Higher expression of PcG in tumors is associated with suppression of PcG target genes p16INK4a and p19ARF (39–40, 44). In transformation processes, direct BMI1 up-regulation induced by c-Myc leads to repression of p16INK4a and p19ARF expression and promotes tumorigenesis and oncogene-induced senescence (45–46). We found that exposure to arsenic dramatically repressed tumor suppressor p16INK4a and p19ARF expression in arsenic-induced carcinogenesis (Fig. 6). Knockdown of PcG proteins, including BMI1 and SUZ12, blocked the cell transformation induced by arsenic through the restoration of p16INK4a and p19ARF expression (Fig. 7). Finally, we suggest that PcG complexes mediate the repression of tumor suppressor p16INK4a and p19ARF expression required for cell transformation induced by arsenic exposure.
In summary, we suggest that PcG complexes are required for cell transformation induced by low dose exposure to arsenic. Exposure to arsenic significantly inhibited tumor suppressor p16INK4a and p19ARF expression by increasing PcG proteins, BMI1 and SUZ12 thus promoting tumorigenesis. Although, evidence is lacking that can explain precisely how arsenic activates the function of PcG proteins, this study suggests that they could be an attractive target for cancer chemotherapy or gene therapy in arsenic-induced carcinogenesis.
Acknowledgment
We thank Tonya Poorman for help submitting our manuscript.
This work was supported by The Hormel Foundation and National Institutes of Health Grants ES016548, R37 CA081064, CA120388, and CA077646.

This article contains supplemental Fig. S1.
- PcG
- polycomb group
- sh
- small hairpin
- EED
- embryonic ectoderm development
- PI
- propidium iodide.
REFERENCES
- 1. Gebel T. W. (1999) Arsenic and drinking water contamination. Science 283, 1458–1459 [DOI] [PubMed] [Google Scholar]
- 2. Li Y., Ling M., Xu Y., Wang S., Li Z., Zhou J., Wang X., Liu Q. (2010) The repressive effect of NF-kappaB on p53 by mot-2 is involved in human keratinocyte transformation induced by low levels of arsenite. Toxicol. Sci. 116, 174–182 [DOI] [PubMed] [Google Scholar]
- 3. Pi J., Diwan B. A., Sun Y., Liu J., Qu W., He Y., Styblo M., Waalkes M. P. (2008) Arsenic-induced malignant transformation of human keratinocytes: involvement of Nrf2. Free Radic. Biol. Med. 45, 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wen G., Calaf G. M., Partridge M. A., Echiburú-Chau C., Zhao Y., Huang S., Chai Y., Li B., Hu B., Hei T. K. (2008) Neoplastic transformation of human small airway epithelial cells induced by arsenic. Mol. Med. 14, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benbrahim-Tallaa L., Waterland R. A., Styblo M., Achanzar W. E., Webber M. M., Waalkes M. P. (2005) Molecular events associated with arsenic-induced malignant transformation of human prostatic epithelial cells: aberrant genomic DNA methylation and K-ras oncogene activation. Toxicol. Appl. Pharmacol. 206, 288–298 [DOI] [PubMed] [Google Scholar]
- 6. Benbrahim-Tallaa L., Webber M. M., Waalkes M. P. (2007) Mechanisms of acquired androgen independence during arsenic-induced malignant transformation of human prostate epithelial cells. Environ. Health Perspect. 115, 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang C., Ma W. Y., Li J., Goranson A., Dong Z. (1999) Requirement of Erk, but not JNK, for arsenite-induced cell transformation. J. Biol. Chem. 274, 14595–14601 [DOI] [PubMed] [Google Scholar]
- 8. Liu J., Benbrahim-Tallaa L., Qian X., Yu L., Xie Y., Boos J., Qu W., Waalkes M. P. (2006) Further studies on aberrant gene expression associated with arsenic-induced malignant transformation in rat liver TRL1215 cells. Toxicol. Appl. Pharmacol. 216, 407–415 [DOI] [PubMed] [Google Scholar]
- 9. Gil J., Bernard D., Peters G. (2005) Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 24, 117–125 [DOI] [PubMed] [Google Scholar]
- 10. Haupt Y., Alexander W. S., Barri G., Klinken S. P., Adams J. M. (1991) Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell 65, 753–763 [DOI] [PubMed] [Google Scholar]
- 11. Lessard J., Sauvageau G. (2003) Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255–260 [DOI] [PubMed] [Google Scholar]
- 12. Ringrose L., Paro R. (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38, 413–443 [DOI] [PubMed] [Google Scholar]
- 13. Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K. (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23, 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kirmizis A., Bartley S. M., Farnham P. J. (2003) Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol. Cancer Ther. 2, 113–121 [PubMed] [Google Scholar]
- 15. Tie F., Furuyama T., Prasad-Sinha J., Jane E., Harte P. J. (2001) The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128, 275–286 [DOI] [PubMed] [Google Scholar]
- 16. Gil J., Peters G. (2006) Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat. Rev. Mol. Cell Biol. 7, 667–677 [DOI] [PubMed] [Google Scholar]
- 17. Maertens G. N., El Messaoudi-Aubert S., Racek T., Stock J. K., Nicholls J., Rodriguez-Niedenführ M., Gil J., Peters G. (2009) Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One 4, e6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim H. G., Lee K. W., Cho Y. Y., Kang N. J., Oh S. M., Bode A. M., Dong Z. (2008) Mitogen- and stress-activated kinase 1-mediated histone H3 phosphorylation is crucial for cell transformation. Cancer Res. 68, 2538–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morey L., Helin K. (2010) Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 35, 323–332 [DOI] [PubMed] [Google Scholar]
- 20. Bracken A. P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Mönch K., Minucci S., Porse B. T., Marine J. C., Hansen K. H., Helin K. (2007) The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abernathy C. O., Liu Y. P., Longfellow D., Aposhian H. V., Beck B., Fowler B., Goyer R., Menzer R., Rossman T., Thompson C., Waalkes M. (1999) Arsenic: health effects, mechanisms of actions, and research issues. Environ. Health Perspect. 107, 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ouyang W., Luo W., Zhang D., Jian J., Ma Q., Li J., Shi X., Chen J., Gao J., Huang C. (2008) PI-3K/Akt pathway-dependent cyclin D1 expression is responsible for arsenite-induced human keratinocyte transformation. Environ. Health Perspect. 116, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang Y., Zhang J., McHenry K. T., Kim M. M., Zeng W., Lopez-Pajares V., Dibble C. C., Mizgerd J. P., Yuan Z. M. (2008) Induction of cytoplasmic accumulation of p53: a mechanism for low levels of arsenic exposure to predispose cells for malignant transformation. Cancer Res. 68, 9131–9136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kakunaga T. (1973) A quantitative system for assay of malignant transformation by chemical carcinogens using a clone derived from BALB-3T3. Int. J. Cancer 12, 463–473 [DOI] [PubMed] [Google Scholar]
- 25. Muramatsu D., Sasaki K., Kuroda S., Hayashi K., Tanaka N., Sakai A. (2009) Comparison of sensitivity to arsenic compounds between a Bhas 42 cell transformation assay and a BALB/c 3T3 cell transformation assay. Mutat. Res. 675, 66–70 [DOI] [PubMed] [Google Scholar]
- 26. Cui X., Wakai T., Shirai Y., Hatakeyama K., Hirano S. (2006b) Chronic oral exposure to inorganic arsenate interferes with methylation status of p16INK4a and RASSF1A and induces lung cancer in A/J mice. Toxicol. Sci. 91, 372–381 [DOI] [PubMed] [Google Scholar]
- 27. Cui X., Wakai T., Shirai Y., Yokoyama N., Hatakeyama K., Hirano S. (2006a) Arsenic trioxide inhibits DNA methyltransferase and restores methylation-silenced genes in human liver cancer cells. Hum. Pathol. 37, 298–311 [DOI] [PubMed] [Google Scholar]
- 28. Reichard J. F., Puga A. (2010) Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics 2, 87–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou X., Sun H., Ellen T. P., Chen H., Costa M. (2008) Arsenite alters global histone H3 methylation. Carcinogenesis 29, 1831–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kennison J. A. (1995) The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29, 289–303 [DOI] [PubMed] [Google Scholar]
- 31. Oktaba K., Gutiérrez L., Gagneur J., Girardot C., Sengupta A. K., Furlong E. E., Müller J. (2008) Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 15, 877–889 [DOI] [PubMed] [Google Scholar]
- 32. Leung C., Lingbeek M., Shakhova O., Liu J., Tanger E., Saremaslani P., Van Lohuizen M., Marino S. (2004) Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428, 337–341 [DOI] [PubMed] [Google Scholar]
- 33. Sparmann A., van Lohuizen M. (2006) Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6, 846–856 [DOI] [PubMed] [Google Scholar]
- 34. Hussain M., Rao M., Humphries A. E., Hong J. A., Liu F., Yang M., Caragacianu D., Schrump D. S. (2009) Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 69, 3570–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pasini D., Bracken A. P., Hansen J. B., Capillo M., Helin K. (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell Biol. 27, 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Herranz N., Pasini D., Díaz V. M., Francí C., Gutierrez A., Dave N., Escrivà M., Hernandez-Muñoz I., Di Croce L., Helin K., García de Herreros A., Peiró S. (2008) Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell Biol. 28, 4772–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Villa R., Pasini D., Gutierrez A., Morey L., Occhionorelli M., Viré E., Nomdedeu J. F., Jenuwein T., Pelicci P. G., Minucci S., Fuks F., Helin K., Di Croce L. (2007) Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell 11, 513–525 [DOI] [PubMed] [Google Scholar]
- 38. Pizzatti L., Binato R., Cofre J., Gomes B. E., Dobbin J., Haussmann M. E., D'Azambuja D., Bouzas L. F., Abdelhay E. (2010) SUZ12 is a candidate target of the non-canonical WNT pathway in the progression of chronic myeloid leukemia. Genes Chromosomes Cancer 49, 107–118 [DOI] [PubMed] [Google Scholar]
- 39. Pietersen A. M., Horlings H. M., Hauptmann M., Langerød A., Ajouaou A., Cornelissen-Steijger P., Wessels L. F., Jonkers J., van de Vijver M. J., van Lohuizen M. (2008) EZH2 and BMI1 inversely correlate with prognosis and TP53 mutation in breast cancer. Breast Cancer Res. 10, R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Becker M., Korn C., Sienerth A. R., Voswinckel R., Luetkenhaus K., Ceteci F., Rapp U. R. (2009) Polycomb group protein Bmi1 is required for growth of RAF driven non-small-cell lung cancer. PLoS One 4, e4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee M. O., Lee H. J., Kim M. A., Kim E. K., Lee J. H., Heo J. H., Lee S. H., Cho S. H., Fornace A. J., Jr., Jeong H. C., Cha H. J. (2011) p16Ink4a suppression of lung adenocarcinoma by Bmi-1 in the presence of p38 activation. J. Thorac. Oncol. 6, 423–431 [DOI] [PubMed] [Google Scholar]
- 42. Srivastav R. K., Schwede S., Klaus M., Schwermann J., Gaestel M., Niedenthal R. (2011) Monitoring protein-protein interactions in mammalian cells by trans-SUMOylation. Biochem. J. 438, 495–503 [DOI] [PubMed] [Google Scholar]
- 43. Kotake Y., Cao R., Viatour P., Sage J., Zhang Y., Xiong Y. (2007) pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4α tumor suppressor gene. Genes Dev. 21, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reynolds P. A., Sigaroudinia M., Zardo G., Wilson M. B., Benton G. M., Miller C. J., Hong C., Fridlyand J., Costello J. F., Tlsty T. D. (2006) Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J. Biol. Chem. 281, 24790–24802 [DOI] [PubMed] [Google Scholar]
- 45. Jacobs J. J., Kieboom K., Marino S., DePinho R. A., van Lohuizen M. (1999b) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397, 164–168 [DOI] [PubMed] [Google Scholar]
- 46. Jacobs J. J., Scheijen B., Voncken J. W., Kieboom K., Berns A., van Lohuizen M. (1999a) Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 13, 2678–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]