Background: CD151 associates with integrins and regulates integrin-dependent small GTPase-mediated cellular behaviors.
Results: CD151-expressing cells exhibited increased Ras, Rac, and Cdc42 complexes with integrins and active forms of small GTPases, as compared with CD151-deficient cells.
Conclusion: CD151 contributes to integrin signaling to small GTPases by facilitating association of integrins with small GTPases.
Significance: Investigating integrin interactions with small GTPases is critical for understanding adhesion signaling.
Keywords: Adhesion, Adhesion Receptor, Integrin, Signaling, Small GTPases, Tetraspanins, CD151
Abstract
Tetraspanin CD151 associates with laminin-binding α3β1/α6β1 integrins in epithelial cells and regulates adhesion-dependent signaling events. We found here that CD151 plays a role in recruiting Ras, Rac1, and Cdc42, but not Rho, to the cell membrane region, leading to the formation of α3β1/α6β1 integrin-CD151-GTPases complexes. Furthermore, cell adhesion to laminin enhanced CD151 association with β1 integrin and, thereby, increased complex formation between the β1 family of integrins and small GTPases, Ras, Rac1, and Cdc42. Adhesion receptor complex-associated small GTPases were activated by CD151-β1 integrin complex-stimulating adhesion events, such as α3β1/α6β1 integrin-activating cell-to-laminin adhesion and homophilic CD151 interaction-generating cell-to-cell adhesion. Additionally, FAK and Src appeared to participate in this adhesion-dependent activation of small GTPases. However, engagement of laminin-binding integrins in CD151-deficient cells or CD151-specific siRNA-transfected cells did not activate these GTPases to the level of cells expressing CD151. Small GTPases activated by engagement of CD151-β1 integrin complexes contributed to CD151-induced cell motility and MMP-9 expression in human melanoma cells. Importantly, among the four tetraspanin proteins that associate with β1 integrin, only CD151 exhibited the ability to facilitate complex formation between the β1 family of integrins and small GTPases and stimulate β1 integrin-dependent activation of small GTPases. These results suggest that CD151 links α3β1/α6β1 integrins to Ras, Rac1, and Cdc42 by promoting the formation of multimolecular complexes in the membrane, which leads to the up-regulation of adhesion-dependent small GTPase activation.
Introduction
Tetraspanins (also known as the transmembrane 4 superfamily) are a large group of ubiquitously expressed cell surface transmembrane proteins that play an important role in a variety of cellular functions, including cell proliferation, activation, differentiation, migration, and cancer cell invasion and metastasis (1, 2). Although the biochemical function(s) of tetraspanins is not fully defined, the ability of tetraspanin proteins to form multimolecular complexes with other membrane proteins involved in signaling pathways, such as integrins and signaling enzymes, suggests that tetraspanins may participate in transmembrane signaling events by integrating several signaling components into a single functional signaling complex and/or modulating the signaling properties of associated receptor proteins with lateral cross-talk (2–4).
CD151 (PETA-3/SFA-1) is a tetraspanin member that is expressed in various cell types, including epidermal basal cells, epithelial cells, endothelial cells, Schwann cells, muscle, and platelets (5). Although the physiological function of CD151 is largely unknown, much attention has been brought to the role of CD151 in malignant cancer progression because its expression level correlates with a poor prognosis in various human cancers, and its proinvasive and prometastatic activity has been revealed in several experimental models (6–11). CD151 is predominantly localized on the cell surface in contact with basement membranes and to a lesser extent at cell-cell junctions in epithelial cells (5, 12) and forms multiprotein clusters on the cell surface with laminin receptor type integrins, E-cadherin, and other tetraspanins (2, 4, 13–15). Because most cellular functions modulated by CD151, such as cell adhesion, motility, and spreading, are integrin-dependent adhesive behaviors, it has been proposed that CD151 participates in integrin signaling by regulating adhesion receptor activity of integrins and modifying integrin signaling pathways as well. Indeed, CD151 association was found to enhance the binding activity of α3β1 integrin to laminin through stabilizing its activated conformation (16). It was also reported that CD151 affects subcellular localization and molecular organization of α3β1 and α6β4 integrins (17, 18). Moreover, CD151 has been shown to modulate the outside-in signaling activity/pathways of αIIbβ3 and α3β1/α6β4 integrins in platelet and breast cancer cells, respectively (18, 19). Association of CD151 with phosphatidylinositol 4-kinase, protein kinase C, and PTPμ protein-tyrosine phosphatase (14, 20, 21) suggests the role of CD151 in linking integrins to intracellular signaling enzymes, which do not regularly participate in integrin-mediated signaling cascades. However, integrin signaling pathways modified and/or modulated by CD151 association have not been established.
Small GTPase proteins, such as Ras and Rho family members, play an important role in controlling adhesion-dependent signaling events (22–24). Several members of Ras GTPases were shown to control integrin function by modulating integrin affinity with extracellular matrix (ECM)2 ligands (25, 26). Conversely, many studies have demonstrated that integrins affect the activation of Rho, Rac, Cdc42, and Ras, either alone or in conjunction with other classes of receptors (22, 27–34). For instance, cell adhesion to laminin through α3β1 and α6β4 integrins, which are associated with CD151, induced and sustained Rac1 activation in keratinocytes, leading to formation of stable lamellipodia at the leading edge (33, 34). Activated GTPases in turn trigger not only actin cytoskeleton reorganization but also downstream signaling cascades, including MAPK pathways (35–37). In these integrin signaling pathways, FAK and Src in focal adhesion contacts are known to be crucial mediators regulating the activation of small GTPases (38, 39). However, it is unclear how small GTPases orient in close proximity to FAK and Src in focal contacts upon integrin engagement.
It was previously shown that engagement of tetraspanin CD81 activates the Rho GTPase family (40). We presumed that tetraspanin CD151 also plays a role in regulating small GTPases, because adhesion-dependent cell behaviors regulated by CD151, such as cell motility and spreading, are known to be modulated by small GTPases (28, 32, 41). The present study demonstrates that CD151 induces subcellular translocation of the small GTPases, Ras, Rac1, and Cdc42, to the membrane through formation of multimolecular complexes containing the β1 family of integrins, CD151, and small GTPases. Adhesion signals initiated by β1 integrin interactions with laminin and homophilic CD151 interactions activated these adhesion receptor complex-associated GTPases through the FAK-Src pathways. However, sole engagement of β1 integrins in CD151-deficient cells or CD151-specific siRNA-transfected cells was not sufficient to activate these GTPases. Additional β1 integrin-associated tetraspanins, such as CD9, CD81, and CD82, were unable to promote molecular association of integrins with small GTPases and did not induce activation of small GTPases upon engagement of the associated integrins. Thus, CD151 contributes to integrin-dependent activation of small GTPases by facilitating their association with the associated integrins.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies, and Reagents
C8161 and MelJuSo human melanoma cell lines were cultured in DMEM/F-12 medium supplemented with 10% fetal bovine serum (FBS) in 5% CO2 at 37 °C. For the culture of Rat-1 rat fibroblast cells and 293 human embryonic kidney cells, DMEM/high glucose medium containing 10% FBS was used. The stable CD151 transfectant clones of MelJuSo cells were previously generated (42). Antibodies against CD151 were purchased from BD Biosciences (14A2H1) and Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) (H-80, N-20). Anti-TM4SF2 and anti-EWI-2 antibodies were purchased from Abcam (Cambridge, UK) and R&D Systems (Minneapolis, MN), respectively. Anti-β1 integrin (JB1A, 6S6) and anti-CD63 (RFAC4) antibodies were obtained from Millipore (Billerica, MA). Antibodies to CD9 (C-4), CD81 (H-121), CD82 (G-2), pan-Ras (FL-189), Rho A/B/C (H-70), Rac1 (C-14), Cdc42 (P1), phospho-FAK(Tyr-925), FAK (A-17), Src (B-12), caveolin-1 (N-20), and MC1-R/melanocyte-stimulating hormone receptor (MSH-R) (H-60) were purchased from Santa Cruz Biotechnology, Inc. An antibody specific to phospho-Src(Tyr-416) was obtained from Cell Signaling (Beverly, MA). Expression vectors of dominant negative and constitutively active mutants of H-ras, K-ras RhoA, Rac1, and Cdc42 were purchased from the Missouri S&T cDNA Resource Center. All other reagents were from Sigma unless indicated otherwise.
Transfection of Small Interfering RNA (siRNA)
siRNAs for CD151 and FAK were designed and synthesized using the software and SilencerTM siRNA construction kit from Ambion (Austin, TX) according to the manufacturer's instructions. Specific oligonucleotide sequences for each target gene were as follows: 5′-GUUGGAGACCUUCAUCCAGdTdT-3′ (sense) and 5′-CUGGAUGAAGGUCUCCAACdTdT-3′ (antisense) targeting CD151; 5′-GAGAAGGCUCAGCAAGAAGdTdT-3′ (sense) and 5′-CUUCUUGCUGAGCCUUCUCdTdT-3′ (antisense) targeting FAK. The siRNA control was 5′-UUCUCCGAACGUGUCACGUdTdT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAAdTdT-3′ (antisense), which bears no homology with relevant human genes (43). Transfection of siRNA was carried out using Lipofectamine reagent as described previously (42).
Immunofluorescence Analysis
Immunocytochemistry was performed on glass coverslips (Fisherbrand), coated with or without 0.1 mg/ml poly-l(+)-lysine or laminin. Cells grown on coverslips were fixed with 3.7% paraformaldehyde for 10 min, permeabilized with 0.2% Triton X-100 for 15 min, and blocked with 5% BSA in PBS for 1 h at room temperature. Cells were then stained for 1 h at 37 °C with the primary antibody diluted in the blocking solution, washed three times with TBS containing 0.1% Triton X-100, and incubated with AlexaFluor488 (green)-conjugated anti-mouse IgG or AlexaFluor555 (red)-conjugated anti-rabbit IgG (Molecular Probes) for 45 min at 37 °C. Fluorescence photomicrographs for mounted coverslips were taken using a confocal laser-scanning microscope (Olympus FV1000).
Small GTPase Protein Pull-down Assays
Raf1-RBD-agarose was obtained from Upstate Biotechnology, Inc. (Lake Placid, NY). GST-rhotekin-RBD and GST-PAK-PBD fusion proteins bound to glutathione-Sepharose 4B beads were prepared as described previously (44, 45). Cells (1–2 × 106) were lysed in GST lysis buffer (50 mm Tris, pH 7.4, 200 mm NaCl, 1% Nonidet P-40, 10% glycerol, 2 mm MgCl2, 100 μm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 20 μg/ml leupeptin). The cleared cell lysates were subsequently incubated with the Raf1-RBD-agarose, GST-rhotekin-RBD, and GST-PAK-PBD beads for 45 min at 4 °C with end-over-end mixing. After washes with GST lysis buffer, proteins bound to beads were eluted into Laemmli buffer and resolved in 12% SDS-PAGE. The small GTPase proteins were detected by immunoblotting with the appropriate antibodies.
Cellular Fractionation by Sucrose Density Gradient Ultracentrifugation
Cells were lysed in HBSE buffer (20 mm HEPES, pH 7.2, 150 mm NaCl, 1 mm EDTA) supplemented with protease inhibitors (EDTA-free tablets; Roche Applied Science) in the presence of 1% Brij 97 for 1 h on ice and homogenized in a loose fitting Dounce homogenizer using 15 strokes. Postnuclear supernatants (1 ml) were obtained by centrifugation (2,500 × g, 10 min, 4 °C), mixed with an equal volume of 90% sucrose (w/v in HBSE), and placed in the bottom of Beckman SW55Ti ultracentrifuge tubes and then overlaid with discontinuous sucrose gradients (2 ml of 35% sucrose and 1 ml of 5% sucrose in HBSE). The gradient was centrifuged for 18 h at 150,000 × g at 4 °C. Twelve fractions of 0.4 ml were collected from the top of the gradient and analyzed by SDS-PAGE and immunoblotting.
Membrane Fractionation and Membrane Fragment Treatment
Detergent-free purification of membrane fragments from empty vector- and CD151-transfected MelJuSo cells was performed as described in previous studies (42, 46). Briefly, cells were washed with ice-cold PBS and then scraped into buffer A (20 mm Tris-HCl, pH 7.5, 2 mm EDTA, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 20 μg/ml leupeptin, and 2 mm benzimidine). The cells were homogenized using a tight fitting Dounce homogenizer (20–25 strokes). Postnuclear supernatants were adjusted to 10% sucrose and loaded onto a 30% sucrose cushion in an ultracentrifuge tube. After centrifugation for 60 min at 150,000 × g in a tabletop ultracentrifuge (Beckman Instruments) with T-1270 rotor, a light-scattering band confined to a 10–30% sucrose interface was collected and stored at −70 °C until use. For cell treatments, membrane fragments were evenly suspended in serum-free medium by passage 5–6 times through a 26-gauge needle and immediately added to cells.
Other Analyses/Assays
Immunoprecipitation and immunoblotting analyses, gelatin zymography, and a wound-healing migration assay were carried out as described previously (42).
RESULTS
CD151 Associates with Ras, Rac1, and Cdc42 Together with β1 Integrins
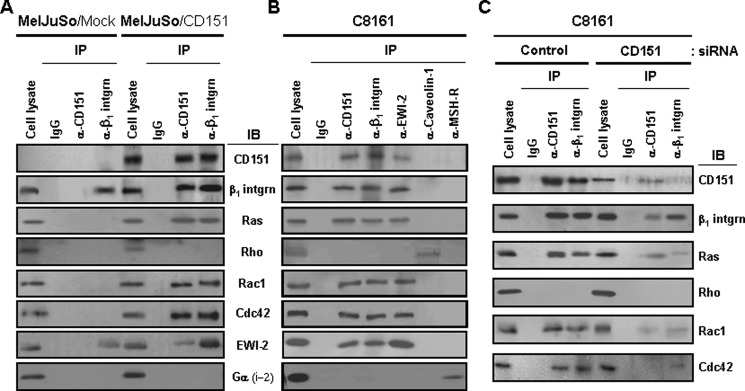
We previously found that CD151 associates with α3β1 and α6β1 laminin receptor type integrins in human melanoma cells (42). In the present study, we first examined whether α3β1/α6β1 integrin-CD151 complexes are physically associated with small GTPases in human melanoma cell lines, MelJuSo cells transfected with exogenous CD151, and C8161 parental cells with endogenous CD151. Transfection of exogenous CD151 into MelJuSo cells resulted in greater expression of CD151 as compared with C8161 cells, but not greater than the A375SM melanoma cell line (supplemental Fig. S1), indicating that the effect of CD151 in MelJuSo CD151 transfectant cells reflects the physiological situation in melanoma cells. Following lysis of empty vector- or CD151-transfected MelJuSo cells with the nonionic detergent Brij 97, a mild detergent preserving tetraspanin-integrin interactions (47, 48), the small GTPases, Ras, Rac1, and Cdc42, were co-precipitated with an anti-CD151 antibody in CD151 transfectant cells but not in mock transfectant cells (Fig. 1A). Co-immunoprecipitation of CD151 with EWI-2, which associates with tetraspanins, including CD151 (49), but not with Gα(i-2), a G protein subunit associated with a variety of membrane receptors, including MSH-R, supports the association of CD151 with Ras, Rac1, and Cdc42. These small GTPases were also detected in the CD151 immunoprecipitates of C8161 cells with endogenous CD151 (Fig. 1B). Furthermore, these small GTPases were found in the immunoprecipitates of CD151-associated EWI-2 in C8161 cells but not in those of caveolin-1 and MSH-R, which do not associate with CD151. Importantly, the β1 integrin immunoprecipitates of CD151-transfected MelJuSo cells and C8161 cells also contained Ras, Rac1, and Cdc42 as well as CD151. However, as shown in MelJuSo mock transfectant cells, β1 integrins lacking CD151 association were not able to form molecular complexes with these GTPases. Also, siRNA-mediated knockdown of CD151 in C8161 cells significantly diminished the amount of Ras, Rac1, and Cdc42 co-immunoprecipitated with β1 integrin (Fig. 1C). With confocal microscopy, the fluorescent images of Ras, Rac1, and Cdc42 in C8161 cells were very similar to the CD151 images (supplemental Fig. S2). Overlaid images between CD151 and these GTPases illustrated co-localization of CD151 with small GTPases in the cell membrane. Meanwhile, among the four major small GTPases examined, the Rho protein was the only GTPase absent in the immunoprecipitates of CD151 and β1 integrin (Fig. 1). Furthermore, Rho was not found in the membrane of C8161 cells with endogenous CD151 (supplemental Fig. S2). These results suggest molecular association of CD151-β1 integrin complexes with small GTPases, such as Ras, Rac1, and Cdc42, in the cell membrane.
FIGURE 1.
CD151 facilitates molecular association of CD151-β1 integrin complexes with Ras, Rac1, and Cdc42. A and B, stable MelJuSo mock and CD151 transfectant cells (A) and parental C8161 cells (B) were lysed with Brij 97, and immunoprecipitation (IP) was performed with mouse normal IgG, anti-CD151, anti-β1 integrin, anti-EWI-2, anti-caveolin-1, or anti-MSH-R Abs. The immunoprecipitates were analyzed by immunoblotting (IB) using anti-pan-Ras, anti-RhoA/B/C, anti-Rac1, anti-Cdc42, anti-EWI-2, or anti-Gαi-2 Abs. C, C8161 cells were transiently transfected with control or CD151-targeting siRNAs, and the CD151 and β1 integrin immunoprecipitates were examined for the presence of Ras, Rho, Rac1, and Cdc42 by immunoblotting analysis.
CD151 Induces Subcellular Translocation of Ras and Rac1 to the Membrane Region Containing CD151-β1 Integrin Complexes
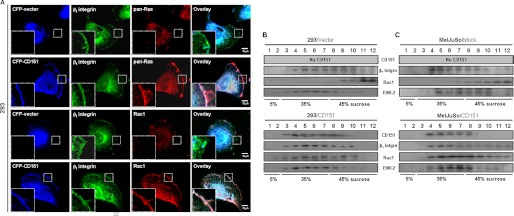
To confirm CD151-dependent co-localization of small GTPases with β1 integrins in the cell membrane, we examined the subcellular localization of Ras and Rac1 in CD151-deficient and -expressing cells with confocal microscopy. In 293 embryonic kidney cells deficient in CD151 expression, Ras and Rac1 were not found in the membrane region, and, thereby, they did not display fluorescent images overlapped with β1 integrins in the cell membrane (Fig. 2A). In contrast, CD151-transfected 293 cells showed Ras and Rac1 present in the cell membrane, where these GTPases displayed overlapped images with CD151 and β1 integrins. Thus, CD151 expression in 293 cells results in membrane translocation of Ras and Rac1. We next examined the sucrose density distribution of Rac1 in the postnuclear fraction of Brij 97 detergent lysates to compare subcellular compartmentalization of Rac1 with β1 integrins in CD151-deficient and CD151-expressing cells. In CD151-deficient 293 and MelJuSo cells, Rac1 was predominantly recovered in the 45% sucrose fractions but not in the 35% sucrose fractions (Fig. 2, B and C). However, in CD151-transfected cells, the bulk of Rac1 was recovered in the 35% sucrose fractions, where CD151, β1 integrins, and EWI-2 were enriched. Thus, consistent with their physical association, Rac1 in CD151-expressing cells exhibited very similar sucrose density distribution patterns to β1 integrins and CD151. Because CD151, β1 integrins, and EWI-2 are known to be abundant in the tetraspanin-associated microdomain of the membrane (48, 50), these data provide another line of evidence for CD151-mediated subcellular co-localization of Rac1 with β1 integrins in the cell membrane.
FIGURE 2.
CD151 induces subcellular translocation of Rac1 and Ras to a membrane region containing β1 integrins. A, 293 cells were transiently transfected with expression vectors encoding CFP or CFP-fused CD151. After fixation, the cells were permeabilized and probed with mouse anti-β1 integrin Ab or rabbit Abs against pan-Ras and Rac1. Following incubation with fluorescent secondary antibodies described under “Experimental Procedures,” the cells were analyzed by confocal microscopy. Images in the far right column represent overlapped images between CFP (blue), β1 integrin (green), and small GTPases (red). Magnified images of the membrane area are shown in insets in fluorescent images. B and C, transiently empty vector- and CD151-transfected 293 cells (B) and stable MelJuSo mock and CD151 transfectant cells (C) were lysed in buffer containing 1% Brij 97, and the subcellular fraction was separated by ultracentrifugation using a discontinuous sucrose density gradient. Twelve fractions were collected from the top of the gradient and analyzed by immunoblotting using Abs against CD151, integrin β1, Rac1, or EWI-2. Data shown are representative of three separate experiments with similar results.
Engagement of β1 Integrins by Laminin Binding Augments Molecular Association of CD151-β1 Integrin Complexes with Small GTPases
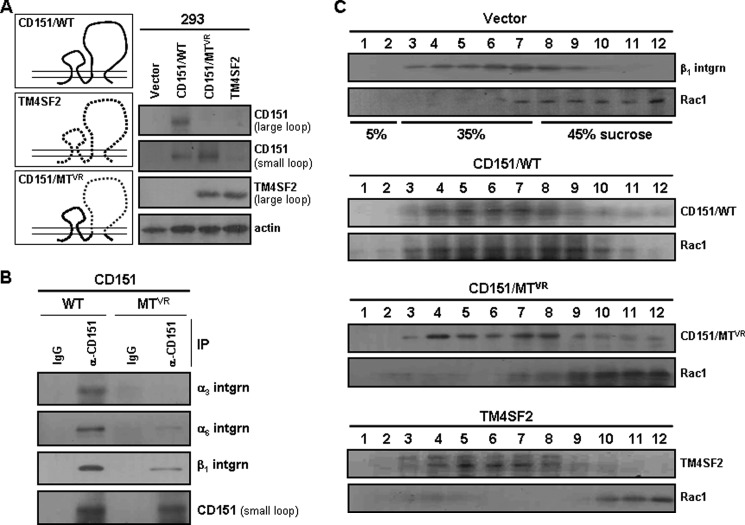
We examined whether CD151-associated α3β1 and α6β1 integrins also play a role in promoting physical association of CD151-β1 integrin complexes with small GTPases. We first constructed a CD151 mutant, CD151VR, in which the large extracellular loop region (serine 158 to glycine 207) of CD151 was replaced with the corresponding region from another tetraspanin, TM4SF2 (Fig. 3A). This region, named as the “variable region” of CD151, was previously shown to contain sequences critical for the interaction of CD151 with its major partner, α3β1 integrin (15). In a mild detergent lysis condition, preserving interactions between wild-type CD151 and α3β1/α6β1 integrins, the CD151VR mutant did not co-precipitate with the α3 integrin subunit (Fig. 3B). Also, only a trace amount of α6 and β1 integrin subunits was detected in the immunoprecipitate of the CD151VR mutant, indicating impaired interaction between the CD151VR mutant and α3β1/α6β1 integrins (Fig. 3B). Unlike wild type, the CD151VR mutant did not display a similar sucrose density distribution pattern to Rac1, despite its localization in the cell membrane (Fig. 3C). Because intracellular regions/domains of the CD151VR mutant are identical to the wild type, CD151 is not likely to interact with Rac1 directly in the intracellular side of the membrane. Instead, these results suggest that CD151 associated with β1 integrins, but not CD151 alone, is able to associate with Rac1. Therefore, it seems likely that the ability of CD151 to associate with α3β1/α6β1 integrins might be important for its ability to facilitate the formation of CD151-β1 integrin-small GTPase complexes.
FIGURE 3.
A CD151 mutant with impaired α3β1 integrin association fails to recruit Rac1 to the membrane. A, CD151VR mutant cDNA that encodes CD151 with a large extracellular loop substituted with that of TM4SF2, as illustrated, was generated by PCR and subcloned into a pcDNA3 vector. The wild-type (WT) and mutant (MTVR) CD151 expression vectors were transiently transfected into 293 cells, and their expression levels were examined by immunoblotting analysis using mAbs H-80 and N-20, which recognize large and small extracellular loops of CD151, respectively. B, following transfection with the wild-type and mutant CD151 expression constructs, Brij 97 detergent lysates were subjected to immunoprecipitation (IP) with anti-CD151 mAb (N20) followed by immunoblotting analysis with Abs against integrin α3, α6, or β1. C, following sucrose density gradient ultracentrifugation, 12 fractions were examined for the presence of β1 integrin, CD151, and Rac1 by immunoblotting analysis.
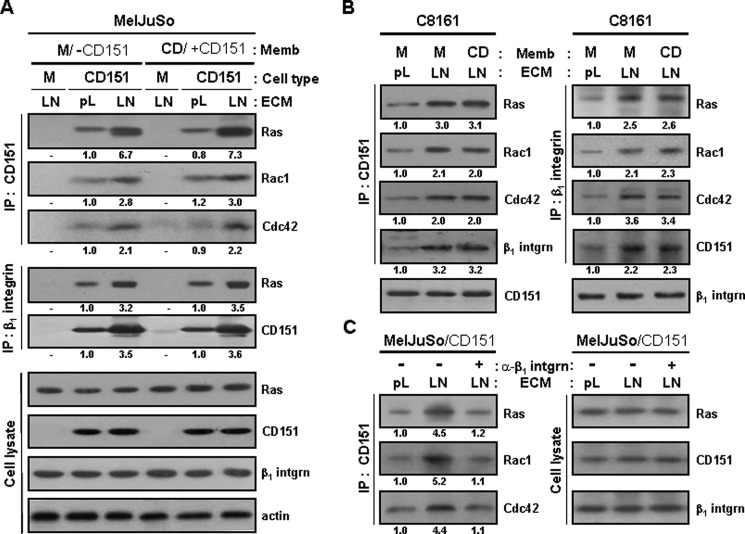
Results in Figs. 1–3 were obtained from cells grown on normal tissue culture polystyrene plates, in which the surface was coated with electrically charged plasma. Cell adhesion to these plates is known to stimulate a broad range of integrin types nonspecifically to moderate levels (51, 52). To examine whether specific activation of CD151-associated β1 integrins affects association between CD151-β1 integrin complexes and small GTPases, we precoated the culture plate surface with either poly-l-lysine or laminin before seeding the cells. Compared with cell binding to poly-l-lysine, a cationic polyelectrolyte that minimally activates any types of integrins (51–53), the attachment of CD151-transfected MelJuSo cells to laminin, which strongly activates CD151-associated α3β1/α6β1 integrins, significantly increased the levels of small GTPases associated with CD151 and β1 integrins (Fig. 4A). Notably, α3β1 and α6β1 integrins activated by laminin binding were shown to associate with CD151 more than integrins bound to poly-l-lysine, which might result in increased association of Ras, Rac1, and Cdc42 with CD151-β1 integrin complexes. C8161 melanoma cells with endogenous CD151 also exhibited a positive effect of β1 integrin engagement with laminin in the formation of CD151-β1 integrin-small GTPases complexes (Fig. 4B). However, when CD151-transfected cells were pretreated with an anti-β1 integrin blocking antibody before seeding onto laminin-coated plates, cell adhesion to laminin did not result in increased association of CD151 with the GTPases (Fig. 4C). We have previously shown that homophilic interactions of CD151 through homotypic cell-to-cell adhesion simulates CD151-β1 integrin complex-dependent signaling (42). However, we here found that, without β1 integrin engagement with laminin, homophilic CD151 interactions resulted in little or no effect on molecular association of CD151 with the small GTPases and β1 integrins as well (Fig. 4, A and B). Activated β1 integrin-induced association of GTPases with CD151-β1 integrin complexes in the cell membrane was also observed in immunofluorescent images of C8161 cells with endogenous CD151 (supplemental Fig. S3). When C8161 cells were attached to poly-l-lysine, Ras was minimally detected in the membrane, and, consequently, no association of Ras with CD151 and β1 integrins was observed. In contrast, β1 integrins activated by laminin binding induced Ras translocation to the membrane, where Ras was co-localized with CD151 and β1 integrins. It thus appears that subcellular localization of Ras is influenced by the activation status of CD151-associated β1 integrins. Taken together, these results strongly suggest that association of Ras, Rac1, and Cdc42 with CD151-β1 integrin complexes in the membrane is facilitated not only by CD151 expression but also by the adhesion-dependent activation of CD151-associated β1 integrins.
FIGURE 4.
Cell adhesion to laminin enhances physical association of CD151-β1 integrin complexes with Ras, Rac1, and Cdc42. A and B, stable MelJuSo mock (M) and CD151 transfectant cells (A) and parental C8161 cells (B) were seeded onto plates precoated with laminin (LN) or poly-l-lysine (pL) for 24 h. Following 60 min of serum starvation, the cells were treated with membrane fragments of mock (M) or CD151 (CD) transfectant MelJuSo cells for 60 min. Immunoprecipitation and immunoblotting analyses were performed in the same manner as in Fig. 1A. C, the CD151-transfected MelJuSo cells were suspended into serum-free medium and mixed with normal IgG or anti-β1 integrin blocking mAb (6S6; 20 μg/ml) in end-over-end fashion for 60 min. Twenty-four hours after seeding onto laminin- or poly-l-lysine-coated plates, cell lysate with Brij 97 was subjected to immunoprecipitation with anti-CD151 Ab followed by immunoblotting analysis with Abs against Ras, Rac1, or Cdc42. Numbers below the immunoblot indicate the relative band intensity and are the mean of three immunoblots obtained from separate experiments.
CD151-β1 Integrin Complex-mediated Adhesion Signaling Activates Ras, Rac1, and Cdc42
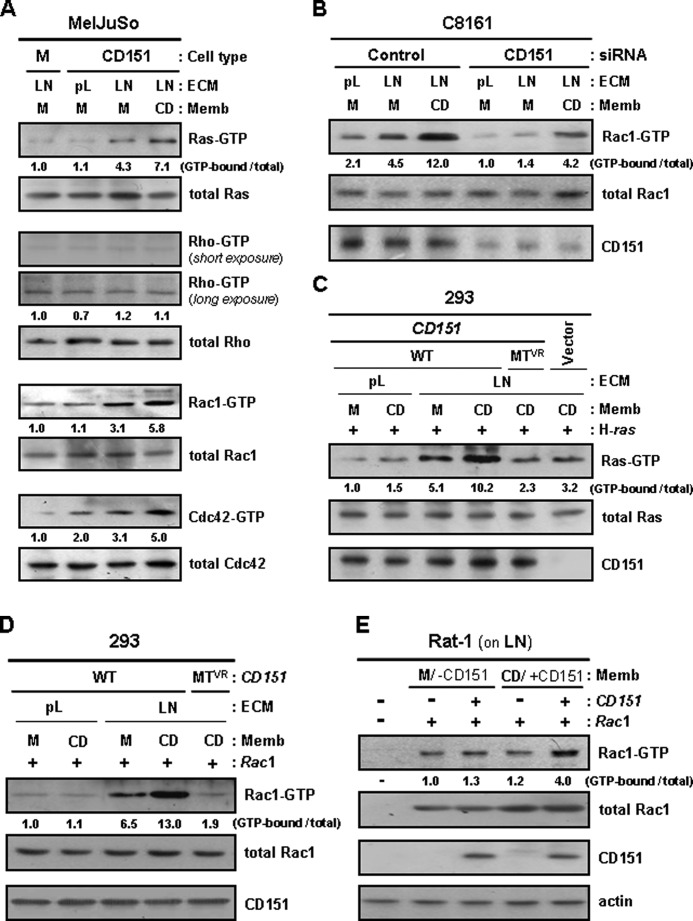
We further examined whether small GTPases associated with CD151-β1 integrin complexes are activated by adhesion events mediated by these complexes. CD151-transfected MelJuSo cells seeded on laminin-coated plates exhibited higher levels of the GTP-loaded Ras, Rac1, and Cdc42 than empty vector-transfected cells (Fig. 5A). Furthermore, siRNA-mediated knockdown of CD151 in C8161 cells significantly suppressed Rac1 activation by cell adhesion to laminin (Fig. 5B). Additionally, CD151-β1 integrin complex-mediated signaling did not activate Rho, the small GTPase not associated with CD151-β1 integrin complexes. Thus, CD151 appears to play an important role in laminin receptor integrin-mediated activation of Ras, Rac1, and Cdc42. However, the positive effect of CD151 expression on the activation of these GTPases was not found in cells attached to poly-l-lysine, indicating that specific activation of CD151-assocated β1 integrin is essential for CD151-dependent activation of Ras, Rac1, and Cdc42. The CD151VR mutant that was unable to associate with the α3β1 integrin was also shown to interfere with the adhesion-dependent activation of Ras and Rac1 (Fig. 5, C and D). Collectively, these data strongly suggest that CD151-mediated physical association of β1 integrins with small GTPases contributes to adhesion-dependent activation of small GTPases. Moreover, in CD151-expressing cells with laminin adhesion, the levels of GTP-loaded Ras, Rac1, and Cdc42 were further increased by treatment with membrane fragments obtained from CD151-transfected cells but not by treatment with empty vector-transfected cell membrane fragments deficient in CD151 (Fig. 5, A and B). CD151-transfected 293 human embryonic kidney and Rat-1 rat fibroblast cells also responded to CD151-containing membrane fragments for Ras and Rac1 activation (Fig. 5, C–E). However, in CD151-deficient cells, treatment with CD151-containing membrane fragments did not induce Ras and Rac1 activation. Also, cell adhesion to poly-l-lysine abolished the positive effect of CD151-containing membrane fragment treatment on the activation of Ras and Rac1 in CD151-transfected 293 cells (Fig. 5, C and D). Thus, CD151-β1 integrin adhesion receptor complexes, primarily activated by interaction of β1 integrins with laminin, could be further stimulated for intracellular signaling by homophilic interactions of CD151 on the surface of two contacting cells. These results implicate CD151-mediated positive signaling cross-talk between cell-to-ECM and cell-to-cell adhesions.
FIGURE 5.
Integrin interaction with laminin and homophilic interactions of CD151 both contribute to activation of Ras, Rac1, and Cdc42. A and B, stable MelJuSo mock (M) and CD151 transfectant cells (A) and control or CD151-targeting siRNA-transfected C8161 cells (B) were cultured onto plates precoated with laminin (LN) or poly-l-lysine (pL) for 24 h. Following 60 min of serum starvation, the cells were treated with membrane fragments of MelJuSo mock (M) or CD151 (CD) transfectant cells for 60 min. GTP-loaded Ras, Rho, Rac1, and Cdc42 proteins were precipitated from cell lysates and detected as described under “Experimental Procedures.” C and D, 293 cells were transfected with CD151 expression vectors encoding the wild-type (WT) or mutant (MTVR) form illustrated in Fig. 3A, together with H-ras (C) or Rac1 (D) expression constructs, and then cultured on laminin (LN)- or poly-l-lysine (pL)-coated plates for 24 h. Following cell treatment with membrane fragments of MelJuSo mock (M) or CD151 (CD) transfectant cells, GTP-loaded Ras (C) and Rac1 (D) proteins were examined in the same manner as in A. E, Rat-1 cells grown on laminin were transfected with CD151 and/or Rac1 expression vectors. After treating cells with membrane fragments of MelJuSo transfectant cells, GTP-loaded Rac1 proteins were examined in the same manner as in A. Numbers below the immunoblot indicate the relative band intensity of GTP-bound form normalized to the total protein level and are the mean of three immunoblots obtained from separate experiments.
Integrin-mediated FAK-Src Pathway Contributes to Adhesion-dependent Activation of GTPases
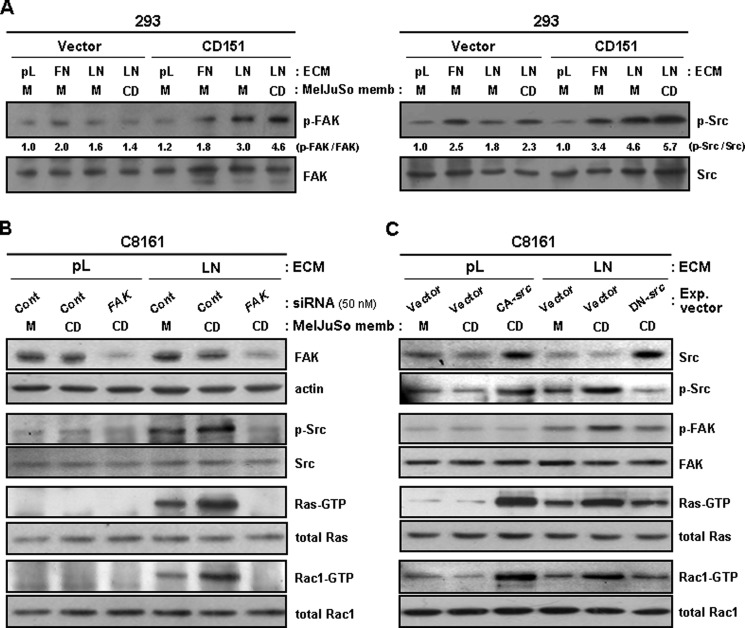
We previously found that FAK and Src, downstream signal transducers of integrins, are involved in integrin-dependent CD151 signaling events in human melanoma cells (42). The present study also demonstrated that CD151 expression in 293 cells resulted in increased phosphorylation of FAK and Src upon integrin engagement (Fig. 6A). Also, treatment with CD151-containing membrane fragments further increased phosphorylation levels of FAK and Src in CD151-transfected 293 cells but not in empty vector-transfected 293 cells. The stimulatory effect of homophilic CD151 interactions on integrin-dependent activation of FAK and Src indicates that CD151 plays a role in up-regulating integrin signaling to FAK and Src. We next examined whether FAK and Src participate in CD151-β1 integrin complex-mediated adhesion signaling to small GTPases. Similar to the same finding in Fig. 5, the levels of GTP-loaded Ras and Rac1 were increased by both β1 integrin-laminin interactions and homophilic CD151 interactions (Fig. 6B); however, CD151-β1 integrin complex-induced activation of Ras and Rac1 was abrogated by siRNA knockdown of FAK. Expression of a dominant negative Src mutant also interfered with Ras and Rac1 activation by CD151-β1 integrin complex-mediated adhesion signals, whereas a constitutively active Src mutant increased the levels of GTP-bound Ras and Rac1 regardless of integrin engagement (Fig. 6C). Taken together, these data suggest that the canonical integrin pathway, involving FAK and Src activation, is responsible for CD151-β1 integrin adhesion receptor complex-mediated activation of Ras and Rac1.
FIGURE 6.
CD151-mediated activation of Ras and Rac1 is dependent on FAK and Src activities involved in integrin signaling. A, empty vector- and CD151-transfected 293 cells were cultured onto plates precoated with laminin (LN) or poly-l-lysine (pL) for 24 h. After treating cells with membrane fragments of MelJuSo mock (M) or CD151 (CD) transfectant cells for 60 min, phosphorylation levels of FAK and Src in cell lysates were assessed by immunoblotting analysis using Abs recognizing phospho-FAK(Tyr-925) and phospho-Src(Tyr-416). Numbers below the immunoblot indicate relative ratio of phospho-FAK/total FAK or phospho-Src/total Src. B and C, C8161 cells cultured on laminin (LN) or poly-l-lysine (pL) were transfected with control or FAK-targeting siRNA (B) and the expression vectors encoding the constitutively active (CA) or dominant negative (DN) mutant of Src (C). Following treatment of cells with membrane fragments from mock (M) or CD151 (CD) transfectant MelJuSo cells, GTP-loaded Ras and Rac1 proteins were examined in the same manner as in Fig. 5.
CD151-induced Cell Motility and MMP-9 Expression Require Ras, Rac1, and Cdc42 Activity
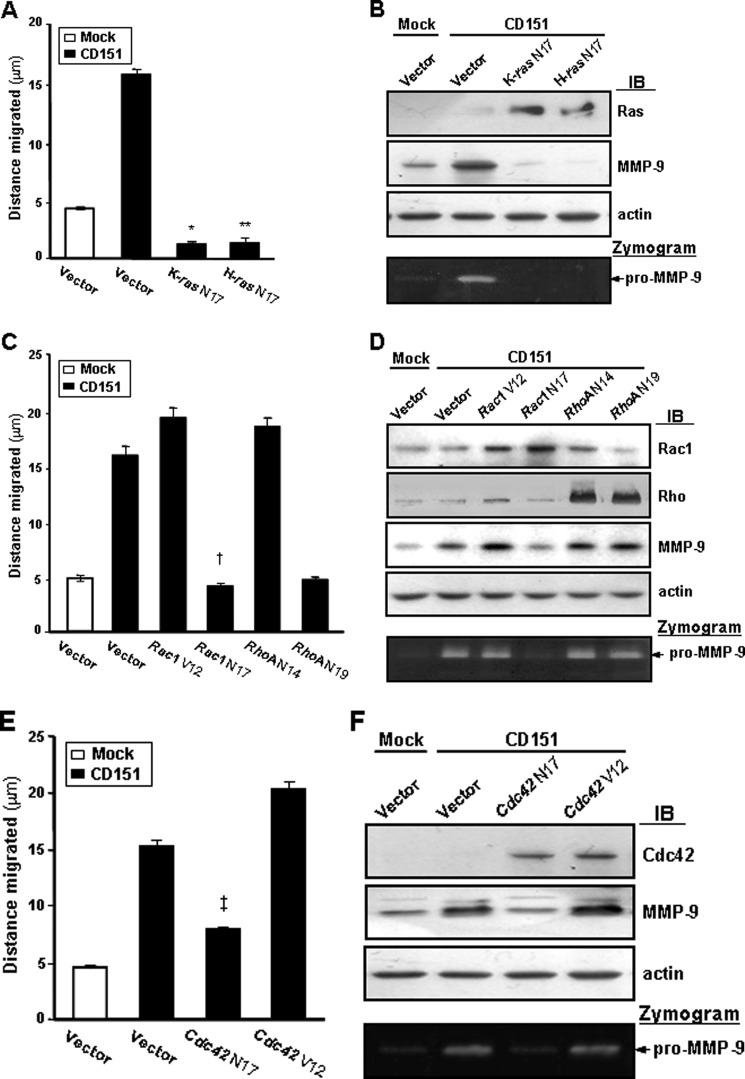
Our previous report showed that CD151 expression increases cell motility and matrix metalloproteinase-9 (MMP-9) expression in MelJuSo melanoma cells (42). To verify the functional involvement of small GTPases in CD151-mediated cellular events, we transiently transfected expression constructs encoding various mutant forms of small GTPases into CD151-transfected MelJuSo cells. Transfection of dominant inhibitory ras mutants, such as K-ras N17 and H-ras N17, decreased the migrating ability and MMP-9 expression of CD151 transfectant cells to levels below those of mock transfectant cells (Fig. 7, A and B). Dominant-negative Rac1 (Rac1 N17) and Cdc42 (Cdc42 N17) also interfered with CD151-induced cell motility and MMP-9 expression in MelJuSo cells, whereas the constitutively active forms of Rac1 (Rac1 V12) and Cdc42 (Cdc42 V12) further increased motility and MMP-9 expression of CD151 transfectant cells (Fig. 7, C–F). Meanwhile, a dominant inhibitory RhoA mutant (RhoA N19) did not suppress the positive effect of CD151 expression on MMP-9 expression (Fig. 7D), indicating that Rho activity is irrelevant to CD151-induced MMP-9 expression. Although RhoA N19 significantly inhibited the migrating ability of CD151 transfectant cells (Fig. 7C), this may result from the malfunction of the general cell migration machinery, which has been known to require Rho-mediated focal adhesion formation. Because Ras, Rac1, and Cdc42, but not Rho, were associated with and activated by CD151-β1 integrin complexes (Figs. 1 and 5), these results strongly suggest that these small GTPases participate in adhesion signaling pathways leading to increased motility and MMP-9 expression of melanoma cells.
FIGURE 7.
CD151-induced cell motility and MMP-9 expression in melanoma cells are mediated by RasH/K, Rac1, and Cdc42. MelJuSo mock and CD151 transfectant cells were transiently transfected with the expression vectors encoding a dominant inhibitory mutant of RasK (K-ras N17) or RasH (H-ras N17) (A and B); constitutively active Rac1 (Rac1 V12) or RhoA (RhoA N14), dominant-negative Rac1 (Rac1 N17) or RhoA (RhoA N19) (C and D); and constitutively active (Cdc42 V12) or dominant inhibitory (Cdc42 N17) mutant of Cdc42 (E and F). A, C, and E, following transfection, cell migration was measured at 48 h after wounding as described previously (42). B, D, and F, protein levels and activities of MMP-9 in the conditioned media obtained from cells cultured in serum-free medium for 3 days were assessed by immunoblotting analysis (IB) and gelatin zymography, respectively. Asterisks and daggers indicate that the differences are statistically significant (*, **, †, and ‡, p < 0.01 versus control vector-transfected cells, Student's t test). Error bars, S.D.
Distinct Role of CD151 in Tetraspanin-mediated Integrin Complex Formation with Small GTPases
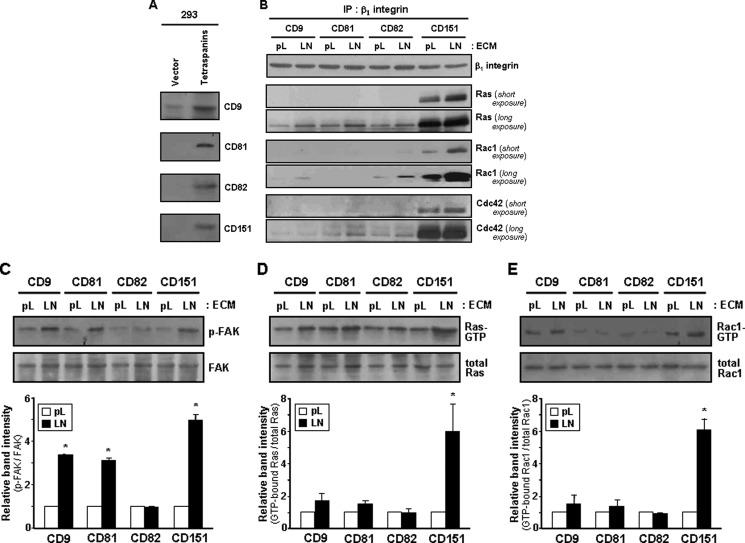
Because many tetraspanin proteins have been known to form complexes with integrins in the membrane, we examined whether members of the tetraspanin family other than CD151 could also facilitate complex formation between integrins and small GTPases. When CD9, CD81, CD82, and CD151, tetraspanin proteins that have been known to associate with β1 integrins (2), were individually overexpressed in 293 cells by gene transfection (Fig. 8A), Ras, Rac1, and Cdc42 were found in the β1 immunoprecipitate of CD151-transfected cells at a significantly higher level than that of other tetraspanin-transfected cells (Fig. 8B). Furthermore, among the four tetraspanins, only CD151 was found to co-precipitate with these GTPases (data not shown). Compared with cell adhesion to poly-l-lysine, adhesion of CD151-expressing cells to laminin significantly increased the amount of Ras, Rac1, and Cdc42 associated with β1 integrins (Fig. 8B), possibly due to the increased association of CD151 with activated β1 integrins (Fig. 4). Notably, CD151 was able to induce physical association of Rac1 and Cdc42 with β1 integrins in cells adhered to poly-l-lysine, whereas CD9, CD81, and CD82 did not (Fig. 8B). These results strongly suggest that CD151 might be a unique tetraspanin capable of promoting molecular association between β1 integrins and small GTPases regardless of integrin activation. Meanwhile, integrin-dependent activation of FAK was observed not only in the CD151-transfected cells, but also in the CD9- and CD81-transfected cells (Fig. 8C). However, an increase in the level of GTP-loaded Ras and Rac1 by cell adhesion to laminin was observed in the CD151-transfected cells but not in the CD9- and CD81-transfected cells (Fig. 8, D and E). These results provide strong evidence for a distinct role of CD151 in mediating physical association of Ras, Rac1, and Cdc42 with integrin adhesion receptor complexes and, thereby, stimulating integrin-dependent activation of small GTPases.
FIGURE 8.
Complex formation between β1 integrins and small GTPases and adhesion-dependent activation of the GTPases occurs in CD151-expressing cells but not in CD9-, CD81-, and CD82-expressing cells. 293 cells were transiently transfected with expression vectors encoding CD9, CD81, CD82, or CD151. A, expression level of each tetraspanin in the transfectant cells was assessed by immunoblotting analysis. B, C, D, and E, the transfectant cells were seeded onto plates precoated with laminin (LN) or poly-l-lysine (pL) for 24 h. B, cell lysate with Brij 97 was subjected to immunoprecipitation with an anti-β1 integrin Ab followed by immunoblotting analysis using Abs against Ras, Rac1, or Cdc42. C, phosphorylation levels of FAK in cell lysates were compared by immunoblotting analysis using anti-phospho-FAK(Tyr-925) Ab. D and E, GTP-loaded Ras (D) and Rac1 (E) proteins in cell lysates were examined in the same manner as in Fig. 5. Bar graphs indicate relative band intensity of the GTP-bound form normalized to total protein level and show the mean ± S.D. (error bars) of three immunoblots obtained from separate experiments.
DISCUSSION
Cell adhesion to ECM initiates outside-in integrin signaling pathways leading to various adhesion-dependent cellular events. Among the intracellular signal-transducing molecules, small GTPase proteins have been known to play a central role in modulating integrin-dependent cell behaviors, including cell adhesion, migration, and spreading. Following cell adhesion to ECM, integrin-mediated Rho activation induces focal adhesion assembly and actin stress fiber formation (54). By regulating the actin cytoskeleton, Rac and Cdc42 also induce the formation of lamellipodia and filopodia, respectively, which are necessary for cell migration and spreading (55). Although the mechanisms by which integrin signaling activates small GTPases are still unclear, there is no doubt that small GTPases take part in integrin signaling pathways. In addition to integrin regulation of small GTPases, Ras and Rho family members were found to regulate integrin functions by modulating affinity and avidity of integrins to ECM ligands (25, 26, 56), implying a feedback regulation loop between integrins and small GTPases. Integrins and GTPases were therefore proposed to be organized into complex signaling cascades that regulate adhesion-dependent cellular activities.
Tetraspanin proteins have been known to be involved in integrin signaling events through complex formation with integrins in the membrane (4, 57, 58). Among the tetraspanin family members, CD151 has been well characterized as a molecular adapter facilitating the assembly of functional signaling complexes in the membrane and is strongly associated with laminin-binding integrins, such as α3β1, α6β1, α6β4, and α7β1 (2, 13). We have also previously demonstrated the association of CD151 with α3β1 and α6β1 integrins in human melanoma cell lines (42). Although CD151 was reported to be involved in adhesion-dependent regulation of Ras (59), no evidence for the molecular association of integrins with small GTPases has been found to date. In the present study, we found that CD151 associates with several small GTPases, along with α3β1 and α6β1 integrins, in human melanoma and embryonic kidney cells. Among the GTPases examined, Ras, Rac1, and Cdc42, but not Rho, were found to associate with CD151-β1 integrin complexes in the membrane. In contrast, other tetraspanin members that associate with β1 integrins, such as CD9, CD81, and CD82 (2), did not exhibit strong activity to promote the association of β1 integrins with Ras, Rac1, and Cdc42 to the same extent as CD151. It thus appears that CD151 plays a distinct role in linking β1 integrins to the small GTPases, Ras, Rac1, and Cdc42, leading to the formation of a multimolecular adhesion signaling unit consisting of β1 integrins, CD151, and small GTPases in the membrane.
Previously, it was reported that the short C-terminal cytoplasmic region of CD151 is important for determining the outside-in signaling functions of α6β1 integrins and CD151 activity for Ras regulation (59, 60). Therefore, we speculated that CD151 might directly interact with Ras through its C-terminal cytoplasmic tail. However, we found here that a CD151 mutant deleted for six amino acids in the C-terminal region was still capable of forming complexes with Ras and Rac1 in the membrane, similar to a CD151 mutant with a deleted N-terminal cytoplasmic region (supplemental Fig. S4). Because no cytoplasmic region(s) of CD151 other than the N- and C-terminal regions has a length sufficient to interact with cellular proteins at the cytoplasmic face, we excluded the possibility of direct interaction of CD151 with small GTPases in the intracellular side of the membrane. Instead, we found here that a CD151 mutant incapable of associating with α3β1/α6β1 integrins was not able to recruit Rac1 to the membrane region, despite its localization in the membrane. These data suggest that the ability of CD151 to associate with α3β1/α6β1 integrins might be important for its ability to induce membrane translocation of Rac1. Recently, it was reported that CD151-integrin association and CD151 association with other tetraspanins were both important for laminin-binding integrin-dependent motility of A431 epidermoid carcinoma cells (61). Therefore, it can be speculated that a membrane structure formed by CD151-α3β1/α6β1 integrin complexes, such as tetraspanin-enriched microdomains (48, 50) and/or some adaptor/scaffold protein(s) in association with CD151-α3β1/α6β1 integrin complexes, plays a role in recruiting small GTPases to the membrane region.
α3β1 integrin-dependent cell adhesion to laminin was previously shown to activate Rac, but not Rho, through the p130CAS-Crk-Dock180 pathway, whereas α5β1 integrin binding to fibronectin selectively activates Rho (62). Because CD151 predominantly associates with α3β1 and α6β1 integrins in epithelial cells rather than the α5β1 integrin, CD151 expression was expected to preferentially affect Rac signaling over Rho signaling. Indeed, a previous study showed that CD151 overexpression resulted in activation of Rac and Cdc42, but not Rho, in A431 cells (63). In the present study, we also found that CD151-transfected melanoma cells responded to cell adhesion to laminin for activation of Rac1 and Cdc42, but not Rho activation, indicating that CD151 contributes to preferential transduction of laminin-binding integrin signals to Rac and Cdc42 over Rho. The present study implicates that this differential activation of Rac and Cdc42 over Rho by laminin-binding integrin signaling is attributable to CD151-mediated selective association of α3β1/α6β1 integrins with Rac and Cdc42 among Rho family members. Additionally, the present study demonstrates that CD151-dependent activation of FAK and Src in integrin signaling occurs in cells adhered to laminin to a greater extent than fibronectin, which may be due to preferential association of CD151 with laminin-binding integrins.
Meanwhile, our current data seem to disagree with a previous report (59), where CD151 was identified as a negative regulator of Ras GTPase. Sawada et al. (59) showed that expression of CD151 in Rat-1 fibroblast cells did not only attenuate Ras activation but also diminished activation of Akt and ERK1/2, downstream targets in the Ras signaling pathway. Importantly, they observed a marked effect of CD151 expression on Ras inactivation only when integrins were disengaged from their ECM ligands, implying that cell detachment renders CD151 unavailable for Ras activation. In contrast, our present study demonstrates CD151-induced activation of Ras upon engagement of laminin-binding β1 integrins. In previous (42) and present studies, we also found that CD151 up-regulated integrin-mediated activation of FAK and Src, upstream effectors for Ras activation, in integrin signaling cascades. These opposite activities of CD151 toward Ras in terms of cell adhesion and de-adhesion may together be responsible for CD151-induced cell motility, because cell migration requires dynamic adhesion and de-adhesion repeats at the leading edge of extending lamellae. Therefore, both adhesion-dependent activation and de-adhesion-dependent inactivation of Ras by CD151 are expected to contribute to the migratory process by facilitating up- and down-regulation cycles of small GTPase-mediated development of the cell migration machinery, such as lamellipodia and filopodia.
We previously showed that homophilic interactions between CD151 proteins on the surface of neighboring cells provoke transmembrane signaling pathways in human melanoma cells, leading to increased MMP-9 expression through c-Jun activation (42). These CD151 signaling pathways were further stimulated by engagement of laminin-binding β1 integrins that are associated with CD151. These previous results strongly suggest that both cell-to-laminin adhesion, which activates α3β1/α6β1 integrins, and homotypic cell-to-cell adhesion, which generates homophilic CD151 interactions, contribute to CD151-β1 integrin adhesion receptor complex-mediated signaling cascades. In the present study, we found that cell adhesion to laminin resulted in activation of Ras, Rac1, and Cdc42 in various cell types with ectopically or endogenously expressed CD151. Importantly, the GTP-loaded levels of these GTPases in CD151-expressing cells with adhesion to laminin were further increased by treatment with CD151-containing membrane fragments but not by treatment with CD151-deficient membrane fragments. However, this positive effect of homophilic CD151 interactions on small GTPase activation was not observed in cells adhered to poly-l-lysine. Considering the data in our previous and present studies, we propose two functional roles of CD151 in outside-in adhesion signaling events. First, CD151 facilitates integrin-mediated signaling by linking β1 integrins to intracellular signaling molecules in the membrane, leading to efficient transduction of the cell-to-ECM adhesion signals into the cell. Second, CD151 integrates homotypic cell-to-cell adhesion signal into cell-to-ECM adhesion signaling cascades at the most upstream step by functioning as an adhesion co-receptor responding to cell-to-cell contacts that generate homophilic CD151 interactions. This proposed role of CD151 as an adhesion co-receptor that recognizes cell-to-cell adhesion is in agreement with its location on the cell surface at cell-cell junctions, although CD151 is mainly localized on the cell surface in contact with basement membranes in epithelial cells (5, 12). Several studies on CD81-mediated signaling events have already provided an example for tetraspanin proteins functioning as membrane receptors capable of provoking intracellular signaling cascades. When CD81 was cross-linked with an immobilized antibody or bound to hepatitis C virus E2 protein, the Rho GTPase family members Rac, Rho, and Cdc42 were shown to be activated in hepatocytes, along with the Raf/MEK/ERK signaling cascades (40). T lymphocytes also responded to CD81 cross-linking for Lck activation, leading to enhanced T-cell receptor/CD3 signaling (64). Interestingly, ERK activation by CD81 cross-linking also occurred in a fibroblast cell line lacking the β1 family of integrins (65), implying that CD81 signaling cascades occur independently of integrin engagement. Thus, tetraspanin CD81 has already been found to be capable of initiating its own intracellular signaling cascades, although cellular ligand(s) for CD81 are unknown. Taken altogether, we propose that the co-stimulatory effects of CD151 on integrin-mediated adhesion signaling depend not only on its ability to promote molecular association of integrins with intracellular signaling molecules, including small GTPases, but also on its role as an adhesion co-receptor that converges the cell-to-cell adhesion signal(s) on integrin-mediated cell-to-matrix adhesion signaling.
In summary, we here demonstrated for the first time that CD151 links the β1 family of integrins to small GTPases Ras, Rac1, and Cdc42 by facilitating the formation of CD151-α3β1/α6β1 integrin-GTPases complexes in the membrane. These β1 integrin-associated GTPases are able to receive cell-to-laminin adhesion signals quickly and efficiently. CD151 also participates in integrin-dependent activation of small GTPases by adding cell-to-cell adhesion signal(s) through its homophilic interactions to integrin-FAK-Src pathways upstream of the GTPases. Thus, both integrin interaction with laminin and homophilic interactions of CD151 contribute to CD151-β1 integrin complex-mediated adhesion signaling. Collectively, the data in this study suggest a novel mechanism by which integrins transduce adhesion signals to downstream small GTPases to induce GTPase-mediated cell adhesive behaviors. Moreover, the findings of CD151-β1 integrin complexes as an adhesion receptor signaling unit and interdependent signaling networks between CD151 and integrins provide a new insight into the cross-talk between cell-to-cell and cell-to-matrix adhesions.
Acknowledgments
We thank Elaine Por for corrections of the manuscript. We also thank Dr. Jae-Bong Park (Hallym University, Korea) for providing the RhoA, Rac1, and Cdc42 expression vectors.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST) (Grant 2011-0024380). This work was also supported by the Mid-career Researcher Program through an NRF grant funded by the MEST (Grant 2009-0054632).

This article contains supplemental Figs. S1–S4.
- ECM
- extracellular matrix
- Ab
- antibody
- FAK
- focal adhesion kinase
- MMP
- matrix metalloproteinase
- MSH-R
- melanocyte-stimulating hormone receptor
- RBD
- Rho-binding domain.
REFERENCES
- 1. Maecker H. T., Todd S. C., Levy S. (1997) The tetraspanin superfamily. Molecular facilitators. FASEB J. 11, 428–442 [PubMed] [Google Scholar]
- 2. Boucheix C., Rubinstein E. (2001) Tetraspanins. Cell Mol. Life Sci. 58, 1189–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sugiura T., Berditchevski F. (1999) Function of α3β1-tetraspanin protein complexes in tumor cell invasion. Evidence for the role of the complexes in production of matrix metalloproteinase 2 (MMP-2). J. Cell Biol. 146, 1375–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hemler M. E. (2001) Specific tetraspanin functions. J. Cell Biol. 155, 1103–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sincock P. M., Mayrhofer G., Ashman L. K. (1997) Localization of the transmembrane 4 superfamily (TM4SF) member PETA-3 (CD151) in normal human tissues. Comparison with CD9, CD63, and α5β1 integrin. J. Histochem. Cytochem. 45, 515–525 [DOI] [PubMed] [Google Scholar]
- 6. Testa J. E., Brooks P. C., Lin J. M., Quigley J. P. (1999) Eukaryotic expression cloning with an antimetastatic monoclonal antibody identifies a tetraspanin (PETA-3/CD151) as an effector of human tumor cell migration and metastasis. Cancer Res. 59, 3812–3820 [PubMed] [Google Scholar]
- 7. Tokuhara T., Hasegawa H., Hattori N., Ishida H., Taki T., Tachibana S., Sasaki S., Miyake M. (2001) Clinical significance of CD151 gene expression in non-small cell lung cancer. Clin. Cancer Res. 7, 4109–4114 [PubMed] [Google Scholar]
- 8. Kohno M., Hasegawa H., Miyake M., Yamamoto T., Fujita S. (2002) CD151 enhances cell motility and metastasis of cancer cells in the presence of focal adhesion kinase. Int. J. Cancer 97, 336–343 [DOI] [PubMed] [Google Scholar]
- 9. Hashida H., Takabayashi A., Tokuhara T., Hattori N., Taki T., Hasegawa H., Satoh S., Kobayashi N., Yamaoka Y., Miyake M. (2003) Clinical significance of transmembrane 4 superfamily in colon cancer. Br. J. Cancer 89, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ang J., Lijovic M., Ashman L. K., Kan K., Frauman A. G. (2004) CD151 protein expression predicts the clinical outcome of low-grade primary prostate cancer better than histologic grading. A new prognostic indicator? Cancer Epidemiol. Biomarkers Prev. 13, 1717–1721 [PubMed] [Google Scholar]
- 11. Sadej R., Romanska H., Baldwin G., Gkirtzimanaki K., Novitskaya V., Filer A. D., Krcova Z., Kusinska R., Ehrmann J., Buckley C. D., Kordek R., Potemski P., Eliopoulos A. G., Lalani el-N., Berditchevski F. (2009) CD151 regulates tumorigenesis by modulating the communication between tumor cells and endothelium. Mol. Cancer Res. 7, 787–798 [DOI] [PubMed] [Google Scholar]
- 12. Sterk L. M., Geuijen C. A., Oomen L. C., Calafat J., Janssen H., Sonnenberg A. (2000) The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin α6β4 and may regulate the spatial organization of hemidesmosomes. J. Cell Biol. 149, 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sterk L. M., Geuijen C. A., van den Berg J. G., Claessen N., Weening J. J., Sonnenberg A. (2002) Association of the tetraspanin CD151 with the laminin-binding integrins α3β1, α6β1, α6β4, and α7β1 in cells in culture and in vivo. J. Cell Sci. 115, 1161–1173 [DOI] [PubMed] [Google Scholar]
- 14. Chattopadhyay N., Wang Z., Ashman L. K., Brady-Kalnay S. M., Kreidberg J. A. (2003) α3β1 integrin-CD151, a component of the cadherin-catenin complex, regulates PTPmu expression and cell-cell adhesion. J. Cell Biol. 163, 1351–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stipp C. S., Kolesnikova T. V., Hemler M. E. (2003) Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28, 106–112 [DOI] [PubMed] [Google Scholar]
- 16. Nishiuchi R., Sanzen N., Nada S., Sumida Y., Wada Y., Okada M., Takagi J., Hasegawa H., Sekiguchi K. (2005) Potentiation of the ligand-binding activity of integrin α3β1 via association with tetraspanin CD151. Proc. Natl. Acad. Sci. U.S.A. 102, 1939–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hasegawa M., Furuya M., Kasuya Y., Nishiyama M., Sugiura T., Nikaido T., Momota Y., Ichinose M., Kimura S. (2007) CD151 dynamics in carcinoma-stroma interaction. Integrin expression, adhesion strength, and proteolytic activity. Lab. Invest. 87, 882–892 [DOI] [PubMed] [Google Scholar]
- 18. Yang X. H., Richardson A. L., Torres-Arzayus M. I., Zhou P., Sharma C., Kazarov A. R., Andzelm M. M., Strominger J. L., Brown M., Hemler M. E. (2008) CD151 accelerates breast cancer by regulating α6 integrin function, signaling, and molecular organization. Cancer Res. 68, 3204–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lau L. M., Wee J. L., Wright M. D., Moseley G. W., Hogarth P. M., Ashman L. K., Jackson D. E. (2004) The tetraspanin superfamily member CD151 regulates outside-in integrin αIIbβ3 signaling and platelet function. Blood 104, 2368–2375 [DOI] [PubMed] [Google Scholar]
- 20. Yauch R. L., Berditchevski F., Harler M. B., Reichner J., Hemler M. E. (1998) Highly stoichiometric, stable, and specific association of integrin α3β1 with CD151 provides a major link to phosphatidylinositol 4-kinase and may regulate cell migration. Mol. Biol. Cell 9, 2751–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X. A., Bontrager A. L., Hemler M. E. (2001) Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific β1 integrins. J. Biol. Chem. 276, 25005–25013 [DOI] [PubMed] [Google Scholar]
- 22. Clark E. A., King W. G., Brugge J. S., Symons M., Hynes R. O. (1998) Integrin-mediated signals regulated by members of the rho family of GTPases. J. Cell Biol. 142, 573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwartz M. A., Ginsberg M. H. (2002) Networks and cross-talk. Integrin signaling spreads. Nat. Cell Biol. 4, E65–68 [DOI] [PubMed] [Google Scholar]
- 24. Yamada K. M., Even-Ram S. (2002) Integrin regulation of growth factor receptors. Nat. Cell Biol. 4, E75–E76 [DOI] [PubMed] [Google Scholar]
- 25. Kinbara K., Goldfinger L. E., Hansen M., Chou F. L., Ginsberg M. H. (2003) Ras GTPases. Integrins' friends or foes? Nat. Rev. Mol. Cell Biol. 4, 767–776 [DOI] [PubMed] [Google Scholar]
- 26. Bos J. L. (2005) Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17, 123–128 [DOI] [PubMed] [Google Scholar]
- 27. DeMali K. A., Balciunaite E., Kazlauskas A. (1999) Integrins enhance platelet-derived growth factor (PDGF)-dependent responses by altering the signal relay enzymes that are recruited to the PDGF β receptor. J. Biol. Chem. 274, 19551–19558 [DOI] [PubMed] [Google Scholar]
- 28. Price L. S., Leng J., Schwartz M. A., Bokoch G. M. (1998) Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell 9, 1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kjoller L., Hall A. (1999) Signaling to Rho GTPases. Exp. Cell Res. 253, 166–179 [DOI] [PubMed] [Google Scholar]
- 30. Ren X. D., Kiosses W. B., Schwartz M. A. (1999) Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18, 578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Danen E. H., Sonneveld P., Sonnenberg A., Yamada K. M. (2000) Dual stimulation of Ras/mitogen-activated protein kinase and RhoA by cell adhesion to fibronectin supports growth factor-stimulated cell cycle progression. J. Cell Biol. 151, 1413–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Etienne-Manneville S., Hall A. (2001) Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCζ. Cell 106, 489–498 [DOI] [PubMed] [Google Scholar]
- 33. Russell A. J., Fincher E. F., Millman L., Smith R., Vela V., Waterman E. A., Dey C. N., Guide S., Weaver V. M., Marinkovich M. P. (2003) α6β4 integrin regulates keratinocyte chemotaxis through differential GTPase activation and antagonism of α3β1 integrin. J. Cell Sci. 116, 3543–3556 [DOI] [PubMed] [Google Scholar]
- 34. Choma D. P., Pumiglia K., DiPersio C. M. (2004) Integrin α3β1 directs the stabilization of a polarized lamellipodium in epithelial cells through activation of Rac1. J. Cell Sci. 117, 3947–3959 [DOI] [PubMed] [Google Scholar]
- 35. Boudreau N. J., Jones P. L. (1999) Extracellular matrix and integrin signaling. The shape of things to come. Biochem. J. 339, 481–488 [PMC free article] [PubMed] [Google Scholar]
- 36. Arthur W. T., Noren N. K., Burridge K. (2002) Regulation of Rho family GTPases by cell-cell and cell-matrix adhesion. Biol. Res. 35, 239–246 [DOI] [PubMed] [Google Scholar]
- 37. Hall A. (2005) Rho GTPases and the control of cell behavior. Biochem. Soc. Trans. 33, 891–895 [DOI] [PubMed] [Google Scholar]
- 38. Choma D. P., Milano V., Pumiglia K. M., DiPersio C. M. (2007) Integrin α3β1-dependent activation of FAK/Src regulates Rac1-mediated keratinocyte polarization on laminin-5. J. Invest. Dermatol. 127, 31–40 [DOI] [PubMed] [Google Scholar]
- 39. Huveneers S., Danen E. H. (2009) Adhesion signaling. Cross-talk between integrins, Src, and Rho. J. Cell Sci. 122, 1059–1069 [DOI] [PubMed] [Google Scholar]
- 40. Brazzoli M., Bianchi A., Filippini S., Weiner A., Zhu Q., Pizza M., Crotta S. (2008) CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 82, 8316–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berrier A. L., Mastrangelo A. M., Downward J., Ginsberg M., LaFlamme S. E. (2000) Activated R-Ras, Rac1, PI 3-kinase, and PKCϵ can each restore cell spreading inhibited by isolated integrin β1 cytoplasmic domains. J. Cell Biol. 151, 1549–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hong I. K., Jin Y. J., Byun H. J., Jeoung D. I., Kim Y. M., Lee H. (2006) Homophilic interactions of Tetraspanin CD151 up-regulate motility and matrix metalloproteinase-9 expression of human melanoma cells through adhesion-dependent c-Jun activation signaling pathways. J. Biol. Chem. 281, 24279–24292 [DOI] [PubMed] [Google Scholar]
- 43. Duxbury M. S., Ito H., Zinner M. J., Ashley S. W., Whang E. E. (2004) CEACAM6 gene silencing impairs anoikis resistance and in vivo metastatic ability of pancreatic adenocarcinoma cells. Oncogene 23, 465–473 [DOI] [PubMed] [Google Scholar]
- 44. Reid T., Furuyashiki T., Ishizaki T., Watanabe G., Watanabe N., Fujisawa K., Morii N., Madaule P., Narumiya S. (1996) Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the Rho-binding domain. J. Biol. Chem. 271, 13556–13560 [DOI] [PubMed] [Google Scholar]
- 45. Benard V., Bohl B. P., Bokoch G. M. (1999) Characterization of Rac and Cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J. Biol. Chem. 274, 13198–13204 [DOI] [PubMed] [Google Scholar]
- 46. Lee H., Ghose-Dastidar J., Winawer S., Friedman E. (1993) Signal transduction through extracellular signal-regulated kinase-like pp57 blocked in differentiated cells having low protein kinase C β activity. J. Biol. Chem. 268, 5255–5263 [PubMed] [Google Scholar]
- 47. Charrin S., Manié S., Billard M., Ashman L., Gerlier D., Boucheix C., Rubinstein E. (2003) Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 304, 107–112 [DOI] [PubMed] [Google Scholar]
- 48. Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F. (2009) Tetraspanin-enriched microdomains. A functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 [DOI] [PubMed] [Google Scholar]
- 49. Stipp C. S., Kolesnikova T. V., Hemler M. E. (2003) EWI-2 regulates α3β1 integrin-dependent cell functions on laminin-5. J. Cell Biol. 163, 1167–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Charrin S., le Naour F., Silvie O., Milhiet P. E., Boucheix C., Rubinstein E. (2009) Lateral organization of membrane proteins. Tetraspanins spin their web. Biochem. J. 420, 133–154 [DOI] [PubMed] [Google Scholar]
- 51. Vleggeert-Lankamp C. L., Pêgo A. P., Lakke E. A., Deenen M., Marani E., Thomeer R. T. (2004) Adhesion and proliferation of human Schwann cells on adhesive coatings. Biomaterials 25, 2741–2751 [DOI] [PubMed] [Google Scholar]
- 52. Chen G., Kawazoe N., Tateishi T. (2008) Effects of ECM proteins and cationic polymers on the adhesion and proliferation of rat islet cells. Open Biotech. J. 2, 133–137 [Google Scholar]
- 53. Yarwood S. J., Woodgett J. R. (2001) Extracellular matrix composition determines the transcriptional response to epidermal growth factor receptor activation. Proc. Natl. Acad. Sci. U.S.A. 98, 4472–4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ridley A. J., Hall A. (1992) The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 55. Nobes C. D., Hall A. (1995) Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62 [DOI] [PubMed] [Google Scholar]
- 56. Schwartz M. A., Shattil S. J. (2000) Signaling networks linking integrins and Rho family GTPases. Trends Biochem. Sci. 25, 388–391 [DOI] [PubMed] [Google Scholar]
- 57. Yánez-Mó M., Mittelbrunn M., Sánchez-Madrid F. (2001) Tetraspanins and intercellular interactions. Microcirculation 8, 153–168 [DOI] [PubMed] [Google Scholar]
- 58. Berditchevski F. (2001) Complexes of tetraspanins with integrins. More than meets the eye. J. Cell Sci. 114, 4143–4151 [DOI] [PubMed] [Google Scholar]
- 59. Sawada S., Yoshimoto M., Odintsova E., Hotchin N. A., Berditchevski F. (2003) The tetraspanin CD151 functions as a negative regulator in the adhesion-dependent activation of Ras. J. Biol. Chem. 278, 26323–26326 [DOI] [PubMed] [Google Scholar]
- 60. Zhang X. A., Kazarov A. R., Yang X., Bontrager A. L., Stipp C. S., Hemler M. E. (2002) Function of the tetraspanin CD151-α6β1 integrin complex during cellular morphogenesis. Mol. Biol. Cell 13, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zevian S., Winterwood N. E., Stipp C. S. (2011) Structure-function analysis of tetraspanin CD151 reveals distinct requirements for tumor cell behaviors mediated by α3β1 versus α6β4 integrin. J. Biol. Chem. 286, 7496–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gu J., Sumida Y., Sanzen N., Sekiguchi K. (2001) Laminin-10/11 and fibronectin differentially regulate integrin-dependent Rho and Rac activation via p130Cas-CrkII-DOCK180 pathway. J. Biol. Chem. 276, 27090–27097 [DOI] [PubMed] [Google Scholar]
- 63. Shigeta M., Sanzen N., Ozawa M., Gu J., Hasegawa H., Sekiguchi K. (2003) CD151 regulates epithelial cell-cell adhesion through PKC- and Cdc42-dependent actin cytoskeletal reorganization. J. Cell Biol. 163, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Soldaini E., Wack A., D'Oro U., Nuti S., Ulivieri C., Baldari C. T., Abrignani S. (2003) T cell costimulation by the hepatitis C virus envelope protein E2 binding to CD81 is mediated by Lck. Eur. J. Immunol. 33, 455–464 [DOI] [PubMed] [Google Scholar]
- 65. Carloni V., Mazzocca A., Ravichandran K. S. (2004) Tetraspanin CD81 is linked to ERK/MAPKinase signaling by Shc in liver tumor cells. Oncogene 23, 1566–1574 [DOI] [PubMed] [Google Scholar]