Background: Three-dimensional architecture of the CFTR channel is not well defined.
Results: Different patterns of disulfide cross-linking between cysteine residues introduced into two transmembrane segments are observed in open and closed channels.
Conclusion: The alignment of transmembrane segments changes during gating, consistent with translational movement of these segments.
Significance: These findings illuminate three-dimensional structural rearrangements during CFTR channel opening.
Keywords: ABC Transporter, Anion Transport, CFTR, Chloride Channels, Ion Channels, Ion Channel Structure and Function
Abstract
Multiple transmembrane (TM) segments line the pore of the cystic fibrosis transmembrane conductance regulator Cl− channel; however, the relative alignment of these TMs and their relative movements during channel gating are unknown. To gain three-dimensional structural information on the outer pore, we have used patch clamp recording to study the proximity of pairs of cysteine side chains introduced into TMs 6 and 11, using both disulfide cross-linking and Cd2+ coordination. Following channel activation, disulfide bonds could apparently be formed between three cysteine pairs (of 15 studied): R334C/T1122C, R334C/G1127C, and T338C/S1118C. To examine the state dependence of cross-linking, we combined these cysteine mutations with a nucleotide-binding domain mutation (E1371Q) that stabilizes the channel open state. Investigation of the effects of the E1371Q mutation on disulfide bond formation and Cd2+ coordination suggests that although R334C/T1122C and T338C/S1118C are closer together in the channel open state, R334C/G1127C are close together and can form disulfide bonds only when the channel is closed. These results provide important new information on the three-dimensional structure of the outer mouth of the cystic fibrosis transmembrane conductance regulator channel pore: TMs 6 and 11 are close enough together to form disulfide bonds in both open and closed channels. Moreover, the altered relative locations of residues in open and in closed channels that we infer allow us to propose that channel opening and closing may be associated with a relative translational movement of TMs 6 and 11, with TM6 moving “down” (toward the cytoplasm) during channel opening.
Introduction
Cystic fibrosis is caused by genetic mutations that result in loss of function of the cystic fibrosis transmembrane conductance regulator (CFTR)3 protein (1). CFTR is a member of the ATP-binding cassette (ABC) family of membrane proteins, most of which function as ATP-dependent active transporters (2). CFTR is unique within the ABC family in functioning as a phosphorylation-regulated, ATP-gated Cl− channel (3). CFTR is essential for salt and water transport in many epithelial tissues, consistent with the pleiotropic symptoms associated with cystic fibrosis (1).
In common with other ABC proteins, CFTR has a modular architecture, consisting of two membrane-spanning domains (MSDs) each followed by a cytoplasmic nucleotide-binding domain (NBD). CFTR also has a unique cytoplasmic regulatory domain (or R domain) linking the two MSD-NBD halves of the protein. Physiological regulation of CFTR is by phosphorylation and dephosphorylation of the R domain, predominantly at protein kinase A phosphorylation sites, with phosphorylation being an absolute prerequisite for channel activation (3). Once phosphorylated, channel opening and closing are controlled by ATP interactions with the two NBDs (4, 5) (see supplemental materials). The transmembrane channel pore is formed by the two MSDs, each of which contains six transmembrane α-helices (TMs 1–12) (6). Opening and closing of the channel pore is thought to reflect conformational rearrangements of the TMs that are controlled by ATP interactions with the cytoplasmic NBDs (7–9) that are analogous to the conformational changes in the substrate translocation pathway that underlie ATP-driven transport in canonical ABC protein transport mechanisms (2, 10, 11).
The structure of the MSDs in CFTR has been observed only at low resolution (12). However, functional evidence has indicated that TMs 1, 5, 6, 11, and 12 contribute to the lining of the pore and interact with Cl− ions (6, 7, 13–16). Extracellular loops between TMs 1 and 2 (ECL1) and TMs 11 and 12 (ECL6) have also been proposed to contribute to the outer mouth of the channel pore (15, 17). However, for the most part the structure and function of these parts of the protein have been studied in isolation, and there is very little direct functional information relating to the three-dimensional arrangement of different TMs (18, 19).
Consistent with the idea that channel opening and closing reflect conformational changes in the channel pore, previous studies have identified changes in the accessibility of individual amino acid side chains in the TMs to either the cytoplasm or the extracellular solution. These accessibility changes may be associated either with activation/phosphorylation (16, 19–21) or with ATP-dependent channel gating (7–9, 22–24) (see supplemental materials). Our recent work has suggested that ATP-dependent channel opening is associated with (i) opening of a “gate” between the pore inner vestibule and narrow regions (8), and (ii) constriction of the outer mouth of the pore (9). However, because previous studies have been limited to changes in the accessibility of single sites, relative motion of different parts of the protein during these putative rearrangements of the MSDs has not previously been identified.
The outer mouth of the CFTR pore is lined both by TM6 (6, 23, 25) and the TM11-ECL6-TM12 loop (15). In the present study, we have sought to understand the three-dimensional arrangement of these two parts of the protein by engineering disulfide cross-links between introduced cysteine residues. Furthermore, we have investigated the dependence of cross-link formation on different channel protein conformations, associated both with channel activation (by phosphorylation) as well as ATP-dependent channel opening (see supplemental materials). In addition to identifying the proximity and alignment of TMs 6 and 11, our results suggest that opening and closing of the channel pore are associated with a change in alignment, suggestive of a relative translational movement of these two TMs.
EXPERIMENTAL PROCEDURES
Experiments were carried out on baby hamster kidney cells transiently transfected with CFTR. In this study we have used a human CFTR variant in which all cysteines had been removed by mutagenesis (as described in Ref. 26) and that includes a mutation in the first NBD (V510A) to increase protein expression in the cell membrane (27). This Cys-less variant, which we have used in previous studies of pore conformational change (8, 9), has functional properties that are very similar to those of wild-type CFTR (28). Additional mutations were introduced into this Cys-less background using the QuikChange site-directed mutagenesis system (Agilent Technologies, Santa Clara, CA) and verified by DNA sequencing. To engineer disulfide cross-links between different TMs, we combined individual cysteine substitutions at sites that have previously been shown to line the channel pore and to react with externally applied cysteine-reactive reagents. In TM6, we selected three functionally important residues: Arg-334 and Lys-335, which act to attract extracellular Cl− ions electrostatically to the pore (17, 29), and Thr-338, which lines the narrowest part of the pore (6) and contributes to a region of high resistance to Cl− flux (30). Each of these three residues have been proposed to show conformation-dependent access from the extracellular solution (9, 22, 23). The three TM6 mutants R334C, K335C and T338C were combined with cysteine substitution of five externally accessible TM11-ECL6-TM12 residues (S1118C, T1121C, T1122C, G1127C, and T1134C) (15, 25) to give a total of 15 cysteine pair mutants (see Figs. 1 and 7). In some cases, cysteine mutants were combined with the NBD2 mutation E1371Q, which we (8, 9) and others (7, 23) have used to abolish ATP-dependent channel gating and lock CFTR channels in the open state (see supplemental materials).
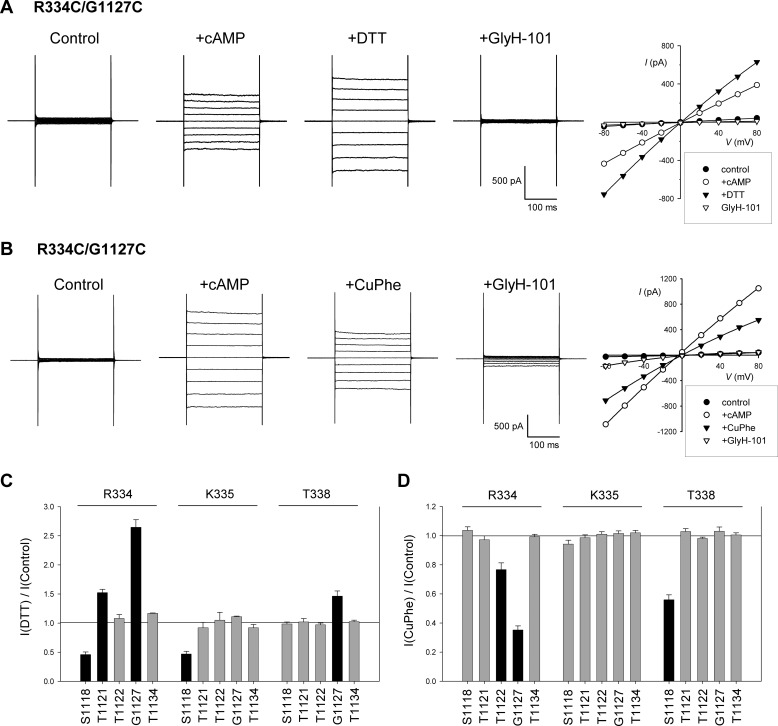
FIGURE 1.
Spontaneous and oxidant-induced disulfide bond formation between cysteine side chains in TM6 and TM11. A and B, example whole cell current recordings for R334C/G1127C during voltage steps (−80 mV to +80 mV in 20-mV increments from a holding potential of 0 mV). Currents were recorded after attainment of a stable whole cell configuration (control), after application of cAMP stimulatory mixture (see “Experimental Procedures”) (+cAMP), after application of DTT (A) or CuPhe (B), and after CFTR channel inhibition by addition of 50 μm GlyH-101. Current-voltage relationships from these two cells, measured from the final 100 ms of the voltage steps for each of the four sets of current traces shown, are illustrated to the right. C and D, mean effect of DTT (C) or CuPhe (D) application on whole cell current amplitude for each of the 15 double cysteine mutants studied. In both cases, the TM6 residue substituted by cysteine (Arg-334, Lys-335, or Thr-338) is named above the panel, and the TM11-ECL6-TM12 residue substituted is below the panel. In both C and D, the mutants in which treatment with DTT or CuPhe resulted in a significant change in current amplitude (p < 0.05) are represented by black bars, and those mutants for which there was no significant change are represented by gray bars. The means of data from three to six cells are shown in C and D.
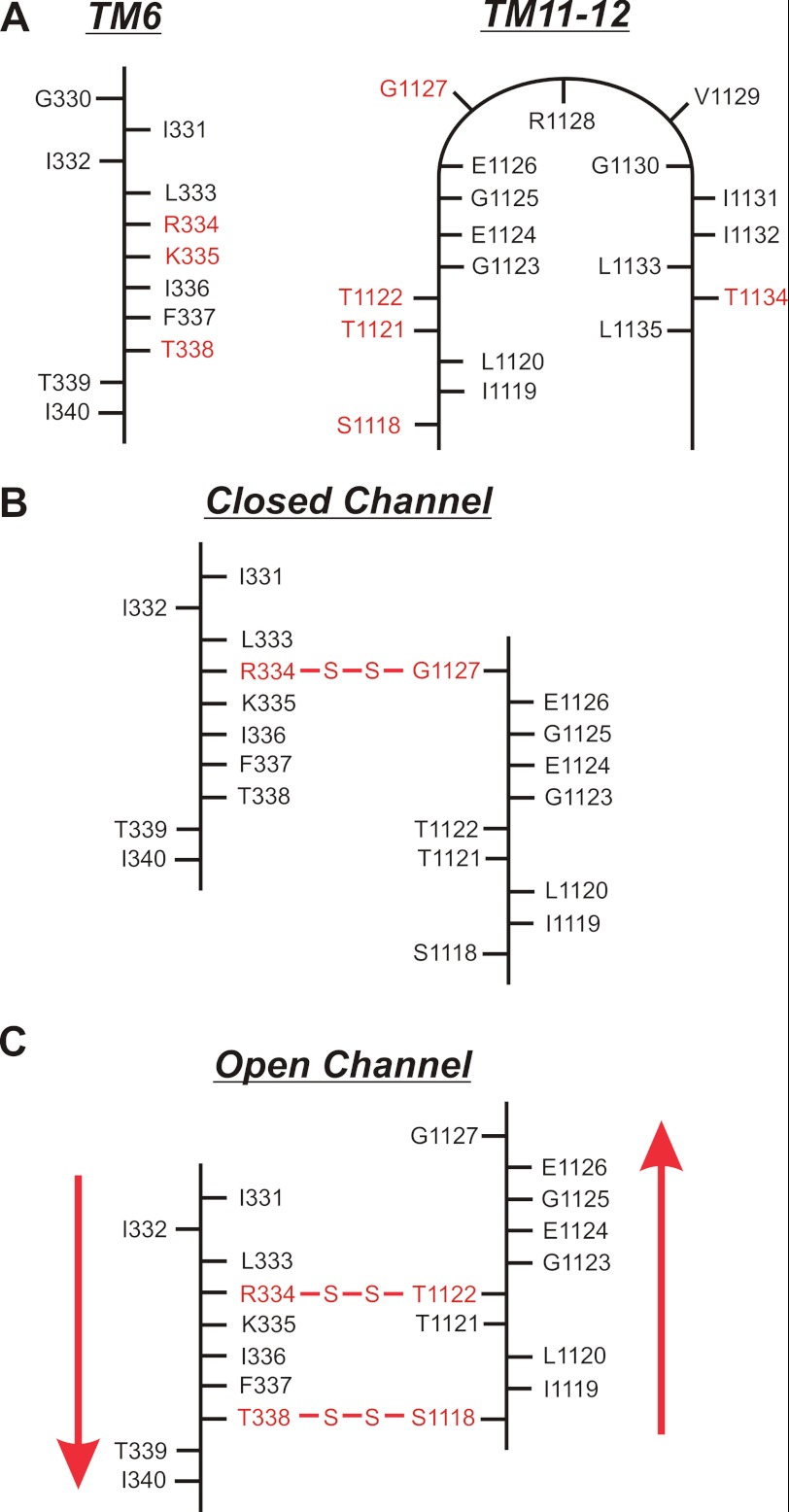
FIGURE 7.
Proposed alignment and relative movement of TMs 6 and 11 during channel opening and closing. A, two-dimensional location of mutated residues in TM6 (left panel) and TM11-ECL6-TM12 (right panel). For TM6, side chains accessible to the extracellular solution are to the right, and nonaccessible side chains are to the left (21, 23, 29, 40). For TM11–12, accessible side chains are outside the loop, and nonaccessible side chains are inside (15, 25). Residues mutated in the present study are in red. Functional evidence suggests that other accessible TM11–12 residues Val-1129, Ile-1131, and Ile-1132 are located at more superficial positions in the pore outer mouth (15). B and C, proposed location of disulfide bonds formed in closed (B) and open (C) channels and inferred relative alignment of TMs 6 and 11 in these two states (Table 1). The suggested relative movement of these two TMs is indicated by the red arrows in C; when the channel opens, TM6 moves relatively “downward” (toward the cytoplasm) and/or TM11 moves relatively “upward” (toward the extracellular solution).
The proximity of cysteine side chains introduced into different parts of the protein was identified functionally using patch clamp recording. Formation of disulfide bonds between two cysteine side chains was inferred from changes in channel function resulting from treatment with the oxidizing agent copper(II)-o-phenanthroline (CuPhe) and/or the reducing agent DTT. The proximity of two cysteine side chains was also determined by tight binding of externally applied Cd2+ ions, resulting in inhibition of channel function. The functional consequences of both CuPhe-induced cross-linking and Cd2+ coordination have previously been used to infer distances between pairs of cysteine side chains of <10 Å in other channel types (31–35). Furthermore, both CuPhe-induced cysteine cross-linking (18, 19) and Cd2+ inhibition of single cysteine mutants (23) have been investigated in the CFTR channel pore using electrophysiological recording.
Whole cell patch clamp recordings were made as described in our recent study (9). Bath (extracellular) solution contained 145 mm NaCl, 15 mm sodium glutamate, 4.5 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 10 mm HEPES, 5 mm glucose, pH 7.4, and pipette (intracellular) solution contained 139 mm CsCl, 2 mm MgCl2, 5 mm EGTA, 10 mm HEPES, 5 mm glucose, 1 mm ATP, 0.1 mm GTP, pH 7.2. Whole cell currents were monitored using voltage steps from a holding potential of 0 mV (Figs. 1 and 4) or continuously at a membrane potential of +30 mV (Figs. 2, 3, 5, and 6). Following attainment of the whole cell configuration and recording of stable baseline currents, CFTR channels were activated by extracellular application of a cyclic AMP stimulatory mixture containing forskolin (FSK; 10 μm), 3-isobutyl-1-methylxanthine (100 μm), and 8-(4-chlorophenylthio) cyclic AMP (100 μm). At the end of the experiment, remaining currents were confirmed as being carried by CFTR by their sensitivity to the specific CFTR inhibitor GlyH-101. As described previously (9, 18), channels bearing the E1371Q mutation were constitutively active, and whole cell currents carried by such channels were not further increased in amplitude by application of cAMP mixture, although they were sensitive to GlyH-101. CuPhe was prepared freshly before each experiment by mixing stock solutions of CuSO4 (200 mm in distilled water) with 1,10-phenanthroline (200 mm in ethanol) in a 1:4 molar ratio. The final concentration used to treat cells was 100 μm Cu2+, 400 μm phenanthroline (18). In preliminary experiments Cd2+ was applied to the extracellular solution at different concentrations to generate concentration-inhibition relationships; in all cases, these were consistent with a simple, reversible inhibition of the channel by Cd2+ ions. However, for clarity in the experiments illustrated, only results with a single, representative concentration of Cd2+ are shown.
FIGURE 4.
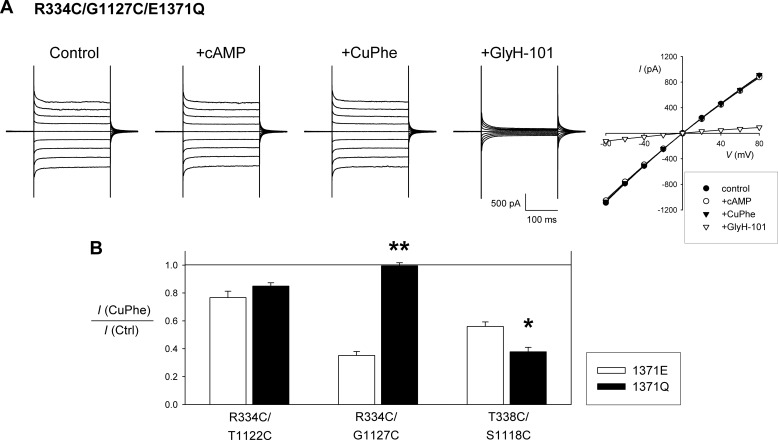
Disulfide bond formation in constitutively open channels. A, example whole cell current recording and corresponding current-voltage relationship for R334C/G1127C/E1371Q, illustrating a lack of inhibition by CuPhe. Compare with R334C/G1127C in Fig. 1 under identical experimental conditions. Note the overlapping current-voltage relationships for control (●), +cAMP (○), and +CuPhe (▾). B, mean effect of CuPhe treatment on whole cell current amplitude for the three CuPhe-sensitive double mutants named, with either the native Glu present at position 1371 (1371E, white bars) or with Gln substituted at this position (1371Q, black bars). Asterisks indicate a significant difference from the effect in a 1371E background. *, p < 0.05; **, p < 0.00001. The means of data from three to six cells are shown. Ctrl, control.
FIGURE 2.
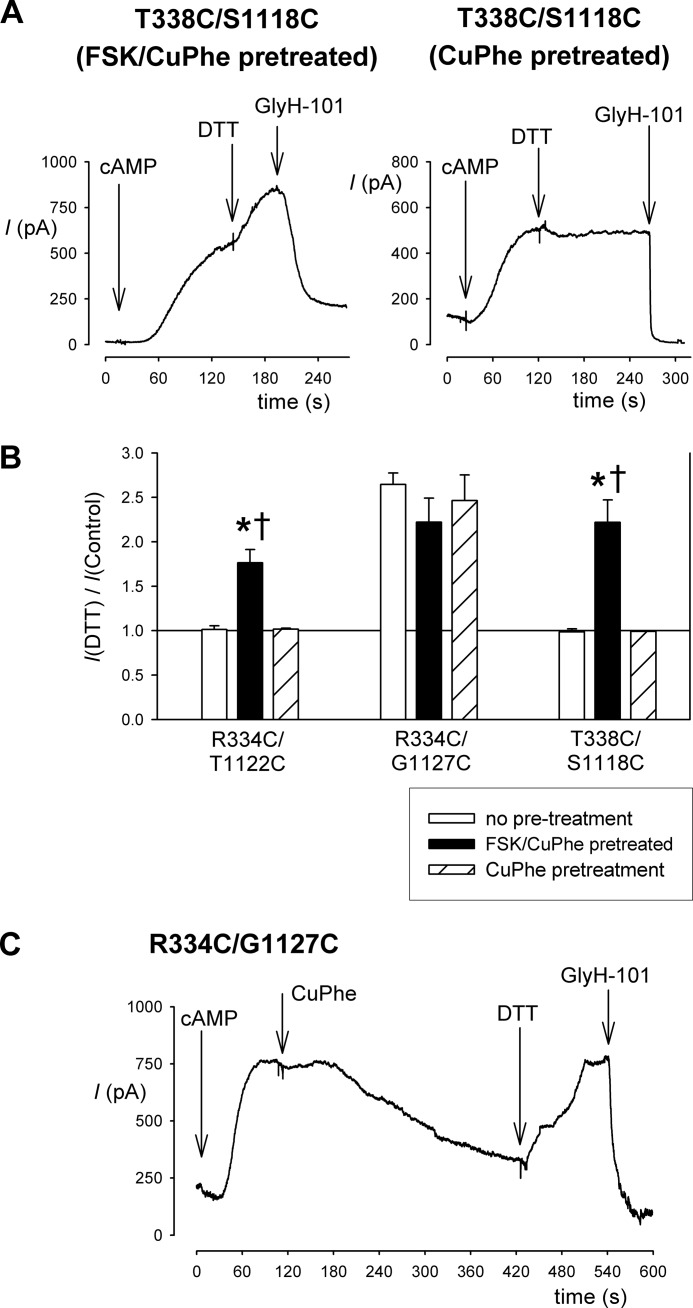
Irreversibility of CuPhe inhibition in double cysteine mutants. A, example whole cell currents recorded at +30 mV for T338C/S1118C from cells that had been pretreated with CuPhe either together with FSK (left panel) or without FSK (right panel) for 5 min. Following extensive washing and attainment of the whole cell patch configuration, channel activation by application of cAMP stimulatory mixture revealed currents that were increased by DTT application (5 mm) only if CuPhe was applied together with FSK. In each case, the identity of CFTR currents was confirmed using the specific inhibitor GlyH-101 (50 μm). B, mean effect of DTT application on whole cell current amplitude for the three double cysteine mutants named, following different pretreatment conditions. Asterisks indicate a significant difference from no pretreatment conditions (p < 0.01), and daggers indicate a significant difference from pretreatment with CuPhe in the absence of FSK (p < 0.01). The means of data from three to six cells are shown. C, stability of CuPhe inhibition of R334C/G1127C and reversibility by excess DTT. Example whole cell current from a cell treated sequentially with cAMP stimulatory mixture, CuPhe, DTT, and GlyH-101.
FIGURE 3.
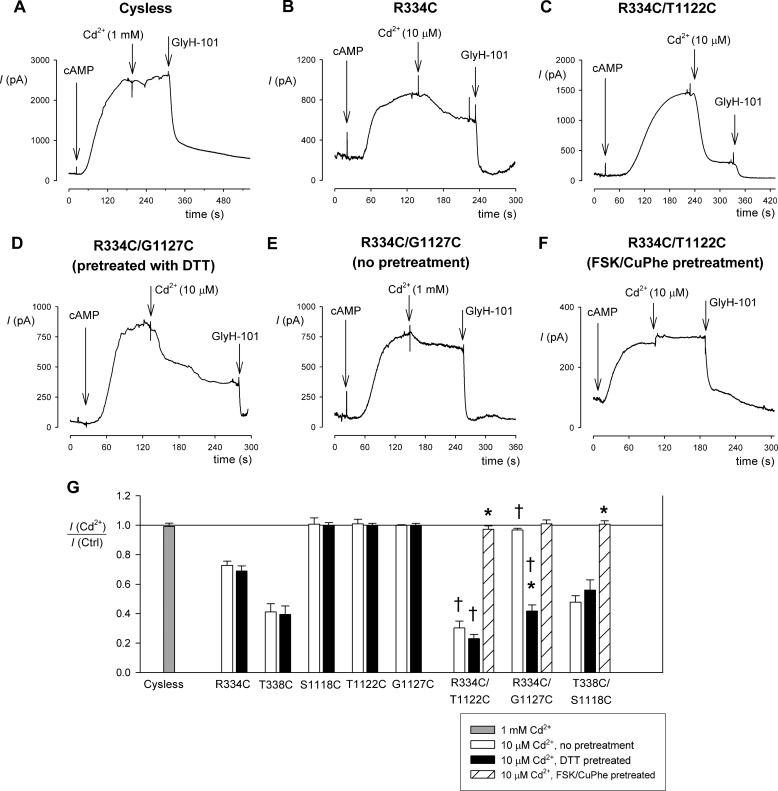
Cadmium inhibition of single and double cysteine mutants. A–F, example whole cell currents recorded at +30 mV for Cys-less (A), R334C (B), R334C/T1122C (C and F), and R334C/G1127C (D and E). CFTR currents were activated by application of cAMP stimulatory mixture in each case, prior to the addition of Cd2+ to the extracellular solution at the concentrations illustrated. The cells were pretreated with DTT (5 mm) except where stated otherwise (E and F). G, mean inhibitory effect of Cd2+ on different channel variants under different conditions as illustrated. Asterisks indicate a significant difference from the same channel variant without pretreatment (p < 0.0001), and daggers indicate a significant difference from the corresponding single TM6 mutant (R334C or T338C) under the same pretreatment conditions (p < 0.005). Note that CuPhe-induced disulfide formation prevented strong inhibition by Cd2+ ions in all double cysteine mutants tested. The means of data from three to five cells are shown. Ctrl, control.
FIGURE 5.
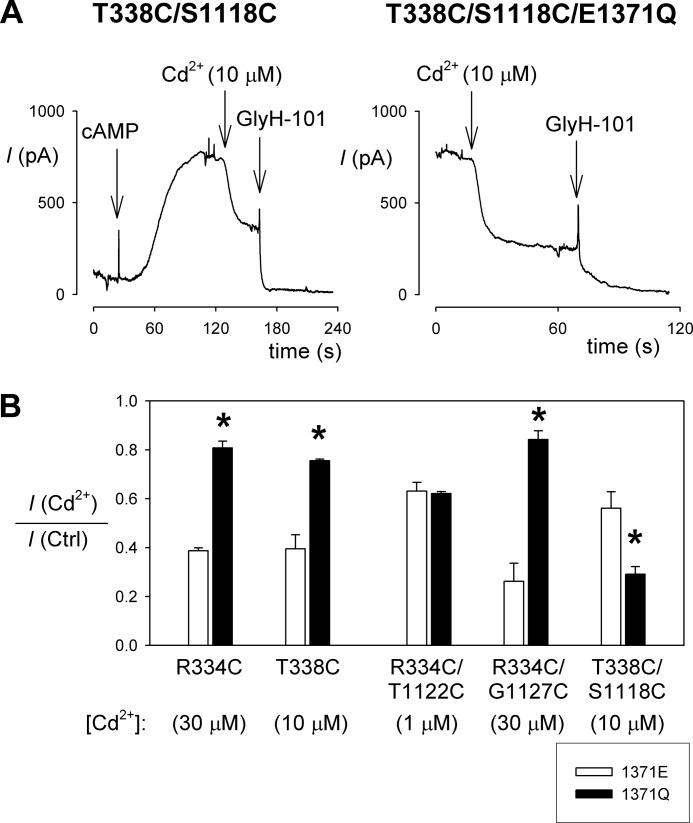
Cadmium inhibition in constitutively open channels. A, example whole cell currents recorded at +30 mV for T338C/S1118C (left panel) and T338C/S1118C/E1371Q (right panel), showing the increased sensitivity to inhibition by external Cd2+ (10 μm) in E1371Q-containing channels. B, effect of the E1371Q mutation on Cd2+ sensitivity in single and double cysteine mutants. Cadmium concentrations, chosen to give ∼50% inhibition for each mutant, are given below the name of the mutant used. Asterisks indicate a significant difference from the effect in a Glu-1371 background (p < 0.05). The means of data from three to four cells are shown. Ctrl, control.
FIGURE 6.
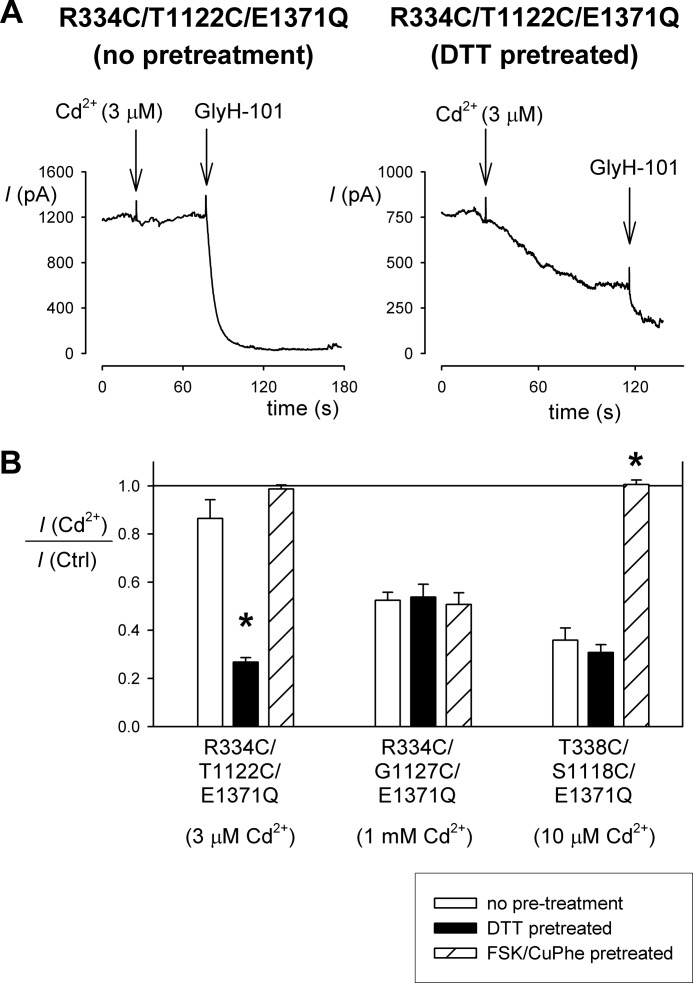
Evidence for spontaneous disulfide bond formation in constitutively open channels. A, example whole cell currents recorded at +30 mV for R334C/T1122C/E1371Q, showing that inhibition by external Cd2+ (3 μm) is dependent on prior treatment with DTT to reduce spontaneous disulfide bonds. B, dependence of Cd2+ inhibition on different pretreatment conditions in different cysteine double mutants in an E1371Q background. Cadmium concentrations, chosen to give ∼50% inhibition for each mutant, are given below the name of the mutant used. Asterisks indicate a significant difference from the effect without pretreatment (p < 0.0005). The means of data from three to six cells are shown. Note that the Cd2+ sensitivity of the single cysteine mutants R334C/E1371Q and T338C/E1371Q was independent of DTT pretreatment (not shown), as shown in Fig. 3 for R334C and T338C. Ctrl, control.
Experiments were carried out at room temperature (21–24 °C). The values are presented as the means ± S.E. Unless stated otherwise, tests of significance were carried out using an unpaired t test, with p < 0.05 being considered statistically significant. All chemicals were from Sigma-Aldrich, except for GlyH-101 (EMD Chemicals, Gibbstown, NJ).
RESULTS
Disulfide Cross-linking between Different TMs
To investigate the potential for disulfide bonds to form between different TMs, we introduced pairs of cysteines into TM6 and TM11/12 (see Fig. 7A). The ability of these pairs of cysteines to form disulfide bonds was investigated functionally using whole cell recording (Fig. 1). Two different protocols were used to identify disulfide formation. First, channels were activated using cAMP, following which DTT was applied to reduce any disulfide bonds that might have formed spontaneously (Fig. 1A). Second, after cAMP stimulation, the cells were treated with CuPhe to catalyze the formation of disulfide bonds (Fig. 1B). In most cases, neither DTT nor CuPhe affected whole cell current amplitude (Fig. 1, C and D), suggesting either that disulfide bond formation did not occur or that these bonds were without significant functional consequence. However, DTT treatment caused a significant change in current amplitude in five mutants: R334C/G1127C (Fig. 1, A and C), R334C/S1118C, R334C/T1121C, K335C/S1118C, and T338C/G1127C (Fig. 1C). This suggests that these pairs of cysteine residues were cross-linked prior to channel activation and that although the presence of the cross-links did not interfere with cAMP activation, they altered channel function in some way such that breaking the disulfide bond with DTT led to either a decrease (R334C/S1118C, K335C/S1118C) or an increase (R334C/T1121C, R334C/G1127C, and T338C/G1127C) in overall current amplitude. In the absence of single channel recordings, it is not clear whether these disulfide bonds affect channel open probability, channel conductance, or both. The fact that so many paired mutants (with at least one pair including each individual cysteine substituted) failed to respond to DTT effectively rules out DTT effects on individual cysteine side chains not involved in disulfide bond formation.
To investigate disulfide bond formation after channel activation, cells were treated with CuPhe (Fig. 1B). Note that in this series of experiments, the cells were pretreated with DTT (5 mm) to ensure reduction of any spontaneously formed disulfide bonds. In most cases, CuPhe treatment failed to affect whole cell current amplitude (Fig. 1D), again suggesting that, in active channels, disulfide bonds could not readily form, or they were without functional effect. However, CuPhe treatment led to a rapid inhibition of current amplitude in R334C/G1127C (Fig. 1, B and D), R334C/T1122C, and T338C/S1118C (Fig. 1D), suggesting that disulfide bond formation somehow impeded normal channel function; again, this may reflect changes in channel open probability and/or conductance. As with DTT treatment (see above), the preponderance of negative results (Fig. 1D) indicates that CuPhe specifically induced disulfide bond formation rather than exerting its functional effects by interactions with any specific individual cysteine side chain. For example, CuPhe inhibited R334C/T1122C and T338C/S1118C channel function, but had no effect on R334C/S1118C or T338C/T1122C.
To demonstrate the irreversibility and stability of CuPhe-induced disulfide bonds in R334C/T1122C. R334C/G1127C and T338C/S1118C, we pretreated cells to induce disulfide bond formation and then tested their reversibility by DTT treatment (Fig. 2). The cells were exposed to CuPhe with or without FSK (10 μm) for 5 min to promote disulfide bond formation. Following pretreatment, the cells were washed with normal bath solution for at least 5 min to remove the CuPhe and FSK before being transferred to the patch clamp recording chamber. Following attainment of the whole cell patch configuration, the cells were stimulated with cAMP stimulatory mixture as usual, and the presence of disulfide bonds was determined by sensitivity to application of DTT. For each of the three cysteine double mutant channels tested, pretreatment with FSK and CuPhe resulted in currents that were stimulated by exposure to DTT (Fig. 2, A and B), indicating the presence of disulfide bonds. Both for R334C/T1122C and T338C/S1118C, this DTT sensitivity was in contrast to cells that had been pretreated by CuPhe alone or had received no pretreatment, suggesting that channel activation with FSK was a necessary precondition for disulfide bond formation (Fig. 2, A and B). In contrast, DTT sensitivity of R334C/G1127C channel currents was independent of the pretreatment conditions (Fig. 2B), most likely reflecting the presence of spontaneously formed disulfide bonds in this mutant (Fig. 1C). Because of this spontaneous DTT sensitivity, we were unable to use this pretreatment approach to demonstrate stable CuPhe-induced inhibition of R334C/G1127C channel function. For this mutant alone, we therefore applied both CuPhe and DTT sequentially following channel activation with cAMP (Fig. 2C). Under these conditions, DTT was able to reverse 106 ± 4% (n = 3) of the inhibitory effect of CuPhe treatment.
Interestingly, the pattern of spontaneous disulfide bond formation identified in Fig. 1C is quite distinct from that of CuPhe-induced disulfide bond formation identified in Fig. 1D; only R334C/G1127C was sensitive to both DTT and CuPhe. Treatment with DTT identifies disulfide bonds that have formed spontaneously, presumably in resting, unstimulated channels. Furthermore, because we have no way of knowing the rate at which these spontaneous cross-links formed prior to the experiment, it is possible that some of these represent gradual accumulation or “trapping” of channels in a relatively rare conformation in which the two cysteine side chains in question were close enough together to form a disulfide bond. In contrast, CuPhe treatment induced rapid (within minutes) disulfide bond formation in stimulated (phosphorylated) channels that were presumably actively opening and closing. We therefore focused on the three CuPhe-sensitive paired cysteine mutants identified in Fig. 1D.
Channel Inhibition by External Cd2+ Ions
Cysteine side chains can bind metal ions like Cd2+, and indeed, current inhibition by externally applied Cd2+ ions has previously been used to identify individual pore-lining cysteine side chains in CFTR (23). Furthermore, coordination of Cd2+ ions by multiple cysteine side chains can lead to stronger binding if these side chains are separated by <10 Å (see “Experimental Procedures”). As shown in Fig. 3A, Cys-less CFTR was insensitive to even a high concentration of Cd2+ (1 mm). Single TM6 cysteine mutants R334C (Fig. 3, B and G) and T338C (Fig. 3G) were inhibited by a lower concentration of Cd2+ (10 μm). TM11 mutants S1118C, T1122C, and G1127C were insensitive to this concentration of Cd2+ (Fig. 3G), although 1 mm Cd2+ did inhibit both S1118C (38 ± 4% inhibition, n = 3) and T1122C (70 ± 5%, n = 3) but not G1127C (1 ± 2%, n = 3). As shown in Fig. 3C, the double mutant R334C/T1122C was more strongly blocked by 10 μm Cd2+ than the corresponding individual mutants. The double mutant R334C/G1127C was also strongly inhibited by 10 μm Cd2+ (Fig. 3, D and G), but only in cells that had been pretreated with DTT; in nonpretreated cells, inhibition was weak even when [Cd2+] was increased to 1 mm (Fig. 3E). This finding is further evidence for spontaneous disulfide bond formation between R334C and G1127C; presumably these two cysteine side chains are unable to bind Cd2+ ions until the disulfide bond has been reduced. In contrast, Cd2+ inhibition in R334C/T1122C was independent of DTT pretreatment (Fig. 3G), again consistent with a lack of spontaneous disulfide formation between these two cysteines. The third cysteine pair, T338C/S1118C, was inhibited by Cd2+ to a similar extent as the single T338C mutant, and inhibition of this double mutant was unaffected by DTT pretreatment (Fig. 3G). In all three double cysteine mutants, pretreatment with FSK and CuPhe to induce disulfide bond formation led to Cd2+-insensitive currents (Fig. 3, F and G), providing further evidence for stable disulfide bond formation in these mutants and demonstrating the lack of strong Cd2+ binding following forced disulfide bond formation.
Isolating Effects on the Channel Open State
The functional effects of disulfide bond formation (Fig. 1) and Cd2+ binding (Fig. 3) described above occur in activated, phosphorylated channels that are opening and closing normally and so may reflect a mixture of effects on open channels and on closed channels. To separate the effects on open channels, we have used the E1371Q mutant, which results in a constitutively open channel in baby hamster kidney cells (8, 9, 18, 36) (see supplemental materials). Previous studies have used this mutation to isolate the open state and thereby to infer that access to individual introduced cysteine residues is state-dependent (7–9, 23) (see supplemental materials). An example of constitutive, cAMP-independent currents carried by channels bearing the E1371Q mutation is shown in Fig. 4A (R334C/G1127C/E1371Q). Interestingly, this recording shows that the R334C/G1127C/E1371Q mutant, unlike R334C/G1127C (Fig. 1), is not affected by CuPhe (Fig. 4). This suggests that R334C and G1127C cannot form disulfide bonds in open channels, implying that the disulfides formed by CuPhe treatment in R334C/G1127C occur when the channel is closed. Treatment with CuPhe did cause current inhibition in R334C/T1122C/E1371Q and in T338C/S1118C/E1371Q (Fig. 4B), suggesting that these cysteine pairs are capable of forming disulfide bonds in open channels. In fact, CuPhe caused a significantly stronger inhibition of T338C/S1118C/E1371Q than of T338C/S1118C (Fig. 4B), hinting that disulfide bond formation between these two cysteines might occur preferentially in open channels. The ability of CuPhe to inhibit the function of locked open E1371Q channels might suggest that this inhibition reflects reduced channel conductance rather than open probability, although this was not tested directly by single channel recording.
We also investigated the effect of the E1371Q mutation on Cd2+ inhibition. This experiment is complicated by the fact that the E1371Q mutation alters the Cd2+ sensitivity of single cysteine mutants (23), perhaps because channel opening is associated with constriction of the pore outer mouth and decreased accessibility from the extracellular solution (9). Indeed, we found that the Cd2+ sensitivity of the Cd2+-sensitive TM6 mutants R334C and T338C were significantly decreased in an E1371Q background (Fig. 5B), consistent with decreased access by Cd2+. Similarly, the Cd2+ sensitivity of R334C/G1127C was decreased in an E1371Q background, although it is difficult to discriminate between a decrease in Cd2+ coordination by the two cysteine side chains versus a decrease in Cd2+ access as suggested by results with R334C/E1371Q; the Cd2+ sensitivity of R334C/G1127C/E1371Q was similar to that of R334C/E1371Q (Fig. 5B). In contrast, the apparent Cd2+ sensitivity of T338C/S1118C was significantly increased in an E1371Q background (Fig. 5), suggesting stronger coordination of Cd2+ ions in open channels. This apparent increase in Cd2+ sensitivity is all the more remarkable given the decrease in Cd2+ sensitivity in T338C/E1371Q (Fig. 5B), implying reduced Cd2+ accessibility in open channels. As a result, whereas Cd2+ inhibition is similar in T338C/S1118C as in T338C (Fig. 3), it is significantly stronger in T338C/S1118C/E1371Q compared with T338C/E1371Q (p < 0.00005) (Fig. 5B), suggesting that the cysteine side chains in T338C/S1118C are able to coordinate Cd2+ ion binding in open channels. R334C/T1122C was able to bind Cd2+ ions tightly (note use of 1 μm Cd2+ in Fig. 5B), irrespective of the presence of the E1371Q mutation (Fig. 5B), suggesting that Cd2+ coordination by these cysteines can occur in open channels.
The results shown in Fig. 5 are from cells that had been pretreated with DTT to reduce any spontaneous disulfide bonds. Note that the results of Cd2+ inhibition experiments shown in Fig. 3 suggested spontaneous disulfide bond formation between R334C and G1127C, but not between R334C and T1122C or between T338C and S1118C. However, in these experiments disulfide bonds are presumed to have formed spontaneously in resting, unstimulated channels. To test whether disulfide bonds could form spontaneously in constitutively open channels, we therefore tested the DTT sensitivity of Cd2+ inhibition in double cysteine mutants bearing the E1371Q mutation (Fig. 6). These experiments show that R334C/T1122C/E1371Q channels are potently inhibited by Cd2+ only following DTT pretreatment; in the absence of pretreatment, the channels appeared much less sensitive to Cd2+ (Fig. 6). This suggests that disulfide bonds can form spontaneously between R334C and T1122C in constitutively open channels and that these disulfide bonds prevent the cysteine side chains in question from tightly binding Cd2+ ions. In contrast, in both R334C/G1127C/E1371Q and T338C/S1118C/E1371Q Cd2+ inhibition appeared independent of DTT pretreatment (Fig. 6B), arguing against spontaneous disulfide bond formation between these cysteine pairs. Furthermore, in both R334C/T1122C/E1371Q and T338C/S1118C/E1371Q, but not in R334C/G1127C/E1371Q, Cd2+ sensitivity was greatly impaired by pretreatment with FSK and CuPhe to induce disulfide bond formation (Fig. 6B), suggesting that disulfide bonds cannot be induced between R334C and G1127C under these conditions. These results are noteworthy in comparison with R334C/G1127C itself, which did appear to form spontaneous disulfide bonds that interfered with Cd2+ binding (Fig. 3). This suggests that although these cysteine side chains can come close enough to form a disulfide bond spontaneously in closed, nonactivated channels, they do not form a disulfide bond spontaneously in the open state.
DISCUSSION
Of the 12 TMs in CFTR, TM6 makes the most important functional contribution to the pore (6, 13, 21, 24, 25). At the outer mouth of the pore, the positive charges of Arg-334 and Lys-335 act to attract extracellular Cl− ions to the pore electrostatically (17, 29, 37, 38). Slightly further into the pore from the outside, Thr-338 contributes to a physical constriction in the pore (6) that also acts as a major part of the resistance to Cl− flow (30). In the present study we have probed the proximity of these important amino acids to pore-lining residues coming from another part of the protein, TM11-ECL6-TM12 (Fig. 7A). Functional evidence suggests that other pore-lining side chains in this region (Val-1129, Ile-1131, and Ile-1132) exist at a more superficial part of the pore outer vestibule (15). Our evidence for the existence of disulfide bridges between cysteine side chains introduced into these two parts of CFTR, either spontaneously formed (Fig. 1C) or induced by oxidant (Fig. 1D), suggests that TM6 is physically close to TM11 in the three-dimensional structure of the pore. Furthermore, the fact that all three sites mutated in TM6 (Arg-334, Lys-335, and Thr-338) and all four in TM11 (Ser-1118, Thr-1121, Thr-1122, and Gly-1127) were able to form disulfide cross-links (Fig. 1, C and D) offers independent verification that the amino acid side chains at these positions are accessible to the outer pore vestibule (15, 23). In contrast, we did not find any evidence for disulfide bond formation for the TM12 mutation T1134C (Fig. 1), even though this residue has been proposed to line a narrow region in the pore (25). Our previous work has failed to identify an important role for TM12 in formation of the narrow pore region (16, 39).
Interestingly, the pattern of disulfide bond formation between TMs 6 and 11 is different when identified by DTT sensitivity (Fig. 1C) compared with CuPhe sensitivity (Fig. 1D); only R334C/G1127C was positive for disulfide bond formation in both experimental assays. We assume that DTT identifies cross-links that have formed spontaneously, presumably in unstimulated channels under resting conditions, whereas CuPhe applied after channel activation promotes cross-linking between cysteine side chains in channels undergoing normal gating. If this is the case, then the difference between cross-links identified in Fig. 1 (C and D) might reflect differences in the proximity of different cysteine side chains in resting, unstimulated (unphosphorylated) CFTR channels (Fig. 1C) and activated, phosphorylated channels (Fig. 1D) (see supplemental materials). It is interesting to speculate, then, that this different pattern might reflect a conformational rearrangement in the outer pore mouth that is associated with phosphorylation, possibly independent of that associated with the ATP-dependent opening and closing of phosphorylated channels. Assuming further that the cross-linking pattern identified by DTT (Fig. 1C) reflects nonactivated channels, then there may be some structural flexibility in the protein under these conditions that allows, for example, R334C to form disulfide bonds with either S1118C, T1121C, and G1127C and that also allows K335C to form a disulfide with S1118C while at the same time allowing the more deeply located T338C to form a disulfide with the purportedly more superficially located G1127C (Figs. 1C and 7A).
Because CuPhe was applied after channel activation and resulted in a rapid change in channel function, we assume that it identifies cysteine pairs that are close together in active channels, at least at some point during the gating cycle (see supplemental materials). We have focused on three cysteine pairs that formed disulfide bonds readily under these conditions: R334C/T1122C, R334C/G1127C, and T338C/S1118C (Fig. 1D). No CuPhe-induced changes in channel current were observed for any K335C-containing mutants (Fig. 1D); although there could be several reasons for this apparent negative result and although an apparent lack of cross-link formation is not evidence for a lack of physical proximity between different amino acids, this could reflect the side chain of Lys-335 facing somewhat away from the main axis of the pore, as proposed previously (40). There were also no CuPhe-induced effects in T1121C-containing mutants (Fig. 1D).
For the strongly CuPhe-reactive cysteine pairs R334C/T1122C, R334C/G1127C, and T338C/S1118C, our results suggest different conformations in open channels and in closed channels. This suggestion is based on results using the ATP hydrolysis-defective E1371Q mutant which, as described previously (8, 9), effectively isolates the open state when expressed in baby hamster kidney cells (see supplemental materials). As a result, we expect results from channels bearing the E1371Q mutation to reflect the effects on the channel open state, whereas results from channels without this NBD2 mutation (after activation by cAMP agonists) reflect a mixture of effects on open and closed channels (see supplemental materials). Where different results are obtained with and without the E1371Q mutation, therefore, we can discriminate between effects that occur preferentially in open channels (effects that are enhanced in E1371Q mutants) and effects that occur preferentially in closed channels (effects that are diminished in E1371Q mutants). Comparison of results for these three reactive cysteine pairs with and without E1371Q are summarized in Table 1 and in the following paragraphs.
TABLE 1.
Summary of results for disulfide cross-linked cysteine pairs R334C/T1122C, R334C/G1127C, and T338C/S1118C
Cysteine pairs were studied with either the native Glu present at position 1371 (Glu-1371) or with Gln substituted at this position (Gln-1371). As described under “Discussion,” Glu-1371 is expected to reflect both open and closed channels, and Gln-1371 reflects almost exclusively open channels. In parentheses are indicated the figures in which the quantitative evidence justifying each nonquantitative statement in the table is presented.
| R334C/T1122C |
R334C/G1127C |
T338C/S1118C |
||||
|---|---|---|---|---|---|---|
| Glu-1371 | Gln-1371 | Glu-1371 | Gln-1371 | Glu-1371 | Gln-1371 | |
| Spontaneous cross-linking | No (Figs. 1 and 3) | Yes (Fig. 6) | Yes (Figs. 1 and 3) | No (Fig. 6) | No (Figs. 1 and 3) | No (Fig. 6) |
| CuPhe-induced cross-linking | Yes (Fig. 1) | Yes (Fig. 4) | Yes (Fig. 1) | No (Fig. 4) | Yes (Fig. 1) | Enhanced (Fig. 4) |
| Cd2+ inhibition | Strong (Fig. 3) | Strong (Fig. 5) | Moderate (Fig. 3) | Abolished (Fig. 5) | Weak (Fig. 3) | Strengthened (Fig. 5) |
For R334C/T1122C, both CuPhe-induced disulfide bonds and strong Cd2+ coordination can occur in E1371Q-containing channels, suggesting that these two cysteine side chains are close together in open channels. Similar effects seen without E1371Q may therefore also reflect effects on open channels, although we have no evidence to rule out effects also on closed channels. Interestingly, these two cysteines can form disulfide bonds spontaneously only in the presence of the E1371Q mutation, supporting a close proximity in open channels.
For R334C/G1127C, the E1371Q mutation has a strongly negative effect on the apparent proximity of the two cysteines; it abolished both spontaneous and CuPhe-induced disulfide bond formation and also drastically weakened inhibition by Cd2+ ions. Based on this, we suggest that these two cysteines are only close enough together to form disulfide bonds and to coordinate Cd2+ binding when the channel is closed; the effects observed in non-E1371Q-containing channels therefore presumably reflect effects that occur during channel closures.
For T338C/S1118C, evidence for proximity is actually strengthened by the E1371Q mutation. Thus, E1371Q both enhanced CuPhe-induced disulfide bond formation and also strengthened Cd2+ inhibition. This latter effect is particularly noteworthy because E1371Q strengthened Cd2+ inhibition in T338C/S1118C, even though it weakened Cd2+ inhibition in the T338C single mutant (Fig. 5B), suggesting reduced Cd2+ accessibility from the extracellular solution. The facilitation of disulfide formation and Cd2+ binding seen in T338C/S1118C/E1371Q suggests that these two cysteine side chains are closer together in open channels than in closed channels.
The apparent state dependence of cross-linking (as well as Cd2+ coordination) suggested above is summarized graphically in Fig. 7 (B and C). Based on these results, we suggest that channel opening and closing may be associated with a relative translational movement of TMs 6 and 11; as shown by the red arrows in Fig. 7C, we propose that channel opening may be associated with a relative “downward” (toward the cytoplasm) movement of TM6 and/or a relative “upward” (toward the extracellular solution) movement of TM11. This suggested that translational movement is an addition to previously suggested movements of the TMs during CFTR channel opening and closing, such as “inward facing/outward facing” TM-tilting movements (9), constrictive/dilational-type pore closing and opening (8, 21), and rotational movements of individual TMs (7, 24). Of course, these different kinds of rearrangements are not mutually exclusive; the TMs may move translationally, as well as tilt and rotate, causing localized constrictions or dilations at different points in the transmembrane permeation pathway during pore opening and closing.
Our results provide important structural constraints for the CFTR channel pore in the open state and in the closed state. Outer pore residues in TMs 6 and 11 are in close proximity in both the open and the closed states, as well as in unstimulated, presumably unphosphorylated channels. Furthermore, these two TMs appear to move relative to one another during pore opening and closing. We propose that the outer mouth of the pore undergoes both an overall constriction (9) and a translation of individual TMs during pore opening. Because conformational rearrangement of the MSDs driven by ATP interactions with the NBDs (such as those that control CFTR channel opening and closing) are a conserved aspect of the mechanism of action of all ABC transporters, the rearrangements of the TMs that we propose may also have structural and functional implications for the transport mechanism of ABC proteins in general.
This work was supported by the Canadian Institutes of Health Research.

This article contains supplemental text and Fig. S1.
- CFTR
- cystic fibrosis transmembrane conductance regulator
- ABC
- ATP-binding cassette
- CuPhe
- copper(II)-o-phenanthroline
- ECL
- extracellular loop
- FSK
- forskolin
- MSD
- membrane-spanning domain
- NBD
- nucleotide-binding domain
- TM
- transmembrane α-helix.
REFERENCES
- 1. O'Sullivan B. P., Freedman S. D. (2009) Cystic fibrosis. Lancet 373, 1891–1904 [DOI] [PubMed] [Google Scholar]
- 2. Rees D. C., Johnson E., Lewinson O. (2009) ABC transporters. The power to change. Nat. Rev. Mol. Cell Biol. 10, 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gadsby D. C., Vergani P., Csanády L. (2006) The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature 440, 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hwang T. C., Sheppard D. N. (2009) Gating of the CFTR Cl− channel by ATP-driven nucleotide-binding domain dimerisation. J. Physiol. 587, 2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muallem D., Vergani P. (2009) ATP hydrolysis-driven gating in cystic fibrosis transmembrane conductance regulator. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Linsdell P. (2006) Mechanism of chloride permeation in the cystic fibrosis transmembrane conductance regulator chloride channel. Exp. Physiol. 91, 123–129 [DOI] [PubMed] [Google Scholar]
- 7. Bai Y., Li M., Hwang T. C. (2011) Structural basis for the channel function of a degraded ABC transporter, CFTR (ABCC7). J. Gen. Physiol. 138, 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang W., Linsdell P. (2012) Conformational change opening the CFTR chloride channel pore coupled to ATP-dependent channel gating. Biochim. Biophys. Acta 1818, 851–860 [DOI] [PubMed] [Google Scholar]
- 9. Wang W., Linsdell P. (2012) Alternating access to the transmembrane domain of the ATP-binding cassette protein cystic fibrosis transmembrane conductance regulator (ABCC7). J. Biol. Chem. 287, 10156–10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kos V., Ford R. C. (2009) The ATP-binding cassette family. A structural perspective. Cell Mol. Life Sci. 66, 3111–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Locher K. P. (2009) Review. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosenberg M. F., O'Ryan L. P., Hughes G., Zhao Z., Aleksandrov L. A., Riordan J. R., Ford R. C. (2011) The cystic fibrosis transmembrane conductance regulator (CFTR). Three-dimensional structure and localization of a channel gate. J. Biol. Chem. 286, 42647–42654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ge N., Muise C. N., Gong X., Linsdell P. (2004) Direct comparison of the functional roles played by different membrane spanning regions in the cystic fibrosis transmembrane conductance regulator chloride channel pore. J. Biol. Chem. 279, 55283–55289 [DOI] [PubMed] [Google Scholar]
- 14. Aubin C. N., Linsdell P. (2006) Positive charges at the intracellular mouth of the pore regulate anion conduction in the CFTR chloride channel. J. Gen. Physiol. 128, 535–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fatehi M., Linsdell P. (2009) Novel residues lining the CFTR chloride channel pore identified by functional modification of introduced cysteines. J. Membr. Biol. 228, 151–164 [DOI] [PubMed] [Google Scholar]
- 16. Qian F., El Hiani Y., Linsdell P. (2011) Functional arrangement of the 12th transmembrane region in the CFTR chloride channel pore based on functional investigation of a cysteine-less CFTR variant. Pflügers Arch. 462, 559–571 [DOI] [PubMed] [Google Scholar]
- 17. Zhou J. J., Fatehi M., Linsdell P. (2008) Identification of positive charges situated at the outer mouth of the CFTR chloride channel pore. Pflügers Arch 457, 351–360 [DOI] [PubMed] [Google Scholar]
- 18. Zhou J. J., Li M. S., Qi J., Linsdell P. (2010) Regulation of conductance by the number of fixed positive charges in the intracellular vestibule of the CFTR chloride channel pore J. Gen. Physiol. 135, 229–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang W., El Hiani Y., Linsdell P. (2011) Alignment of transmembrane regions in the cystic fibrosis transmembrane conductance regulator chloride channel pore. J. Gen. Physiol. 138, 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fatehi M., Linsdell P. (2008) State-dependent access of anions to the cystic fibrosis transmembrane conductance regulator chloride channel pore. J. Biol. Chem. 283, 6102–6109 [DOI] [PubMed] [Google Scholar]
- 21. El Hiani Y., Linsdell P. (2010) Changes in accessibility of cytoplasmic substances to the pore associated with activation of the cystic fibrosis transmembrane conductance regulator chloride channel. J. Biol. Chem. 285, 32126–32140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Z. R., Song B., McCarty N. A. (2005) State-dependent chemical reactivity of an engineered cysteine reveals conformational changes in the outer vestibule of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 280, 41997–42003 [DOI] [PubMed] [Google Scholar]
- 23. Beck E. J., Yang Y., Yaemsiri S., Raghuram V. (2008) Conformational changes in a pore-lining helix coupled to cystic fibrosis transmembrane conductance regulator channel gating. J. Biol. Chem. 283, 4957–4966 [DOI] [PubMed] [Google Scholar]
- 24. Bai Y., Li M., Hwang T. C. (2010) Dual roles of the sixth transmembrane segment of the CFTR chloride channel in gating and permeation. J. Gen. Physiol. 136, 293–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Norimatsu Y., Ivetac A., Alexander C., Kirkham J., O'Donnell N., Dawson D. C., Sansom M. S. (2012) Cystic fibrosis transmembrane conductance regulator. A molecular model defines the architecture of the anion conduction path and locates a “bottleneck” in the pore. Biochemistry 51, 2199–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mense M., Vergani P., White D. M., Altberg G., Nairn A. C., Gadsby D. C. (2006) In vivo phosphorylation of CFTR promotes formation of a nucleotide-binding domain heterodimer. EMBO J. 25, 4728–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li M. S., Demsey A. F., Qi J., Linsdell P. (2009) Cysteine-independent inhibition of the CFTR chloride channel by the cysteine-reactive reagent sodium (2-sulphonatoethyl) methanethiosulphonate (MTSES). Br. J. Pharmacol. 157, 1065–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holstead R. G., Li M. S., Linsdell P. (2011) Functional differences in pore properties between wild-type and cysteine-less forms of the CFTR chloride channel. J. Membr. Biol. 243, 15–23 [DOI] [PubMed] [Google Scholar]
- 29. Smith S. S., Liu X., Zhang Z. R., Sun F., Kriewall T. E., McCarty N. A., Dawson D. C. (2001) CFTR. Covalent and noncovalent modification suggests a role for fixed charges in anion conduction. J. Gen. Physiol. 118, 407–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fatehi M., St Aubin C. N., Linsdell P. (2007) On the origin of asymmetric interactions between permeant anions and the CFTR chloride channel pore. Biophys. J. 92, 1241–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holmgren M., Shin K. S., Yellen G. (1998) The activation gate of a voltage-gated K+ channel can be trapped in the open state by an intersubunit metal bridge. Neuron 21, 617–621 [DOI] [PubMed] [Google Scholar]
- 32. Neale E. J., Elliott D. J., Hunter M., Sivaprasadarao A. (2003) Evidence for intersubunit interactions between S4 and S5 transmembrane segments of the Shaker potassium channel. J. Biol. Chem. 278, 29079–29085 [DOI] [PubMed] [Google Scholar]
- 33. Sobolevsky A. I., Yelshansky M. V., Wollmuth L. P. (2004) The outer pore of the glutamate receptor channel has 2-fold rotational symmetry. Neuron 41, 367–378 [DOI] [PubMed] [Google Scholar]
- 34. Clarke C. E., Veale E. L., Wyse K., Vandenberg J. I., Mathie A. (2008) The M1P1 loop of TASK3 K2P channels apposes the selectivity filter and influences channel function. J. Biol. Chem. 283, 16985–16992 [DOI] [PubMed] [Google Scholar]
- 35. Bell D. C., Turbendian H. K., Valley M. T., Zhou L., Riley J. H., Siegelbaum S. A., Tibbs G. R. (2009) Probing S4 and S5 segment proximity in mammalian hyperpolarization-activated HCN channels by disulfide bridging and Cd2+ coordination. Pflügers Arch. 458, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li M. S., Holstead R. G., Wang W., Linsdell P. (2011) Regulation of CFTR chloride channel macroscopic conductance by extracellular bicarbonate. Am. J. Physiol. Cell Physiol. 300, C65–C74 [DOI] [PubMed] [Google Scholar]
- 37. Gong X., Linsdell P. (2003) Molecular determinants and role of an anion binding site in the external mouth of the CFTR chloride channel pore. J. Physiol. 549, 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou J. J., Fatehi M., Linsdell P. (2007) Direct and indirect effects of mutations at the outer mouth of the CFTR chloride channel pore. J. Membr. Biol. 216, 129–142 [DOI] [PubMed] [Google Scholar]
- 39. Gupta J., Evagelidis A., Hanrahan J. W., Linsdell P. (2001) Asymmetric structure of the cystic fibrosis transmembrane conductance regulator chloride channel pore suggested by mutagenesis of the twelfth transmembrane region. Biochemistry 40, 6620–6627 [DOI] [PubMed] [Google Scholar]
- 40. Cheung M., Akabas M. H. (1996) Identification of CFTR channel-lining residues in and flanking the M6 membrane-spanning semgent. Biophys. J. 70, 2688–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]