Background: Constitutively active mutants (CAM) of G-protein-coupled receptors are often related to human diseases.
Results: A novel type of CAM mimicking the ligand revealed a double binding mode of the PrRP receptor and its binding pocket.
Conclusion: The applied modeling-guided mutagenic approach discloses distinct insights into the molecular mechanisms of GPCR ligand recognition and activation.
Significance: The concept can be adopted to study hereditary harmful CAMs and assist GPCR-based drug development.
Keywords: 7-Helix Receptor, Computer Modeling, G-protein-coupled Receptors (GPCR), Neuropeptide, Peptide Hormones, Receptor Structure-Function
Abstract
The prolactin-releasing peptide receptor and its bioactive RF-amide peptide (PrRP20) have been investigated to explore the ligand binding mode of peptide G-protein-coupled receptors (GPCRs). By receptor mutagenesis, we identified the conserved aspartate in the upper transmembrane helix 6 (Asp6.59) of the receptor as the first position that directly interacts with arginine 19 of the ligand (Arg19). Replacement of Asp6.59 with Arg19 of PrRP20 led to D6.59R, which turned out to be a constitutively active receptor mutant (CAM). This suggests that the mutated residue at the top of transmembrane helix 6 mimics Arg19 by interacting with additional binding partners in the receptor. Next, we generated an initial comparative model of this CAM because no ligand docking was required, and we selected the next set of receptor mutants to find the engaged partners of the binding pocket. In an iterative process, we identified two acidic residues and two hydrophobic residues that form the peptide ligand binding pocket. As all residues are localized on top or in the upper part of the transmembrane domains, we clearly can show that the extracellular surface of the receptor is sufficient for full signal transduction for prolactin-releasing peptide, rather than a deep, membrane-embedded binding pocket. This contributes to the knowledge of the binding of peptide ligands to GPCRs and might facilitate the development of GPCR ligands, but it also provides new targeting of CAMs involved in hereditary diseases.
Introduction
Identification of direct receptor-ligand interactions for the ∼800 identified G-protein-coupled receptors (GPCRs)4 is as challenging as it is important for drug discovery (1); 50% of all currently available drugs target the specific manipulation of GPCR activity (2, 3). The PrRP receptor superfamily is expressed in almost all cells/tissues, is involved in a plethora of different signaling pathways, and plays an important role in a large variety of physiological processes.
The prolactin-releasing peptide receptor (PrRPR) was originally isolated from rat hypothalamus (4). PrRPR has been detected widely throughout the human and rat brain (5) and most commonly activates the Gq protein-coupled signaling pathway (6). Its eponymous endogenous ligand, the prolactin-releasing peptide (PrRP), was identified in 1998 by a reverse pharmacology approach (7, 8). PrRP features two equipotent isoforms, PrRP31 (31 residues) and an N-terminally truncated PrRP20 (20 residues) (6, 8). PrRP is an RF-amide peptide, consisting of a common C-terminal arginine and an amidated phenylalanine motif. Further, it plays a role in energy metabolism, stress responses, circadian rhythm, analgesia, and anorexigenic effects (7, 9). Structure-activity relationship studies of PrRP using N-terminally truncated mutants and alanine substitution within these constructs (10–12) demonstrated the biological significance of the C-terminal Arg and Phe residues and the amidation of the C terminus.
Site-directed mutagenesis is a powerful and widely used tool to study receptor activation. This approach alone can provide insight into the function of GPCRs, but it is often used in combination with information provided by other techniques, such as crystallography or molecular modeling, to relate receptor function to a tertiary structure (13). The conserved Asp6.59 residue of the Y receptor family was shown to interact with a specific Arg of either human pancreatic polypeptide or neuropeptide Y (NPY) in a subtype-specific manner (14, 15). The numbering of receptor residues has been performed as suggested by Ballesteros and Weinstein (16). PrRPR shares its phylogenic origin with Y receptors (17), leading to sequence similarities (Fig. 1A) and a number of conserved residues, including Asp6.59 (Fig. 1C). Furthermore, the ligands of these receptors are structurally similar (18) and share a similar C-terminal sequence (Fig. 1B). Although the RF-amide motif was previously identified as a major requirement for PrRP-induced agonist activity (10, 11), the critical residues on the receptor remain unknown, and the ligand binding mode is still poorly understood.
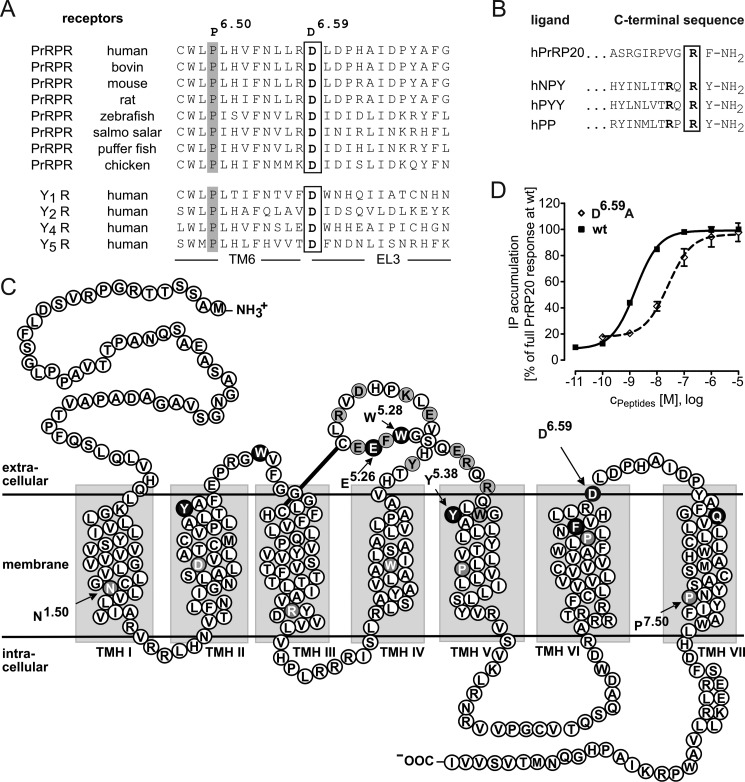
FIGURE 1.
Identification of the conserved Asp6. 59 residue in the hPrRPR sequence as potential point of interaction. A, conservation of Asp6.59 shown in the amino acid sequence alignment. The region of upper TMH6 and the beginning of the subsequent EL3 of the four human Y receptor subtypes and the PrRPR are presented. B, comparison of the C-terminal amino acids of the Y receptor ligands and the PrRP20. C, snake plot representing the sequence of the human PrRPR. Residues highlighted in black were investigated as double mutants in the D6.59R construct. Selective alanine scanning was performed on residues pictured in gray, resulting in no functional alteration. Residues with white letters on gray correspond to the X.50 nomenclature (16). D, IP accumulating signal transduction assay performed for 1 h with COS-7 cells in a concentration-response dependent manner reveals an impact of D6.59A PrRPR in comparison with the WT PrRP receptor. Data represent the mean ± S.E. of multiple independent experiments (n = 32 for hPrRPR and n = 12 for D6.59A PrRPR). Receptor activity is expressed as percentage of the full response of PrRP20 at the WT PrRP receptor.
Here, we describe the first mutagenesis study of the human PrRP receptor (PrRPR). We used the extracellular region to elucidate the binding site and the molecular mechanism of GPCR activation. Considering the relevance of the C-terminal Arg and Phe residues of PrRP for receptor binding, we applied the concept of the double-cycle mutagenesis approach (15, 19, 20) and identified the first direct contact point between PrRP20 and the PrRPR, consisting of the conserved Asp6.59 and the Arg19 residue of PrRP20. To prove the existence of this interaction, we switched the residues involved in the salt bridge formation and created D6.59R PrRPR and Asp19PrRP20. This newly introduced Arg in the receptor variant D6.59R might serve as surrogate for the absent Arg19 of the ligand, as it led to a new type of constitutive activity. Given the lack of data of experimentally determined structures of peptide GPCRs, we developed a comparative model of the human PrRPR. By combining molecular modeling with double-cycle mutagenesis experiments in the framework of this constitutively active mutant (CAM), we conceived an effective strategy to explore structural determinants of ligand recognition on a molecular level. More specifically, we were able to identify Tyr5.38, Trp5.28, Glu5.26, and to some extent, Phe6.54 to be involved in receptor activation and ligand binding. This combinatory approach enabled us to clarify the double binding mode of Arg19 of the peptide ligand, which has two putative interaction partners within the PrRPR, Glu5.26 and Asp6.59. The assembled experimental data were used to generate a model of the PrRP-receptor interaction in molecular detail. Furthermore, our data describe the binding mode of a peptide ligand to GPCR by solely interacting with residues localized in the extracellular domain or upper part of the transmembrane helices (TMHs). In our approach, we identified a receptor mutant with constitutive activity, which most likely relies on mimicking a direct ligand-receptor interaction. This provides knowledge on the function of an active mode of GPCRs and may be applied to other peptide GPCRs.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
Rink-amide resin (NovaBiochem; Laüfelfingen, Switzerland) was used to synthesize PrRP20, Ala19PrRP20, Asp19PrRP20, and Ala20PrRP20 by automated solid phase peptide synthesis (Syro; MultiSynTech, Bochum, Germany) as described previously, using the orthogonal 9-fluorenyl-methoxycarbonyl-tert-butyl strategy (21). Purification and verification of the peptides were achieved as described previously (supplemental Table S1) (22).
DNA Extraction from SMS-KAN
To obtain genomic DNA from SMS-KAN cells (human neuroblastoma cells, DSMZ, Braunschweig, Germany), ∼1 million cells were digested overnight at 55 °C with 500 μl of lysis buffer (1 m NaCl, 20% SDS, 0.5 m EDTA, 1 m Tris, pH 8.5, was adjusted using hydrochloric acid (HCl)) containing 50 μg of proteinase K (Promega, Mannheim, Germany). Genomic DNA was extracted using phenol/chloroform and precipitated from the aqueous phase with isopropyl alcohol, washed with ethanol, and then dissolved in water.
Cloning and Mutagenesis of the PrRP Receptors in Eukaryotic Expression Vectors
The coding sequence of the human PrRPR was obtained by PCR amplification from the isolated genomic DNA of SMS-KAN cells and cloned into the eukaryotic expression vector peYFP-N1 (Clontech) C-terminally fused to eYFP, using the XhoI and BamHI restriction site to result in the construct phPrRPR_eYFP-N1. The correctness of the entire coding sequence was confirmed by DNA sequencing using the dideoxynucleotide (ddNTP) termination method developed by Sanger et al. (23). Plasmids encoding single point mutations (Tables 1 and 2) were prepared by using the QuikChangeTM site-directed mutagenesis method (Stratagene, CA) with the desired mutagenic primers. For intermolecular double-cycle mutagenesis approaches, the single alanine mutated receptor constructs were investigated, using single alanine-modified PrRP20 analogs. Plasmids encoding double mutations containing Y2.64A, W2.71A, E5.26A, E5.26R; W5.28A, D6.59A, F6.54A, or Q7.35A as a second mutation, respectively, were prepared by using the QuikChangeTM site-directed mutagenesis approach with the D6.59R or D6.59A construct as template. In addition, all PrPR receptor constructs were also generated N-terminally fused to the coding sequence of the hemagglutinin (HA) tag. The entire coding sequence of each resulting receptor mutant was proven by sequencing.
TABLE 1.
Functional characterization of wild type and Asp6.59 PrRP receptor mutants with different PrRP analogs
IP accumulating signal transduction assay was performed for 1 h with different concentrations of modified PrRP20 peptides to determine EC50 values from concentration-response curves. NT represents not tested; NR indicates no response after stimulation with 10 μm, and n displays the number of individual experiments.
| PrRPR mutants | PrRP20 |
Ala19PrRP20 |
Asp19PrRP20 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| EC50 (nm)a (pEC50 ± S.E.) | EC50 ratiob (mutant/WT) | Emax ± S.E.c | n | EC50 (nm)a (pEC50 ± S.E.) | EC50 ratiob (analog/WT) | n | EC50 (nm)a (pEC50 ± S.E.) | n | |
| WT | 1.66 (8.78 ± 0.04) | 1 | 100 | 32 | 1202 (5.92 ± 0.08) | 736 | 11 | 1318 (5.88 ± 0.12) | 5 |
| D6.59A | 26 (7.59 ± 0.15) | 15 | 98 ± 7 | 12 | 166 (6.78 ± 0.17) | 0.16 | 3 | 6456 (5.19 ± 0.16) | 4 |
| D6.59R | NDd | NDe | 60 ± 13 | 4 | >10 000 (<5) | NDe | 2 | 138 (6.86 ± 0.23) | 3 |
| D6.59K | 1380 (5.86 ± 0.20) | 847 | 90 ± 10 | 3 | NT | 115 (6.94 ± 0.17) | 2 | ||
| D6.59E | 3.98 (8.4 ± 0.19) | 2 | 106 ± 10 | 2 | NT | NT | |||
| D6.59N | 36.3 (7.44 ± 0.25) | 22 | 105 ± 20 | 2 | NT | NT | |||
| E5.26A | 537 (6.27 ± 0.09) | 361 | 81 ± 6 | 8 | > 10 000 (< 5) | 21 | 3 | NT | |
| E5.26R | > 10 000 (< 5) | NDe | 70 ± 6 | 2 | NDd | NDe | 2 | >10 000 (<5) | 2 |
| E5.26A/D6.59A | NDd | NDe | 58 ± 7 | 2 | NDd | NDe | 2 | NT | |
| E5.26R/D6.59R | NR | NDe | 8 ± 2 | 2 | NR | NDe | 2 | NDd | 2 |
a EC50/pEC50 values were calculated from the mean ± S.E. of n independent experiments, measured in duplicate.
b The ratio was determined using the Prism 5.03 global fitting function for EC50 shift determination.
c Efficacy was determined as percentage compared with full PrRP20 response at WT.
d ND means not determined because of the lack of efficacy. The plateau of the curve was not reached.
e ND means not determinable.
TABLE 2.
Signal transduction of selected alanine of PrRP receptor mutants from extracellular loop 2 and top TMH5
The IP accumulating signal transduction assay was performed for 1 h with different concentrations of modified PrRP20 peptides to determine EC50 values from concentration-response curves. n represents the number of independent experiments.
| PrRPR mutant | Emax ± S.E.a | pb | pEC50 ± S.E.c | EC50c | EC50 ratio (mutant/WT)d | n |
|---|---|---|---|---|---|---|
| % | nm | |||||
| WT | 100 | 8.78 ± 0.04 | 1.66 | 1 | 32 | |
| Y4.65A | 63 ± 22 | NS | 8.03 ± 0.32 | 9.3 | 6 | 2 |
| E4.68A | 93 ± 8 | NS | 8.19 ± 0.19 | 6.4 | 4 | 3 |
| K4.70A | 111 ± 35 | NS | 8.41 ± 0.41 | 3.9 | 2 | 2 |
| D4.73A | 146 ± 41 | NS | 8.75 ± 0.49 | 1.78 | 1 | 2 |
| R4.75A | 87 ± 15 | NS | 8.32 ± 0.37 | 4.8 | 3 | 3 |
| E5.25A | 124 ± 10 | NS | 7.99 ± 0.13 | 10 | 6 | 3 |
| E5.26A | 81 ± 5 | 0.0094 | 6.26 ± 0.10 | 549 | 331 | 8 |
| F5.27A | 122 ± 50 | NS | 8.14 ± 0.49 | 7.2 | 4 | 2 |
| W5.28A | 48 ± 5 | <0.0001 | 6.02 ± 0.14 | 954 | 580 | 7 |
| E5.32A | 114 ± 11 | NS | 8.62 ± 0.14 | 2.4 | 1 | 2 |
| R5.33A | 115 ± 15 | NS | 8.57 ± 0.20 | 2.7 | 2 | 2 |
| R5.35A | 81 ± 4 | 0.0122 | 8.35 ± 0.32 | 4.5 | 3 | 2 |
| Y5.38A | 46 ± 6 | <0.0001 | 6.99 ± 0.14 | 102 | 61 | 5 |
| W5.40A | 101 ± 38 | NS | 8.78 ± 0.49 | 1.7 | 1 | 2 |
| D6.59A | 97 ± 6 | NS | 7.59 ± 0.15 | 26 | 15 | 12 |
| F6.54A | 101 ± 3 | NS | 7.61 ± 0.10 | 25 | 15 | 3 |
a Efficacy was determined as percentage compared with full PrRP20 response at WT.
b Significance p was estimated using the unpaired t test (NS represents no significantly different means with p ≥ 0.05).
c EC50/pEC50 values were calculated from the mean ± S.E. of n independent experiments, measured in duplicate.
d The ratio was determined using the prism 5.03 function of dose-response EC50 shift determination by global fitting.
Cell Culture
Cell culture material was supplied by PAA Laboratories GmbH (Pasching, Austria). Culture of COS-7 (African green monkey, kidney), HEK293 (human embryonic kidney), and SMS-KAN cells was done as recommended by the supplier (DSMZ, Braunschweig, Germany). Briefly, cells were grown as monolayers at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. COS-7 cells were cultured in Dulbecco's modified Eagle's medium containing 10% (v/v) heat-inactivated fetal calf serum (FCS), 100 units/ml penicillin, and 100 μg/ml streptomycin, and HEK293 cells were grown in DMEM/Ham's F-12 (1:1) without l-glutamine containing 15% (v/v) heat-inactivated FCS as described previously (15, 24). SMS-KAN cells were maintained in nutrient mixture Ham's F-12/Dulbecco's modified Eagle's medium (1:1) with 15% (v/v) FCS, 4 mm glutamine, and 0.2 mm nonessential amino acids (25).
Fluorescence Microscopy
HEK293 cells (1.2 × 105) were seeded onto 8-well chamber slides (ibidi, Munich, Germany). The transient transfection of HEK293 cells were performed using 0.1 to 1 μg of vector DNA and 1 μl of LipofectamineTM 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. The nuclei were visualized with Hoechst 33342 (1 μg/ml; Sigma) for 10 min after 1 h of starving with OPTI®-MEM I reduced serum medium (Invitrogen). Fluorescence images were obtained using an ApoTome Imaging System with an Axio Observer microscope (Zeiss, Jena, Germany). All investigated receptors were correctly integrated in the membrane as confirmed by live-cell microscopy (supplemental Fig. S1A).
Quantification of Receptor Cell Surface Localization by Cell Surface ELISA
To quantify plasma membrane receptors, a cell surface ELISA was performed using an antibody directed against the native 15 N-terminal amino acids of the PrRPR. 50,000 HEK293 cells were grown in 96-well plates and transfected with the PrRP WT receptor or its mutants after reaching 75–85% of confluence. The cells were starved with Opti®-MEM I (30 min) 17 h post-transfection and fixed in 4% paraformaldehyde (30 min). For immune staining, cells were blocked with 2% BSA and permeabilized with 0.5% Triton X-100, 2% BSA in Dulbecco's modified Eagle's medium for 1 h (37 °C) to determine total receptor amounts, whereas surface expressed receptors were quantified without permeabilization. Incubation was performed with the primary antibody (1:2000 dilutions) for 2 h (25 °C) and followed by 1.5-h (25 °C) incubation with the secondary antibody (1:5000). Receptors were detected by using the rabbit anti-N terminus (GPR10 antibody (N1), GTX108137, GeneTex) followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (sc-2004, Santa Cruz Biotechnology, Heidelberg, Germany). The results were fully confirmed in a second independent ELISA setup, using a peroxidase-conjugated anti-HA-antibody (1:1000 dilutions, 12CA5, Roche Applied Science) versus the N-terminally fused HA tag of the generated PrRPR constructs (data not shown). Quantification of the bound peroxidase was performed as described and analysis performed with the GraphPad Prism 5.03 program (14). Values are presented as mean values ± S.E. of four individual experiments, measured in triplicate.
Radioligand Binding Studies
For radioligand binding studies, 1.5 × 106 COS-7 cells were seeded into 25-cm2 flasks. At 60–70% confluency, cells were transiently transfected using 4 μg of vector DNA and 15 μl of MetafecteneTM (Biontex Laboratories GmbH, Martinsried/Planegg, Germany). Approximately 24 h after transfection, binding assays were performed on intact cells using N-[propionyl-3H]hPrRP20. Binding was determined with 1 nm N-[propionyl-3H]hPrRP20 in the absence (total binding) or in the presence (non-specific binding) of 1 μm unlabeled hPrRP20, respectively, as described previously (26, 27). Our former evaluated protocol (28) was used to obtain N-[propionyl-3H]hPrRP20 by selective labeling with a specific activity of 3.52 TBq/mmol and resulting in a Kd value of 0.58 nm. Specific binding of each PrRP receptor mutant was compared with specific binding of the PrRP WT receptor. IC50 values and the Kd value were calculated with GraphPad Prism 5.03 (GraphPad Software, San Diego), fitted to a one-site competition or a one-site binding model, respectively. Triplicates were measured in at least two independent experiments for the determination of IC50 values, whereas one experiment in triplicate was made for Kd value estimation.
Signal Transduction Assay
Signal transduction (inositol phosphate (IP) accumulation) assays were performed as described previously with minor modifications (22). The time of incubation was increased to 3 h for the double mutants of PrRPR and reduced to 1 h for measurement of concentration-response curves. To test for constitutive activity, COS-7 cells were incubated without agonist for 1, 3, and 6 h at 37 °C. Each ligand-receptor interaction was analyzed with the GraphPad Prism 5.03 program by establishing the corresponding data set from different experiments. All signal transduction assays were repeated at least twice independently and measured in duplicate. The global curve fitting function of GraphPad Prism 5.03 was asked to determine given EC50 ratios. The statistical significance of relevant samples was computed by using the unpaired Student's t test, based on the means, and values with p < 0.05 were considered to be significant.
Multiple Sequence Alignment
ClustalW (29) was used to align the primary sequence of the PrRPR with the sequences of mammalian Y and PrRP receptors. Next, the transmembrane regions of six GPCRs of known structure (see below) were structurally aligned with Mustang (30). The profiles resulting from these first two steps were then aligned to one another with ClustalW, and the human PrRPR sequence alignment used for modeling was taken from this final profile-profile alignment. The C-terminal 310 residues of the PrRPR primary sequence were threaded onto the three-dimensional coordinates of six available GPCR experimental structures; PDB codes are as follows: 1U19 (31), 3CAP (32), 3DQB (33), 2RH1 (34), 2VT4 (35), and 3EML (36).
Construction of the Comparative Models
Extracellular loop (EL) regions were reconstructed using kinematic loop closure (37) and cyclic coordinate descent (38), as implemented in the Rosetta version 3 software suite. The models were refined with the Rosetta version 3 all-atom energy function. Energetically favorable models were grouped into 15 structurally similar groups by k-means clustering, and the lowest scoring models of each cluster were analyzed. Models based on the template PDB 3DQB had the lowest energy and were used to inform the mutagenesis studies.
Model Refinement and Peptide Docking
The comparative model constructed in light of the new mutagenesis data presented herein was generated using the original multiple sequence alignment. To model the PrRPR-ligand complex, an iterative peptide docking and loop remodeling procedure was performed. Energetically favorable changes in orientation were determined using the RosettaMembrane all-atom energy function (39). The PrRP8-20 model was docked into the putative binding site of the receptor while allowing remodeling of EL1, EL2, and EL3. Using the RosettaDock protocol (40), translational movements of the peptide of up to 4 Å were allowed in three dimensions, and the peptide was allowed to rotate along its x, y, and z axes by up to 10°. Loop regions were constructed using cyclic coordinate descent (38). The conformational search was enhanced by conducting the modeling in the presence of loose distance restraints, where models that placed Asp6.59, Glu5.26, Trp5.28, and Tyr5.38 within 10 Å of Arg19 of the peptide were more energetically favorable than those that did not. The PrRP8-20 model was generated by de novo folding the peptide using RosettaNMR with sparse NMR chemical shift and distance data (41). Of 19,241 PrRP receptor complex docked models, the top 10 by total score were analyzed. Two of these models were considered structurally redundant, leaving eight unique models that agree with the experimental data presented herein (Fig. 8).
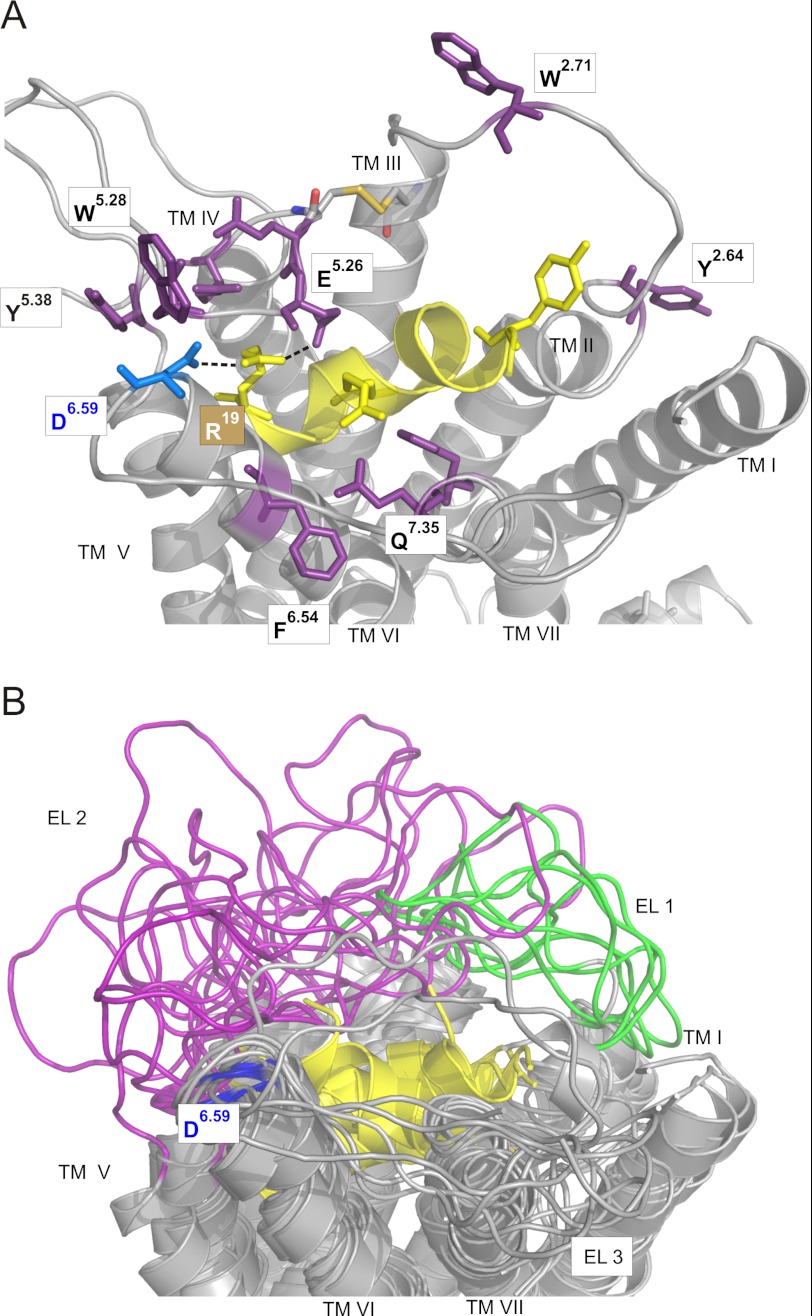
FIGURE 8.
Comparative model of PrRPR docked to the 13 C-terminal residues of PrRP20. A, selected comparative model generated by Rosetta in the presence of the PrRP ligand to support experimental data. The same color code used in Fig. 5A is used here. The figure displays an ensemble of low energy PrRP/receptor models generated in Rosetta that agrees well with experimental data. Residue Asp6.59 is colored in blue; the peptide is presented in yellow, and residues in vicinity to PrRP are in purple. B, eight nonredundant low energy comparative models of the PrRP-receptor complex. These eight models were generated in the presence of structural constraints derived from the mutagenesis data described (see main text) and are considered energetically favorable according to the Rosetta version 3 all-atom scoring function. The peptide is highlighted in yellow; Asp6.59 of the receptor is in blue; EL1 of the receptor is in green, and EL2 of the receptor is in magenta.
RESULTS
Arg19 of the Endogenous Ligand PrRP20 Interacts with the Asp6.59 of PrRPR
Based on the data of the NPY/Y receptor system (14, 15), we hypothesized Asp6.59 to be the interaction partner of Arg19 in the PrRP/PrRPR system. To test this hypothesis, charge and size prerequisites in position Asp6.59 were elucidated by systematic substitution to D6.59A, D6.59E, D6.59N, D6.59R, and D6.59K (Table 1). The expected impact on function was confirmed by the right-shifted concentration-response curve of D6.59A, compared with the wild type (WT) receptor after stimulation with PrRP20 (Fig. 1D). The increased EC50 value (26 nm) of the D6.59A mutant confirms the importance of the Asp6.59 side chain. In addition, the results obtained for the other Asp6.59 single mutants support the hypothesis of an ionic interaction; D6.59E behaves similarly to WT. The oppositely charged D6.59K shows strong effects in potency and the bulkier, more positively charged D6.59R is not tolerated (Table 1). The impact of the substitutions increased as follows: Glu < Ala < Lys < Arg, showing that the lack of charge is a first critical component. This is followed by necessities in space and strength of the opposing charged Lys and Arg at position 6.59, showing different and increasing repulsion of the substitutions by PrRP20 stimulation (Table 1). Therefore, the charge seems to be a major prerequisite at position 6.59.
The signal transduction results obtained for PrRPR stimulation with peptide analogs Ala19PrRP20 and Ala20PrRP20 confirmed the essential influence of the formerly described RF-amide motif with respect to binding and signaling (Table 1 and supplemental Tables S1 and S2) (10–12). Circular dichroism (CD) spectroscopy showed that these variations have no influence on the PrRP20 overall structure, at least not detectable by CD (data not shown).5
Double Cycle Mutagenesis Suggests Additional Receptor Region “X” Critical for Peptide Binding
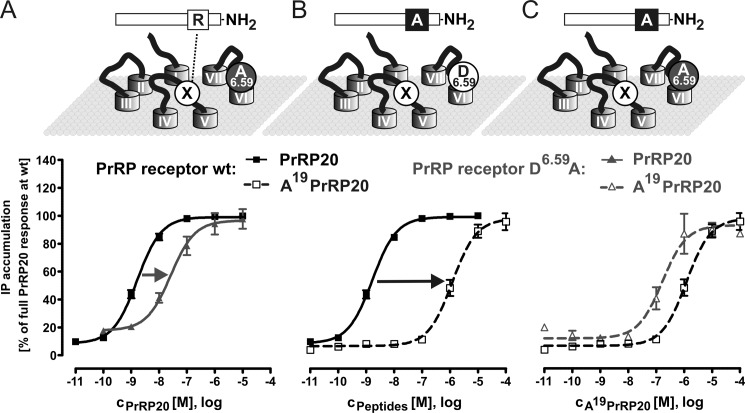
The concentration-response curve of the D6.59A receptor with PrRP20 reveals a 15-fold elevated EC50 value (Fig. 2A and Table 1), whereas the WT receptor stimulated with Ala19PrRP20 results in a 736-fold elevated EC50 value (Fig. 2B and Table 1). This finding suggests that Arg19 has one or more additional interaction partners, X, which explains the increased importance of Arg19 for receptor activity. Stimulation of the D6.59A receptor with Ala19PrRP20 resulted in a 0.16-fold elevated EC50 value, compared with PrRP20 stimulation. This non-additive effect of the double-cycle mutagenesis experiment implies that the effects of the individual replacements are not independent of each other. Among more complicated mechanisms, such as indirect interactions of the two residues, the effect may also be due to a direct interaction between Asp6.59 of PrRPR and Arg19 of PrRP20 (Fig. 2C and Table 1).
FIGURE 2.
Functional characterization of PrRP receptor mutant D6. 59A with PrRP20 and the modified ligand Ala19PrRP20. Schemes representing the postulated mode of ligand binding. Because of the significant difference in effect on EC50 of Asp6.59 and Arg19 mutants, a second contact point for Arg19 can be assumed. Complementary mutagenesis approach was used in combination with the signal transduction assay on cells, expressing the WT PrRPR or the D6.59A mutant to observe concentration-response curves. Data represent the means ± S.E. of multiple independent experiments (n = 32 for hPrRPR with PrRP20, n = 12 for D6.59A PrRPR with PrRP20, n = 11 for hPrRPR with Ala19PrRP20, and n = 3 for D6.59A PrRPR with Ala19PrRP20). Receptor activity is expressed as percentage of full PrRP20 response at the WT PrRP receptor. A, modification of receptor side. D6.59A PrRPR in comparison with WT receptor was stimulated with PrRP20. B, exploring the ligand side. Both PrRP20 and Ala19PrRP20 were investigated using WT PrRPR. C, complementary approach. Ala19PrRP20 stimulation of WT and mutant receptor resulted almost matching concentration-response curves, indicating an interaction between Asp6.59 of the receptor and Arg19 of the ligand.
Reciprocal Mutagenesis Leads to a Constitutively Active Receptor Mutant
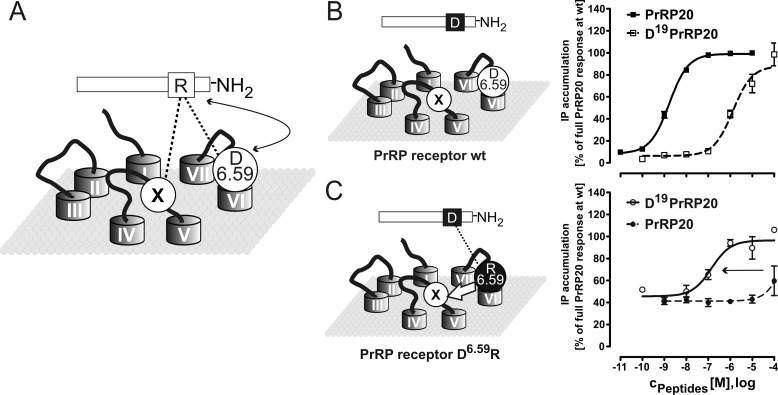
To confirm the direct interaction between Arg19 and Asp6.59, the corresponding residues were swapped (Fig. 3A). The herein performed reciprocal mutagenesis approach assumes that a lost interaction between two residues induced by single mutation to the counter amino acid can partly be recovered by a second mutation that establishes the interaction in a reverse manner. We used this method to verify the salt bridge between Asp6.59 and Arg19 in the PrRP/PrRPR system by using the peptide Asp19PrRP20 and the D6.59R receptor mutant (Fig. 3C). The single mutant peptide Asp19PrRP20 shows a similar effect as Ala19PrRP20, with an increased EC50 value of 1,318 nm (Table 1) without impact on the efficacy (Fig. 3B). We conclude that all peptide-receptor interactions that involve position Arg19 have been disrupted (Figs. 2B and 3B). In the reverse experiment, PrRP20 barely stimulated the D6.59R receptor mutant with no determinable EC50 value (Fig. 3C). In comparison with both single mutant experiments, the activation of the D6.59R, as well as D6.59K mutants, with Asp19PrRP20 revealed a gain of function (EC50 values: D6.59R = 138 nm and D6.59K = 115 nm, Table 1; Figs. 3C and 4C), confirming the direct interaction of Arg19 and Asp6.59. At the same time, the experiment provides further evidence in support of a second interaction site, X for D6.59R, as the EC50 value is still elevated by a factor of 84 compared with the WT interaction.
FIGURE 3.
Reciprocal mutagenesis of the PrRPR. A, this scheme displays the assumed WT situation with the direct interaction of ligand Arg19PrRP20 and receptor Asp6.59PrRPR, as well as the second unknown interaction of the Arg19 to the receptor. B, stimulation of WT receptor by Asp19PrRP20 and the corresponding concentration-response curves of the signal transduction assay. C, reciprocal mutagenesis scheme is shown with related concentration-response curves. Interestingly, D6.59R mutant is partially basally active and can be activated by Asp19PrRP20. The latter is due to the established Asp-Arg interaction. IP accumulation presented in B and C represent the means ± S.E. of multiple independent experiments (n = 32 for hPrRPR with PrRP20, n = 5 for D6.59R PrRPR with PrRP20, n = 4 for hPrRPR with Asp19PrRP20, and n = 3 for D6.59R PrRPR with Asp19PrRP20). Receptor activity is expressed as percentage of full PrRP20 response at the WT PrRP receptor.
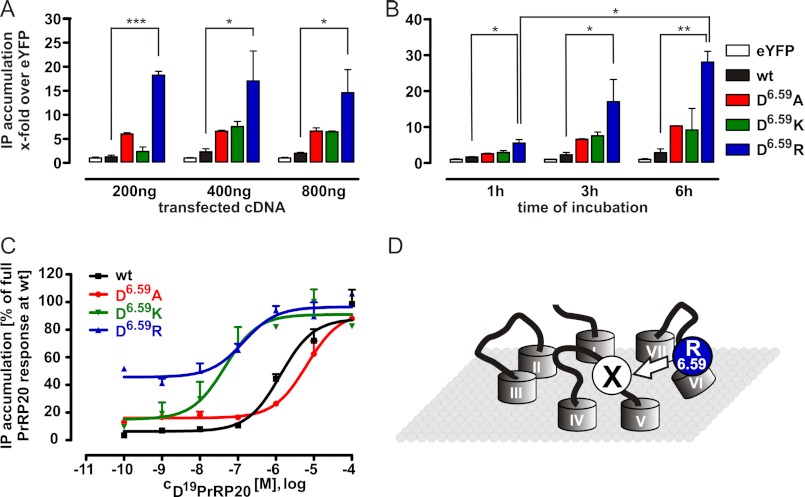
FIGURE 4.
Investigation of the constitutive activity of D6.59R PrRPR mutant. A, test of influence of transfection upon constitutive activity of WT PrRPR and Asp6.59 constructs. The IP accumulation of differently transient transfected COS-7 cells expressing the various PrRPR mutants was measured without any agonist after 3 h (given as x-fold over eYFP-expressing cells). (Each bar represents the mean ± S.E. of two different experiments; at least in triplicate; *, p < 0.05; **, p < 0.01; ***, p < 0.001.) B, constitutive activity of WT PrRPR and Asp6.59 mutant was investigated in a time-dependent manner. The IP accumulation of COS-7 cells expressing the different PrRPR variants was measured without any agonist after different times (given as x-fold over eYFP-expressing cells). C, concentration-response curves of Asp6.59 PrRP receptor mutants. Data represent the mean ± S.E. of multiple independent experiments (n = 5 for hPrRPR, n = 4 for D6.59A PrRPR, n = 3 for D6.59R PrRPR, and n = 2 for D6.59K PrRPR). Receptor activity is expressed as percentage of full PrRP20 response at the WT PrRP receptor. D, scheme of assumed explanation for the agonist-independent activity of the D6.59R receptor mutant. We postulate that the D6.59R is a CAM because D6.59R mimics Arg19 of PrRP20 by intramolecular interaction with a receptor region X, inducing a partially active receptor conformation.
A novel possibility to identify the missing interaction site arose because the D6.59R receptor mutant presented a strongly increased basal activity, which is indicated by curves with higher initial IP accumulation (Figs. 3C and. 4C). In contrast, D6.59A and D6.59K solely reveal a slight elevated basal activity. This can be explained by looser steric and electrostatic constraints at this position, thus making it more susceptible for induced basal activity, whereas for D6.59K, the spatial and more charged prerequisites are missing. The observed effect of constitutive activity is independent of transient transfection, which is a critical component. Different amounts of transfected DNA resulted in essentially similar cellular responses (Fig. 4A). Finally, the constitutive activity of the D6.59R receptor mutant was confirmed by an increased time-dependent IP accumulation compared with WT (Fig. 4B; 1 and 3 h = p < 0.05; 6 h = p < 0.01). All investigated receptors were correctly integrated in the membrane as confirmed by live cell microscopy (supplemental Fig. S1A) and revealed similar cell surface levels as determined by surface ELISA (supplemental Fig. S1, B and C).
Identification of X by Modeling-Guided Double Mutant Analysis
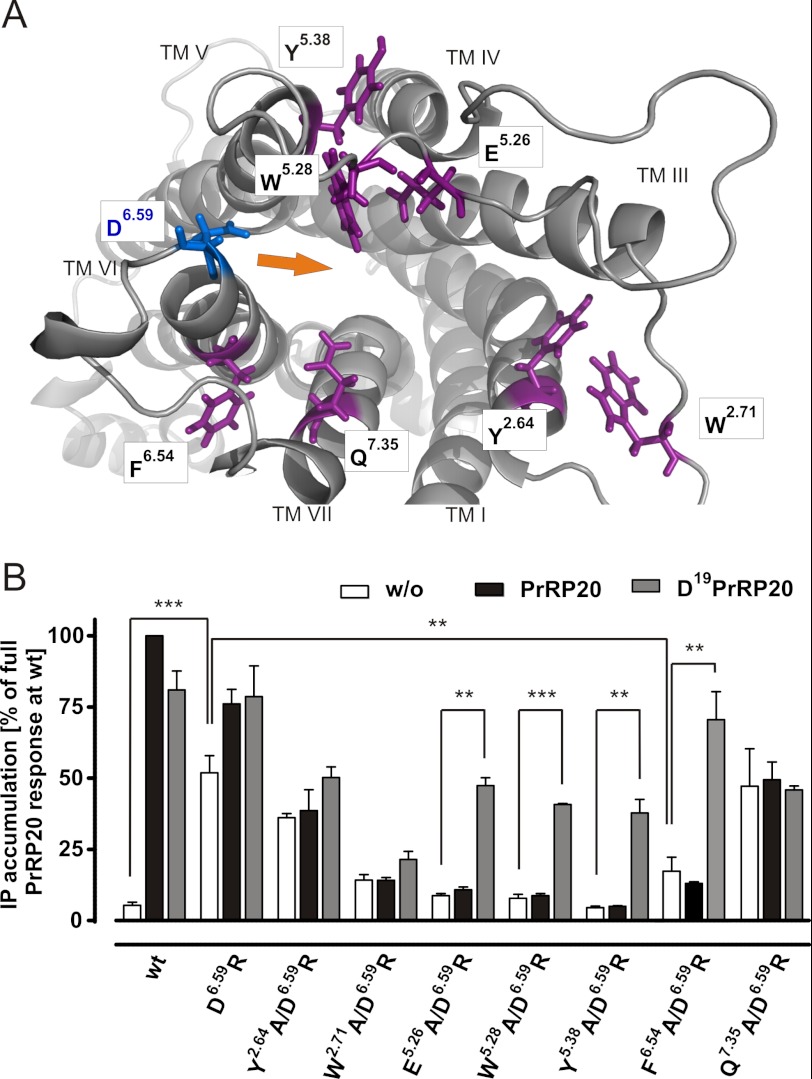
We hypothesize that D6.59R PrRPR is a CAM caused by the interaction of D6.59R with residue X. D6.59R mimics Arg19 of PrRP20, inducing a partially active receptor conformation (Fig. 4D). We further hypothesize that D6.59R/XX.XA double mutants will lose constitutive activity and, most importantly, retain activation by Asp19PrRP20. To determine likely positions for X, a comparative model of the PrRPR was constructed using the Rosetta molecular modeling software suite. Details of the modeling protocol are given under “Experimental Procedures.” According to the lowest energy model based on the semi-active opsin structure (PDB code 3DQB (32)), Glu5.26, Trp5.28, Tyr5.38, Phe6.54, and Gln7.35 were found proximal to Asp6.59 and were proposed to be potential interaction partners for D6.59R (Fig. 5A) or for Arg19PrRP20 when testing the WT receptor. The more distant residues, Tyr2.64 and Trp2.71, were chosen for control experiments.
FIGURE 5.
Molecular model of the PrRPR based on PDB code 3DQB and resulting double mutations based on the D6.59R PrRPR construct. A, residues in proximity to the extracellular side are shown in purple. These were investigated in double mutational analysis with D6.59R PrRPR. The Asp6.59 on top of TMH4 is colored in blue, and the suggested inward movement of the extracellular helical part of TMH6 is indicated by an orange arrow. B, new approach to identify the missing interaction site, X, arose by insertion of a second alanine substitution of assumed interacting residues to the D6.59R PrRPR. The second mutation is expected to diminish the basal activity but retain the capability to be activated by Asp19PrRP20. IP accumulation assay of COS-7 cells transfected with eYFP as control and the following constructs of PrRPR: WT; D6.59R; Y2.64A/D6.59R; W2.71A/D6.59R; E5.26A/D6.59R; W5.28A/D6.59R; Y5.38A/D6.59R; F6.54A/D6.59R; and Q7.35A/D6.59R; respectively. Incubation was performed for 3 h without ligand, PrRP20, or Asp19PrRP20, and results are presented in IP accumulation as percentage of full PrRP20 response at the WT PrRP receptor. (Each bar represents the mean ± S.E. of at least duplicates of four different experiments; **, p < 0.01; ***, p < 0.001.)
With guidance from the receptor modeling data (Fig. 5A), we generated and tested the double mutants Y2.64A/D6.59R, W2.71A/D6.59R, E5.26A/D6.59R, W5.28A/D6.59R, Y5.38A/D6.59R, F6.54A/D6.59R, and Q7.35A/D6.59R of PrRPR. Interestingly, E5.26A/D6.59R, W5.28A/D6.59R, and Y5.38A/D6.59R receptor mutants completely lost their constitutive activity in a ligand-independent signal transduction assay (Fig. 5B). The IP accumulation after 3 h of these unstimulated receptors dropped to a PrRPR WT level. The F6.54A/D6.59R dropped as well but remained partially constitutively active (Fig. 5B). These effects could be due to disruption of the hypothesized interaction to the Arg6.59 residue or to decisive structural alterations, resulting in generally nonfunctional mutants. The latter situation was excluded after activation of these constructs using 10 μm Asp19PrRP20 as an agonist (Fig. 5B; p < 0.01). In concentration-response experiments, the EC50 values were determined to be higher than 100 μm (Fig. 6A). The fact that Asp19PrRP20, not WT PrRP20, was able to activate these constructs re-emphasizes the direct interaction of Asp19 with D6.59R.
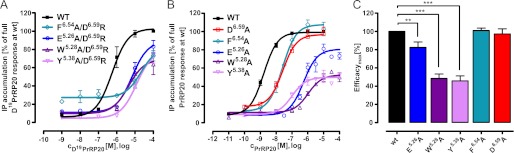
FIGURE 6.
Functional characterization of PrRPR mutants with impact on receptor activation and ligand binding. A, COS-7 cells transfected with WT PrRPR or E5.26A/D6.59R, W5.28A/D6.59R, Y5.38A/D6.59R, F6.54A/D6.59R receptor mutants, were stimulated for 3 h with different Asp19PrRP20 concentrations using a signal transduction assay. Data represent the mean ± S.E. of five (PrRPR), three (E5.26A/D6.59R, W5.28A/D6.59R, and Y5.38A/D6.59R) or two (F6.54A/D6.59R) independent experiments, measured in duplicate. B, COS-7 cells transfected with WT (n = 32) and E5.26A (n = 8), W5.28A (n = 7), Y5.38A (n = 5), D6.59A (n = 12), and F6.54A (n = 3) PrRPR mutants, respectively, were investigated in signal transduction assay, and data are presented in concentration-response curves as percentage of full PrRP20 response at WT PrRP receptor. Stimulation was performed for 1 h. The height of the curves correlates with the efficacy of the mutants. Potency is given by the degree of shift to the right and its resulting EC50 value. C, COS-7 cells transfected with the mentioned constructs in B were incubated for 1 h in a signal transduction assay with 1 × 10−5 m (mutants) or 1 × 10−7 m (WT) PrRP20, and without stimulus. Results are expressed as percentage of IP accumulation compared with the PrRPR, with lowest mean of value being 0% and highest 100%. (Bars represent the mean ± S.E. of duplicates of at least three different experiments; **, p < 0.05; ***, p < 0.001.)
Other double mutants, such as Y2.64A/D6.59R and Q7.35A/D6.59R, showed slightly reduced constitutive activity but seemed to be trapped in that state, as no further activation/stimulation was achieved. W2.71A/D6.59R appears to have structural restrictions because no significant receptor activation could be observed. From the plethora of residues in the upper TMHs and ELs of PrRPR, which may interact with D6.59R, the initial comparative models and mutational studies clearly suggested seven residues to potentially interact with D6.59R. Of these potential interaction sites, we hypothesize Glu5.26, Trp5.28, Tyr5.38, and Phe6.54 to be engaged in D6.59R-induced basal activity. Therefore, we postulate the latter residues to be involved in ligand binding and/or receptor activation. The combination of mutagenesis and comparative modeling enabled us to extract three residues of relevance from the plethora of residues in the upper TMHs and ELs of the PrRPR.
Confirmation of Binding and Activation Site Using Single Mutants
To clarify the exact impact of the identified positions Glu5.26, Trp5.28, Tyr5.38, and Phe6.54, single alanine mutants at these positions were generated. Signal transduction studies of the single alanine mutants E5.26A (331-fold over WT), W5.28A (580-fold over WT), Y5.38A (61-fold over WT), and F6.54A (15-fold over WT) confirm the impact of residues Glu5.26, Trp5.28, Tyr5.38, and Phe6.54 on ligand binding (Table 2 and Fig. 6B). Their distribution in EL2 and TMH5 suggests that this region plays a significant role in ligand binding. Therefore, EL2 and TMH5 were studied systematically to identify additional interaction sites that might have been missed due to inaccuracies of the comparative model. All charged (Arg, Lys, Glu, and Asp) and aromatic (Trp, Phe, and Tyr) residues between positions 4.65 and 5.40 were substituted to alanine (Table 2). None of the tested mutants resulted in significantly increased EC50 values (Fig. 6B and Table 2). This demonstrates that the model-guided intramolecular mutagenesis experiment, at least in this setting, was more effective than alanine scanning in selecting the critical interaction partners.
Because a constitutive internalization of the PrRP receptor has recently been reported (42), the cellular expression levels in the plasma membrane were investigated to verify our results of potency of the PrRP WT receptor and its mutants. Binding studies of transiently transfected COS-7 cells revealed a sufficient number of surface WT receptors per cell (∼95,000), calculated from the obtained Bmax value (445 Bq), the specific activity (3.52 × 1015 Bq/mol), and cell number (6.6 × 105). All PrRP receptor constructs with impact on potency were shown to be surface-exposed and quantified by surface ELISA (supplemental Fig. S1). The deviation from the WT PrRPR surface expression levels (WT = 39.6 ± 1.1%) varies from 16.3% (W5.28A) to 59.6% (F6.54A/D6.59A). However, these differences, basically resulting from transient transfection, reveal minor effects in the IP accumulation signaling assay setup, as the receptor mutant F6.54A (20.9 ± 3.7%) shows reduced total surface expression levels (supplemental Fig. S1B) but full WT-like efficacy (Fig. 5, B and C). Additionally, all PrRPR mutants are properly exported to the cell surface in comparable amounts as the WT receptor (39.6%, supplemental Fig. S1C). Therefore, the herein obtained results of potency of agonists at their receptor constructs do not result from altered expression or export levels.
A reduced efficacy was observed in the concentration-response-dependent signal transduction assay for W5.28A and Y5.38A (p < 0.001) and, with decreased impact, also for E5.26A (p < 0.0094, Fig. 6C and Table 2). In summary, our findings support a binding mechanism in which Glu5.26, in addition to Asp6.59, directly engage Arg19 of PrRP20 through ionic interactions. Phe6.54 might contribute to the overall global conformation of the binding pocket and positioning of TMH6, as its single mutation is less invasive but still is in distance for direct ligand interactions. We further suggest that Trp5.28 and Tyr5.38 are possibly in direct contact with the ligand and are indeed critical for receptor activation and the transmission of an external signal into the cell.
Exploration of Second Interaction Partner and Dual Binding Mode at Arg19
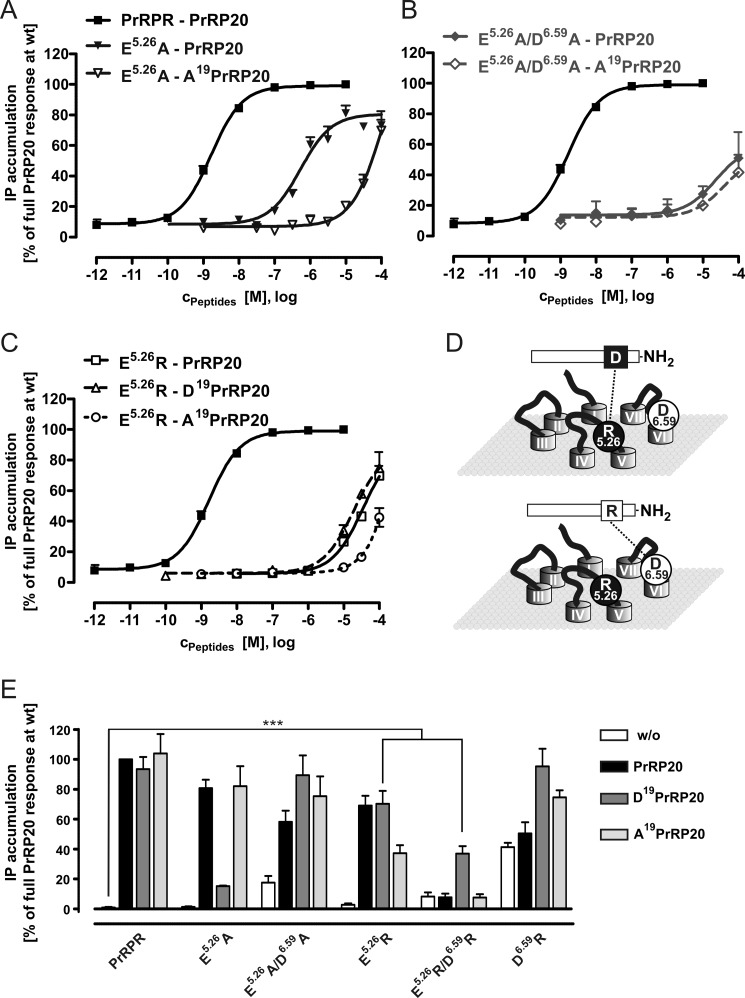
We generated the E5.26A/D6.59A double mutant of the receptor, which lacks both putative binding partners to the Arg19. In addition, the reciprocal PrRPR mutants, E5.26R/D6.59R and E5.26R, were generated to test the interaction by swapping the putative binding residues. The E5.26A and the E5.26A/D6.59A receptor mutants were investigated in a double-cycle mutagenesis study, where they were stimulated with Ala19PrRP20 and WT PrRP20 (Table 1 and Fig. 7A). The E5.26A mutant stimulated with Ala19PrRP20 resulted in a strongly increased EC50 value higher than 10 μm, 21-fold shifted compared with PrRP20 stimulation (537 nm). The enhanced EC50 value can be explained by the disruption of the second Arg19 interaction to receptor residue Asp6.59. Indeed, this effect agrees with a similar impact of the D6.59A mutation (15-fold shifted; Table 1), which also diminished the direct interaction to the Arg19 of the ligand to a similar extent (Figs. 2A and 7A). Furthermore, the stimulation of the E5.26A/D6.59A receptor mutant with either PrPR20 or Ala19PrRP20 resulted in matching curves. As no additional loss in potency was observed compared with the E5.26A mutant tested with Ala19PrRP20 (Fig. 7B), the experiment provides evidence that Glu5.26 is involved in binding to Arg19.
FIGURE 7.
Stimulation analysis of Glu5. 26 mutants reveals a preferential activation of Arg mutants by the reciprocal ligand Asp19PrRP20. Functional investigation of PrRPR mutants E5.26A, E5.26R, and E5.26A/D6.59A with the ligands PrRP20, Ala19PrRP20, or Asp19PrRP20 is shown. The signal transduction assay was performed in COS-7 cells expressing the WT PrRPR or E5.26A, E5.26R, or E5.26A/D6.59A mutants to observe concentration-response curves. Results of two independent experiments, each performed in duplicate, are presented as mean ± S.E. of duplicates. A, E5.26A PrRPR was stimulated with both PrRP20 and Ala19PrRP20 and demonstrated an equipotent loss in potency compared with the D6.59A PrRPR mutation (Fig. 2A). Additionally, this panel highlights the direct interaction between Arg19 and Asp6.59. B, stimulation with of the E5.26A/D6.59A receptor with Ala19PrRP20 or PrRP20 revealed no further loss in potency and a slightly decreased efficacy compared with the E5.26A PrRPR. This indicates that Glu5.26 might be the second binding partner of Arg19. C, functional characterization of the reciprocal E5.26R PrRPR mutant using Arg19-modified PrRP20 analogs. D, scheme shows the assumed interplay of attraction and repulsion for the reciprocal interaction of the ligands Arg19PrRP20 and Asp19PrRP20 with the E5.26R PrRP receptor mutant from C. E, IP accumulation assay of COS-7 cells transfected with eYFP as control and the following constructs of PrRPR: WT; E5.26A; E5.26A/D6.59A; E5.26R; E5.26R/D6.59R; D6.59R, respectively. Incubation was performed for 1 h using 100 μm of PrRP20, Asp19PrRP20, Ala19PrRP20, and without ligand. (Each bar represents the mean ± S.E. of at least duplicates of two different experiments; ***, p < 0.001.)
Next, the capability of receptor mutants E5.26A, E5.26A/D6.59A, E5.26R, E5.26R/D6.59R, D6.59A, and WT PrRPR to transmit signaling was tested (Fig. 7E). Importantly, the reciprocal receptor mutants E5.26R and E5.26R/D6.59R were significantly and best activated by Asp19PrRP20 (both: p < 0.001). In fact, E5.26R/D6.59R was solely activated by Asp19PrRP20. Finally, the E5.26R mutant was stimulated with PrRP20, Ala19PrRP20, and Asp19PrRP20 in a concentration-response experiment (Fig. 7C). This receptor mutant behaved similarly, when stimulated by PrRP20 and Asp19PrRP20 (both: EC50 value >10 μm). Along with the experiments testing Asp19PrRP20 stimulation of WT PrRPR, we demonstrate an approximately equal repulsive effect of Arg19 to E5.26R or Asp19 to Asp6.59 (Fig. 7D). This strengthens our hypothesis of a dual binding mode of Arg19 to Glu5.26 and Asp6.59.
Comparative Model of PrRP-Receptor Complex Provides Structural Information on Mode of Binding
Using the Arg19/Glu5.26 and Arg19/Asp6.59 contacts as restraints, a de novo-folded model of PrRP8-20 based on reported NMR data (18) was docked into an ensemble of comparative models of the PrRPR. The conformation of the EL regions was constructed simultaneously with ligand docking to accurately capture conformational changes induced by the peptide. Details of the modeling procedures are given in the supplemental Materials and Methods. The lowest energy Rosetta model features salt bridges between Asp6.59, Glu5.26, and Arg19. Trp5.28 and Tyr5.38 form π-stacking interactions that may be indicative of a “toggle-switch” mechanism (Fig. 8A) (43). Phe6.54 appears to be further apart from Arg19 but might contribute to the positioning of TMH6 via intra-molecular interactions and is in distance for π-stacking interactions with the Phe20 of PrRP20. Additional interactions between peptide and receptor hold the peptide in an optimal binding conformation deeply buried in the upper TMH segments and supported by the ELs from above.
DISCUSSION
We have evolved a strategy to interrogate detailed molecular mechanisms of GPCR activation by combining reciprocal, double-cycle, and intramolecular double mutagenesis with computational modeling. We apply this technique effectively to PrRPR and its CAM, D6.59R PrRPR, identifying distinct receptor residues involved in activation and/or ligand binding.
This is the first comprehensive mutational study of the extracellular and transmembrane regions of the PrRPR. The double-cycle mutagenic approach suggests the interaction (direct or indirect) between residues Asp6.59 and Arg19 and provides a first anchor point for receptor/ligand investigations. Interacting residues can be characterized by reciprocal mutagenesis, as shown before in an intramolecular study with the D2.61R/R7.39D swap in the gastrin-releasing peptide receptor (44) or the Asp6.44/Asn7.49 residues of the thyrotropin receptor (45). By applying this method to the PrRP/PrRPR system, the salt bridge of Asp6.59 to Arg19 was verified, and more importantly, by generating the D6.59R receptor, we identified the first CAM of the PrRPR. Up to now, numerous CAMs were generated and investigated in a plethora of previous studies, emphasizing their increasing importance. For example, CAMs of the human angiotensin II type 1 receptor with N3.35G (46), the β1B (47)/β2-adrenergic receptor (48, 50), the cannabinoid receptor 1 (51), muscarinic m1 (52), and m5 receptors (53), among others, have been found. Interestingly, more than 60 naturally occurring CAM GPCRs are known so far (54) and are often related to human disorders (55). Consequently, GPCRs activated in an agonist-independent manner are of emerging importance for drug development (3).
CAMs more readily undergo transition between active and inactive conformations due to removed conformational constraints of the inactive form (56). Because D6.59R in PrRPR is located at the top of TMH6, we hypothesize that this helix is involved in receptor activation via an inward movement of the upper helical region (Fig. 4D). Similarly to the PrRPR D6.59R CAM, mutant-induced receptor activity was observed in the S6.58Y/T6.59P double mutant of m5 muscarinic receptors (57). These data indicate that the top of TMH6 is directly involved in the switch between the active and the inactive state of several GPCRs and that the interaction with the ligand stabilizes the receptor in this active conformation, a notion that supports the “global toggle switch model” (58, 60). This model suggests that activation results from an inward movement of the extracellular ends of TMH6 and -7 toward TMH3, concomitant with a movement of the intracellular part of the TMHs in the opposite direction, which enables signaling via G-protein coupling. PrRPR represents an excellent model system to further investigate this hypothesis and gain insights to receptor activating mechanisms.
Previous work on the thyrotropin receptors showed the effects of spatially distant double mutants on constitutive activity (61, 62). However, we focused on the investigation of the molecular vicinity surrounding Asp6.59, as we suggest that specific inter-residue interactions of the generated CAM occur. To take advantage of the D6.59R CAM to elucidate the mechanism of ligand binding and PrRPR activation, we established an effective combination of intramolecular double- and inter-molecular reciprocal mutagenic approaches to study PrRPR activation by WT PrRP20, Ala19PrRP20, and Asp19PrRP20. With guidance from the initial PrRPR comparative model, possible interacting residues were considered (Fig. 5A), and the double mutants E5.26A/D6.59R, W5.28A/D6.59R, Y5.38A/D6.59R, and F6.54A/D6.59R revealed an involvement of these residues in receptor activation. Importantly, these receptor mutants were significantly activated by Asp19PrRP20 but not by WT PrRP20 (Fig. 5B), proving that the receptor mutants were not mis-folded and that Asp19 on the ligand is still able to interact with D6.59R. CAMs are thought to mimic, at least partially, the active conformation of the WT receptor and to spontaneously adopt a structure able to activate G-proteins (63). Therefore, we hypothesize that in Asp19PrRP20, residue Asp19 takes over the role of the destroyed intramolecular interaction of the double mutants, reactivating the “silenced” CAM. The conformation of a basally silenced GPCR might impair its intrinsic capacity for signaling compared with the WT receptor. Notably, further mutations within EL2/TMH5 had no considerable impact on receptor potency, in contrast to all three positions identified via intramolecular interactions (Table 2). This demonstrates the precision and usefulness of the modeling-guided double mutational approach to identify interacting residues in close proximity to the ligand.
In contrast, the W2.71A/D6.59R control turned out to be deficient in signaling. This is expected and in agreement with the high conservation of Trp2.70/Trp2.71 in most peptide GPCRs, e.g. in the NPY receptor system (14). Furthermore, Trp2.71 is located in the structurally relevant WXGF motif, which is suggested to be a key component in the activation mechanism in many GPCRs in the rhodopsin family (64). Recent investigations on TMH2 of the CAM N3.35G hAT1 suggested TMH2 to pivot, bringing the top of TMH2 closer to the binding pocket (65). Our results obtained for the conserved Tyr2.64 on top of TMH2 do not support such a spatial approach to Asp6.59 and thus to the binding pocket. This reflects the divergence of GPCR activation and accentuates that the mode of activation is not a common mechanism.
The results obtained from studies of the E5.26A mutation lead to the conclusion that this residue is predominantly responsible for ligand binding. Our initial double cycle mutagenic experiments at Asp6.59 support a more complex double binding role for Arg19 of PrRP20, which appears to be in contact with two sites on PrRPR. Accordingly, we suggest Glu5.26 to be the second binding partner for peptide residue Arg19 (Fig. 7D). The extensive mutagenic studies of residue Glu5.26 strongly indicate the participation in binding to Arg19, and the constitutive activity of D6.59R supports the hypothesis of a second Arg-specific interaction site in PrRPR that can be satisfied by the D6.59R but not the D6.59K mutant. A similar dual binding mode for arginine was recently reported for gonadotropin-releasing hormone receptor (66). This has been supported by other studies, where substitution of Arg19 to lysine, citrulline, α-amino-4-guanidinobutyric acid, or α-amino-3-guanidinopropionic acid on the peptide lead to reduced binding affinities (12). Interestingly, the tight ensemble of models that is in agreement with the experimental data presented herein exhibits variability in EL1 and -2 while still maintaining the contacts between Asp6.59 and Glu5.26 with Arg19. Given this structural variability in our models, we emphasize that the presented approach is an iterative process, where initial models can be used to guide experimental design, and the resulting data allow for model refinement. The current PrRP receptor model can only be considered valid in the light of the functional data. However, it provides insight into possible structural mechanisms of peptide-receptor interactions and receptor activation.
W5.28A and Y5.38A also showed lowered ligand potency, but both mutants revealed a strongly decreased ability to transmit signals compared with the WT receptor (Table 2). This effect may result from intramolecular structural alteration due to the lack of aromaticity at the Y5.38A site. Mutational studies reported for the nearby Tyr5.39 residue in both cannabinoid receptors (CB1 and CB2) revealed that the aromaticity at this position is crucial (67). The PrRP receptor model places Trp5.28 in close proximity to Tyr5.38 (Fig. 8A). In this model, the residues form stacking interactions, but this remains to be proven experimentally. We speculate that, due to the effects observed for potency and efficacy, Trp5.28 and Tyr5.38 are related to receptor activation. In contrast, F6.54A mutant reveals full WT efficacy accompanied with reduced potency. From the docked modeling data, we speculate that this residue contributes to the correct conformation of the binding pocket and might interact with the Phe20 of the PrPR20.
Evolutionary and structural studies revealed that the PrRPR belongs to the family of RF-amide peptide receptors, consisting of five discovered groups as follows: the neuropeptide FF group, the PrRP group, the gonadotropin-inhibitory hormone group, the kisspeptin group, and the 26RFa group (68–70). However, further phylogenic investigations revealed that the PrPRR shares an ancient receptor with the NPY receptors (17). The human PrRPR possesses high sequence identity with the human NPY2R, particularly in the upper and middle regions of TMH4, TMH5, and TMH6. It is suggested that the PrRPR family began co-evolving with ancestral PrRP/C-RF-amide peptide with a redundant NPY binding receptor (17). This explains the importance of the conserved Asp6.59 residue and in turn might have been responsible for the development of a double binding mode for Arg19 in the PrRPR/PrRP system. It could be speculated that other RF-amide receptors evolved similar binding modes for the crucial arginine within the RF-amide motif, especially for the closely related 26RF-amide receptor. In contrast, for the well investigated Y-receptor family, a double binding mode was not identified, neither for Arg33 at Y2- and Y5-receptor nor for Arg35 at Y1- and Y4 receptor (14, 15). However, the second interaction might occur via the second arginine 33 or 35, respectively.
Regarding medical and physiological implications, the expression of CAMs can entail oncogenic effects, such as tumor formation in nude mice (71). A variety of diseases is known to be triggered by elevated basal activity, including autosomal dominant hypocalcemia (49) and ovarian hyperstimulation syndrome (59). Our findings provide insight into the harmful potential of CAMs and demonstrate the need for applicable drugs that are able to diminish mutation-induced receptor activity. We are confident that our technique is a promising tool to investigate residues relevant for ligand binding and receptor activation because a CAM is used as a template. Our approach paves the way for obtaining specific structure/function information on a molecular level, which is of indispensable value, as no crystal structure for a peptide GPCR currently exists. This method will hopefully contribute to the elucidation of the structural mechanisms of harmful CAMs and help to develop and increase the number of inverse-agonist drugs that target these receptors.
Acknowledgments
We thank Kristin Löbner and Christina Dammann for their technical assistance in peptide synthesis, Janet Schwesinger for sequencing, and Regina Reppich-Sacher for recording mass spectra. We also thank members of the RosettaCommons, Elizabeth Dong, David Nannemann, Steven Combs, and Anette Kaiser, for their assistance and insight provided concerning the molecular modeling.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 MH090192 and R01 GM080403 (to J. M.) and Deutsche Forschungsgemeinschaft Grant SFB 610, BE 1264-11 (to A. B. S.).

This article contains supplemental Materials and Methods, Fig. S1, and Tables S1 and S2.
S. H. DeLuca, D. Rathmann, A.G. Beck-Sickinger, and J. Meiler, manuscript in preparation.
- GPCR
- G-protein-coupled receptor
- PrRPR
- prolactin-releasing peptide receptor
- CAM
- constitutively active mutant
- NPY
- neuropeptide Y
- IP
- inositol phosphate
- eYFP
- enhanced yellow fluorescence protein
- EL
- extracellular loop
- TMH
- transmembrane helix
- GnRH
- gonadotropin-releasing hormone
- PrRP
- prolactin-releasing peptide
- PDB
- Protein Data Bank.
REFERENCES
- 1. Hopkins A. L., Groom C. R. (2002) The druggable genome. Nat. Rev. Drug Discov. 1, 727–730 [DOI] [PubMed] [Google Scholar]
- 2. Lagerström M. C., Schiöth H. B. (2008) Structural diversity of G-protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7, 339–357 [DOI] [PubMed] [Google Scholar]
- 3. Bond R. A., Ijzerman A. P. (2006) Recent developments in constitutive receptor activity and inverse agonism, and their potential for GPCR drug discovery. Trends Pharmacol. Sci. 27, 92–96 [DOI] [PubMed] [Google Scholar]
- 4. Welch S. K., O'Hara B. F., Kilduff T. S., Heller H. C. (1995) Sequence and tissue distribution of a candidate G-coupled receptor cloned from rat hypothalamus. Biochem. Biophys. Res. Commun. 209, 606–613 [DOI] [PubMed] [Google Scholar]
- 5. Fujii R., Fukusumi S., Hosoya M., Kawamata Y., Habata Y., Hinuma S., Sekiguchi M., Kitada C., Kurokawa T., Nishimura O., Onda H., Sumino Y., Fujino M. (1999) Tissue distribution of prolactin-releasing peptide (PrRP) and its receptor. Regul. Pept. 83, 1–10 [DOI] [PubMed] [Google Scholar]
- 6. Langmead C. J., Szekeres P. G., Chambers J. K., Ratcliffe S. J., Jones D. N., Hirst W. D., Price G. W., Herdon H. J. (2000) Characterization of the binding of 125I-human prolactin releasing peptide (PrRP) to GPR10, a novel G-protein-coupled receptor. Br. J. Pharmacol. 131, 683–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fukusumi S., Fujii R., Hinuma S. (2006) Recent advances in mammalian RF-amide peptides. The discovery and functional analyses of PrRP, RFRPs, and QRFP. Peptides 27, 1073–1086 [DOI] [PubMed] [Google Scholar]
- 8. Hinuma S., Habata Y., Fujii R., Kawamata Y., Hosoya M., Fukusumi S., Kitada C., Masuo Y., Asano T., Matsumoto H., Sekiguchi M., Kurokawa T., Nishimura O., Onda H., Fujino M. (1998) A prolactin-releasing peptide in the brain. Nature 393, 272–276 [DOI] [PubMed] [Google Scholar]
- 9. Lin S. H. (2008) Prolactin-releasing peptide. Results Probl. Cell Differ. 46, 57–88 [DOI] [PubMed] [Google Scholar]
- 10. Boyle R. G., Downham R., Ganguly T., Humphries J., Smith J., Travers S. (2005) Structure-activity studies on prolactin-releasing peptide (PrRP). Analogs of PrRP-(19–31)-peptide. J. Pept. Sci. 11, 161–165 [DOI] [PubMed] [Google Scholar]
- 11. Roland B. L., Sutton S. W., Wilson S. J., Luo L., Pyati J., Huvar R., Erlander M. G., Lovenberg T. W. (1999) Anatomical distribution of prolactin-releasing peptide and its receptor suggests additional functions in the central nervous system and periphery. Endocrinology 140, 5736–5745 [DOI] [PubMed] [Google Scholar]
- 12. Danho W., Swistok J., Khan W., Truitt T., Kurylko G., Fry D., Greeley D., Sun H., Dvorozniak M., Machie G., Spence C., Goodnow R. (2003) Structure-activity relationships and bioactive conformations of prolactin-releasing peptides. Ligands for a potential obesity target. 18th American Peptide Symposium, Boston, MA, July 19–23, 2003, Kluwer Academic, Norwell, MA [Google Scholar]
- 13. Conner A. C., Barwell J., Poyner D. R., Wheatley M. (2011) The use of site-directed mutagenesis to study GPCRs. Methods Mol. Biol. 746, 85–98 [DOI] [PubMed] [Google Scholar]
- 14. Lindner D., van Dieck J., Merten N., Mörl K., Günther R., Hofmann H. J., Beck-Sickinger A. G. (2008) GPC receptors and not ligands decide the binding mode in neuropeptide Y multireceptor/multiligand system. Biochemistry 47, 5905–5914 [DOI] [PubMed] [Google Scholar]
- 15. Merten N., Lindner D., Rabe N., Römpler H., Mörl K., Schöneberg T., Beck-Sickinger A. G. (2007) Receptor subtype-specific docking of Asp6.59 with C-terminal arginine residues in Y receptor ligands. J. Biol. Chem. 282, 7543–7551 [DOI] [PubMed] [Google Scholar]
- 16. Ballesteros J. A., Weinstein H. (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 [Google Scholar]
- 17. Lagerström M. C., Fredriksson R., Bjarnadóttir T. K., Fridmanis D., Holmquist T., Andersson J., Yan Y. L., Raudsepp T., Zoorob R., Kukkonen J. P., Lundin L. G., Klovins J., Chowdhary B. P., Postlethwait J. H., Schiöth H. B. (2005) Origin of the prolactin-releasing hormone (PRLH) receptors. Evidence of coevolution between PRLH and a redundant neuropeptide Y receptor during vertebrate evolution. Genomics 85, 688–703 [DOI] [PubMed] [Google Scholar]
- 18. D'Ursi A. M., Albrizio S., Di Fenza A., Crescenzi O., Carotenuto A., Picone D., Novellino E., Rovero P. (2002) Structural studies on Hgr3 orphan receptor ligand prolactin-releasing peptide. J. Med. Chem. 45, 5483–5491 [DOI] [PubMed] [Google Scholar]
- 19. Carter P. J., Winter G., Wilkinson A. J., Fersht A. R. (1984) The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell 38, 835–840 [DOI] [PubMed] [Google Scholar]
- 20. Krylov D., Mikhailenko I., Vinson C. (1994) A thermodynamic scale for leucine zipper stability and dimerization specificity. e and g interhelical interactions. EMBO J. 13, 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lang M., Bufe B., De Pol S., Reiser O., Meyerhof W., Beck-Sickinger A. G. (2006) Structural properties of orexins for activation of their receptors. J. Pept. Sci. 12, 258–266 [DOI] [PubMed] [Google Scholar]
- 22. Findeisen M., Rathmann D., Beck-Sickinger A. G. (2011) Structure-activity studies of RF-amide peptides reveal subtype-selective activation of neuropeptide FF1 and FF2 receptors. ChemMedChem. 6, 1081–1093 [DOI] [PubMed] [Google Scholar]
- 23. Sanger F., Nicklen S., Coulson A. R. (1992) DNA sequencing with chain-terminating inhibitors. 1977. Biotechnology 24, 104–108 [PubMed] [Google Scholar]
- 24. Böhme I., Stichel J., Walther C., Mörl K., Beck-Sickinger A. G. (2008) Agonist induced receptor internalization of neuropeptide Y receptor subtypes depends on third intracellular loop and C terminus. Cell. Signal. 20, 1740–1749 [DOI] [PubMed] [Google Scholar]
- 25. Reynolds C. P., Biedler J. L., Spengler B. A., Reynolds D. A., Ross R. A., Frenkel E. P., Smith R. G. (1986) Characterization of human neuroblastoma cell lines established before and after therapy. J. Natl. Cancer Inst. 76, 375–387 [PubMed] [Google Scholar]
- 26. Höfliger M. M., Castejón G. L., Kiess W., Beck Sickinger A. G. (2003) Novel cell line selectively expressing neuropeptide Y-Y2 receptors. J. Recept. Signal. Transduct. Res. 23, 351–360 [DOI] [PubMed] [Google Scholar]
- 27. Böhme I., Mörl K., Bamming D., Meyer C., Beck-Sickinger A. G. (2007) Tracking of human Y receptors in living cells. A fluorescence approach. Peptides 28, 226–234 [DOI] [PubMed] [Google Scholar]
- 28. Koglin N., Lang M., Rennert R., Beck-Sickinger A. G. (2003) Facile and selective nanoscale labeling of peptides in solution by using photolabile protecting groups. J. Med. Chem. 46, 4369–4372 [DOI] [PubMed] [Google Scholar]
- 29. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Konagurthu A. S., Whisstock J. C., Stuckey P. J., Lesk A. M. (2006) MUSTANG. A multiple structural alignment algorithm. Proteins 64, 559–574 [DOI] [PubMed] [Google Scholar]
- 31. Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., Buss V. (2004) The retinal conformation and its environment in rhodopsin in light of a new 2.2-Å crystal structure. J. Mol. Biol. 342, 571–583 [DOI] [PubMed] [Google Scholar]
- 32. Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 33. Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 34. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) High resolution crystal structure of an engineered human β2-adrenergic G-protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) The 2.6-Å crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mandell D. J., Coutsias E. A., Kortemme T. (2009) Sub-Å accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 6, 551–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Canutescu A. A., Dunbrack R. L., Jr. (2003) Cyclic coordinate descent. A robotics algorithm for protein loop closure. Protein Sci. 12, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barth P., Schonbrun J., Baker D. (2007) Toward high resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. U.S.A. 104, 15682–15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 41. Rohl C. A. (2005) Protein structure estimation from minimal restraints using Rosetta. Methods Enzymol. 394, 244–260 [DOI] [PubMed] [Google Scholar]
- 42. Madsen K. L., Thorsen T. S., Rahbek-Clemmensen T., Eriksen J., Gether U. (2012) Protein interacting with C kinase 1 (PICK1) reduces reinsertion rates of interaction partners sorted to Rab11-dependent slow recycling pathway. J. Biol. Chem. 287, 12293–12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 44. Donohue P. J., Sainz E., Akeson M., Kroog G. S., Mantey S. A., Battey J. F., Jensen R. T., Northup J. K. (1999) An aspartate residue at the extracellular boundary of TMII and an arginine residue in TMVII of the gastrin-releasing peptide receptor interact to facilitate heterotrimeric G-protein coupling. Biochemistry 38, 9366–9372 [DOI] [PubMed] [Google Scholar]
- 45. Govaerts C., Lefort A., Costagliola S., Wodak S. J., Ballesteros J. A., Van Sande J., Pardo L., Vassart G. (2001) A conserved Asn in transmembrane helix 7 is an on/off switch in the activation of the thyrotropin receptor. J. Biol. Chem. 276, 22991–22999 [DOI] [PubMed] [Google Scholar]
- 46. Arsenault J., Cabana J., Fillion D., Leduc R., Guillemette G., Lavigne P., Escher E. (2010) Temperature-dependent photolabeling of the human angiotensin II type 1 receptor reveals insights into its conformational landscape and its activation mechanism. Biochem. Pharmacol. 80, 990–999 [DOI] [PubMed] [Google Scholar]
- 47. Scheer A., Fanelli F., Costa T., De Benedetti P. G., Cotecchia S. (1996) Constitutively active mutants of the α1B-adrenergic receptor. Role of highly conserved polar amino acids in receptor activation. EMBO J. 15, 3566–3578 [PMC free article] [PubMed] [Google Scholar]
- 48. Lefkowitz R. J., Cotecchia S., Samama P., Costa T. (1993) Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol. Sci. 14, 303–307 [DOI] [PubMed] [Google Scholar]
- 49. Okazaki R., Chikatsu N., Nakatsu M., Takeuchi Y., Ajima M., Miki J., Fujita T., Arai M., Totsuka Y., Tanaka K., Fukumoto S. (1999) A novel activating mutation in calcium-sensing receptor gene associated with a family of autosomal dominant hypocalcemia. J. Clin. Endocrinol. Metab. 84, 363–366 [DOI] [PubMed] [Google Scholar]
- 50. Samama P., Cotecchia S., Costa T., Lefkowitz R. J. (1993) A mutation-induced activated state of the β2-adrenergic receptor. Extending the ternary complex model. J. Biol. Chem. 268, 4625–4636 [PubMed] [Google Scholar]
- 51. D'Antona A. M., Ahn K. H., Wang L., Mierke D. F., Lucas-Lenard J., Kendall D. A. (2006) A cannabinoid receptor 1 mutation proximal to the DRY motif results in constitutive activity and reveals intramolecular interactions involved in receptor activation. Brain Res. 1108, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Högger P., Shockley M. S., Lameh J., Sadée W. (1995) Activating and inactivating mutations in N- and C-terminal i3 loop junctions of muscarinic acetylcholine Hm1 receptors. J. Biol. Chem. 270, 7405–7410 [DOI] [PubMed] [Google Scholar]
- 53. Spalding T. A., Burstein E. S., Brauner-Osborne H., Hill-Eubanks D., Brann M. R. (1995) Pharmacology of a constitutively active muscarinic receptor generated by random mutagenesis. J. Pharmacol. Exp. Ther. 275, 1274–1279 [PubMed] [Google Scholar]
- 54. Seifert R., Wenzel-Seifert K. (2002) Constitutive activity of G-protein-coupled receptors. Cause of disease and common property of wild type receptors. Naunyn-Schmiedeberg's Arch. Pharmacol. 366, 381–416 [DOI] [PubMed] [Google Scholar]
- 55. Spiegel A. M. (1996) Defects in G-protein-coupled signal transduction in human disease. Annu. Rev. Physiol. 58, 143–170 [DOI] [PubMed] [Google Scholar]
- 56. Gether U., Ballesteros J. A., Seifert R., Sanders-Bush E., Weinstein H., Kobilka B. K. (1997) Structural instability of a constitutively active G-protein-coupled receptor. Agonist-independent activation due to conformational flexibility. J. Biol. Chem. 272, 2587–2590 [DOI] [PubMed] [Google Scholar]
- 57. Ford D. J., Essex A., Spalding T. A., Burstein E. S., Ellis J. (2002) Homologous mutations near the junction of the sixth transmembrane domain and the third extracellular loop lead to constitutive activity and enhanced agonist affinity at all muscarinic receptor subtypes. J. Pharmacol. Exp. Ther. 300, 810–817 [DOI] [PubMed] [Google Scholar]
- 58. Schwartz T. W., Frimurer T. M., Holst B., Rosenkilde M. M., Elling C. E. (2006) Molecular mechanism of 7TM receptor activation. A global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 46, 481–519 [DOI] [PubMed] [Google Scholar]
- 59. Gromoll J., Simoni M., Nordhoff V., Behre H. M., De Geyter C., Nieschlag E. (1996) Functional and clinical consequences of mutations in the FSH receptor. Mol. Cell. Endocrinol. 125, 177–182 [DOI] [PubMed] [Google Scholar]
- 60. Elling C. E., Frimurer T. M., Gerlach L. O., Jorgensen R., Holst B., Schwartz T. W. (2006) Metal ion site engineering indicates a global toggle switch model for seven-transmembrane receptor activation. J. Biol. Chem. 281, 17337–17346 [DOI] [PubMed] [Google Scholar]
- 61. Kleinau G., Jaeschke H., Mueller S., Worth C. L., Paschke R., Krause G. (2008) Molecular and structural effects of inverse agonistic mutations on signaling of the thyrotropin receptor. A basally active GPCR. Cell. Mol. Life Sci. 65, 3664–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grüters A., Schöneberg T., Biebermann H., Krude H., Krohn H. P., Dralle H., Gudermann T. (1998) Severe congenital hyperthyroidism caused by a germ line neo mutation in the extracellular portion of the thyrotropin receptor. J. Clin. Endocrinol. Metab. 83, 1431–1436 [DOI] [PubMed] [Google Scholar]
- 63. Cotecchia S., Fanelli F., Costa T. (2003) Constitutively active G-protein-coupled receptor mutants. Implications on receptor function and drug action. Assay Drug Dev. Technol. 1, 311–316 [DOI] [PubMed] [Google Scholar]
- 64. Klco J. M., Nikiforovich G. V., Baranski T. J. (2006) Genetic analysis of the first and third extracellular loops of the C5a receptor reveals an essential WXFG motif in the first loop. J. Biol. Chem. 281, 12010–12019 [DOI] [PubMed] [Google Scholar]
- 65. Domazet I., Holleran B. J., Martin S. S., Lavigne P., Leduc R., Escher E., Guillemette G. (2009) The second transmembrane domain of the human type 1 angiotensin II receptor participates in the formation of the ligand binding pocket and undergoes integral pivoting movement during the process of receptor activation. J. Biol. Chem. 284, 11922–11929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Flanagan C. A., Rodic V., Konvicka K., Yuen T., Chi L., Rivier J. E., Millar R. P., Weinstein H., Sealfon S. C. (2000) Multiple interactions of the Asp(2.61(98)) side chain of the gonadotropin-releasing hormone receptor contribute differentially to ligand interaction. Biochemistry 39, 8133–8141 [DOI] [PubMed] [Google Scholar]
- 67. McAllister S. D., Tao Q., Barnett-Norris J., Buehner K., Hurst D. P., Guarnieri F., Reggio P. H., Nowell Harmon K. W., Cabral G. A., Abood M. E. (2002) A critical role for a tyrosine residue in the cannabinoid receptors for ligand recognition. Biochem. Pharmacol. 63, 2121–2136 [DOI] [PubMed] [Google Scholar]
- 68. Ukena K., Vaudry H., Leprince J., Tsutsui K. (2011) Molecular evolution and functional characterization of the orexigenic peptide 26RFa and its receptor in vertebrates. Cell Tissue Res. 343, 475–481 [DOI] [PubMed] [Google Scholar]
- 69. Osugi T., Ukena K., Sower S. A., Kawauchi H., Tsutsui K. (2006) Evolutionary origin and divergence of PQRF-amide peptides and LPXRF-amide peptides in the RF-amide peptide family. Insights from novel lamprey RF-amide peptides. FEBS J. 273, 1731–1743 [DOI] [PubMed] [Google Scholar]
- 70. Findeisen M., Rathmann D., Beck-Sickinger A. G. (2011) RF-amide peptides. Structure, function, mechanisms, and pharmaceutical potential. Pharmaceuticals 4, 1248–1280 [Google Scholar]
- 71. Allen L. F., Lefkowitz R. J., Caron M. G., Cotecchia S. (1991) G-protein-coupled receptor genes as protooncogenes. Constitutively activating mutation of the α1B-adrenergic receptor enhances mitogenesis and tumorigenicity. Proc. Natl. Acad. Sci. U.S.A. 88, 11354–11358 [DOI] [PMC free article] [PubMed] [Google Scholar]