Background: Regulation of protein function by phosphorylation is an important mechanism to control many cellular processes.
Results: We found that the mRNA-binding protein HuR is phosphorylated by Cdk5 at the serine 202 residue.
Conclusion: The aberrant phosphorylation of HuR at Ser-202 affects centrosome function and induces arrest of cell cycle progression.
Significance: This work emphasizes HuR phosphorylation as a novel molecular target in cancer.
Keywords: Cancer, Centrosome, mRNA, Phosphorylation, RNA-binding Protein
Abstract
Hu antigen R (HuR) is an mRNA-binding protein belonging to the ELAV family. It is highly expressed in cancer and involved in cell survival and proliferation. The impact of post-translational regulation of HuR and resulting cellular effects are poorly understood. In the current report, we describe a direct interaction between HuR and Cdk5 in glioma. We determined that Cdk5 specifically phosphorylates HuR at the serine 202 residue in the unique hinge region. The molecular consequences of this interaction are an altered HuR ability to bind, stabilize, and promote translation of mRNAs. At the cellular level, the anomalous HuR phosphorylation at this site evokes robust defects in centrosome duplication and cohesion as well as arrest of cell cycle progression. Subcellular fractionation and immunofluorescence technique confirm a direct integration of HuR and Cdk5 with centrosomes. We propose that HuR stores mRNA in the centrosome and that HuR phosphorylation by Cdk5 controls de novo protein synthesis in near proximity to centrosomes and, thus, impacts centrosome function.
Introduction
The cyclin-dependent kinase (Cdk)2 family is implicated in cell cycle progression, cell differentiation, and migration. The Cdk family contains two types of members: first a the common one, which must bind to cyclin proteins to become active (Cdk1, -2, -3, -4, -6, -7, -8, and -11), and a unique, cyclin-independent member (Cdk5) activated by p39, p35, and p25 co-factors (1–4). The expression of cyclin-dependent Cdk members is not tissue-specific; however, the unique Cdk5 is mainly expressed in the central nervous system (CNS) (4, 5). Cdk5 is regulated by both growth factor (PDGF, EGF/βFGF)-dependent and -independent mechanisms. Phosphorylation-independent Cdk5 activation by p35 and p25 is strictly controlled during development and aberrant in tumor tissue (6–8).
Many CNS functions, including memory, movements, and plasticity, have been reported as controlled by Cdk5 (9). As a result, a number of CNS diseases, such as Huntington disease, Alzheimer disease, and Parkinson disease have been associated with aberrant expression or regulation of Cdk5 (10). During early stages of development, the depletion of Cdk5 from neuronal stem cells abolishes neurogenesis (11, 12). The homozygote Cdk5 gene deletion results in global abnormalities of neuronal migration, affects differentiation, and is lethal for the mouse embryo (13). In mature neurons, Cdk5 subcellular localization controls cell cycle initiation and consequent neuronal death (14, 15). Cdk5 is required for synapses formation and is important in local protein synthesis and synaptic transmission (16, 17). In tumor cells, Cdk5 overexpression or activation by p35 and growth factors promotes an aggressive metastatic behavior, resistance to the chemotherapeutic drugs, and, in some cases, alterations of cell cycle progression (7, 8, 18, 19). The mechanisms implicated in Cdk5 function include phosphorylation and activation of transcriptional factors, ion channels, cytoskeleton proteins, and kinases (19–21).
In the present paper, we report a novel Cdk5 target, the mRNA-binding protein HuR. HuR is highly conserved during development, is required for angiogenesis and neurogenesis in normal tissues, and is involved in inflammation, migration, proliferation, and survival of different types of transformed cells (22–28). HuR is essential for cell cycle progression and may influence cytoskeleton architecture (22–28). At the molecular level, HuR directs mRNA stabilization, localization, splicing, and translation. Several kinases (Cdk1, PKCα, PKCδ, and Chk2) phosphorylate HuR and influence HuR function and subcellular localization (29, 30).
This study provides evidence of HuR and Cdk5 interaction in cancer cells. We determine that HuR is phosphorylated by Cdk5 at serine 202. Centrosomes were discovered as sites of functional significance of HuR and Cdk5 interaction. At the cellular level, the prolonged disruption of the balance between phosphorylated and dephosphorylated HuR at serine 202 provokes an arrest of cell cycle progression in glioma cells.
EXPERIMENTAL PROCEDURES
DNA Constructs and Stable Cell Line Creation
mRNA was isolated from primary GBM samples with TRIzol reagent and converted to cDNA by RT-PCR for cloning. Cdk5 was cloned by using CDK5-FORW (5′-GCGATGCAGAAATACGAGAAAC) and CDK5-Rev (5′-CTAGGGCGGACAGAAGTCGG) primers (Sigma) and ligated in pCR-2.1TOPO vector (Invitrogen). Cdk5 was recloned in pDsRed2-C1 (Clontech) (HindIII/BamHI) by using Forw/Cdk5/Hind3 (5′-GAGCTCAAGCTTCGCAGAAATACGAGAAACTGGAAAGGATTG) and Rev/Cdk5/BamHI (5′-GCCGGTGGATCCCTAGGGCGGACAGAAGTCGGAG) primers. HuR S202A, HuR S221A, and HuR S202D constructs were achieved by point mutagenesis on FLAG-HuR wild type (WT) in pTre2Hyg vector. HuR WT, HuR S202A, and HuR S221A were cloned in pGEX-6P vector for recombinant protein expression and purification from bacteria. The PACT domain of pericentrin attached to the red fluorescence protein (mKO1) under the Ta promoter was a gift from Dr. Akira Sakakibara. PACT-mKO1 was recloned under the CMV promoter. All constructs were confirmed by sequencing. The human p35 and cyclin A clones were purchased from OriGene. The stable cell lines with conditional expression of FLAG-HuR constructs were created by using the TET-ON system in U251 cells. During experiments, cells were expanded in DMEM/F-12 medium supplemented with FBS (10%) or FBS (0%) for synchronization, l-glutamine, and penicillin/streptomycin antibiotic. The stable clone's medium was supplemented with G418 and hydromycin B to ensure selection. Construct induction was achieved with doxycycline (DOX) at 0.5 μg/ml.
Cell cycle analysis by PI and EDU incorporation was performed by using the Click-iT EDU 647 flow cytometry assay kit (Invitrogen). Construct expression was induced by cell treatment with DOX (for HuR) or by transfection (for p35) (transfection kit from LONZA) for 1–2 days, and then 10 μm EDU was incorporated for 5–8 h, and cells were washed and fixed in 100% ethanol and stored at −20 ºC. The following week, EDU was developed and PI was incorporated by using the Invitrogen protocol, and cells were analyzed by a flow cytometry technique. The parental TETON cell line without construct expression was used as a control.
Fluorescence microscopy was performed by using an Olympus DP71 fluorescence microscope with a DP controller 3.3.1.294 system and Nikon Eclipse Ti fluorescence microscope with NIS-Elements software. Polyclonal DYKDDDDK tag (Alexa® 488) antibody (Cell Signaling Technology) was employed for subcellular detection of FLAG-HuR S202A, FLAG-HuR S202D, and FLAG-HuR WT proteins. Cells were fixed and permeabilized by using a protocol from Invitrogen, Universal Buffer was used to block nonspecific binding, and cells were incubated with antibody (1:500) overnight at 4 ºC and washed in PBS. HuR-EGFP, Cdk5-DsRed, and PACT-mKO1 constructs were transfected to cells by using a transfection kit from LONZA.
Protein/mRNA Pull-down Assay
FLAG-tagged HuR was immunoprecipitated with A/G beads coated with FLAG antibodies, co-precipitated mRNA was purified by using TRIzol reagent, and mRNA targets (GAPDH, hey1, cyclin A1, and jagged-1; primer/probes Hs00266705_g1, Hs01114115_m1, Hs00171105_m1, Hs00164982_m1, respectively; Applied Biosystems) were analyzed by using TAQMAN. Cdk5/HuR co-immunoprecipitation was performed using the immunoprecipitation method as described (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), ExtraCruz C, dilution in kinase buffer) with monoclonal Cdk5 and HuR 3A2 (Santa Cruz Biotechnology, Inc.) antibodies for immunoprecipitation and polyclonal Cdk5 (Cell Signaling Technology) and HuR-H280 (Santa Cruz Biotechnology, Inc.) antibodies for immunoblotting. Protein fractionation was performed using the nuclear/cytoplasm fractionation kit (Pierce).
RNA kinetics were analyzed following 12 h of actinomycin D (ACMD) treatment. Each graph point (see Fig. 3C) represents mRNA values normalized to the corresponding initial mRNA level at the 0 h time point. A one-phase exponential decay equation was fitted using Prism 5 (GraphPad, La Jolla, CA).
FIGURE 3.
Mutations of serine 202 to alanine or aspartate in HuR alter the HuR/mRNA interaction. A, an example of conditional FLAG-HuR WT, FLAG-HuR S202A, and FLAG-HuR S202D protein expression in U251 detected by Western blot. B, kinetics of hey1, jagged-1, and cyclin A mRNA degradation in control TETON and HuR S202A/D-induced and non-induced clones following ACMD treatments for 12 h (n = 6). Triangles and squares represent decay of mRNA degradation in induced and non-induced clones, respectively. C, average values of GAPDH, hey1, jagged-1, and cyclin A mRNAs co-immunoprecipitated with FLAG-HuR S202A/D mutants (by using FLAG antibody) normalized to the values of corresponding mRNA targets co-immunoprecipitated with control mouse IgG-coated beads from induced clones (n = 3). Error bars, S.D.
In vitro recombinant HuR protein phosphorylation by recombinant Cdk5/p35 (Invitrogen) was performed at 30 °C for 30 min in kinase buffer (Cell Signaling Technology) supplemented with ATP. Phosphorylated HuR-GST or HuR (GST was truncated with thrombin) was detected in Western blot by using p-S-Cdk substrate-specific antibody (Cell Signaling Technology). For in vitro experiments, recombinant HuR-GST proteins was purified by using 5-ml GSTrap HP columns (HiTrap), dialyzed against PBS in 3–12-ml Slide-A-Lyzer dialysis cassettes (Thermo Scientific) and verified by Western blot.
The α screen assay was developed using glutathione α donor beads (PerkinElmer Life Sciences) (which bind to HuR-GST protein) and protein A acceptor beads (PerkinElmer Life Sciences) (which bind to phospho-Cdk substrate antibody). In vitro phosphorylation of recombinant HuR-GST by Cdk5/p35 was performed at 24–28 °C (room temperature) for 1 h, and beads were added for the next 1 h. Acceptor beads were preincubated with phospho-Cdk substrate antibody at room temperature for 1 h before the addition to the reaction. The signal was recorded on a PerkinElmer Life Sciences Envision Multilabel Plate Reader. The titrations of Cdk5/p35, HuR-GST, and Cdk family inhibitor alsterpaullone (Alexis Biochemicals) were performed at room temperature.
In Vitro mRNA/HuR Binding
High protein binding 96-well plates (Nunc) were loaded with equal amounts (1 μg) of GST, HuR-GST, and HuR-GST phosphorylated by Cdk5/p35. Cyclin A RNA was in vitro synthesized (Ambion T7 kit) and loaded in binding buffer. Ribogreen was added, and the fluorescence signal was read. All fluorescence readings were compared with signal from corresponding protein wells with 0 nm RNA, normalized to the maximum signal representing HuR-GST/cyclin A binding, and fit with a dose-response function using OriginPro software. Binding buffer contained 25 mm KCl, 5 mm Hepes, 1.25 MgCl2, 3.8% glycerol, 0.02 mm DTT, 1 mm EDTA.
Centrosome purification and fractionation was performed as described by Meigs and Kaplan (31). The purity of the centrosome fractions was confirmed by staining with γ-tubulin. Briefly, the centrosome fractions for each clone were purified from cells induced or non-induced by DOX. The initial step of purification included separation of nuclear fraction from centrosome/cytoplasmic fractions and was verified by lamin A/C distribution in fractions by Western blot (supplemental Fig. 5A). The crude centrosome fractions were isolated and collected following centrifugation in Ficoll cushion solution. A total of 15–17 fractions were collected for each clone after the second step of purification (centrifugation and fractionation in a discontinuous 62.5–20% sucrose gradient). The purity of the centrosomal fractions was determined by the appearance of γ-tubulin, which was observed in 2–3 of 15 fractions at ∼48% of sucrose gradient level for each clone (disregarding the first two fractions, which probably contain cell debris) (31). The presence of two specific centrosomal proteins Cdk5rap2 and γ-tubulin and the absence of non-centrosomal protein GAPDH in centrosome fractions confirm the purity of the purification (supplemental Fig. 5B).
HuR/mRNA Co-immunoprecipitate from Cytoplasmic Fraction
The cells were electroporated with constructs as control, p35 cDNA for p35 overexpression, p35 for p35 overexpression, and siRNA of Cdk5 for Cdk5 inhibition. Following p35 overexpression, cells were expanded in the absence or presence of the Cdk5 inhibitor alsterpaullone (0.9 μm). Cells were collected after 48–72 h of growth, and nuclear/cytoplasmic fractionation of cell lysate was performed by using a nuclear/cytoplasmic kit (Thermo Scientific). Cytoplasmic lysates were precleared, and immunoprecipitations were performed by using protein A/G beads-agarose coated with HuR (4 μg) or IgG (4 μg). Half of the beads-antibody-protein/mRNA bound complexes for each condition were washed three times and analyzed in Western blot, and the other half were washed and used for mRNA isolation. The mRNA was isolated by using the RNeasy minikit (Qiagen). De novo protein analysis was performed by using the Click-iT 488 labeling kit for protein assays (Invitrogen), l-homoproparglycine (Invitrogen), and RPMI (l-methionine-free) (Invitrogen) medium.
Statistical analysis was performed by using Excel and OriginPro data analysis and graphing software. The statistical significance was determined by Student's t test and cut off at the p > 0.05 level. The statistically significant data are labeled by an asterisk in the graphs.
RESULTS
The cytoplasmic localization of Cdk5 has been reported as crucial for cell cycle initiation and proliferation (36). An increase in the cytoplasmic localization of HuR is noted in highly aggressive cancers and is essential for cell survival, migration, and proliferation (22–28). To support our hypothesis that unique proline-directed serines in HuR serve as points of regulatory control by Cdk5, we provide the following results.
Cdk5 Interacts with HuR
To determine if HuR serves as a substrate for Cdk5, we examined the expression, interaction, and subcellular distribution of both proteins in glioma tumor samples and established cell lines. Fig. 1A illustrates the expression of HuR and Cdk5 in all samples of brain tumors with a clear increase of HuR in the most aggressive forms (WHO IV). The established glioma cell lines also exhibited expression of both Cdk5 and HuR proteins uniformly (Fig. 1, bottom). The co-immunoprecipitation of HuR and Cdk5 in two primary glioma xenolines confirmed a direct interaction between these proteins (Fig. 1B). The RNase treatment of cell lysates prior to co-immunoprecipitation did not diminish the HuR and Cdk5 interaction (supplemental Fig. 1). The details of Cdk5/HuR protein interaction were investigated in the established U251 line. Nuclear/cytoplasmic fractionation revealed that Cdk5 and its co-activator p35 are localized mainly in the cytoplasmic fraction, with HuR levels greatest in the nucleus and less in the cytoplasm (Fig. 1C (a)). A direct interaction between Cdk5 and HuR was confirmed in total and cytoplasmic fractions by using a co-immunoprecipitation assay (Fig. 1C (b)). We did not observe a significant Cdk5/HuR interaction in the nuclear fraction. Expression of HuR-EGFP and Cdk5-DS-Red constructs in U251 cells verified a mostly cytoplasmic localization of Cdk5 (Fig. 1D (a)), nuclear and cytoplasmic localization of HuR (Fig. 1D (b)), and co-localization of HuR and Cdk5 in discrete cytoplasmic areas (Fig. 1D (c)). Our data confirm a direct interaction between HuR and Cdk5 in glioma tumor samples and established cell lines.
FIGURE 1.
Cdk5 is expressed and interacts with HuR in glioma tumor samples and cell lines. A, expression of Cdk5 and HuR in samples of brain tumors. A (a), lanes 1–4, control brain; lane 5, pilocytic astrocytoma (WHO I); lanes 6–8, newly diagnosed GBM (WHO IV); lanes 9 and 10, recurrent GBM (WHO IV); lane 11, U251 cell line. A (b), established glioma cell lines. Lane 1, U251; lane 2, U118; lane 3, U138; lane 4, SF188; lane 5, LN319, lane 6, SNB191; lane 7, MD59K; lane 8, D37. B, co-immunoprecipitation (IP) of Cdk5 and HuR from two primary GBM xenolines. C, expression (a) and co-immunoprecipitation (b) of Cdk5 and HuR from total (T), nuclear (N), and cytoplasmic (C) fractions of U251. Lamin C1/C2 and α-tubulin were chosen as indicators of nuclear and cytoplasmic fractions, respectively. D, localization of Cdk5-DsRed (a) and HuR-EGFP (b) and co-localization of Cdk5-DsRed and HuR-EGFP constructs (c) in U251. Nuclei were stained with DAPI.
Cdk5 Directly Phosphorylates the Serine 202 Residue in HuR Hinge Region
The hinge region of HuR contains two proline-directed serine residues (amino acids 202 and 221) that may serve as potential Cdk5 phosphorylation sites. Recombinant wild type, S202A, and S221A HuR-GST proteins were created, and an in vitro Cdk5/HuR phosphorylation assay was employed to evaluate HuR phosphorylation by Cdk5. Fig. 2A illustrates HuR-wild type (GST-cleaved) phosphorylation by Cdk5 detected by p-S-Cdk substrate-specific antibody in a kinase assay. Substitution of the serine 221 with alanine had no significant effect on Cdk5-dependent HuR phosphorylation (Fig. 2B). However, the substitution of serine 202 to alanine in HuR completely abolished HuR phosphorylation by Cdk5 (Fig. 2C). To check if Cdk5/p35 may phosphorylate alternative sites to serine Ser-202 in HuR, we examined the phosphorylation of recombinant GST, GST-HuR wild type, and GST-HuR S202A (2.5 μg of each) proteins by Cdk5/p35 (50 ng) with an in vitro assay to detect the generation of phosphoproteins (Pro-Q phosphoprotein gel staining; Invitrogen) by Western blot (supplemental Fig. 2). The experiment was performed three times, suggesting that Cdk5/p35 preferentially phosphorylates the Ser-202 residue of HuR because the mutation of Ser-202 to alanine in HuR abolished the phosphofluorescence signal induced by Cdk5/p35 phosphorylation. This finding suggests that the serine 202 in the HuR hinge region is a unique phosphorylation site regulated by Cdk5.
FIGURE 2.
Serine 202 is the unique residue in HuR phosphorylated by Cdk5. A, phosphorylation of recombinant HuR WT protein (GST cleaved) by Cdk5 detected in Western blot by p-S-Cdk substrate-specific antibody. B, phosphorylation of HuR S221A-GST at two different concentrations (150 and 300 ng) by Cdk5 detected by p-S-Cdk substrate-specific antibody. C, phosphorylation of recombinant HuR WT-GST and HuR S202A-GST mutant are exhibited at two different concentrations (200 and 400 ng). D (a), an example of α screen signal from recombinant HuR WT-GST and HuR S202A-GST mutant proteins phosphorylated by Cdk5. Inset, average signal from phosphorylated HuR WT-GST (0 and 500 ng) and HuR-202A-GST mutant (0 and 500 ng) normalized to the maximum of HuR WT-GST phosphorylated signal (n = 5). D (b), Cdk5/p35 titration in HuR WT-GST (500 ng) phosphorylation reaction. Inset, average data normalized to the maximum of phosphorylation signal in each experiment (n = 3). D (c), the Cdk family ATP competitive inhibitor, alsterpaullone, blocks Cdk5 phosphorylation of HuR in a dose- and ATP-dependent manner.
An α screen assay was developed and employed to analyze details of the Cdk5/HuR interaction. Fig. 2D (a) illustrates a concentration-dependent phosphorylation of HuR wild type and HuR S202A mutant by Cdk5. Phosphorylation of HuR S202A was not detectable over a wide concentration range. The ratio of luminescence signals of phosphorylated HuR-GST wild type (500 ng) to phosphorylated HuR S202A-GST mutant (500 ng) by Cdk5 was 14 ± 1-fold greater (n = 8 experiments) (Fig. 2D (a), inset), suggesting specific serine 202 phosphorylation by Cdk5. Fig. 2D (b) illustrates titration of Cdk5 in a phosphorylation reaction performed with a constant HuR wild type amount (500 ng). The K½ of Cdk5 for HuR phosphorylation was 17 ± 6 ng (n = 5). The ATP competitive inhibitor of the Cdk family (alsterpaullone) induced a decline of HuR phosphorylation by Cdk5 in an ATP-dependent manner (Fig. 2D (c)). Thus, our data confirms that Cdk5 specifically phosphorylates serine 202 in HuR.
Phosphorylation of HuR at Ser-202 Alters HuR/mRNA Interaction
U251 clones were engineered to conditionally express WT, S202A, or S202D FLAG-tagged HuR proteins in order to evaluate the cellular and molecular consequences of HuR phosphorylation at the Ser-202 residue (Fig. 3A). Previously, it has been reported that HuR binds to the 3′-UTRs of mRNAs for the transcriptional regulator hey1, angiogenesis-promoting factor jagged-1, and cell cycle regulator cyclin A. To investigate the impact of HuR Ser-202 phosphorylation on stabilization of mRNA targets, we evaluated hey1, jagged-1, and cyclin A mRNA degradation following ACMD treatment in cells with conditional expression of HuR Ser-202 mutants. Mutants were induced to the same level for HuR S202A and HuR S202D clones by the same DOX concentration (0.5 μg/ml) (data not shown). Fig. 3B illustrates the kinetics of hey1, jagged-1, and cyclin A mRNAs in TETON control, HuR S202A, and HuR S202D clones before and after construct induction with DOX (two experiments in triplicate). Table 1 provides a summary of the RNA half-lives for the above targets and c-myc. c-myc was included due to its rapid decay and as a positive control for the inhibition of transcription. The declines in mRNA signals of TETON, HuR S202A, and HuR S202D cell lines following ACMD treatment were fitted by a single phase exponential decay function for all mRNA targets. The control TETON clones demonstrated similar RNA half-lives with or without DOX induction, supporting a lack of influence for DOX. The non-induced mutant clones for both S202A and S202D demonstrated half-lives greater than the TETON controls but similar to each other, suggesting consistency with the system but a possible effect of the antibiotics utilized to maintain the clone stability. Following induction of the mutant clones with DOX, a significant alteration in target RNA half-life was noted. With induction of the S202D mutant, the half-lives were increased 2–3 fold; however, following induction of the S202A mutant, a half-life was determined only for the c-myc target due to lack of decay within the experimental time window. These data suggest that the phosphorylation state of HuR influences target RNA binding affinity.
TABLE 1.
RNA half-life values for potential HuR targets following induction of Ser-202 mutants
| Teton DOX− | Teton DOX+ | S202A DOX− | S202A DOX+ | S202D DOX− | S202D DOX+ | |
|---|---|---|---|---|---|---|
| h | h | h | h | h | h | |
| hey1 | 4.68 | 5.67 | 8.5 | NDa | 7.56 | 16.47 |
| jagged-1 | 11.24 | 9.24 | 15.02 | ND | 8.82 | 20.24 |
| Cyclin A | 8.26 | 9.53 | 16.25 | ND | 11.46 | 35.29 |
| c-myc | 0.38 | 0.40 | 0.49 | 6.7 | 0.39 | 10.0 |
a ND, not determined.
To assess the effect of serine Ser-202 phosphorylation on HuR/mRNA binding, a HuR/mRNA co-immunoprecipitation assay was employed on HuR S202A and HuR S202D induced clones. The mRNAs co-precipitated with HuR mutants (FLAG antibody-coated beads) were specified and quantified by the TAQMAN technique and normalized to the corresponding mRNA values precipitated with normal mouse IgG-coated beads (Fig. 3C). These values were also normalized to the relative HuR expression for each clone to remove any bias based on induction differences. Note that HuR S202A mutant precipitates significantly larger amounts of hey1, jagged-1, and cyclin A mRNAs (187 ± 19 (n = 3), 130 ± 30 (n = 3), and 79 ± 20 (n = 2)) compared with the corresponding mRNA targets precipitated with HuR S202D mutant (103 ± 24, 44 ± 32, and 40 ± 6, respectively; n = 3 experiments). The difference is significant at p = 0.02 and p = 0.03 levels correspondingly for Jagged-1 and hey1 mRNA targets. The p value for cyclin A did not reach significance, which was felt to be due to fewer repetitions and greater variability. These data indicate that HuR phosphorylation at Ser-202 alters HuR/mRNA binding and that a significantly more favorable mRNA interaction exists with the HuR S202A mutant compared with HuR S202D mutant. To verify a role for p35/Cdk5 in the HuR/mRNA interaction at the cellular level, the mRNA/HuR co-immunoprecipitations were performed from cytoplasmic fractions of control U251 cells (control construct-expressing cells), cells overexpressing p35, cells overexpressing p35 in the presence of Cdk5 siRNA, and cells overexpressing p35 in the presence of a common Cdk5 inhibitor alsterpaullone (0.9 μm) (supplemental Fig. 3). Alsterpaullone reduces Cdk5/p35 activity in an ATP-dependent manner (Fig. 2D (c)) as well as reducing Cdk5 expression (supplemental Fig. 3A). Supplemental Fig. 3, A and B, illustrates HuR protein immunoprecipitated from cytoplasmic fractions and averaged values of mRNA co-immunoprecipitated with HuR from cytoplasmic fractions normalized to the mRNA value co-immunoprecipitated with HuR in the control condition. The averages of three experiments confirm that p35 overexpression significantly diminished the HuR/mRNA interaction (74 ± 8%) and that this path could be overcome by the Cdk5 inhibitor alsterpaullone or Cdk5 siRNA, suggesting a significant p35/Cdk5 role in HuR/mRNA interaction in the cell cytoplasm.
In Vitro mRNA/HuR Binding
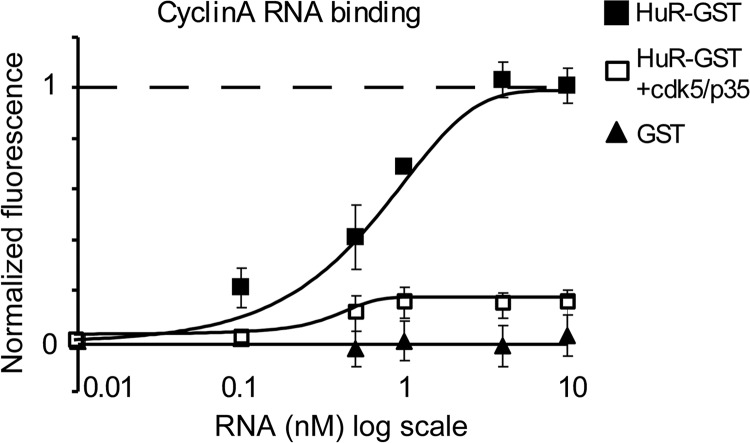
To clarify the mechanism of HuR/mRNA interaction under regulation by Cdk5/p35, we performed cyclin A RNA binding to recombinant HuR-GST under non-phosphorylated and phosphorylated by Cdk5/p35 conditions in vitro (Fig. 4). Cyclin A RNA containing 3′-UTR was synthesized in vitro from human cyclin A clone. Fig. 4 illustrates the binding signal of cyclin A RNA (from 0 to 10 nm) to non-phosphorylated HuR-GST, phosphorylated HuR-GST, and GST. The averaged EC50 of cyclin A RNA to HuR was 0.8 ± 0.1 nm. The maximum of cyclin A binding to phosphorylated HuR reached only 15–20% of maximum cyclin A binding to non-phosphorylated HuR-GST. GST alone did not exhibit any binding ability to cyclin A. These data suggest that HuR phosphorylated by Cdk5/p35 has an unfavorable conformation for cyclin A binding.
FIGURE 4.
In vitro HuR binding to cyclin A RNA. Cyclin A mRNA binding curves to HuR-GST in non-phosphorylated and phosphorylated by Cdk5/p35 conditions are illustrated in the graph. The binding values for each mRNA concentration have been normalized to the maximum HuR-GST/cyclin A binding observed at 10 nm in vitro synthesized cyclin A RNA. The maximum cyclin A RNA binding to HuR-GST in a phosphorylated by Cdk5/p35 condition was diminished by 85 ± 8% (n = 4) compared with the maximum binding to non-phosphorylated HuR-GST. No significant mRNA binding was detected to GST alone (n = 4). The averaged EC50 = 0.8 ± 0.1 nm (n = 4) for the cyclin A RNA/HuR binding affinity. Error bars, S.D.
Prolonged Alteration of HuR Phosphorylation at Ser-202 Induces Centrosome Abnormalities and Arrest of Cell Cycle Progression
HuR is involved in proliferation of several types of cancer (25–28, 32, 33). An EDU incorporation assay was employed to evaluate the influence of HuR phosphorylation at Ser-202 on cell cycle progression. A significant decrease in EDU incorporation was observed in 43 ± 11% and 42 ± 25% (n = 4 experiments) of cells when HuR S202A and HuR S202D clones were induced, respectively, compared with the parental non-induced clones. The overexpression of HuR WT exhibited a slight, non-significant effect on EDU incorporation (the increase was 4 ± 8% (n = 4) compared with the parental non-induced clone) (Fig. 5A (a)). Plots of EDU incorporation versus DNA content (PI incorporation) (n = 4) and PI incorporation alone (n = 5) revealed cell cycle arrest in HuR Ser-202 mutant clones associated with an accumulation of cells in the G2-M phases (Fig. 5A (b)). This finding suggests that consecutive phosphorylation and dephosphorylation of serine 202 in HuR are essential for cell cycle progression.
FIGURE 5.
Alteration of serine 202 phosphorylation in HuR affects cell cycle progression and centrosome duplication and cohesion. A (a), averaged percentage of cells incorporating EDU from induced FLAG-HuR WT and FLAG-HuR Ser-202 mutant clones (48 h of induction) normalized to the corresponding values from non-induced clones during the same period of EDU incorporation (n = 4). b, plots of EDU versus PI incorporation in induced and non-induced HuR clones. The HuR mutants and wild type HuR were equally (+15%) induced in our experiments by the same amount of DOX (inset represents correspondingly HuR WT-FLAG (lanes 1 and 2), HuR S202A-FLAG (lanes 3 and 4), and HuR S202D-FLAG (lanes 5 and 6) clones before and after induction with 0.5 μg/ml Dox). B, distribution (in percentage) of cells with different centrosome numbers in non-induced and induced clones. Gray, cells with less then 2 centrosomes per nuclei; blue, cells with amplified (more than two centrosomes per nuclei) number; red and green, cells with 2 centrosomes per nuclei and with lateral or central localization during cytokinesis, respectively. Each diagram illustrates analysis of at least 100 cells obtained from the sum of three experiments from the same clone. Nuclei were stained with DAPI, and centrosomes were marked with the PACT domain of pericentrin attached to the red fluorescence protein. C, HuR localization in centrosomes confirmed by immunostaining (a) and by subcellular fractionation (b). Bar in a, 50 μm. Fractions 1–9 represent protein content corresponding to the 60–48% range (left to right) of sucrose gradient levels, which were achieved by fractionation of proteins from discontinuous 60–20% sucrose gradient after nuclear/cytoplasmic and Ficoll gradient purifications (48). γ-Tubulin marks pure centrosome fraction. D (a and b), protein fractions from 60–48% (left to right) sucrose gradient levels from FLAG-HuR S202A and FLAG-HuR S202D induced clones. Note that HuR mutants appear in pure centrosome fractions, which are marked with γ-tubulin.
We hypothesized that abnormalities in cell cycle progression resulting from induced HuR S202A and S202D may be tied to defects in centrosome duplication and cohesion caused by aberrant HuR-dependent regulation of cyclin A levels. We marked centrosomes with the conserved PACT domain of pericentrin attached to the red fluorescence protein and analyzed their structure in HuR WT-, S202A-, or S202D-overexpressing cells. Fig. 5B represents distribution (in percentage) of cells with different centrosome number per nuclei in control and induced clones. Robust defects in centrosome duplication and cohesion were observed in HuR mutant clones (Fig. 5B, diagrams). The majority of HuR S202A-overexpressing cells exhibited amplification of centrosome number (Fig. 5B); however, the majority of HuR S202D-overexpressing cells displayed a decrease in centrosome number as well as abnormal centrosome cohesion (Fig. 5B).
To detail the HuR influence on centrosome function, we employed an immunostaining technique and immunoblotting analysis of purified centrosome fractions (31). The immunostaining approach revealed HuR co-localization with centrosomes marked by PACT-mko.1 fluorescence construct (Fig. 5C (a)). The centrosome purification by subcellular fractionation confirmed that HuR and Cdk5 proteins were integrated with centrosomes (Fig. 5C (b) and supplemental Fig. 4). The centrosome protein fraction obtained by using a discontinuous (60–20%) sucrose gradient was identified with γ-tubulin at ∼48–53% sucrose level and exhibited a significant amount of endogenous HuR and Cdk5 proteins (Fig. 5C (b)). The analysis of purified centrosome fractions from HuR Ser-202 mutant clones revealed that the mutations of serine 202 to A or D did not prohibit the ability of HuR to co-localize with the centrosome fraction marked with γ-tubulin (Fig. 5D (a and b)). Our data suggest that HuR may integrate in centrosomes and that the unbalanced HuR phosphorylation at serine 202 distorts centrosome function and cell cycle progression.
Phosphorylation of Serine 202 in HuR and Overactivation of Cdk5 Have a Common Impact on Cyclin A Protein Level
Cyclin A oscillation in centrosomes is essential for normal duplication and cohesion (34, 35). We hypothesized that abnormal serine 202 phosphorylation in HuR may affect cyclin A levels in the centrosomes. The comparison of crude centrosome content from different clones revealed that the level of cyclin A protein correlates positively with the amount of HuR S202A mutant and inversely correlates with the mimicking serine S202 phosphorylation of HuR S202D mutant (Fig. 6A). The average value of cyclin A was 2.1 ± 0.5 (n = 3)-fold higher in crude centrosome fractions from the induced HuR S202A clone compared with the cyclin A value in crude centrosomes of the HuR S202D induced clone; the value of cyclin A in crude centrosome fractions of non-induced HuR S202A and HuR S202D clones were not significantly different (1 ± 0.2, n = 3). The difference was statistically significant for the induced HuR S202A compared with S202D (p = 0.04). To check if HuR phosphorylation influences cyclin A translation, we performed fluorescence labeling of de novo synthesizing proteins by using modified glycine in U251 cells. Fluorescence labeling confirms spots of de novo protein synthesis in proximity (0–3 μm) to centrosomes, suggesting possible attachment of translational apparatus to centrosomes (supplemental Fig. 5). The flow cytometry analysis of de novo protein synthesis revealed that overexpression of HuR S202D mutant was associated with a decrease of de novo protein synthesis compared with protein synthesis in the HuR wild type- or HuR S202A mutant-overexpressing cells. (Fig. 6B, n = 4). The immunoprecipitation of de novo synthesized cyclin A (6 h of labeled translation) confirmed a decrease in cyclin A protein level in HuR S202D-overexpressing cells compared with HuR S202A-overexpressing cells (Fig. 6B (b)). To further support the role of HuR phosphorylation on cyclin A expression, a decrease of cyclin A protein level was observed following p35 overexpression in U251 cells (Fig. 6C (a)). The total decrease of protein synthesis reached 19 ± 5% (n = 4) compared with control (vector) overexpressing cells (Fig. 6C (b and c)). This finding highlights a common impact of HuR Ser-202 phosphorylation and Cdk5 activation by p35 on cyclin A protein translation in U251 cells.
FIGURE 6.
Serine 202 phosphorylation in HuR affects protein translation. A, the protein content of crude centrosome fractions from induced and non-induced HuR Ser-202 mutant clones. The chemiluminescence values of cyclin A bands are normalized to the corresponding values of γ-tubulin bands. B (a), averages of de novo protein synthesis in induced clones normalized to the corresponding values from non-induced clones obtained by flow cytometry (n = 4). Note the significant reduction of protein synthesis in HuR S202D induced clone. b, de novo synthesized cyclin A protein (during 6 h of labeled translation) immunoprecipitated (IP) from HuR S202D and HuR S202A induced clones. C, overexpression of p35 was associated with a decrease in cyclin A protein levels (a) and a reduction of de novo protein synthesis (b and c). De novo protein synthesis is illustrated in the flow cytometry plots (b) and in quantified graph (c) (n = 4) following 24 and 48 h of p35 overexpression. Error bars, S.D.
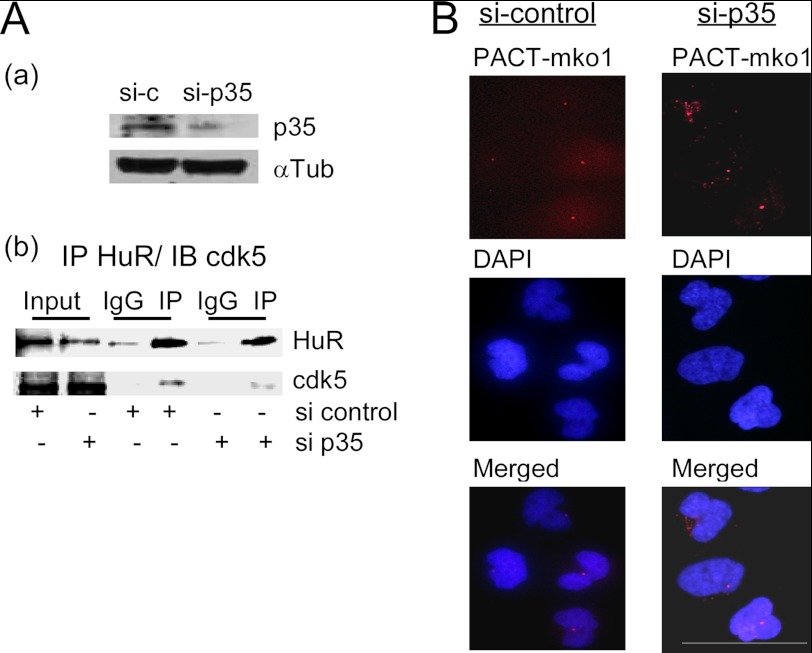
Further confirmation of centrosome regulation by HuR/Cdk5 interaction was achieved by centrosome analysis following inhibition of the Cdk5/HuR interaction. The knockdown of Cdk5 co-activator p35 (Fig. 7A) provoked a decrease in the HuR/Cdk5 interaction confirmed by a co-immunoprecipitation assay (Fig. 7B) and was associated with robust defects in centrosome duplication. The amplification in centrosome number was observed in more than 50% of cells after p35 inhibition (Fig. 7C) versus only 5–10% of control cells following transfection with scrambled siRNA.
FIGURE 7.
Knockdown of p35 by siRNA reduces HuR/Cdk5 interaction and provokes amplification of centrosome number. A, reduction of p35 expression by siRNA (a) and reduction of Cdk5 co-immunoprecipitation with HuR (b). B, illustration of amplification of centrosome number per nuclei following p35 knockdown. Nuclei were stained with DAPI, and centrosomes were marked with the PACT domain of pericentrin attached to the red fluorescence protein. Bar, 50 μm. IP, immunoprecipitation; IB, immunoblot.
DISCUSSION
In the current report, we have provided several lines of evidence supporting a direct Cdk5/HuR interaction: 1) co-immunoprecipitation of both proteins from several glioma lines; 2) co-localization by fluorescence microscopy; and 3) direct interaction in an in vitro phosphorylation assay. We confirm HuR integration in centrosomes and a phosphorylation-dependent influence on centrosome function and cell cycle progression.
The role of HuR in cell cycle progression via regulation of cyclins and growth factors is well documented (25–27, 32). The phosphorylation-dependent HuR shuttling between nucleus and cytoplasm, mRNA stabilization, and up- and down-regulation of protein synthesis underlie HuR function during cell cycle progression (30, 37–39). Several kinases (Cdk1 and PKCα) have been reported to regulate HuR function during the cell cycle (29, 30). We found that HuR phosphorylation by Cdk5 may influence cell function via alteration of cyclin A protein level. The oscillation of cyclin A protein levels is essential for mitosis progression through activation of DNA replication and for centrosome duplication through cyclin A/Cdk2 phosphorylation-dependent regulation of Msp1 kinase (34, 35, 40). The range of cyclin A oscillation is determined by both translation and degradation and may have positive and negative impacts on cell cycle progression. We observed an inverse correlation between HuR phosphorylation at Ser-202 and cyclin A protein level in centrosomes. In agreement with previous observations, we found that elevation of cyclin A level in centrosomes is associated with amplification of centrosome number in HuR S202A induced clone, and a decline of cyclin A protein level in HuR S202D induced clone is linked to a decrease in centrosome duplication and aberrant cohesion (35). The centrosome's role in mitotic orientation of progenitors is well documented (41). Recently, centrosomes have emerged as key regulators of mitosis in cancer cells with abnormalities in centrosome function tightly related to genomic instability and aberrant cell cycle progression (42–44). The arrest of cell cycle progression observed in HuR S202A/D induced clones could be well explained by 1) disruption of centrosome duplication, 2) inadequate cyclin A protein level during G2-M phase of cell cycle progression, or 3) both.
The direct integration and regulation of centrosome function by HuR highlights a new, active contribution for this mRNA-binding protein in cell cycle progression. Numerous key proteins that are involved in centrosome function, including cyclins, growth factors, β-actin, β-catenin, and connexin, are controlled by HuR at the mRNA and translational levels (28, 45–48). We hypothesize that HuR may store mRNA in centrosomes and facilitate local translation during mitosis. In support of our hypothesis, we observed spots of de novo protein synthesis attached to the centrosomes and the presence of translation initiation factors in crude centrosomes (supplemental Fig. 1). In agreement with our theory, several authors have reported a correlation between local de novo protein synthesis and centrosome formation, supporting a dependence of centrosome structure on components of the translational machinery (49, 50). We found that HuR phosphorylation by Cdk5 influences cyclin A mRNA stabilization and translation in proximity to the centrosome. However, other proline-directed kinases, such as Aurora A, Nek2, SIK2, and Polo-like kinase 1, have also been reported in proximity to centrosomes and may play an active function in promoting cell cycle progression (51–55). The particular type of proline-directed kinases involved in HuR phosphorylation at Ser-202 may depend on the cell line or cellular fate.
The function of HuR is heavily dependent on the phosphorylation status of its hinge region (29, 30, 56–58). The hinge domain is directly or indirectly responsible for HuR subcellular localization, binding affinity with mRNA, and regulation of translational efficiency (29, 30, 56–58). The main consequence of aberrant serine Ser-202 phosphorylation in the HuR hinge region is the arrest of cell cycle progression. Overall, our data suggest that HuR S202A has a more favorable role in mRNA stabilization and processing compared with HuR S202D, specifically in the regulation of cyclin A mRNA delivery and processing in proximity to centrosomes. In addition, the alterations of mRNA levels and the decrease in protein synthesis following induction of HuR Ser-202 mutants point to an important role of hinge region phosphorylation in HuR RNA binding function. The mechanism of this effect is not clear and could be partially explained by different HuR Ser-202 mutant affinities to mRNA, alterations in subcellular location or trafficking ability of HuR Ser-202 mutants, or direct and indirect HuR influences on the translational machinery. This certainly situates the HuR hinge domain as a potential novel therapeutic target in cancer. Given these results, HuR and Cdk5 with emphasis on the phosphoregulation of the hinge region may be considered as unique molecular targets of a novel class in cancer therapeutics.
Acknowledgments
We greatly appreciate the space and support provided by the Southern Research Institute. We thank Dr. Lynn Rasmussen for great advice and Robert E McHugh for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA112397 (to L. B. N.).

This article contains supplemental Figs. 1–5.
- Cdk
- cyclin-dependent kinase
- HuR
- Hu antigen R
- DOX
- doxycycline
- ACMD
- actinomycin D
- PI
- propidium iodide
- EDU
- 5-ethyl-2′-deoxyuridine
- GBM
- glioblastoma.
REFERENCES
- 1. Satyanarayana A., Kaldis P. (2009) Mammalian cell-cycle regulation. Several Cdks, numerous cyclins, and diverse compensatory mechanisms. Oncogene 28, 2925–2939 [DOI] [PubMed] [Google Scholar]
- 2. Lew J., Huang Q. Q., Qi Z., Winkfein R. J., Aebersold R., Hunt T., Wang J. H. (1994) A brain-specific activator of cyclin-dependent kinase 5. Nature 371, 423–426 [DOI] [PubMed] [Google Scholar]
- 3. Nebreda A. R. (2006) Cdk activation by non-cyclin proteins. Curr. Opin. Cell Biol. 18, 192–198 [DOI] [PubMed] [Google Scholar]
- 4. Tsai L. H., Delalle I., Caviness V. S., Jr., Chae T., Harlow E. (1994) p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature 371, 419–423 [DOI] [PubMed] [Google Scholar]
- 5. Tsai L. H., Takahashi T., Caviness V. S., Jr., Harlow E. (1993) Activity and expression pattern of cyclin-dependent kinase 5 in the embryonic mouse nervous system. Development 119, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 6. Sasaki Y., Cheng C., Uchida Y., Nakajima O., Ohshima T., Yagi T., Taniguchi M., Nakayama T., Kishida R., Kudo Y., Ohno S., Nakamura F., Goshima Y. (2002) Fyn and Cdk5 mediate semaphorin-3A signaling, which is involved in regulation of dendrite orientation in cerebral cortex. Neuron 35, 907–920 [DOI] [PubMed] [Google Scholar]
- 7. Feldmann G., Mishra A., Hong S. M., Bisht S., Strock C. J., Ball D. W., Goggins M., Maitra A., Nelkin B. D. (2010) Inhibiting the cyclin-dependent kinase CDK5 blocks pancreatic cancer formation and progression through the suppression of Ras-Ral signaling. Cancer Res. 70, 4460–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goodyear S., Sharma M. C. (2007) Roscovitine regulates invasive breast cancer cell (MDA-MB231) proliferation and survival through cell cycle regulatory protein Cdk5. Exp. Mol. Pathol. 82, 25–32 [DOI] [PubMed] [Google Scholar]
- 9. Lalioti V., Pulido D., Sandoval I. V. (2010) Cdk5, the multifunctional surveyor. Cell Cycle 9, 284–311 [DOI] [PubMed] [Google Scholar]
- 10. Dhariwala F. A., Rajadhyaksha M. S. (2008) An unusual member of the Cdk family. Cdk5. Cell Mol. Neurobiol. 28, 351–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jessberger S., Aigner S., Clemenson G. D., Jr., Toni N., Lie D. C., Karalay O., Overall R., Kempermann G., Gage F. H. (2008) Cdk5 regulates accurate maturation of newborn granule cells in the adult hippocampus. PLoS Biol. 6, e272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lagace D. C., Benavides D. R., Kansy J. W., Mapelli M., Greengard P., Bibb J. A., Eisch A. J. (2008) Cdk5 is essential for adult hippocampal neurogenesis. Proc Natl. Acad Sci. U.S.A. 105, 18567–18571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hirasawa M., Ohshima T., Takahashi S., Longenecker G., Honjo Y., Veeranna, Pant H. C., Mikoshiba K., Brady R. O., Kulkarni A. B. (2004) Perinatal abrogation of Cdk5 expression in brain results neuronal migration defects. Proc. Natl. Acad. Sci. U.S.A. 101, 6249–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Hare M. J., Kushwaha N., Zhang Y., Aleyasin H., Callaghan S. M., Slack R. S., Albert P. R., Vincent I., Park D. S. (2005) Differential roles of nuclear and cytoplasmic cyclin-dependent kinase 5 in apoptotic and excitotoxic neuronal death. J. Neurosci. 25, 8954–8966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J., Li H., Yabut O., Fitzpatrick H., D'Arcangelo G., Herrup K. (2010) Cdk5 suppresses the neuronal cell cycle by disrupting the E2F1-DP1 complex. J. Neurosci. 30, 5219–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng K., Ip N. Y. (2003) Cdk5. A new player at synapses. Neurosignals 12, 180–190 [DOI] [PubMed] [Google Scholar]
- 17. Samuels B. A., Hsueh Y. P., Shu T., Liang H., Tseng H. C., Hong C. J., Su S. C., Volker J., Neve R. L., Yue D. T., Tsai L. H. (2007) Cdk5 promotes synaptogenesis by regulating the subcellular distribution of the MAGUK family member CASK. Neuron 56, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strock C. J., Park J. I., Nakakura E. K., Bova G. S., Isaacs J. T., Ball D. W., Nelkin B. D. (2006) Cyclin-dependent kinase 5 activity controls cell motility and metastatic potential of prostate cancer cells. Cancer Res. 66, 7509–7515 [DOI] [PubMed] [Google Scholar]
- 19. Courapied S., Sellier H., de Carné Trćesson S., Vigneron A., Bernard A. C., Gamelin E., Barré B., Coqueret O. (2010) The Cdk5 kinase regulates the STAT3 transcription factor to prevent DNA damage upon topoisomerase I inhibition. J. Biol. Chem. 285, 26765–26778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hawasli A. H., Benavides D. R., Nguyen C., Kansy J. W., Hayashi K., Chambon P., Greengard P., Powell C. M., Cooper D. C., Bibb J. A. (2007) Cyclin-dependent kinase 5 governs learning and synaptic plasticity via control of NMDAR degradation. Nat. Neurosci. 10, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schalm S. S., Tee A. R., Blenis J. (2005) Characterization of a conserved C-terminal (RSPRR) motif in S6K1 required for its mTOR-dependent regulation. J. Biol. Chem. 280, 11101–11106 [DOI] [PubMed] [Google Scholar]
- 22. Brennan C. M., Steitz J. A. (2001) HuR and mRNA stability. Cell Mol. Life Sci. 58, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hinman M. N., Lou H. (2008) Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 65, 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim H. H., Gorospe M. (2008) Phosphorylated HuR shuttles in cycles. Cell Cycle 7, 3124–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. López de Silanes I., Lal A., Gorospe M. (2005) HuR. Post-transcriptional paths to malignancy. RNA Biol. 2, 11–13 [DOI] [PubMed] [Google Scholar]
- 26. Nabors L. B., Gillespie G. Y., Harkins L., King P. H. (2001) HuR, an RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′-untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 61, 2154–2161 [PubMed] [Google Scholar]
- 27. Nabors L. B., Suswam E., Huang Y., Yang X., Johnson M. J., King P. H. (2003) Tumor necrosis factor α induces angiogenic factor up-regulation in malignant glioma cells. A role for RNA stabilization and HuR. Cancer Res. 63, 4181–4187 [PubMed] [Google Scholar]
- 28. Dormoy-Raclet V., Ménard I., Clair E., Kurban G., Mazroui R., Di Marco S., von Roretz C., Pause A., Gallouzi I. E. (2007) The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the β-actin mRNA in a U-rich element-dependent manner. Mol. Cell Biol. 27, 5365–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim H. H., Abdelmohsen K., Lal A., Pullmann R., Jr., Yang X., Galban S., Srikantan S., Martindale J. L., Blethrow J., Shokat K. M., Gorospe M. (2008) Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 22, 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doller A., Huwiler A., Müller R., Radeke H. H., Pfeilschifter J., Eberhardt W. (2007) Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cδ elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Biol. Cell 18, 2137–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meigs T. E., Kaplan D. D. (2008) Isolation of centrosomes from cultured mammalian cells. CSH Protoc. 2008, pdb.prot5039 [DOI] [PubMed] [Google Scholar]
- 32. Wang W., Caldwell M. C., Lin S., Furneaux H., Gorospe M. (2000) HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 19, 2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Filippova N., Yang X., Wang Y., Gillespie G. Y., Langford C., King P. H., Wheeler C., Nabors L. B. (2011) The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol. Cancer Res. 9, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Boer L., Oakes V., Beamish H., Giles N., Stevens F., Somodevilla-Torres M., Desouza C., Gabrielli B. (2008) Cyclin A/Cdk2 coordinates centrosomal and nuclear mitotic events. Oncogene 27, 4261–4268 [DOI] [PubMed] [Google Scholar]
- 35. Kasbek C., Yang C. H., Yusof A. M., Chapman H. M., Winey M., Fisk H. A. (2007) Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol. Biol. Cell 18, 4457–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang J., Cicero S. A., Wang L., Romito-Digiacomo R. R., Yang Y., Herrup K. (2008) Nuclear localization of Cdk5 is a key determinant in the postmitotic state of neurons. Proc. Natl. Acad. Sci. U.S.A. 105, 8772–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu L., Rao J. N., Zou T., Xiao L., Wang P. Y., Turner D. J., Gorospe M., Wang J. Y. (2009) Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol. Biol. Cell 20, 4885–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gallouzi I. E., Brennan C. M., Stenberg M. G., Swanson M. S., Eversole A., Maizels N., Steitz J. A. (2000) HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad Sci. U.S.A. 97, 3073–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen Chi M., Chalmel F., Agius E., Vanzo N., Khabar K. S., Jégou B., Morello D. (2009) Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. PLoS One 4, e4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. John P. C., Mews M., Moore R. (2001) Cyclin-Cdk complexes. Their involvement in cell cycle progression and mitotic division. Protoplasma 216, 119–142 [DOI] [PubMed] [Google Scholar]
- 41. Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. (2008) Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 10, 93–101 [DOI] [PubMed] [Google Scholar]
- 42. Duensing S., Münger K. (2001) Centrosome abnormalities, genomic instability and carcinogenic progression. Biochim. Biophys. Acta 1471, M81–88 [DOI] [PubMed] [Google Scholar]
- 43. Doxsey S., Zimmerman W., Mikule K. (2005) Centrosome control of the cell cycle. Trends Cell Biol. 15, 303–311 [DOI] [PubMed] [Google Scholar]
- 44. Lai P. Y., Wang C. Y., Chen W. Y., Kao Y. H., Tsai H. M., Tachibana T., Chang W. C., Chung B. C. (2011) Steroidogenic factor 1 (NR5A1) resides in centrosomes and maintains genomic stability by controlling centrosome homeostasis. Cell Death Differ. 18, 1836–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor S. M., Nevis K. R., Park H. L., Rogers G. C., Rogers S. L., Cook J. G., Bautch V. L. (2010) Angiogenic factor signaling regulates centrosome duplication in endothelial cells of developing blood vessels. Blood 116, 3108–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ale-Agha N., Galban S., Sobieroy C., Abdelmohsen K., Gorospe M., Sies H., Klotz L. O. (2009) HuR regulates gap junctional intercellular communication by controlling β-catenin levels and adherens junction integrity. Hepatology 50, 1567–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bahmanyar S., Kaplan D. D., Deluca J. G., Giddings T. H., Jr., O'Toole E. T., Winey M., Salmon E. D., Casey P. J., Nelson W. J., Barth A. I. (2008) β-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 22, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bahmanyar S., Guiney E. L., Hatch E. M., Nelson W. J., Barth A. I. (2010) Formation of extra centrosomal structures is dependent on β-catenin. J. Cell Sci. 123, 3125–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Müller H., Schmidt D., Steinbrink S., Mirgorodskaya E., Lehmann V., Habermann K., Dreher F., Gustavsson N., Kessler T., Lehrach H., Herwig R., Gobom J., Ploubidou A., Boutros M., Lange B. M. (2010) Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 29, 3344–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vijayakumar S., Shah N., Fernadez-Garcia I., Barcellos-Hoff M. (2011) Origins and consequences of radiation in induced centrosome aberrations. Low Dose Radiation Research Investigators' Workshop, Bethesda, MD, May 9–11, 2011, United States Department of Energy, Washington, D. C [Google Scholar]
- 51. Dutertre S., Descamps S., Prigent C. (2002) On the role of aurora-A in centrosome function. Oncogene 21, 6175–6183 [DOI] [PubMed] [Google Scholar]
- 52. Qin L., Tong T., Song Y., Xue L., Fan F., Zhan Q. (2009) Aurora-A interacts with cyclin B1 and enhances its stability. Cancer Lett. 275, 77–85 [DOI] [PubMed] [Google Scholar]
- 53. Rosales J. L., Rattner J. B., Lee K. Y. (2010) The primary microcephaly 3 (MCPH3)-interacting protein, p35, and its catalytic subunit, Cdk5, are centrosomal proteins. Cell Cycle 9, 618–620 [DOI] [PubMed] [Google Scholar]
- 54. Kishi K., van Vugt M. A., Okamoto K., Hayashi Y., Yaffe M. B. (2009) Functional dynamics of Polo-like kinase 1 at the centrosome. Mol. Cell Biol. 29, 3134–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fry A. M. (2002) The Nek2 protein kinase. A novel regulator of centrosome structure. Oncogene 21, 6184–6194 [DOI] [PubMed] [Google Scholar]
- 56. Kim H. H., Yang X., Kuwano Y., Gorospe M. (2008) Modification at HuR(S242) alters HuR localization and proliferative influence. Cell Cycle 7, 3371–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li H., Park S., Kilburn B., Jelinek M. A., Henschen-Edman A., Aswad D. W., Stallcup M. R., Laird-Offringa I. A. (2002) Lipopolysaccharide-induced modification of HuR. J. Biol. Chem. 277, 44623–44630 [DOI] [PubMed] [Google Scholar]
- 58. Mazroui R., Di Marco S., Clair E., von Roretz C., Tenenbaum S. A., Keene J. D., Saleh M., Gallouzi I. E. (2008) Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J. Cell Biol. 180, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]