The genome sequence of Ustilago hordei revealed that transposable elements were involved in restructuring of the genome, which affected fungal reproductive biology and evolution of genes encoding effector proteins. Comparison to other smuts indicated loss of genome defense components in Ustilago maydis and control of repetitive sequences by repeat-induced point mutation in U. hordei.
Abstract
Ustilago hordei is a biotrophic parasite of barley (Hordeum vulgare). After seedling infection, the fungus persists in the plant until head emergence when fungal spores develop and are released from sori formed at kernel positions. The 26.1-Mb U. hordei genome contains 7113 protein encoding genes with high synteny to the smaller genomes of the related, maize-infecting smut fungi Ustilago maydis and Sporisorium reilianum but has a larger repeat content that affected genome evolution at important loci, including mating-type and effector loci. The U. hordei genome encodes components involved in RNA interference and heterochromatin formation, normally involved in genome defense, that are lacking in the U. maydis genome due to clean excision events. These excision events were possibly a result of former presence of repetitive DNA and of an efficient homologous recombination system in U. maydis. We found evidence of repeat-induced point mutations in the genome of U. hordei, indicating that smut fungi use different strategies to counteract the deleterious effects of repetitive DNA. The complement of U. hordei effector genes is comparable to the other two smuts but reveals differences in family expansion and clustering. The availability of the genome sequence will facilitate the identification of genes responsible for virulence and evolution of smut fungi on their respective hosts.
INTRODUCTION
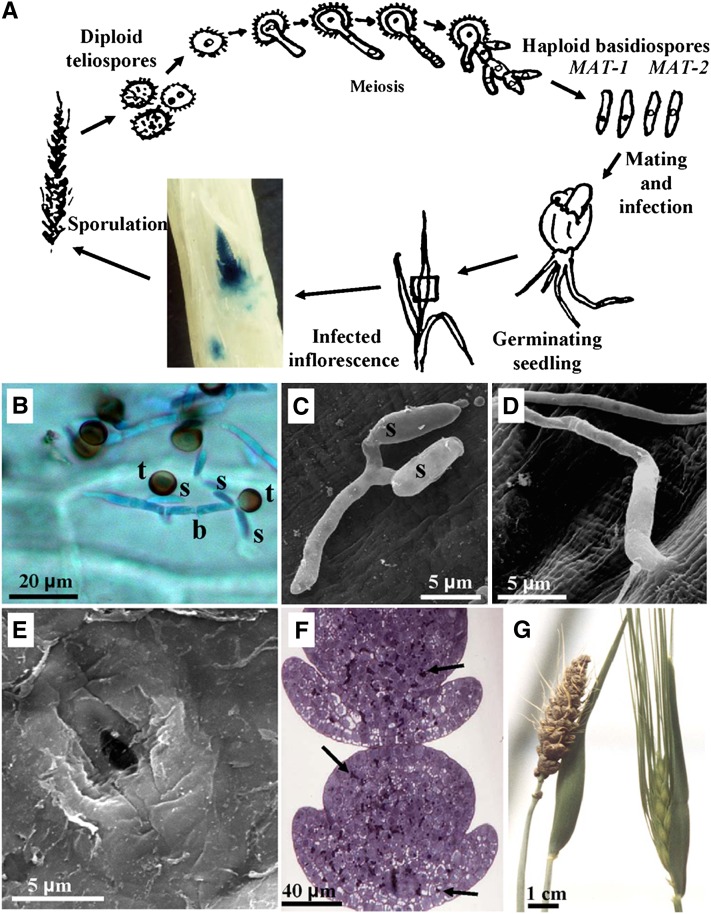
The smut fungus Ustilago hordei causes the economically significant disease of covered smut on barley (Hordeum vulgare) and oats (Avena sativa) and has a worldwide distribution. Diploid teliospores, survival propagules from infected seed heads, germinate with the seed to produce four meiotic, haploid basidiospores. Fusion of two basidiospores of opposite mating type results in an obligate parasitic, mycelial dikaryon (Bakkeren and Kronstad, 1994). The dikaryon can infect the barley plant at the seedling stage and needs the host for survival and spore formation. During plant colonization, the dikaryotic mycelium grows in or just below the shoot meristem. The infection proceeds mostly symptomless until differentiation of the colonized meristem into floral tissue, which cues the fungus to proliferate and sporulate in the spikelets of the inflorescence (Figure 1; Hu et al., 2002).
Figure 1.
Infection Process of U. hordei on Barley.
(A) Schematic representation of the U. hordei life cycle. Dispersed teliospores become lodged under seed hulls and germinate together with the seed. Mating precedes infection and occurs on the young coleoptile. The photographic insert depicts a light microscopy picture of an immature inflorescence at 47 d, colonized after infection by a compatible, mated mixture of U. hordei strain Uh359 stably transformed with a β-glucuronidase–expressing construct and strain Uh362. Fungal hyphae were colored blue following treatment of the inflorescence with glucuronide (Hu et al., 2002).
(B) Light microscopy picture of germinated teliospores (t) stained with cotton blue on the surface of a barley coleoptile 17 h after inoculation having produced a basidium (b), from which haploid basidiospores (s) are emerging. Microscopy procedure as described by Gaudet et al. (2010).
(C) Scanning electron micrograph of two mated basidiospores (s) of opposite mating type fused through conjugation hyphae to produce the dikaryotic infection filament on a barley coleoptile.
(D) Scanning electron micrograph of a dikaryotic infection hypha entering the barley coleoptile wall by direct penetration.
(E) Scanning electron micrograph showing a penetration site, from which the hypha was dislodged during sample preparation. Scanning electron microscopy in (C) to (E) as described by Hu et al. (2002).
(F) Light microscopy picture of an immature inflorescence showing extensive, early teliospore formation (arrows).
(G) Emerged barley inflorescence where all kernels have been replaced by black teliospores (left) next to a healthy head (right).
Mating in U. hordei is controlled by a single mating-type locus (MAT) with two known allelic specificities, MAT-1 and MAT-2 (Bakkeren and Kronstad, 1994; Bakkeren et al., 2008). Each MAT locus is over 450 kb in length, and MAT-1 is composed of 47 genes dispersed between large stretches of long terminal repeats (LTRs) and transposable elements (TEs) occupying ∼50% of the region. The loci are delimited by a pheromone and pheromone receptor pair (a locus) and a pair of homeodomain-containing transcription factors (b locus) (Lee et al., 1999; Bakkeren et al., 2006). While a bipolar mating system seems to be present in most smut fungi, the maize-infecting smut fungi Ustilago maydis and Sporisorium reilianum have a tetrapolar mating system with both a and b loci occurring at two separate chromosomal locations (Bölker et al., 1992; Kämper et al., 1995; Schirawski et al., 2005).
Bipolarity in U. hordei results from a tight linkage of the a and b mating type loci on the same chromosome due to suppression of recombination in the heavily TE-populated intervening sequence (Bakkeren and Kronstad, 1994; Bakkeren et al., 2006). Large stretches of TEs are not seen in either the U. maydis or S. reilianum genomes, indicating species differences in TE activity and possibly genomic defenses against parasitic DNA elements (Kämper et al., 2006; Schirawski et al., 2010). Moreover, RNA silencing has been shown to be functional in U. hordei but not in U. maydis (Laurie et al., 2008). For S. reilianum, genes encoding RNA silencing components have been identified, but it is unknown whether the mechanism is functional (Schirawski et al., 2010). RNA silencing is instrumental in controlling TEs and facilitating repressive chromatin at repetitive loci and is considered to have been present in the last common ancestor to all eukaryotes (Cerutti and Casas-Mollano, 2006; Shabalina and Koonin, 2008).
The elucidation of its genome sequence promoted U. maydis to a model species for the study of biotrophic basidiomycete plant pathogens (Kämper et al., 2006). The U. maydis genome now serves as a reference to study genome and virulence factor evolution in smut fungi (Bakkeren et al., 2006; Schirawski et al., 2010). In particular, genome comparison between U. maydis and S. reilianum revealed a very high degree of synteny. This allowed identification of gene clusters with low sequence conservation, of which some could be experimentally shown to contain virulence factors (Schirawski et al., 2010). Such factors may also account for differences among U. hordei, S. reilianum, and U. maydis, including infection strategy, site of sporulation, and host selection. U. hordei and S. reilianum can infect their respective hosts systemically, initially without the induction of obvious macroscopic symptoms. Sporulation of both species occurs almost exclusively in inflorescences of the respective host plants. By contrast, U. maydis can cause a local infection of any aboveground part of the maize (Zea mays) plant, resulting in induction of tumors in which copious fungal proliferation and sporulation take place. Race- and strain-specific virulence/compatibility interactions exist in U. hordei toward barley (Linning et al., 2004; Grewal et al., 2008), but no dominant avirulence functions that genetically interact with dominant host resistance genes on a gene-for-gene basis have been identified in the U. maydis– or the S. reilianum–maize interaction (Lubberstedt et al., 1998, 1999; Baumgarten et al., 2007).
To study genome evolution in smut fungi and to understand the molecular basis of the different smut fungal life styles, we sequenced, assembled, and annotated the genome of the barley smut fungus U. hordei. Assembly of the so far largest known smut fungal genome has been hampered by the extensive presence of repetitive elements. In spite of generally conserved synteny to other smut fungal genomes, the presence of repetitive elements appears to have shaped genome evolution at several loci with profound effects on fungal biology.
RESULTS AND DISCUSSION
Sequencing of the U. hordei Genome
Nuclear DNA of the U. hordei strain Uh4875-4 (MAT-1) (Linning et al., 2004) was sequenced to 25-fold coverage by a combination of a genomic and a 10-kb paired-end library on the GS FLX 454 platform and end-sequencing of a tiled BAC library on an ABI3730 sequencer (Table 1). Assembly of the sequence reads proved challenging due to the extensive presence of repetitive sequences. Integrating the paired-end information of both the 10-kb and BAC library sequences allowed computer-assisted manual assembly of 71 supercontigs. To support the assembly, optical mapping data was generated allowing tentative assignment of supercontigs to 23 chromosomes (see Supplemental Data Set 1 online). The total genome size is estimated to be 26.1 Mb. Approximately 81% of the genome is represented by sequence in scaffolds and nonassembled contigs.
Table 1. U. hordei Genome Statistics Compared to That of Other Sequenced Smut Fungi.
| Genome Specifics | U. hordei | U. maydis | S. reilianum |
|---|---|---|---|
| Assembly statistics | |||
| Total contig length (Mb) | 20.7 | 19.7 | 18.2 |
| Total scaffold length (Mb) | 21.2a | 19.8 | 18.4 |
| Average base coverage | 20× | 10× | 20× |
| N50 contig (kb)b | 48.7 | 127.4 | 50.3 |
| N50 scaffold (kb) | 307.7 | 817.8 | 738.5 |
| Chrs | 23 | 23 | 23 |
| GC content (%) | 52.0 | 54.0 | 59.7 |
| Coding | 54.3 | 56.3 | 62.6 |
| Noncoding | 43.4 | 50.5 | 54.3 |
| Nonassembled contigs (<500bp) | 48.0 | ||
| Coding sequence | |||
| Percentage coding (%) | 57.5 | 61.1 | 65.9 |
| Average gene size (bp) | 1,708 | 1,836 | 1,858 |
| Average gene density | 3.0 kb/gene | 2.9 kb/gene | 2.8 kb/gene |
| Protein-coding genes | 7,113 | 6,786 | 6,648 |
| Homologous proteinsc | 6,325 (89%) | 6,214 (92%) | 6,394 (96%) |
| Predicted proteinsd | 788 (11%) | 572 (8%) | 254 (4%) |
| Exons | 10,907 | 9,783 | 9,776 |
| Average exon size | 1,107 | 1,230 | 1,221 |
| Exons/gene | 1.53 | 1.44 | 1.47 |
| tRNA genes | 110 | 111 | 96 |
| Noncoding sequence | |||
| Introns | 3,161 | 2,997 | 3,103 |
| Introns/gene | 0.44 | 0.44 | 0.46 |
| Average intron length (base) | 141 | 142 | 144 |
| Average intergenic distance (bp) | 1,186 | 1,127 | 929 |
This excludes 4.9 Mb present in 866,023 small (<500 bp), nonassembled contigs.
Size in kilobases of ≥50% of the assembled contigs.
Proteins with support through matched similarities with other organisms, with an identity of more than 20% in SIMAP (Rattei et al., 2010).
Predicted proteins not supported by similarity in other organisms.
Analysis of all scaffolds and contigs allowed the prediction of 7113 nuclear protein coding genes, which together covered 57.5% of the available sequence (Table 1). Several tests indicated that most if not all of the gene space was covered (see Methods). The number of predicted proteins is almost 5% larger than the complement of 6786 proteins in U. maydis and contains 3023 conserved hypothetical proteins and 779 hypothetical proteins. Twenty-seven proteins have predicted functions related to TEs and show homology to retrotransposon HobS hobase or nucleocapsid proteins, transposase, or gag-pol-type polyproteins. However, they seem to represent nonfunctional TEs with pseudogenes since the open reading frames are interrupted by in-frame stop codons. In addition, the genome contains numerous noncalled pseudogenes with functions related to TEs, and these are located in the abundant small contigs (see below) that could not be placed in the genome assembly.
Comparative Chromosome Analysis
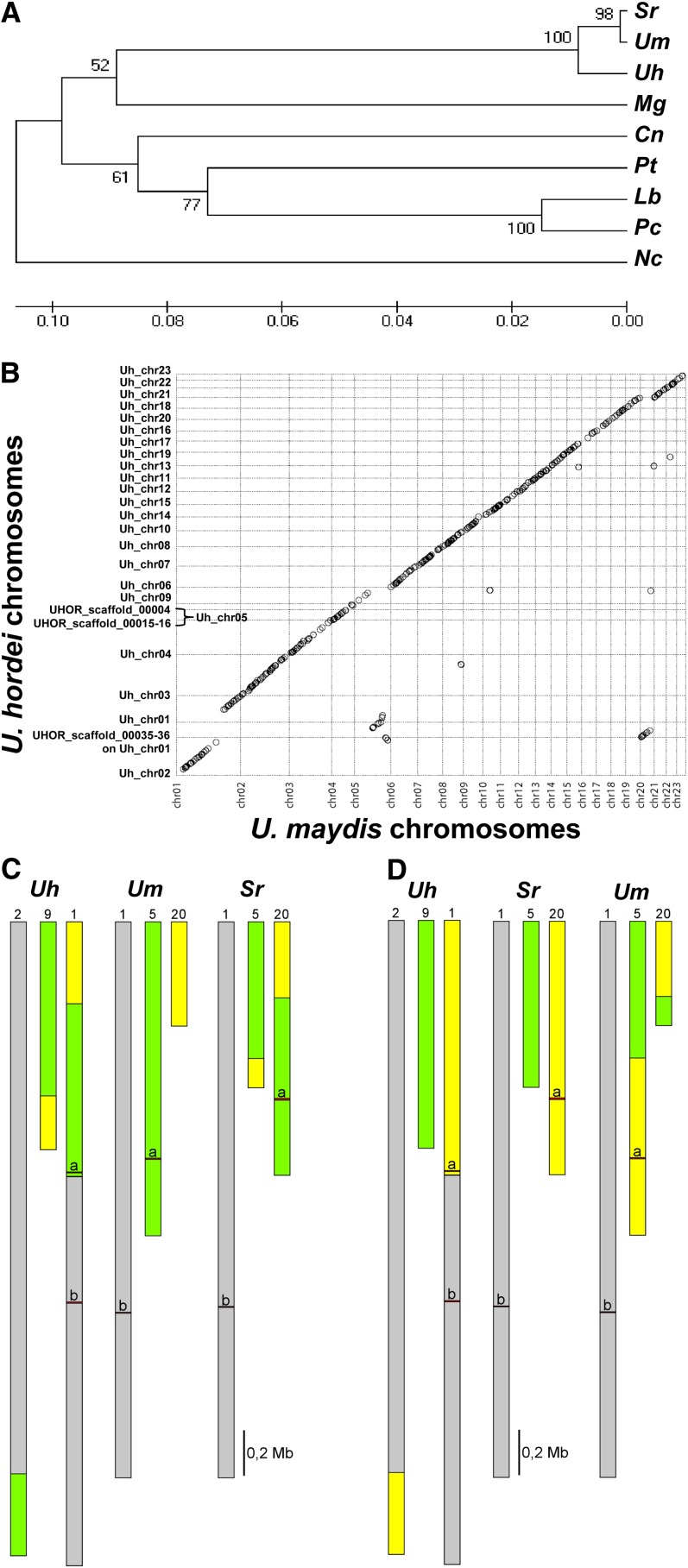
U. hordei is a relative of both U. maydis and S. reilianum and was estimated to have diverged from U. maydis between 21 and 27 million years ago (Bakkeren et al., 2006; Figure 2A). Of the 7113 U. hordei proteins, 69% show high (>57.1%) amino acid identity to homologs in U. maydis, and more than 98% of these also show high amino acid identity to homologs in S. reilianum, highlighting the relatedness of the three organisms. Overall, the genomes of U. hordei and U. maydis showed a high degree of synteny (Figure 2B; see Supplemental Data Set 1 online) that was only slightly lower than the synteny between the genomes of U. maydis and S. reilianum (Schirawski et al., 2010). In addition to a few smaller translocation events, three major rearrangements involving U. hordei chromosomes (Chrs) 1, 2, and 9 and U. maydis Chrs 1, 5, and 20 were evident (Figure 2B). One of these rearrangements corresponded exactly to a previously described translocation event, in which parts of Chrs 5 and 20 were exchanged in S. reilianum relative to U. maydis (Figure 2C; Schirawski et al., 2010). Indeed, chromosomal colinearity between the three species is much higher relative to S. reilianum than relative to U. maydis (Figures 2C and 2D). This suggests that the Chr arrangement in S. reilianum may represent the Chr arrangement of the ancestor of the three species. In S. reilianum, the a mating type locus resides on the left arm of Chr 20, while the b mating-type locus is on the left arm of Chr 1 (Figure 2D). Assuming the arrangement in S. reilianum to be ancestral, the relocation events that happened moved the a locus from Chr 20 to Chr 5 in the U. maydis lineage, while the b locus was transferred from Chr 1 to Chr 20 in the U. hordei lineage. This relocation event in the U. hordei lineage had fundamental biological consequences because it placed the a and b mating type loci on the same Chr (now designated Chr 1), thereby enabling development of a bipolar mating behavior on U. hordei (Figure 2D; Bakkeren et al., 2006).
Figure 2.
Taxonomic Placement of U. hordei and Genome Synteny with U. maydis and S. reilianum.
(A) Evolutionary placing of U. hordei in relation to other fungi. Consensus tree using the unweighted pair group method with arithmetic mean with bootstrap support (1000 replicates) based on β-tubulin protein sequences (Cn, C. neoformans CNC03260; Lc, Laccaria bicolor XP_001883306; Mg, M. globosa MGL_1528; Nc, N. crassa NCU04054; Pc, Phanerochaete chrysosporium e_gwh2.10.156.1; Pt, Puccinia. triticina PTTG_02454; Sr, S. reilianum sr16450; Uh, U. hordei UHOR_08314; Um, U. maydis um05828). All positions containing gaps and missing data were eliminated from the data set prior to computing evolutionary distances using the Poisson correction method (Zuckerkandl and Pauling, 1965) conducted in MEGA5 (Tamura et al., 2011).
(B) Dot plot of the synteny occurring between the chromosomes of U. maydis and the assembled supercontigs of U. hordei, created using the program nucmer from the MUMmer program package (http://mummer.sourceforge.net/).
(C) Synteny comparison of the indicated chromosomes of U. hordei and S. reilianum to those of U. maydis.
(D) Synteny comparison of the indicated chromosomes of U. hordei and U. maydis to those of S. reilianum. Syntenic parts are indicated by the same color.
Accumulation of Repetitive and TEs in the U. hordei Genome
The sequence assembly made use of a tiled BAC fingerprint map (Bakkeren et al., 2006), imposing constraints and requiring the introduction of gaps (stretches of N) of various lengths. A high number (2088) of small gaps within the supercontigs, ranging in size from 4 to 2615 bp and covering a total length of 1016 kb, was identified. Since 1267 flanks on either side of gaps and 681 flanks on both ends of gaps (in total 63% of flanks) mapped to previously identified TEs and repeats of U. hordei (Bakkeren et al., 2006), these sequence gaps likely resulted because of failure to assemble repetitive DNA at these sites. In support of this, of 3669 small contigs of <500 bp that could not be embedded in the assembly, 1118 (30%) matched a TE or LTR element and had a low GC content (48%), a feature associated with TEs in U. hordei. For comparison, an average GC content of 52% was found in all assembled contigs, including non-TE, gene coding (54%) and noncoding regions (43%; Table 1).
A comprehensive search of the U. hordei genome sequence for repetitive elements identified a large number of DNA transposons, a large array of LTR and unclassified retrotransposons, as well as LINE elements (see Supplemental Table 1 online). In total, repetitive sequences occupy almost 8% of the assembled U. hordei genome, which currently largely exceeds the repetitive genomic DNA content of other sequenced pathogenic basidiomycetes with the exception of possibly atypical rust fungi (see Supplemental Table 1 online; Duplessis et al., 2011). The similarity found among the repeats in U. hordei, U. maydis, and S. reilianum is quite low: >95% (1078/1120) of U. maydis repeats are not similar to any U. hordei repeat and 99% (5411/5441) of the U. hordei repeats are not similar to any S. reilianum repeat. In addition, 95% (1066/1120) of U. maydis repeats are not similar to any S. reilianum repeat (all calculated at >e−10, 30% alignment length and 30% identity threshold). Among the repeats showing similarity with the parameters used, just four families of small 100- to 115-bp elements could be identified that seemed evolutionarily related between all three species. However, no syntenic conservation of the repeats with respect to gene positions was detected among the three genomes. This suggests that the majority of these elements were acquired after separation of these species.
When relatedness of 5441 sequences representing TEs and repeat elements in U. hordei was analyzed, a limited number of families could be identified: 258 at 90% similarity and 142 at 80% or 65% similarity (see Supplemental Table 2 online). Among these 142 families, 41 represented only one element, but 4601 elements (>84%) could be assigned to only seven families of more than 50 repeat elements, with the largest two families holding 2751 Ty1/copia-type elements (row f in Figure 3; see Supplemental Figure 1 online) and 1376 Ty3/Gypsy class elements (row g in Figure 3; see Supplemental Figure 1 online). When diversity among 1120 U. maydis, 1226 Sporobolomyces roseus, and 110 Malassezia globosa repeat element sequences was compared, clustering was much reduced in these species compared with U. hordei, indicating more diverged within-species elements (see Supplemental Table 1 online). With the exception of M. globosa, which has a small genome with little reported repeat content (Xu et al., 2007), genome sizes of the other four species are comparable, yet U. hordei contains at least 4 times more highly related repeat elements (see Supplemental Table 1 online). These combined data suggest a recent expansion of a few related elements newly introduced in the U. hordei lineage after separation from a common ancestor. This is reminiscent of the genome of Magnaporthe grisea where many similar repeat elements were also contained in a few clusters of highly related repeats (>65% similarity) (Dean et al., 2005). Organisms have been shown to go through bursts of TE activity when new elements are introduced into naïve genomes or when repression is relieved, possibly through epigenetic changes, caused by stress such as a changing (plant host) environment (reviewed in Slotkin and Martienssen, 2007).
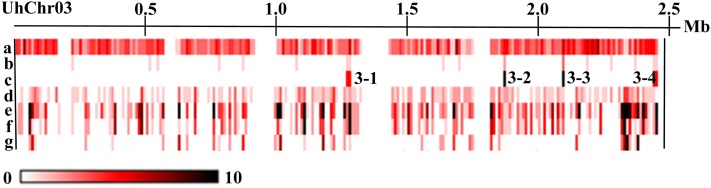
Figure 3.
Distribution of Genes, Sequencing Gaps, and Repetitive Elements on U. hordei Chr 3 Represented as a Heat Map.
Feature frequency was determined in a 10-kb sliding window. Rows indicate (a) all annotated gene models (interruptions indicate lack of sequence information between supercontigs), (b) small CSEP genes, (c) CSEP cluster homology regions, (d) small sequencing gaps within supercontigs, (e) all repetitive and TE sequences, (f) family of Ty1/copia-type elements, and (g) family of Ty3/Gypsy class elements. See Supplemental Figure 1 online for a complete genome-wide distribution.
Overall, the repetitive sequences are evenly distributed over the genome except for the MAT-1 mating-type region on Chr 1 where the repeat content reaches a value of ∼45% (Bakkeren et al., 2006; see Supplemental Figure 1 online), similar to the overall load found in several recently sequenced genomes of obligate basidiomycete rust pathogens (Duplessis et al., 2011). This accumulation in U. hordei is thought to result from a lack of a purifying recombination ability in the MAT region due to sequence dissimilarity compared with MAT-2, which likely fixed the a and b mating-type gene complexes in a bipolar organization (Lee et al., 1999). Repetitive sequences have also accumulated at other, smaller regions, in particular five regions on Chrs 3, 9, 17, 18, and 23 (row e in Figure 3; see Supplemental Figure 1 online). In these regions, no obvious clustering of genes with similar functions that may bestow a selective advantage when kept together is apparent, such as producing secondary metabolites. Rather, some of these regions harbor candidate-secreted effector protein (CSEP) genes, and the higher density of TEs and repeats may indicate a higher rate of genome evolution, possibly due to selection pressure (see further).
Genes Encoding RNA Interference and DNA Remodeling Components Present in U. hordei Are Cleanly Excised in U. maydis
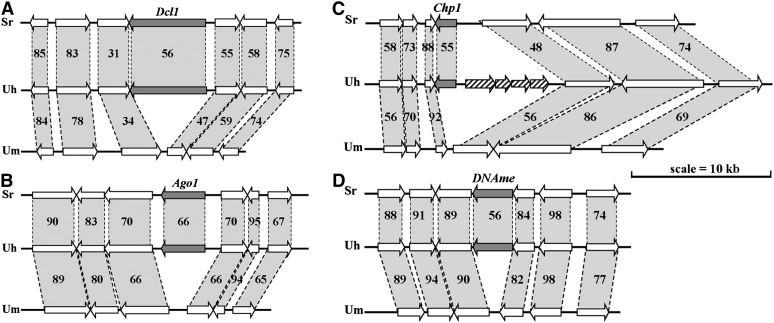
Among genes that occur in the U. hordei genome but do not have homologs in U. maydis are those putatively coding for components of the RNA interference (RNAi) machinery, such as ARGONAUTE (Uh-Ago1, UHOR_06256), three RNA-dependent RNA polymerases (Uh-RdRP1, UHOR_08874; Uh-RdRP2, UHOR_01631; and Uh-RdRP3, UHOR_15740), and DICER (Uh-Dcl1, UHOR_08937) as well as genes implicated in heterochromatin formation, such as chromodomain-coding HP1-like (Uh-Chp1, UHOR_05116) (Zofall and Grewal, 2006) and C5-cytosine methyltransferase (Uh-DNAme, UHOR_08509) (Smith and Shilatifard, 2007). These genes are widely represented in the fungal kingdom, although some are missing from several species and their copy number varies (Table 2; see Supplemental Figure 2 online). Whereas these seven genes are lacking from the U. maydis genome, they are present in S. reilianum (see Supplemental Table 3 online). Most baffling, all of these genes appear cleanly deleted from otherwise conserved syntenous regions in the U. maydis genome (Figure 4; see Supplemental Figure 3 online). Comparison of orthologous loci in S. reilianum and U. hordei revealed conserved synteny for the Uh-Dcl1, Uh-Ago1, Uh-RdRP3, and Uh-DNAme loci. However, in the Uh-Chp1, Uh-RdRP1, and Uh-RdRP2 loci, U. hordei carried additional genes potentially encoding proteins with functions in retrotransposition (Figure 4; see Supplemental Figure 3 online). This suggests that these loci, possibly under selection pressure, evolved differently in the three species and were active sites of TE activity and recombination, allowing gene insertions in U. hordei and gene deletions in U. maydis. Indeed, numerous copies of two variants of a 10-bp repeat sequence were found seemingly randomly distributed over the U. maydis genome (see Supplemental Figure 4 online; Kämper et al., 2006), occurring on average 1.76 times in the 1-kb regions flanking all 6786 protein-coding genes (sd = 1.88). However, these repeats accumulated at the former U. maydis Dcl1 and Ago1 loci (eight repeats) as well as at the former RdRP1 and RdRP2 loci (six and nine repeats, respectively; see Supplemental Figure 5 online). This suggests that intrachromosomal recombination led to loss of the RNAi and heterochromatin genes in U. maydis. Intrachromosomal recombination leaving small repeats was proposed to be the most frequent mechanism operating in maize for single gene deletions (Woodhouse et al., 2010).
Table 2. Number of RNAi Genes in Selected Fungal Species.
| RNAi Gene | Ascomycetes |
Basidiomycetes |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nc | Ca | Sc | Sp | Pc | Cc | Lb | Cn | Mg | Pgta | Sr | Uh | Um | |
| Ago | 2 | 1 | 0 | 1 | 7 | 8 | 6b | 2 | 0 | 3 | 1 | 1 | 0 |
| Dicer | 2 | 0 | 0 | 1 | 3 | 3 | 2 | 2 | 0 | 3 | 1 | 1 | 0 |
| RdRP | 3 | 0 | 0 | 1 | 9 | 7 | 6b | 1 | 0 | 5 | 3 | 3 | 0 |
Ca, Candida albicans; Cc, Coprinopsis cinerea; Cn, C. neoformans; Lb, L. bicolor; Mg, M. globosa; Nc, N. crassa; Pc, Phanerochaete chrysosporium; Pgt, P. graminis f. sp tritici; Sc, S. cerevisiae; Sp, S. pombe; Sr, S. reilianum; Uh, U. hordei; Um, U. maydis. Table adapted from Nakayashiki et al. (2006). Note that the number of genes is reduced in the pathogenic basidiomycetes compared to other basidiomycetes.
Comparative BLAST searches identified one more possible (partial) Ago sequence and two more matching Dicer sequences in the currently available Pgt genome (Duplessis et al., 2011; http://www.broadinstitute.org/annotation/genome/puccinia_group/MultiHome.html; March, 2012); comparative phylogenetic analysis suggests the likely presence of allelic variants in the available genome information that is derived from dikaryotic material. Fewer genes were found in the currently available Pt genome that is likely still partial: one Ago, four Dicer, and two RdRp sequences. Even fewer were found in the partial Pst genome sequence currently available (Cantu et al., 2011).
Database: Laccaria bicolor Best Models (proteins) Joint Genome Institute.
Figure 4.
Synteny around Loci Present in U. hordei and S. reilianum but Missing in U. maydis.
RNA silencing genes Dicer Uh-Dcl1 (UHOR_08937) (A), Argonaute Uh-Ago1 (UHOR_06256) (B), chromodomain gene Uh-Chp1 (UHOR_05116) (C), and a Cytosine 5-specific methyltransferase Uh-DNAme (UHOR_08509) (D) are depicted as dark arrows. Synteny is indicated by shaded bars with percentage of amino acid identity between the predicted flanking proteins. Note the four genes to the right of Uh-Chp1 (C), one of which (UHOR_15241) is related to two genes, UHO_0374 and UHO_0261, possibly coding a DNA polymerase III ε-subunit and previously found embedded in repeats in the U. hordei MAT-1 region (GenBank accession number AM118080; Bakkeren et al., 2006), and three genes (UHOR_17012, UHOR_17013, and UHOR_17014) that match many related genes in the genome and likely code for reverse transcriptase, gag-pol/copia-type proteins (pfam07727) usually indicative of a retrotransposon.
Genome Defenses Related to TE Control and Heterochromatin Formation
To control the negative consequences of proliferating TEs, such as gene inactivation, ectopic recombination, and genome instability, U. hordei may use mechanisms similar to those described for Schizosaccharomyces pombe. In S. pombe, RNAi facilitates heterochromatin formation adjacent to centromers, affecting centromere function and chromosome segregation (Chen et al., 2008; Kloc and Martienssen, 2008). HP1-like chromodomain proteins (CHP1, CHP2, and SWI6) associate with methylated histones in pericentromeric repetitive DNA when transcription during S-phase of the cell cycle is targeted by RNAi (Fischer et al., 2009). U. hordei (and S. reilianum, but not U. maydis) contains the necessary genes Chp1-, Chp2-, and Swi6-like needed for this mechanism (see Supplemental Table 3 online). Direct control of TE activity by RNAi in fungi has also been demonstrated in Saccharomyces species, where DICER and ARGONAUTE proteins silence endogenous retrotransposons (Drinnenberg et al., 2009), and in Cryptococcus neoformans, where RNAi controls TE activity during sexual development (Janbon et al., 2010; Wang et al., 2010). Therefore, fungi seem to use RNAi for protection against TEs similar to other eukaryotes.
Alternatively, TE activity can be controlled by the mechanism of repeat-induced point mutation (RIP). First discovered in Neurospora crassa (Selker and Stevens, 1985), RIP was noticed to operate during sexual reproduction of several ascomycete fungi. RIP results in C-to-T transitions at repetitive loci rendering TEs inactive through mutation (Galagan and Selker, 2004). In this study, analysis of homologous TEs clearly revealed that U. hordei employs RIP to control TE activity: Numerous point mutations representing C-to-T (G-to-A) transitions were discovered when similar TEs were aligned (see Supplemental Figure 6 online). Many occurred at TCG triplets (or CGA on the opposite strand) as found in a related smut fungus Microbotryum violaceum (Hood et al., 2005), though they were not strictly limited to these triplets as in M. violaceum. This could point to a more complex RIP mechanism than found to date in ascomycetes (Clutterbuck, 2011) or indicate a different mechanism. RIP targets repeats causing deamination of cytosine residues at CpG dinucleotides. This is evident by a decrease/depletion of CpG dinucleotides and a concomitant increase in both CpA and TpG (Zemach et al., 2010). A genome-wide analysis found a clear decrease in CpG dinucleotides only in U. hordei repeats (see Supplemental Figure 7 online). When comparing TE repeats in S. reilianum, C-to-T (G-to-A) and C-to-G or -A transitions were found, indicative of a RIP mechanism in this fungus as well (see Supplemental Figure 8 online). However, this was not easily corroborated when analyzing a potential genome-wide reduction in CpG dinucleotides (see Supplemental Figure 7 online). The low abundance of repeats could compromise a proper analysis in this fungus; it was concluded previously that S. reilianum and U. maydis likely do not possess a RIP mechanism (Horns et al., 2012).
In contrast with U. hordei, U. maydis lacks RNAi, HP1-like, and putative DNA methyltransferase genes, and no RIP mutation mechanism has been identified thus far. The lack of a homolog to U. hordei DNA methyltransferase, UHOR_08509, possibly a functional homolog of Rid (for RIP-defective) in N. crassa (Freitag et al., 2002), would be consistent with this finding. Therefore, U. maydis lacks universal mechanisms for TE control and heterochromatin formation and likely uses alternative means for genome maintenance. The high frequency of homologous recombination reported for U. maydis (Holloman et al., 2008) may compensate for lack of conserved defense mechanisms.
Several diverse fungal species lacking RNAi components have now been described, and possible consequences for lifestyle and biology are emerging. Recently, in Saccharomyces cerevisiae, incompatibility between RNAi and double-stranded RNA killer viruses has been described (Drinnenberg et al., 2011), suggesting that in the environment where U. maydis is found, it could be advantageous to possess toxin-producing killer virus while the presence of an RNAi silencing mechanism would preclude the persistence of such viruses in U. hordei and S. reilianum. This could be related to differences in lifestyle, with U. maydis having to compete with other microbes on all aboveground parts of the plant, while U. hordei and S. reilianum, which cause a systemic infection, may be less prone to having to fight competitors. It may be that the presence of advantageous double-stranded RNA viruses drove the loss of RNAi components or, vice versa, that the loss of the silencing mechanism allowed viruses to occupy this fungal niche, now having dropped its antiviral defenses. Another interesting example concerns two Cryptococcus gattii isolates, one of which lacks Ago1 and Ago2 sequences and is virulent toward healthy humans, whereas the other isolate only infects immunocompromised individuals (D’Souza et al., 2011). It is currently unknown whether there is any correlation between loss of silencing components and changes in virulence and/or TE content in these evolved subspecies in the C. gattii complex.
U. hordei has a bipolar mating system that, by nature, promotes inbreeding. Inbreeding leads to increased homozygosity, reducing the potential for ectopic recombination between TEs. Evolution of bipolar mating may thus have been beneficial for the fungus because it promoted inbreeding and stabilization of the genome under TE stress after expansion of the TE repertoire. The same process could also have been beneficial for the TEs since inbreeding helps fix TEs within a population (Blumenstiel, 2011). In tetrapolar species, such as U. maydis and S. reilianum, outcrossing increases heterozygosity. This would raise the risk of ectopic recombination between related TEs that could eliminate TEs, but at the same time promote genome instability. The combination of bipolar mating, RNAi, and repressive chromatin features of HP1-like proteins and DNA methylation supported by RIP in U. hordei would provide a robust means by which to manage the abundant TEs. However, unless eliminated, as appears to be the case in U. maydis, TEs would likely occasionally escape even the tightest of controls and accumulate in loci subsequently suppressed in recombination. This could influence the expression of genes at these loci and affect their evolution within a population. Of considerable interest in this context are loci where repetitive DNA/TEs coincide with effectors that interact with the plant host.
CSEPs
Of the 515 U. hordei proteins predicted to be secreted, 333 were judged to be CSEPs (see Supplemental Data Set 2 online). Interestingly, these proteins matched 36 additional related proteins with an amino acid identity above 20% (see Supplemental Data Set 2 online). These paralogs have no identifiable signal peptide and may have either lost or mutated this feature during recombination/duplication and selection or they may still be secreted through an alternate mechanism. When comparing U. hordei and U. maydis proteins, CSEPs seem to be more diverged than the rest of the proteome (see Supplemental Figure 9 online), suggesting that they evolve faster, possibly as a result of interactions with host components selecting for diversification. Only 47 (14%) of the 333 U. hordei CSEPs are U. hordei specific. The vast majority has a homolog in either U. maydis or S. reilianum, and of those, 112 (34%) are even highly conserved in all three organisms (see Supplemental Data Set 2 online and Methods for definition of specific or conserved). This may indicate the existence of two functional groups of CSEPs: Those of high conservation may target essential plant processes, while those that are weakly conserved may target the highly dynamic plant defense systems (Doehlemann et al., 2009; Schirawski et al., 2010).
In several fungi and oomycetes, effectors have been described that are rich in Cys and, although not necessarily similar in primary amino acid sequence, many have Cys residues in similar patterns and spacing, potentially involved in disulphide bridge formation and secondary structure. In U. hordei, 60 CSEPs and six related paralogs could tentatively be placed in 18 arbitrary classes of which 11 had members with Cys patterns equivalent to U. maydis effectors (see Supplemental Data Set 3 online). For comparison, in U. maydis, 62 such effectors were placed in 13 classes (Mueller et al., 2008). The largest class of U. hordei effectors contained 19 Mig1-related proteins having a characteristic and conserved pattern of Cys residues. In U. maydis, Mig1 was shown to be highly induced during biotrophic growth in maize (Basse et al., 2002). U. maydis has only three copies of Mig1-like genes, while eight copies exist in S. reilianum (see Supplemental Figure 10 online; Schirawski et al., 2010). Some of the U. hordei Mig1-related genes do not have a predicted signal peptide sequence and could have been inactivated to avoid secretion and recognition by the host. Interestingly, whereas the Mig1-related genes occur in one locus on Chr 8 in both S. reilianum and U. maydis, they are dispersed over the genome in U. hordei. It is plausible that TE activity contributed to the dispersal of the Mig1-related genes over the genome in U. hordei since 15 out of the 19 genes have flanking DNA of which one side (nine) or both sides (six) match repeats or TE sequences. It is presently unknown if any of these dispersed copies could represent avirulence (avr) genes; only six avr genes have been genetically distinguished of which only Uh-Avr1 has been mapped (Linning et al., 2004).
Many predicted secreted effectors in U. maydis (Kämper et al., 2006) and S. reilianum (Schirawski et al., 2010) are arranged in clusters. In U. hordei, a subset consisting of 135 CSEPs (40%) and three paralogs lacking a recognizable secretion signal could be assigned to 45 regions with two or more CSEPs. At least 16 regions contained Uh CSEPs that matched homologs identified in the U. maydis clusters (Kämper et al., 2006) and/or S. reilianum diversity clusters (Schirawski et al., 2010). However, in general, the 45 Uh regions were less compact than the clusters in U. maydis and S. reilianum, and to recognize them, we had to allow CSEPs and/or paralogs to be interrupted by up to 10 unrelated proteins (see Supplemental Data Set 4 online). This could indicate that ancestral clustering may have disappeared during adaptation to a new host. Just one region (6-1) contains only Uh-specific CSEPs that are clustered, although at least 13 cluster homology regions have Uh-specific CSEPs as well as ones with weak homology to either U. maydis or S. reilianum effectors (see Supplemental Data Set 4 online). Apart from some clustering, the distribution of the majority of Uh CSEPs over the genome appears random, although some chromosomes carry few CSEPs, such as Chrs 13, 16, and 19.
Interestingly, some identified Uh CSEP cluster homology regions coincide with areas where higher densities of TEs are found (see Supplemental Figure 1 online, cf. rows b, c, and e). Examples are the CSEP regions 1-2, 3-4, 7-2, 8-2, 9-3, and 18-1 on the respective chromosomes (see Supplemental Data Set 4 online). Overall, TE and repeat sequences were found ∼5 times more often in 1-kb flanks of 308 CSEP genes (43% on either side and 8% on both sides) than of 6411 other Uh genes (9% on either side and 2% on both sides), and the CSEPs in the TE-rich regions mentioned above were indeed often flanked by TE and repeat sequences (see Supplemental Data Set 4 online). Moreover, 18 of the 43 Um and Sr diversity regions (mentioned in Kämper et al., 2006 and Schirawski et al., 2010) contained Uh CSEPs with flanking TE and repeat sequences, including five of the Uh-Mig1 genes. TE activity may contribute to rearrangements and activation or inactivation of effectors, driving their evolution and creating a selective advantage with respect to overcoming host resistance. Effector genes have been detected in repeat-rich regions in several pathogens, and it has been speculated that their presence in highly fluid parts of genomes may allow the organism to adapt quickly to a hostile host environment (Haas et al., 2009; Baxter et al., 2010; Raffaele et al., 2010; Spanu et al., 2010; Duplessis et al., 2011). U. maydis and S. reilianum, two related successful obligate pathogens with streamlined genomes largely devoid of TEs, may have evolved so far unknown but efficient mechanisms to mutate and adapt.
In conclusion, the U. hordei genome provides clues to the biology and evolution of smut fungi. Of significant importance is how TEs have seemingly restructured the U. hordei genome affecting reproductive biology and possibly evolution of genes encoding effector proteins. A shift in favor of inbreeding helped maintain TEs even under the likely repressive forces of RNAi and RIP. This work reveals striking differences in TE load and genome defense strategies and provides a starting point for future studies aimed at addressing the relationship between RNAi and TEs, and TEs and effector genes.
METHODS
Ustilago hordei Genome Sequencing
U. hordei haploid strain Uh4857-4 (alias Uh364, mating type MAT-1; Linning et al., 2004) was chosen for sequencing because a BAC library was available (Bakkeren et al., 2006). Preparation of genomic DNA for pyrosequencing followed a described method (Schirawski et al., 2010). Essentially, U. hordei was grown in full medium with shaking to an OD600 of 0.5. Cells were pelleted and treated with cell wall–digesting enzymes for the generation of protoplasts. Protoplasts were lysed, and genomic DNA was prepared using a genomic DNA isolation kit (Q-tip 100; Qiagen). To reduce the content of mitochondrial DNA, Q-tip 100–purified DNA of U. hordei was subjected to CsCl density gradient centrifugation. Fractions containing genomic DNA were pooled and dialyzed, and the genomic DNA was precipitated and solved to a concentration of 1 μg/μL in TE buffer. Subsequent library construction and genome sequencing of U. hordei was done at 454 Life Sciences. As a first step, a whole-genome shotgun random library was constructed and sequenced using the Genome Sequencer FLX (Roche). A total of 2,149,477 high-quality filtered sequence reads with an average read length of 236 bases were generated. Total sequence output was 658,792,120 bases, which corresponded to almost 25-fold coverage, and 21,196,105 bases had a quality score of at least 40 (i.e., accuracy was at least 99.99%). Individual reads were de novo assembled using the 454 Newbler assembler. Unfortunately, the assembler was not able to assemble the complete data set. The unfinished assembly resulted in 3219 contigs of more than 500 bases with a total length of 21,271,904 bases. The N50 contig size was 16,017 bases. To improve the assembly, a new library was constructed as a 10-kb paired-end library that was sequenced at 454 Life Sciences using the Genome Sequencer FLX (Roche). A total of 603,649 paired-end reads with a mean length of 250 bp were generated and added to the assembly. In addition, sequencing information of an available BAC library consisting of 2304 clones was used, for which a fingerprint map had been generated, yielding 60,429 HindIII fragments with a calculated length of 212,744,135 bp or ∼8 times the estimated 26-Mb genome (Bakkeren et al., 2006). BAC end sequencing was performed on 2304 BAC clones at the Michael Smith Genome Sciences Centre (Vancouver, Canada) using standard M13 reverse primer (5′-CAGGAAACAGCTATGAC-3′), T7 primer (5′-AATACGACTCACTATAG-3′), and BigDye v3.1 chemistry on ABI3730 sequencers (Applied Biosystems). The CAP3 program (Huang and Madan, 1999) was used to search for overlap, resulting in 2597 sequences with an average length of 786 bases, covering a total of 2,042,392 quality bases (∼10% of the estimated genome size). With this additional sequence information and a new version of the Newbler software (version 2.0.0), the assembly could be completed, which resulted in 6618 contigs, of which 2949 were at least 500 bases in length. In total, 3,030,000 reads covering more than 634,000,000 high-quality bases were used for the final assembly, which corresponded to more than 24-fold coverage. Integrating the pairing information of paired ends and paired BAC reads, 1268 scaffolds were further assembled into 27 supercontigs. Reassessment placed 71 contigs (4.01024, 4.00815, 4.01124, 4.00842, 4.00047, and 5.00001 to 5.00066) on these supercontigs. Constraints required the introduction of sequence gaps (stretches of NNNs) of various lengths. Finally, assembly of the 27 supercontigs was verified by generating an EcoRI-based Optical Map using MapSolver (www.OpGen.com). Some scaffolds were remaining (641 having sizes of between 0.5 and 20 kb and seven that were larger than 20 kb) that could not be placed on the supercontigs because of lack of support from BAC end reads or optical mapping data. Comparative analysis to the Ustilago maydis genome resulted in the tentative assignment of the 71 contigs to 23 potential U. hordei chromosomes with synteny to (parts of) the 23 recognized U. maydis chromosomes; Uh Chr 5 is a likely homolog of Um Chr 4 (Figure 2; see Supplemental Data Set 1 online). The total genome size based on the sum of the contigs is 21.2 Mb, and based on optical mapping is estimated to be 26.1 Mb. Thus, assembled sequence information covers 81.1% of the total estimated genome size (76.7% without the small, nonassigned contigs).
Repeat Content
Repeat content was computed using RepeatMasker based on its default library, a RepeatScout library, and custom annotated repeats library. The RepeatScout library was generated using seed lengths of 10 to 21 and default parameters. Elements of at least 50 bp in length were included. Low-complexity repeats and tandem repeats were removed as part of the RepeatScout algorithm, using Nseg (Wootton and Federhen, 1993) and Tandem Repeat Finder (Benson, 1999). TEclass, a tool for automated classification of unknown eukaryotic TEs (Abrusán et al., 2009), was run using a 2010/01/20 version of RepBase. The larger value was reported when RepeatMasker and TEclass predicted different counts for certain TE classes. Low complexity and simple repeats and small RNAs were classified exclusively by RepeatMasker using the default library. If repeats overlapped by more than 50 nucleotides, only the larger one was counted. To search for similarity among TEs from the various species, the similarity searching program ssearch36 from the FASTA package (Pearson et al., 1997) was used. To visualize the distribution of the various repeats and TEs (Figure 3; see Supplemental Figure 1 online), overall protein density, and CSEPs, a heat map of genomic feature density was prepared from optical map data, feature coordinates, and assembled sequence. Custom Perl scripts were written to carry out the following. Optical map data were used to order and group the genome assembly’s supercontigs into chromosomes. These chromosomes were, in turn, divided into 10-kb bins. Genes (including a subset of 308 secretory proteins), secretory gene cluster regions, and intrasupercontig sequencing gaps were assigned to appropriate bins and counted. Repeats and TE sequences were similarly allocated. In addition, the repeat/TE sequences were enumerated within four subgroups based on descending frequency of membership. The frequency counts for the chromosomal bins were maintained in a MySQL database, and the results of appropriate queries of this database for each chromosome were input into a Web interface for the matrix2png heat map program (www.bioinformatics.ubc.ca/matrix2png/bin/matrix2png.cgi). Since many of the TEs and repeat elements were located on many small contigs that could not be assembled into the larger genome contigs, only a partial distribution could be revealed. Genome entropies reflecting informative (coding) DNA were computed as described by Lauc et al. (1992) by counting the number of times different k-mers occurred in the genome and applying the formula for entropy H = −Σ (pi * log (pi), where pi is the probability of k-mer i occurring in the genome; i = 1 to n, where n is the number of possible k-mers. For trinucleotides, n = 43 = 64. The entropy value was lower for U. hordei compared with the other three genomes, which correlated with the repeat content (see Supplemental Table 1online). The trend in Supplemental Table 1 online did not change for 1 to 10mers.
Ribosomal Repeats
rRNA sequences (18S-ITS1-5.8S-ITS2-25S) were previously shown to be contained in 8.6-kb repeats (Bakkeren et al., 2006). By hybridization of pulse field gel-separated chromosomes, probes representing the ribosomal gene repeats revealed a chromosome that was called IV of ∼2 Mbp in the same U. hordei strain (Bakkeren et al., 2006). In that study, sizeable differences in the length of this chromosome (from 2.25 to 2.0 Mb or from 1.9 to 1.7 Mb) were uncovered by hybridization, even between clonal but transformed lines of the same basidiospore (Figure 7A, panel 2, in Bakkeren et al., 2006; cf. lanes 1 to 4 and 5 to 7) and among basidiospore progeny from the same teliospore (cf. lanes 1 to 4 versus 5 to 7); no hybridization was found to other chromosomes. This variation in chromosome length additionally suggested that this chromosome contained the rDNA repeats by revealing frequent mitotic and meiotic recombination occurring between them. From the BAC library analysis in that study, the copy number of rRNA repeats was estimated at 140, occupying ∼1.2 Mbp of the estimated 2 Mbp chromosome. In this study, three contigs (5.00015, 5.00016, and 5.00004) totaling 807 kb, were tentatively fitted to chromosome 5 for which a size of ∼1.5 Mbp was calculated based on optical mapping data (see Supplemental Data Set 1 online). In U. maydis, Chr IV holds the ribosomal repeats, estimated to occupy ∼600 kb (Kämper et al., 2006), and this chromosome was found syntenous to U. hordei Chr 5 in this study. Ribosomal repeat sequences were compared by BLASTn to the first set of 454-generated 603,649 sequences (mean length 250 bp covering 150,389,109 bp), yielding 12,000 matches to mainly unassembled reads and small contigs, as expected for highly repetitive sequences. In 532,042 paired-end reads (mean length of 100 bp covering 56,323,004 bp), 6950 matches were found. These two sets provided an estimate of the number of ribosomal repeats of 61 and 41, occupying 525 and 352 kb, respectively. The discrepancy might result from these sequencing approaches/techniques not being random (the paired ends generated from 10-kb clones). Because of many assumptions and uncertainties about exact match length and coverage, this calculation likely provides an underestimate of the number of repeat units.
Open Reading Frames and the Secretome
Gene models were predicted through use of specifically trained programs GeneMark (Besemer and Borodovsky, 2005; Ter-Hovhannisyan et al., 2008) and Fgenesh (http://softberry.com) and were compared with open reading frames and predicted proteins in the U. maydis genome using BLAST (Altschul et al., 1990). In all available genome reads, 7113 protein-encoding gene models were identified. In addition, 110 tRNA-encoding genes were predicted using tRNAscan-SE (Lowe and Eddy, 1997). To assess the genome completeness, a BLAST search was performed with highly conserved core eukaryotic genes present in higher eukaryotes (Aguileta et al., 2008; Parra et al., 2009). From the expected 246 single-copy orthologs extracted from 21 genomes, 246 were present in all three species (Uh, Sr, and Um), indicating that the gene space was likely completely covered by the assembly. From numerous genome sequencing projects, including from several Ustilaginales, it has become apparent that the 454 sequencing strategy is particularly well suitable for coding regions but has drawbacks in dealing with repetitive material (Wicker et al., 2006). A BLASTx search of all 4.9 Mb contained in small (<500 bp) contigs that could not be assembled into the larger genome contigs did not reveal any genes that may have been missed by the gene calling programs; a number of them only revealed partial sequence homologies to transposases, etc. To predict a set of secreted effector proteins in U. hordei, all 7113 predicted proteins were first analyzed by the SignalP 3.0 program (http://www.cbs.dtu.dk/services/SignalP/) to predict a signal peptide based on SignalP-HMM results. This resulted in 1142 signal peptide–predicted proteins, which were subsequently run through TMHMM 2.0 (Krogh et al., 2001) to identify transmembrane proteins. Proteins with one transmembrane domain were kept in the data set if the transmembrane domain was located close to the predicted signal peptide, since this could indicate a function related to the translocation and not necessarily predict membrane retention. All other proteins with predicted transmembrane domains were removed. The resulting 931 proteins were subsequently screened by TargetP v1.1 (Emanuelsson et al., 2007) to identify and remove proteins that were predicted to be mitochondrial, resulting in a set of 540 predicted secreted proteins. These 540 protein sequences were then analyzed with ProtComP 9.0 (http://linux1.softberry.com/berry.phtml), which compares them to proteins in the LocDB and PotLocDB databases that hold proteins with known or reliably predicted localization. Proteins with predicted extracellular localization or with no database correlation were kept in the data set, resulting in 515 predicted secreted proteins. Subsequently, proteins annotated with known functions, such as cell wall–modifying enzymes, were removed; the remaining set contained a number of sequences matching the TEs and repeats that were removed as well, although intriguingly these could include chimeric TE-effector sequences, indicating inactivated effectors. The final set consisted of 333 proteins. This analysis was performed essentially as described for the 6786 U. maydis gene models where 386 potential secreted effector proteins were identified (Mueller et al., 2008). In Sporisorium reilianum, a similar analysis of the 6648 gene models (Schirawski et al., 2010) yielded 400 potential secreted effector proteins. In addition, a comprehensive paralog search within each species using a similarity matrix of proteins (SIMAP) analysis (Rattei et al., 2010) pulled in 36, 98, and 95 unique paralogs with a SIMAP value of ≥20% for U. hordei, U. maydis, and S. reilianum, respectively. SIMAP calculates amino acid identities: Amino acid identities of homologous stretches are multiplied by the length of the homologous region and divided by the length of the longer protein (http://webclu.bio.wzw.tum.de/portal/web/simap/). The paralogs do not have predicted signal peptides and were therefore not included in the initial set based on SignalP and TargetP searches. However, by definition they are related to the predicted effectors. Examples are the four additional Mig1- and two additional Mig2-related proteins that have a similar Cys pattern. Interestingly, >96% of the weakly conserved genes present in the previously identified U. maydis–S. reilianum divergence clusters (Schirawski et al., 2010) have weakly conserved homologs in U. hordei (see Supplemental Data Set 2 online). This supports the notion (Schirawski et al., 2010) that these highly diversified proteins encode virulence functions that likely play a role in smut fungal adaptation to different hosts and lifestyles.
Proteome Comparison
The 7113 predicted proteins of U. hordei strain Uh4857-4 (http://mips.helmholtz-muenchen.de/genre/proj/MUHDB) and 6786 proteins from U. maydis strain 521 (http://mips.helmholtz-muenchen.de/genre/proj/ustilago; Kämper et al., 2006; Schirawski et al., 2010) were compared with each other using SIMAP (Rattei et al., 2010). The average amino acid identity of all pairs of genes occurring in both organisms was 63%. Genes and gene pairs were grouped in one of three categories depending on their amino acid identities of the proteins they code for. Uh-Um gene pairs representing an amino acid identity above 57.2% (4898 of the U. hordei genes or 69%) were considered highly conserved, while those with an amino acid identity between 57.2 and 20.1% were considered low conserved (1162 U. hordei genes). Any genes representing amino acid identities of 20.0% or below to their best hits were regarded as species specific. However, among the latter group, 23 genes coded for proteins matching Um or Sr proteins with SIMAP values slightly below the cutoff value of 20% but were in syntenic positions flanking ones showing higher SIMAP values and are therefore possibly very diverged orthologs. Therefore, there are 1185 (17%) low conserved genes and 1030 (14%) U. hordei–specific genes (Uh); the reciprocal comparison yielded 608 U. maydis–specific genes (Um).
Phylogeny
Amino acid sequences for the RNaseIII domain of Dicer, the PIWI domain of Ago, and the RdRP domain of RdRP genes were described previously for other fungi (Nakayashiki et al., 2006). Using these sequences, BLAST searches (tBLASTx) were performed on the U. hordei genome. Retrieved sequences were used to search the National Center for Biotechnology Information for hits supporting their identity. Genes were predicted using FGENESH and domains predicted using the CDART program at the National Center for Biotechnology Information. Phylogenies were done using MEGA5 (Tamura et al., 2011) with sequences described previously (Nakayashiki et al., 2006), but with the addition of Laccaria bicolor dicers Lb1 (scaffold_4, 1619948-1616065) and Lb2 (scaffold_6, 1342360-1348160) from the Department of Energy Joint Genome Institute (http://genome.jgi-psf.org/Lacbi1/Lacbi1.home.html) and related sequences from Puccinia graminis f. sp tritici from the Broad Institute (http://www.broadinstitute.org/annotation/genome/puccinia_group/MultiHome.html). U. maydis and S. reilianum sequences were obtained from the databases at the Munich Information Center for Protein Sequences (MUMDB and MSRDB, respectively, at http://www.helmholtz-muenchen.de/en/mips/projects/fungi/index.html) and the Broad Institute (http://www.broad.mit.edu/annotation/genome/ustilago_maydis). Alignments used to produce the phylogenetic trees shown in Supplemental Figures 2 and 10 online are presented in Supplemental Data Sets 5 and 6 online, respectively.
Accession Numbers
The genome, the nonassigned contigs, and gene models are accessible through the Munich Information Center for Protein Sequences at http://mips.helmholtz-muenchen.de/genre/proj/MUHDB and were deposited at the European Nucleotide Archive (http://www.ebi.ac.uk/ena/data/view/) under accession numbers CAGI01000001 to CAGI01000713.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Heat Map Revealing Distribution of Various Features on the U. hordei Chrs.
Supplemental Figure 2. Phylogenetic Relationship of Selected Dicer and Argonaute Protein Sequences.
Supplemental Figure 3. Comparative Microsynteny around Loci Present in U. hordei (Uh) and S. reilianum (Sr) but Missing in U. maydis (Um).
Supplemental Figure 4. Distribution of the 10-bp Repeats in the U. maydis Genome.
Supplemental Figure 5. Sequences of the Loci in U. maydis Deleted for RNA Silencing Genes.
Supplemental Figure 6. Indication of RIP in U. hordei.
Supplemental Figure 7. Analysis of Dinucleotide Frequency in Three Smut Fungi.
Supplemental Figure 8. Indication of RIP in S. reilianum.
Supplemental Figure 9. Diversity among U. hordei and U. maydis Proteins.
Supplemental Figure 10. Molecular Phylogeny Tree Depicting the Relatedness among All Mig1-Related Proteins Found in the Genomes of U. hordei, U. maydis, and S. reilianum.
Supplemental Table 1. TEs and Repeat Content among Five Basidiomycete Genomes.
Supplemental Table 2. Number of Clusters of TEs and Repeat Elements in Four Basidiomycete Genomes.
Supplemental Table 3. Conservation between U. hordei and S. reilianum Proteins Involved in Transcriptional Gene Silencing and Chromatin Remodeling but Absent from U. maydis.
Supplemental Data Set 1. U. hordei Contigs Matching Chrs of U. maydis.
Supplemental Data Set 2. List of 333 U. hordei Candidate-Secreted Effector Proteins and 36 Paralogs, Their Sr and Um Homologs.
Supplemental Data Set 3. U. hordei Candidate-Secreted Effector Proteins with Similar Patterns of Cys (C) Residue Occurrence and Spacing (X Number of Amino Acid Residues).
Supplemental Data Set 4. Clustering of U. hordei Candidate-Secreted Effector Proteins and Their Homologs in U. maydis and S. reilianum.
Supplemental Data Set 5. Alignment Used to Produce Phylogeny Presented in Supplemental Figure 2.
Supplemental Data Set 6. Alignment Used to Produce Phylogeny Presented in Supplemental Figure 10.
Supplementary Material
Acknowledgments
We are grateful for special funds from the Max Planck Society for sequencing, optical mapping, and manual annotation. R.K. acknowledges support through SFB593 and the LOEWE center (SYNMIKRO). G.B. acknowledges support from the Natural Sciences and Engineering Research Council of Canada and the Agriculture and Agri-Food Canada Canadian Crop Genomics Initiative. We thank the sequencing team at the Michael Smith Genome Sciences Centre for BAC end sequencing, Julien Dutheil for critical comments on the article, Elmar Meyer for technical assistance, Denis Gaudet and Guanggan Hu for photographs in Figures 1B and 1C to 1F, respectively, and Xiangfeng Wang for the analysis in Supplemental Figure 4 online.
AUTHOR CONTRIBUTIONS
J.D.L., S.A., R.L., P.W., U.G., M.M., J.S., and G.B. analyzed data. G.M. assembled and annotated the sequence. G.B. and R.M. provided the BAC end sequences. J.S. provided the sequenced DNA. R.K. and G.B. obtained funding for the sequencing. G.B., J.S., J.D.L., S.A., and R.K. wrote the article.
Glossary
- LTR
long terminal repeat
- TE
transposable element
- Chr
chromosome
- CSEP
candidate-secreted effector protein
- RNAi
RNA interference
- RIP
repeat-induced point mutation
- SIMAP
similarity matrix of proteins
References
- Abrusán G., Grundmann N., DeMester L., Makalowski W. (2009). TEclass—A tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25: 1329–1330 [DOI] [PubMed] [Google Scholar]
- Aguileta G., Marthey S., Chiapello H., Lebrun M.H., Rodolphe F., Fournier E., Gendrault-Jacquemard A., Giraud T. (2008). Assessing the performance of single-copy genes for recovering robust phylogenies. Syst. Biol. 57: 613–627 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bakkeren G., et al. (2006). Mating factor linkage and genome evolution in basidiomycetous pathogens of cereals. Fungal Genet. Biol. 43: 655–666 [DOI] [PubMed] [Google Scholar]
- Bakkeren G., Kämper J., Schirawski J. (2008). Sex in smut fungi: Structure, function and evolution of mating-type complexes. Fungal Genet. Biol. 45 (suppl. 1): S15–S21 [DOI] [PubMed] [Google Scholar]
- Bakkeren G., Kronstad J.W. (1994). Linkage of mating-type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc. Natl. Acad. Sci. USA 91: 7085–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basse C.W., Kolb S., Kahmann R. (2002). A maize-specifically expressed gene cluster in Ustilago maydis. Mol. Microbiol. 43: 75–93 [DOI] [PubMed] [Google Scholar]
- Baumgarten A.M., Suresh J., May G., Phillips R.L. (2007). Mapping QTLs contributing to Ustilago maydis resistance in specific plant tissues of maize. Theor. Appl. Genet. 114: 1229–1238 [DOI] [PubMed] [Google Scholar]
- Baxter L., et al. (2010). Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330: 1549–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. (1999). Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J., Borodovsky M. (2005). GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33(Web Server issue): W451–W454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel J.P. (2011). Evolutionary dynamics of transposable elements in a small RNA world. Trends Genet. 27: 23–31 [DOI] [PubMed] [Google Scholar]
- Bölker M., Urban M., Kahmann R. (1992). The a mating type locus of U. maydis specifies cell signaling components. Cell 68: 441–450 [DOI] [PubMed] [Google Scholar]
- Cantu D., Govindarajulu M., Kozik A., Wang M., Chen X., Kojima K.K., Jurka J., Michelmore R.W., Dubcovsky J. (2011). Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS ONE 6: e24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H., Casas-Mollano J.A. (2006). On the origin and functions of RNA-mediated silencing: From protists to man. Curr. Genet. 50: 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.S., Zhang K., Nicolas E., Cam H.P., Zofall M., Grewal S.I. (2008). Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Clutterbuck A.J. (2011). Genomic evidence of repeat-induced point mutation (RIP) in filamentous ascomycetes. Fungal Genet. Biol. 48: 306–326 [DOI] [PubMed] [Google Scholar]
- Dean R.A., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- Doehlemann G., van der Linde K., Assmann D., Schwammbach D., Hof A., Mohanty A., Jackson D., Kahmann R. (2009). Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5: e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I.A., Fink G.R., Bartel D.P. (2011). Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333: 1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I.A., Weinberg D.E., Xie K.T., Mower J.P., Wolfe K.H., Fink G.R., Bartel D.P. (2009). RNAi in budding yeast. Science 326: 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza C.A., et al. (2011). Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. MBio. 2: e00342–e00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis S., et al. (2011). Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 108: 9166–9171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fischer T., Cui B., Dhakshnamoorthy J., Zhou M., Rubin C., Zofall M., Veenstra T.D., Grewal S.I.S. (2009). Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl. Acad. Sci. USA 106: 8998–9003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M., Williams R.L., Kothe G.O., Selker E.U. (2002). A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa. Proc. Natl. Acad. Sci. USA 99: 8802–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J.E., Selker E.U. (2004). RIP: The evolutionary cost of genome defense. Trends Genet. 20: 417–423 [DOI] [PubMed] [Google Scholar]
- Gaudet D.A., Wang Y., Penniket C., Lu Z.X., Bakkeren G., Laroche A. (2010). Morphological and molecular analyses of host and nonhost interactions involving barley and wheat and the covered smut pathogen Ustilago hordei. Mol. Plant Microbe Interact. 23: 1619–1634 [DOI] [PubMed] [Google Scholar]
- Grewal T.S., Rossnagel B.G., Bakkeren G., Scoles G.J. (2008). Identification of resistance genes to barley covered smut and mapping of the Ruh1 gene using Ustilago hordei strains with defined avirulence genes. Can. J. Plant Pathol. 30: 277–284 [Google Scholar]
- Haas B.J., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398 [DOI] [PubMed] [Google Scholar]
- Holloman W.K., Schirawski J., Holliday R. (2008). The homologous recombination system of Ustilago maydis. Fungal Genet. Biol. 45 (suppl. 1): S31–S39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M.E., Katawczik M., Giraud T. (2005). Repeat-induced point mutation and the population structure of transposable elements in Microbotryum violaceum. Genetics 170: 1081–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horns F., Petit E., Yockteng R., Hood M.E. (2012). Patterns of repeat-induced point mutation in transposable elements of basidiomycete fungi. Genome Biol. Evol. 4: 240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G.G., Linning R., Bakkeren G. (2002). Sporidial mating and infection process of the smut fungus, Ustilago hordei, in susceptible barley. Can. J. Bot. 80: 1103–1114 [Google Scholar]
- Huang X., Madan A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G., Maeng S., Yang D.-H., Ko Y.-J., Jung K.-W., Moyrand F., Floyd A., Heitman J., Bahn Y.-S. (2010). Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fungal Genet. Biol. 47: 1070–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101 [DOI] [PubMed] [Google Scholar]
- Kämper J., Reichmann M., Romeis T., Bölker M., Kahmann R. (1995). Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81: 73–83 [DOI] [PubMed] [Google Scholar]
- Kloc A., Martienssen R. (2008). RNAi, heterochromatin and the cell cycle. Trends Genet. 24: 511–517 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Lauc G., Ilić I., Heffer-Lauc M. (1992). Entropies of coding and noncoding sequences of DNA and proteins. Biophys. Chem. 42: 7–11 [DOI] [PubMed] [Google Scholar]
- Laurie J.D., Linning R., Bakkeren G. (2008). Hallmarks of RNA silencing are found in the smut fungus Ustilago hordei but not in its close relative Ustilago maydis. Curr. Genet. 53: 49–58 [DOI] [PubMed] [Google Scholar]
- Lee N., Bakkeren G., Wong K., Sherwood J.E., Kronstad J.W. (1999). The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl. Acad. Sci. USA 96: 15026–15031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linning R., Lin D., Lee N., Abdennadher M., Gaudet D., Thomas P., Mills D., Kronstad J.W., Bakkeren G. (2004). Marker-based cloning of the region containing the UhAvr1 avirulence gene from the basidiomycete barley pathogen Ustilago hordei. Genetics 166: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. (1997). tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubberstedt T., Klein D., Melchinger A.E. (1998). Comparative QTL mapping of resistance to Ustilago maydis across four populations of European flint-maize. Theor. Appl. Genet. 97: 1321–1330 [Google Scholar]
- Lubberstedt T., Xia X.C., Tan G., Liu X., Melchinger A.E. (1999). QTL mapping of resistance to Sporisorium reiliana in maize. Theor. Appl. Genet. 99: 593–598 [DOI] [PubMed] [Google Scholar]
- Mueller O., Kahmann R., Aguilar G., Trejo-Aguilar B., Wu A., de Vries R.P. (2008). The secretome of the maize pathogen Ustilago maydis. Fungal Genet. Biol. 45 (suppl. 1): S63–S70 [DOI] [PubMed] [Google Scholar]
- Nakayashiki H., Kadotani N., Mayama S. (2006). Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 63: 127–135 [DOI] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Ning Z., Keane T., Korf I. (2009). Assessing the gene space in draft genomes. Nucleic Acids Res. 37: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W.R., Wood T., Zhang Z., Miller W. (1997). Comparison of DNA sequences with protein sequences. Genomics 46: 24–36 [DOI] [PubMed] [Google Scholar]
- Raffaele S., et al. (2010). Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330: 1540–1543 [DOI] [PubMed] [Google Scholar]
- Rattei T., Tischler P., Götz S., Jehl M.A., Hoser J., Arnold R., Conesa A., Mewes H.W. (2010). SIMAP—A comprehensive database of pre-calculated protein sequence similarities, domains, annotations and clusters. Nucleic Acids Res. 38(Database issue): D223–D226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J., Heinze B., Wagenknecht M., Kahmann R. (2005). Mating type loci of Sporisorium reilianum: Novel pattern with three a and multiple b specificities. Eukaryot. Cell 4: 1317–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J., et al. (2010). Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330: 1546–1548 [DOI] [PubMed] [Google Scholar]
- Selker E.U., Stevens J.N. (1985). DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc. Natl. Acad. Sci. USA 82: 8114–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina S.A., Koonin E.V. (2008). Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. (Amst.) 23: 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Smith E., Shilatifard A. (2007). The A, B, Gs of silencing. Genes Dev. 21: 1141–1144 [DOI] [PubMed] [Google Scholar]
- Spanu P.D., et al. (2010). Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330: 1543–1546 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V., Lomsadze A., Chernoff Y.O., Borodovsky M. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18: 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hsueh Y.-P., Li W., Floyd A., Skalsky R., Heitman J. (2010). Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 24: 2566–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Schlagenhauf E., Graner A., Close T.J., Keller B., Stein N. (2006). 454 sequencing put to the test using the complex genome of barley. BMC Genomics 7: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M.R., Schnable J.C., Pedersen B.S., Lyons E., Lisch D., Subramaniam S., Freeling M. (2010). Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 8: e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton J.C., Federhen S. (1993). Statistics of local complexity in amino acid sequences and sequence databases. Comput. Chem. 17: 149–163 [Google Scholar]
- Xu J., et al. (2007). Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA 104: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., McDaniel I.E., Silva P., Zilberman D. (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zofall M., Grewal S.I. (2006). Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell 22: 681–692 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, V. Bryson and H.J. Vogel, eds (New York: Academic Press), pp. 97–166 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.