This research identifies a previously unknown clade of plant-specific proteins whose founding member is shown to facilitate meiotically heritable trans-homologue repression in maize. The findings highlight a diversification of epigenomic control mechanisms in higher plants.
Abstract
Meiotically heritable epigenetic changes in gene regulation known as paramutations are facilitated by poorly understood trans-homolog interactions. Mutations affecting paramutations in maize (Zea mays) identify components required for the accumulation of 24-nucleotide RNAs. Some of these components have Arabidopsis thaliana orthologs that are part of an RNA-directed DNA methylation (RdDM) pathway. It remains unclear if small RNAs actually mediate paramutations and whether the maize-specific molecules identified to date define a mechanism distinct from RdDM. Here, we identify a novel protein required for paramutation at the maize purple plant1 locus. This required to maintain repression2 (RMR2) protein represents the founding member of a plant-specific clade of predicted proteins. We show that RMR2 is required for transcriptional repression at the Pl1-Rhoades haplotype, for accumulation of 24-nucleotide RNA species, and for maintenance of a 5-methylcytosine pattern distinct from that maintained by RNA polymerase IV. Genetic tests indicate that RMR2 is not required for paramutation occurring at the red1 locus. These results distinguish the paramutation-type mechanisms operating at specific haplotypes. The RMR2 clade of proteins provides a new entry point for understanding the diversity of epigenomic control operating in higher plants.
INTRODUCTION
Paramutation describes both the process and the result of heritable epigenetic changes in gene regulation that are typically facilitated by trans-homolog interactions (Brink, 1958). Such events lead to apparent exceptions to the familiar modes of Mendelian inheritance (Brink, 1956) with profound implications for our understanding of population genetics and evolutionary biology (Jablonka and Raz, 2009). Although it remains unknown how paramutations occur and what common structural features mediate these events, such information promises novel approaches to germplasm improvement (Hollick and Springer, 2009).
Paramutation-like behaviors are found in diverse eukaryotes, including mouse (reviewed in Chandler and Stam, 2004). The best-known examples occur among specific haplotype variants of the maize (Zea mays) loci red1 (r1), booster1 (b1), and purple plant1 (pl1), which provide transcription factors needed for anthocyanin pigment production (Dooner et al., 1991; Chandler and Stam, 2004). Somatic, germinal, and trans-generational changes are easily detected and tracked with these haplotypes because they provide a visual readout of gene expression (Hollick, 2010; Erhard and Hollick, 2011). Genetic studies have begun to identify cis-linked and trans-acting components affecting paramutation; however, the specific mechanism remains unclear (Erhard and Hollick, 2011).
It has been proposed that small RNAs (sRNAs) acting in an RNA-directed DNA methylation (RdDM) pathway (Haag and Pikaard, 2011; Zhang and Zhu, 2011) mediate the trans-homolog interactions that typify paramutation behaviors (Chandler, 2010; Teixeira and Colot, 2010). This model is attractive because both RNA polymerase IV (Pol IV) catalytic subunits (RPD1 and RPD2a) and a putative RNA-dependent RNA polymerase (RDR2), presumably orthologous to the Arabidopsis thaliana RdDM-related proteins NUCLEAR RNA POLYMERASE D1 (NRPD1), NRPD2, and RNA-DEPENDENT RNA POLYMERASE2 (RDR2), respectively, have been identified by mutations affecting paramutation at pl1 (Dorweiler et al., 2000; Hollick et al., 2005; Alleman et al., 2006; Woodhouse et al., 2006; Erhard et al., 2009; Stonaker et al., 2009). However, paramutation events still occur at pl1 and b1 in the absence of the subset of 24-nucleotide sRNAs dependent on required to maintain repression1 (RMR1), a Rad54-like putative ATPase related to A. thaliana CLASSY1 and DEFECTIVE IN RNA-DIRECTED DNA METHYLATION1 (Hale et al., 2007). These findings cast doubt on a simple model in which sRNAs themselves mediate paramutations (Erhard and Hollick, 2011).
Despite the common requirement for RPD1 (Hollick et al., 2005) and RDR2 (Dorweiler et al., 2000), it also remains unclear if all of the known examples of paramutation in maize are mechanistically identical. Details of the paramutation behaviors at pl1, b1, and r1 reveal both similarities and differences, and the interacting haplotypes appear to use distinct cis-linked sequences at each locus (Hollick, 2010).
Paramutation at pl1 occurs specifically among Pl1-Rhoades (Pl1-Rh) haplotypes (Hollick et al., 1995; Gross and Hollick, 2007) responsible for seedling, plant, and anther pigmentation. Pl1-Rh can persist in a continuum of regulatory states described by a 1 to 7 graded series of anther color scores (ACSs). States conferring ACS 1 to 4 phenotypes (variegated) are denoted Pl′ and represent transcriptionally and posttranscriptionally repressed derivatives of the so-called Pl-Rh state that confers fully pigmented (ACS 7) phenotypes (Hollick et al., 2000; Hale et al., 2007).
When Pl1-Rh haplotypes of contrasting Pl-Rh and Pl′ states are combined by sexual crosses, the pigment phenotype of the sporophytic tissues (Pl-Rh/Pl′) resembles that of Pl′/Pl′ genotypes, and only Pl1-Rh haplotypes having a Pl′ identity are transmitted to progeny plants (Hollick et al., 1995). This behavior is pl1 locus dependent, parent-of-origin independent, and not the result of chromosomal transmission ratio distortions (Hollick et al., 1995, 2000, 2005). Therefore, the Pl-Rh state invariably changes to a Pl′ state at some point, either during somatic development of the Pl-Rh/Pl′ sporophyte, at meiosis, or during subsequent gametophyte development. The cis-linked sequences mediating this behavior have yet to be described, but they are clearly distinct from those required at b1, which are unique in the genome (Stam et al., 2002a).
Paramutation at b1 occurs specifically among B1-Intense (B1-I) haplotypes (Coe, 1966) responsible for plant pigmentation. B1-I can persist in a fully active state denoted B-I and a transcriptionally repressed derivative state denoted B′ (Patterson et al., 1993). Similar to Pl1-Rh, only B1-I haplotypes of B′ state are transmitted from B-I/B′ sporophytes (Coe, 1966). Paramutation behavior at b1 is dependent on the repeated nature of seven tandem 853-bp units found ∼100 kb 5′ of the B1-I coding region that act as a transcriptional enhancer (Stam et al., 2002a, 2002b). Because sRNAs representing these repeated sequences are found in both B-I/B-I and B′/B′ plants as well as other plants that only contain a single unit of this 853-bp sequence (Arteaga-Vazquez et al., 2010), these sRNAs by themselves appear insufficient to mediate b1 paramutation.
Paramutation at r1 occurs among specific components of R-r:standard (R-r) haplotypes responsible for kernel pigmentation. R-r is multigenic (Eggleston et al., 1995; Walker et al., 1995) with an inverted repeat of seed color genes and a separate plant color gene. Similar to both Pl1-Rh and B1-I, R-r haplotypes in a relatively repressed state, denoted R-r′, are transmitted from R-r/R-r′ plants (Brown and Brink, 1960). Structurally distinct r1 haplotypes, R-stippled (R-st) and R-marbled (R-mb), composed of four and three tandem R1-coding genes, respectively, are able to elicit the same behavior at R-r: Only R-r haplotypes having R-r′ states are transmitted from R-st/R-r and R-mb/R-r plants (Brink, 1956; Brink and Weyers, 1957). The cis-linked sequences mediating these events are the duplicated r1 genes (and potentially the intergenic sequences) themselves (Kermicle et al., 1995; Panavas et al., 1999) and a small promoter region of R-r responsible for kernel expression (Kermicle, 1996; Walker, 1998).
The so-called paramutant state of B1-I (B′) is extremely stable and has not been reported to change back to a B-I-like state under any circumstance (Coe, 1966). By contrast, both R-r′ and Pl′ states can revert, at a relatively high frequency, to fully active R-r and Pl-Rh states after being hemizygous or after being heterozygous with other haplotypes that do not show paramutation properties (Styles and Brink, 1969; Hollick and Chandler, 1998; Gross and Hollick, 2007).
Pl1-Rh can also be transmitted in a Pl-Rh state, at various frequencies, from Pl′/Pl′ plants deficient for RPD1 (Hollick et al., 2005), RDR2 (Dorweiler et al., 2000), or RMR1 (Hollick and Chandler, 2001). Although all these mutants have pigment phenotypes indistinguishable from plants with a Pl-Rh/Pl-Rh genotype, not all the Pl1-Rh haplotypes adopt a meiotically heritable Pl-Rh state. In the absence of RPD2a, one of three second largest catalytic subunits of Pol IV found in maize, the pigment phenotype of all Pl′/Pl′ plants appears identical to that of Pl-Rh/Pl-Rh plants, but only Pl′ states are sexually transmitted (Stonaker et al., 2009). These findings are consistent with the proposal that the repression seen in both Pl-Rh/Pl′ and Pl′/Pl′ sporophytes is mechanistically distinct, though not necessarily unrelated, from that responsible for maintaining meiotically heritable Pl′ states (Erhard and Hollick, 2011).
Forward genetic screens using ethyl methanesulfonate (EMS) as a mutagen identify at least 12 independent loci whose functions are required to maintain repression of the Pl′ state (Dorweiler et al., 2000; Hollick and Chandler, 2001; Hollick et al., 2005; Stonaker et al., 2009; J.B. Hollick, unpublished data). The four loci described to date encode proteins with likely A. thaliana orthologs associated with Pol IV complexes containing NRPD1, NRPD2, RDR2, and RMR1-related proteins encoded by AT3G24340 and AT1G05490 (Law et al., 2011). All four maize proteins are required for accumulation of 24-nucleotide RNA species (Nobuta et al., 2008; Erhard et al., 2009; Hale et al., 2009; Stonaker et al., 2009), yet only RPD1 is clearly required to mediate acquisition of a meiotically heritable Pl′ state in Pl-Rh/Pl′ plants (Hollick et al., 2005).
In maize and other grasses, diverse Pol IV isoforms defined by distinct second largest subunits appear to target different genomic targets (Sidorenko et al., 2009; Stonaker et al., 2009). Maize mutants deficient for RPD1 have unique developmental abnormalities likely caused by expanded expression domains of specific developmental regulators (Parkinson et al., 2007; Erhard et al., 2009). It has been proposed that, in large genomes such as that found in maize, Pol IV is responsible for regulating specific alleles or haplotypes through RdDM-independent interactions with neighboring transposons (Hale et al., 2009). This idea is supported by observations in which polyadenylated RNAs from certain retrotransposons accumulate in the absence of RPD1 but are not found in the absence of RPD2a, RDR2, or RMR1 (Hale et al., 2009; Stonaker et al., 2009). Presumably, a Pol IV complex involving one of the other two potential RPD2 proteins is responsible for this retrotransposon inhibition.
To date, all the components required to maintain the Pl′ state, either somatically or germinally, identify Pol IV–related proteins. Loss-of-function rpd1, rdr2, and rmr1 mutants have coincident hypomethylation of certain cytosine residues found within a CACTA-type transposon fragment related to doppia found immediately 5′ of the Pl1-Rh coding region (Hale et al., 2007). This finding is consistent with the doppia region being targeted by an RdDM-type mechanism. However, other pl1 alleles that do not show paramutation-type behaviors, such as Pl1-Blotched, have an identical doppia fragment (Gross and Hollick, 2007) and the Pl′ and Pl-Rh states are not distinguished by any such differences in doppia 5-methylcytosine (5meC) patterns (Hale et al., 2007). The doppia sequences may be necessary for paramutation behaviors, but they are clearly insufficient and their 5meC status appears to be immaterial. It thus remains unclear if an RdDM-type effect of Pol IV plays a role in the paramutation mechanisms or whether Pol IV acts more directly through as-yet-uncharacterized functions.
Here, we identify a pioneer protein encoded by the rmr2 locus; this protein has not been associated with Pol IV or an RdDM-type mechanism in any species. Rmr2 is required for somatic maintenance of Pl′ (Hollick and Chandler, 2001) and for at least two examples of transgene silencing (McGinnis et al., 2006). RMR2 represents the founding member of a small clade of plant-specific proteins whose molecular function is not obvious. Phylogenetic analyses indicate that the RMR2 subclade is grass specific. Genetic test results indicate that RMR2 is required for the establishment of paramutation at pl1 but not at r1. Molecular assays show that RMR2 is required for the accumulation of 24-nucleotide RNAs from both repetitive and unique genomic regions. However, these RMR2-dependent sRNAs are not absolutely required to promote paramutation at either pl1 or r1. Curiously, specific 5meC patterns affected by RMR2 are distinct from those affected by RPD1. These results indicate that RMR2 plays a role in the establishment of paramutation specifically at pl1 and that it has both Pol IV–overlapping functions (sRNAs) and functions distinct from Pol IV (specific 5meC patterns). RMR2 represents a novel component of the increasingly diverse set of nuclear systems available to generate and maintain heritable epigenetic variation in higher plants.
RESULTS
Rmr2 Maintains Transcriptional Repression of Paramutant Pl1-Rh
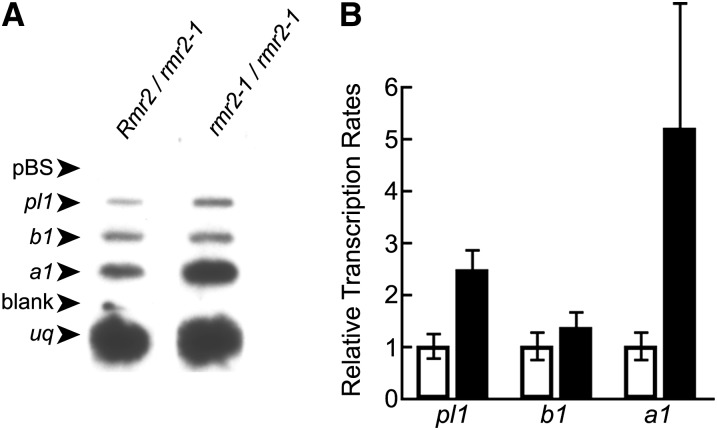
RNA expression from the Pl′ state is repressed at both transcriptional and posttranscriptional levels relative to Pl-Rh (Hollick et al., 2000, 2005; Hale et al., 2007). RNase protection experiments showed that pl1 RNA levels are increased ∼14-fold in the anthers of homozygous rmr2-1 mutants (Hollick and Chandler, 2001), but it remained unknown if this difference was related to changes in transcription rates, RNA stability, or both.
We used in vitro transcription reactions with nuclei isolated from husk leaves to determine whether pl1 transcription rates were increased in rmr2-1 mutants relative to heterozygous siblings. Source plants contained B1-I (B-I) so that activation of the anthocyanin pathway could be monitored with both visual and molecular assays. Radiolabeled nascent RNAs produced from isolated nuclei were hybridized to plasmid DNA samples of cDNAs from the pl1, b1, anthocyaninless1 (a1), and ubiquitin2 (uq) genes (Figure 1A). Normalized to uq, the transcription rate of pl1 was increased ∼2.4-fold in rmr2-1 mutants relative to heterozygous siblings (Figure 1B). This increase is similar to the 2.8-fold difference measured between Pl′ and Pl-Rh states (Hollick et al., 1995).
Figure 1.
In Vitro Transcription Analysis of Rmr2 Function.
(A) Slot blots of cDNA clones from the pl1, b1, a1, and uq genes hybridized with radiolabeled nascent RNAs derived from husk nuclei isolated from mutant and nonmutant siblings. pBS is a plasmid control.
(B) Quantification of mean relative transcription rates of the indicated genes normalized to uq (±se). Measurements for rmr2-1/rmr2-1 genotypes (closed bars, n = 5) are displayed relative to data of Rmr2/rmr2-1 genotypes (open bars, n = 5) set at unit value.
Transcription rates at b1 remained unchanged (Figure 1A), indicating the rmr2-1 mutation has no effect on a nonparamutant state like B-I. However, the relative transcription rate of a1 was increased ∼5.2-fold consistent with the increased plant pigment seen in rmr2-1 mutants. This result comports with the expected increase in PL1 transcription factors, the paralog of which (C1, for colored aleurone1) is known to bind to the a1 promoter and activate a1 transcription (Sainz et al., 1997).
The in vitro transcription data show that Rmr2 function is required to maintain transcriptional repression of the Pl′ state. However, the data do not exclude the possibility that posttranscriptional control of pl1 RNA is also affected in rmr2-1 mutants. Genetic deficiencies for the largest subunit of Pol IV (RPD1) also increase the transcription rate of Pl′ (approximately fourfold) (Hollick et al., 2005), but it remains unknown if posttranscriptional control is similarly affected.
Rmr2 Maintains Somatic Repression of Paramutant Pl1-Rh
Previous genetic tests indicated that Pl′ could revert to a heritable Pl-Rh state in both rmr1-1 and rmr2-1 homozygotes albeit at low (∼0.1) frequencies (Hollick and Chandler, 2001). We reasoned that multigenerational deficiencies of either function would enhance the frequency of heritable reversions. To test this prediction, single recombinant inbred lines (RILs) of single seed descent were generated for both rmr1-1 (Hale et al., 2009) and rmr2-1 mutations and then representative plants were crossed to a common tester line to monitor transmission of a Pl′-like state. Anther color remained dark in all RILs, indicating that the Pl1-Rh haplotype was being maintained in a Pl-Rh-like state. For the rmr1-1 lineage, all four S9 plants sampled exclusively transmitted nonparamutagenic Pl1-Rh haplotypes (Table 1) consistent with full reversions of Pl′ to Pl-Rh states occurring during inbreeding. By contrast, only Pl′-like states were transmitted from rmr2-1 plants (n = 3) at the S7 generation (Table 1). This result indicates that Rmr2 is required to maintain somatic repression of Pl′ states but, similar to RPD2a (Stonaker et al., 2009), is not required to maintain the meiotically heritable features that facilitate paramutation. This result stands in contrast with the previously reported reversion rates (2 and 9%) seen among rmr2-1 homozygotes (Hollick and Chandler, 2001) and thus indicates a trans-generational effect of Pl1-Rh behaviors.
Table 1. Paramutation Tests following Inbreeding.
|
rmr/rmr; Pl′/Pl′ × Rmr/Rmr; Pl-Rh/Pl-Rh | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| rmr Allele | Inbred Generation | No. of Test Crosses | No. of Progeny Individuals with Specific ACS Phenotypes |
||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| rmr1-1 | 9 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 61 |
| rmr2-1 | 7 | 3 | 7 | 27 | 0 | 0 | 0 | 0 | 0 |
Rmr2 Is Partially Required to Establish Paramutation at pl1
When haplotypes of both Pl′ and Pl-Rh form are combined through sexual crosses, the resulting sporophyte exclusively transmits Pl′-like haplotypes (Hollick et al., 1995, 2000); acquisition of a paramutant Pl′ state is efficiently facilitated and then maintained through meiosis. In specific mutants, however, one or both of these two requirements for establishment may be impaired (Dorweiler et al., 2000; Hollick et al., 2005). By independently tracking the segregation and inheritance of parental haplotypes, one can potentially distinguish a mutant’s failure to maintain repression from its failure to facilitate paramutation. Using such genetic assays, it was evident that RMR1 is not required to promote paramutation at pl1 or b1 (Hale et al., 2007).
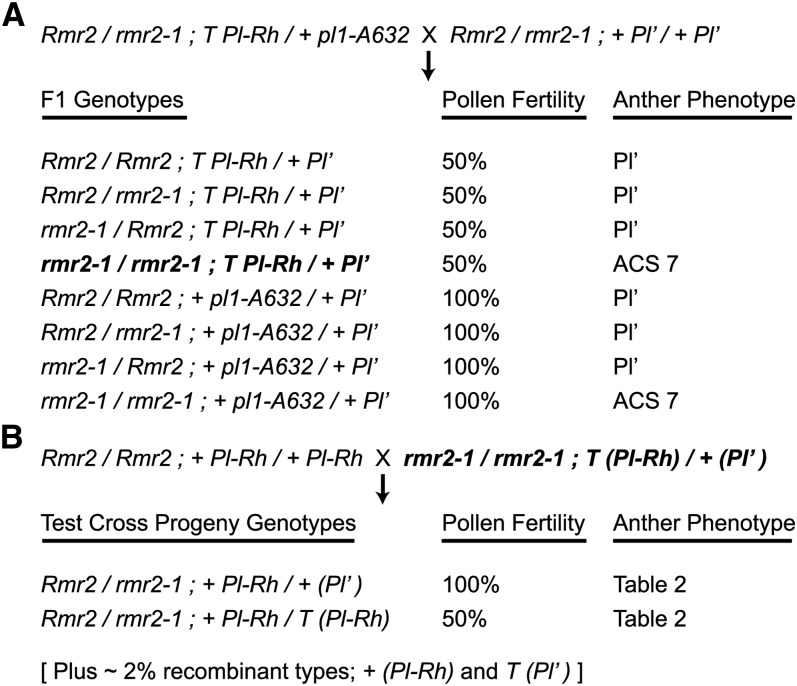
We asked whether Rmr2 was required to establish a Pl′ state by combining Pl1-Rh haplotypes of both Pl′ and Pl-Rh states in rmr2-1 mutants and in Rmr2/rmr2-1 siblings and then assaying the potential of the segregating Pl1-Rh haplotypes to facilitate paramutation in test cross progenies (Figure 2). The parental haplotype of the Pl-Rh form was tightly linked (∼2 centimorgans [cM]) to a T6-9 (043-1) interchange breakpoint (T), and the haplotype of the Pl′ type was resident on a structurally normal chromosome (+). Plants heterozygous for this interchange chromosome (T/+) are semisterile and are easily identified by examining fresh pollen samples with a pocket microscope (Hollick et al., 2005). Five semisterile plants (+ Pl′/T Pl-Rh) having fully colored anthers (rmr2-1/rmr2-1) and five semisterile plants having Pl′-like anthers (Rmr2/rmr2-1 or Rmr2/Rmr2) were crossed to + Pl-Rh/+ Pl-Rh testers, and the progeny were scored for both pollen fertility and anther colors (Table 2; see Supplemental Table 1 online). All 80 test cross progeny individuals derived from nonmutant plants had clear Pl′-like anther phenotypes (see Supplemental Table 1 online), indicating that paramutation at pl1 was efficiently established and maintained in nonmutant plants.
Figure 2.
Crossing Scheme Used to Test Paramutation in the Absence of Rmr2 Function.
(A) Parental genotypes and F1 characters of plants used to combine Pl′ and Pl-Rh states in rmr2-1 homozygotes. “T” refers to the T6-9 (043-1) interchange breakpoint, and “+” refers to a normal chromosome 6. Anther phenotypes are either nonmutant (Pl′) or the ACS 7 displayed by rmr2-1 mutants. Plants having the F1 genotype in bold were subjected to the test cross shown in (B).
(B) Parental genotypes and test cross progeny characters used to evaluate whether or not Pl1-Rh haplotypes are transmitted from rmr2-1 mutants with the ability to facilitate paramutation (paramutagenicity). Progeny anther phenotypes are listed in Table 2.
Table 2. Paramutation Occurring in rmr2-1/rmr2-1; + (Pl′)/T Pl-Rh Plantsa.
| Test Cross Parent | Progeny Structural Genotype | No. of Progeny Individuals with Specific ACSs |
||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| 03-1194-11 | +/T | 0 | 2 | 0 | 2 | 0 | 2* | 0 |
| 03-1196-10 | +/T | 0 | 0 | 0 | 4 | 1 | 0 | 1* |
| 03-1196-15 | +/T | 0 | 2 | 0 | 2 | 0 | 1* | 3 |
| 03-1196-17 | +/T | 0 | 0 | 0 | 2 | 0 | 2* | 5 |
| 03-1196-18 | +/T | 0 | 1 | 0 | 2 | 0 | 0 | 1* |
| 03-1194-11 | +/+ | 0 | 7 | 0 | 2 | 0 | 0 | 0 |
| 03-1196-10 | +/+ | 1 | 8 | 0 | 1 | 0 | 0 | 0 |
| 03-1196-15 | +/+ | 6 | 3 | 0 | 1 | 0 | 0 | 1* |
| 03-1196-17 | +/+ | 0 | 5 | 5 | 0 | 0 | 1* | 1 |
| 03-1196-18 | +/+ | 0 | 8 | 0 | 1 | 0 | 0 | 0 |
| Totals | +/T | 0 | 5 | 0 | 12 | 1 | 5* | 8 (2*) |
| Totals | +/+ | 7 | 31 | 5 | 5 | 0 | 1* | 1 (1*) |
If paramutation absolutely requires Rmr2 function, then the Pl1-Rh resident on the translocation chromosome (T Pl1-Rh) should remain in an unaltered Pl-Rh state despite being exposed to a normal chromosome carrying a Pl1-Rh haplotype of Pl′ state in the F1 (Figure 2A, bold genotype). This scenario would be reflected in the test cross progeny if all the semisterile individuals inheriting the translocation chromosome had Pl-Rh–like anthers (Table 2, +/T rows). Reciprocally, all fully fertile test cross progeny individuals inheriting the normal chromosome would have Pl′-like anthers (Table 2, +/+ rows). However, if paramutation does not require Rmr2 function, then both F1 Pl1-Rh haplotypes will be of Pl′ state and result in both semisterile (Table 2, +/T rows) and fully fertile (Table 2, +/+ rows) test cross progeny individuals having Pl′-like anthers.
We found that paramutation at pl1 was partially impaired in rmr2-1 mutants. The semisterile (+/T) test cross progeny plants had anther colors representing both Pl′ and Pl-Rh–like types (Table 2). Half (17/33) of these plants had clear Pl′-like phenotypes, indicating that many of the parental haplotypes of Pl-Rh form had acquired and maintained a Pl′ identity. Even in the absence of Rmr2 function, paramutation had occurred. However, nearly one-third of the semisterile progeny plants had fully colored anthers, indicating that the parental haplotype of Pl-Rh form could also remain unchanged in rmr2-1/rmr2-1, + Pl′/T Pl-Rh plants; paramutation was impaired. Six other progeny plants had ACSs intermediate of typical Pl′ and Pl-Rh–like phenotypes, further indicating that paramutation was impaired in rmr2-1 mutants. By contrast, of all the 51 fully fertile (+/+) progeny plants, only two had fully colored anthers, indicating that a Pl-Rh–like state had been transmitted on a normal chromosome from rmr2-1/rmr2-1; + Pl′/T Pl-Rh plants (Table 2). It was not possible to distinguish whether these two examples represented recombinants between the parental haplotype of the Pl-Rh form and the structurally normal chromosome or if they represented a low level of reversion of Pl′ to Pl-Rh occurring in the rmr2-1 mutants. Because reversions of Pl′ to Pl-Rh in the absence of Rmr2 are infrequent (Hollick and Chandler, 2001), these genetic segregation data indicate that establishment of paramutation at the pl1 locus is partially dependent on Rmr2 function.
Rmr2 Is Not Required to Establish Paramutation at r1
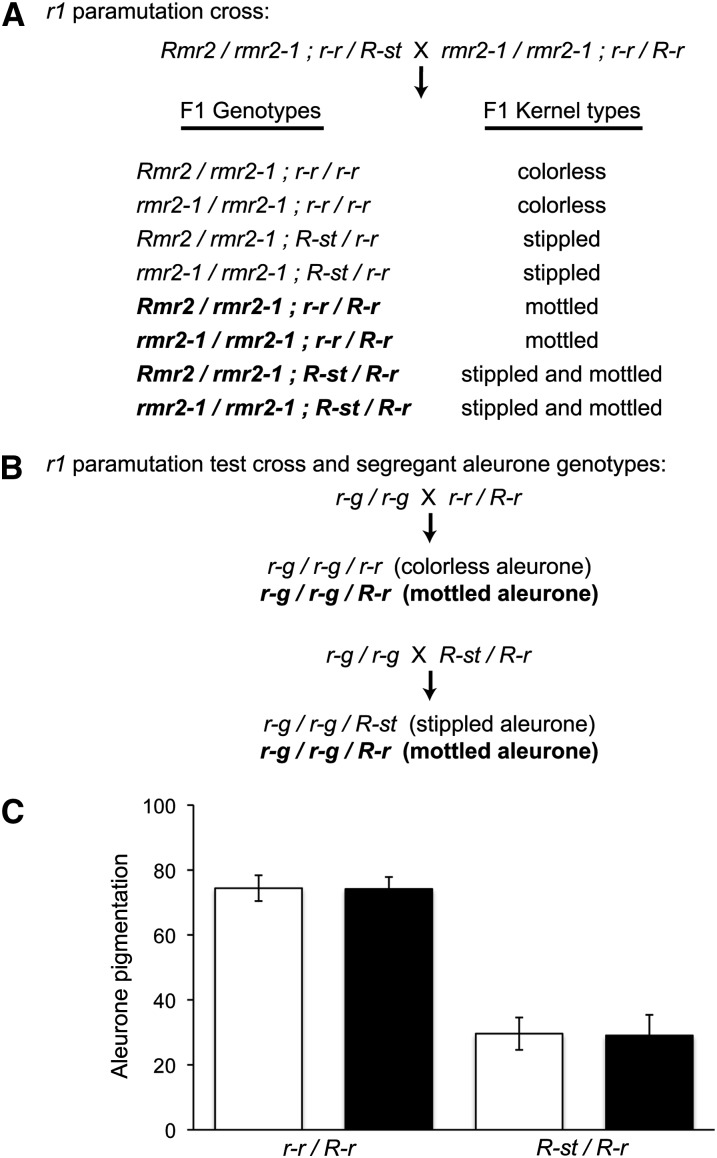
Pigment levels conferred by R-r are evaluated in kernels resulting from crosses to recessive r-g/r-g testers. Previous genetic tests showed that RPD1 is required for R-r to establish a heritable R-r′ state in R-r/R-st sporophytes (Hollick et al., 2005). Using a nearly identical scheme, we found that Rmr2 function was unnecessary for the establishment of R-r′ states (Figure 3). Guided by unique anther and kernel color patterns, we synthesized and identified both mutant (rmr2-1/rmr2-1) and nonmutant (Rmr2/rmr2-1) siblings in which R-r was heterozygous with either R-st or r-r (Figure 3A). These plants were crossed to r-g/r-g testers, and kernels were sorted by diagnostic pigment patterns (Figure 3B). Pigment levels were measured in groups of kernels that inherited the R-r haplotype and compared between the contrasting rmr2 genotypes (Figure 3C). The data showed that R-r action was near identical following transmission from the rmr2-1 mutant and nonmutant genotypes regardless of whether R-r was heterozygous with r-r or R-st. This result indicates that Rmr2 is not required to establish paramutation occurring at r1.
Figure 3.
r1 Paramutation Analysis.
(A) Parental genotypes and F1 characters of plants used to combine specific r1 haplotypes in rmr2-1 homozygotes. Plants having the F1 genotypes in bold were subjected to the test crosses shown in (B). Plants from three distinct F1 progeny sets were evaluated.
(B) Parental genotypes and test cross aleurone types used to evaluate whether or not R-r haplotypes are transmitted from rmr2-1 mutants with the ability to facilitate paramutation (paramutagenicity).
(C) Histogram of mean pigment levels for kernels having mottled aleurones quantified using a reflectometer (±sd). Data represent materials generated from both Rmr2/rmr2-1 (open bars) and rmr2-1/rmr2-1 (closed bars) genotypes (n = 21, 18, 14, and 15 individual test crosses for the respective genotypes).
A similar genetic test was initiated to ask if Rmr2 was required for paramutation occurring at b1. This test was complicated by the relatively tight linkage between the two loci (see below) and by inherent instability of the B-I state. We were unable to maintain a B1-I rmr2-1 haplotype in B-I form in many lineages; therefore, any potential requirement for Rmr2 function could not be reliably determined.
rmr2 Maps to Chromosome 2S
Given the unique genetic role of rmr2 in the paramutation process, we sought to identify the molecular nature of this locus by first mapping its genomic location. Genetic segregation frequencies between plant color (controlled by the B1-I haplotype) and anther color (determined by Rmr2 function) showed that rmr2 was tightly linked to b1 on chromosome 2S. When colored plants (B1-I/b1) with variegated anthers (Rmr2/rmr2-1) were crossed to or by plants having no plant color (b1/b1) but darkly pigmented anthers (rmr2-1/rmr2-1), individuals from the resulting progeny sets (n = 7) displayed the expected segregation ratios for both plant (178 colored:170 colorless) and anther colors (159 ACS 5-7:189 ACS 1-4). However, the frequency of individuals having both plant and anther color (23 of 348; 0.066) deviated significantly (χ2 = 51; P < < 0.01) from the expectation of Mendelian segregation for unlinked factors (0.25). These data placed rmr2 ∼6.6 cM from b1 on chromosome 2S.
rmr2 was mapped centromere-proximal to b1 using specific simple sequence length polymorphism (SSLP) markers. To construct an appropriate mapping population, individuals from an rmr2-1 RIL (F2S7) were crossed to a color-converted A632 inbred and the resulting F1 progeny plants were self-pollinated. Genomic DNA was isolated from F2 individuals having darkly colored anthers (rmr2-1/rmr2-1) and assayed for parental SSLPs found on 2S. Strong cosegregation was seen with the ordered umc1845, umc1185, and bnlg1064 loci, which are all centromere-proximal to b1. A632 polymorphisms at these loci were found at frequencies indicating tight genetic linkage (umc1845, 20 of 167 samples, ∼6 cM; umc1185, 3 of 59 samples, ∼2.5 cM; bnlg1064, 17 of 172 samples, ∼4.9 cM). In the three samples having an A632 SSLP at umc1185, there was also an A632 SSLP at bnlg1064. These data placed rmr2 between umc1845 and umc1185, proximal to b1.
We searched for candidate genes potentially related to chromatin biology or the RdDM pathway within the umc1845 to umc1185 interval and identified a single gene model encoding a SET domain protein with possible histone methyltransferase function (SDG104). We sequenced this candidate gene from an rmr2-1 mutant (data not shown) and did not identify any lesions that would result in obvious protein dysfunction. EST evidence and associated gene models representing 46 transcribed features are found in this interval, yet BLAST searches did not identify any other candidates that might encode proteins known to be involved in epigenetic modifications. We thus set about generating and isolating transposon-induced rmr2 alleles with which to functionally identify the locus and provide a molecular tag for cloning.
Transposon Tagging and High-Throughput Sequencing Identify an rmr2 Candidate Gene
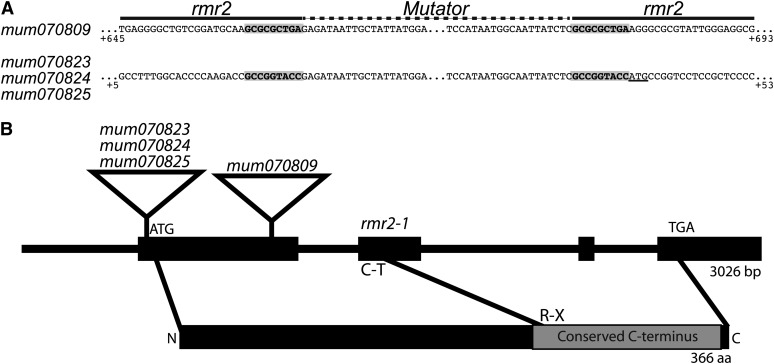
A field-based screen for Mutator (Mu) transposon-induced rmr2 alleles (rmr2-mum) identified four potential isolates from ∼12,000 M1 plants. Pollen bulked from plants carrying an active Mu system (see Methods) was applied to the silks of plants heterozygous for the rmr2-1 reference allele, and the resulting M1 progeny were grown to maturity. Four M1 progeny having Pl-Rh–like anthers were crossed to both color-converted A619 and A632 lines to isolate the putative rmr2-mum alleles from rmr2-1. Because rmr2-1 was linked to a white tip1 (wt1) locus mutation (∼4.5 cM centromere proximal; see Methods), all F2 progenies inheriting rmr2-1 segregated seedlings having a chlorotic white tip on their first leaves. Reciprocally, all F2 progenies inheriting an rmr2-mum allele would not have seedlings with white-tipped leaves. F2 plants not carrying the wt1 mutation, yet having Pl-Rh-like anthers, were backcrossed to the respective A619 and A632 lines, and the rmr2-mum alleles were recovered in homozygous condition in BC1F2 families. One of these families had plants displaying mutable dark-color phenotypes characteristic of transposon-derived alleles. The four new alleles were designated rmr2-mum070809, rmr2-mum070823, rmr2-mum070824, and rmr2-mum070825.
We used a recently developed method of Illumina-based sequencing of Mu insertion sites (Williams-Carrier et al., 2010) to identify possible rmr2 gene models. Six genomic DNA samples were assayed in total: both A619 and A632 BC1F2 individuals homozygous for the rmr2-mum070809, 070823, and 070825 alleles. This analysis identified only a single gene model (GRMZM2G009208) that shared Mu insertions among the set of sampled genomes. This model resides at a position on chromosome 2S (∼22 Mb) consistent with the mapped position of rmr2 and thus represented the likely candidate.
Further molecular assays supported this candidate gene as the identity of the rmr2 locus. We designed reverse primers to the GRMZM2G009208 gene model and used these in combinations with a degenerate Mu-end primer to validate the insertion sites identified by the Illumina data and to identify another Mu insertion representing the rmr2-mum070824 allele (Figure 4A). All these Mu insertions were coincident with 9-bp target-site duplications, a hallmark of Mu element integrations (Figure 4A). We then sequenced this candidate gene from DNA isolated from plants homozygous for the EMS-induced rmr2-1 reference allele and identified a C-to-T transition mutation resulting in a nonsense codon in the second exon. This lesion is predicted to eliminate translation of a highly conserved C-terminal portion of the predicted protein. These data strongly indicate that the rmr2 locus corresponds to a relatively small protein-coding gene identified in the maize gene build assembly 5b.60 as GRMZM2G009208, hereafter referred to as rmr2.
Figure 4.
Mu Insertion Site Sequences and rmr2 Gene Model.
(A) Mu insertion sites are flanked by direct sequence duplications (gray boxes). Positions of the rmr2 sequence relative to the predicted transcription start site are indicated below the sequences. The start codon is underlined. Mu insertion allele abbreviations removed the rmr2- prefix from each allele name.
(B) Schematic of gene (above) and protein (below) models highlighting the four exons and conserved CTR used for phylogenetic analysis (Figure 5). Lines connecting the models mark the start, stop, and rmr2-1 nonsense codons. Relative positions of the Mu and EMS lesions are indicated. aa, amino acids.
rmr2 Encodes a Novel Pioneer Protein
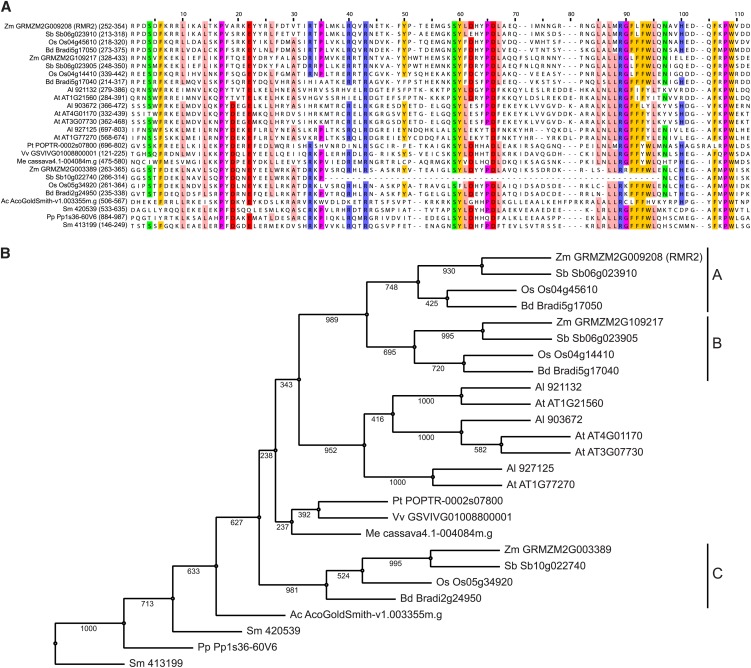
Bioinformatic tools and available nucleotide sequences were used to better understand the potential molecular function of RMR2. EST and full-length cDNA sequences support a 366 amino acid–encoding rmr2 gene model (Figure 4B) (http://maizesequence.org). TBLASTN searches of the predicted RMR2 protein identified two putative paralogs (GRMZM2G109217 and GRMZM2G003389) and hypothetical or predicted proteins in other multicellular plants, all lacking confirmed functional annotations. Multiple sequence alignments of these potentially related proteins (Figure 5A; see Supplemental Data Set 1 online) highlighted a conserved C-terminal region (CTR) corresponding to approximately one-third (amino acids 252 to 354) of the predicted RMR2 protein. We used this conserved region in a position-specific iterated BLAST search (Altschul et al., 1997) to identify more distantly related proteins, but this analysis returned only hypothetical and similarly unconfirmed annotations exceeding an expected = 0.01 threshold.
Figure 5.
RMR2 Conserved Sequence Alignments and Phylogeny.
(A) Alignment of the RMR2 conserved CTR in selected multicellular plants. Each gene name is followed by the amino acid residue positions in parentheses. Residues are colored if they have >60% identity across the sampled proteins.
(B) Maximum likelihood relationships based on alignments shown in (A) for representative grasses: maize (Zm), sorghum (Sorghum bicolor; Sb), rice (Oryza sativa; Os), and Brachypodium distachyon (Bd); eudicots: Aquilegia coerulea (Ac), Vitis vinifera (Vv), Manihot esculenta (Me), Populus trichocarpa (Pt), A. lyrata (Al), and A. thaliana (At); and outgroup: P. patens (Pp) and S. moellendorffii (Sm). Branch lengths correspond to the indicated bootstrap values (n = 1000). Three clades of grass proteins are labeled A, B, and C.
In silico translation predicts that rmr2 encodes a 40.5-kD, negatively charged protein (theoretical pI = 4.70) (http://web.expasy.org/protparam; Gasteiger et al., 2005). The Plant-mLoc protein sorting algorithm (http://www.csbio.sjtu.edu.cn/cgi-bin/PlantmPLoc.cgi; Chou and Shen, 2010) predicts a nuclear localization for RMR2, but comparisons to existing nuclear localization signals (https://www.predictprotein.org; Rost et al., 2004) did not identify any related nuclear localization signal. PONDR-FIT (http://www.disprot.org/pondr-fit.php; Xue et al., 2010) predicted two regions with high probability of being disordered in the N terminus and the middle of the polypeptide. Pfam searches did not find any conserved motifs in the presumed RMR2 primary structure matching their databases (http://www.sanger.ac.uk/resources/databases/pfam.html; Finn et al., 2010). However, sequence-structure comparison software (FUGUE; http://tardis.nibio.go.jp/fugue; Shi et al., 2001) identified structural similarities of the RMR2 conserved region to F93 (FUGUE z-score = 3.87), a predicted winged helix-turn-helix (HTH) DNA binding protein from hyperthermophilic sulfolobus turreted icosahedral virus (Larson et al., 2007) and to human saposin b (FUGUE z-score = 3.80). The protein homology/analogy recognition engine (PHYRE2; http://www.sbg.bio.ic.ac.uk/~phyre2; Kelley and Sternberg, 2009) predicted structural similarity between residues 255 and 305 (the first half of the conserved CTR) and most of the HTH DNA binding domain (amino acids 3 to 60 of the 80 residue domain) of the Mycobacterium tuberculosis EspR transcription factor (Rosenberg et al., 2011).
Representative alignments of proteins sharing the conserved CTR of RMR2 (Figure 5A; see Supplemental Data Set 1 online) defined a unique arrangement of amino acid residues conserved from mosses through eudicots. Residues corresponding to RMR2 amino acids 308 to 315 follow the conserved pattern of Ser-Tyr-Leu(or Phe)-acidic(Glu or Asp)-X-X-Pro-Asp-Leu(or Phe) and a later motif (329 to 339) has a four-residue Leu-rich region followed by a nearly ubiquitous Arg-Gly, a four-residue aromatic-rich region and a final Leu (or other hydrophobic residue). Finally, residues corresponding to 350 and 351 are always Pro-Trp usually following Phe-X (348 and 349). In addition to these RMR2-family motifs, there are also nearly invariant residues (Phe-257, Leu-261, Leu-265, Pro-268, Glu-273, and Arg-294). Phylogenetic surveys of existing genome sequences indicated that all grasses have three distinct proteins containing this conserved region. These proteins separate into three clades (Figure 5B) distinct from the clade containing the multiple A. thaliana representatives. Despite the A. thaliana proteins sharing the conserved C terminus, none appear to be orthologous to the proteins representing grass clade A that contains RMR2. The grass clades also correspond with physical locations of each gene in the maize genome: Clade A and B genes are present on chromosome 2 (also in tandem for two of the three other grass genomes analyzed), while the clade C gene resides on chromosome 9. RT-PCR amplification of cDNAs indicated that all three maize A, B, and C clade representatives are expressed in immature ear, immature tassel, and shoot apical meristems. However, only rmr2 mRNA appeared to be present in embryo and endosperm tissues (see Supplemental Figure 1 online).
RMR2 Is Required for Accumulation of 24-Nucleotide RNAs
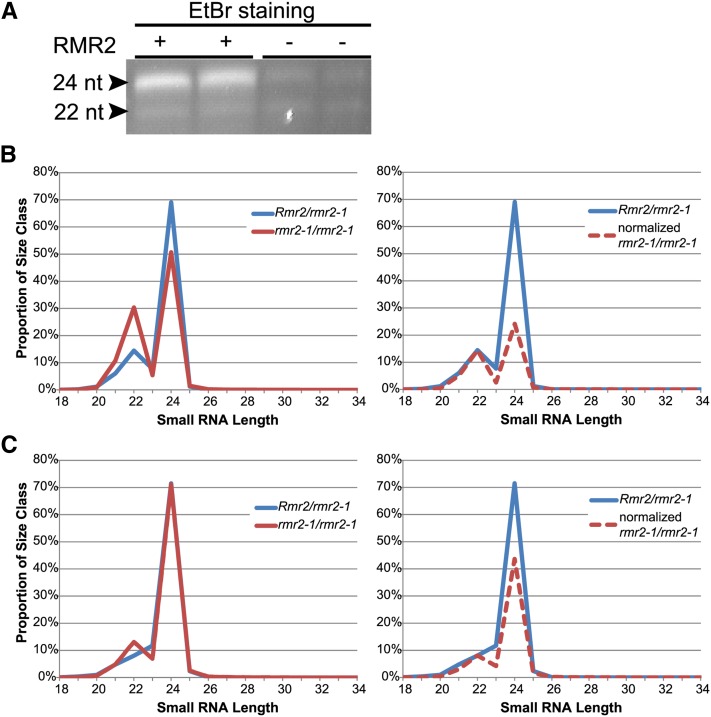
All RMR proteins identified to date are required for the biogenesis and/or maintenance of 24-nucleotide RNA species (Erhard et al., 2009; Hale et al., 2009; Stonaker et al., 2009). This requirement is obvious from comparing ethidium bromide–stained sRNAs from mutant and nonmutant siblings following PAGE fractionation. To evaluate whether RMR2 is similarly required, total RNA was extracted from developing cobs (4 cm in length) of both rmr2-1 homozygotes (n = 2) and Rmr2/rmr2-1 (n = 2) siblings. Cobs were collected prior to flowering and processed after the rmr2 genotype was determined by diagnostic anther colors. sRNA fractions were enriched from total RNA samples, separated on a polyacrylamide gel, and stained (Figure 6A). Two readily visible bands are routinely seen in such fractionations: one predominant band corresponding to the 24-nucleotide-size class (Erhard et al., 2009; Hale et al., 2009; Stonaker et al., 2009) and a less pronounced band below representing a 22-nucleotide maize-specific size class (Nobuta et al., 2008). Among the four biological samples we evaluated, the 22-nucleotide RNA band stained with remarkable uniformity, indicating that similar levels of this sized species are found in both genotypes. By contrast, the 24-nucleotide RNA population was markedly reduced in the absence of RMR2 relative to nonmutant siblings (Figure 6A).
Figure 6.
RMR2-Dependent sRNAs.
(A) Ethidium bromide (EtBr) staining of PAGE fractionated sRNAs from Rmr2/rmr2-1 (+) and rmr2-1/rmr2-1 (−) siblings. Sizes of the abundant sRNA classes are indicated. nt, nucleotides.
(B) Plots comparing the proportion of total, genome-matched sRNA sequences versus their size. The two graphs represent the total percentages (left) and the percentage of normalized abundances of maize-specific 22-mers found in the nonmutant samples (right). Blue and red lines correspond to the profiles from Rmr2/rmr2-1 and rmr2-1/rmr2-1 genotypes, respectively. Total genome-matched reads mapped to one or more sites in the entire maize genome.
(C) Plots comparing the proportion of distinct, genome-matched sRNA sequences versus their size. The two graphs represent the total percentages (left) and the percentage of normalized abundances of maize-specific 22-mers found in the nonmutant samples (right). Blue and red lines correspond to the profiles from Rmr2/rmr2-1 and rmr2-1/rmr2-1 genotypes, respectively. Distinct genome matched reads map uniquely to the entire maize genome.
We then used sequencing-by-synthesis on the Illumina platform to deeply sequence sRNA libraries made from the 4-cm cobs of rmr2 mutant and nonmutant siblings (see Supplemental Table 4 online). The sRNA size and abundance profile revealed there was a ∼65% loss of 24-nucleotide RNA species in the absence of RMR2 function, relative to normalized levels of 22-nucleotide sRNAs (Figure 6B). Among the sRNA reads mapping only to unique regions of the maize genome, there was still ∼40% loss of the 24-nucleotide species (Figure 6C). These data show that RMR2 is required for the accumulation of sRNAs representing both repetitious and unique regions of the genome in developing maize cobs, and this requirement appears to be preferential, if not exclusive, for the 24-nucleotide size class.
RMR2 Is Required for Cytosine Methylation Patterns at the 3′ End of Pl1-Rh
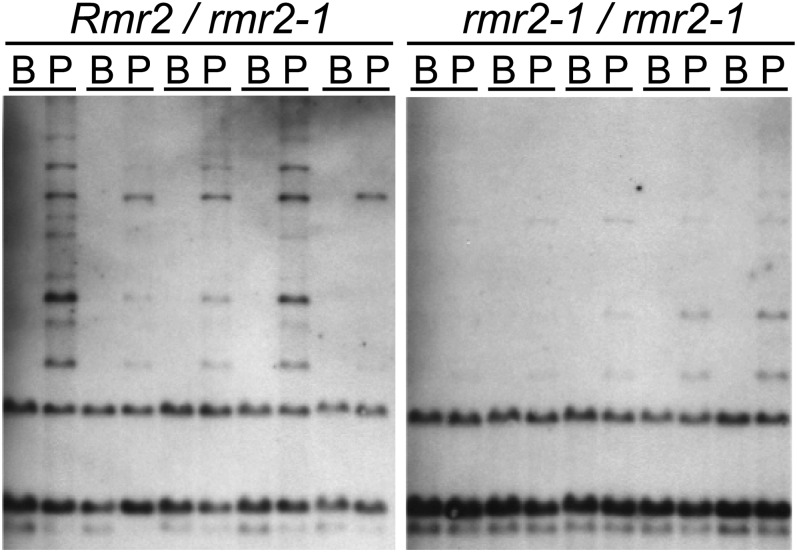
Pol IV, RDR2, and RMR1 are also required to establish and/or maintain specific 5meC profiles (Hale et al., 2007; Parkinson et al., 2007). While RPD1 is required to maintain and/or establish 5meC in all sequence contexts at the fractured doppia element 5′ of Pl1-Rh (Hale et al., 2007), it has no obvious effect on the 5meC at specific CHG sites found in the 3′ region nor at other sites evaluated by differential BstNI-PspGI digestions at centromeric repeats and 45S rDNA clusters (Parkinson et al., 2007). Using identical digestion, gel blotting, and hybridization conditions, we found that RMR2 is required to maintain high levels of 5meC at these CHG sites found 3′ of Pl1-Rh in genomic DNA samples isolated from flag leaves (Figure 7). This difference in 5meC patterns did not appear to extend to centromeric repeats as, similar to RPD1, we found no compelling indication that RMR2 was required to maintain 5meC at these assayed sites (see Supplemental Figure 2 online). Although this survey was limited, the data clearly show that RMR2 is required for certain 5meC patterns distinct from those maintained and/or established by RPD1. This places RMR2 in an RdDM-like pathway unaffected by RPD1, the sole largest subunit of Pol IV in maize (Erhard et al., 2009).
Figure 7.
RMR2-Dependent Methylation of the Pl1-Rh 3′ Region.
Genomic DNA samples from sibling Rmr2/rmr2-1 and rmr2-1/rmr2-1 plants (five each) digested with methylation-insensitive (BstNI; B) or -sensitive (PspGI; P) endonucleases and probed with a radiolabeled genomic clone specific to 3′ pl1 sequences.
DISCUSSION
The RMR2 family represents apparently diverse proteins in multicellular plants related by a novel amino acid motif embedded in a conserved CTR. The corresponding gene models encode hypothetical proteins ranging from 278 (Selaginella moellendorffii gene model: 413199) to 1123 amino acids (Physcomitrella patens gene model: Pp1s36-60V6). The number of RMR2 family members per organism also varies. Compared with eudicots, which typically have single RMR2-type proteins, the grasses appear to have conserved additional diversity. However, further improvements to plant genome assemblies may identify additional gene models containing a similar CTR. Unlike other eudicots, A. thaliana and Arabidopsis lyrata genomes both contain several (four and three, respectively) gene models predicting proteins with identity to the RMR2 CTR. In every replicate phylogram (n = 1000, bootstrap frequencies of 1.0), these A. thaliana proteins group into three subclades, but these representatives are all most closely related to the grass clade C proteins. Analysis of the A. thaliana developmental expression atlas (Schmid et al., 2005) indicates that all of the RMR2 family genes are expressed throughout development (see Methods for details). However, AT1G21560 RNA is typically present at 10-fold higher levels than the other three members. These mRNA levels decrease to the level of the other two subclade representatives in mature pollen and in stage 8, 9, and 10 embryos. Importantly, there is no evidence of subclade-specific expression in embryo and endosperm tissues as seen with the maize RMR2-type genes. These comparisons raise the possibility that RMR2 has evolved to carry out grass-specific functions.
The molecular function of the RMR2 protein family remains unknown; however, bioinformatic analyses hint at a possible role in nucleic acid binding. Both FUGUE and PHYRE2 algorithms predicted a series of α-helices in the RMR2 CTR that have the highest structural similarity to HTH DNA binding folds. The HTH folds of F93 (FUGUE result) and EspR (PHYRE2 result) both have unique traits. EspR in particular was shown by Rosenberg et al. (2011) to create large DNA loops because of the angle of the two DNA binding helices within an EspR dimer. PHYRE2 analysis of the RMR2 clade C maize member (GRMZM2G003389) identified a more significant similarity between its C-terminal helices and the EspR HTH fold (confidence of 82% versus 49% for RMR2). Pfam queries also identified zinc-finger DNA binding motifs in P. patens (gene model: Pp1s36_60V6) and S. moellendorffii (gene model: 420539). The other S. moellendorffii gene model (413199) sharing identity with the RMR2 CTR has two tandem AT-hook motifs. AT-hooks can bind minor grooves of DNA and commonly function with other DNA binding motifs to alter DNA structure (Aravind and Landsman, 1998).
None of the RMR2 family members have obvious catalytic domains, leading to the prediction of a structural rather than enzymatic role for these generally small (300 to 600 amino acids) proteins. The secondary structure of the RMR2 N-terminal region is predicted to be intrinsically disordered. In combination with the low sequence conservation and variable sizes of the RMR2 family N termini, these observations support the idea that these proteins have rapidly evolving protein–protein interaction domains (Dunker et al., 2005). Perhaps the CTR of RMR2 directs nucleic acid binding, while the poorly conserved N-terminal region coordinates contacts with other peptides. These speculative assignments remain to be tested, but interestingly, a DNA binding protein associating with the B1-I upstream repeats appears to play a role in facilitating paramutation at b1 (Brzeska et al., 2010). In A. thaliana, a DNA binding protein (SSH1/DTF1) required for both DNA methylation-dependent and methylation-independent gene silencing (Liu et al., 2011) was recently found in coimmunoprecipitations with NRPD1 (Law et al., 2011). These DNA binding proteins may act to specify genomic targeting of specific RNA polymerase complexes.
Because accumulation of 24-nucleotide RNAs and certain 5meC patterns require RMR2 function, it seems likely that RMR2 functions in an RdDM-like pathway. In A. thaliana, the 24-nucleotide RNA class directs methylation of repetitive sequences via ARGONAUTE4-mediated association with nascent Pol V or Pol II transcripts and recruitment of de novo cytosine methyltransferases (Haag and Pikaard, 2011; Zhang and Zhu, 2011). The hypomethylation of 3′ Pl1-Rh CHG sequences in rmr2 mutants presumably results from the disruption of an RdDM-like pathway in the absence of the 24-nucleotide RNAs that require RMR2. Recently, Law et al. (2011) found several A. thaliana proteins, including RDR2 and the RMR1 orthologs, physically associated with Pol IV. If maize has similar RNA polymerase complexes, then RMR2 association with these, either directly or indirectly, would agree with predictions of an adapter-like function for RMR2. However, the 3′ Pl1-Rh hypomethylation seen in rmr2 mutants is not found in rpd1 mutants (Parkinson et al., 2007), indicating that RMR2 can maintain specific 5meC patterns independently of Pol IV. Whether the Pol IV-independent function represents a subclass of RMR2-dependent sRNAs acting through an alternate RNA polymerase or a function unrelated to sRNAs remains to be determined.
Paramutation requires trans-homolog interactions currently thought to be mediated by a diffusible substance (Hollick, 2010). The 24-nucleotide RNAs, whose accumulation requires several trans-acting proteins identified in our genetic screen (Pol IV, RMR1, RDR2, and now RMR2), could potentially act as such a diffusible substance. However, here, we found that paramutations at both r1 and pl1 can occur in the absence of RMR2. This result indicates that neither the RMR2-dependent sRNA profiles nor any RMR2-dependent cytosine methylation patterns mediate paramutation. Similarly, absence of RMR1 does not inhibit paramutations, despite the loss of ∼65% of the 24-nucleotide RNAs (Hale et al., 2007). The extent of coincident sRNA signatures absent in rmr1 and rmr2 mutants remains to be seen. So far, mutations of rmr1, rmr2, and rpd2a appear to deplete subsets of the total sRNA pool requiring RPD1 (Erhard et al., 2009; Hale et al., 2009; Stonaker et al., 2009). Whether the RMR2-dependent 24-nucleotide sRNA profile defines a global reduction in sRNA levels or the loss of a specific subpopulation remains unknown. It remains possible that specific sRNAs responsible for mediating paramutations at r1 and pl1 still exist in the absence of RMR1 and RMR2. The alternate possibility is that paramutation requires RPD1-dependent functions that are distinct from the sRNAs and any downstream effects on cytosine methylation patterns.
We also found that RMR2 was required for paramutations at the pl1 locus in at least one-third of the examples tested. This finding indicates that RMR2 plays a role in either facilitating the trans-homolog interactions taking place between Pl1-Rh haplotypes or in maintaining the newly acquired repressed state resulting from such interactions. Our observations that RMR2 did not appear to be required for paramutations occurring at r1 implicate a molecular distinction between paramutation mechanisms operating at these different loci. A key goal is to determine which features make a haplotype susceptible to paramutations. Generally, cis-linked, repetitive structural features related to transcription seem to be necessary. RMR2 represents a trans-acting protein having effects on only a subset of paramutable loci, perhaps by sensing distinctions in cis-linked elements.
The genetic behaviors at pl1 seen in rmr2 mutants further support a mechanistic distinction between the repression of Pl1-Rh in plant tissues of both Pl-Rh/Pl′ and Pl′/Pl′ sporophytes and the maintenance of a meiotically heritable feature responsible for facilitating paramutations in the next generation (Erhard and Hollick, 2011). Transcriptional repression of Pl′ in the sporophyte requires RMR2 function, yet darkly pigmented rmr2 mutants pass paramutagenic Pl′ states to their progeny. The darkly colored tissues seen in rmr2 mutants mirror a derepressed transcription state of Pl′ in the sporophyte. However, a reversion to a fully active Pl-Rh–like state can only be scored if that plant transmits Pl-Rh–like haplotypes to its progeny. In two previous genetic tests, 2 and 9% of progeny individuals from darkly pigmented rmr2-1/rmr2-1; Pl′/Pl′ plants transmitted Pl-Rh states (Hollick and Chandler, 2001). Here, no reversions were found when Pl1-Rh haplotypes of original Pl′ state were maintained in homozygous condition for seven generations in the absence of RMR2. This breeding scheme reinforced the stability of Pl′ states despite continually presenting Pl-Rh–like sporophytes. These contrasting observations might indicate a variable requirement for RMR2 in different genetic backgrounds, or perhaps they reflect different depths of repression in the Pl′ states between haplotypes with newly acquired Pl′ states and those carried through several generations in a repressed Pl′ state. Regardless, Pl′ states remain heritably repressed even after passage through multiple generations in the absence of RMR2 and RMR2-dependent sRNA profiles. Therefore, just as RMR2-dependent 24-nucleotide RNA profiles are not required to acquire a paramutant state, they are also not required to maintain such states through meiosis.
RMR2 also maintains repressed states of features found elsewhere in the maize genome. McGinnis et al. (2006) found that two structurally distinct heritably silenced transgenes were reactivated in the absence of RMR2. Loss of RMR1 and RDR2 similarly reactivated the complex transgenes, indicating that all three proteins function together to maintain the repression (McGinnis et al., 2006). We also observed evidence of developmental defects diagnostic of a “Mu syndrome” (Walbot, 1991) and high Mu-induced mutation rates, after maintaining a weakly active Mu line in the absence of RMR2 (see Methods). Both empirical phenotypes indicate a general derepression of previously silenced autonomous Mu elements (Walbot, 1991). However, all of our non-Mu lines lacking RMR2 do not display developmental defects characteristic of rpd1 mutants (Parkinson et al., 2007; Erhard et al., 2009). The one rmr2-1 RIL advanced to the S9 generation displays evidence of inbreeding depression but otherwise appears normal (J.B. Hollick, unpublished data). Like the genome-wide effects on 24-nucleotide RNAs, the RMR2 requirement to maintain repression is not limited to isolated loci; however, it remains unknown if all of these silencing roles are mediated through an RdDM-like pathway. Further characterizations of RMR2-dependent sRNA profiles and the RMR2-dependent transcriptome promise to clarify these more global roles in epigenomic regulation.
The maize proteins representing the three grass RMR2 family subclades may functionally overlap, possibly accounting for paramutations still occurring at r1 and pl1 in rmr2 mutants. Because embryo and endosperm tissues appear to exclusively express rmr2 RNA, any functions in those tissues presumably rely solely on RMR2. If the putative paralogs have similar roles, perhaps the GRMZM2G109217 or GRMZM2G003389 protein acts in the acquisition of paramutation at r1. The three possible second largest Pol IV subunits represent another case of increased diversity of epigenetic control mechanisms in grasses as rpd2a mutants only share a subset of phenotypes with the rpd1 mutants that must remove all Pol IV activity (Erhard et al., 2009; Stonaker et al., 2009). At this point, partially redundant or completely distinct roles cannot be distinguished for the grass RMR2-related proteins.
The plant-specific origins of Pol IV, together with the apparent grass-specific nature of RMR2, raise the possibility that paramutations occurring in maize are mechanistically distinct from similar behaviors described in other eukaryotes. Paramutations in maize require Pol IV function, but it is increasingly likely that the sRNAs presumably derived from Pol IV transcripts are not causative agents (Erhard and Hollick, 2011). It is possible that many components of a presumed maize RdDM pathway are identified in our mutational screens because of the intimate and pervasive juxtaposition of repetitive sequences targeted by RdDM and attendant genic regions found in the maize genome (Baucom et al., 2009). This opinion focuses attention on features of transcription and perhaps nascent RNA processing at or nearby such juxtapositions that might facilitate trans-homolog interactions and subsequent heritable changes to the chromosome (Erhard and Hollick, 2011). Identification of the proteins that functionally interact with RMR2 will provide a novel avenue to understanding the particular paramutation mechanism occurring at pl1. A broader understanding of the role(s) played by all RMR2-related proteins promises to open new areas of investigation into the diversification of epigenomic control mechanisms in higher plants.
METHODS
Genetic Materials and Stock Syntheses
The genetic nomenclature used follows established guidelines for maize (Zea mays) and has been fully described previously (Hollick et al., 2005). Hand pollinations were used for all stock syntheses and genetic assays. Detailed pedigree information is available upon request. All stocks contain functional alleles required for anthocyanin pigmentation in the anthers unless otherwise noted. The b1, pl1, and r1 haplotypes are indicated as these encode the transcription factors affecting the pigment patterns used in the stock syntheses and analyses. All plants are homozygous for the Pl1-Rh haplotype unless otherwise described. Details regarding most of the specific source stocks, pollen and anther scoring, and paramutation analyses are provided by Hollick et al. (2005).
The rmr2-1 reference allele was isolated from an EMS mutagenesis as previously described (Hollick and Chandler, 2001). The rmr2-1 mutation was recovered in homozygous condition from an F2 family (Hollick and Chandler, 2001) and inbred by single-seed descent to the F2S9 generation. All test crosses measuring the paramutagenic potential of Pl1-Rh haplotypes were made using a Pl1-Rh (Pl-Rh)–converted A619 inbred as previously described (Hollick et al., 2005). Experimental designs and specific stocks used for testing whether or not paramutation at pl1 and r1 could occur in the absence of Rmr2 function were nearly identical to those described for the rmr6-1 mutation (Hollick et al., 2005) except that the original rmr2-1 donor derived from an A632 F2 rather than an A619 F2 as described and three progeny sets (2252, 2256, and 2283) were evaluated for R-r pigmenting activity.
For in vitro transcription reactions, progenies were generated that segregated 1:1 for Rmr2/rmr2-1 and rmr2-1/rmr2-1 siblings; plants with these genotypes are clearly identified by either Pl′-like (variegated) or Pl-Rh-like (darkly colored) anther phenotypes, respectively. These progenies also segregated B1-I haplotypes of B-I state as the rmr2-1/rmr2-1 parent was heterozygous B-I/b1. Plants with a B-I/b1 genotype have obvious plant pigmentation in both Rmr2/rmr2-1 and rmr2-1/rmr2-1 conditions. The necessary B1-I (B-I) rmr2-1 recombinant chromosome was isolated from a darkly pigmented plant having Pl-Rh–like anthers in the progeny of parents having B1-I (B-I) Rmr2/b1 rmr2-1 and b1 rmr2-1/b1 rmr2-1 genotypes. A W23 inbred full-color derivative served as the source of the B1-I (B-I) haplotype (Hollick et al., 1995).
For the sRNA libraries, sibling Rmr2/rmr2-1 and rmr2-1/rmr2-1 plants were generated by crossing a BC1 parent (Rmr2/rmr2-1) by an F2 (rmr2-1/rmr2-1) mutant. Both F2 and BC1 plants derive from crosses to a largely K55 stock (b1, Pl1-Rh, and r-r). Prior to the initial cross to the K55 stock, the rmr2-1 allele had been maintained in homozygous condition for three generations by sib crosses.
For the DNA gel blot analyses, sibling Rmr2/rmr2-1 and rmr2-1/rmr2-1 plants were derived from crossing an rmr2-1 heterozygous parent by an rmr2-1 mutant (F2S1) recovered from initially crossing an original rmr2-1 M2 isolate to the A632 inbred line. The heterozygous parent (F3) was produced from crossing rmr2-1 heterozygotes by rmr2-1 mutant siblings for two successive generations following the initial outcross of an original rmr2-1 M2 isolate to a Pl′/Pl′ tester (Hollick and Chandler, 2001).
Materials used for molecular mapping derived from F2 families using F2S7 rmr2-1/rmr2-1 plants as described above and a Pl1-Rh–converted A632 inbred (Hollick et al., 2005) as parents.
A specific stock was synthesized to genetically mark the rmr2-1 reference allele used in the Mu mutagenesis experiment. Two b1 rmr2-1 wt recombinant chromosomes were isolated in summer 2000 from a single F2 progeny plant having white-tipped seedling leaves and dark Pl-Rh–like anthers. F1 plants derived from parents of genotypes glossy2 (gl2) b1 Rmr2 wt (Patterson et al., 1991) and Gl2 b1 rmr2-1 Wt1 were self-pollinated to obtain the b1 rmr2-1 wt/b1 rmr2-1 wt double recombinant; two plants of 1319 progeny had this genotype.
The Mu lines used derive from those described previously (Dorweiler et al., 2000). Specifically, these source lines were crossed by rmr2-1/rmr2-1 plants and then propagated through a single plant with darkly colored anthers that appeared among ∼11,000 progeny. This plant was considered to be heterozygous for a new, induced rmr2 mutation provisionally designated rmr2-mum1 (rmr2-1/rmr2-mum1). Efforts to isolate this new allele from the reference allele proved unsuccessful given the lack of available parental polymorphisms. Following two generations of selfing plants that retained darkly colored anthers, a line was recovered that had a severely stunted plant phenotype and a single plant from this was used as a pollen source to an A619 inbred (b1, pl1-A619, and R-r). Plants in the subsequent progenies displayed a variety of abnormal developmental phenotypes characteristic of active Mu lines (Walbot, 1991). Plants from the F2 progeny were both backcrossed to the Pl1-Rh–converted A619 inbred and selfed. Progenies from the individual self pollinations were evaluated for anther colors, and only those that did not segregate plants with darkly colored anthers (rmr2 mutants) were maintained as lines for Mu-based mutagenesis. For the Mu-based mutagenesis, pollen from the Mu lines was bulked from at least 200 individual plants (tassel quality is poor) and applied to the silks of plants having a B1-I (B-I) Rmr2 Wt1/b1 rmr2-1 wt genotype. Approximately 12,000 progeny were screened at flowering in summer 2008 for plants having darkly colored anthers.
The plants containing putative Mu-induced rmr2 alleles (rmr2-mum) were crossed to both A619 and A632 Pl1-Rh-converted inbreds and then recovered in the F2 progenies that failed to segregate seedlings having white-tipped (rmr2-1 wt1/rmr2-1 wt1) seedling leaves. These were subsequently backcrossed to the respective inbred and again recovered in the BC1F2 progenies by their dark seedling color phenotype. These seedlings served as the source for genomic DNA provided for the Illumina sequencing of Mu insertion sites.
In Vitro Transcription Reactions
Nuclei isolations from internal husk leaves, transcription reactions, and specific slot blot hybridizations were performed as described previously (Hollick et al., 2005).
Illumina Mu-End Sequencing
An Illumina G1 analyzer was used with the specific library preparations and downstream analyses described previously (Williams-Carrier et al., 2010).
Phylogenetic Analysis
Sequences were obtained through protein BLAST (BLASTP, BLOSUM62 comparison matrix, allowing gaps and a word length of 3) (Altschul et al., 1990) queries with the maize RMR2 protein sequence on http://phytozome.net (Goodstein et al., 2012). A core ClustalX (Larkin et al., 2007) alignment of the top three protein matches for each grass, plus all Arabidopsis thaliana matches and the Physcomitrella patens top match was used to delimit the conserved CTR. Additional homologs (from Phytozome or National Center for Biotechnology Information BLASTP searches) were then manually added with Jalview (Waterhouse et al., 2009) to the conserved C-terminal alignment. The final alignment (Figure 5A) used for the phylogram was chosen for breadth and depth of coverage. The second P. patens protein (Pp1s114_65V6) was excluded to avoid an ∼15–amino acid insertion in the middle of the conserved region. Manually adjusted ClustalX alignment was analyzed with PHYLIP to create a maximum likelihood bootstrapped phylogram with 1000 randomized data sets (proml subprogram: Jones-Taylor-Thornton probability model with each data set randomly input 10 times and global rearrangements for each data set) (Felsenstein, 2005).
sRNA Libraries and Analyses
sRNA libraries were constructed from total RNA isolated from 4-cm developing cobs of Rmr2/rmr2-1 and rmr2/rmr2-1 using TRIzol reagent (Invitrogen). The sRNA library preparation protocol is based on Illumina’s sRNA Sequencing Sample Preparation Guide. The libraries were sequenced on an Illumina GAII at the Delaware Biotechnology Institute.
The adapter sequences were removed using a Perl script, generating sRNA sequences plus abundances. To account for differences in library depth, the library abundance for each signature was normalized to 10 million (units of transcripts per 10 million). The data were matched to the maize genome, annotation version 4a.53 AGPv1. Library summary statistics are provided in Supplemental Table 4 online.
RT-PCR and DNA Gel Blots
Details of the DNA gel blot analyses, including the specific probes used, are provided by Parkinson et al. (2007).
Previously generated cDNA (Erhard et al., 2009) and genomic A632 DNA templates were amplified with primer pairs specific for either GRMZM2G009208, GRMZM2G109217, GRMZM2G003389, or alanine aminotransferase (Aat) (see Supplemental Table 2 online). The RT-PCR products were separated on a 3% agarose gel and stained with ethidium bromide. See Supplemental Table 3 online for the predicted amplicon sizes from cDNA and genomic DNA templates.
A. thaliana Expression Profile Analysis
The AtGenExpress Visualization Tool (jsp.weigelworld.org/expviz/expviz.jsp) was used to query the AtGE Development microarray database (Schmid et al., 2005) for the A. thaliana RMR2 family members (AT3G07730, AT1G21560, AT1G77270, and AT4G01170). AAT1 (AT1G17290) was used as a control for expression across samples and LEAFY3 (AT5G61850) as a tissue-specific control primarily to indicate the background noise when not expressed. AT3G07730 and AT4G01170 were not differentiated by the microarray hybridizations.
Primers Used in This Work
The sequences are listed in Supplemental Table 2 online.
Accession Numbers
DNA sequence data from this article can be found in the GenBank/EMBL database under accession number JQ682647 (rmr2-1). RNA sequences have been deposited in the Gene Expression Omnibus database under accession number GSE37204. The raw and normalized Illumina data are also available at http://mpss.udel.edu/maize.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RNA Profile of Maize rmr2 Paralogs.
Supplemental Figure 2. Methylation Profile of Centromere Repeats.
Supplemental Table 1. Paramutation Occurring in Rmr2/rmr2-1; + (Pl′)/T Pl-Rh Plants.
Supplemental Table 2. List of Primers Used in the Work.
Supplemental Table 3. List of Expected RT-PCR Product Sizes.
Supplemental Table 4. Summary Statistics of SBS Libraries.
Supplemental Data Set 1. Text File of Alignment Corresponding to the Phylogenetic Tree in Figure 5.
Acknowledgments
We thank Vicki Chandler (University of Arizona, Tucson, AZ) for providing the Mu line germplasm, Stephen Gross (Joint Genome Institute, Walnut Creek, CA) for Pl1-Rh sequence data, and Karl F. Erhard Jr. (University of California, Berkeley) for helpful comments. This research was funded in part by a National Institutes of Health National Research Service Award Trainee appointment on Grant T32_GM_007127. This work was supported by the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (99-35301-7753, 2001-35301-10641, and 2005-35301-15891) and the National Science Foundation (MCB-0419909 and MCB-0920623). The views expressed are solely those of the authors and are not endorsed by the sponsors of this work.
AUTHOR CONTRIBUTIONS
J.K., B.C.M., A.B., and J.B.H. designed the research. J.-E.R.B., I.T.L., J.L.S., J.P.L., C.C.L., S.E.P., J.K., S.A.S., R.W.-C., and J.B.H. performed research. J.-E.R.B., I.T.L., S.E.P., J.K., S.A.S., B.C.M., R.W.-C., and J.B.H. analyzed data. J.-E.R.B. and J.B.H. wrote the article.
Glossary
- sRNA
small RNA
- RdDM
RNA-directed DNA methylation
- ACS
anther color score
- EMS
ethyl methanesulfonate
- 5meC
5-methylcytosine
- RIL
recombinant inbred line
- cM
centimorgan
- SSLP
simple sequence length polymorphism
- CTR
C-terminal region
References
- Alleman M., Sidorenko L., McGinnis K., Seshadri V., Dorweiler J.E., White J., Sikkink K., Chandler V.L. (2006). An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442: 295–298 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Landsman D. (1998). AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26: 4413–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga-Vazquez M.A., Sidorenko L., Rabanal F.A., Shrivistava R., Nobuta K., Green P.J., Meyers B.C., Chandler V.L. (2010). RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc. Natl. Acad. Sci. USA 107: 12986–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom R.S., Estill J.C., Chaparro C., Upshaw N., Jogi A., Deragon J.-M., Westerman R.P., San Miguel P.J., Bennetzen J.L. (2009). Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink R.A. (1956). A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41: 872–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink R.A. (1958). Paramutation at the R locus in maize. Cold Spring Harb. Symp. Quant. Biol. 23: 379–391 [DOI] [PubMed] [Google Scholar]
- Brink R.A., Weyers W.H. (1957). Invariable genetic change in maize plants heterozygous for marbled aleurone. Proc. Natl. Acad. Sci. USA 43: 1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.F., Brink R.A. (1960). Paramutagenic action of paramutant R-r and R-g alleles in maize. Genetics 45: 1313–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska K., Brzeski J., Smith J., Chandler V.L. (2010). Transgenic expression of CBBP, a CXC domain protein, establishes paramutation in maize. Proc. Natl. Acad. Sci. USA 107: 5516–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V.L. (2010). Paramutation’s properties and puzzles. Science 330: 628–629 [DOI] [PubMed] [Google Scholar]
- Chandler V.L., Stam M. (2004). Chromatin conversations: Mechanisms and implications of paramutation. Nat. Rev. Genet. 5: 532–544 [DOI] [PubMed] [Google Scholar]
- Chou K.-C., Shen H.-B. (2010). Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 5: e11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E.H., Jr (1966). The properties, origin, and mechanism of conversion-type inheritance at the B locus in maize. Genetics 53: 1035–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., Robbins T.P., Jorgensen R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199 [DOI] [PubMed] [Google Scholar]
- Dorweiler J.E., Carey C.C., Kubo K.M., Hollick J.B., Kermicle J.L., Chandler V.L. (2000). mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12: 2101–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., Cortese M.S., Romero P., Iakoucheva L.M., Uversky V.N. (2005). Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 272: 5129–5148 [DOI] [PubMed] [Google Scholar]
- Eggleston W.B., Alleman M., Kermicle J.L. (1995). Molecular organization and germinal instability of R-stippled maize. Genetics 141: 347–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard K.F., Jr, Stonaker J.L., Parkinson S.E., Lim J.P., Hale C.J., Hollick J.B. (2009). RNA polymerase IV functions in paramutation in Zea mays. Science 323: 1201–1205 [DOI] [PubMed] [Google Scholar]
- Erhard K.F., Hollick J.B. (2011). Paramutation: A process for acquiring trans-generational regulatory states. Curr. Opin. Plant Biol. 14: 1–7 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (2005). PHYLIP (Phylogeny Inference Package) Version 3.6. (Seattle, WA: University of Washington) [Google Scholar]
- Finn R.D., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38(Database issue): D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. (2005). Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, J. Walker, ed (Totowa, NJ: Humana Press), pp. 571–607 [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40(Database issue): D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.M., Hollick J.B. (2007). Multiple trans-sensing interactions affect meiotically heritable epigenetic states at the maize pl1 locus. Genetics 176: 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J.R., Pikaard C.S. (2011). Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12: 483–492 [DOI] [PubMed] [Google Scholar]
- Hale C.J., Erhard K.F., Jr, Lisch D., Hollick J.B. (2009). Production and processing of siRNA precursor transcripts from the highly repetitive maize genome. PLoS Genet. 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C.J., Stonaker J.L., Gross S.M., Hollick J.B. (2007). A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in Zea mays. PLoS Biol. 5: e275
- Hollick J.B. (2010). Paramutation and development. Annu. Rev. Cell Dev. Biol. 26: 557–579
- Hollick J.B., Chandler V.L. (1998). Epigenetic allelic states of a maize transcriptional regulatory locus exhibit overdominant gene action. Genetics 150: 891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J.B., Chandler V.L. (2001). Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics 157: 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J.B., Kermicle J.L., Parkinson S.E. (2005). Rmr6 maintains meiotic inheritance of paramutant states in Zea mays. Genetics 171: 725–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J.B., Patterson G.I., Asmundsson I.M., Chandler V.L. (2000). Paramutation alters regulatory control of the maize pl locus. Genetics 154: 1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J.B., Patterson G.I., Coe E.H., Jr, Cone K.C., Chandler V.L. (1995). Allelic interactions heritably alter the activity of a metastable maize pl allele. Genetics 141: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J.B., Springer N. (2009). Epigenetic phenomena and epigenomics in maize. In Epigenomics, A.C. Ferguson-Smith, J.M. Greally, and R.A. Martienssen, eds (Houten, The Netherlands: Springer Media B.V.), pp. 119–147 [Google Scholar]
- Jablonka E., Raz G. (2009). Transgenerational epigenetic inheritance: Prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84: 131–176 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J.E. (2009). Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kermicle J.L. (1996). Epigenetic silencing and activation of a maize r gene. In Epigenetic Mechanisms of Gene Regulation, V.E.A. Russo, R.A. Martienssen, and A.D. Riggs, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 267–287 [Google Scholar]
- Kermicle J.L., Eggleston W.B., Alleman M. (1995). Organization of paramutagenicity in R-stippled maize. Genetics 141: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Larson E.T., Eilers B., Menon S., Reiter D., Ortmann A., Young M.J., Lawrence C.M. (2007). A winged-helix protein from Sulfolobus turreted icosahedral virus points toward stabilizing disulfide bonds in the intracellular proteins of a hyperthermophilic virus. Virology 368: 249–261 [DOI] [PubMed] [Google Scholar]
- Law J.A., Vashisht A.A., Wohlschlegel J.A., Jacobsen S.E. (2011). SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bai G., Zhang C., Chen W., Zhou J., Zhang S., Chen Q., Deng X., He X.-J., Zhu J.-K. (2011). An atypical component of RNA-directed DNA methylation machinery has both DNA methylation-dependent and -independent roles in locus-specific transcriptional gene silencing. Cell Res. 21: 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K.M., Springer C., Lin Y., Carey C.C., Chandler V.L. (2006). Transcriptionally silenced transgenes in maize are activated by three mutations defective in paramutation. Genetics 173: 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K., et al. (2008). Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl. Acad. Sci. USA 105: 14958–14963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T., Weir J., Walker E.L. (1999). The structure and paramutagenicity of the R-marbled haplotype of Zea mays. Genetics 153: 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson S.E., Gross S.M., Hollick J.B. (2007). Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev. Biol. 308: 462–473 [DOI] [PubMed] [Google Scholar]
- Patterson G.I., Harris L.J., Walbot V., Chandler V.L. (1991). Genetic analysis of B-Peru, a regulatory gene in maize. Genetics 127: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G.I., Thorpe C.J., Chandler V.L. (1993). Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics 135: 881–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg O.S., Dovey C., Tempesta M., Robbins R.A., Finer-Moore J.S., Stroud R.M., Cox J.S. (2011). EspR, a key regulator of Mycobacterium tuberculosis virulence, adopts a unique dimeric structure among helix-turn-helix proteins. Proc. Natl. Acad. Sci. USA 108: 13450–13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Yachdav G., Liu J. (2004). The PredictProtein Server. Nucleic Acids Res. 32(Web Server issue): W321–W326
- Sainz M.B., Grotewold E., Chandler V.L. (1997). Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Shi J., Blundell T.L., Mizuguchi K. (2001). FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310: 243–257 [DOI] [PubMed] [Google Scholar]
- Sidorenko L., Dorweiler J.E., Cigan A.M., Arteaga-Vazquez M., Vyas M., Kermicle J., Jurcin D., Brzeski J., Cai Y., Chandler V.L. (2009). A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 5: e1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M., Belele C., Dorweiler J.E., Chandler V.L. (2002a). Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16: 1906–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M., Belele C., Ramakrishna W., Dorweiler J.E., Bennetzen J.L., Chandler V.L. (2002b). The regulatory regions required for B′ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics 162: 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonaker J.L., Lim J.P., Erhard K.F., Jr, Hollick J.B. (2009). Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet. 5: e1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles E.D., Brink R.A. (1969). The metastable nature of paramutable R alleles in maize. IV. Parallel enhancement of R action in heterozygotes with r and in hemizygotes. Genetics 61: 801–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F.K., Colot V. (2010). Repeat elements and the Arabidopsis DNA methylation landscape. Heredity 105: 14–23 [DOI] [PubMed] [Google Scholar]
- Walbot V. (1991). The Mutator transposable element family of maize. In Current Topics in Genetic Engineering. J.K. Setlow, ed (New York: Plenum Press), pp. 1–37 [Google Scholar]
- Walker E.L. (1998). Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148: 1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E.L., Robbins T.P., Bureau T.E., Kermicle J., Dellaporta S.L. (1995). Transposon-mediated chromosomal rearrangements and gene duplications in the formation of the maize R-r complex. EMBO J. 14: 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. (2009). Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R., Stiffler N., Belcher S., Kroeger T., Stern D.B., Monde R.-A., Coalter R., Barkan A. (2010). Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J. 63: 167–177 [DOI] [PubMed] [Google Scholar]
- Woodhouse M.R., Freeling M., Lisch D. (2006). Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 4: e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. (2010). PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804: 996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhu J.-K. (2011). RNA-directed DNA methylation. Curr. Opin. Plant Biol. 14: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]