Reconstruction of the grass genome paleohistory revealed subgenome partitioning of microRNA (miRNA) genes during post-whole-genome duplication diploidization. The evolutionary scenario of miRNAs from the ancestral founder pool to the modern complements displayed dosage balance constrictions on the deletion/retention of miRNAs and associated target genes wherein transposable elements may play a major role in miRNA gene synteny disruption.
Abstract
The recent availability of plant genome sequences, combined with a robust evolutionary scenario of the modern monocot and eudicot karyotypes from their diploid ancestors, offers an opportunity to gain insights into microRNA (miRNA) gene paleohistory in plants. Characterization and comparison of miRNAs and associated protein-coding targets in plants allowed us to unravel (1) contrasted genome conservation patterns of miRNAs in monocots and eudicots after whole-genome duplication (WGD), (2) an ancestral miRNA founder pool in the monocot genomes dating back to 100 million years ago, (3) miRNA subgenome dominance during the post-WGD diploidization process with selective miRNA deletion complemented with possible transposable element–mediated return flows, and (4) the miRNA/target interaction-directed differential loss/retention of miRNAs following the gene dosage balance rule. Together, our data suggest that overretained miRNAs in grass genomes may be implicated in connected gene regulations for stress responses, which is essential for plant adaptation and useful for crop variety innovation.
INTRODUCTION
Genome sequences from flowering plants that are derived from a common ancestor 135 to 250 million years ago (mya) are increasingly available for evolutionary studies. Numerous paleogenomics efforts aiming at reconstructing genome paleohistory from founder ancestors have been reported and demonstrated that monocots (i.e., mainly grasses), including Panicoideae (sorghum [Sorghum bicolor], Paterson et al., 2009; maize [Zea mays], Schnable et al., 2010), Ehrhartoideae (rice [Oryza sativa], International Rice Genome Sequencing Project, 2005), and Pooideae (Brachypodium distachyon; International Brachypodium Initiative, 2010), were derived from n = 5 to 12 ancestral grass karyotypes (AGKs) containing 6045 ordered protogenes with a physical size of 33 Mb (Salse et al., 2008, 2009a, 2009b; Bolot et al., 2009; Salse, 2012). Modern grass genomes were shaped from this AGK through whole-genome duplication (WGD) and ancestral chromosome fusion events. Likewise, the recent comparison of numerous eudicot genomes (i.e., mainly eurosid), including grape (Vitis vinifera; Jaillon et al., 2007), poplar (Populus trichocarpa; Tuskan et al., 2006), Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000), soybean (Glycine max; Schmutz et al., 2010), and cacao (Theobroma cacao; Argout et al., 2011), revealed that modern eudicot genomes were evolved from an n = 7 ancestor that went through a paleohexaploidization event to reach an n = 21 intermediate followed by numerous WGD and chromosome fusion events (Jaillon et al., 2007; Abrouk et al., 2010). During the last 135 to 250 million years of evolution, the protein-coding gene families have been then shaped by various gene duplication mechanisms, including WGD (or polyploidization), segmental duplication, and tandem duplication. It is now well established that almost all modern diploid plant species are paleopolyploids (Paterson et al., 2004; Tang et al., 2008a, 2008b; Van de Peer et al., 2009; Wang et al., 2011).
Polyploidization is followed by a genome-wide diploidization (also referred to as partitioning or fractionation) process in which one or the other gene duplicate is lost. It has been shown that protein-coding genes belonging to different functional categories behave differently during this process. Diploidization-resistant genes, mainly transcription factors (TFs) or regulators (TRs), are often retained as duplicated copies following WGDs, whereas others are progressively deleted back to a single copy (singleton) state (Thomas et al., 2006; Sankoff et al., 2010; Pont et al., 2011). This diploidization phenomenon is affected by overall differential expression of the progenitor genomes to the newly formed tetraploid (Chang et al., 2010; Schnable et al., 2010). Gene dosage relations remain balanced after WGD with preferential retention of dosage-sensitive genes due to unbalanced states when deleted (Birchler et al., 2005; Freeling and Thomas, 2006). Fates of duplicated genes in this hypothesis are considered based on their roles in macromolecular complexes or networks. Diploidization-resistant genes are considered dosage sensitive because they are part of product–product interaction (referred as connected genes such as TFs and TRs), and their deletion or modification of product concentration will impact or imbalance the whole network (Blanc and Wolfe, 2004; Maere et al., 2005a; Seoighe and Gehring, 2004). Other classes of genes return to the singleton state following WGD. In this context, the connection of miRNAs and their targets, mainly TRs, is a good model to investigate the gene dosage balance hypothesis (Birchler et al., 2005; Freeling and Thomas, 2006; Birchler and Vietia, 2007; Edger and Pires, 2009) as well as subgenome dominance phenomena following ancient polyploidization in plants.
It is now well known that noncoding RNAs, especially small endogenous RNA molecules, play important roles in a wide range of biological processes in plant development, metabolism, cell cycle and differentiation, and stress response (Reinhart et al., 2002; Lim et al., 2003; Axtell and Bartel, 2005; Bagga and Pasquinelli, 2006). Posttranscriptional gene silencing, via repression or cleavage of mRNAs by small RNAs, has been well studied during the last decade (Axtell and Bartel, 2005). Small interfering RNAs and microRNAs (miRNAs) have been shown to mediate posttranscriptional gene regulation (Tomari and Zamore, 2005). Plant miRNAs, 20 to 22 nucleotides long, are products of noncoding genes (Bartel, 2004). miRNA biogenesis pathways involve primary miRNA, precursor miRNA (pre-miRNA), and mature miRNA that recognizes the target mRNAs (mostly protein coding genes; Reinhart et al., 2002) by sequence complementarity to exonic regions favoring RNA cleavage processes (Bagga and Pasquinelli, 2006). In plants, the presence of miRNAs in single cell alga and Bryophytes supports the early emergence of the miRNA/target interactome (algae, T. Zhao et al., 2007; moss, Arazi et al., 2005), preceding and facilitating the developmental patterning needed for multicellular differentiation and adaptation. On the other hand, novel miRNAs are continuously being generated from tandemly inverted target genes (Allen et al., 2004) or by recruitment from repetitive sequences (Li et al., 2011).
Besides miRNA conservation between species and lineages, the precise impact of genome evolutionary events, such as WGD, on miRNAs is poorly investigated. In Arabidopsis, for example, WGD, segmental duplication, and tandem duplication events were found as the major mechanisms for the expansion of conserved miRNA families from their traceable ancestral copies (Maher et al., 2006; Zhang et al., 2009). Despite this, a systematic and detailed study of miRNAs during paleohistorical genome evolution across the plant subfamily is still missing. Particularly, how miRNAs behave following polyploidization events in plants is not established. The recently achieved, robust, and detailed plant genome paleohistorical scenario, especially for grass genomes, offers the opportunity to follow precisely the evolutionary paths of miRNA genes during this process. Based on the extensive identification and characterization of miRNAs and associated targets in nine plant genomes, we address in this analysis especially grass-relevant conclusions regarding (1) the conservation of miRNAs and the deduced ancestral or core miRNA gene pool, (2) the impact of genome duplication on the elaboration of miRNA gene families, (3) the consequences of the diploidization process of paleopolyploids on miRNA retention, and (4) the mechanisms of the transposable element (TE)–mediated miRNA transposition. Together, we propose that miRNA/target interaction may underpin the differential retention of miRNAs following the gene dosage balance hypothesis during the formation of subgenome dominance in grass paleopolyploids.
RESULTS
Contrasted miRNA Conservation Patterns between Monocot and Eudicot Genomes
Plant miRNAs from four monocots (rice, maize, sorghum, and Brachypodium) and five eudicots (grape, Arabidopsis, soybean, poplar, and cacao) were identified using MIReNA software (Mathelier and Carbone, 2010) with the 22 conserved families of miRNAs from rice and grape in the miRBase (Kozomara and Griffiths-Jones, 2011) as reference sequences. These miRNAs were then positioned on the chromosome pseudomolecules for synteny analysis (see Methods; see Supplemental Data Set 1 and Supplemental Tables 1 and 2 online). A total of 150, 192, 177, and 112 miRNAs were obtained for rice, sorghum, maize, and Brachypodium, respectively (Table 1), more than those reported in the miRBase (141, 116, 145, and 75, respectively). Comparison of the two data sets (our MIReNA-detected miRNA set versus annotated miRNAs from miRBase) revealed 73% consistency in miRNA discovery rates for all four monocot species studied. The remaining 27% MIReNA-specific miRNAs were found to be GC poor, shorter, and generally located in centromeric and pericentromeric regions (see Supplemental Figure 1 online). Such characteristics may contribute to their escape from most commonly used miRNA gene prediction pipelines. Therefore, our MIReNA-based detection provides a large (i.e., ∼24% greater than miRBase) and robust (73% miRNAs identical to miRBase) miRNA data set that should complement and refine the miRBase catalog. Overall, between 20 (i.e., sorghum and Brachypodium) and 22 (i.e., rice) miRNA families were identified and mapped to the corresponding genomes (Table 1; see Supplemental Data Set 1 and Supplemental Table 1 online).
Table 1.
miRNA Families and Evolutionary Data of Studied Plant Genomes
| Species | Sequence Data |
Evolutionary Data |
|||
|---|---|---|---|---|---|
| # miRNA | # Family | OrthoSeq | ParaSeq | ConserFam | |
| Monocot | |||||
| Rice (12chr- 372Mb- 41046genes-1R) | 150 | 22 | 93 (62%) | 17 (23%) | 19 |
| Sorghum (10chr- 659Mb - 34008genes-1R) | 192 | 20 | 87 (45%) | 14 (15%) | 18 |
| Maize (10chr- 2365Mb- 32540genes-2R) | 177 | 21 | 73 (41%) | 21 (24%) | 18 |
| B. distachyon (5chr- 271Mb-25504genes1R) | 112 | 20 | 63 (56%) | 8 (14%) | 15 |
| Total | 631 miRNAs | – | 316 Conserved, 51% average | 60 Duplicates,19% average | – |
| Eudicot | |||||
| Grape (19chr- 302Mb- 21189genes-1R) | 147 | 19 | 28 (19%) | 8 (11%) | 13 |
| Arabidopsis [(5chr- 119Mb- 33198genes-3R) | 90 | 20 | 11 (12%) | 13 (29%) | 7 |
| Poplar (19chr- 294Mb- 30260genes-2R) | 132 | 20 | 15 (11%) | 25 (38%) | 6 |
| Soybean (20chr- 949Mb- 46194genes-3R) | 274 | 20 | 14 (5%) | 67 (49%) | 7 |
| Cacao (10chr- 218Mb- 27814genes-1R) | 84 | 19 | 10 (12%) | 4 (10%) | 5 |
| Total | 727 miRNAs | – | 78 Conserved, 12% average | 117 Duplicates, 27% average | – |
miRNAs and their family numbers from four monocots and five eudicots are listed here. ConserFam, conserved families among species; OrthoSeq, miRNAs located at the orthologous regions between species; ParaSeq, miRNAs located at intragenomic duplication regions; R, round of duplications (i.e., WGD); –, not available. The corresponding percentages based on total numbers are shown in parentheses.
By studying the locations of miRNAs relative to the syntenic blocks on the chromosomes of the four species, we found that miRNAs of 19 out of 22 families were represented in orthologous relationships. Unexpectedly, miR168, miR397, and miR535 families with the lowest number of family members were never identified at orthologous positions among rice, sorghum, maize, and Brachypodium genomes (see Supplemental Table 1 online). Using the orthology and paralogy criteria described in Methods, we were able to conclude that 62, 45, 41, and 56% (i.e., ∼50% on average) of the miRNAs fell in the syntenic blocks and 17, 14, 21, and 8 miRNAs were found in the duplicated genomic regions, respectively, in rice, sorghum, maize, and Brachypodium (Table 1). We also observed three miRNA families that were particularly impacted by genome duplications: miR156/157, miR167, and miR169 families that are associated with the highest number of duplicated copies in the four considered grass genomes (see Supplemental Data Set 1 and Supplemental Table 1 online). Moreover, we determined that seven families have no paralogous copies, and 12 families have at least one sister pair in the respective genomes (Table 1). Therefore, we show that for the monocots, the miRNAs evolved with an average ∼50% of conserved genes between species, similar to protein-coding genes (Murat et al., 2010). In other words, half of miRNA genes are either unique in a species or have largely diverged. The mechanism driving such noncolinearity of miRNAs will be investigated in the following sections.
By contrast, the colinearity of miRNA genes in the eudicots is significantly eroded compared with monocots where only 19, 12, 5, 11, and 12% (i.e., 12% on average) are conserved respectively in grape, Arabidopsis, soybean, poplar, and cacao genomes (Table 1). These conserved miRNAs involved only five to 13 families among the 22 surveyed, suggesting that WGD events that took place in eudicot evolution have greatly reduced the syntenic conservation of miRNAs (see Supplemental Table 2 online). Except for the ancestral paleotetraploidization and hexaploidization that were identified in monocots (50 to 70 mya) and eudicots (130 to 150 mya), respectively, the eudicots experienced numerous additional recent WGDs (Salse, 2012; referenced as ‘R’ in Table 1). Here, we provide evidence that WGDs in monocots but more significantly in eudicots reduced the conservation of miRNAs at the orthologous positions between modern species (Lyons et al., 2008). For instance, during a similar evolutionary period of 50 to 70 million years since they diverged from their common ancestor, rice/sorghum for the monocots and poplar/soybean for the eudicots show 51 and 5%, respectively, of conserved miRNAs. Whereas the two considered monocot genomes did not experience any lineage-specific WGD, poplar and soybean experienced one and two lineage-specific WGDs, respectively, then driving the observed loss of syntenic conservation of miRNA genes.
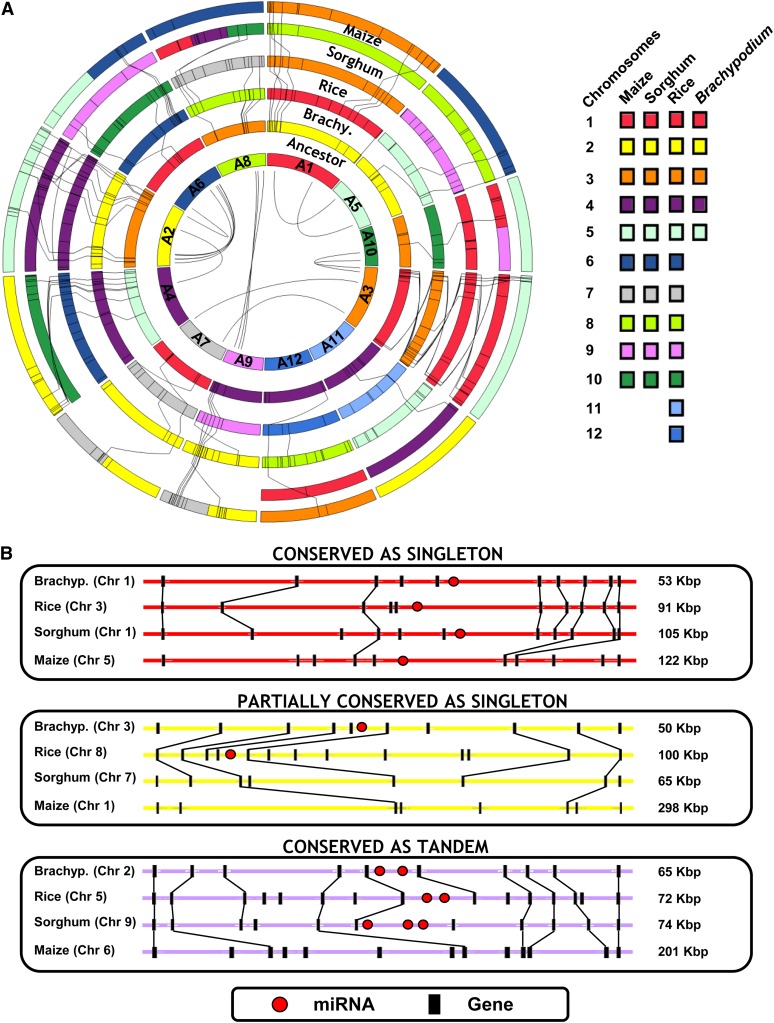
Thus, the monocot genomes are better places to study the structural evolution rules for miRNA genes. We aligned miRNA genes to the chromosome circles of the four species (Salse, 2012). Figure 1 represents the miRNA conservation in grasses at the genome-wide level (Figure 1A) as well as the microsynteny level (Figure 1B). The orthologous chromosomes are shown with a color code that illuminates their common origin from the founder ancestors of 12 protochromosomes (inner circle). miRNA genes are positioned on the chromosomes as thin black bars, and conserved miRNAs are linked with black thin lines between circles. This representation shows that conserved miRNA genes tend to be located at telomeric or subtelomeric regions (i.e., 31.1% of conserved miRNAs) of the plant chromosomes, whereas nonconserved miRNAs are mainly within centromeric or pericentromeric locations (i.e., 23.8% of conserved miRNAs) of the plant chromosomes. The biased distribution of the miRNAs is in agreement with what has already been established for the protein-coding genes in grasses in general (Murat et al., 2010). On the microscale level, miRNA genes are conserved as singletons between all the species (Figure 1B, top; representing 20% of the conserved miRNAs) or partially conserved between two or three species (48%; Figure 1B, center) or as tandem clusters (32%; Figure 1B, bottom).
Figure 1.
Genome-Wide miRNA Conservation in Grasses.
(A) Grass genome synteny is illustrated as concentric circles. The chromosome number is highlighted with a color code (at right) that illuminates the n = 12 monocot ancestral genome structure. Any radius of the circle shows orthologous chromosomes from Brachypodium, rice, sorghum, and maize genomes. Maize genome is depicted as a double circle originating from the maize-specific recent WGD. Colinear miRNAs are linked with black lines between circles, and ancestral duplicated miRNAs are linked with black lines at the center of the circle.
(B) Illustration of conservation fate from the miRNA (red dots) evolutionary analysis as (1) conservation between the four species (top inset), (2) conservation between two species (middle inset), and (3) conservation as clusters (bottom inset). Colinear genes are shown as black boxes and linked with lines.
Grass miRNA Gene Evolutionary Scenario Suggests Subgenome Dominance
In grass genomes, miRNA families appear to expand differently during evolution, resulting in variable family sizes. The miR156/157 family was the largest, with 23 members on average for the four species studied, followed by miR169 and miR395 families (17.75 members on average; Figure 2; see Supplemental Data Set 1 and Supplemental Table 1 online). Then, there were three miRNA families with an average size greater than 10 members (miR172-399-167), seven families comprising between five and 10 (miR159-160-164-166-170/171-396-529), and nine families associated with an average size below five (miR168-319-390-393-394-397-398-528-535). However, even though the numbers of miRNAs varied for each family within a species, the size of the same miRNA family was often similar in different species (Table 1; see Supplemental Table 1 online). Figure 2 (bottom) illustrates the distribution of the considered 22 miRNA families in the four grass genomes. On a statistical basis (i.e., 5% P value threshold) miR159, miR160, and miR172 are overrepresented in maize, rice, and sorghum, respectively, which may indicate that these repeat-amplified copies (see next sections) are no longer functional and, therefore, no longer under selective constraint. Oddly, members of the miR168, miR397, miR398, and miR535 families were not readily detectable simultaneously in all four grass genomes, but identified only in two or three of the species studied. This may be caused by the limitation of the computational algorithm or such miRNA loci are missing on the pseudomolecules, which may not entirely cover the genomes of interest.
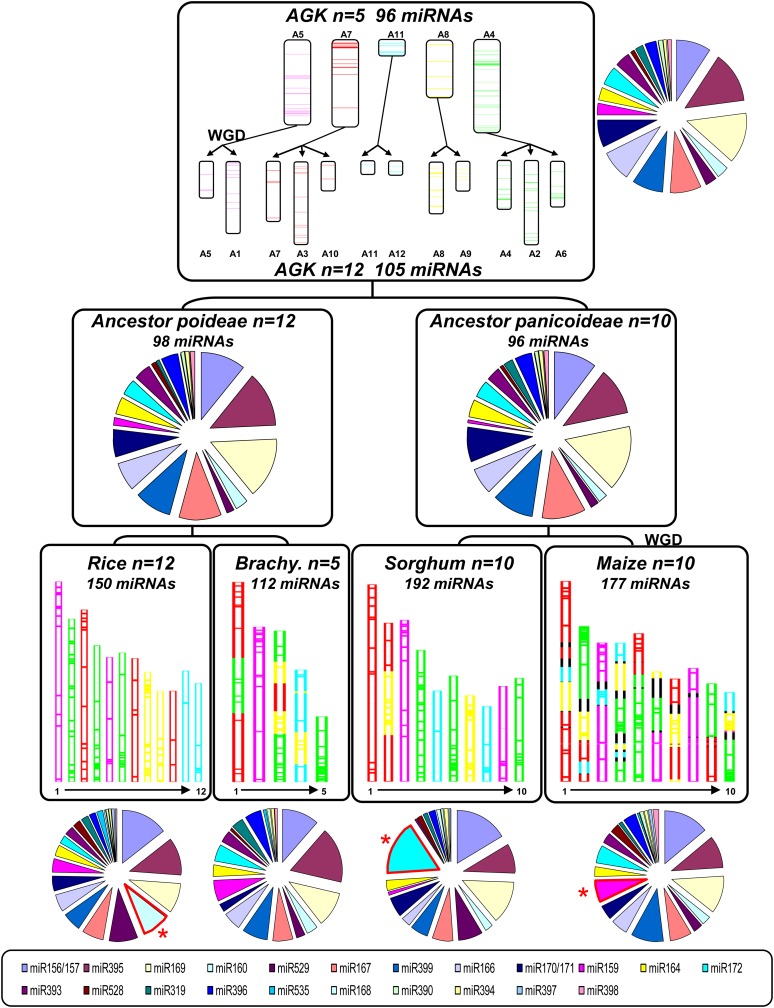
Figure 2.
Paleohistory of miRNA Evolution in Plants.
The modern grass genome structures (bottom) are depicted with a five-color code that illuminates their relationship with the n = 5 AGK (top). The percentage of miRNAs per family (reference color code legend at the bottom) are shown with circular distribution for the four monocot genomes (bottom), the rice/Brachypodium and sorghum/maize ancestral genome intermediates (center), as well as for the AGK (top), according to the paleogenomic study in the monocots from Murat et al. (2010).
Modern grass genomes have been shown to be derived from five protochromosomes via a WGD event at ∼50 to 70 mya, followed by ancestral chromosome translocations and fusions, as well as family-based and lineage-specific shuffling (Figure 2; Salse et al., 2008, 2009a; Bolot et al., 2009; Abrouk et al., 2010; Murat et al., 2010; Salse, 2012). Precise identification of the ancestral genome structure allowed us to investigate here the ancestral miRNA content in these five protochromosomes (A5, A4, A7, A8, and A11) from which miRNAs of the four modern species (150, 192, 177, and 112 for rice, sorghum, maize, and Brachypodium, respectively) were derived, giving a comparable net birth and death rate of 0.74 to ∼1.02, 1.32 to ∼1.81, 1.11 to ∼1.52, and 0.3 to ∼0.72 conserved miRNAs per million years for rice, sorghum, maize, and Brachypodium, respectively. Through the integration of paralogous and orthologous analysis described in the previous section, we defined 18 ancestral miRNAs for A5, 16 miRNAs for A7, five miRNAs for A11, 18 miRNAs for A8, and 39 miRNAs for A4. In total, we identified 96 ancestral miRNAs (i.e., conserved at orthologous position between the four genomes) covering 19 of the 22 miRNA families considered (Figure 2).
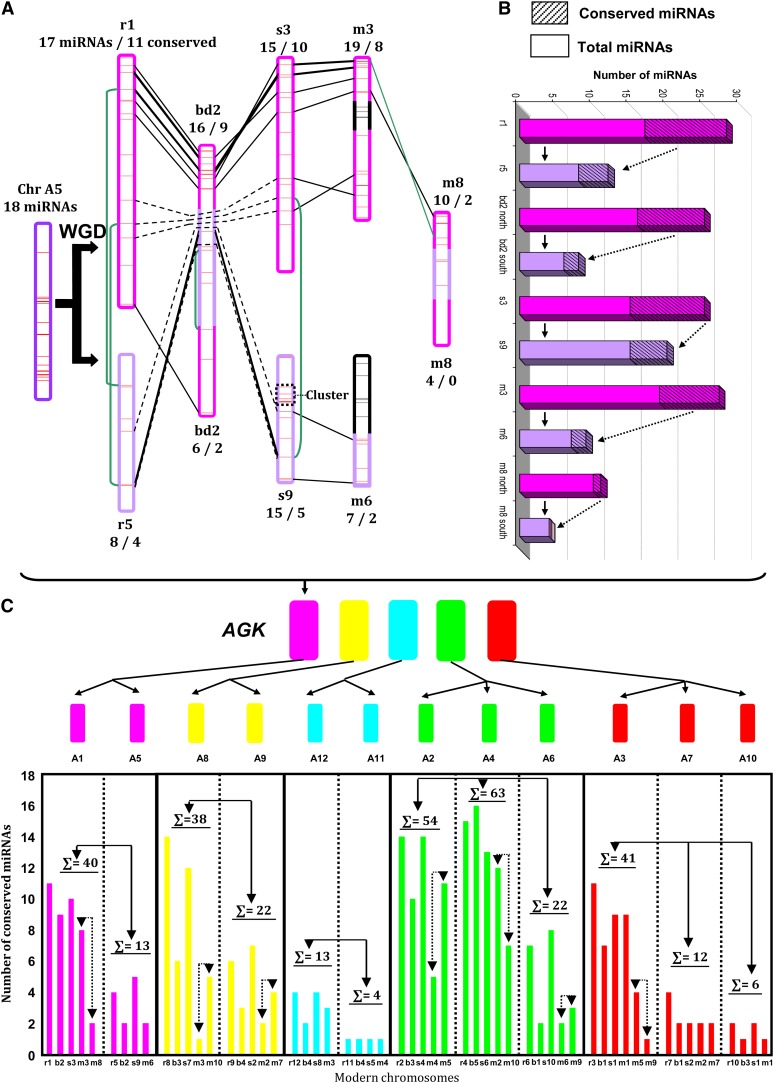
Therefore, our results confirm that grass miRNAs are ancient and were already present in the genomes of the ancestral species or so-called paleogenomes. However, we observe a clear difference of evolutionary trends between paralogous regions in terms of the miRNA content. Figure 3A provides the detailed evolutionary path of the ancestral protochromosome A5 in modern species (i.e., rice chromosomes 1/5, Brachypodium chromosome 2, sorghum chromosomes 3/9, and maize chromosomes 3/6/8). We characterized 18 ancient miRNAs from A5 protochromosome, and we know that A5 is derived from the ancestral duplication shared between the chromosome groups: group 1 involving r5-bd2-s9-m6/8 and group 2 involving r1-bd2-s3-m3/8 (Figure 3A). Normally, we should have observed an equal distribution of miRNAs in both chromosomal groups that derived from a unique protochromosome (referenced as Chr A5). However, we observed a higher number (i.e., greater than twofold differences in miRNA gene content) of miRNAs in group 2 (cf. Figure 3A top with 17, 16, 15, 19, and 10 miRNAs, respectively, on chromosomes r1, bd2S, s3, m3, and m8S) compared with group 1 (cf. Figure 3A bottom with 8, 6, 15, 7, and 4 miRNAs, respectively, on chromosomes r5, bd2L, s9, m6, and m8L). Overall, 77 miRNAs were identified in the group 2 of orthologous chromosomes compared with 40 for group 1, demonstrating unbalanced distribution of miRNAs on the duplicated chromosomes. This is also true for the number of orthologous (i.e., conserved) miRNAs on these two chromosomes (40 in group 2 versus 13 in group 1; Figure 3A). Figure 3B illustrates such biased distribution for each of the duplicated chromosomes regarding the total number of miRNAs (plain bars) and conserved miRNAs (dashed bars). One exception involves the total number of miRNAs (15) observed for both sorghum chromosomes 3 and 9. The enrichment of miRNA content on sorghum chromosome 9 is due to lineage-specific miR169 and miR399 clusters as highlighted (cf. dashed box in Figure 3A and Supplemental Data Set 1 online). Finally, we confirmed that the differences in miRNA content as well as conservation were statistically significant between the duplicated chromosomes deriving from the same ancestral protochromosome (plain arrows; Student’s t test P value = 0.026) as well as between the recent duplicated chromosomes in maize (dashed arrows; Student’s t test P value = 0.038; Figure 3C). Thus, the differences in miRNA content as well as conservation between ancestrally duplicated chromosomes suggest that miRNAs on one of the duplicated segments were subject to biased deletion and/or transposition. The biased partitioning of miRNAs during the genome diploidization process provides additional evidence for the subgenome dominance phenomenon following WGD.
Figure 3.
Differential Conservation Patterns of miRNA Duplicates.
(A) Illustration of paralogous chromosomes in rice (Chr 1-5), Brachypodium (Chr 2), sorghum (Chr 3-9), and maize (Chr 3-6-8) originating from the single ancestral preduplication chromosome A5. miRNAs are shown with horizontal colored bars on the chromosomes, colinear miRNAs are linked with black lines, and paleo-duplicated miRNAs are linked with green lines. The numbers of miRNAs and associated number of orthologous miRNAs are mentioned at the extremity of each chromosome.
(B) Illustration of the number (y axis) in total miRNA content (solid bars) and conservation (dashed bars) between duplicated chromosomes (x axis) in rice (chr 1-5), Brachypodium (chr 2 north-south), sorghum (chr 3/9), and maize (chr 3/6/8).
(C) Illustration of the conservation of duplicated miRNAs (y axis) in the modern genomes (bottom) Brachypodium (b), rice (r), sorghum (s), and maize (m) derived from a single paleo-tetraploid event from ancestral chromosomes A5, A8, A11, A4, and A7 (top). Differences in miRNA content in one of the paleo-duplicated (black arrows) or neo-duplicated (dotted arrows for maize) chromosomes are shown.
miRNA/Target Conservation Patterns Support the Gene Dosage Balance Hypothesis
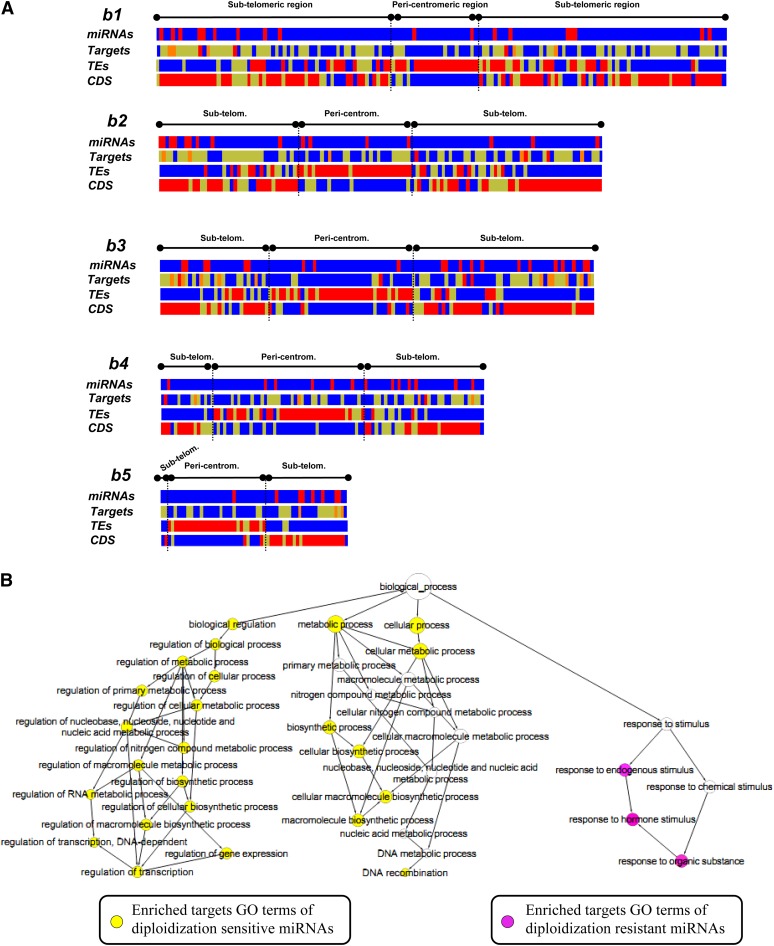
Recent studies on the retention of duplicate genes following diploidization of ancient polyplodization events suggest that protein-coding genes for signal transduction and transcription regulation (i.e., TFs and TRs) are preferentially maintained in a dosage-sensitive manner (Tang et al., 2008b; Salse et al., 2009a). To investigate the diploidization phenomenon upon miRNAs and associated targets, we first characterized the genomic distribution of miRNAs and their targets in the four grass genomes. miRNAs and associated targets have been precisely identified based on the strategy described in Methods and are illustrated in the case of the Brachypodium genome as a detailed example in the next section. Figure 4A depicts, as an example, the five Brachypodium chromosomes through heat maps for miRNAs, miRNA targets, TEs, and protein coding sequence (CDS). Genes (107.7 genes/Mb in subtelomeric region versus ∼50% [59.1 genes/Mb] in pericentromeric regions) and TEs (21.5% of subtelomeric regions covered by TEs versus greater than twofold in pericentromeric regions [51.4%]) are not randomly distributed on the chromosomes, and miRNAs and their associated targets appear more biased in their chromosome-wide distribution. Pericentromeric regions known to be poor in protein-coding genes (20.1%) are depleted in miRNAs (i.e., 17% of the Brachypodium miRNAs in pericentromeric regions) and associated targets (i.e., 16.3% of the Brachypodium miRNA targets in pericentromeric regions). Similar distributions regarding CDS, TEs, miRNAs, and miRNA targets have been observed for the rice, maize, and sorghum compared with the detailed illustration from the Brachypodium genome previously. These data suggest a distinct organization and conservation of miRNAs and protein-coding genes in grass genomes.
Figure 4.
Chromosomal Distribution and Conservation of miRNAs and Their Targets.
(A) Brachypodium heat map is illustrated for the miRNAs (first heat map color code blue = 0 and red ≤1 of miRNAs in intervals), miRNA targets (second heat map, color code color blue = 0, yellow <4, red >5 of miRNA targets in intervals), TEs (third heat map, color code blue <80%, yellow >80%, red ∼100% of TEs in intervals), and CDS (last heat map, color code blue <40, yellow = 40 to 50, red >50 of CDS in intervals) for the five pseudomolecules.
(B) A Cytoscape view of enriched GO biological processes among targets of overretained (i.e., duplicated) and underretained (i.e., nonduplicated) miRNAs in rice. The size of the node is proportional to the number of targets in the GO category. Purple solid nodes represent GO terms enriched in targets of overretained miRNAs; yellow solid nodes represent GO terms enriched in targets of underretained miRNAs; white nodes represent nonenriched GO terms to show the hierarchical relationship between enriched ontology branches. Enrichment significance level P value < 0.05 and FDR < 0.05.
We then studied the genome distribution of miRNAs as well as their targets in the context of the diploidization process after the ancestral WGD event in monocots (see the case example illustrated in Figure 3B) by mapping them to the ancient chromosomal duplication pairs and searching the paralogs between these duplicated pairs within the genomes of the four grass species. We observed that on average, there is ∼15% divergence (∼3- to ∼4-nucleotide difference) among mature miRNA sequences in the same families in four grass species (14.30, 14.97, 15.46, and 14.23% for rice, sorghum, maize, and Brachypodium, respectively). In addition, current miRNA functional modes require high sequence similarity between the miRNA and the targets, making it feasible for miRNAs from multiple precursors to target the same genes should they be expressed in the same spatial condition. This is in contrast with the divergence among TFs where each individual TF of the same family often carries a distinctive function. Therefore, we consider a miRNA family as a whole when studying their member retention in the context of dosage balance. Under this definition, an average of 45.3% miRNA families (9/17, 6/18, 6/15, and 11/20 for rice, sorghum, maize, and Brachypodium, respectively, each family with minimum two members; see Supplemental Tables 3 to 5 online) contained paralogous copies that are retained after WGD, close to the retention rate of TFs (60% in rice; Xiong et al., 2005) and suggesting possible dosage sensitivity.
Furthermore, we found a positive correlation between overretained miRNA families and the number of their target genes (see Supplemental Figure 2 and Supplemental Table 6 online), indicating that overretained miRNAs indeed confer more complex regulatory networks. As expected, most paralog-containing families were associated with overretained target genes in all four species. In rice, for example, seven out of nine (77.8%) of these retained miRNA families confer retained genes among their targets, of which 55.9% (19 out of 34) were TFs, demonstrating that coretained miRNA/target pairs were highly resistant to diploidization, probably due to the connected functions of miRNAs and their targets. Therefore, it appears that the genomic evolution of miRNAs also follows the gene balance hypothesis (Birchler and Veitia, 2010).
To understand better the roles of miRNAs retained after the paleohistorical WGD, we compared the functions of the targets of miRNA families with and without intraspecific syntenic paralogs using Gene Ontology (GO) enrichment analysis. The results showed that genes for biological regulation and metabolic processes in biological process (BP) terms were enriched (false discovery rate [FDR] < 0.05) with targets regulated by underretained miRNA families, whereas targets of overretained miRNA families (e.g., miR-159-160-167-169; see Supplemental Table 4 online) were significantly enriched (FDR < 0.05) in BP terms “response to stimulus” (cf. pink enrichment color code in Figure 4B; see Supplemental Table 7 online). As mentioned previously, most of these targets are TFs, such as auxin response factors, NAC domain proteins, MYB proteins, and NFYA (for nuclear transcription factor Y, subunit α), and the regulating miRNAs have been shown to be involved in responses to salt stress, water deficit, mechanical stress, nutrient deficiency, and reactive oxygen species (Ding et al., 2009; Reyes and Chua, 2007; B. Zhao et al., 2007, 2009, 2011). These observations therefore indicate a possible correlation between the overretained miRNAs and their targets for trait improvement and/or adaptation.
TE-Mediated miRNA Gene Reshuffling
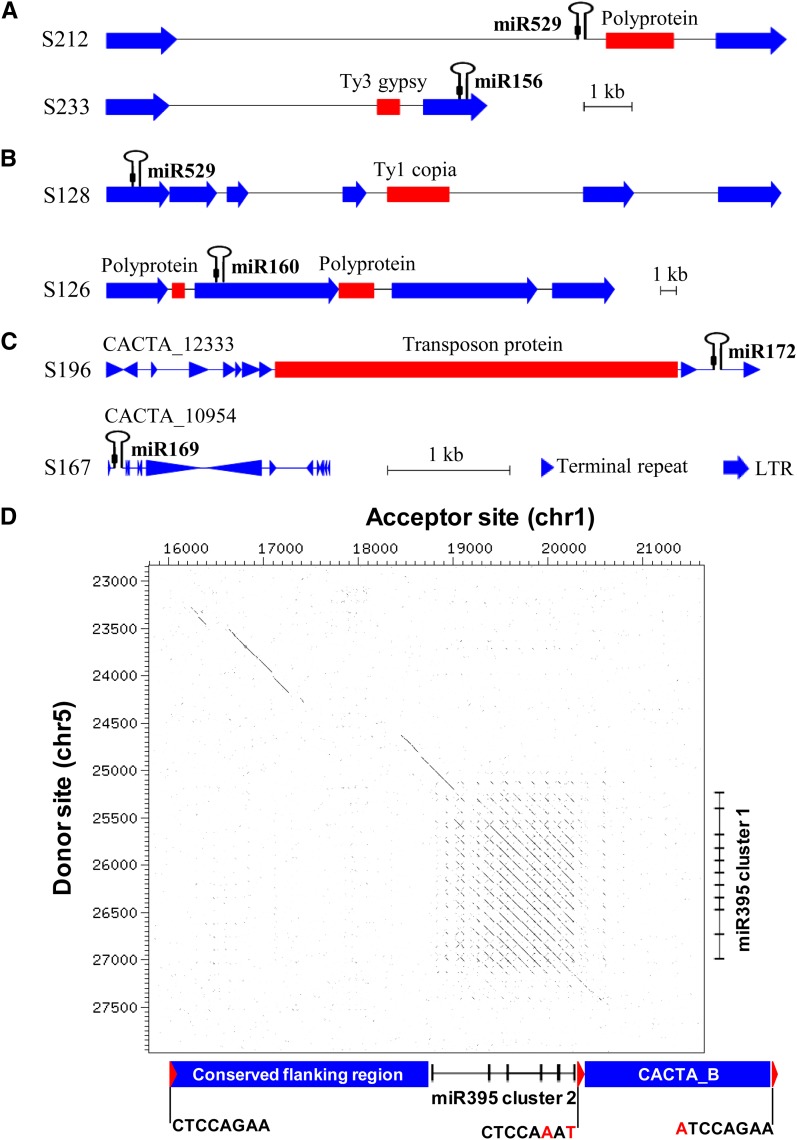
Our data indicate that the conservation of miRNA colinearity was negatively correlated with the number of WGD events as shown by the contrasting genome conservation patterns between monocot and eudicot genomes. The diploidization process resulted in nearly half of the miRNAs locating at nonsyntenic genomic regions. In light of their significant roles in reshuffling protein coding genes (Jiang et al., 2004; Morgante et al., 2005; Paterson et al., 2009; Murat et al., 2010; Wicker et al., 2010), it is conceivable that TEs should contribute substantially to the erosion of miRNA colinearity in grass genomes. To study the effects of TEs on the miRNA genome organization, we extracted the flanking sequences of these miRNAs and searched against TE sequences (see Methods). In sorghum, for example, a total of 66 miRNAs from 15 families were found to be located within TEs (see Supplemental Tables 8 and 9 online). Many of these TEs were retrotransposons, some nested, while others belonged to the DNA transposons, such as CACTA elements. A number of miRNAs may have been carried to their current genomic locations by TEs because they were identified as located either between the two LTRs or even on the LTRs of retrotransposons (Figures 5A to 5C). We found that miRNAs may also be transposed via TE-mediated double-strand break (DSB) events (Kapitonov and Jurka, 2007; Agmon et al., 2009). For instance, two regions of ∼3 kb encompassing two miR395 clusters on Brachypodium chromosomes 1 and 5 were ∼80% identical at the sequence level. The chromosome 5 region in Brachypodium was syntenic between rice, sorghum, and maize (the so-called donor site), whereas the chromosome 1 region was not (the so-called acceptor site; Figure 5D). A typical CACTA element was found immediately adjacent to the 3′ region of the acceptor site. The duplicated region was flanked by three characteristic target sequence duplications (TSDs), indicating an external sequence insertion that was achieved by a DSB event (Wicker et al., 2010).
Figure 5.
Mechanisms of TE-Mediated miRNA Gene Transposition.
(A) to (C) Potential TE-mediated miRNA transposition.
(A) miRNAs located inside the retrotransposon polyprotein region.
(B) miRNAs located on LTRs of nested retrotransposons.
(C) miRNAs carried by CACTA transposon elements.
(D) Blue arrows indicated the long terminal repeat DotPlot alignment showing high similarity between the donor and the acceptor sites that contain two miR395 clusters on Brachypodium chromosomes 5 and 1, respectively. Red arrows indicate TSDs produced upon CACTA insertion. Imperfectly matched bases are in red. The length of CACTA is not in proportion.
Among transposed miRNAs, 17% (11 out of 66 from 15 families) were indeed located in the syntenic regions across all four grasses, indicating that TE-transposed miRNAs already existed in the ancestral genomes (see Supplemental Table 9 online). TEs should contribute significantly to the expansion of underretained miRNA families since the transposition frequency of underretained miRNA families was much higher than those overretained. In sorghum, for example, on average 43.5% underretained miRNAs and 16.3% overretained miRNAs were associated with TEs (Student’s t test P value = 0.024; see Supplemental Figure 3 and Supplemental Table 10 online). The sorghum miR529 and miR172 families are two largest underretained families (15 and 32 genes, respectively) and contained more than 66% (10 out of 15) and 71% (23 out of 32) of TE-transposed members, respectively. One interesting observation was the significantly biased distribution of TE-transposed miRNAs on the paleo-chromosomes. We found more transposed miRNAs on chromosomes A5, A9, A11, A6, and A7/A10 (4, 8, 10, 5, and 0/4, respectively) than on chromosomes A1, A8, A12, A2/A4, and A3 (2, 5, 5, 3/11, and 1, respectively; paired t test P value = 0.0051; see Supplemental Table 11 online). As shown in Figure 3, A1, A8, A12, A2/A4, and A3 belong to the dominant subgenome, whereas A5, A9, A11, A6, and A7/A10 were nondominant in miRNA content. The preferred distribution of transposed miRNAs on the nondominant subgenomes implies a TE-mediated miRNA return flow, probably for counter balancing/compensating for the lost miRNAs on these chromosomes.
DISCUSSION
Erosion of miRNA Gene Synteny by WGD
Most of the investigated eudicots (grape, Arabidopsis, soybean, poplar, and cacao) experienced up to three WGD events, whereas the investigated monocots (rice, maize, sorghum, and Brachypodium) went through only one shared ancestral WGD during their speciation except for maize, which has a recent extra WGD (Salse, 2012). Selective gene elimination and significant genome reorganization are readily detected after each genome polyploidization (Thomas et al., 2006; Schnable et al., 2009; Sankoff et al., 2010; Woodhouse et al., 2010). We observed that on average ∼50% of miRNAs are conserved in orthologous relationships, involving 19 out of 22 families in monocots, significantly higher than those in eudicotyledonous species (∼11% on average), probably resulting from the diploidization-promoted genome reorganization. Conserved miRNAs were in the form of singletons or tandem clusters between species, demonstrating a preexisting pool of miRNAs that have been amplified in the ancestor of the modern grasses. Nevertheless, WGD seems to not significantly affect the total number of conserved miRNA genes, indicating fast miRNA gene death after their birth. This is in contrast with the ever-growing number of novel miRNAs. In Arabidopsis, for instance, within 10 million years of evolution, 33% of the detected species-specific miRNA families have been acquired between two Arabidopsis lineages (thaliana versus lyrata; Khraiwesh et al., 2010), leading to a net gene flux (birth/death) rate estimated at 1.2 to 3.3 miRNAs per million years (Fahlgren et al., 2007). Conserved miRNAs maintained a relatively slow but steady pace of birth and death rate of 0.87 to 1.27 per million years in all four grass species.
miRNA Gene Subgenome Partitioning during Grass Genome Evolution
The inference of ancestral gene content based on the integration of orthologous and paralogous relationships suggests an original founder pool of almost ∼100 miRNAs for the monocot lineage covering 19 of the 22 considered miRNA families. Moreover, reconstruction of the evolutionary scenario that shaped the modern genome miRNA gene content from the founder pool shows a significant difference in miRNA content as well as conservation between the duplicated chromosomes derived from the same ancestral protochromosome. Our observation about the miRNA retention following WGD reinforces the polyploidy subgenome dominance hypothesis, where the retention of duplicated genes is considered not random, but structurally favored on one subgenome that then became dominant in term of gene content compared with the other. These conclusions complement recent studies describing biased patterns in duplicate gene retention (some are miRNA targets) following genome duplications in multiple eukaryotic genomes, including several grass genomes, such as rice (Freeling, 2009; Throude et al., 2009) and maize (Schnable et al., 2010). We found in this study potential return flows of TE-mediated miRNAs to the nondominant subgenomes that may compensate for the loss of miRNAs caused by diploidization. Such a counterbalance in miRNA content may be selected to improve adaptation.
miRNA Paleohistory Supports the Gene Balance Hypothesis
Numerous miRNAs discovered in plants have been shown to target TFs/TRs as well as genes involved in abiotic and biotic stress response and hormone signaling (Willmann and Poethig, 2007). The observed biased retention of duplicated miRNAs, depending on the function of the targeted genes, is in favor of the gene dosage balance hypothesis in which maintaining proper network balance/relative dosage (e.g., regulatory or signaling networks) or stoichiometry of protein complexes is considered to be vital for normal cellular function. Loss of such a duplicated gene would likely result in a regulatory network imbalance and consequently is preferentially maintained after WGD. Similarly, miRNA families with overretained members are important transcription/translation regulators that should be dosage sensitive like TFs and are hence resistant to elimination during diploidization. The high percentage of TFs among overretained targets of overretained miRNA families suggests that such coretention of miRNAs and their targets is important for gene regulation during genome diploidization and is indeed sensitive to gene dosage changes. Thus, the miRNA paleohistory is in support of the gene balance hypothesis (Birchler and Veitia, 2010). In other words, the existence of coloss or coretention of miRNAs and target genes may result in innovative miRNA–target interactions that are pivotal for adaptive response to various environmental stimuli, in addition to maintaining a constant set of miRNAs for basic biological functions.
TEs as One of the Major Forces for Genomic Reorganization of miRNA Genes
Although WGD, segmental, and tandem gene duplication dominate the expansion mechanisms of miRNA families (Maher et al., 2006), our data show that TE-mediated transpositional mechanisms may play important roles in shaping the genome organization of miRNAs. A significant number of miRNAs were found to be closely associated with TEs and may be carried around in the grass genomes. Noncolinear miRNAs may also be transposed by additional approaches such as repeat and/or TE-mediated DSB mechanisms. For example, CACTA-like elements are the predominant DNA transposons in sorghum, constituting ∼4.7% of the genome, and are frequently observed to relocate genes and gene fragments (Paterson et al., 2009). Although infrequently, the involvement of a CACTA in translocating a cluster of miR395 genes into a nonsyntenic region through the DSB mechanism was observed in Brachypodium. TE-mediated miRNA transposition may also contribute to the expansion of miRNA gene families, especially in plants with large genome sizes where TEs are the major component of repetitive sequences. Furthermore, TEs may mediate miRNA return flows to the nondominant subgenomes derived from the selective deletion during diploidization. Such compensation may be taken to establish novel gene dosage balance under particular adaptation conditions. Therefore, TE-associated translocation is one of the major forces for miRNA mobilization and amplification. Since the percentage of nonsyntenic miRNAs reached nearly 50% of the total miRNA genes, it is conceivable that additional mechanisms (e.g., rearrangements, inversions, nonreciprocal translocations, etc.) may also contribute to the miRNA gene reshuffling in the grass genomes, which may need further investigation.
Evolutionary Manipulation of miRNA Genes is Essential for Plant Adaptation
The functions of miRNA/target networks in determining the plant developmental plasticity have been widely reported (Rubio-Somoza et al., 2009). Our analysis suggests that conserved, duplication-resistant miRNA families were associated with targets enriched in GO BP terms “transcription regulation” and “metabolic processes,” suggesting that they are involved in essential biological pathways. For instance, the miR172/AP2 (APETALA2) interaction has been shown to play important roles in the vegetative/flowering transition phase (Aukerman and Sakai, 2003; Mlotshwa et al., 2006; Usami et al., 2009). In addition, miR164-319 are essential for senescence through ETHYLENE INSENSITIVE, NAC, and TCP (for Teosinte branched 1, Cycloideae, Pcf) gene expression regulation (Guo et al., 2005; Schommer et al., 2008; Kim et al., 2009). Also, miR159-160-164-166-167-390 seem to acquire new functions through target genes (such as MYB and auxin response factor) from tip growth in mosses to leaf and root vascular patterning and organ polarity in eudicots (Mallory et al., 2004a, 2004b; Kim et al., 2005; Rubio-Somoza et al., 2009; Yoon et al., 2010). Most of these miRNA genes are prone to diploidization and are mainly involved in the basic regulation of plant development, such as rice architecture in Jiao et al. (2010), maize leaf polarity in Juarez et al. (2004), Arabidopsis vascular development in Kim et al. (2005), and leaf margin serration in Paterson et al. (2004). By contrast, conserved and maintained/retained miRNA/target networks may play crucial roles in adaptation to biotic and/or abiotic stresses, such as nutrient deficiency response (miR169 versus NFYA TF; Zhao et al., 2011), sulfate accumulation and allocation (miR395 versus APS/SULTR2-1; Liang et al., 2010), phosphate starvation response (miR399 versus a ubiquitin conjugating enzyme; Fujii et al., 2005), oxidative stress tolerance (miR398 versus Cu/Zn superoxide dismutase; Sunkar et al., 2006), and defense response (miR393 versus auxin-mediated pathogen elicitors; Navarro et al., 2006). Thus, these overretained miRNAs render plants with more robust adaptive traits and the mechanism may be useful for modern crop variety improvements.
Model of miRNA/Target Network Evolution and Conservation in Plants
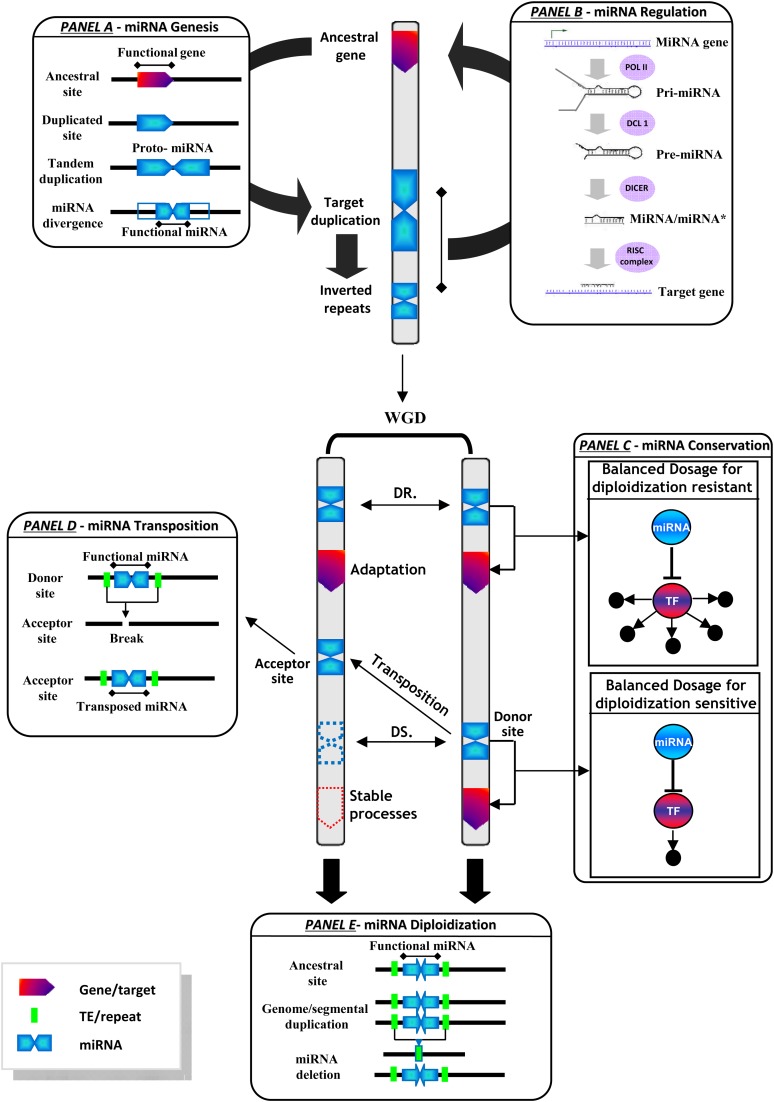
Based on our data together with previous studies, we propose a model in Figure 6 illustrating the conclusions raised in this article regarding miRNA evolution via conservation/mobility, polyploidization, and derived impact upon targets and associated traits. miRNAs are known to be first generated randomly either through gene duplication, transcriptionally based exonization, or TE-based origin (gene duplication exemplified in Figure 6A; Allen et al., 2004). Once such inverted repeat structure obtains the capacity to be expressed, the resultant hairpin product acts as a substrate for Dicer-like enzyme complexes to generate a mature miRNA and finally regulates target gene expression (Figure 6B). Whereas miRNAs in plants have an ancestral origin (Figure 6A), the identification of nonconserved or even species-specific miRNAs suggests that miRNA biogenesis is a constant phenomenon during evolution. We propose that WGD should be the major mechanism for generation of miRNA genes that are differentially conserved or lost during the diploidization process depending on the target gene function (Figure 6C), compounded by TE-mediated transposition (Figure 6D) or deletion (Figure 6E), to improve plant adaptation. Meanwhile, species-specific, or non-conserved miRNAs (Figure 6C), arise continually probably in response to adaptation to changing environments, although they are often weakly expressed, processed imprecisely, and highly divergent, suggesting an on-going maturation process under neutral evolution and, maybe, selection (Cuperus et al., 2011). Therefore, the modern plant miRNA repertoire should consist of ancestrally conserved, transposed, as well as recently spawned novel miRNA loci. Overall, our analysis of miRNA paleohistory demonstrates miRNA gene evolution by a WGD event following the subgenome dominance and gene dosage balance theories. The functional partition mechanism of overretained miRNAs may provide valuable references for miRNA-related modern grass genome manipulation that will lead to more robust crop varieties keeping in mind the important roles of miRNAs to plants.
Figure 6.
Evolutionary Model of miRNA Genesis, Regulation, Conservation, and Transposition in Plants.
Major conclusions are schematically illustrated in five panels regarding miRNA (1) genesis through gene duplication and sequence divergence (A), (2) regulation of targeted gene via the formation of primary miRNA and pre-miRNA and miRNA/miRNA* complexes (B), (3) conservation (C) or transposition (D) following the gene dosage hypothesis, and (4) evolution via diploidization following the subgenome dominance process (E). miRNAs (blue arrow), protein-coding genes (red arrow), and TEs (green box) are shown on a single ancestral chromosome (top black bar) and derived duplicated (bottom black bars) chromosomes after a WGD. DR, diploidization resistant; DS, diploidization sensitive.
METHODS
Plant Genome Sequences
Genome pseudomolecules of four monocot and four eudicot genomes were downloaded from the Phytozome (http://www.phytozome.net/) website of the Joint Genome Institute, including rice (Oryza sativa; 12 chromosomes, 372 Mb, 41,046 genes; International Rice Genome Sequencing Project, 2005), sorghum (Sorghum bicolor; 10 chromosomes, 659 Mb, 34,008 genes; Jackson et al., 2009), maize (Zea mays; 10 chromosomes, 2365 Mb, 32,540 genes; Schnable et al., 2009), Brachypodium distachyon (five chromosomes, 271 Mb, 25,504 genes; International Brachypodium Initiative, 2010), grape (Vitis vinifera; 19 chromosomes, 302 Mb, 21,189 genes; Jaillon et al., 2007), poplar (Populus trichocarpa; 19 chromosomes, 294 Mb, 30,260 genes; Tuskan et al., 2006), Arabidopsis thaliana (five chromosomes, 119 Mb, 33,198 genes; Arabidopsis Genome Initiative, 2000), soybean (Glycine max; 20 chromosomes, 949 Mb, 46,195 genes; Schmutz et al., 2010). The cacao gene (10 chromosomes, 218 Mb, 27,814 genes) was used as described in Argout et al., 2011.
Genome Synteny and Duplication Analysis
Plant genome synteny and duplication patterns were used as described (Salse et al., 2008, 2009a; Abrouk et al., 2010; Murat et al., 2010). Briefly, three sequence alignment parameters were used for the synteny and duplication analysis. These parameters are alignment length (AL), cumulative identity percentage, and cumulative alignment length percentage, which can improve the stringency and significance of BLAST (Altschul et al., 1990) sequence alignment by parsing BLASTN results and rebuilding high scoring pairs (HSPs). The AL parameter corresponds to the sum of all HSP lengths. The cumulative identity percentage [(Σ ID by HSP/AL) × 100] corresponds to the cumulative percentage of identity observed for all HSPs divided by cumulative AL. The cumulative alignment length percentage [AL/query length], is the sum of the HSPs lengths (AL) for all HSPs divided by the length of the query sequence. These parameters allow the identification of the best alignment with the highest cumulative percentage of identity in the longest cumulative length for improving the conservation between two compared sequences.
Then, the orthologous or paralogous pairs identified were statistically validated based on two criteria, which were density ration (DR) and the cluster ration (CR), taking into account the physical size (Size), number of annotated genes (Gnumber), and number of orthologous or paralogous couples (Cnumber) in the regions. DR [((Size1 + Size2)/(2 × Cnumber)) × 100] considers the number of links between two regions (duplicated or syntenic) as a function of the size of the considered blocks. CR [(2 × Cnumber)/(Gnumber1 + Gnumber2)] considers the number of the links between two regions as a function of the number of annotated genes available in considered blocks. The remaining collinear or duplicated regions were considered as artificially obtained at random considering the number of links between two regions characterized by a physical size and number of annotated genes available.
miRNA genes and Target Detection
miRNAs were identified in nine plant genomes using MIReNA software (Mathelier and Carbone, 2010) using a total of 259 mature miRNAs (141 from rice and 118 from grape) from the 22 most conserved plant miRNA families in the miRBase database (release 16, September, 2010) as reference. We set the parameters in MIReNA for detecting miRNAs following the criteria established for the plant miRNA annotation (Ambros, 2004; Meyers et al., 2008), including fewer than four mismatches between miRNA/miRNA*, no bulges in miRNA/miRNA* larger than two bases, and fewer than four mismatches between detected miRNA and the miRNA reference. MIReNA software considers the previous criteria and is structured in four main steps for the identification of novel miRNAs: (1) align known miRNAs on the pseudomolecules and extract the positions of the different matches, (2) extend sequence on each side of the match and secondary structure prediction by RNAfold, and (3) calculate the percentage of unmatched nucleotides between miRNA and miRNA*, the adjusted minimum folding energy (AMFE), and minimum free energy index (MFEI) (Zhang et al., 2006). AMFE [(MFE / L) × 100] was calculated with minimum free energy and the pre-miRNA sequence length (L), while the MFEI was the quotient of AMFE divided by %GC (the percentage of GC composition of the pre-miRNA). For plant miRNA detection, the parameters were set as the percentage of mismatch between miRNA and miRNA* <26%, AMFE <−32, and MFEI <−0.85. (4) The fourth step is to remove sequences overlapping with coding sequences or TEs (using RepeatMasker). miRNA targets have been identified and associated with a single related miRNA using miRanda software (http://www.microrna.org), when both sequences aligned with >90% identity and >90% of the considered miRNA length (i.e., one to two mismatches allowed).
Determination of miRNA gene Orthology/Paralogy and Ancestral Content
Gene synteny and duplication between and within species were determined previously (Salse et al., 2008, 2009a; Abrouk et al., 2010; Murat et al., 2010). After locating the detected miRNAs to the corresponding pseudomolecules, the gene synteny flanking the miRNAs was referenced between the species. miRNA genes of the same family on the same syntenic region were considered interspecific orthologs or intraspecific paralogs. Identified miRNAs were considered as ancestral when they have been identified as conserved between at least two genomes at orthologous positions. Then, information about the ancestral shared duplications (Salse et al., 2008, 2009a; Abrouk et al., 2010; Murat et al., 2010) was used to eliminate redundancy and to deduce the ancestral miRNA gene content.
Target Gene GO Enrichment Analysis
The functional enrichment of the miRNA targets was performed using the BinGO software (Maere et al., 2005b), and the Cytoscape plugin (Kohl et al., 2011) was used to display the GO hierarchy tree. For enrichment P value calculation (at a significance level of <0.05), the hypergeometric test method was applied. For multiple hypothesis testing, FDR correction of the Benjamini and Hochberg method was used to reduce false negatives (Benjamini et al., 2001). The GO annotation of target genes is available from the agriGO website (http://bioinfo.cau.edu.cn/agriGO/).
Identification of TE-Mediated miRNA Transposition
To locate the miRNA position relative to TEs, miRNA precursors were searched against repeat sequences using BLASTN. miRNAs located inside a TE were considered “TE carried.” Sorghum repeats were downloaded from the Munich Information Center for Protein Sequences (ftp://ftpmips.helmholtz-muenchen.de/plants/sorghum/). Maize repeat sequences were downloaded from http://ftp.maizesequence.org/current/repeats/ and rice from The Institute for Genomic Research. The Brachypodium repeat data set was used as described in the International Brachypodium Initiative (2010). For DSB analysis, miRNA genes with >70% identity were considered duplicates with each other. Interspecific syntenic regions were considered as donor sites, whereas nonsyntenic regions were acceptor sites. miRNA genes were considered to be transferred from a donor sites to an acceptor sites by a copy-and-paste process if the flanking regions also exhibited high sequence similarity (Wicker et al., 2010). To identify the precise border of the duplicated regions, sequences of 20 kb from each side of a miRNA gene were extracted and aligned using DOTTER (Sonnhammer and Durbin, 1995). In the meantime, they were searched for repeat elements. A duplicated region containing a miRNA or miRNA cluster with a juxtaposed TE encompassed by two TSDs was considered as a TE-mediated DSB event.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of miRBase and MIReNA Data Sets.
Supplemental Figure 2. Positive Correlation between Overretained miRNA Families and the Number of Target Genes.
Supplemental Figure 3. Transposition Frequency of miRNA Families.
Supplemental Table 1. Conserved and Duplicated miRNA Genes Identified in Monocots.
Supplemental Table 2. Conserved and Duplicated miRNA Genes Identified in Eudicots.
Supplemental Table 3. Overretention Frequency of miRNA Families in Rice, Brachypodium, Sorghum, and Maize.
Supplemental Table 4. Overretained miRNA Families in Rice, Brachypodium, Sorghum, and Maize.
Supplemental Table 5. Underretained miRNA Families in Rice, Brachypodium, Sorghum, and Maize.
Supplemental Table 6. Number of miRNA Targets in Rice, Brachypodium, Sorghum, and Maize.
Supplemental Table 7. miRNA Target Gene Enrichment Analysis in GO Biological Process Categories.
Supplemental Table 8. Sorghum miRNAs Colocalizing with CACTA TEs.
Supplemental Table 9. Sorghum miRNAs Colocalizing with Retrotransposons.
Supplemental Table 10. Overretained miRNA Families with Lower Transposition Frequency.
Supplemental Table 11. Preferential Distribution of Transposed miRNAs between Duplicated Ancient Chromosome Pairs.
Supplemental Data Set 1. miRNA Catalog Characterized in Monocots (First Sheet) and Eudicots (Second Sheet).
Supplementary Material
Acknowledgments
This work has been supported by grants from the Agence Nationale de la Recherche (Program ANRjc-PaleoCereal, reference: ANR-09-JCJC-0058-01 and Program ANR Blanc-PAGE, reference: ANR-2011-BSV6-00801) to J.S. and the National High Tech Program (863) from the Ministry of Science and Technology of China (2009AA02Z307 and 2012AA10A308) to L.M.
AUTHOR CONTRIBUTIONS
J.S. and L.M. designed the experiments. M.A., R.Z., F.M., A.L., and C.P. performed the analysis and contributed to the article preparation. J.S. and L.M. wrote the article and contributed equally to this work.
Glossary
- AGK
ancestral grass karyotype
- WGD
whole-genome duplication
- mya
million years ago
- TF
transcription factor
- TR
transcription regulator
- miRNA
microRNA
- pre-miRNA
precursor miRNA
- TE
transposable element
- CDS
coding sequence
- GO
Gene Ontology
- BP
biological process
- FDR
false discovery rate
- DSB
double-strand break
- TSD
target sequence duplication
- AL
alignment length
- HSP
high scoring pair
- AMFE
adjusted minimum folding energy
- MFEI
minimum free energy index
References
- Abrouk M., Murat F., Pont C., Messing J., Jackson S., Faraut T., Tannier E., Plomion C., Cooke R., Feuillet C., Salse J. (2010). Palaeogenomics of plants: Synteny-based modelling of extinct ancestors. Trends Plant Sci. 15: 479–487 [DOI] [PubMed] [Google Scholar]
- Agmon N., Pur S., Liefshitz B., Kupiec M. (2009). Analysis of repair mechanism choice during homologous recombination. Nucleic Acids Res. 37: 5081–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36: 1282–1290 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ambros V. (2004). The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arazi T., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C. (2005). Cloning and characterization of micro-RNAs from moss. Plant J. 43: 837–848 [DOI] [PubMed] [Google Scholar]
- Argout X., et al. (2011). The genome of Theobroma cacao. Nat. Genet. 43: 101–108 [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17: 1658–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S., Pasquinelli A.E. (2006). Identification and analysis of microRNAs. Genet. Eng. (N.Y.) 27: 1–20 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125: 279–284 [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. (2007). The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 19: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. (2010). The gene balance hypothesis: Implications for gene regulation, quantitative traits and evolution. New Phytol. 186: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Riddle N.C., Auger D.L., Veitia R.A. (2005). Dosage balance in gene regulation: Biological implications. Trends Genet. 21: 219–226 [DOI] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolot S., Abrouk M., Masood-Quraishi U., Stein N., Messing J., Feuillet C., Salse J. (2009). The ‘inner circle’ of the cereal genomes. Curr. Opin. Plant Biol. 12: 119–125 [DOI] [PubMed] [Google Scholar]
- Chang P.L., Dilkes B.P., McMahon M., Comai L., Nuzhdin S.V. (2010). Homoeolog-specific retention and use in allotetraploid Arabidopsis suecica depends on parent of origin and network partners. Genome Biol. 11: R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Fahlgren N., Carrington J.C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D., Zhang L., Wang H., Liu Z., Zhang Z., Zheng Y. (2009). Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. (Lond.) 103: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger P.P., Pires J.C. (2009). Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17: 699–717 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Carrington J.C. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (2009). Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60: 433–453 [DOI] [PubMed] [Google Scholar]
- Freeling M., Thomas B.C. (2006). Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16: 805–814 [DOI] [PubMed] [Google Scholar]
- Fujii H., Chiou T.J., Lin S.I., Aung K., Zhu J.K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Guo H.S., Xie Q., Fei J.F., Chua N.H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jaillon O., et al. French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., Qian Q., Li J. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544 [DOI] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. (2007). Helitrons on a roll: Eukaryotic rolling-circle transposons. Trends Genet. 23: 521–529 [DOI] [PubMed] [Google Scholar]
- Khraiwesh B., Arif M.A., Seumel G.I., Ossowski S., Weigel D., Reski R., Frank W. (2010). Transcriptional control of gene expression by microRNAs. Cell 140: 111–122 [DOI] [PubMed] [Google Scholar]
- Kim J., Jung J.H., Reyes J.L., Kim Y.S., Kim S.Y., Chung K.S., Kim J.A., Lee M., Lee Y., Narry Kim V., Chua N.H., Park C.M. (2005). microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 42: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Woo H.R., Kim J., Lim P.O., Lee I.C., Choi S.H., Hwang D., Nam H.G. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kohl M., Wiese S., Warscheid B. (2011). Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 696: 291–303 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39(Database issue): D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li C., Xia J., Jin Y. (2011). Domestication of transposable elements into microRNA genes in plants. PLoS ONE 6: e19212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Yang F., Yu D. (2010). MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 62: 1046–1057 [DOI] [PubMed] [Google Scholar]
- Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P. (2003). Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y. (2005a). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005b). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Maher C., Stein L., Ware D. (2006). Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 16: 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Dugas D.V., Bartel D.P., Bartel B. (2004a). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14: 1035–1046 [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. (2004b). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A., Carbone A. (2010). MIReNA: Finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics 26: 2226–2234 [DOI] [PubMed] [Google Scholar]
- Meyers B.C., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S., Yang Z., Kim Y., Chen X. (2006). Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol. Biol. 61: 781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Brunner S., Pea G., Fengler K., Zuccolo A., Rafalski A. (2005). Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 37: 997–1002 [DOI] [PubMed] [Google Scholar]
- Murat F., Xu J.H., Tannier E., Abrouk M., Guilhot N., Pont C., Messing J., Salse J. (2010). Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 20: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Paterson A.H., Bowers J.E., Chapman B.A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Pont C., Murat F., Confolent C., Balzergue S., Salse J. (2011). RNA-seq in grain unveils fate of neo- and paleopolyploidization events in bread wheat (Triticum aestivum L.). Genome Biol. 12: R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. (2002). MicroRNAs in plants. Genes Dev. 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.L., Chua N.H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49: 592–606 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I., Cuperus J.T., Weigel D., Carrington J.C. (2009). Regulation and functional specialization of small RNA-target nodes during plant development. Curr. Opin. Plant Biol. 12: 622–627 [DOI] [PubMed] [Google Scholar]
- Salse J. (2012). In silico archeogenomics unveils modern plant genome organisation, regulation and evolution. Curr. Opin. Plant Biol. 15: 122–130 [DOI] [PubMed] [Google Scholar]
- Salse J., Abrouk M., Bolot S., Guilhot N., Courcelle E., Faraut T., Waugh R., Close T.J., Messing J., Feuillet C. (2009a). Reconstruction of monocotelydoneous proto-chromosomes reveals faster evolution in plants than in animals. Proc. Natl. Acad. Sci. USA 106: 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J., Abrouk M., Murat F., Quraishi U.M., Feuillet C. (2009b). Improved criteria and comparative genomics tool provide new insights into grass paleogenomics. Brief. Bioinform. 10: 619–630 [DOI] [PubMed] [Google Scholar]
- Salse J., Bolot S., Throude M., Jouffe V., Piegu B., Quraishi U.M., Calcagno T., Cooke R., Delseny M., Feuillet C. (2008). Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D., Zheng C., Zhu Q. (2010). The collapse of gene complement following whole genome duplication. BMC Genomics 11: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M. (2010). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C., Gehring C. (2004). Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 20: 461–464 [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L., Durbin R. (1995). A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–GC10 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Kapoor A., Zhu J.K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H. (2008a). Synteny and collinearity in plant genomes. Science 320: 486–488 [DOI] [PubMed] [Google Scholar]
- Tang H., Wang X., Bowers J.E., Ming R., Alam M., Paterson A.H. (2008b). Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18: 1944–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throude M., et al. (2009). Structure and expression analysis of rice paleo duplications. Nucleic Acids Res. 37: 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y., Zamore P.D. (2005). MicroRNA biogenesis: Drosha can’t cut it without a partner. Curr. Biol. 15: R61–R64 [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Usami T., Horiguchi G., Yano S., Tsukaya H. (2009). The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Fawcett J.A., Proost S., Sterck L., Vandepoele K. (2009). The flowering world: A tale of duplications. Trends Plant Sci. 14: 680–688 [DOI] [PubMed] [Google Scholar]
- Wang X., et al. Brassica rapa Genome Sequencing Project Consortium (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Wicker T., Buchmann J.P., Keller B. (2010). Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res. 20: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann M.R., Poethig R.S. (2007). Conservation and evolution of miRNA regulatory programs in plant development. Curr. Opin. Plant Biol. 10: 503–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M.R., Schnable J.C., Pedersen B.S., Lyons E., Lisch D., Subramaniam S., Freeling M. (2010). Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 8: e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu T., Tian C., Sun S., Li J., Chen M. (2005). Transcription factors in rice: A genome-wide comparative analysis between monocots and eudicots. Plant Mol. Biol. 59: 191–203 [DOI] [PubMed] [Google Scholar]
- Yoon E.K., Yang J.H., Lim J., Kim S.H., Kim S.K., Lee W.S. (2010). Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 38: 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pan X., Cannon C.H., Cobb G.P., Anderson T.A. (2006). Conservation and divergence of plant microRNA genes. Plant J. 46: 243–259 [DOI] [PubMed] [Google Scholar]
- Zhang L., Chia J.M., Kumari S., Stein J.C., Liu Z., Narechania A., Maher C.A., Guill K., McMullen M.D., Ware D. (2009). A genome-wide characterization of microRNA genes in maize. PLoS Genet. 5: e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ge L., Liang R., Li W., Ruan K., Lin H., Jin Y. (2009). Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Liang R., Ge L., Li W., Xiao H., Lin H., Ruan K., Jin Y. (2007). Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 354: 585–590 [DOI] [PubMed] [Google Scholar]
- Zhao M., Ding H., Zhu J.K., Zhang F., Li W.X. (2011). Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 190: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Li G., Mi S., Li S., Hannon G.J., Wang X.-J., Qi Y. (2007). A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 21: 1190–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.