An Arabidopsis thaliana abscisic acid (ABA) overly sensitive mutant, abo6, was identified, and ABO6 was found to encode a DEXH box RNA helicase that regulates the splicing of genes of complex I in mitochondria. ABA and auxin signaling are shown to regulate primary root growth and seed germination via reactive oxygen species produced in mitochondria.
Abstract
It is well known that abscisic acid (ABA) promotes reactive oxygen species (ROS) production through plasma membrane–associated NADPH oxidases during ABA signaling. However, whether ROS from organelles can act as second messengers in ABA signaling is largely unknown. Here, we identified an ABA overly sensitive mutant, abo6, in a genetic screen for ABA-mediated inhibition of primary root growth. ABO6 encodes a DEXH box RNA helicase that is involved in regulating the splicing of several genes of complex I in mitochondria. The abo6 mutant accumulated more ROS in mitochondria, as established using a mitochondrial superoxide indicator, circularly permuted yellow fluorescent protein. Two dominant-negative mutations in ABA insensitive1 (abi1-1) and abi2-1 greatly reduced ROS production in mitochondria. The ABA sensitivity of abo6 can also be compromised by the atrbohF mutation. ABA-mediated inhibition of seed germination and primary root growth in abo6 was released by the addition of reduced GSH and exogenous auxin to the medium. Expression of auxin-responsive markers ProDR5:GUS (for synthetic auxin response element D1-4 with site-directed mutants in the 5′-end from soybean):β-glucuronidase) and Indole-3-acetic acid inducible2:GUS was greatly reduced by the abo6 mutation. Hence, our results provide molecular evidence for the interplay between ABA and auxin through the production of ROS from mitochondria. This interplay regulates primary root growth and seed germination in Arabidopsis thaliana.
INTRODUCTION
One of the most important traits of a crop plant is the root system. The root system determines the efficiency of nutrient and water uptake from soil and influences the distribution of photosynthates throughout the plant. Root growth is regulated by hormones and environmental conditions (Teale et al., 2008; Fukaki and Tasaka, 2009). The phytohormone abscisic acid (ABA) plays crucial roles in regulating root growth and also helps regulate seed dormancy and germination, stomatal movement, vegetative growth, and responses to biotic and abiotic stress (Zhu, 2002; Ton et al., 2009; Cutler et al., 2010; Kim et al., 2010). Although ABA is required for root growth and ABA-deficient mutants have reduced root systems (Xiong and Zhu, 2003), high concentrations of exogenous ABA inhibit root growth. Our understanding of how ABA stimulates or inhibits seed germination, seedling growth, and stomatal movement has been greatly enhanced by genetic and molecular analyses; however, our understanding remains incomplete (Zhu, 2002; Chinnusamy et al., 2008; Cutler et al., 2010; Kim et al., 2010).
ABA stimulates the production of reactive oxygen species (ROS; including O2− and hydrogen peroxide [H2O2]) through plasma membrane–localized NADPH oxidases (Kwak et al., 2003). These ROS act as important second messengers in regulating root growth, stomatal movement, and seed germination in the ABA signaling pathway (Kwak et al., 2003). Mutations in two NADPH oxidases, respiratory burst oxidase homologs D and F (RBOHD/F), reduce ROS production, and Arabidopsis thaliana rbohF single or rbohD rbohF double mutants are insensitive to ABA-mediated inhibition of root growth (Kwak et al., 2003). The ABA-activated SnRK2 protein kinase OPEN STOMATA1 (OST1) interacts with and phosphorylates RBOHF, suggesting that RBOHF is regulated by OST1 (Sirichandra et al., 2009). The activities of ABA INSENSITIVE1 (ABI1) and ABI2, two critical negative regulators in the ABA signaling pathway, are inhibited by the H2O2 in vitro (Meinhard and Grill, 2001; Meinhard et al., 2002). ABI2 interacts with GPX3 (the glutathione peroxidase 3 that regulates the redox state of guard cells) (Miao et al., 2006), and ABI1 binds to phosphatidic acid produced by phospholipase Dα1 (Zhang et al., 2004; Zhang et al., 2009). Phosphatidic acid interacts with and stimulates RBOHD/F for ROS production (Zhang et al., 2009). A mutation in RBOHC results in ROS reduction, inhibition of root hair growth, and reduced activities of Ca2+ channels in root hairs, suggesting that ROS regulate plant cell expansion by activating Ca2+ channels (Foreman et al., 2003). A recent study on a plasma membrane–associated Pro-rich extensin-like receptor kinase 4 also suggests that Ca2+ is important for root growth in the ABA signaling pathway (Bai et al., 2009). In both the rbohC mutant and the rbohD rbohF double mutant, the root lengths are greatly reduced, suggesting that ROS are required for plant root growth (Foreman et al., 2003; Kwak et al., 2003). A recent study on ROS production as mediated by the basic helix-loop-helix transcriptional factor UPBEAT1 indicates that ROS control the transition from cell proliferation to differentiation in roots via a separate pathway that involves auxin signaling (Tsukagoshi et al., 2010). ROS are also produced by fatty acid β-oxidation and glycolate oxidases in peroxisomes and by electron transport in the chloroplasts and mitochondria (Laloi et al., 2004). The biological roles of these ROS in the ABA signaling pathway are largely unknown.
In the mitochondrial electron transport chain, complex I and III are major sites for ROS production in darkness and in tissues lacking chloroplasts (Laloi et al., 2004). Impairment of the components in this chain leads to the accumulation of ROS in mitochondria. Because the mitochondrial electron transport chain also contains an alternative oxidase that can be activated under stress to remove toxic ROS (Maxwell et al., 1999), ROS produced from mitochondria are present only in low concentrations in green tissues (Apel and Hirt, 2004).
Most mitochondrial proteins are encoded by nuclear genes, and only a small number are encoded by mitochondrial genes. The mitochondrial pre-RNAs require processing to become mature RNAs. The processing of mitochondrial RNAs, including intron splicing and RNA editing, requires the help of many nuclear proteins, such as DEAD box RNA helicases (Köhler et al., 2010) and pentatricopeptide repeat (PPR) proteins (Liu et al., 2010). DEAD/DExH box RNA helicases, which require the help of other proteins and ATP hydrolysis, act as molecular motors that rearrange inter- or intramolecular RNA secondary structures or that dissolve RNA-protein complexes. Only a few RNA helicases have been analyzed in Arabidopsis. LOW EXPRESSION OF OSMOTICALLY RESPONSIVE GENES4 is required for mRNA export from the nucleus to the cytoplasm (Gong et al., 2002, 2005), and STRESS-RESPONSE SUPPRESSOR1 (STRS1) and STRS2 are involved in the response to many abiotic stresses (Kant et al., 2007). Another RNA helicase, PUTATIVE MITOCHONDRIAL RNA HELICASE2, is involved in group II intron splicing in mitochondria (Köhler et al., 2010). Mutation in the RNA helicase SIZE EXCLUSION LIMIT1 leads to increased ROS- and plasmodesmata-mediated cell–cell transport and embryo death (Stonebloom et al., 2009). However, the molecular roles of most of these RNA helicases are unknown.
To study the molecular mechanisms by which ABA regulates primary root growth, we performed a genetic screen in which we identified mutants that are sensitive or insensitive to ABA inhibition of primary root growth. As reported previously, we identified several ABA-overly sensitive (abo) mutants for ABA inhibition of primary root growth (Yin et al., 2009; Zhou et al., 2009; Liu et al., 2010; Ren et al., 2010; Wang et al., 2011). Among them, ABO5 encodes a PPR protein required for cis-splicing of mitochondrial nad2 intron 3 (Liu et al., 2010). In this study, we characterized another abo mutant, abo6. ABO6 encodes a DEXH box RNA helicase that is localized in the mitochondria and is required for splicing of several genes in complex I. abo6 mutants accumulated more ROS than the wild type in the mitochondria, while ABA treatment enhanced ROS accumulation, and impairing ABA signaling by the abi1-1 or abi2-1 dominant-negative mutation reduced ROS accumulation, as indicated by a mitochondrial superoxide indicator circularly permuted yellow fluorescent protein (cpYFP) (Wang et al., 2008). The addition of reduced GSH (a reducing chemical) to the medium rescued ABA inhibition of primary root growth, while addition of the GSH inhibitor buthionine sulphoximine (BSO) enhanced the ABA inhibition of primary root growth. Further analyses suggested that ABA-mediated inhibition of primary root growth involves auxin homeostasis. Together, these results indicate that, like the ROS produced by plasma membrane NADPH oxidases, the ROS produced by mitochondria are important second messengers in the ABA signaling pathway.
RESULTS
Isolation of abo6
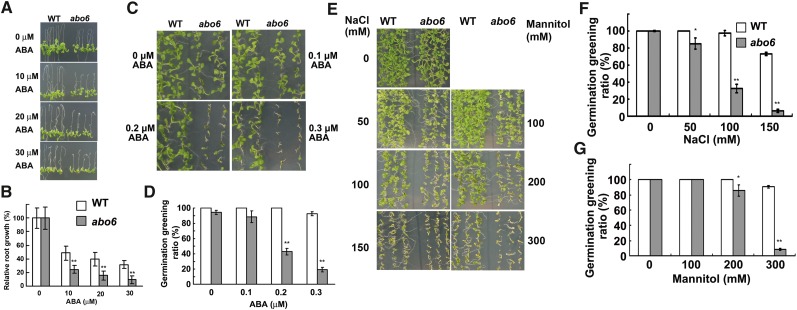
abo6 was isolated during a genetic screen for ABA-sensitive mutants; the screen used a root-bending assay on Murashige and Skoog (MS) medium containing 30 μM ABA (Yin et al., 2009). We compared the root growth of 4-d-old wild-type and abo6 seedlings transferred onto MS medium supplemented with different concentrations of ABA for 7 d. As shown in Figures 1A and 1B, although abo6 had shorter primary roots than the wild type on MS medium without ABA (see Supplemental Figure 1 online), primary root growth was more sensitive in abo6 than in the wild type to ABA (Figure 1A). In addition, seed germination greening (greening cotyledon ratio) was more sensitive in abo6 than in the wild type to ABA (Figures 1C and 1D) and to NaCl and mannitol (Figures 1E to 1G). abo6 plants were later flowering than the wild type (see Supplemental Figure 1 online).
Figure 1.
Primary Root Growth and Seed Germination of abo6 and the Wild Type.
(A) Primary root growth of seedlings grown on MS medium or MS medium supplemented with 10, 20, and 30 μM ABA. Four-day-old seedlings were transferred to the different media and grown for 7 d before being photographed. WT, the wild type.
(B) Relative primary root growth of abo6 or wild-type seedlings grown on medium containing different concentrations of ABA (plates were oriented vertically with root tips down). Values are means ± se (growth without ABA was set to 100%). About 10 roots were measured in each of three plates, and three independent experiments were performed. Data are means ± se of three biological repetitions. **P < 0.01.
(C) Germination greening of abo6 and the wild type on MS medium containing different concentrations of ABA. Plates were removed from 4°C and kept in a growth chamber for 7 d before they were photographed.
(D) Statistical analysis of the seed germination greening ratio in (C).
(E) Seed germination greening as affected by different concentrations of NaCl or mannitol. The plates were kept in a growth chamber for 7 d before they were photographed.
(F) and (G) Statistical analysis of seed germination greening ratio in (E).
For seed germination greening in (D), (F), and (G), ∼30 seeds were counted in each of three plates for one experiment, and three independent experiments were performed with similar results. Data are means ± se of three biological repetitions. *P < 0.05; **P < 0.01.
[See online article for color version of this figure.]
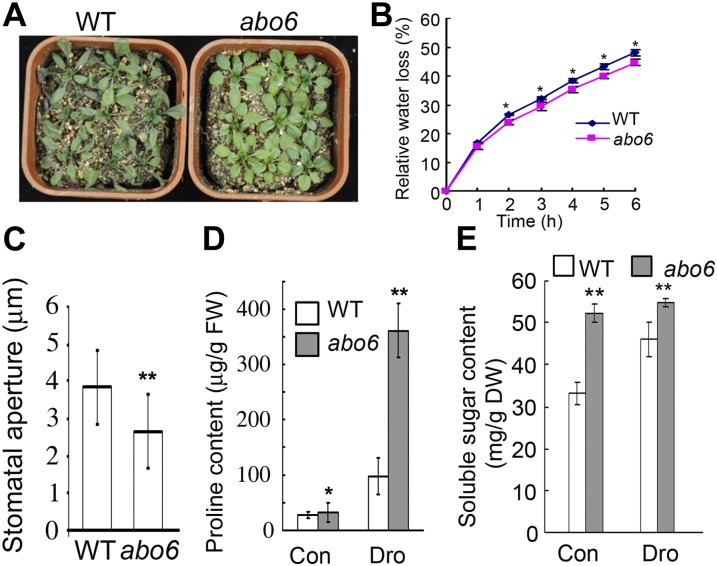
We analyzed whether abo6 was more drought resistant than the wild type. abo6 and wild-type plants were grown in pots under water sufficient conditions. After 2 weeks, the plants were subjected to drought stress by withholding water. After 10 d of drought treatment, the wild-type leaves showed clear wilting signs, but abo6 leaves remained turgid (Figure 2A). Because the abo6 mutants were slightly smaller than the wild-type plants, and because leaf surface area affects the rate of transpiration from leaves, we measured the water loss of detached leaves so as to obtain a direct measure of transpiration. The rate of water loss was slower from detached abo6 leaves than from detached wild-type leaves (Figure 2B), which was consistent with the drought-tolerant phenotype of abo6 plants growing in soil. Under water-sufficient conditions, the stomatal apertures of abo6 were smaller than those of the wild type (Figure 2C). We also found that Pro and soluble sugar contents were greater in abo6 than in the wild type under both water-sufficient and drought conditions (Figures 2D and 2E). These results suggest that the abo6 mutant suffers from constitutive stress and also partially explain its drought-tolerant phenotype.
Figure 2.
abo6 Is More Drought Tolerant Than the Wild Type.
(A) Phenotypes of seedlings grown in soil and subjected to drought treatment. Seedlings were grown in soil under short-day (12 h light/12 h dark) conditions for 2 weeks before water was withheld. Seedlings were photographed after water had been withheld for 10 d. WT, the wild type.
(B) Relative water loss in detached leaves. Detached leaves from nine seedlings grown in soil under short-day conditions were weighed at the time of detachment (=100% water content or 0% water loss), kept uncovered on the greenhouse bench, and weighed each hour for 6 h. Three independent experiments were performed with the similar results. Data are means ± se of three biological repetitions, n = 3. *P < 0.05.
(C) Stomatal aperture of seedlings growing in soil under water sufficient conditions. At least 90 apertures in total were measured. Data are means ± se of three biological repetitions. **P < 0.01.
(D) Pro content of seedlings grown under water-sufficient (control [Con]) and drought (Dro) conditions. FW, fresh weight.
(E) Soluble sugar content of seedlings grown under water-sufficient and drought conditions. DW, dry weight.
In (D) and (E), leaves of 4-week-old seedlings grown in soil under short-day conditions were detached and kept uncovered on the greenhouse bench. After 3 h, Pro (D) and soluble sugar (E) content in the detached leaves were measured. Three independent experiments were performed. Data are means of three biological repetitions ±se, n = 3. *P < 0.05; **P < 0.01.
[See online article for color version of this figure.]
ABO6 Encodes a DEXH RNA Helicase That Is Localized to Mitochondria
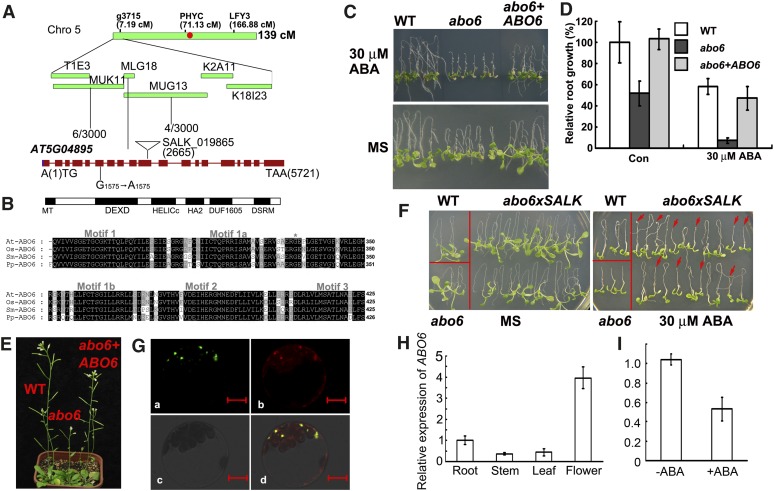
The abo6 mutant (in the Columbia gl1 accession) was crossed with the Landsberg accession. The F2 seedlings that showed ABA sensitivity in root growth (on MS medium supplemented with 30 μM ABA) were selected. ABO6 was initially mapped to the upper arm of chromosome 5 and was precisely mapped to the BAC clones MUK11 and MUG13 (Figure 3A). We sequenced all of the open reading frames in this region and identified a G1575-to-A1575 (counting from the first putative ATG) mutation in the sixth exon of AT5G04895; this mutation caused a Gly334-to-Glu334 change in AT5G04895.
Figure 3.
Cloning of ABO6.
(A) Map-based cloning of ABO6. ABO6 was mapped to the top of chromosome 5. A total of 1500 samples were used to narrow the location of ABO6 to BAC clones MUK11 and MUG13. A G-to-A mutation in position 1575 and a T-DNA insertion in position 2665 in AT5G04895 are indicated. Red dot, centromere; cM, centimorgans.
(B) The structure of the predicted ABO6 protein and DEXD domain. MT, mitochondrial signal peptide; DEXD, DEXD motif; HELICc, a helicase C-terminal domain; HA2, a helicase-associated domain; DUF1605, domain-of-unknown function; DSRM, double-stranded RNA binding domain. The DEXD domains of ABO6 homologs from Oryza sativa AAL58955 (Os-ABO6), Selaginella moellendorffii EFJ13473 (Sm-ABO6), and Physcomitrella patens EDQ53672 (Pp-ABO6) were aligned with ABO6 using ClustalX 2.0.5, and the conserved DEXH domains was selected. The asterisk refers to the conserved amino acid Gly-334 of ABO6 that was mutated to Glu in abo6 in motif1a.
(C) ABA sensitivity of abo6 complemented with Pro super:ABO6. The wild type (WT), abo6, and transgenic line 9 (abo6+ABO6) were grown on MS medium containing 0 or 30 μM ABA.
(D) Relative primary root growth (root length) of abo6, abo6 complemented with Pro super:ABO6, and the wild type on MS medium with and without ABA. Primary root length of the wild type on MS medium without ABA was taken as 100%.
(E) Growth phenotypes of abo6, abo6 complemented with Pro super:ABO6, and wild-type seedlings growing in soil.
(F) ABA-sensitive phenotype of F1 seedlings from abo6 crossed with a heterozygous T-DNA line disrupting AT5G04895 (SALK_019865, see [A] for insertion position). Four-day-old seedlings were transferred to MS medium containing 0 or 30 μM ABA, and seedlings were photographed 7 d later. Red arrows point to abo6/SALK_019865.
(G) Localization of the ABO6 signal peptide fused with GFP. The signal peptide of ABO6 was fused with GFP and transiently expressed in protoplasts of Arabidopsis leaves. The protoplasts were stained with MitoTracker Red and observed with a confocal laser microscope. (a) GFP image; (b) MitoTracker Red image; (c) bright-field images; (d) merged images of GFP and MitoTracker Red. Bars = 10 μm.
(H) Relative expression of ABO6 in roots, stems, leaves, and flowers as determined by qRT-PCR. ACTIN2/8 was used as a control. Three independent experiments were performed, each with three technical replicates. Results shown are from one representative experiment.
(I) Expression of ABO6 is reduced by ABA treatment. Seedlings were treated with 50 μM ABA for 5 h, and total RNAs were used for qRT-PCR. ACTIN2/8 was used as a control. Three independent experiments were performed. Data are means of three biological repetitions ± se, n = 3.
ABO6 was annotated as a DEAD box RNA helicase that contains a DEXH box (Figure 3B). There are 58 DEAD box proteins in Arabidopsis, most of which belong to the sf2 family, a family that is divided into DEAD, DEAH, and DExD/H subfamilies (Boudet et al., 2001; Mingam et al., 2004). The abo6 mutation Gly-334 to Glu-334 occurs in the DEXH region. Gly-334 is a well-conserved amino acid in the DEXH region of ABO6 homologs from different species (Figure 3B). The cDNA of AT5G04895 was amplified and overexpressed under the control of a super promoter in abo6. We obtained 22 independent transgenic lines and selected several T3 homozygous lines to test for ABA sensitivity of root growth: All were rescued to the wild-type phenotype on MS medium containing 30 μM ABA. As an example, the homozygous transgenic line 9 complemented both the ABA-sensitive phenotype (Figures 3C and 3D) and the retarded-growth phenotype (Figure 3E), indicating that the ABA-sensitive phenotype of abo6 was caused by the mutation in AT5G04895. We obtained T-DNA lines but failed to obtain homozygous T-DNA insertion plants, suggesting that ABO6 is required for plant survival. We crossed abo6 with a heterozygous T-DNA mutant (Figure 3A) and analyzed the F1 plants for root sensitivity to ABA. We found that the ratio of normal plants to sensitive plants was near 1:1 (36:38) (Figure 3F). The ABA-sensitive plants were heterozygous in that they carried both the abo6 mutation and the T-DNA insertion. Together, these results confirmed that AT5G04895 is ABO6.
ABO6 is predicted to localize to mitochondria, according to the program TargetP (http://www.cbs.dtu.dk/services/TargetP). To verify this result, we fused the N terminus (nucleotides 1 to 1227) of the abo6 cDNA fragment (including a putative signal peptide) in frame with green fluorescent protein (GFP) under the control of a super promoter. The constructs were transiently transformed into Arabidopsis protoplasts stained with MitoTracker Red, a dye that specifically accumulates in mitochondria. The green fluorescence of the fusion protein was colocalized with the fluorescence of the MitoTracker Red, indicating that ABO6 is targeted to mitochondria (Figure 3G).
To determine the expression of ABO6, we performed quantitative RT-PCR. We found that ABO6 transcripts were more abundant in roots and flowers than in leaves and stems (Figure 3H). Interestingly, the expression of ABO6 was reduced by ABA treatment (Figure 3I), which is consistent with the public microarray data (Nakashima et al., 2009), suggesting that it has a role in the ABA response pathway.
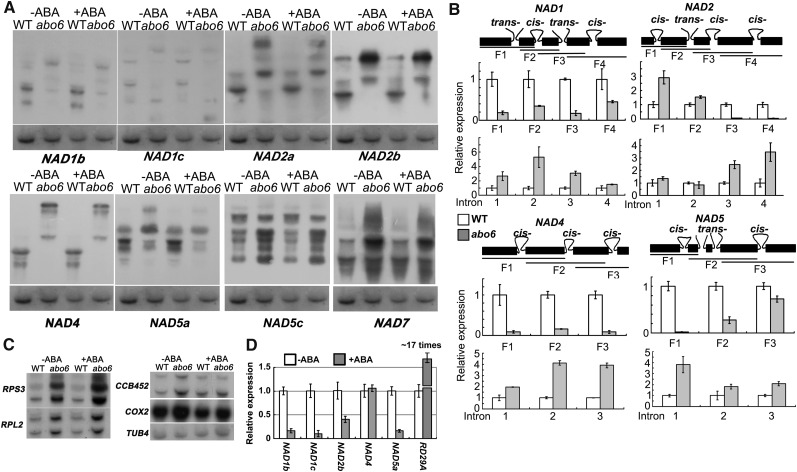
ABO6 Affects the Splicing of Complex I Genes in Mitochondria
DEAD box RNA helicases have been implicated in a variety of RNA metabolic processes, including RNA synthesis, RNA modification, RNA cleavage, and RNA degradation (Gong et al., 2002). Because ABO6 is targeted to mitochondria, we wanted to determine whether it participates in mitochondrial RNA metabolism. Among the nine mitochondria-encoded nad genes (nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, and nad9, encoding subunits of the NADH dehydrogenase [complex I]), nad4 and nad7 require cis-splicing, and nad1, nad2, and nad5 require both cis- and trans-splicing (Unseld et al., 1997). We found that the abo6 mutation impaired RNA splicing in NAD1b, NAD1c, NAD2a, NAD2b, NAD4, and NAD5a but not in NAD5c and NAD7 (Figure 4A) or in COX2 (a subunit of cytochrome c oxidase [complex IV]), RPL2, RPS3 (encode subunits of ribosomal proteins), and CCB452 (involved in cytochrome c biogenesis), genes that are required for splicing but not in complex I (Figure 4C). The transcripts of NAD5c, NAD7, COX2, RPL2, RPS3, and CCB452 were more abundant in abo6 than in the wild type. The high accumulation of mitochondrial transcripts is also found in other mutants, such as slo1, in which a PPR gene required for RNA editing of nad4 and nad9 is mutated. However, the reason for this is not known (Sung et al., 2010). We further used quantitative RT-PCR (qRT-PCR) to determine the effect of the abo6 mutation on splicing sites. For NAD1, the splicing in all four introns was greatly reduced in abo6, and the transcripts of each intron were more abundant in abo6 than in the wild type (Figure 4B). For NAD2, splicing was not affected in the first (cis-) and second (trans-) intron but was impaired in the third (cis-) and fourth (cis-) intron; the abundance of transcripts of the first and second introns was not changed, but the abundance of transcripts of the third and fourth introns was increased. For NAD4, the cis-splicing in all three introns was significantly reduced, and the intron expression was higher in abo6 than in the wild type (Figure 4B). For NAD5, the third exon is very short and the primers were designed to cover the second and fourth exons. Both the cis- and trans-splicing (introns 2 and 3) were reduced in NAD5, and transcripts of the corresponding introns were increased (Figure 4B). These results indicate that ABO6 is involved in both cis- and trans-splicing of some nad genes. We also found that ABA treatment reduced the expression of NAD1, NAD2, and NAD5 but had no effect on the expression of NAD4 (Figure 4D).
Figure 4.
RNA Gel Blot of nad Genes.
(A) RNA gel blot analysis of the expression of nad genes or gene fragments using specific probes. Probes: NAD1b, the third exon; NAD1c, the fifth exon; NAD2a, the second exon; NAD2b, the forth exon; NAD4, the third exon; NAD5a, the second exon; NAD5c, the forth exon; NAD7, the third exon (see [B] for the exon fragments used). TUBULIN4 was used as a loading control. WT, the wild type.
(B) qRT-PCR analysis of the transcripts of different exons and introns. In each gene, trans- or cis-splicing is indicated. The amplified fragment is labeled F1, F2, F3, or F4 or is indicated by intron number.
(C) RNA gel blots of COX2 (a subunit of cytochrome c oxidase [complex IV]), RPL2, RPS3 (encoding subunits of ribosomal proteins), and CCB452 (involved in cytochrome c biogenesis) genes that are required for splicing. TUBULIN4 was used as a loading control.
(D) qRT-PCR analysis of the transcripts of some NAD genes by ABA treatment. The transcripts without ABA treatment were compared with those under ABA treatment. Three independent experiments were done with similar results, each with three biological replicates. Results shown are from one representative experiment. RD29A was used as a positive control for effective ABA induction.
ROS from Mitochondria Are the Second Messengers for ABA Inhibition of Primary Root Growth and Seed Germination in abo6
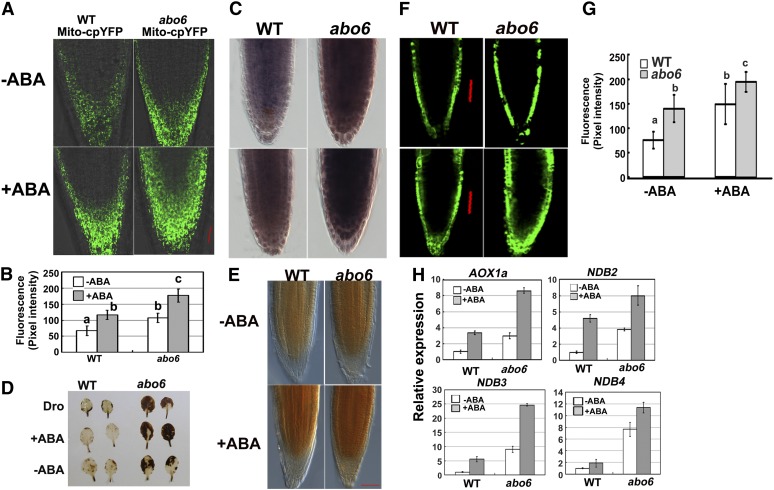
Impairment of complex I in mitochondria would result in respiratory dysfunction and would elicit the production of excess ROS. To detect ROS production from mitochondria, we generated transgenic Arabidopsis plants that harbored a mitochondrial matrix-targeted superoxide indicator, Mito-cpYFP, that emits strong fluorescence in the presence of the superoxides found in mitochondria with superoxide in mitochondria, but not with other ROS or not in other cell organelles (Wang et al., 2008). In the absence of ABA treatment, abo6/Mito-cpYFP emitted stronger fluorescence than the wild type/Mito-cpYFP (Figure 5A), indicating that abo6 accumulated a higher level of ROS than the wild type. ABA treatment induced higher levels of fluorescence in both abo6/Mito-cpYFP and wild type/Mito-cpYFP than no ABA treatment (Figure 5A), and abo6/Mito-cpYFP emitted stronger fluorescence than wild type/Mito-cpYFP (Figure 5A). Quantification of intensities verified that abo6 produced more ROS than did the wild type with or without ABA treatment (Figure 5B). Nitroblue tetrazolium (NBT) staining (for detection of superoxides) of root tips supported these results under both normal conditions and ABA treatment (Figure 5C, also see Figures 7C, 7D, 8I, and 8J for quantification). These results indicate that ABA induces the accumulation of ROS in mitochondria, and abo6 mutation leads to increased ROS accumulation in mitochondria.
Figure 5.
abo6 Accumulates More ROS Than the Wild Type.
(A) Fluorescence analysis of mitochondrial cpYFP in wild-type and abo6 roots without or with ABA treatment. Bar = 50 μm.
(B) Intensity of cpYFP quantified using AxioVision Rel. 4.8 software. About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.01. WT, the wild type.
(C) NBT staining (detection of superoxide) of root tips of the wild type and abo6 under both normal conditions and ABA treatment. Bar = 50 μm.
(D) DAB staining of leaves under normal conditions, ABA treatment, or drought treatment. Leaves from seedlings grown in soil under short-day conditions for 4 weeks were detached and treated with or without 50 μM ABA for 3 h or left uncovered on a laboratory bench for 3 h for drought treatment.
(E) DAB staining of primary roots treated or not with 50 μM ABA for 5 h. Bar = 50 μm.
(F) DCFH-DA staining of primary roots treated or not treated with 50 μM ABA for 5 h. Bar = 50 μm.
(G) Intensity of DCFH-DA staining quantified using AxioVision Rel. 4.8 software. About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.01 (a and b, a and c) or P < 0.05 (b and c).
(H) The expression of AOX1a, NDB2, NDB3, and NDB4 in seedlings treated or not treated with 50 μM ABA for 5 h. Three independent experiments were done with similar results, each with three biological replicates. Results shown are from one representative experiment. Data are means of three biological repetitions ± se, n = 3.
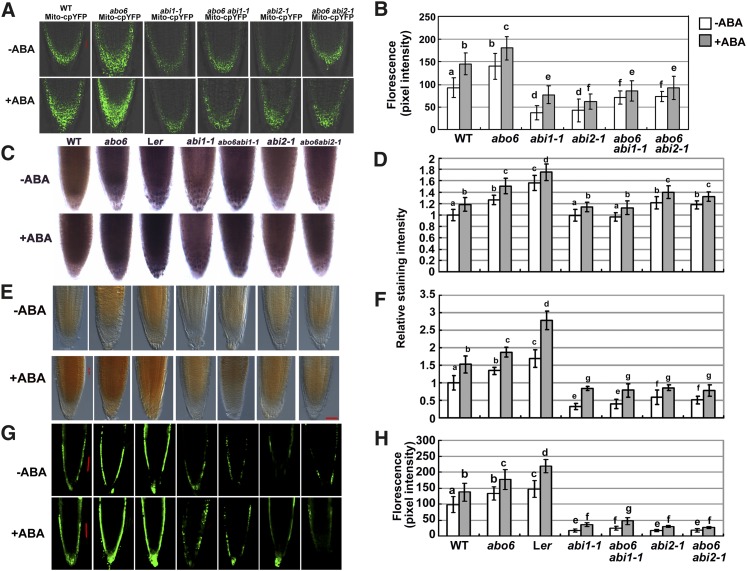
Figure 7.
Genetic Analysis of abo6 with abi1-1 and abi2-1.
(A) Fluorescence analysis of mitochondrial cpYFP in the roots of abo6, abi1-1, abi2-1, abo6 abi1-1, abo6 abi2-1, and wild-type (WT) plants without or with ABA treatment. Bar = 50 μm.
(B) Intensity of Mito-cpYFP fluorescence quantified using AxioVision Rel. 4.8 software. About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.05.
(C) NBT staining (detection of superoxides) of root tips of the wild type (Columbia), abo6, Ler, abi1-1, abi2-1, abo6 abi1-1, and abo6 abi2-1 under both normal conditions and ABA treatment.
(D) Relative intensity of NBT staining in (C). About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.05.
(E) DAB staining of the primary roots of the wild type, abo6, Ler, abi1-1, abi2-1, abo6 abi1-1, and abo6 abi2-1 plants treated or not with 30 μM ABA for 5 h. Bar = 50 μm.
(F) Relative intensity of DAB staining in (E). About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.05.
(G) DCFH-DA staining assay of the primary roots in abo6, wild-type, Ler, abi1-1, abi2-1, abo6 abi1-1, and abo6 abi2-1 plants treated or not with 30 μM ABA for 5 h. Bar = 50 μm.
(H) Intensity of DCFH-DA staining quantified using AxioVision Rel. 4.8 software. About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.05.
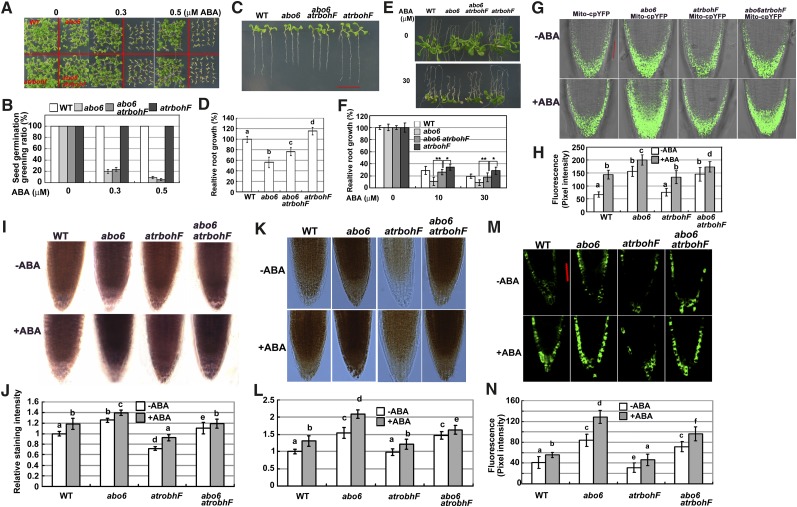
Figure 8.
ABA Inhibition of Primary Root Growth of abo6 Can Be Compromised by the rbohF Mutation.
(A) and (B) Seed germination greening of the wild type (WT), abo6, rbohF, and abo6 rbohF on MS medium containing different concentrations of ABA. About 30 seeds were counted in each of three plates for one experiment, and three independent experiments were performed with similar results in (B). Data are means ± se of three biological repetitions.
(C) and (D) Root growth comparison of the wild type, abo6, rbohF, and abo6 rbohF (7-d-old seedlings) on MS medium. Three independent experiments were performed, each with triple replicates (more than 10 roots for each replicate) in (D). Samples with different letters are significantly different: P < 0.01.
(E) and (F) ABA inhibition of primary root growth in wild-type, abo6, rbohF, and abo6 rbohF seedlings. Four-day-old seedlings were transferred to MS medium containing different concentrations of ABA and grown for 14 d. Primary root length of the wild type or abo6 on MS alone was set at 100%. Three independent experiments were performed, each with triple replicates (more than 10 roots for each replicate). *P < 0.05; **P < 0.01.
(G) Fluorescence analysis of mitochondrial cpYFP in the roots of wild-type, abo6, rbohF, and abo6 rbohF plants treated or not with 50 μM ABA for 5 h.
(H) Intensity of Mito-cpYFP quantified using AxioVision Rel. 4.8 software. About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.05 (b and d) or 0.01.
(I) NBT staining of root tips of wild-type, abo6, rbohF, and abo6 rbohF plants treated or not with 50 μM ABA for 5 h.
(J) Relative intensity of NBT staining in (I). About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.05.
(K) DAB staining assay of the primary roots in the wild type abo6, rbohF, and abo6 rbohF treated or not with 50 μM ABA for 5 h. Bar = 50 μm.
(L) Relative intensity of DAB staining in (K). About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.05.
(M) DCFH-DA staining of the primary roots of wild-type, abo6, rbohF, and abo6 rbohF plants treated or not with 50 μM ABA for 5 h. Bar = 50 μm.
(N) Fluorescence intensity of DCFH-DA staining quantified using AxioVision Rel. 4.8 software. About 20 roots were analyzed for each experiment. Values are means ± se of three biological repetitions. Samples with different letters are significantly different: P < 0.05.
We used 3′, 3′- diaminobenzidine (DAB; for detection of H2O2) staining to examine ROS in leaves and found that the concentration of ROS was higher in abo6 leaves than in wild-type leaves (Figure 5D). ABA and drought treatment induced more ROS in abo6 than in the wild type (Figure 5D). Similar results were obtained when we used DAB staining to measure ROS accumulation with and without ABA treatment in primary roots (Figure 5E). 2′,7′-Dichlorodihydrofluorescin diacetate (DCFH-DA) staining (a fluorescent probe for detection of H2O2) and its quantification in primary root tips confirmed the higher accumulation of ROS in abo6 than in the wild type with and without ABA treatment (Figures 5F and 5G). These results indicate that abo6 mutants accumulate more ROS than the wild type in both leaves and roots and that ABA treatment enhances ROS production.
ROS produced in mitochondria usually act as retrograde signals to induce the expression of some nuclear genes, including alternative oxidase and type II NAD(P)H dehydrogenases (Clifton et al., 2005). We tested the gene expression of AOX1a and the B class NADH dehydrogenase gene family (NDB2, NDB3, and NDB4) in abo6 and the wild type. As shown in Figure 5H, the transcripts of AOX1a, NDB2, NDB3, and NDB4 were several times more abundant in abo6 than in the wild type under normal growth conditions. ABA treatment increased the transcripts of these four genes in both abo6 and the wild type (Figure 5H).
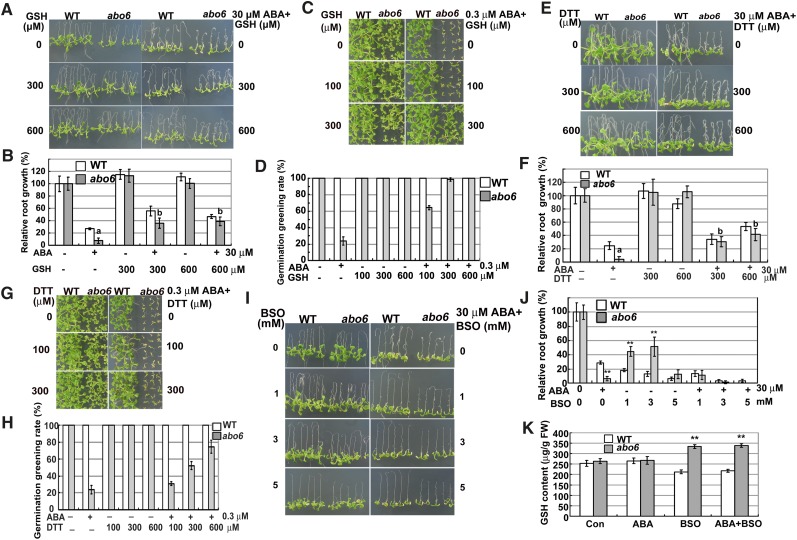
To determine whether the ROS produced from mitochondria can act as second messengers that inhibit primary root growth, we compared the primary root growth of the abo6 mutant with that of the wild type on MS medium supplemented with a ROS scavenging reagent, reduced GSH. As shown in Figures 6A and 6B, relative primary root growth was similar for the wild type and abo6 on MS medium containing 300 μM GSH or 600 μM GSH. As shown above, 30 μM ABA greatly inhibited the primary root growth of abo6, while the relative primary root growth of abo6 was comparable with that of the wild type on MS medium supplemented with 30 μM ABA plus 300 μM GSH or 600 μM GSH. The addition of different concentrations of GSH to the medium also released the ABA inhibition of abo6 seed germination (Figures 6C and 6D).
Figure 6.
Exogenous Addition of GSH or DTT Can Partially Rescue the ABA-Sensitive Phenotypes of abo6.
(A) Primary root growth of seedlings on MS medium or MS medium containing ABA, GSH, or ABA plus GSH, respectively. Four-day-old seedlings were transferred to MS medium or MS medium containing different concentrations of GSH, ABA, or GSH plus ABA for 7 d before being photographed. WT, the wild type.
(B) Statistical analysis of relative primary root length in (A). Samples of abo6 with different letters are significantly different: P < 0.01.
(C) Germination greening of seeds on MS medium or MS medium supplemented with ABA, GSH, or ABA plus GSH. Plates were kept in a growth chamber for 14 d before they were photographed.
(D) Statistical analysis of the seed germination greening rate in (C).
(E) The primary root growth of seedlings on MS medium or MS medium supplemented with ABA, DTT, or ABA plus DTT.
(F) Statistical analysis of the relative primary root length in (E). Samples of abo6 with different letters are significantly different: P < 0.01.
(G) Germination greening of seeds on MS medium or MS medium supplemented with ABA, DTT, or ABA plus DTT. Plates were kept in a growth chamber for 14 d before they were photographed.
(H) Statistical analysis of seed germination greening rate in (G).
(I) Primary root growth of seedlings on MS medium or MS medium supplemented with ABA, BSO, or ABA plus BSO.
(J) Statistical analysis of relative primary root length in (I). **P < 0.01.
(K) GSH content in the roots of wild type and abo6 under different treatments. **P < 0.01.
In (B), (F), and (J), primary root length of the wild type or abo6 on MS alone was set at 100%. Values are means ± se in (B), (D), (F), (H), (J), and (K). Three independent experiments were performed, each with triple replicates (more than 30 seeds or 10 roots for each replicate).
[See online article for color version of this figure.]
Previous studies suggested that a GSH pool is important for mediating plant growth (Bashandy et al., 2010; Koprivova et al., 2010a). To determine whether the response of abo6 to GSH was specific or was representative of its response to other ROS scavenging agents, we investigated whether another ROS scavenging agent, DTT, could also recover the ABA-sensitive phenotypes of abo6. The addition of 300 μM DTT did not apparently affect the primary root growth of the wild type and abo6 but clearly released the ABA inhibition of primary root growth in both abo6 and the wild type and made the relative root growth of abo6 comparable to that of the wild type (Figures 6E and 6F). The addition of 600 μM DTT weakly inhibited primary root growth of the wild type but not of abo6 and released the ABA inhibition of primary root growth of abo6 and the wild type. The addition of DTT also reduced the inhibition of seed germination greening caused by ABA treatment in abo6 (Figures 6G and 6H). We further compared two other mutants, ndufs4 (for NADH dehydrogenase [ubiquinone] fragment S subunit 4 of complex I) and abo5 (cis-splicing of mitochondrial nad2 intron 3 in complex I), on MS medium supplemented with GSH or DTT. Two mutants are sensitive to ABA in seed germination and root growth (Liu et al., 2010). The addition of GSH or DTT to the MS medium released the ABA inhibition of both seed germination and primary root growth in the two mutants to a similar degree as in abo6 (see Supplemental Figure 2 online). These results suggest that the oxidation status is important for ABA inhibition of seed germination and primary root growth.
We further tested whether a specific nontoxic inhibitor of γ-glutamylcysteine synthetases, BSO, could impact the growth of abo6 and the wild type. The root growth of abo6 was more resistant to BSO than that of the wild type (Figures 6I and 6J), which is consistent with other complex I defective mutants, such as bir6 (for BSO-insensitive roots6), which is defective in splicing of intron 1 of the nad7 transcript, css1, which is disrupted in the splicing of an intron in maturase, and a γ-carbonic anhydrase mutant (At1g47260) (Koprivova et al., 2010b). The resistance to BSO is consistent with the higher accumulation of GSH under BSO treatment in abo6 than in the wild type (Figure 6K), but the mechanism explaining why complex I mutants with higher GSH are resistant to BSO is unknown (Koprivova et al., 2010b). These results suggest that at least the GSH pool in these mutants is not lower than that in the wild type under ABA or BSO treatment. Increased BSO combined with ABA had an additive effect on the inhibition of root growth in both abo6 and the wild type (Figures 6I and 6J), indicating that BSO enhancement of oxidative stress increases ABA-mediated inhibition of root growth.
Dominant-Negative Mutations in ABI1 and ABI2 Reduce ROS Production and Release the ABA Sensitivity of abo6 in Root Growth
ABI1 and ABI2 encode protein phosphatases 2C (Leung et al., 1994, 1997; Meyer et al., 1994), which are important components in early ABA signaling. The dominant-negative mutations in abi1-1 and abi2-1 block the interaction of PYR/PYL/RCAR ABA receptors with ABI1 and ABI2, resulting in the inhibition of ABA signal transduction (Cutler et al., 2010). In the abi1-1 mutant, ABA-induced ROS accumulation is greatly reduced in guard cells, while in the abi2-1 mutant, ABA-induced ROS accumulation is normal in guard cells, but the ROS signal cannot be transmitted to downstream targets to regulate stomatal movement (Murata et al., 2001). We used cpYFP superoxide indicator to compare ROS accumulation in abo6, abi1-1, abi2-1, and the wild type. As shown in Figures 7A and 7B, both abi1-1/Mito-cpYFP and abi2-1/Mito-cpYFP emitted less fluorescence than abo6/Mito-cpYFP or wild type/Mito-cpYFP. abo6 abi1-1/Mito-cpYFP and abo6 abi2-1/Mito-cpYFP emitted more fluorescence than abi1-1/Mito-cpYFP or abi2-1/Mito-cpYFP but much less than abo6/Mito-cpYFP or wild type/Mito-cpYFP. ABA treatment resulted in more fluorescence than no ABA treatment in abi1-1/Mito-cpYFP, abi2-1/Mito-cpYFP, abo6 abi1-1/Mito-cpYFP, or abo6 abi2-1/Mito-cpYFP plants. NBT staining of root tips confirmed these results, both under normal conditions and ABA treatment (Figures 7C and 7D). Because abi1-1 and abi2-1 are in the Landsberg erecta (Ler) accession, we also included Ler in our analysis. The primary roots of Ler produced more ROS than those of Columbia gl1 (here used as the wild-type control) (Figures 7C and 7D), which might partially explain the greater ABA sensitivity of Ler than Columbia in both seed germination greening and primary root growth.
Next, we performed both DAB and DCFH-DA staining to analyze ROS (H2O2) production. Both abi1-1 and abi2-1 accumulated less ROS (H2O2) in roots than the wild type under ABA treatment or no ABA treatment (Figures 7E and 7F for DAB staining and Figures 7G and 7H for DCFH-DA), suggesting that ROS production regulated by ABI2 in roots is different from that in guard cells (Murata et al., 2001). ABA treatment increased ROS accumulation in all plants. The abi1-1 abo6 and abi2-1 abo6 double mutants produced more ROS than abi1-1 or abi2-1, respectively, which all accumulated less ROS than the wild type or abo6 under ABA treatment or no ABA treatment. The primary roots of Ler accumulated more H2O2 than those of Columbia.
The abi1-1 abo6 and abi2-1 abo6 mutants were resistant to ABA inhibition of primary root growth and seed germination greening (see Supplemental Figure 3 online), which was not caused by a change in the accumulation of GSH in roots (see Supplemental Figure 4 online). As previous studies have indicated that ABA regulates ROS production at the plasma membrane (Kim et al., 2010), our results suggest that the early ABA signaling components, ABI1 and ABI2, also regulate ROS production from mitochondria.
ABA-Mediated Inhibition of abo6 Primary Root Growth Is Alleviated by the rbohF Mutation
The Arabidopsis genome contains 10 NADPH oxidase catalytic subunit genes. A previous study indicated that ABA evaluates ROS production by regulating the plasma membrane NADPH oxidases RBOHD and RBOHF (Kwak et al., 2003). To determine the contribution of the mitochondrial ROS made by abo6 to the cellular response, we performed a phenotypic analysis of abo6 and the rbohF mutant, which is insensitive to ABA in terms of primary root growth (Kwak et al., 2003). abo6 rbohF showed similar ABA sensitivity as abo6 in terms of seed germination, indicating that abo6 dominates the seed germination (Figures 8A and 8B). The primary root growth of abo6 rbohF was faster than that of abo6 but slower than the wild type and rbohF under normal conditions (Figures 8C and 8D). The primary root growth of abo6 rbohF was less inhibited by ABA than was abo6 (Figures 8E and 8F), indicating that the rbohF mutant could alleviate the ABA inhibition of primary root growth of abo6. Under both ABA and no ABA treatment, rbohF accumulated similar levels of superoxide in mitochondria as the wild type, as detected by Mito-cpYFP fluorescence (Figures 8G and 8H). abo6 rbohF accumulated a similar level of superoxides in mitochondria as abo6 in normal conditions and less than abo6 upon ABA treatment (Figures 8G and 8H). These results indicate that RBOHF affects superoxide accumulation in mitochondria under ABA treatment. NBT staining showed similar superoxide accumulation patterns as Mito-cpYFP fluorescence (Figures 8I and 8J). Upon DAB staining (Figures 8K and 8L), rbohF accumulated less H2O2 in root tips than the wild type in both the presence and absence of exogenously applied ABA, while abo6 rbohF accumulated similar levels of H2O2 as abo6 in the absence of ABA treatment, less than abo6 under ABA treatment, more than rbohF in both the presence and absence of ABA treatment, more than the wild type under normal conditions, and similar as the wild type under ABA treatment. DCFH-DA staining (Figures 8M and 8N) and showed similar H2O2 accumulation patterns in all phenotypes as DAB staining. Taken together, these results suggest that ROS homeostasis can be affected by the ROS produced at both the plasma membrane and mitochondria in Arabidopsis.
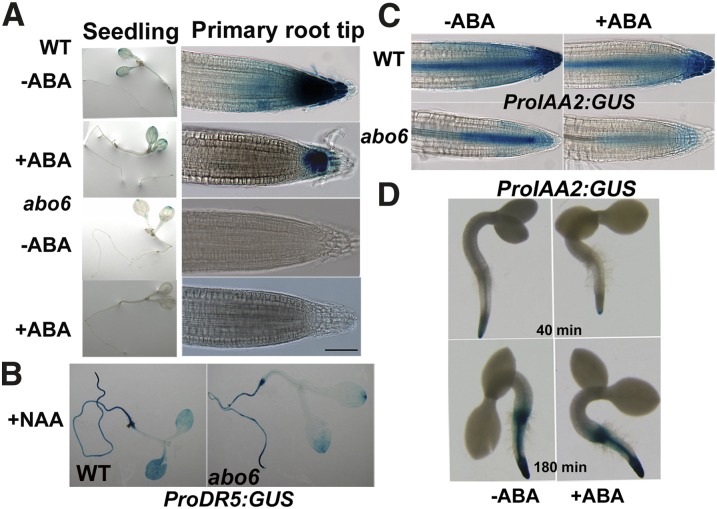
ProDR5:GUS and ProIAA2:GUS Expression Are Reduced in the Primary Root Tips of abo6
DR5 is a synthetic promoter that contains an auxin response element whose expression provides reliable information concerning auxin distribution in roots and other tissues (Ulmasov et al., 1997; Benková et al., 2003). We introduced ProDR5:GUS (for synthetic auxin response element D1-4 with site-directed mutants in the 5′-end from soybean:β-glucuronidase) into the abo6 mutant. GUS staining in the cotyledon was less in abo6 than in the wild type. In the primary root tips, GUS staining was faint in abo6 but strong in the wild type (Figure 9A). ABA treatment reduced ProDR5:GUS expression in both primary root tips and leaves of the wild type and also in leaves of abo6 (Figure 9A). The addition of 1-napthalene acetic acid (NAA) to the medium induced the expression of GUS to higher levels in both abo6 and the wild type (Figure 9B), indicating that the auxin signaling pathway is not defective in abo6. However, ProDR5:GUS expression in abo6 was still lower than that in the wild type upon NAA treatment, suggesting that ROS are negative regulators of auxin response (Nakagami et al., 2006). IAA2 is another auxin marker whose expression is rapidly induced by auxin (Shibasaki et al., 2009). We introduced Indole-3-acetic acid inducible2 (ProIAA2):GUS into the abo6 mutant. The expression of ProIAA2:GUS in primary root tips was lower in abo6 than in the wild type (Figure 9C). ABA treatment (30 μM) reduced ProIAA2:GUS expression in both abo6 and the wild type. As a previous study indicates that a low concentration of ABA (2 μM) likely increases ProIAA2:GUS expression (Belin et al., 2009), we performed similar experiments to test the effect of ABA treatment on ProIAA2:GUS expression. As shown in Figure 9D, GUS staining was greatly reduced by ABA treatment when staining was performed for a short time (40 min). When samples were stained for a longer period (180 min), the strength of GUS staining was hard to compare, suggesting that the conditions used by our lab and by Belin et al. (2009) might have differed. These results suggest that auxin accumulation or responsiveness is less in abo6 than in the wild type.
Figure 9.
abo6 Reduces Auxin Accumulation or the Auxin Signal.
(A) ProDR5:GUS expression in abo6 and the wild type (WT). Four-day-old seedlings were transferred to MS medium with or without 30 μM ABA for 2 d and were then stained for 24 h before being photographed. Bar = 100 μm.
(B) Exogenous addition of NAA restores the expression of ProDR5:GUS in abo6.
(C) ProIAA2:GUS expression in abo6 and the wild type. Four-day-old seedlings were transferred to MS medium with or without 30 μM ABA for 2 d and were then stained for 40 min before being photographed.
(D) Comparison of ProIAA2:GUS expression in the wild type for different staining times without or with 2 μM ABA treatment. Upon endosperm rupture (after ∼36 h on MS medium at 22°C), wild-type seeds harboring ProIAA2:GUS were transferred to MS medium with or without 2 μM ABA for 10 h and stained for 40 min or (180 min) before being photographed.
Exogenous Auxins Restore the ABA Inhibition of Seed Germination Greening and Primary Root Growth of abo6
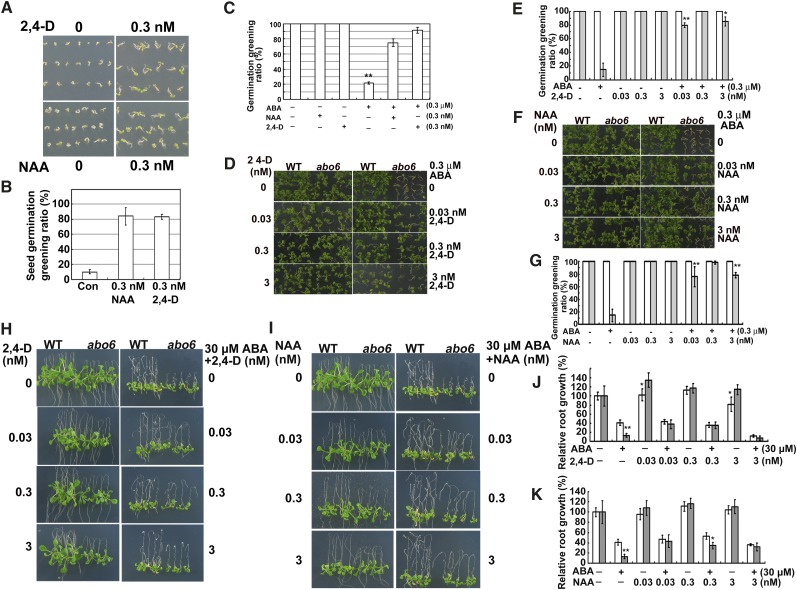
Previous studies indicate that ROS degrade or oxidize auxin and that ROS are negative regulators of the auxin response (Gazarian et al., 1998; Nakagami et al., 2006). The high accumulation of ROS in abo6 may perturb auxin homeostasis and thereby affect root growth. We tested whether the exogenous addition of auxins could restore the ABA sensitivity of abo6. The two auxins used were NAA, which is able to cross the plasma membrane without the help of auxin facilitators, and 2,4-D, which requires auxin facilitators to cross the plasma membrane. Wild-type seeds germinated earlier in the presence of 0.3 nM 2,4-D or 0.3 nM NAA than in the absence of 2,4-D or NAA (Figures 10A and 10B), and the addition of 0.3 nM 2,4-D or 0.3 nM NAA could release the ABA-mediated inhibition of seed germination of the wild type (Figure 10C), indicating that auxin is able to promote seed germination of wild-type Arabidopsis. An increase from 0.03 to 3 nM NAA or 2,4-D to the medium restored the ABA-mediated inhibition of seed germination greening of abo6 (Figures 10D to 10G). Similarly, the addition of low concentrations of NAA or 2,4-D to the medium also released the inhibition of primary root growth by 30 μM ABA in abo6 (Figures 10H to 10K).
Figure 10.
Auxins Promote Seed Germination and Restore ABA-Inhibited Primary Root Growth.
(A) 2,4-D and NAA promote seed germination of the wild type. Wild-type seeds were germinated on MS medium supplemented with 0.3 nM NAA or 2,4-D and kept in a growth chamber for 3 days before being photographed.
(B) Seed germination greening ratio in (A). Con, MS medium.
(C) 2,4-D and NAA release the ABA-inhibited seed germination of the wild type. Wild-type seeds were germinated on MS medium supplemented with 0.3 nM 2,4-D or NAA, plus 0.3 μM ABA and kept in a growth chamber for 5 d. Seed germination greening was quantified.
(D) Germination greening of seeds on MS medium or MS medium supplemented with ABA, 2,4-D, or ABA plus 2,4-D. Plates were kept in a growth chamber for 14 d before being photographed. WT, the wild type.
(E) Statistical analysis of seed germination greening in (D).
(F) Germination greening of seeds on MS medium or MS medium supplemented with ABA, NAA, or ABA plus NAA. Plates were kept in a growth chamber for 14 d before being photographed.
(G) Statistical analysis of the seed germination greening in (F).
(H) Primary root growth of seedlings on MS medium or MS medium supplemented with ABA, 2,4-D, or ABA plus 2,4-D. Four-day-old seedlings were transferred to MS medium containing 0 to 3 nM 2,4-D, ABA, or 2,4-D plus ABA for 7 d before being photographed.
(I) Primary root growth of seedlings on MS medium or MS medium supplemented with ABA, NAA, or ABA plus NAA. Four-day-old seedlings were transferred to MS medium containing 0 to 3 nM NAA, ABA, or NAA plus ABA for 7 d before being photographed.
(J) Statistical analysis of relative primary root length in (H).
(K) Statistical analysis of the relative primary root length in (I).
In (B), (C), (E), (G), (J), and (K), germination greening or primary root length of the wild type or abo6 on MS alone was set at 100%. For statistical analysis, three independent experiments were performed, each with triple replicates (more than 30 seeds or 10 roots for each replicate). Values are means ± se, n = 3. *P < 0.05; **P < 0.01. In (E), (G), (J), and (K), the white column is the wild type and the gray column is abo6.
[See online article for color version of this figure.]
The Expression of Auxin Carriers Is Less in abo6 Than in the Wild Type
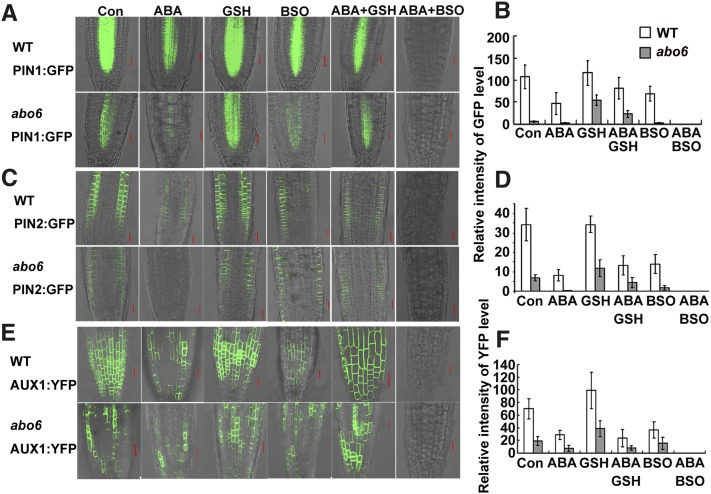
The expression of auxin transport proteins is regulated by auxin homeostasis. The reduced auxin response in abo6 may be due to perturbed polarized auxin transport, reduced auxin levels, or both. We compared the expression of auxin transport proteins using PIN-FORMED1 (PIN1), PIN2, or AUXIN RESISTANT1 (AUX1) promoter-derived PIN1-GFP, PIN2-GFP, or AUX1-YFP in abo6 and the wild type. As shown in Figures 11A and 11B, the PIN1-GFP level was lower in abo6 than in the wild type. ABA treatment reduced PIN1-GFP in both abo6 and the wild type, but the relative level of PIN1-GFP was reduced more in abo6 than in the wild type. Similar differences were observed between ProPIN2:PIN2-GFP and ProAUX1:AUX1-YFP roots of abo6 and the wild type (Figures 11C to 11F). The addition of GSH to the medium increased the relative levels of PIN1-GFP, PIN2-GFP, or AUX1-YFP more in abo6 than in the wild type (Figures 11A to 11F). The addition of BSO to the medium consistently decreased the relative amount of PIN1-GFP, PIN2-GFP, or AUX1-YFP in both abo6 and the wild type (Figures 11A to 11F). The addition of GSH recovered the relative amount of PIN1-GFP, PIN2-GFP, or AUX1-YFP in both ABA-treated abo6 and ABA-treated wild-type plants. However, a combination of ABA and BSO substantially decreased the amount of PIN1-GFP, PIN2-GFP, or AUX1-YFP so that the GFP/YFP signals were hardly detectably (Figure 11). These results suggest that ROS production in abo6 contributes to the decreased abundance of auxin transport proteins, which would decrease the auxin transport capacities and auxin levels.
Figure 11.
The Expression of PIN1, PIN2, and AUX1 in abo6 and the Wild Type under Different Treatments.
Four-day-old seedlings were transferred to MS medium (Con) or MS medium containing 30 μM ABA, 600 μM GSH, 2.5 mM BSO, 30 μM ABA plus 600 μM GSH, or 30 μM ABA plus 2.5 mM BSO for 2 d before being photographed with a confocal microscope with the same settings. GFP/YFP signal strength was quantified using AxioVision Rel. 4.8 software. At least 20 roots were analyzed. Values are means ± se. WT, the wild type. Bar = 20 μm.
(A) ProPIN1:PIN1-GFP expression.
(B) Statistical analysis of GFP signal strength in (A).
(C) ProPIN2:PIN2-GFP expression.
(D) Statistical analysis of GFP signal strength in (C).
(E) ProAUX1:AUX1-YFP expression.
(F) Statistical analysis of YFP signal strength in (E).
DISCUSSION
It has been well documented that ROS can be enhanced when plants are exposed to various abiotic stresses, such as drought and salt stress, indicating that plants have developed efficient defense systems against oxidative stress. Drought stress quickly induces the accumulation of the phytohormone ABA, which plays crucial roles in regulating stomatal movement and in modulating the expression of stress-inducible genes. ABA stimulates the production of ROS by activating plasma membrane–localized NADPH oxidases (Kwak et al., 2003; Zhang et al., 2009), but the roles of ROS from mitochondria in the ABA signaling pathway are not well understood. Here, we provide evidence that ABO6 encodes a mitochondria-localized DEXH RNA helicase that is involved in both trans- and cis-splicing of several, but not all, nad genes in mitochondrial complex I. Impairment of ABO6 leads to increased oxidative stress in mitochondria, which is further enhanced by ABA treatment.
In plant mitochondria, the electron transfer system consists of four complexes, including NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), cytochrome bc1 (complex III), and cytochrome c oxidase (complex IV). Complex I consists of more than 35 proteins encoded by both nuclear and mitochondrial DNA. Complex I is not essential in plants because plant mitochondria have four additional NAD(P)H dehydrogenases and an alternative oxidase that could bypass complex I, III, and IV. Some mutants defective in complex I, such as Arabidopsis fro1 (Lee et al., 2002) and tobacco (Nicotiana tabacum) CMS I and II (Gutierres et al., 1997), are not lethal. However, ABO6 is an essential gene, suggesting that besides having roles in intron splicing of nad genes in complex I, ABO6 may have other unidentified roles in RNA metabolism.
The abo6 mutant exhibits various ABA-sensitive phenotypes (i.e., exposure of abo6 to increased levels of ABA reduces seed germination, primary root growth, and stomatal opening). Interestingly, the expression of ABO6 is downregulated by ABA treatment, suggesting that ABA modulates ROS production partially by regulating ABO6 expression. However, we could not detect the splicing changes of nad genes under ABA treatment, suggesting that the reduction in ABO6 by ABA treatment might have some undetectable effects on nad genes. Blocking earlier steps in the ABA signaling pathway by two dominant-negative regulators, abi1-1 and abi2-1, also blocks ROS production in the mitochondria of the abo6 mutant, indicating that ROS produced from mitochondria are regulated by the ABA signaling pathway. These results suggest that mitochondria are key players in ROS homeostasis through ABA-mediated ROS production. The results also indicate that production of ROS by mitochondria may contribute to plant adaptation to drought stress and is important in the ABA signaling pathway.
abo6 accumulated more ROS than the wild type with and without ABA treatment, suggesting that abo6 suffers from constitutive oxidative stress. When the ROS scavenging reagent GSH was added to the growth medium to reduce ROS, the ABA-sensitive phenotypes of abo6 were recovered to a level similar to that of the wild type. Previous studies indicate that a defect of complex I in bir6 (encoding a PPR protein for splicing of intron 1 of nad7) and css1 (impaired in intron maturase for nad4 splicing) leads to resistance to the GSH biosynthesis inhibitor BSO (Koprivova et al., 2010b). abo6 exhibits similar resistance as bir6 and css1 to BSO in terms of root growth. These results suggest that complex I mutants maintain higher GSH levels under BSO treatment. A similar result was observed in cell cultures treated with the complex I inhibitor rotenone (Garmier et al., 2008). These results indicate that impairment of complex I enhances ROS production, which in turn increases the GSH levels for antioxidation so that cellular redox homeostasis is maintained. Because of the high ROS levels in abo6, addition of GSH or DTT can restore the ABA sensitivity in both seed germination and primary root growth by reducing ROS accumulation. Previous studies indicate that GSH is an important regulator of root growth because a mutation of γ-glutamylcysteine synthetase in the root meristemless (rml1) mutant leads to root growth arrest (Vernoux et al., 2000). Although ROS is required for root growth (Foreman et al., 2003; Kwak et al., 2003), overaccumulation of ROS by ABA treatment of abo6 inhibits seed germination and primary root growth.
We found that the expression of two auxin-responsive marker genes, ProDR5:GUS and ProIAA2:GUS, was greatly reduced by abo6 mutation even under normal conditions. Exogenous addition of both NAA and 2,4-D restored the ABA-sensitive phenotype of abo6. The high accumulation of ROS in abo6 should be the main cause of low auxin contents because auxin can be degraded or oxidized by ROS in plant cells (Gazarian et al., 1998). Furthermore, the interaction between ROS and auxin has been suggested by studies of auxin homeostasis and auxin-inducible gene regulation through increased H2O2 levels (Nakagami et al., 2006; Potters et al., 2007). We also observed that the auxin facilitators PIN1, PIN2, and AUX1 are more reduced in abo6 than in the wild type by ABA treatment. Because 2,4-D but not NAA requires auxin carriers to pass through the membrane but both auxins had similar effects on ABA inhibition of seed germination and primary root growth, auxin carriers are not required for the response of abo6 to ABA. These results suggest that reduced auxin content is the most likely explanation for the ABA-sensitive phenotype in abo6. However, reduced auxin carriers might also contribute to the ABA-sensitive phenotype of abo6 because auxin transport and auxin metabolism have feedback control (Benjamins and Scheres, 2008). Based on these results, we suspect that a reduction in both auxin content and auxin carriers contributes to the effects of ABA-mediated ROS production on the seed germination and primary root growth. The auxin-responsive phenotypes of abo6 by overaccumulation of ROS are consistent with those observed in the ntra ntrb cad2 triple mutants that are deficient in cytosolic NADPH thioredoxin reductases (NTRA and NTRB) and glutathione biosynthesis (CAD2) (Bashandy et al., 2010). These results demonstrate that either reducing GSH or increasing ROS will disrupt the redox homeostasis that is important for auxin transport and auxin homeostasis in the regulation of seed germination and root growth.
Crosstalk between ABA and auxin has been suggested by several studies. A study of ABA-potentiating auxin-mediated repression of embryonic axis elongation suggests that ABA enhances auxin signaling rather than increasing auxin flow or auxin levels (Belin et al., 2009). The auxin signaling-defective mutants axr1 and axr2 have reduced sensitivity to ABA during root elongation (Wilson et al., 1990; Tiryaki and Staswick, 2002; Monroe-Augustus et al., 2003). In another study, transgenic plants overexpressing the H2O2-inducible gene UGT74E2, which encodes an indole-3-butyric acid (IBA) glucosyltransferase, had elevated levels of both IBA-Glc and free IBA (IAA precursor) and a modified IAA pattern (Tsukagoshi et al., 2010); the transgenic plants exhibited enhanced ABA sensitivity, which was further enhanced by adding IBA, IBA-Glc, or IAA. Auxin can also enhance ABA inhibition of seed germination (Brady et al., 2003; Liu et al., 2007). Mutations in INDOLE-3-BUTYRIC ACID RESPONSE5 dual specificity phosphatase and RCN1 protein phosphatase 2A lead to auxin insensitivity but also to ABA insensitivity in germination and root growth. These studies highlight coenhancement between ABA and auxin signaling or auxin homeostasis during germination and seedling development (Liu et al., 2007; Belin et al., 2009; Tognetti et al., 2010). However, our results demonstrate that abo6 is sensitive to ABA mainly because of the reduced auxin levels caused by ROS overproduction. Because auxin is necessary for primary root elongation, the addition of auxin complements the lack of auxin in the primary root tips and releases the ABA inhibition of primary root growth. We also showed that low concentrations of auxin promote seed germination, while other researchers showed that high concentrations of auxin do not affect seed germination (Brady et al., 2003). Seed germination of abo6 is delayed and more sensitive to ABA than seed germination of the wild type; however, these responses of abo6 can be reversed by the addition of low concentrations of auxin to the medium. These results indicate that a certain threshold level of auxin determines whether auxin will enhance or reduce ABA signaling and that crosstalk between ABA and auxin is complex.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (gl1) seeds were germinated and grown on MS medium (M5519; Sigma-Aldrich) with 3% (w/v) Suc and 0.8% agar in glass plates in a light homoeothermic incubator (22°C) with 23 h light/1 h dark. For the root-bending assay, 4-d-old seedlings were moved to MS medium containing ABA, GSH, BSO, or other chemicals as indicated. For germination, ∼30 seeds were placed on MS medium containing different concentrations of ABA, NaCl, mannitol, GSH, and other reagents, with three plates for each treatment. The seedlings were grown in forest soil and vermiculite (1:1) under 50 µmol m−2 s−1 light at 22°C and 16 h light/8 h dark in a greenhouse.
Mutant Screening
The M2 ethyl methanesulfonate-mutagenized seeds of gl1 were used for screening ABA-sensitive mutants by the root-bending assay (Yin et al., 2009) because growth of gl1 did not differ on MS and MS supplemented with 30 μM ABA. Seedlings (4 d old) were transferred onto MS agar medium supplemented with 30 μM ABA and grown with the roots pointing upward for an additional 7 d; putative mutants with inhibited root growth were selected. The root growth phenotypes of putative mutant seedlings were rechecked in the second and third generations. Approximately 30,000 ethyl methanesulfonate–mutated M2 seedlings were screened, and one abo6 mutant allele was obtained.
Drought Treatments, Water Loss Assay, Stomatal Aperture Measurement, and Pro and Carbohydrate Analyses
Plants grown under short-day conditions (12 h light/12 h dark) were used in these analyses. For drought treatment, 7-d-old mutant and wild-type seedlings were transferred to small pots, grown for 2 weeks under normal conditions, and then subjected to drought treatment by withholding watering for 10 d.
For water loss treatment of detached leaves, shoots of 4-week-old mutant and wild-type plants were cut with a pair of scissors, placed on a piece of weighing paper, and kept at 20°C and 75% humidity in the light. The shoots were weighed immediately after they were cut and periodically thereafter. The percentage of water loss was calculated on the basis of the initial weight of the plants, with three replicate shoots for each assay.
For stomatal aperture assays, epidermal strips from rosette leaves of 4-week-old seedlings were examined with a light microscope (B5-223 IEP; Motic China Group). Stomatal apertures were measured with Motic software for three replicates, with three epidermal strips per replicate and 30 stomata per strip.
For Pro and sugar analysis, the shoots of 4-week-old seedlings were detached and kept on the greenhouse bench for 3 h. They were then ground in liquid nitrogen before Pro and carbohydrate contents were determined according to Chen et al. (2005).
Genetic Mapping and Mutant Complementation
abo6 (ecotype Columbia gl1) was crossed to Ler accession, and 1500 mutant plants were selected from the F2 population based on the ABA-sensitive phenotype on MS medium supplemented with 30 μM ABA. Simple sequence length polymorphism markers were used to map ABO6 first to chromosome 5 between the BAC clones F8L15 and T20L5. Markers in MED24, T1E3, MUK11, MUG13, and K18I23 were then used to narrow the location of the abo6 mutation to a region between the BAC clone MUK11 and MUG13. All candidate genes in this region were sequenced and a G–A point mutation was found in AT5G04895.
For complementation of the abo6 mutant, ABO6 cDNA was amplified from reverse-transcribed cDNA with the primer pair 5′-CTGCAGATGCGTTTCACAAAAAGAATAAGCCTCTTC-3′ and 5′-GGTACCTTACTTGCCCTTGGATCGCCTGC-3′. The PCR products were cloned in the T-easy vector (Promega). The DNA fragments were then cloned in the PstI and KpnI sites of a modified vector pCAMBIA1300 under the control of a super promoter. The cloned construct was transformed into Agrobacterium tumefaciens GV3101 cells and transferred into plants using the floral dip method (Clough and Bent, 1998). Transgenic homozygous lines in the T3 generation were used for complementation.
A Salk T-DNA insertion line SALK_019865, obtained from the Arabidopsis Information Resource, was lethal in the homozygous state. abo6 was crossed with the heterozygous T-DNA lines to obtain the F1 seedlings, and their sensitivity to ABA was examined. The genomic DNA fragments isolated from ABA-sensitive lines were examined by PCR analysis, and all lines were determined to be heterozygous mutants that contained both abo6 and the SALK_019865 T-DNA insertion. The primers used for amplification of T-DNA insertion were as follows: LP, 5′-ATTGGTTCTCATGAGTGAC-3′; RP, 5′-GGTGGTCTCTTTTGCTTTTCC-3′; TF, 5′-ATTTTGCCGATTTCGGAAC-3′. The primers used for amplification of abo6 mutation were as follows: abo6-dcaps-F, 5′-TAGAGCGGGATATCTTCGCAATGC-3′; abo6-dcaps-R, 5′-TAACCGTTTCACCAAGAGGAGCT-3′. The PCR products amplified from the wild type were cut with SacI, whereas those amplified from abo6 were not.
abo6 was also crossed with abi1-1and abi2-1 and the double mutants were identified (Allen et al., 1999).
Subcellular Localization of ABO6
Full-length ABO6 cDNA was obtained by PCR from the T-easy vector containing ABO6 cDNA with the primers 5′-CTGCAGATGCGTTTCACAAAAAGAATAAGCCTCTTC-3′ and 5′-GGTACCCTTGCCCTTGGATCGCCTGC-3′. The PCR products were cloned into the pMD18 T-vector (TaKaRa); the DNA fragments were then subcloned into the PstI and KpnI sites of a modified vector pCAMBIA1300 that contains GFP cDNA between the KpnI and SacI sites. The cloned construct was transformed into Agrobacterium GV3101 cells and transferred into plants using the floral dip method (Clough and Bent, 1998). Because GFP fluorescence was not observed in the transgenic plants, the N-terminal part of ABO6 that contains the mitochondrial signal peptide was used in place of full-length ABO6 cDNA using the primers 5′-CTGCAGATGCGTTTCACAAAAAGAATAAGCCTCTTC-3′ and 5′-GGTACCATCTGGACGCCGTGGAAGAAG-3′ in pCAMBIA1300-GFP. After sequence analysis, correct vectors were transformed into isolated Arabidopsis protoplasts, which were stained with the Mito-tracker and examined with a Zeiss LSM 510 META confocal microscope. For confocal microscopy, green (GFP) images were obtained with an excitation at 488 nm and emission at 525 nm, and red images (MitoTracker stain) were obtained with an excitation at 543 nm and emission at 615 nm.
ROS Assays
H2O2 was detected in situ by DAB staining (Thordal-Christensen et al., 1997). The terminal leaflet of the first fully expanded leaf was sampled from 28-d-old wild-type and abo6 plants treated with 50 μM ABA or drought for 3 h. Leaflets were collected and vacuum-infiltrated with DAB solution (1 mg/mL, pH 3.8; Sigma-Aldrich). The sampled leaflets were placed in a plastic box in a light homoeothermic incubator until a reddish-brown precipitate was observed (8 h); they were then fixed in a solution of 3:1:1 ethanol:lactic acid:glycerol and photographed. In the roots, H2O2 was stained with 0.1 mg/mL DAB in 50 mM Tris buffer, pH 5.0 (Benitez-Alfonso et al., 2009), with subsequent steps being the same as those described for leaflets.
The DCFH-DA staining assay was performed as described previously (Liu et al., 2010). Briefly, 4-d-old seedlings were treated with 0 or 50 μM ABA for 5 h, incubated in 50 μM DCFH-DA in 20 mM K-phosphate, pH 6.0, in darkness for 10 min, and washed three times with the same buffer. Fluorescence was detected with a confocal microscope (Zeiss LSM 510 META) with an excitation at 488 nm and emission at 525 nm and collected by scanning different slices the bottom to the top of a sample. The slices were overlaid using the projection function of Zeiss LSM Image Brower software, and the fluorescence intensities were collected using AxioVision Rel. 4.8 software, which was displayed as estimated mean pixel intensities. Approximately 20 images were acquired per sample, and three independent experiments were performed.
We used NBT staining to detect O2− in situ as described previously (Dunand et al., 2007), with the following modifications. Four-day-old seedlings were treated with 0 or 50 μM ABA for 5 h, incubated in 2 mM NBT in 20 mM K-phosphate with 0.1 M NaCl at pH 6.1 for 15 min, and then transferred to distilled water. Root tips were imaged under bright-field illumination on a Zeiss Scope AI microscope, and all of the images in an experiment were taken with the same settings. Approximately 20 images were taken each time, and three independent experiments were performed.
Determination of GSH Content
Four-day-old seedlings were transferred to MS medium containing 30 μM ABA and 1 mM BSO and grown for another 7 d. Then, root tips were collected for quantitative measurement of GSH. The GSH content was measured using a Glutathione Colorimetric Detection Kit (BioVision), with the following minor modifications (Su et al., 2011). The root tips were ground to powder using liquid nitrogen, ∼50 mg of each sample was then dissolved in 400 μL GSH buffer, and 100 μL 5% sulfosalicylic acid was added and mixed gently. The samples were incubated on ice for 5 min and centrifuged at 12,000g at 4°C for 15 min, and the supernatants were collected for GSH detection.
For determination of GSH content, the samples or GSH standards (20 μL) were mixed with 160 μL of reaction solution (20 μL NADPH generating mix and 140 μL glutathione reaction buffer) on a microtiter plate and incubated at room temperature for 10 min. Then, 20 μL of substrate was added to each sample, and the absorbencies at 405 nm were recorded for 5, 10, 20, and 30 min. The GSH content was calculated according to the standard curve.
RNA Gel Blot Analysis and Quantitative RT-PCR
Two-week-old seedlings were submerged into 50 μM ABA for 5 h. Total RNAs were extracted and hybridized with different probes as described previously (Liu et al., 2010). The primers used to amplify the probes for RNA gel blot analysis are listed in Supplemental Table 1 online.
For real-time qRT-PCR, ∼4 μg of total RNAs was digested with RNase-free DNase I. The digested RNA was then reverse transcribed into cDNA using oligo(dT) primers (Promega) and Moloney Murine Leukemia Virus Transcriptase (Promega) in a 20-μL reaction. The cDNAs were used as templates in real-time PCR reactions using SYBR Premix Ex Tag (TaKaRa) with gene-specific primers and the internal control (ACTIN2/8). The primers used for qRT-PCR are listed in Supplemental Table 2 online; the primers were used to amplify NAD1b, NAD1c, NAD2b, NAD4, and NAD5a are listed in Supplemental Table 1 online (Liu et al., 2010). PCR was performed using a PTC-200 DNA engine cycle (MJ Research) with a Chromo4 detector in 20-μL reactions and the following amplification procedure: 5 min at 95°C (one cycle), 15 s at 95°C, 15 s at 57 to 60°C, 20 s at 72°C (40 cycles), and 5 min at 72°C. Next, a standard dissociation protocol was used to examine whether the amplifications were specific. Triple biological replications were performed for each of three independent samples, and the values shown represent the average of three assays for each real-time PCR sample. The results are given as a ratio of the values in the wild type.
For the analysis of NAD splicing, the primers were designed to amplify two different fragments (de Longevialle et al., 2007). One pair of primers was used to amplify the fragment that spans two adjacent exons (to quantify spliced mRNA); another pair of primers was used to amplify the intron-exon fragment (to quantify unspliced mRNA). These primers are listed in Supplemental Table 3 online. If amplification of the intron-exon fragment in abo6 was greater than that in the wild type, it means that the splicing was impaired in abo6.
Marker Gene Analysis
Arabidopsis plants harboring ProDR5:GUS, ProIAA2:GUS, ProAUX1:AUX1-YFP, ProPIN1:PIN1-GFP, ProPIN2:PIN2-GFP (Wang et al., 2011), and Mito-cpYFP were crossed with abo6. Homozygous plants for the mutant and marker lines were separated from the F2 and F3 population before they were subjected to GUS staining or GFP. Four-day-old seedlings were transferred to MS containing 30 μM ABA, 600 μM GSH, or 2.5 mM BSO and grown for another 2 d.
For GUS staining, seedlings were incubated in 0.1 M phosphate buffer (a mixture of KH2PO4 and K2HPO4, pH 7.0) and 5 mg/mL X-Gluc at 37°C for different times and were then incubated in 75% ethanol for several hours to remove chlorophylls. Staining time was 40 min for ProIAA2:GUS and 24 h for ProDR5:GUS. Seedlings were photographed with an Olympus stereofluorescence microscope (SZX16 -DP72) and a Zeiss Scope AI microscope.
For GFP fluorescence, the roots of the plants treated with different reagents were observed with a Zeiss LSM 510 META confocal microscope and photographed under the same settings. The fluorescence images were collected by scanning different slices from one side to the other side, and the images were overlaid using the projection function of Zeiss LSM image browser software. Fluorescence intensities were collected using AxioVision Rel. 4.8 software, which was displayed as estimated mean pixel intensities. Approximately 20 images were taken per sample, and three independent experiments were performed.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ABO6, At5g04895; ABO5, At1g51965; NDUFS4, At1g47260; ATRBOHF, At1g64060; ABI1, At4g26080; ABI2, At5g57050; AOX1a, At3g22370; NDB2, At4g05020; NDB3, At4g21490; NDB4, At2g20800; RD29A, At5g52310; ACTIN2, At3g18780; ACTIN8, At1g49240; TUB4, At5g44340; RPL2, Atmg00560; COX2, Atmg00160; CCB452, Atmg00180; RPS3, Atmg00090; NAD1B, Atmg01120; NAD1C, Atmg00516; NAD2A, Atmg00285; NAD2B, Atmg01320; NAD4, Atmg00580; NAD5A, Atmg00513; NAD5C, Atmg00060; and NAD7, Atmg00510.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Growth Phenotype of abo6.
Supplemental Figure 2. Seed Germination and Root Growth of ndufs4 and abo5 on ABA Containing MS Medium Supplemented with GSH or DTT.
Supplemental Figure 3. Seed Germination and Root Growth of abo6, the Wild Type, Ler, abi1-1, abi2-1, abo6 abi1-1, and abo6 abi2-1 in Response to ABA.
Supplemental Figure 4. GSH Contents in the Roots of Different Phenotypes under ABA and No ABA Treatments.
Supplemental Table 1. Primers Used to Amplify Fragments in RNA Gel Blots.
Supplemental Table 2. Primers Used for qRT-PCR.
Supplemental Table 3. Primers Used for the Splicing qRT-PCR Experiment of Mitochondrial Transcripts.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Nature Science Foundation of China (91117017 and 31121002), the National Basic Research Program of China (973 Program; 2012CB114300), the National Transgenic Research Project (2011ZX08009-002), and the China Postdoctoral Science Foundation–funded project (2011M500453). We thank Miguel Angel Torres (University of North Carolina at Chapel Hill) for providing atrbohF mutant seeds, Chuanyou Li (Institute of Genetics and Developmental Biology) for providing various auxin related materials, and the ABRC for providing the T-DNA insertion lines of ABO6.
AUTHOR CONTRIBUTIONS
J.H. and Z.G. designed the experiment. J.H. performed the majority of experiments and analyzed the data. Y.D., D.H., G.F., L.W., Y.L., Z.C., and L.-J.Q. assisted with the experiment and data analyses. L.H. assisted with the experiments. J.H. and Z.G. wrote the article.
Glossary
- ABA
abscisic acid
- ROS
reactive oxygen species
- H2O2
hydrogen peroxide
- PPR
pentatricopeptide repeat
- cpYFP
to be defined
- BSO
buthionine sulphoximine
- MS
Murashige and Skoog
- GFP
green fluorescent protein
- qRT-PCR
quantitative RT-PCR
- NBT
nitroblue tetrazolium
- DAB
3′3′ diaminobenzidine
- DCFH-DA
2′,7′-dichlorodihydrofluorescin diacetate
- Ler
Landsberg erecta
- NAA
1-napthalene acetic acid
- GUS
β-glucuronidase
- IBA
indole-3-butyric acid
References
- Allen G.J., Kuchitsu K., Chu S.P., Murata Y., Schroeder J.I. (1999). Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11: 1785–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bai L., Zhang G., Zhou Y., Zhang Z., Wang W., Du Y., Wu Z., Song C.P. (2009). Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 60: 314–327 [DOI] [PubMed] [Google Scholar]
- Bashandy T., Guilleminot J., Vernoux T., Caparros-Ruiz D., Ljung K., Meyer Y., Reichheld J.P. (2010). Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin C., Megies C., Hauserová E., Lopez-Molina L. (2009). Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21: 2253–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Cilia M., San Roman A., Thomas C., Maule A., Hearn S., Jackson D. (2009). Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Boudet N., Aubourg S., Toffano-Nioche C., Kreis M., Lecharny A. (2001). Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res. 11: 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., Sarkar S.F., Bonetta D., McCourt P. (2003). The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34: 67–75 [DOI] [PubMed] [Google Scholar]
- Chen Z., Hong X., Zhang H., Wang Y., Li X., Zhu J.K., Gong Z. (2005). Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43: 273–283 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Gong Z., Zhu J.K. (2008). Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant Biol. 50: 1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton R., Lister R., Parker K.L., Sappl P.G., Elhafez D., Millar A.H., Day D.A., Whelan J. (2005). Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679 [DOI] [PubMed] [Google Scholar]
- de Longevialle A.F., Meyer E.H., Andrés C., Taylor N.L., Lurin C., Millar A.H., Small I.D. (2007). The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand C., Crèvecoeur M., Penel C. (2007). Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: Possible interaction with peroxidases. New Phytol. 174: 332–341 [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D., Davies J.M., Dolan L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Tasaka M. (2009). Hormone interactions during lateral root formation. Plant Mol. Biol. 69: 437–449 [DOI] [PubMed] [Google Scholar]
- Garmier M., Carroll A.J., Delannoy E., Vallet C., Day D.A., Small I.D., Millar A.H. (2008). Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol. 148: 1324–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazarian I.G., Lagrimini L.M., Mellon F.A., Naldrett M.J., Ashby G.A., Thorneley R.N. (1998). Identification of skatolyl hydroperoxide and its role in the peroxidase-catalysed oxidation of indol-3-yl acetic acid. Biochem. J. 333: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Dong C.H., Lee H., Zhu J., Xiong L., Gong D., Stevenson B., Zhu J.K. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Lee H., Xiong L., Jagendorf A., Stevenson B., Zhu J.K. (2002). RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99: 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres S., Sabar M., Lelandais C., Chetrit P., Diolez P., Degand H., Boutry M., Vedel F., de Kouchkovsky Y., De Paepe R. (1997). Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc. Natl. Acad. Sci. USA 94: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant P., Kant S., Gordon M., Shaked R., Barak S. (2007). STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 145: 814–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler D., Schmidt-Gattung S., Binder S. (2010). The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol. Biol. 72: 459–467 [DOI] [PubMed] [Google Scholar]
- Koprivova A., des Francs-Small C.C., Calder G., Mugford S.T., Tanz S., Lee B.R., Zechmann B., Small I., Kopriva S. (2010b). Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J. Biol. Chem. 285: 32192–32199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A., Mugford S.T., Kopriva S. (2010a). Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Rep. 29: 1157–1167 [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C., Apel K., Danon A. (2004). Reactive oxygen signalling: The latest news. Curr. Opin. Plant Biol. 7: 323–328 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Lee H., Xiong L., Zhu J.K. (2002). A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14: 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J., Bouvier-Durand M., Morris P.C., Guerrier D., Chefdor F., Giraudat J. (1994). Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J., Merlot S., Giraudat J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.P., Montgomery T.A., Fahlgren N., Kasschau K.D., Nonogaki H., Carrington J.C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52: 133–146 [DOI] [PubMed] [Google Scholar]
- Liu Y., He J., Chen Z., Ren X., Hong X., Gong Z. (2010). ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J. 63: 749–765 [DOI] [PubMed] [Google Scholar]
- Maxwell D.P., Wang Y., McIntosh L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhard M., Grill E. (2001). Hydrogen peroxide is a regulator of ABI1, a protein phosphatase 2C from Arabidopsis. FEBS Lett. 508: 443–446 [DOI] [PubMed] [Google Scholar]
- Meinhard M., Rodriguez P.L., Grill E. (2002). The sensitivity of ABI2 to hydrogen peroxide links the abscisic acid-response regulator to redox signalling. Planta 214: 775–782 [DOI] [PubMed] [Google Scholar]
- Meyer K., Leube M.P., Grill E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Miao Y., Lv D., Wang P., Wang X.C., Chen J., Miao C., Song C.P. (2006). An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingam A., Toffano-Nioche C., Brunaud V., Boudet N., Kreis M., Lecharny A. (2004). DEAD-box RNA helicases in Arabidopsis thaliana: Establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol. J. 2: 401–415 [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M., Zolman B.K., Bartel B. (2003). IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15: 2979–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Pei Z.M., Mori I.C., Schroeder J. (2001). Abscisic acid activation of plasma membrane Ca(2+) channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1−1 and abi2−1 protein phosphatase 2C mutants. Plant Cell 13: 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Soukupová H., Schikora A., Zárský V., Hirt H. (2006). A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Potters G., Pasternak T.P., Guisez Y., Palme K.J., Jansen M.A. (2007). Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci. 12: 98–105 [DOI] [PubMed] [Google Scholar]
- Ren X., Chen Z., Liu Y., Zhang H., Zhang M., Liu Q., Hong X., Zhu J.K., Gong Z. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki K., Uemura M., Tsurumi S., Rahman A. (2009). Auxin response in Arabidopsis under cold stress: Underlying molecular mechanisms. Plant Cell 21: 3823–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C., Gu D., Hu H.C., Davanture M., Lee S., Djaoui M., Valot B., Zivy M., Leung J., Merlot S., Kwak J.M. (2009). Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Stonebloom S., Burch-Smith T., Kim I., Meinke D., Mindrinos M., Zambryski P. (2009). Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc. Natl. Acad. Sci. USA 106: 17229–17234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Xu J., Li Y., Lei L., Zhao L., Yang H., Feng J., Liu G., Ren D. (2011). Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23: 364–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung T.Y., Tseng C.C., Hsieh M.H. (2010). The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J. 63: 499–511 [DOI] [PubMed] [Google Scholar]
- Teale W.D., Ditengou F.A., Dovzhenko A.D., Li X., Molendijk A.M., Ruperti B., Paponov I., Palme K. (2008). Auxin as a model for the integration of hormonal signal processing and transduction. Mol. Plant 1: 229–237 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z., Wei Y., Collinge D.B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11: 1187–1194 [Google Scholar]
- Tiryaki I., Staswick P.E. (2002). An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 130: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti V.B., et al. (2010). Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J., Flors V., Mauch-Mani B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14: 310–317 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld M., Marienfeld J.R., Brandt P., Brennicke A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15: 57–61 [DOI] [PubMed] [Google Scholar]
- Vernoux T., Wilson R.C., Seeley K.A., Reichheld J.P., Muroy S., Brown S., Maughan S.C., Cobbett C.S., Van Montagu M., Inzé D., May M.J., Sung Z.R. (2000). The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hua D., He J., Duan Y., Chen Z., Hong X., Gong Z. (2011). Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 7: e1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2008). Superoxide flashes in single mitochondria. Cell 134: 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.K., Pickett F.B., Turner J.C., Estelle M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222: 377–383 [DOI] [PubMed] [Google Scholar]
- Xiong L., Zhu J.K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Zhang X., Liu J., Wang Y., He J., Yang T., Hong X., Yang Q., Gong Z. (2009). Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell 21: 386–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Qin C., Zhao J., Wang X. (2004). Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu H., Zhang Q., Li M., Yan M., Wang R., Wang L., Welti R., Zhang W., Wang X. (2009). Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21: 2357–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Hua D., Chen Z., Zhou Z., Gong Z. (2009). Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 60: 79–90 [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.