This study identifies a regulatory oxidative pathway, comprised of thioredoxin and peroxiredoxin, in Arabidopsis thaliana chloroplasts. It shows that the pathway is used to sense photosynthetic peroxide formation under low to moderate light intensity and proposes that the oxidative signal adjusts the photosynthetic linear electron flow to fluctuating environmental conditions.
Abstract
The transition from dark to light involves marked changes in the redox reactions of photosynthetic electron transport and in chloroplast stromal enzyme activity even under mild light and growth conditions. Thus, it is not surprising that redox regulation is used to dynamically adjust and coordinate the stromal and thylakoid compartments. While oxidation of regulatory proteins is necessary for the regulation, the identity and the mechanism of action of the oxidizing pathway are still unresolved. Here, we studied the oxidation of a thylakoid-associated atypical thioredoxin-type protein, ACHT1, in the Arabidopsis thaliana chloroplast. We found that after a brief period of net reduction in plants illuminated with moderate light intensity, a significant oxidation reaction of ACHT1 arises and counterbalances its reduction. Interestingly, ACHT1 oxidation is driven by 2-Cys peroxiredoxin (Prx), which in turn eliminates peroxides. The ACHT1 and 2-Cys Prx reaction characteristics in plants further indicated that ACHT1 oxidation is linked with changes in the photosynthetic production of peroxides. Our findings that plants with altered redox poise of the ACHT1 and 2-Cys Prx pathway show higher nonphotochemical quenching and lower photosynthetic electron transport infer a feedback regulatory role for this pathway.
INTRODUCTION
In nature, plants grow under fluctuating environmental conditions and require frequent acclimation for optimal growth. The acclimation is particularly crucial in the morning when photosynthesis commences and chloroplast metabolism is drastically reconfigured. If not regulated properly, the conversion of light energy into photosynthetic linear electron flow can turn in a short period of time into excessive production of deleterious reactive oxygen species (ROS). Perhaps because of the short time response needed, plants use photosynthetic redox signals as a direct and dynamic means to regulate multiple chloroplast phenomena, controlling the production of reducing equivalents and alleviating their inflicted damage (Karpinski et al., 1999; Trebitsh and Danon, 2001; Pfannschmidt, 2003; Rochaix, 2007; Schürmann and Buchanan, 2008). Yet, the mechanisms by which the redox signals are sensed are only beginning to unravel.
While the redox state of the regulatory redox proteins must change uniquely according to their function (Danon, 2002), the exact nature of these redox changes and the mechanism of their regulation is yet to be fully understood. Naturally, counterbalancing reducing and oxidizing activities controls the regulatory redox state changes. The clarification of the details of each of these opposing reactions is important to our understanding of the biological role of the regulatory pathway and to our basic understanding of the principles of redox regulation. In plants growing under optimal conditions, the majority of photosynthetic reducing equivalents are passed from photosystem I (PSI) via ferredoxin to ferredoxin-dependent NADP+ reductase to produce NADPH, providing the reducing power needed for the carbon fixation reactions. Nevertheless, a fraction of the reducing equivalents are transferred, for regulatory purposes, from ferredoxin via ferredoxin-dependent thioredoxin reductase to several chloroplast thioredoxins (Trxs). This reductive signal is then used by the Trxs to reduce disulfide bonds in regulatory sites of target chloroplast proteins, thereby modulating their activity (Schürmann and Buchanan, 2008; Meyer et al., 2009). In contrast with the photosynthetic reductive signal, the identity and mechanism of action of the oxidizing activity of the Trx-regulated proteins are yet to be established. Defining the oxidizing activity and the time at which it occurs could also be important for discerning the biological context of the redox-regulated phenomenon. Trx-regulated proteins are generally oxidized in the dark and are reduced upon illumination (Schürmann and Buchanan, 2008), but a Trx-implicated oxidizing activity was also found in the light (Trebitsh et al., 2000; Martinsuo et al., 2003), indicating a broader regulatory role for protein thiol oxidation.
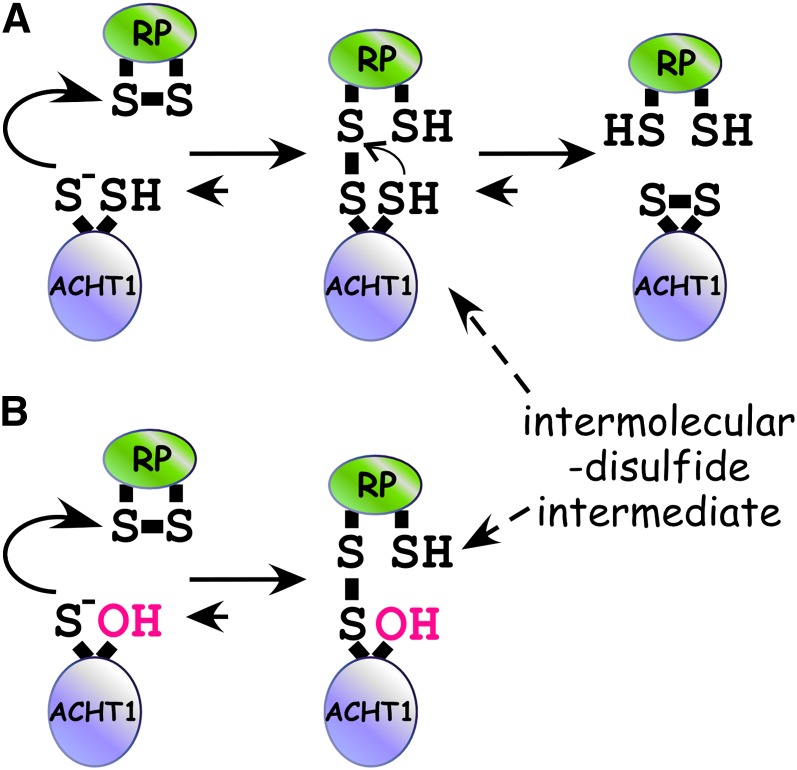
Mechanistically, the dithiol-disulfide exchange reaction between a Trx and its redox partner is a two-step reaction (Figure 1A) (Kallis and Holmgren, 1980). It is initiated by the nucleophilic N-terminal thiol of the Trx dithiol catalytic site. The reaction then goes through a transient intermolecular-disulfide intermediate (i.e., a covalent bond linking the Trx and its target) and is subsequently resolved by the neighboring thiol of the catalytic site. This mechanism allows for the trapping and identification of Trx redox partner proteins. A redox-active site monothiol mutant, in which the resolving Cys is replaced with a Ser or an Ala, is typically used to inhibit the second step and to increase the stability of the reaction intermediate (Figure 1B). This approach facilitated the isolation in vivo (Walker et al., 1996; Frand and Kaiser, 1999; Senkevich et al., 2002) or in vitro (Motohashi et al., 2001; Balmer et al., 2003) of Trx targets and the identification in vitro of the Cys residues participating in the intermolecular disulfide (Alergand et al., 2006). Importantly, protein disulfide isomerase (PDI) and its oxidizing protein, ENDOPLASMIC RETICULUM OXIDOREDUCTIN1 (ERO1), were trapped by this approach in an intermolecular disulfide complex in yeast and human cells (Pollard et al., 1998; Frand and Kaiser, 1999; Mezghrani et al., 2001). In a similar approach, a Cys mutant form of the Saccharomyces cerevisiae AP-1–like transcriptional factor (Yap1) was used to trap and identify a thiol peroxidase as a hydrogen peroxide receptor and redox transducer (Delaunay et al., 2002).
Figure 1.
Reaction Scheme of the Two-Step Dithiol Disulfide Transfer Reaction of ACHT1.
(A) Initially, the nucleophilic N-terminal Cys of the ACHT1 active site (s−) attacks the disulfide bond of its redox protein partner (RP), resulting in an intermolecular-disulfide reaction intermediate between ACHT1 and RP. Next, an attack of the C-terminal Cys of the ACHT1 active site on the intermolecular-disulfide bond releases oxidized ACHT1 and reduced RP.
(B) In a redox-active site mutant of ACHT1, in which the C-terminal Cys is replaced with a Ser, the resolving step is inhibited, resulting in increased stabilization of the intermolecular disulfide intermediate.
[See online article for color version of this figure.]
Compelling data supporting the coexistence of redox regulatory pathways and ROS formation in chloroplasts (Apel and Hirt, 2004; Pitzschke et al., 2006; Miller et al., 2008; Foyer and Noctor, 2009) promoted the notion that ROS might be used for regulatory protein oxidation (Apel and Hirt, 2004; Møller et al., 2007). Studies of bacteria, plants, and animals revealed that ROS signals could be perceived directly by specific sensors (Storz et al., 1990; D'Autréaux and Toledano, 2007) or by the use of antioxidative enzymes, such as peroxiredoxins (Prxs) (D'Autréaux and Toledano, 2007; Fourquet et al., 2008; Dietz, 2011; Fomenko et al., 2011). Prxs are ancient conserved enzymes that catalytically reduce hydrogen peroxide and organic peroxides and in turn oxidize their electron donors, such as Trxs, to regenerate their activity (Hall et al., 2009; Dietz, 2011). Plant Prxs were implicated as redox sensors, linking the redox signaling and ROS networks of cells (Dietz, 2011). Recent findings identified particular 2-Cys Prxs as a circadian rhythmic biomarker in algae cells and mammalian red blood cells (O'Neill and Reddy, 2011; O'Neill et al., 2011) and as a regulator of Drosophila melanogaster embryo development (Degennaro et al., 2011).
Photosynthetic overreduction conditions can lead to the production of deleterious ROS in photosystem II (PSII) and in the reducing side of PSI. Hence, it is not surprising that electron acceptors on the reducing side of each of the two photosystems play key intrachloroplast regulatory roles in the early acclimation to photosynthetic overreduction conditions. Redox state changes of the plastoquinone pool regulate the reversible State I–State II transition phenomenon, redistributing the light capturing capacity between PSII and PSI and the phosphorylation of PSII proteins via the Ser/Thr protein kinases STN7 and STN8 (Allen et al., 1981; Zer et al., 1999; Bellafiore et al., 2005; Bonardi et al., 2005). In comparison, redox states of Trxs and the NADPH pool have been implicated in the coordination of stromal activity and in feedback regulation of the membranal reactions of photosynthesis (Johnson, 2003; Martinsuo et al., 2003; Hald et al., 2008; Schürmann and Buchanan, 2008). Several studies of the state transition phenomenon (Tikkanen et al., 2010), of the translation of the chloroplast psbA mRNA (Trebitsh et al., 2000; Trebitsh and Danon, 2001), and of chloroplast transcription (Pfannschmidt and Liere, 2005; Lemeille et al., 2009; Steiner et al., 2009) further suggested a cooperative regulation of the plastoquinone and the Trx pathways. This cooperative regulation is most likely important for maintaining a critical balance between the production of reducing equivalents by the light reactions and their consumption by stromal activity to avoid generating detrimental ROS. Such disparity could arise temporarily even under moderate light intensity during the transition of plants from night to day (Asada, 2006; Cardol et al., 2010).
The Arabidopsis thaliana ACHT1 (for Atypical Cysteine Histidine rich Trx1) is a member of a small family of chloroplast localized Trxs that are unique to plastid-containing viridiplantae. ACHT members have an atypical redox active site and a less reducing redox midpoint potential than that of the classic chloroplast Trxs (Dangoor et al., 2009). Here, our studies in vivo show that ACHT1 accepts electrons from the linear photosynthetic electron transfer reactions and, in turn, reduces the chloroplast 2-Cys Prx. Notably, we found that ACHT1 undergoes rapid redox state changes during the transition of plants from dark to light and afterwards, such that after a short period of net reduction, the ACHT1 pool is kept in a partially oxidized state in the light. Our collective findings that increased reduction of the 2-Cys Prx by ACHT1 results in increased nonphotochemical quenching (NPQ) and photosynthetic electron transport rate (ETR) and the in vivo properties of the kinetics of ACHT1 and the 2-Cys Prx reaction emphasize a regulatory role rather than a pure antioxidative function for this pathway. Thus, we suggest a mechanism by which ACHT1, together with 2-Cys Prx, forms a regulatory oxidative pathway that feedback regulates photosynthetic electron transport in response to illumination of dark-adapted plants with low to moderate light intensity.
RESULTS
ACHT1 Reoxidizes in Plants after a Short Term of Reduction in the Light
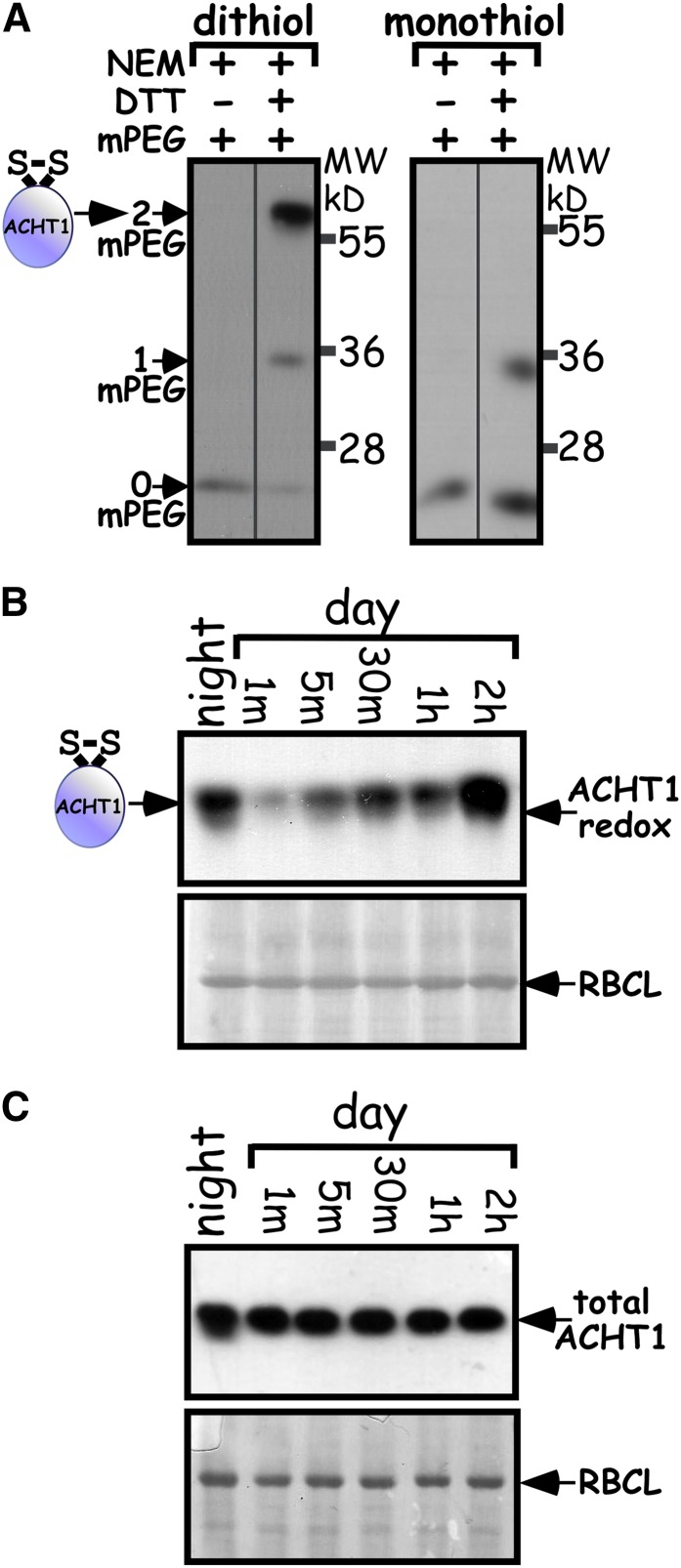
Defining the changes in the redox state of regulatory proteins in vivo is essential to our understanding of their biological role. The highly reductive nature of the intracellular milieu suggests that the regulatory proteins must oxidize for redox signaling (Danon, 2002). Thus, to understand the role of ACHT1 and the mechanism by which it exerts its function, we studied its oxidation in planta. We trapped ACHT1 disulfides in vivo by adapting to plants (Peled-Zehavi et al., 2010) the methodology that was used to study PDI and ERO1 redox state changes and reaction intermediates in yeast cells (Frand and Kaiser, 1999). Transgenic plants expressing the chloroplastic ACHT1 fused with a hemagglutinin (HA) affinity tag at its C terminus under the control of the 35S promoter (Dangoor et al., 2009) were grown under long-day conditions with moderate light intensity, 80 to 100 μE/m2s. Protein disulfides were trapped by extracting whole plants in 10% trichloroacetic acid and then irreversibly blocking the reduced Cys residues with N-ethylmaleimide (NEM). Subsequently, the disulfide-bonded Cys residues, which are protected from NEM modification, were reduced with DTT, labeled with methoxypolyethylene glycol-maleimide (mPEG), and visualized in an immunoblot assay of fractionated denatured proteins using anti-HA specific antibodies. In plants expressing ACHT1 with an active catalytic site (dithiol site; Cys-X-X-Cys), a major high molecular mass band was identified only in extracts reduced with DTT prior to labeling (Figure 2A), as expected for disulfides. The band corresponded to ACHT1 labeled with two mPEGs, likely resulting from capturing ACHT1 in which the catalytic site was disulfide bonded in plants. A second faint band with a molecular mass corresponding to ACHT1 modified by one mPEG and conceivably resulting from capturing ACHT1 engaging in vivo in an intermolecular disulfide bond was identified as well. The 22-kD band that was present in both DTT-treated and nontreated extracts corresponded to a nonlabeled ACHT1, resulting from capturing fully reduced ACHT1 in vivo. In a control experiment with plants expressing ACHT1C35S, containing a mutated catalytic site (monothiol site; Cys-X-X-Ser), the disulfide-bonded form disappeared and was replaced with a single oxidized Cys form (Figure 2A), indicating that only the two Cys residues of the catalytic site of ACHT1 engage in disulfide exchange reactions in planta.
Figure 2.
ACHT1 Reoxidizes in Plants after Illumination.
(A) The two Cys residues of the ACHT1 active site engage in vivo in a disulfide exchange reaction. Immunoblot assay showing the in vivo redox state of ACHT1 Cys residues captured in plants expressing either the catalytic active ACHT1 (dithiol) or the ACHT1C35S mutant (monothiol). The number of oxidized Cys residues was determined by the increased molecular mass of ACHT1 after labeling with mPEG (indicated by arrows). Two Cys residues were oxidized in the catalytic active ACHT1 and one in ACHT1C35S, indicating that the two Cys residues of the ACHT1 active site engage in vivo in disulfide exchange reactions (schematically depicted on the left). NEM treatment blocked reduced thiols. DTT reduced disulfides prior to mPEG labeling.
(B) ACHT1 reoxidizes in plants shortly after illumination. Immunoblot assay showing the in vivo oxidized state of ACHT1 active-site Cys residues captured in plants at the end of the night and at 1 min (m), 5 min, 30 min, 1 h, and 2 h after illumination. Equal loading was verified by the stained level of ribulose-1,5-bis-phosphate carboxylase/oxygenase (RBCL). The results shown are representative of five independent experiments.
(C) ACHT1 amount remains constant throughout the day. Immunoblot assay showing the total amount of ACHT1 in the plants at the indicated time points. Equal loading was verified by the stained level of ribulose-1,5-bis-phosphate carboxylase/oxygenase.
[See online article for color version of this figure.]
Next, we wished to study whether the ACHT1 catalytic site is oxidized during illumination of plants. When the redox state of ACHT1 was determined in vivo at several time points, starting at the end of the night and into the day, we found that the pool of ACHT1 catalytic site was largely disulfide bonded at the end of the night (Figure 2B). Illumination caused rapid reduction, within 1 min into the light period, of the ACHT1 pool in the plants. Thereafter, the atypical Trx ACHT1 catalytic site underwent net reoxidation, although minor fluctuations in its redox state were visible between different time points. A control experiment showed that the total amount of ACHT1 did not change significantly (Figure 2C) and thereby did not interfere with the observed changes in ACHT1 redox state. We concluded that after a short period of net reduction in the light, a significant oxidation reaction of the ACHT1 catalytic site arises and counterbalances its reduction. As a result, the ACHT1 pool is kept thereafter in a partially disulfide-bonded state. The major redox state changes of the ACHT1 pool in plants illuminated with moderate light intensity suggested a regulatory homeostatic role for ACHT1.
ACHT1 Is Reduced in the Light by Photosynthetic Electron Flow
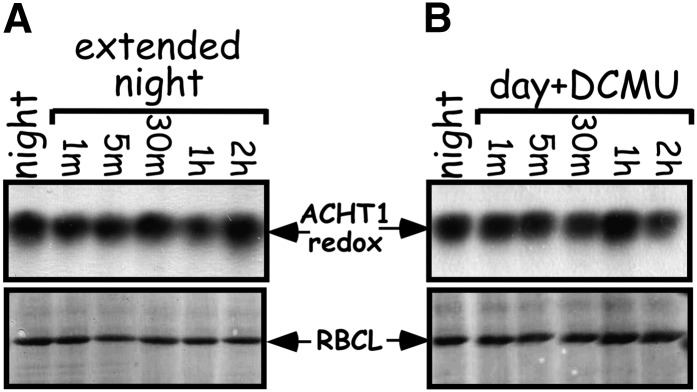
In the light, the classic chloroplastic-type Trxs are typically reduced by photosynthesis through the ferredoxin-Trx system (Schürmann and Buchanan, 2008). Thus, to test whether the atypical Trx ACHT1 is also reduced by photosynthesis, we studied the changes in its redox state in plants that were incubated under an extended dark period (Figure 3A). We found that the rapid reduction of the ACHT1 catalytic site upon illumination was missing in the prolonged dark period, showing that the reduction of ACHT1 is limited to the light period. This result also indicates that the reduction of ACHT1 is not under circadian control.
Figure 3.
ACHT1 Reduction Requires Photosynthetic Reducing Equivalents.
Immunoblot assay showing the in vivo oxidized state of ACHT1 active site Cys residues captured in plants that were left in the dark (A) or illuminated but treated with 50 μM of the photosynthetic transport inhibitor DCMU (B). Equal protein loading was verified as in Figure 2. The results shown are representative of three independent experiments.
We further examined whether inhibition of linear electron flow by the photosynthetic inhibitor DCMU affects ACHT1 redox state in plants. The results in Figure 3B show that DCMU inhibited the typical reduction of ACHT1 in the light (Figure 2B), indicating that the source of electrons for ACHT1 in plants is the linear electron flow of photosynthesis.
ACHT1 Reacts with 2-Cys Prx in Planta
Next, we sought to identify the redox partners with which ACHT1 engages in disulfide transfer reactions in planta and to unravel the source of its oxidation in the light. The disulfide exchange reaction goes through an intermolecular disulfide reaction intermediate that if captured could be used to isolate the authentic ACHT1 redox protein partners (Figure 1). Thus, we used nonreducing denaturing gel electrophoresis to study the intermolecular disulfide complexes formed and trapped by the trichloroacetic acid extraction (Peled-Zehavi et al., 2010) in plants expressing either a catalytic active HA-tagged ACHT1 or the monothiol mutant ACHT1C35S (Figure 4A). The catalytic active ACHT1 captured in planta high molecular mass complexes that were resistant to denaturation by SDS under nonreducing conditions but were sensitive to reduction by DTT (Figure 4A), as expected for authentic reaction intermediates linked by an intermolecular disulfide. As anticipated by the catalytic mechanism of the disulfide transfer reaction (Figure 1), the monothiol ACHT1C35S pulled down higher amounts of similar intermolecular disulfide complexes. Notably, the similar general pattern of the intermolecular disulfide complexes in plants expressing the catalytic active or the monothiol ACHT1 and their increased amounts in the monothiol plants validated the trapping of authentic reaction intermediates. Furthermore, the increased amounts of trapped proteins in ACHT1C35S plants made possible an efficient isolation by affinity purification technique. Hence, to identify the two major ACHT1-intermolecular disulfide complexes (Figure 4A, marked by asterisks), we enriched for SDS-denatured intermolecular disulfide complexes trapped in ACHT1C35S plants by affinity purification using anti-HA matrix and analyzed the corresponding gel slice by mass spectrometry (MS). The analysis identified the nearly identical chloroplastic 2-Cys Prx A and B as candidate targets of ACHT1 (see Supplemental Table 1 online). Reciprocal immunoprecipitation of SDS-denatured intermolecular disulfide complexes with 2-Cys Prx antisera pulled down ACHT1C35S (Figure 4B), authenticating the proteins’ interaction.
Figure 4.
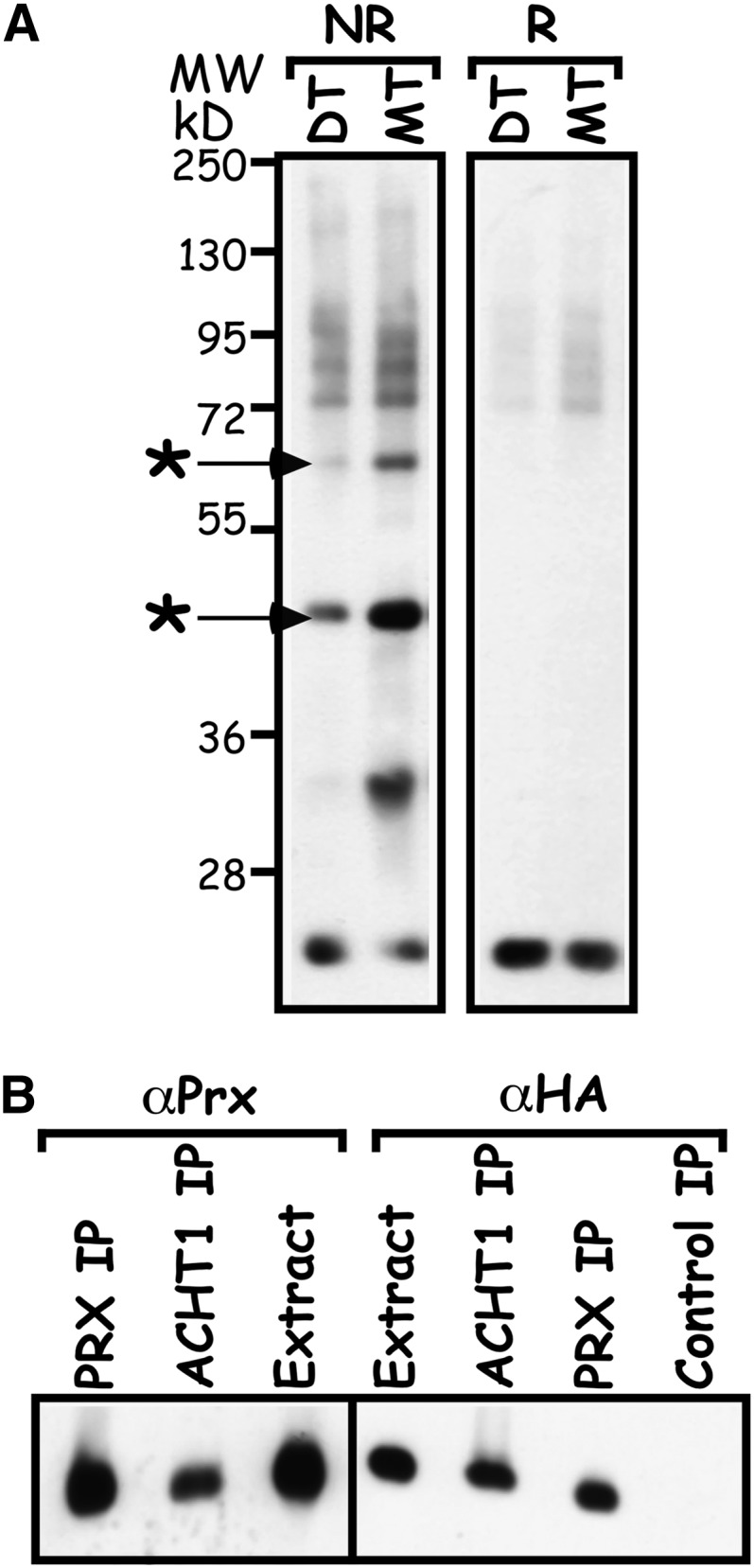
ACHT1 Reacts with 2-Cys Prx in Plants.
(A) Intermolecular disulfide complexes of ACHT1 and its redox protein partners. Immunoblot assay showing the intermolecular disulfide complexes trapped in plants expressing the catalytic active ACHT1 (DT) or the monothiol mutant ACHT1C35S (MT). Proteins were electrophoresed under nonreducing (NR) or reducing (R) conditions, and complexes containing ACHT1 were identified by immunoblot assays using anti-HA-specific antibodies. The major intermolecular disulfide complexes of ACHT1, which were identified by MS, are marked with an asterisk.
(B) Reciprocal immunoprecipitation verified the ACHT1–2-Cys Prx interaction. Immunoblot assay of proteins immunoprecipitated with anti-HA (ACHT1 IP) or anti-2-Cys Prx (Prx IP) affinity matrixes or with nonspecific matrix (Control IP) from plants expressing ACHT1C35S. Purified proteins were run under reducing conditions and blotted with antibodies specific to the HA-tag of ACHT1 (αHA) or to 2-Cys Prx (αPrx).
ACHT1 formed at least three additional intermolecular disulfide complexes with estimated molecular masses ranging between 95 and 130 kD (Figure 4A) and one approximately at 38 kD. The identity and the role of the ACHT1 targets in these intermolecular disulfide complexes are yet to be established.
ACHT1 Reduces the 2-Cys Prx in Planta
The chloroplast 2-Cys Prx is a typical Prx that accepts electrons from Trx and in turn reduces and eliminates hydro- and alkyl-peroxides (Dietz, 2011). As the trapping of ACHT1 2-Cys Prx reaction intermediate (Figure 4) implied that ACHT1 donates electrons to the 2-Cys Prx in plants, we examined the effect of elevated expression of ACHT1 (see Supplemental Figure 1A online) on the redox state in vivo of 2-Cys Prx in the light (Figure 5). The active chloroplast 2-Cys Prx is a head-to-tail homodimer containing two identical catalytic sites. Its reaction with peroxide is initiated by the peroxidatic Cys that forms a Cys sulfenic acid reaction intermediate. Next, the sulfenic acid from one subunit reacts with a resolving Cys of the second subunit to form a stable intersubunit disulfide bond. The 2-Cys Prx is then typically reduced and recycled by Trx (Hall et al., 2009). The mPEG labeling procedure discriminated between the three forms of 2-Cys Prx: fully oxidized, resulting from a 2-Cys Prx dimer in which both the peroxidatic and resolving Cys residues of each 2-Cys Prx molecule are intermolecularly disulfide bonded; semioxidized, resulting from a 2-Cys Prx dimer in which only the peroxidatic Cys of one 2-Cys Prx molecule is intermolecularly disulfide bonded; and a fully reduced form in which all Cys residues of the 2-Cys Prx dimers are reduced. We found that the pool of 2-Cys Prx was more reduced, with less fully and semioxidized forms and more of the fully reduced form, in plants with elevated expression of catalytic active ACHT1 compared with wild-type plants (Figure 5). In addition, the lack of efficient peroxide elimination in vitro in reactions containing ACHT1 but devoid of 2-Cys Prx (see Supplemental Figure 2 online) ruled out a direct catalytic reaction of ACHT1 with peroxides. We concluded that ACHT1 reduces 2-Cys Prx, and in turn is oxidized, in plants in the light.
Figure 5.
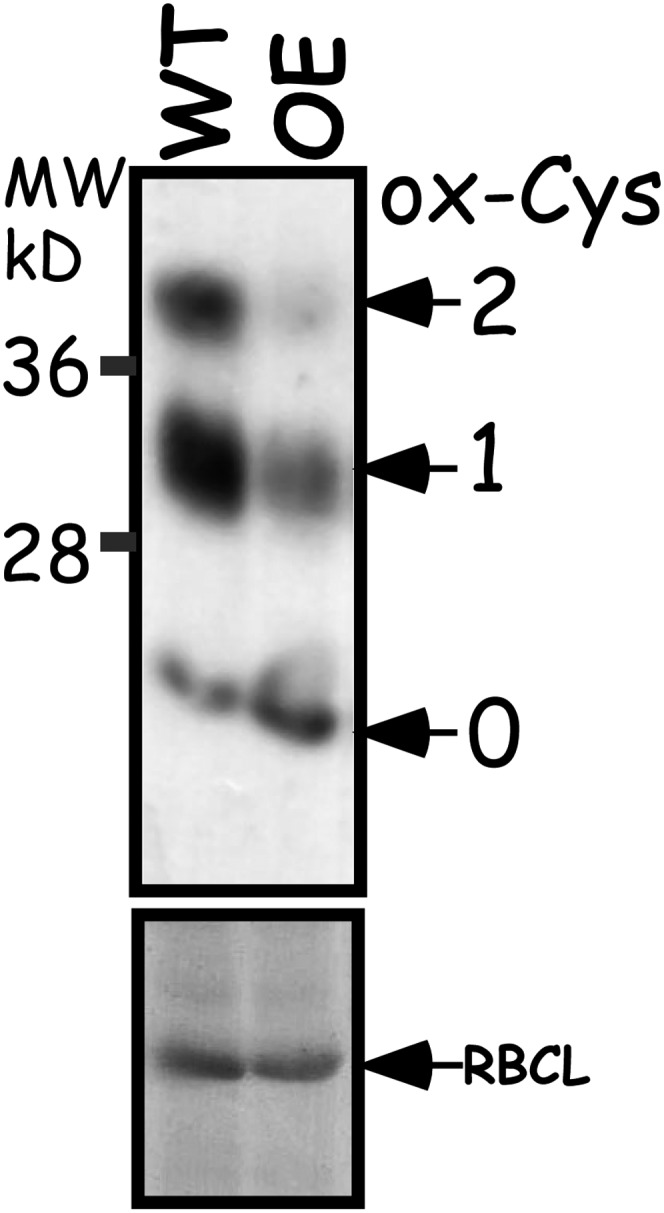
2-Cys Prx Is Reduced by ACHT1 in Planta.
Immunoblot assay showing the oxidized state of 2-Cys Prx Cys residues in plants after 5 min of illumination. The number of oxidized Cys residues (ox-Cys) was determined by the increased molecular mass of 2-Cys Prx after labeling with mPEG (indicated by arrows) as detected by 2-Cys Prx-specific sera. The assay showed that the 2-Cys Prx pool was more reduced in plants with increased expression of ACHT1 (OE) relative to wild-type plants (WT). Equal protein loading was verified as in Figure 2. The results shown are representative of three independent experiments. RBCL, ribulose-1,5-bis-phosphate carboxylase/oxygenase.
2-Cys Prx-ACHT1 Reaction Characteristics in the Light Implies an Oxidative Sensing Role
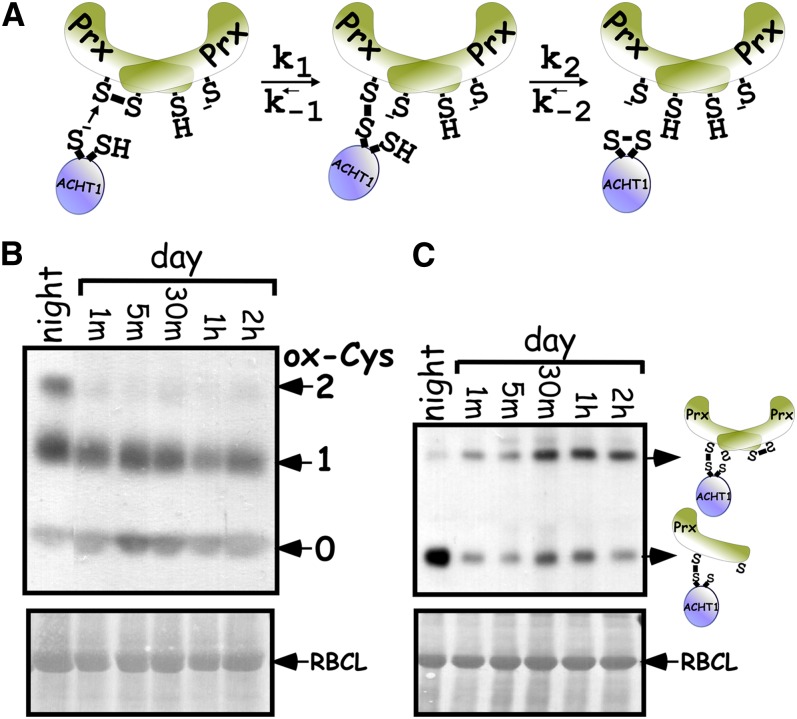
The major redox state changes that ACHT1 underwent during illumination of plants (Figure 2B) suggested a regulatory role for ACHT1. We later found that ACHT1 redox state is determined by the balance between its rate of reduction by photosynthesis and its rate of oxidation by the antioxidative activity of 2-Cys Prx. Notably, the reoxidation of the ACHT1 pool (Figure 2B), which revealed that its rate of oxidation by 2-Cys Prx exceeded its rate of reduction, occurred during mild growth conditions under which peroxide formation was not likely to saturate an antioxidative path, thus corroborating a regulatory role for ACHT1. Hence, to determine whether the ACHT1 reaction with 2-Cys Prx is indeed used to link the ACHT1 redox state with the production of peroxides by photosynthesis, we decided to examine the in vivo characteristics of this pathway in more detail. We specifically focused on the intermolecular disulfide reaction intermediate, as it accumulated in plants expressing a catalytic active ACHT1 to levels higher than anticipated for an efficient reductive activity (Figure 4A, dithiol lane). Theoretical considerations of the reaction kinetics, as depicted in Figure 6A, suggest that the intermolecular-disulfide reaction intermediate of catalytic ACHT1 and 2-Cys Prx (Figure 4A, dithiol lane) could accumulate at times in which concentrations of the final reaction products, oxidized ACHT1 and reduced 2-Cys Prx, build up. As ACHT1 oxidized with prolonged illumination (Figure 2B), we analyzed at the same time points the redox state of 2-Cys Prx (Figure 6B) and the levels of the intermolecular disulfide complexes (Figure 6C). The results showed that the redox state of the 2-Cys Prx pool in wild-type plants in the dark (Figure 6B), assayed by the same methodology used for ACHT1 (Figure 2), was a mixture of fully oxidized and semireduced forms. Within 1 min in the light, the fully oxidized form disappeared and the 2-Cys Prx was detected mainly in the fully reduced and semireduced forms. After this initial stage, the 2-Cys Prx redox state fluctuated, but for most of the time was kept in the reduced and semireduced states. It is also important to note that in contrast with ACHT1 (Figure 2B), the pool of 2-Cys Prx did not go through major redox state changes during illumination of dark-adapted plants as was found for the circadian rhythmic biomarker 2-Cys Prxs (O'Neill and Reddy, 2011; O'Neill et al., 2011), implying that it is not used as a direct regulator.
Figure 6.
Characteristics of the ACHT1-2-Cys Prx Reaction in Plants.
(A) Reaction scheme of 2-Cys Prx reduction by ACHT1. Initially, the N-terminal Cys of the ACHT1 active site (s−) attacks the disulfide bridge linking the two monomers in a 2-Cys Prx dimer, resulting in an intermolecular disulfide reaction intermediate between ACHT1 and 2-Cys Prx. The rate constants of the forward and reverse reactions in this step are K1 and K-1, respectively. Next, an attack of the second Cys of the ACHT1 active site on the intermolecular disulfide bond releases oxidized ACHT1 and a reduced 2-Cys Prx. The rate constants of the forward and reverse reactions for this step are K2 and K-2, respectively. It is generally assumed that K2 is higher than K1 and that K-2 is much smaller than K2, favoring the forward reaction. If K-1 is smaller than K-2, then when the rates of the reverse reactions are high (i.e., when oxidized ACHT1 and reduced 2-Cys Prx concentrations are high), the intermediate will accumulate.
(B) 2-Cys Prx redox state after illumination. Immunoblot assay showing the oxidized state of 2-Cys Prx Cys residues in wild-type plants sampled at the indicated time points. The number of oxidized Cys residues (ox-Cys) was determined by the increased molecular mass of 2-Cys Prx after labeling with mPEG (indicated by numbers and arrows), as detected by 2-Cys Prx-specific sera. The assay showed that, in contrast with ACHT1, the pool of 2-Cys Prx did not go through major redox state changes during illumination of dark-adapted plants. Equal protein loading was verified as in Figure 2. The results shown are representative of four independent experiments. RBCL, ribulose-1,5-bis-phosphate carboxylase/oxygenase
(C) ACHT1-2-Cys Prx intermolecular disulfide complexes accumulate in plants when the ACHT1 pool oxidizes. Immunoblot assay showing the intermolecular disulfide complexes trapped in plants expressing the catalytic active ACHT1. Protein complexes were separated under nonreducing conditions and blotted with antibodies specific to the HA-tag of ACHT1. The heterodimer and heterotrimer ACHT1-2-Cys Prx complexes are indicated by arrows. Equal loading was verified as in Figure 2. The results shown are representative of five independent experiments.
[See online article for color version of this figure.]
Next, we studied the previously identified two ACHT1-2-Cys Prx intermolecular-disulfide reaction intermediates (Figure 4A) trapped at the same time points in plants expressing the catalytic active ACHT1 (Figure 6C). According to their estimated molecular mass and to the known mechanism of action of 2-Cys Prx, the faster migrating disulfide-bonded complex corresponded to an ACHT1-2-Cys Prx heterodimer and the slower migrating complex to a heterotrimer comprised of ACHT1 linked with a 2-Cys Prx disulfide-bridged dimer. In the dark, the amount of the oxidized heterodimer was the highest. Upon illumination, and thereupon, the rapid reduction of ACHT1, the amount of the heterodimer decreased to its lowest level. The subsequent increase in the heterodimer and the heterotrimer corresponded tightly to the timing of ACHT1 reoxidation in the light. Our conclusion that the combined level of the two intermolecular disulfide complexes was highest when ACHT1 was mostly oxidized (Figure 2) further indicated a smaller rate of forward reaction than that required for optimal antioxidative function even under mild light conditions.
Collectively, our findings that oxidized ACHT1 and reduced 2-Cys Prx were high at times at which ACHT1-2-Cys Prx intermolecular disulfide complexes accumulate portray a regulatory pathway in which the oxidation of the ACHT1 pool is favored over forward rates of the antioxidative reaction. Moreover, the significant redox state changes of the ACHT1 pool (Figure 2B) and the relatively minor changes of 2-Cys Prx (Figure 6B) indicate that it is the redox state changes of ACHT1 that might be used for sensing photosynthetic peroxide production.
ACHT1 Redox State depends on Light Intensity
The reaction characteristics of ACHT1 and 2-Cys Prx suggest that redox state changes of ACHT1 might sense via the 2-Cys Prx activity photosynthetic production of peroxides under moderate light. Thus, it was important to determine the changes in the redox state of ACHT1 at higher light intensity, which results in increased production of ROS by the Mehler reaction. As higher light intensity also results in greater photoreductive poise, which might increase the reduction of ACHT1, three possibilities could be envisioned: (1) The reduction rate of ACHT1 by photosynthesis is higher than its oxidation, resulting in a higher level of reduced ACHT1; (2) the increase in ACHT1 reduction equals its oxidation by 2-Cys Prx, resulting in no change in the ACHT1 redox state; and (3) the production of peroxides is proportionally high, leading to a higher rate of ACHT1 oxidation by 2-Cys Prx than its reduction by photosynthesis. To determine which of these occurs, we compared the redox state of ACHT1 in plants illuminated by 200 μE/m2s light intensity to plants illuminated by 80 μE/m2s. We detected an earlier reoxidation of ACHT1 in response to the higher light intensity (Figure 7), indicating that at 2.5 times higher light intensity, the rate of ACHT1 reduction by photosynthesis did not compensate for its increased oxidation by 2-Cys Prx. Thus, the reduction by photosynthetic linear electron flow appears to be limiting, permitting ACHT1 oxidation in response to peroxide production under both low and moderate light intensities. These results further specify that the parameters of the ACHT1-2-Cys Prx system link the redox state changes of ACHT1 with the rate of peroxide elimination by 2-Cys Prx. Notably, ACHT1 oxidation appeared to approach saturation under 200 μE/m2s light intensity, implying that this pathway operates under adequate conditions rather than acute oxidative stress.
Figure 7.
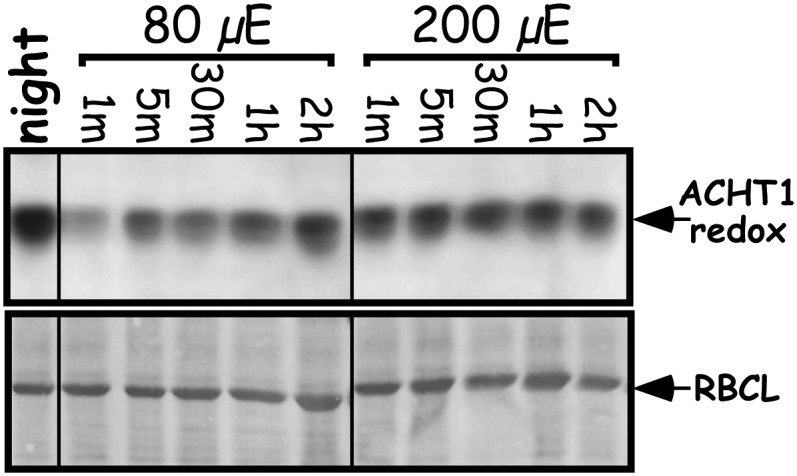
ACHT1 Redox State Is Dependent on Light Intensity.
Immunoblot assay showing the in vivo oxidized state of ACHT1 active site Cys residues captured in plants grown under 80 μE/m2s for 7 d and then illuminated by the same light intensity or by higher light intensity (200 μE/m2s) for the indicated time periods. Equal protein loading was verified as in Figure 1. The results shown are representative of four independent experiments.
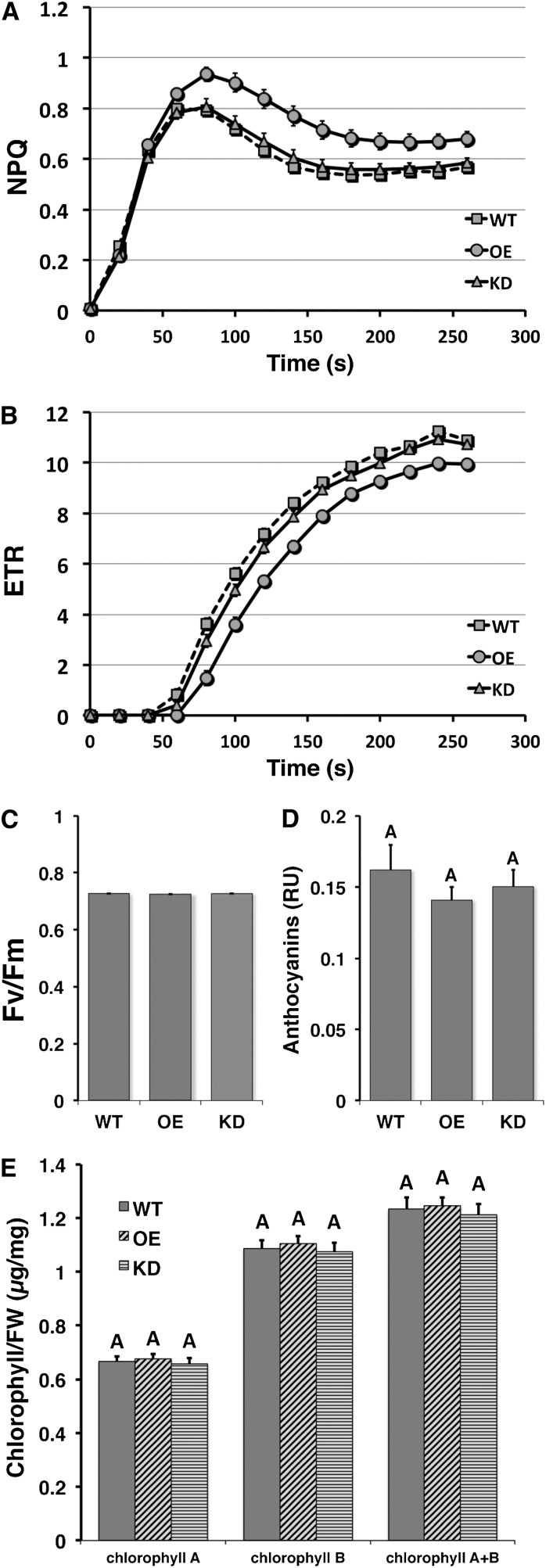
Altered ACHT1 Expression Affects Photosynthetic Electron Flow
Our findings indicated that the redox state of ACHT1 might be used to dynamically regulate a response to redox changes in the acceptor side of PSI. Such conditions could occur daily during illumination of dark-adapted plants when the stromal acceptors of photosynthetic reducing equivalents are being activated (Asada, 2006). Interestingly, ACHT1 was reoxidized under these conditions even when illuminated with moderate light intensity (Figure 2B). Thus, we searched for phenotypic consequences of altered levels of ACHT1 (see Supplemental Figure 1 online) by measuring the photosynthetic parameters of dark-adapted plants during illumination with 80 μE/m2s light intensity (Figure 8). Plants overexpressing (OE) ACHT1 showed increased NPQ (Figure 8A), indicating that a higher fraction of photosynthetic equivalents is channeled into heat rather than into production of reducing equivalents for carbon fixation. The ACHT1 overexpressor plants also showed a corresponding decrease in photosynthetic ETR (Figure 8B), indicating a lower rate of PSII photochemistry. ACHT1 knockdown (KD) plants showed only an insignificant increase in NPQ (Figure 8A) and decrease in ETR values compared to wild-type plants (Figure 8B), suggesting that other ACHT members, coexpressed in Arabidopsis chloroplasts (Dangoor et al., 2009), might partially compensate for the diminished ACHT1 expression. In contrast with these specific photosynthetic defects, the maximum quantum yield values (Fv/Fm) (Figure 8C) and the chlorophyll A and B (Figure 8E) and anthocyanin (Figure 8D) content of the ACHT1 OE and KD plants were all normal. The ACHT1 OE and KD plants also displayed a slightly retarded growth compared to wild-type plants (see Supplemental Figure 3A online) but had normal germination rates (see Supplemental Figure 3B online). While further studies are required to decipher the molecular details of the role of ACHT1, the altered NPQ and ETR values but normal maximum quantum yield and normal chlorophyll and anthocyanin content suggested that the ACHT1 OE plants are particularly defective in a feedback response to light, perhaps in a control of the linear electron flow, and were not under general stress. Additionally, the increased redox poise of the ACHT1 pathway in the overexpresser lines (Figure 5) was expected, in a scenario of pure antioxidative role of this pathway, to confer higher resistance to oxidative stress. Thus, our contrary findings that increased reduction disturbed photosynthetic electron flow further agree with a regulatory role for this pathway.
Figure 8.
Elevated ACHT1 Expression Results in Altered NPQ and ETR but Not in Fv/Fm Values or Chlorophyll and Anthocyanin Content.
(A) Time course of NPQ was calculated according to Maxwell and Johnson (2000) from traces of fluorescence induction in 16-h dark-adapted wild-type (WT), ACHT1 OE, and ACHT1 KD lines illuminated with 80 μE/m2s actinic light.
(B) ETR was calculated from traces of fluorescence induction as in (A).
(C) The maximal quantum yield (Fv/Fm) of the plants was measured before light induction.
(D) Anthocyanins content was determined from the absorbance at 530 and 657 nm of extracts of 14-d-old plants, and the results are expressed as (A530 – A637) × 1000 per milligram of plant weight (relative units [RU]).
(E) Chlorophyll a, chlorophyll b, and chlorophyll a+b concentrations were determined in extracts of 14-d-old plants as described (Porra et al., 1989). FW, fresh weight.
Plants were grown at 20°C/18°C under 8/16 h light/dark cycle using fluorescent white light at approximately 80 μE/m2s. Results from two KD lines (SALK_089128 and SALK_144456) were pooled together as well as the results from two OE lines (2-25 and 2-28). Data represent mean and se (n = 20 for photosynthetic parameter measurements and n = 6 for anthocyanin and chlorophyll determination). Means sharing the same letter are not significantly different based on the Tukey-Kramer honestly significant difference test using a P value cutoff of 0.05.
DISCUSSION
Defining the oxidative and reductive pathways of regulatory proteins is important to our understanding of their biological role. It had been known that illumination of dark-adapted plants results in photosynthesis-mediated reduction of the classical chloroplast-type Trxs that in turn adjust stromal enzyme function to photosynthetic activity (Schürmann and Buchanan, 2008; Meyer et al., 2009). Our study revealed that in contrast with the classical Trxs, the ACHT1 pool was reoxidized in dark-adapted plants shortly after illumination with low to moderate light intensity. This finding indicated that ACHT1 might be involved in oxidative rather than reductive regulation in the daily short-term acclimation of chloroplasts to light. The results also unveiled a Trx oxidative pathway that became active shortly after illumination with low to moderate light intensity. To identify the oxidizing partner of ACHT1, we took advantage of the methodology that trapped in vivo ERO1, the thiol oxidase of PDI, and PDI in intermolecular disulfide complexes (Pollard et al., 1998; Frand and Kaiser, 1999; Mezghrani et al., 2001). The trapping in planta of ACHT1 reaction intermediates, in which ACHT1 is linked to its redox partner by an intermolecular disulfide bridge, was important for maintaining physiological conditions. Indeed, the similar general pattern of the intermolecular disulfide complexes in plants expressing the catalytic active or the monothiol ACHT1 and their increased amounts in the monothiol plants validated the trapping of authentic ACHT1 reaction intermediates. We used MS to identify the 2-Cys Prx as a reaction partner of ACHT1 in dark-adapted plants illuminated by moderate light intensity. We then verified that the two redox proteins react with each other in plants by showing that increased levels of ACHT1 reduced the pool of 2-Cys Prx as predicted by the established reaction mechanism of Prxs. We concluded that ACHT1 reduces 2-Cys Prx and in turn is oxidized shortly after illuminating dark-adapted plants with moderate light intensity.
Several in vivo characteristics of the ACHT1 2-Cys Prx pathway indicated that it is apt for sensing peroxide levels by ACHT1 oxidation under low to moderate light intensity rather than for pure antioxidative function. (1) The oxidation of the ACHT1 pool in the light (Figure 2) revealed that its rate of oxidation by 2-Cys Prx exceeded its rate of reduction by photosynthesis. While ACHT1 oxidation is required if ACHT1 has a regulatory role, it is expected to hinder a pure antioxidative function. (2) ACHT1 oxidation is detected under mild growth conditions under which peroxide formation is not likely to saturate an antioxidative path. (3) Our finding that the intermolecular disulfide complexes of the catalytic active ACHT1 and 2-Cys Prx are most abundant when ACHT1 is largely oxidized (Figures 2 and 6) indicates a smaller rate of forward reaction than that required for optimal antioxidative function. (4) The higher midpoint redox potential of ACHT1 (Dangoor et al., 2009) than that of 2-Cys Prx (Horling et al., 2003) is not consistent with high rates of peroxides detoxification by the ACHT1-2-Cys Prx reaction in chloroplasts under acute oxidative stress conditions. (5) Finally, the detrimental effect on photosynthesis by the increased reduction of the ACHT1 and 2-Cys Prx pathway (Figures 5 and 8) is expected for a regulatory role and not for an antioxidative one. While the above argumentations promote a regulatory role for the ACHT1 and 2-Cys Prx catalytic couple, it is important to remember that the oxidation of ACHT1 indicates an ongoing rate of elimination of peroxides that increases under high light intensities (Figure 7). Thus, functional antioxidative activity of the pathway is essential for its sensing role. Interestingly, the 2-Cys Prx pool did not go through major redox state changes as the ACHT1 pool did during illumination of dark-adapted plants. These findings, which are reminiscent of the Yap1 oxidative sensing pathway (Delaunay et al., 2002) or of the Trx-Prx Tpx1 pathway in Schizosaccharomyces pombe (Day et al., 2012) and not of the circadian rhythmic biomarker 2-Cys Prxs (O'Neill and Reddy, 2011; O'Neill et al., 2011), indicate that ACHT1 redox state changes are used for regulation. Conceivably, the additional proteins trapped in intermolecular disulfide complexes with ACHT1 (Figure 4) could be ACHT1 targets regulated by ACHT1 redox state. Together, our findings portray a peroxide sensing system in which the oxidation of the ACHT1 pool is favored over forward rates of the antioxidative reaction. The oxidation of ACHT1 could, in turn, oxidize regulatory disulfides in its target proteins, thereby linking their activity to changes in the rate of peroxide formation under low to moderate light intensity.
The oxidation of ACHT1 soon after illumination of dark-adapted plants indicated that it is involved in regulation at that period. Also, the saturation of ACHT1 oxidation by moderate light intensity implied that it is involved in daily oxidative regulation under mild growth conditions, rather than in response to acute oxidative stress. Thus, we searched for phenotypic consequences of elevated levels of ACHT1 during the illumination of plants with moderate light intensity after 16 h in darkness (Figure 8). Our findings that increased redox poise of the ACHT1 and 2-Cys Prx pathway decreased the photosynthetic ETR with a concomitant increase in NPQ during the transition from dark to light, but maintained normal levels of chlorophylls and anthocyanins and Fv/Fm values (Figures 5 and 8), imply a redox-based feedback regulation of the thylakoid electron transfer reactions. Numerous studies have assigned a marked role to the intersystem electron carriers, especially the plastoquinone pool and the cytochrome b6f, in the control of the electron flow reactions through regulation of light-harvesting complex II (LHCII) phosphorylation and NPQ (reviewed in Niyogi, 1999; Foyer and Noctor, 2009; Dietz and Pfannschmidt, 2011; Rochaix, 2011). Both changes in the redox state of the plastoquinone pool and ΔpH signals are used to regulate these phenomena. Recent studies of Arabidopsis plants illuminated with low and high light intensity have implicated an additional regulatory path of LHCII phosphorylation involving the ferredoxin-Trx system and hydrogen peroxide (Rintamäki et al., 2000; Piippo et al., 2006; Tikkanen et al., 2010). In this regulatory path, counteracting reductive and oxidative activities control the formation of the regulatory disulfide of LHCII kinase and thereby its activity (Martinsuo et al., 2003).
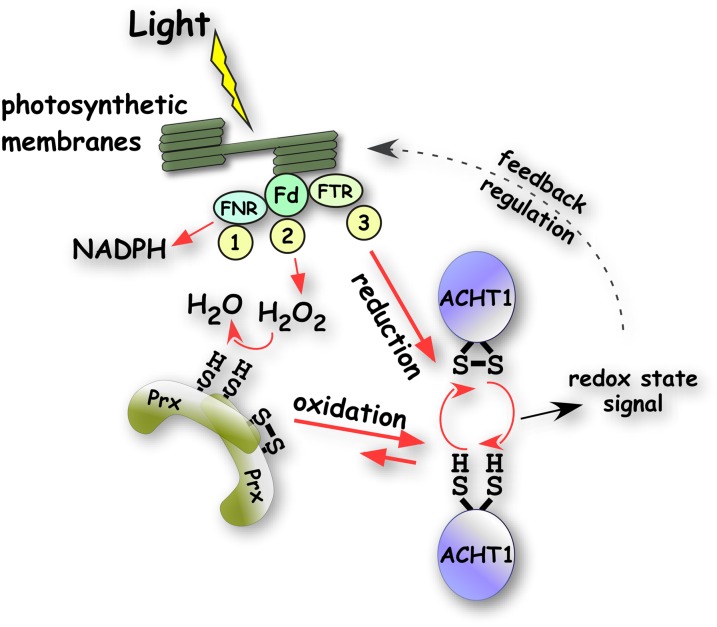
Figure 9 illustrates our working model of the ACHT1 and 2-Cys Prx pathway in plant chloroplasts. The instantaneous ACHT1 redox state is determined by its rate of reduction by photosynthesis and by its rate of oxidation by 2-Cys Prx. The rate of 2-Cys Prx oxidation is driven by peroxides likely formed by the Mehler reaction in PSI, which diverts excessive reducing equivalents from NADPH production to O2 reduction (Mehler, 1951). The association of both ACHT1 and 2-Cys Prx with the thylakoid membranes (König et al., 2002; Dangoor et al., 2009) might argue for localized sensing of peroxide formation. Under moderate light intensity, the reduction of ACHT1 by photosynthesis, and not its oxidation by 2-Cys Prx, is limiting and results in oxidation of ACHT1 (Figure 2). This suggests that the rate of reduction by photosynthesis dictates the dynamic range of ACHT1 function (i.e., the range of peroxide concentrations to which ACHT1, via 2-Cys Prx, dynamically responds). At higher light intensity, a larger proportion of reducing equivalents are transported through the Mehler reaction (Asada, 1999); in turn, the oxidation rate of ACHT1 by 2-Cys Prx increases, resulting in a higher proportion of oxidized ACHT1 (Figure 7). Our results suggest that the oxidation of ACHT1 is close to saturation in plants illuminated with a moderate level of light intensity under a controlled environment (Figure 7). Conceivably, this value is expected to fluctuate in natural conditions, where temperature, CO2, and water availability are not controlled. Similar to the classical type Trxs, which regulate target proteins by reduction (Schürmann and Buchanan, 2008), the oxidation of ACHT1 feedback regulates proteins affecting photosynthetic electron flow and NPQ. The short transient reduced state of ACHT1 precedes ACHT1 reoxidation (Figure 2) and takes place during the transition of the chloroplast from dark to light metabolism. It is possible that ACHT1 might have a transient reductive role in this time frame, during which the Calvin cycle is activated and the oxidative pentose phosphate pathway is inactivated. In case the reduction of additional Trxs, such as other ACHT family members (Dangoor et al., 2009) or the stress-induced chloroplast CDSP32 (Rey et al., 2005), is as limiting as the reduction of ACHT1, they may form a similar oxidative signaling pathway with 2-Cys Prx, which may be used to regulate other plant responses.
Figure 9.
Working Model of ACHT1 in the Light.
Depending on environmental conditions, the photosynthetic reducing equivalents may be diverted from the productive reaction of (1) NADP+ reduction via ferredoxin (Fd) and ferredoxin NADP-reductase (FNR) to the nonproductive (2) Mehler reaction, producing hydrogen peroxide (H2O2) as an intermediate. A portion of the equivalents is used, (3) via ferredoxin Trx reductase (FTR), to reduce chloroplast Trxs, such as ACHT1, for regulatory purposes. The redox state of the chloroplast ACHT1 pool in the light is determined by its reduction rate by ferredoxin Trx reductase (3) and by its oxidation rate by 2-Cys Prx (Prx). 2-Cys Prx itself is oxidized by peroxides produced by nonproductive photosynthesis (2). ACHT1 reduction rate is limiting and leads to net oxidation of ACHT1 shortly after illumination. The redox state of ACHT1 is used for redox-based feedback regulation of the membranal electron transfer reactions.
The chloroplast contains an additional electron donor to 2-Cys Prx, NADPH Thioredoxin Reductase (NTRC) (Pérez-Ruiz et al., 2006). Several differences between NTRC and ACHT1 suggest different roles for the two proteins. NTRC is a soluble stromal protein (Serrato et al., 2004), whereas ACHT1 is mainly associated with the thylakoids (Dangoor et al., 2009). ACHT1 draws its reducing power from photosynthetic linear electron flow, whereas NTRC depends on NADPH, which is produced also in the dark (Pérez-Ruiz et al., 2006). The findings that NTRC-deficient plants lack 2-Cys Prx reduction activity in the night and that the aberrant phenotype is more pronounced when the knockout plants are grown under short-day conditions or prolonged darkness (Pérez-Ruiz et al., 2006) suggested that NTRC is the main reducer of 2-Cys Prx in the night (Kirchsteiger et al., 2009). Yet, there is also evidence for a role for NTRC in the light, since there is decreased activation of ADP-Glc pyrophosphorylase (Michalska et al., 2009) and a lower amount of reduced 2-Cys Prx (Kirchsteiger et al., 2009) in the NTRC-deficient plants than in wild-type plants. Interestingly, our finding that 2-Cys Prx is kept in a more reduced state than what is anticipated from the oxidized state of ACHT1 agrees with the concept that NTRC reduces 2-Cys Prx also in the light.
The fast dynamic response of the ACHT1 redox state under moderate light intensity and the detrimental effect of the increased reduction of ACHT1 and 2-Cys Prx pathway promote the notion that the ACHT1-2-Cys Prx system is adjusting key chloroplast reactions for optimal growth under fluctuating environmental conditions. Yet, the identification of the additional ACHT1 target proteins (Figure 4) and studying their redox state changes are required to unravel its detailed biological role and to further our understanding of the intricate regulation of the redox signaling reactions in the chloroplast.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana var Columbia was grown in ambient air on solid half-strength Murashige and Skoog medium with 0.8% agar. The Arabidopsis T-DNA insertion lines SALK_089128 and SALK_144456 were obtained from the SALK collection. Plants were grown under a 16/8 h light/dark cycle and 20°C/18°C, respectively, at 80 μE/m2s (unless otherwise stated) for 7 d. A 10 mM stock solution of the photosynthetic electron transport inhibitor DCMU (Sigma-Aldrich) was resuspended in 50% ethanol and then diluted to a final concentration of 50 μM in 0.05% Tween 20 and applied by spraying the plants in the dark (30 min before illumination). For light intensity experiments, plants were grown under 80 μE/m2s for 7 d and then either transferred to 200 μE/m2s or kept in 80 μE/m2s. Samples were taken at the indicated times.
Generation of Transgenic Plants
ACHT1 and ACHT1C35S open reading frames were ligated upstream and in frame of the HA3 affinity tag. The fragments were cloned into pART7 vector, under the control of the 35S promoter and 35S terminator. NotI-digested fragments were ligated into the binary vector pBART, which confers BASTA resistance in plants. The binary plasmids were introduced into Agrobacterium tumefaciens through electroporation. Plant transformation was performed by the floral dip method (Clough and Bent, 1998). The chloroplast localization of the HA-tagged ACHT1 protein in these transgenic plants was demonstrated earlier (Dangoor et al., 2009).
Protein Redox State Labeling
The redox state of protein Cys residues in planta was resolved by modification of the oxidized fraction of the Cys residues with 5 kD mPEG (Sigma-Aldrich), a nonreversible thiol label that significantly increases the protein molecular mass as previously described (Peled-Zehavi et al., 2010). Trapping of the redox state of protein Cys residues was achieved by homogenizing the plants in 10% trichloroacetic acid, which protonate Cys residues, thereby preventing nonphysiological thiol exchanges. Following precipitation, pellets were resuspended in denaturing urea buffer (8 M urea, 2% SDS, 100 mM Tris-HCl, pH 7.5, 10 mM EDTA, and protease inhibitor) containing 50 mM NEM to irreversibly block reduced protein thiols. Then, oxidized Cys residues were reduced with DTT (100 mM). A second trichloroacetic acid precipitation was performed; the pellet was resuspended in urea buffer and labeled with 10 mM mPEG (2 h at 27°C). After incubation with mPEG, DTT (50 mM) was added to all the samples, followed by addition of 3× sample buffer (150 mM Tris-HCl, pH 6.8, 6% SDS, 30% glycerol, and 0.3% Pyronin Y). Proteins were then separated by SDS-PAGE and analyzed by immunoblots.
Intermolecular Disulfide Complex Formation
Following trapping of intermolecular disulfide complexes in plants by homogenization in 10% trichloroacetic acid, the proteins were resuspended in denaturing urea buffer containing 50 mM NEM, precipitated again in trichloroacetic acid, and resuspended in buffer (100 mM Tris, pH 7.5, 5 mM EDTA, 1% SDS, 50 mM NEM, and 1% Protease Inhibitor I [Calbiochem 539131]) as previously described (Peled-Zehavi et al., 2010). The protein extract was separated by reducing or nonreducing SDS-PAGE as indicated and analyzed by immunoblots.
Affinity Purification of Complexes and MS Analysis
Affinity purification of in planta–trapped complexes was performed as previously described (Peled-Zehavi et al., 2010). Trapped intermolecular disulfide protein complexes were incubated overnight at 4°C in RIPA buffer (1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 10 mM Tris-HCl, pH 8, and 140 mM NaCl) with either anti-HA resin or anti-2-Cys Prx-conjugated protein G beads. The proteins were eluted with nonreducing 2× sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, and 0.2% Pyronin Y) at 70°C for 10 min. The eluted proteins were loaded on SDS-PAGE gels, and selected areas of the gel were sliced out. The proteins were reduced in gelo with 2.8 mM DTT, modified with 8.8 mM iodoacetamide in 100 mM ammonium bicarbonate, and digested with modified trypsin (Promega) overnight at 37°C. The resulting tryptic peptides were resolved by reverse-phase chromatography. MS was performed at the Smoler Proteomics Center at the Technion (Israel Institute of Technology) by an ion-trap mass spectrometer (Orbitrap; Thermo) in a positive mode using repetitively full MS scan followed by collision-induced dissociation of the seven most dominant ions selected from the first MS scan. The MS data were analyzed using the Protein Discoverer 1.1 (Thermo Fisher) using both Sequest and Mascot search engines, searching against the plant section of the National Center for Biotechnology Information nonredundant database.
Differential comparison of proteins isolated from plants expressing the HA-tagged ACHT1 and proteins from control wild-type plants allowed for the subtraction of nonspecific background proteins. 2-Cys Prx-specific polyclonal antibodies were raised in rabbit using the recombinant mature form of 2-Cys Prx protein (Dangoor et al., 2009) at The Biological Services Unit of The Weizmann Institute of Science.
Determination of Photosynthetic Parameters
Plants were grown in ambient air on solid half-strength Murashige and Skoog medium with 0.8% agar under 8/16 h light/dark cycle and 20°C/18°C, respectively, at 80 μE/m2s. Induction curve kinetics were measured in 14-d-old dark-adapted plants (10 plants per measurement) using an imaging PAM system (Heinz Walz). Actinic light of 80 μE/m2s was switched on 40 s after F0 and Fm determination. Saturating pulses of 10,000 μE/m2s for 600 ms were administered at 20-s intervals. NPQ, ETR, and Fv/Fm values were calculated according to Maxwell and Johnson (2000). All experiments were repeated at least five times. The results from the two KD lines (SALK_089128 and SALK_144456) were pulled together as well as the results from two overexpresser lines (2-25 and 2-28). For characterization of the transgenic lines, see Supplemental Figure 1 online.
Growth Phenotype and Determination of Chlorophyll and Anthocyanin Content
Seeds were surface sterilized, sown on solid half-strength Murashige and Skoog medium with 0.8% agar, and grown under a 8/16 h light/dark cycle and 20°C/18°C, respectively, at 80 μE/m2s. For each plant line, four to six replicates of approximately 50 seedlings were scored at 24 h poststratification for the percentage of germination (determined by radicle emergence) and at 66 h poststratification for the percentage of plantlets with fully opened cotyledons. The results from the two T-DNA insertion lines (SALK_089128 and SALK_144456) were pooled together as well as the results from two overexpresser lines (2-25 and 2-28), and averages and standard error of the mean were calculated by one-way analysis of variance using JMP software (SAS Institute). For characterization of the transgenic lines, see Supplemental Figure 1 online.
Chlorophyll concentration was determined in 14-d-old plants as described (Porra et al., 1989). For anthocyanin content determination, 14-d-old plants were extracted in 300 μL of 1% HCl in methanol for 24 h at 4°C. The samples were diluted with 200 μL water and extracted with 500 μL chloroform. The upper aqueous phase was removed to a clean tube and further diluted with an equal volume of aqueous solution containing 60% methanol and 1% HCl. Absorbance at 657 and 530 nm was measured, and anthocyanin content was calculated from A530 corrected for the background A657. All experiments were repeated at least three times for each plant line, and the results of the two T-DNA and the two overexpresser lines were each pooled together.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACHT1 (At4g26160), 2-Cys PrxA (At3g11630), and 2-Cys PrxB (At5g06290).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of Plant Lines with Altered Expression of ACHT1.
Supplemental Figure 2. ACHT1 Requires 2-Cys Prx for Peroxide Reduction.
Supplemental Figure 3. Growth Phenotype of Plants with Altered ACHT1 Expression.
Supplemental Table 1. 2-Cys Prx Peptides Identified by MS/MS in Immunopurified ACHT1 Complexes.
Supplementary Material
Acknowledgments
We thank Nir Keren and Itzhak Ohad for their guiding advice concerning the chlorophyll fluorescence measurements. We thank Meir Edelman and Ami Navon for their valuable suggestions and Ami Navon for his generous gift of HA-conjugated beads. This study was supported by grants from the Israeli Science Foundation and the Minerva Foundation. A.D. is the incumbent of The Henry and Bertha Benson Chair, Weizmann Institute of Science.
AUTHOR CONTRIBUTIONS
A.D. directed the project. A.D, I.D., and H.P.-Z. designed the research. I.D. and H.P.-Z. conducted the majority of the experiments. A.D. and G.W. performed the photosynthetic measurements. A.D., I.D., and H.P.-Z. wrote the article.
Glossary
- ROS
reactive oxygen species
- PSI
photosystem I
- Trx
thioredoxin
- PDI
protein disulfide isomerase
- Prx
peroxiredoxin
- PSII
photosystem II
- NPQ
nonphotochemical quenching
- ETR
electron transport rate
- HA
hemagglutinin
- NEM
N-ethylmaleimide
- mPEG
methoxypolyethylene glycol-maleimide
- OE
overexpressor
- KD
knockdown
- MS
mass spectrometry
References
- Alergand T., Peled-Zehavi H., Katz Y., Danon A. (2006). The chloroplast protein disulfide isomerase RB60 reacts with a regulatory disulfide of the RNA-binding protein RB47. Plant Cell Physiol. 47: 540–548 [DOI] [PubMed] [Google Scholar]
- Allen J.F., Bennett J., Steinback K.E., Arntzen C.J. (1981). Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291: 25–29 [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Asada K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Asada K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y., Koller A., del Val G., Manieri W., Schürmann P., Buchanan B.B. (2003). Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. USA 100: 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Bonardi V., Pesaresi P., Becker T., Schleiff E., Wagner R., Pfannschmidt T., Jahns P., Leister D. (2005). Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Cardol P., De Paepe R., Franck F., Forti G., Finazzi G. (2010). The onset of NPQ and Deltamu(H)+ upon illumination of tobacco plants studied through the influence of mitochondrial electron transport. Biochim. Biophys. Acta 1797: 177–188 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dangoor I., Peled-Zehavi H., Levitan A., Pasand O., Danon A. (2009). A small family of chloroplast atypical thioredoxins. Plant Physiol. 149: 1240–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A. (2002). Redox reactions of regulatory proteins: Do kinetics promote specificity? Trends Biochem. Sci. 27: 197–203 [DOI] [PubMed] [Google Scholar]
- D’Autréaux B., Toledano M.B. (2007). ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8: 813–824 [DOI] [PubMed] [Google Scholar]
- Day A.M., Brown J.D., Taylor S.R., Rand J.D., Morgan B.A., Veal E.A. (2012). Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol. Cell 45: 398–408 [DOI] [PubMed] [Google Scholar]
- DeGennaro M., Hurd T.R., Siekhaus D.E., Biteau B., Jasper H., Lehmann R. (2011). Peroxiredoxin stabilization of DE-cadherin promotes primordial germ cell adhesion. Dev. Cell 20: 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay A., Pflieger D., Barrault M.B., Vinh J., Toledano M.B. (2002). A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111: 471–481 [DOI] [PubMed] [Google Scholar]
- Dietz K.J. (2011). Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 15: 1129–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K.J., Pfannschmidt T. (2011). Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol. 155: 1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomenko D.E., Koc A., Agisheva N., Jacobsen M., Kaya A., Malinouski M., Rutherford J.C., Siu K.L., Jin D.Y., Winge D.R., Gladyshev V.N. (2011). Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc. Natl. Acad. Sci. USA 108: 2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourquet S., Huang M.E., D’Autreaux B., Toledano M.B. (2008). The dual functions of thiol-based peroxidases in H2O2 scavenging and signaling. Antioxid. Redox Signal. 10: 1565–1576 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2009). Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11: 861–905 [DOI] [PubMed] [Google Scholar]
- Frand A.R., Kaiser C.A. (1999). Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell 4: 469–477 [DOI] [PubMed] [Google Scholar]
- Hald S., Nandha B., Gallois P., Johnson G.N. (2008). Feedback regulation of photosynthetic electron transport by NADP(H) redox poise. Biochim. Biophys. Acta 1777: 433–440 [DOI] [PubMed] [Google Scholar]
- Hall A., Karplus P.A., Poole L.B. (2009). Typical 2-Cys peroxiredoxins—Structures, mechanisms and functions. FEBS J. 276: 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horling F., Lamkemeyer P., König J., Finkemeier I., Kandlbinder A., Baier M., Dietz K.J. (2003). Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol. 131: 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G.N. (2003). Thiol regulation of the thylakoid electron transport chain—A missing link in the regulation of photosynthesis? Biochemistry 42: 3040–3044 [DOI] [PubMed] [Google Scholar]
- Kallis G.B., Holmgren A. (1980). Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J. Biol. Chem. 255: 10261–10265 [PubMed] [Google Scholar]
- Karpinski S., Reynolds H., Karpinska B., Wingsle G., Creissen G., Mullineaux P. (1999). Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284: 654–657 [DOI] [PubMed] [Google Scholar]
- Kirchsteiger K., Pulido P., González M., Cejudo F.J. (2009). NADPH Thioredoxin reductase C controls the redox status of chloroplast 2-Cys peroxiredoxins in Arabidopsis thaliana. Mol. Plant 2: 298–307 [DOI] [PubMed] [Google Scholar]
- König J., Baier M., Horling F., Kahmann U., Harris G., Schürmann P., Dietz K.J. (2002). The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc. Natl. Acad. Sci. USA 99: 5738–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeille S., Willig A., Depège-Fargeix N., Delessert C., Bassi R., Rochaix J.D. (2009). Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 7: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsuo P., Pursiheimo S., Aro E.M., Rintamäki E. (2003). Dithiol oxidant and disulfide reductant dynamically regulate the phosphorylation of light-harvesting complex II proteins in thylakoid membranes. Plant Physiol. 133: 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Mehler A.H. (1951). Studies on reactions of illuminated chloroplasts. II. Stimulation and inhibition of the reaction with molecular oxygen. Arch. Biochem. Biophys. 34: 339–351 [DOI] [PubMed] [Google Scholar]
- Meyer Y., Buchanan B.B., Vignols F., Reichheld J.P. (2009). Thioredoxins and glutaredoxins: Unifying elements in redox biology. Annu. Rev. Genet. 43: 335–367 [DOI] [PubMed] [Google Scholar]
- Mezghrani A., Fassio A., Benham A., Simmen T., Braakman I., Sitia R. (2001). Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 20: 6288–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska J., Zauber H., Buchanan B.B., Cejudo F.J., Geigenberger P. (2009). NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc. Natl. Acad. Sci. USA 106: 9908–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Shulaev V., Mittler R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133: 481–489 [DOI] [PubMed] [Google Scholar]
- Møller I.M., Jensen P.E., Hansson A. (2007). Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58: 459–481 [DOI] [PubMed] [Google Scholar]
- Motohashi K., Kondoh A., Stumpp M.T., Hisabori T. (2001). Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. USA 98: 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 333–359 [DOI] [PubMed] [Google Scholar]
- O’Neill J.S., Reddy A.B. (2011). Circadian clocks in human red blood cells. Nature 469: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J.S., van Ooijen G., Dixon L.E., Troein C., Corellou F., Bouget F.Y., Reddy A.B., Millar A.J. (2011). Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled-Zehavi H., Avital S., Danon A. (2010). Methods of redox signaling by plant thioredoxins. In Methods in Redox Signaling, D. Das, ed (New Rochelle, NY: Mary Ann Liebert), pp. 251–256. [Google Scholar]
- Pérez-Ruiz J.M., Spínola M.C., Kirchsteiger K., Moreno J., Sahrawy M., Cejudo F.J. (2006). Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T. (2003). Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci. 8: 33–41 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T., Liere K. (2005). Redox regulation and modification of proteins controlling chloroplast gene expression. Antioxid. Redox Signal. 7: 607–618 [DOI] [PubMed] [Google Scholar]
- Piippo M., Allahverdiyeva Y., Paakkarinen V., Suoranta U.M., Battchikova N., Aro E.M. (2006). Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol. Genomics 25: 142–152 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Forzani C., Hirt H. (2006). Reactive oxygen species signaling in plants. Antioxid. Redox Signal. 8: 1757–1764 [DOI] [PubMed] [Google Scholar]
- Pollard M.G., Travers K.J., Weissman J.S. (1998). Ero1p: A novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1: 171–182 [DOI] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and chlorophyll B extracted with 4 different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Rey P., Cuiné S., Eymery F., Garin J., Court M., Jacquot J.P., Rouhier N., Broin M. (2005). Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J. 41: 31–42 [DOI] [PubMed] [Google Scholar]
- Rintamäki E., Martinsuo P., Pursiheimo S., Aro E.M. (2000). Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl. Acad. Sci. USA 97: 11644–11649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J.D. (2007). Role of thylakoid protein kinases in photosynthetic acclimation. FEBS Lett. 581: 2768–2775 [DOI] [PubMed] [Google Scholar]
- Rochaix J.D. (2011). Regulation of photosynthetic electron transport. Biochim. Biophys. Acta 1807: 375–383 [DOI] [PubMed] [Google Scholar]
- Schürmann P., Buchanan B.B. (2008). The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 10: 1235–1274 [DOI] [PubMed] [Google Scholar]
- Senkevich T.G., White C.L., Koonin E.V., Moss B. (2002). Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc. Natl. Acad. Sci. USA 99: 6667–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato A.J., Pérez-Ruiz J.M., Spínola M.C., Cejudo F.J. (2004). A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 279: 43821–43827 [DOI] [PubMed] [Google Scholar]
- Steiner S., Dietzel L., Schröter Y., Fey V., Wagner R., Pfannschmidt T. (2009). The role of phosphorylation in redox regulation of photosynthesis genes psaA and psbA during photosynthetic acclimation of mustard. Mol. Plant 2: 416–429 [DOI] [PubMed] [Google Scholar]
- Storz G., Tartaglia L.A., Ames B.N. (1990). Transcriptional regulator of oxidative stress-inducible genes: Direct activation by oxidation. Science 248: 189–194 [DOI] [PubMed] [Google Scholar]
- Tikkanen M., Grieco M., Kangasjärvi S., Aro E.M. (2010). Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol. 152: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T., Danon A. (2001). Translation of chloroplast psbA mRNA is regulated by signals initiated by both photosystems II and I. Proc. Natl. Acad. Sci. USA 98: 12289–12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T., Levitan A., Sofer A., Danon A. (2000). Translation of chloroplast psbA mRNA is modulated in the light by counteracting oxidizing and reducing activities. Mol. Cell. Biol. 20: 1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker K.W., Lyles M.M., Gilbert H.F. (1996). Catalysis of oxidative protein folding by mutants of protein disulfide isomerase with a single active-site cysteine. Biochemistry 35: 1972–1980 [DOI] [PubMed] [Google Scholar]
- Zer H., Vink M., Keren N., Dilly-Hartwig H.G., Paulsen H., Herrmann R.G., Andersson B., Ohad I. (1999). Regulation of thylakoid protein phosphorylation at the substrate level: Reversible light-induced conformational changes expose the phosphorylation site of the light-harvesting complex II. Proc. Natl. Acad. Sci. USA 96: 8277–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.