When young plants emerge from the soil or canopy into direct sunlight, they must rapidly adapt to the new light environment containing a higher ratio of red/far-red light. This work reveals the molecular mechanism of how the photoreceptor phyA differentially transduces red and far-red light signals to light-responsive genes for better plant survival in natural environments.
Abstract
Emerging plants have to adapt to a high ratio of far-red light (FR)/red light (R) light in the canopy before they reach the R-enriched direct sunlight. Phytochrome A (phyA) is the single dominant photoreceptor in young Arabidopsis thaliana seedlings that initiates photomorphogenesis in response to a FR-enriched environment and transduces increasing R signals to early responsive genes. To date, how phyA differentially transmits FR and R signals to downstream genes remains obscure. Here, we present a phyA pathway in which FAR-RED ELONGATED HYPOCOTYL1 (FHY1), an essential partner of phyA, directly guides phyA to target gene promoters and coactivates transcription. Furthermore, we identified two phosphorylation sites on FHY1, Ser-39 and Thr-61, whose phosphorylation by phyA under R inhibits phyA signaling at each step of its pathway. Deregulation of FHY1 phosphorylation renders seedlings colorblind to FR and R. Finally, we show that the weaker phyA response resulting from FHY1 phosphorylation ensures the seedling deetiolation process in response to a R-enriched light condition. Collectively, our results reveal FHY1 phosphorylation as a key mechanism for FR/R spectrum-specific responses in plants and an essential event for plant adaption to changing light conditions in nature.
INTRODUCTION
Light provides a variety of signals, including light quality, intensity, direction, and duration, for plant growth and development (Fankhauser and Casal, 2004; Mathews, 2006). When subterranean seedlings emerge from soil, they obtain positional information, from either under the soil or in direct sunlight or under a canopy, by sensing the light intensity and quality, specifically the proportion of red light (R) and far-red light (FR) wavelengths of light. Among the five phytochrome photoreceptors (phyA to phyE) in Arabidopsis thaliana, phyA mediates most responses to FR, while phyB is primarily responsible for R responses (Whitelam et al., 1993; Neff and Van Volkenburgh, 1994). In etiolated seedlings, phyA accounts for over 85% of all phytochrome (Sharrock and Clack, 2002) and plays a predominant role in early seedling photomorphogenesis and gene expression in response to not only FR, but also R (Tepperman et al., 2004, 2006).
Upon light activation, phyA translocates from the cytoplasm into the nucleus to modulate gene expression (Nagatani, 2004) dependent on FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and its less abundant homolog FHY1-LIKE (FHL) (Hiltbrunner et al., 2005, 2006). FHY1 plays a predominant role compared with FHL, since fhl mutants exhibit a much weaker phenotype, and the transcript level of FHL is only approximately one-fifteenth of that of FHY1 (Zhou et al., 2005). FHY1/FHL also facilitate the association of phyA with the nuclear body (NB; Kircher et al., 2002; Hiltbrunner et al., 2005), a proposed site where light signaling components interact to mediate gene expression (Van Buskirk et al., 2012). Moreover, it bridges the association between phyA and transcription factors LONG AFTER FAR-RED LIGHT1 (LAF1) and LONG HYPOCOTYL IN FAR-RED1 (HFR1) during FR activation (Yang et al., 2009). How the nuclear phyA-FHY1 complex influences gene expression beyond its association with transcription factors remains unknown.
The reversible photoconversion of phytochromes between the inactive R-absorbing Pr form and the active FR-absorbing Pfr form is the molecular basis for plants’ ability to sense the FR/R ratio of the ambient light (Quail, 2010). Under FR, the nuclear Pr-FHY1/FHL complex dissociates readily and sets free FHY1/FHL to continuously transport Pfr phyA into the nucleus. Under R, the high Pfr/Pr ratio results in a higher proportion of the Pfr-FHY1/FHL complex, which has a slower dissociation rate and consequently leads to a lower FHY1/FHL recycling rate (Rausenberger et al., 2011). This model suggests that phyA signaling desensitization under R is not only due to R-induced phyA removal through the ubiquitin-proteasome system (Clough and Vierstra, 1997), but also due to reduced phyA nuclear accumulation compared with the FR condition. However, none of the available models for phyA signaling has taken into account FR/R reversible FHY1 phosphorylation, which is R induced and dependent on Pfr phyA (Shen et al., 2009).
To elucidate how young seedlings differentiate FR and R through phyA, we delineate a complete phyA signaling pathway. Beyond translocating into the nucleus, FHY1 guides phyA to associate with the promoter DNA through association with DNA binding transcription factors and acts as a coactivator for target gene transcription. FR/R reversible phosphorylation of FHY1 is revealed as a part of the plant FR/R sensing mechanism to regulate the level of phyA signaling activity. R-induced phosphorylated FHY1 maintains its association with phyA and actively weakens phyA function at all levels: nuclear translocation, NB targeting, and chromatin association. The weaker phyA response to R is beneficial to the deetiolation process of young seedlings. Compared with the R-responsive reduction of nuclear phyA, FR/R reversible phosphorylation of FHY1 seems to be a relatively rapid and flexible mechanism for desensitization of phyA signaling in plant adaption to the changing light environment.
RESULTS
R Induces FHY1 Phosphorylation at Amino Acid Residues Ser-39 and Thr-61
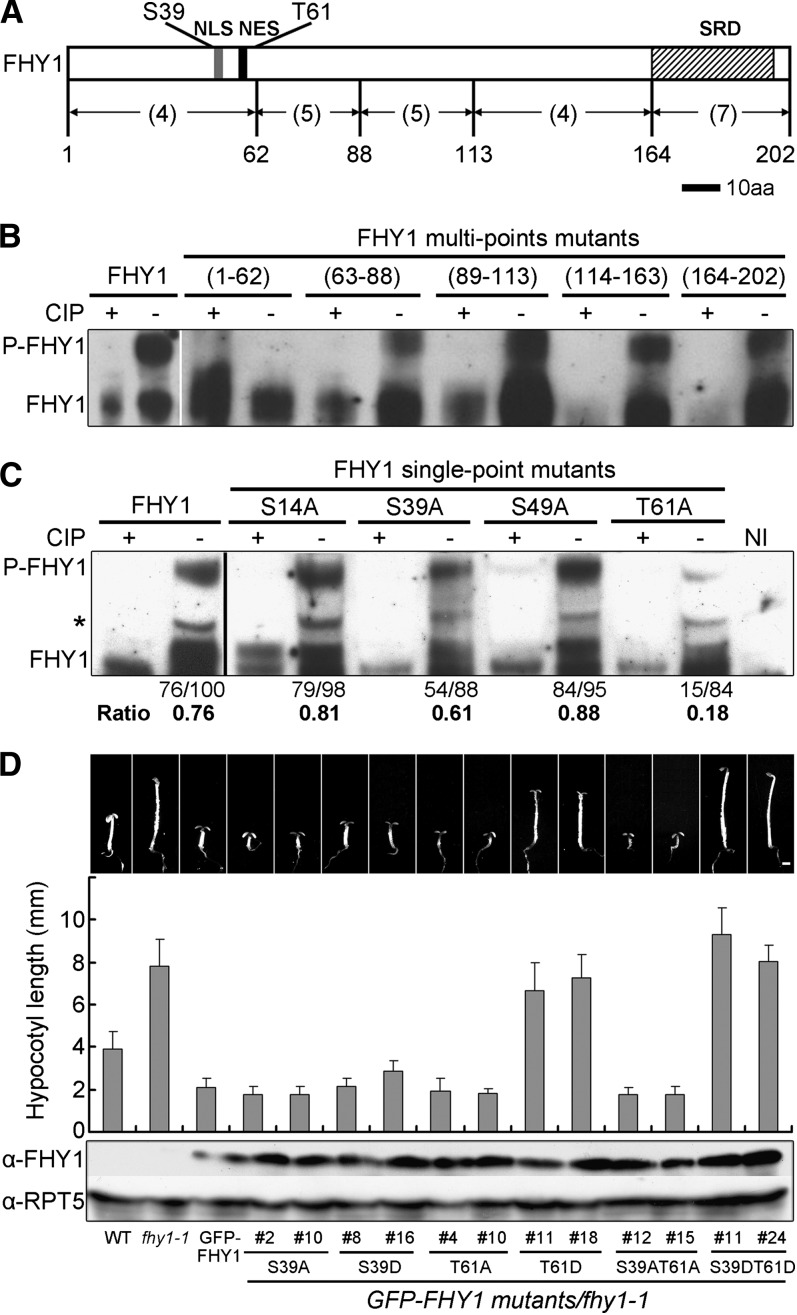
We first set out to identify the amino acid residues in FHY1 that are phosphorylated in R. We divided the 25 putative Ser/Thr (S/T) kinase phosphorylation sites in FHY1 (NetPhos software prediction score >0.5) into five groups (Figure 1A) and mutated the Ser/Thr residues within each group to Ala (A). The resulting five multipoint mutants of FHY1 were examined in a transient assay using Nicotiana benthamiana leaves. We found that the R-induced phosphorylation band disappeared when putative sites in the N-terminal region (1 to 62) were eliminated (Figure 1B), suggesting that FHY1 phosphorylation site(s) were among the four Ser/Thr residues in that region.
Figure 1.
FHY1 Is Phosphorylated at Amino Acid Residues Ser-39 and Thr-61.
(A) Diagram of the FHY1 protein showing the location of Ser-39 and Thr-61 phosphorylation sites. Numbers in parentheses indicate the number of potential phosphorylation sites in the indicated region. aa, amino acids; NES (black bar), nuclear export signal; NLS (gray bar), nuclear localization signal; SRD (hatched bar), Septin-related domain.
(B) and (C) Delineation of FHY1 phosphorylation sites to region 1 to 62 (B) and to Ser-39 and Thr-61 (C). Tobacco leaves infiltrated with constructs expressing myc-tagged multiple-point or single-point FHY1 mutants were incubated in the dark for 2 d and exposed to R for 1 d to induce FHY1 phosphorylation. Protein extracts were incubated with (+) or without (−) calf intestinal alkaline phosphatase (CIP) before immunoblotting using anti-myc (B) or anti-FHY1 (C). Relative band intensities of phosphorylated FHY1 (P-FHY1) over unphosphorylated FHY1, as indicated below the immunoblot, was normalized with the value of the wild-type FHY1 nonphosphorylated band (100). Asterisk indicates a nonspecific band not included in the quantification. NI, noninfiltrated tobacco leaves.
(D) Morphology of FR-grown transgenic seedlings expressing P-deficient or P-mimic FHY1 in fhy1-1 background. Two independent transgenic lines for each mutant were used. Seedlings were grown for 5 d in FR. Measurements of hypocotyl lengths are shown as mean ±sd (n > 20). Immunoblots show expression of mutant GFP-FHY1 proteins in corresponding transgenic lines, with RPT5 as a loading control. Bar = 1mm.
Subsequently, four single-point FHY1 mutants were generated and examined. FHY1T61A lost most of the R-induced phosphorylation, while FHY1S39A lost a moderate amount (Figure 1C), indicating that Ser-39 and Thr-61 are phosphorylated in R with Thr-61 being at higher level. Protein sequence alignments showed that Thr-61 of FHY1 corresponds to Phe-61 in FHL, which cannot be phosphorylated (Zeidler et al., 2004). Consistent with this, R-induced phosphorylation was not detected in FHL (Shen et al., 2009), despite the fact that FHL is highly homologous to FHY1.
Phosphorylation at Ser-39 and Thr-61 Inactivates FHY1 Function in FR
We next generated transgenic plants in the fhy1-1 background expressing either a phosphorylation (P)-deficient mutant by changing Ser and Thr to Ala, or P-mimic mutant by changing Ser and Thr to Asp (D) in green fluorescent protein (GFP)-FHY1. When seedlings were grown under FR, the fhy1-1 phenotype was fully rescued by GFP-FHY1 or GFP-FHY1S39AT61A, but not GFP-FHY1S39DT61D (Figure 1D). Similar results were obtained with plants expressing an FHY1 C-terminal fusion of GFP (see Supplemental Figure 1A online). Remarkably, the S39DT61D double-point mutant displayed a very long hypocotyl and closed cotyledons in FR, identical to that of fhy1-1 null mutant, whereas the T61D single-point mutant caused less but considerable FR deficiency (Figure 1D). In addition, anthocyanin accumulation, another hallmark of the FR response, was deficient in GFP-FHY1S39DT61D transgenic seedlings under FR (see Supplemental Figure 1B online). These data show that phosphorylation on Ser-39 and Thr-61 inactivate FHY1.
Phosphorylation on S39T61 Promotes Its Cytoplasmic Localization and Inhibits Its NB Targeting
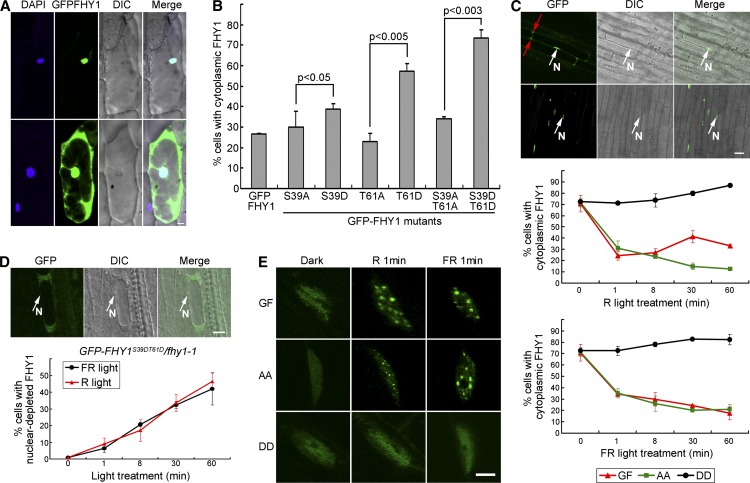
Light-responsive nuclear transport and NB localization are critical for FHY1 function (Hiltbrunner et al., 2005, 2006; Genoud et al., 2008; Rausenberger et al., 2011). We first investigated the effect of FHY1 phosphorylation on its nucleocytoplasmic localization pattern in onion (Allium cepa) cells. GFP-FHY1 protein exhibited either a nuclear-exclusive localization pattern or a ubiquitous localization pattern in which FHY1 was observed in both the cytosol and the nucleus (Figure 2A). We quantified the localization preference of GFP-FHY1 and its phosphorylation mutants based on the percentage of cells containing cytoplasmic localized proteins. We found that all of the P-mimic mutant samples displayed a higher proportion of cytoplasmic FHY1 than the wild type and the corresponding P-deficient mutant samples (Figure 2B). This result shows that phosphorylated FHY1 prefers cytoplasmic localization in onion cells.
Figure 2.
FHY1 Phosphorylation Promotes Cytoplasmic Localization.
(A) FHY1 localizes in the nucleus (top) or stays ubiquitously distributed (bottom). Onion epidermal cells expressing GFP-FHY1 by bombardment were incubated in the dark for 36 h followed by a 2-min white light pulse. The 4′,6-diamidino-2-phenylindole (DAPI)-stained nucleus is shown in blue. DIC, differential interference contrast. Bar = 20 µm.
(B) P-mimic FHY1 prefers cytoplasmic localization. Onion cells expressing the indicated GFP-FHY1 mutants were counted according to their nuclear-cytoplasmic distribution pattern after 36-h dark incubation and a 2-min white light pulse. Mean ± sd (n > 150); P values are from Student’s t tests.
(C) FHY1 phosphorylation mutants exhibit abnormal localization patterns in FR and R in Arabidopsis. Top panel: GFP-FHY1 localizes to the nucleus (top panel) or stays ubiquitously distributed (bottom panel) in hypocotyl cells of a 4-d etiolated GFP-FHY1/fhy1-1 (GF). Red and white arrows indicate cytoplasmic and nuclear localization (N), respectively. Middle and bottom panels: 4-d etiolated GF, GFP-FHYS39AT61A/fhy1-1 (AA), and GFP-FHY1S39DT61D/fhy1-1 (DD) seedlings were exposed to R (middle) or FR (bottom) for indicated time periods. Cells were counted at the end of the indicated light exposure period. Mean ± sd (n > 150). Bar = 20µm.
(D) Nuclear-excluded distribution of P-mimic GFP-FHY1 with extended R or FR treatments. A 4-d etiolated DD seedling exposed to FR for 1 h shows a typical nuclear-excluded localization of the protein (top panels). Proportion of cells containing nuclear-excluded DD protein over the indicated time periods under R (the red line) or FR (the black line) treatment is shown in the bottom. Mean ± sd (n > 150). Bar = 20 µm.
(E) P-mimic FHY1 is unable to localize to nuclear bodies. Etiolated (4 d) GF, AA, and DD seedlings were exposed to R or FR for 1 min, followed by immediate microscopy imaging. Bar = 5µm.
In transgenic Arabidopsis seedlings, we found that GFP-FHY1S39AT61A, like wild-type FHY1, was localized to the NBs in response to R or FR irradiation within as short as 1 min (Figure 2E). However, among a small number of cells that contained nuclear GFP-FHY1S39DT61D, the protein failed to localize to the NBs (Figure 2E), even after 60 min of FR or R irradiation (data not shown). These data suggest that FHY1 phosphorylation prevents its NBs localization.
The R/FR-Specific Localization Profile of FHY1 Is Determined by S39T61 Phosphorylation
We next performed dynamic analyses of FHY1 nuclear localization in transgenic Arabidopsis seedlings to determine the relationship between R/FR light irradiation and FHY1 phosphorylation and subcellular localization. Similar to onion, in Arabidopsis hypocotyl cells, FHY1 protein also exhibited nuclear localization and ubiquitous localization patterns (Figure 2C, top panel). Notably, although R and FR both triggered rapid nuclear translocation of GFP-FHY1 upon light exposure, its sustained localization profile differed in R or FR (Figure 2C, bottom panels). During the first minute of light treatment, before FHY1 phosphorylation was detectable (Shen et al., 2009), GFP-FHY1 started to translocate into the nucleus. Whereas extended FR irradiation led to persistent nuclear translocation of GFP-FHY1, extended R treatment made GFP-FHY1 reach an equilibrium of steady nucleocytoplasmic distribution, with some fluctuations (Figure 2C, bottom panels). The highest proportion of cells containing cytoplasmic GFP-FHY1 was found after 30 min of R, which incidentally corresponds to the time frame when FHY1 phosphorylation level peaks after an R pulse (Shen et al., 2009).
GFP-FHY1S39AT61A and GFP-FHY1S39DT61D mutants were both spectrum blind, but they behaved in opposite ways. GFP-FHY1S39DT61D failed to show the initial burst of nuclear import and largely remained in the cytosol throughout the entire course of either R or FR treatment (Figure 2C, bottom panels). In fact, many cells in GFP-FHY1S39DT61D transgenic seedlings were found to exhibit a nuclear-depleted localization pattern that was not found in GFP-FHY1 or GFPFHY1S39AT61A transgenic seedlings (Figure 2D, top panel). Moreover, the percentage of cells containing nuclear-excluded GFP-FHY1S39DT61D increased steadily following light treatment of R or FR (Figure 2D, bottom panel). This result suggests that phosphorylation of FHY1 promotes cytoplasmic localization of the protein, possibly by reversing its nuclear transportation direction from import to export or by rapid nuclear degradation of FHY1.
GFP-FHY1S39AT61A, on the other hand, displayed an FR-characteristic distribution profile and kinetics regardless of FR or R. In either R or FR, GFP-FHY1S39AT61A displayed a persistent nuclear localization and behaved identically to GFP-FHY1 under FR (Figure 2C, bottom panels). Taken together, we found that FHY1 responds to R or FR irradiation with distinct nucleocytoplasmic distribution dynamics and that mutations at Ser-39 and Thr-61 abolish the ability of FHY1 to distinguish between the two spectra of light.
FHY1 Phosphorylation Inhibits Nuclear Localization of Photoreceptor phyA
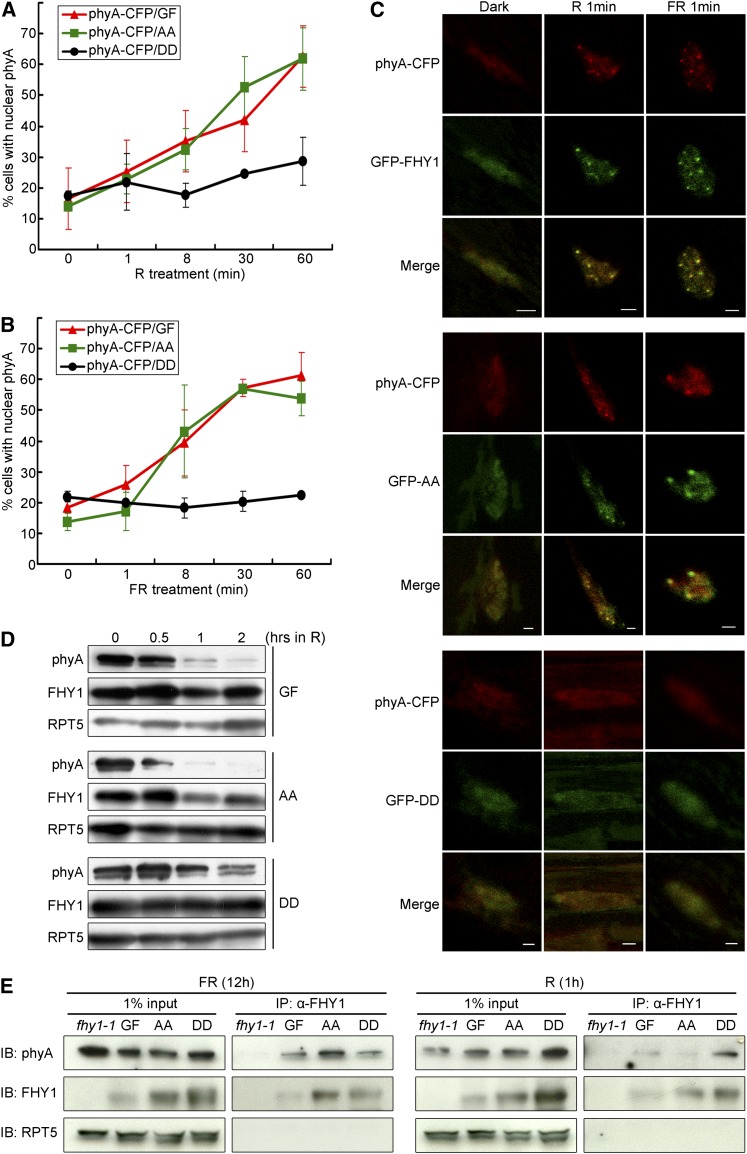
To determine how phyA nuclear localization was affected by FHY1 phosphorylation, the phyA-cyan fluorescent protein (CFP) transgene was crossed into various GFP-FHY1 mutant backgrounds, and its localization profile following R or FR stimulation was examined in F1 plants. Although F1 plants contain one copy of wild-type FHY1, its effect is essentially negligible in the presence of mutant FHY1 transgenes (see Supplemental Figure 2 online). Both R and FR triggered phyA nuclear translocation in GFP-FHY1 and GFP-FHY1S39AT61A background, but not in GFP-FHY1S39DT61D transgenic seedlings (Figures 3A and 3B). Furthermore, phyA was found to colocalize with wild-type FHY1 and FHY1S39AT61A in nuclear bodies in both R and FR, while under the same conditions, no phyA-containing NBs were observed in GFP-FHY1S39DT61D seedlings (Figure 3C). These results indicate that FHY1 phosphorylation inhibits phyA nuclear translocation and abolishes the accumulation of phyA in NBs.
Figure 3.
FHY1 Phosphorylation Promotes phyA Cytoplasmic Localization.
(A) and (B) phyA nuclear translocation is attenuated by P-mimic FHY1 expression in R (A) or FR (B). phyA-CFP/phyA-201 was crossed with GFP-FHY1/fhy1-1 (phyA-CFP/GF), GFP-FHYS39AT61A/fhy1-1 (phyA-CFP/AA), and GFP-FHYS39DT61D/fhy1-1 (phyA-CFP/DD), respectively. Etiolated 4-d-old F1 seedlings were examined for percentage of cells containing nuclear localized phyA-CFP. Mean ± sd (n > 150).
(C) NB localization of phyA in cells expressing the wild type (top panels) or P-deficient FHY1 (middle panels) but not in those expressing P-mimic FHY1 (bottom panels). Etiolated seedlings as described in (A) and (B) were irradiated with R or FR for 1 min before microscopy imaging. Bars = 2 µm.
(D) P-mimic FHY1 stabilizes phyA protein. Four-day etiolated GF, AA, and DD seedlings were exposed to R for indicated time periods. Immunoblots were probed with anti-phyA and anti-FHY1 antibodies. Anti-RPT5 was used as a loading control.
(E) Phosphorylation does not affect the interaction between FHY1 and phyA. Four-day etiolated seedlings were treated with FR or R for the indicated time. Anti-FHY1 was used in immunoprecipitation, and the blots were analyzed with indicated antibodies on the left. Anti-RPT5 was used as an internal control.
We next investigated if FHY1 phosphorylation influences the phyA turnover rate. Immunoblot analysis showed that phyA was more stable in GFP-FHY1S39DT61D than in GFP-FHY1 or GFP-FHY1S39AT61A transgenic seedlings (Figure 3D). Thus, there is a correlation between FHY1 phosphorylation, reduced phyA nuclear localization, and longer phyA half-life. This is consistent with the report that phyA turnover rate is faster in the nucleus (Debrieux and Fankhauser, 2010).
We noticed that FHY1 colocalized with phyA regardless of its phosphorylation status and nucleocytoplasmic partitioning (Figure 3C; see Supplemental Figure 3 online), suggesting that P-FHY1 can still interact with phyA. To address this question directly, we performed immunoprecipitation experiments. PhyA could be coimmunoprecipitated by anti-FHY1 antibody from GFP-FHY1S39DT61D lysate as well as from wild-type FHY1 and GFP-FHY1S39AT61A lysates (Figure 3E), confirming that FHY1-phyA interaction is not abolished by FHY1 phosphorylation at S39T61.
FHY1 and phyA Are Recruited to the CHS Promoter through HY5 or PIF3 in FR
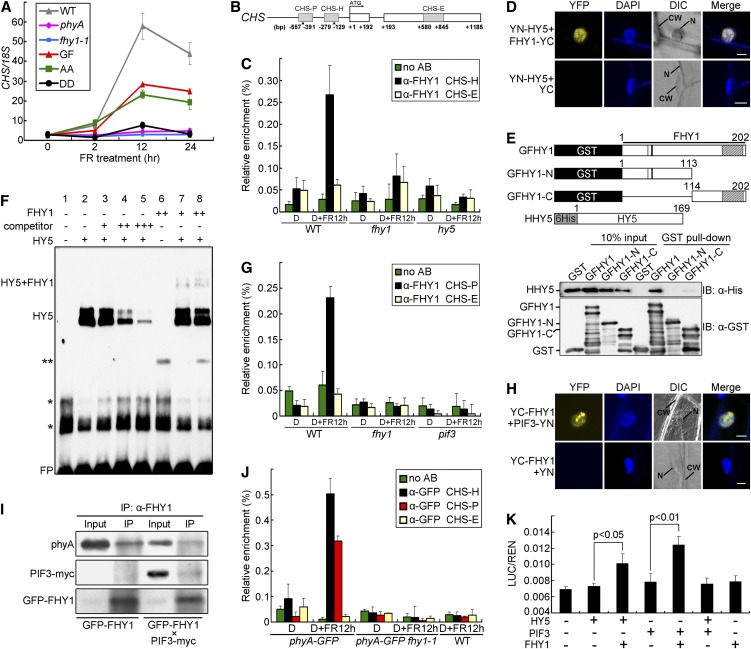
CHS encodes chalcone synthase, a key enzyme in the biosynthesis of anthocyanin (Ferrer et al., 2008). It was chosen as a marker of FR-induced gene expression to investigate the nuclear function of FHY1. CHS was induced by FR in GFP-FHY1 and GFP-FHY1S39AT61A transgenic seedlings, but not in GFP-FHY1S39DT61D seedlings or fhy1-1 or phyA null mutants (Figure 4A), similar to the results on anthocyanin accumulation (see Supplemental Figure 1B online).
Figure 4.
FHY1 and phyA Are Recruited to the CHS Promoter through HY5 and PIF3 in FR.
(A) FHY1 phosphorylation inhibits FR-induced CHS expression. Four-day etiolated seedlings of indicated genotypes were exposed to FR (6 µmol/m2/s) for the indicated time. The values of CHS transcript were normalized to 18S rRNA in the quantitative RT-PCR. WT, wild type.
(B) A diagram of the CHS gene and promoter. CHS-H and CHS-P are HY5- and PIF3 binding regions containing G-boxes (indicated by the triangles). CHS-E is a region on an CHS exon.
(C) FHY1 associates with the CHS-H promoter region in a HY5-dependent manner. Four-day etiolated wild-type (WT), fhy1-1, and hy5 seedlings were left untreated (D) or irradiated with 12 h FR (6 µmol/m2/s) (D+FR12 h). ChIP-qPCR using anti-FHY1 or no antibody (no AB) was followed by amplification of CHS-H and CHS-E (negative control). Data were normalized with corresponding input samples.
(D) BiFC assay showing interaction of FHY1 with HY5. YN (YFP N-terminal)-HY5 and YC (YFP C-terminal)-FHY1 fusion proteins were expressed in onion epidermal cells through cobombardment. CW, cell wall; DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast; N, nucleus. Bar = 20 µm.
(E) FHY1 interacts with HY5 in vitro through its C-terminal domain. Schematic diagrams of GST-tagged FHY1 proteins and His-tagged HY5 (Top). Extracts containing mixtures of HHY5 and specified GFHY1 proteins were subjected to GST pulldown followed by immunoblotting using indicated antibodies (Bottom).
(F) FHY1 and HY5 form a supercomplex on the CHS-H fragment in vitro. The EMSA reactions contained 1 µg (+) or 4 µg (++) indicated proteins and labeled probe (−151 to −193 bp of CHS promoter), without (−) or with 200-fold (+), 1000-fold (++), or 2000-fold (+++) competitor (unlabeled probe). One asterisk represents nonspecific bands; two asterisks represent an unknown band. FP, free probe.
(G) FHY1 associates with the CHS-P promoter region in a PIF3-dependent manner. Etiolated wild-type, fhy1-1, and pif3 seedlings were used in ChIP-qPCR as in (C), except that the CHS-P region was examined.
(H) BiFC assay showing interaction of FHY1 with PIF3. YN-PIF3 and YC-FHY1 fusion proteins were expressed in onion epidermal cells by cobombardment. Bar = 20 µm.
(I) In vivo interaction of FHY1 with PIF3 and phyA. FR-grown GFPFHY1/fhy1-1 or F2 seedlings homozygous for GFPFHY1 and PIF3-myc were used for anti-FHY1 immunoprecipitations. Precipitates were analyzed by immunoblot with indicated antibodies.
(J) phyA is recruited to both CHS-H and CHS-P regions of the CHS promoter under FR. phyA-GFP/phyA-1 and F2 seedlings homozygous for phyA-GFP and fhy1-1 were grown and examined as described in (C) by anti-GFP ChIP-qPCR. Wild-type seedlings were used as negative control for anti-GFP specificity.
(K) FHY1 enhances transcriptional activities of HY5 and PIF3. A luciferase transcription reporter was coinfiltrated into tobacco leaves with constructs expressing HY5, PIF3, or FHY1 as indicated. The reporter activity was measured after incubating the leaves in the dark for 2 d then exposing them to FR for 1 d. Mean ± se (n = 6); P values are from Student’s t tests.
All error bars represent ±sd of triplicate experiments unless otherwise indicated.
Next, we explored the possibility that FHY1 and phyA may regulate CHS transcription at its promoter. The CHS promoter contains two G-boxes at −171 and −516, which can be recognized and bound by transcription factors HY5 and PIF3, respectively (Shin et al., 2007). By chromatin immunoprecipitation–quantitative PCR (ChIP-qPCR) assay, we found that endogenous FHY1 was recruited to the CHS promoter in the HY5 binding region in response to FR (Figures 4B and 4C). Notably, the FHY1 chromatin association was HY5 dependent because it was abolished in the hy5 mutant. Direct interaction of FHY1 with HY5 was confirmed by bimolecular fluorescence complementation (BiFC) assays (Figure 4D). Glutathione S-transferase (GST) pull-down assays further indicated that HY5 bound the C-terminal region of FHY1 (Figure 4E). Given that HY5 could bind to the CHS promoter on chromatin independently of FHY1 (see Supplemental Figure 4A online), whereas FHY1 could not bind the promoter in the absence of HY5 (Figure 4C), we reasoned that FHY1 was probably recruited by HY5 to the CHS promoter. To test this idea, we performed a supershift EMSA assay. As predicted, HY5 alone, but not FHY1 alone, bound the CHS promoter. Moreover, some of the complex containing HY5 and the CHS promoter was supershifted in the presence of FHY1, indicating that FHY1 and HY5 form a supercomplex on the CHS promoter (Figure 4F).
PIF3 also binds to the CHS promoter and activates CHS transcription (Shin et al., 2007). ChIP-qPCR assays showed that FHY1 could associate with the PIF3 binding region on the CHS promoter in FR, and this association was dependent on PIF3 (Figure 4G). In addition, FHY1 interacted with PIF3 in the BiFC assay (Figure 4H) and the in vivo coimmunoprecipitation assay (Figure 4I). Collectively, FHY1 is recruited to the CHS promoter by direct interaction with the corresponding transcription factors HY5 and PIF3.
ChIP-qPCR also showed that phyA-GFP was recruited to the CHS promoter on both the PIF3 and the HY5 binding regions in response to FR in an FHY1-dependent manner (Figure 4J), suggesting that FHY1 tethered both photoreceptor and transcription factor on the chromatin. Interestingly, we found more phyA bound to the CHS promoter under FR than with 5 min of R following the FR treatment (see Supplemental Figure 4B online). This result is consistent with the idea that R-induced FHY1 phosphorylation inactivates phyA functions and with the prediction that nuclear phyA-FHY1 complex is more abundant under FR than R (Rausenberger et al., 2011).
FHY1 Coactivates CHS Transcription with HY5 and PIF3
We used a CHS promoter-driven dual-luciferase reporter assay in N. benthamiana leaves to study the function of FHY1 in transcriptional activation. The CHS promoter activity was only slightly increased upon transient expression of HY5 alone, but markedly increased when FHY1 and HY5 were coexpressed (Figure 4K), indicating that FHY1 can coactivate transcription with HY5. In a transcription activation experiment in yeast cells, we found, likewise, that coexpression of FHY1 and HY5, but not expressing HY5 alone, clearly activated the CHS promoter-driven LacZ reporter (see Supplemental Figure 4C online). Similarly, coinfiltration of FHY1 and PIF3 constructs into tobacco leaves further enhanced the PIF3-promoted transcriptional activity of the CHS promoter (Figure 4K). Interestingly, HY5 and PIF3 did not coactivate with each other when they were coexpressed (Figure 4K), while both required coactivation by FHY1 for high activity in FR. These results show that, besides nuclear transportation of photoreceptor phyA, FHY1 also plays a role in transcription regulation, thereby directly contributing to downstream gene expression. In addition to CHS, other FR-induced genes, such as RBCS1A and PORA, were similarly affected by FHY1 phosphorylation (see Supplemental Figure 5 online).
FHY1 Phosphorylation Abolishes Its Association with the CHS Promoter
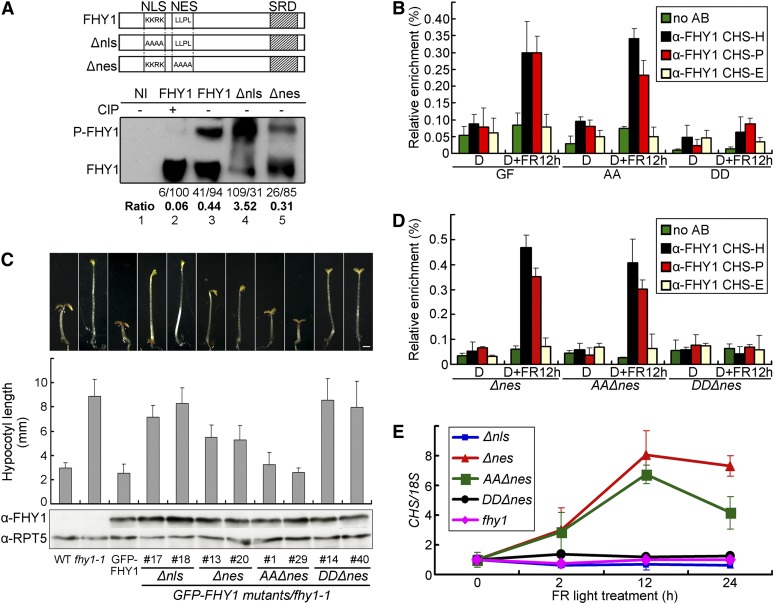
To determine whether FHY1 phosphorylation may affect its promoter association, we performed ChIP-qPCR with FHY1 phosphorylation site mutants. Unlike GFP-FHY1 and GFP-FHY1S39AT61A, GFP-FHY1S39DT61D was not detected on the CHS promoter (Figure 5B). To rule out that this result was caused by reduced nuclear abundance of GFP-FHY1S39DT61D (Figure 2), we generated the NES and NLS mutants of FHY1 (Figure 5A, top panel), identical mutations of which have been shown to restrict the protein to the cytosol (Δnls) or the nucleus (Δnes) (Zeidler et al., 2004). Both Δnls and Δnes mutants of FHY1 proteins could be phosphorylated (Figure 5A, bottom panel), which indicates that FHY1 can be phosphorylated in both nuclear and cytosolic compartments, though phosphorylation of cytoplasmic FHY1 appeared to be more efficient.
Figure 5.
FHY1 Phosphorylation Prevents Its Association with CHS Promoter.
(A) Phosphorylation of FHY1 occurs in both cytoplasm and nucleus. A diagram of FHY1 constructs showing NLS or NES mutations generated (Top). Tobacco leaves infiltrated with indicated FHY1 constructs were exposed to R for 1 d (Bottom). Protein extracts were treated with (+) or without (−)calf intestinal alkaline phosphatase (CIP) before immunoblots with anti-FHY1. The ratios of P-FHY1 over FHY1 are indicated below the immunoblot. Band intensities were normalized to the value (set at 100) of nonphosphorylated FHY1 in lane 2. NI, noninfiltrated tobacco leaves.
(B) P-mimic FHY1 is unable to associate with the CHS promoter. Four-day etiolated seedlings of GFP-FHY1/fhy1-1 (GF), GFP-FHY1S39AT61A/fhy1-1 (AA), and GFP-FHY1S39DT61D/fhy1-1 (DD) were left untreated (D) or treated with 12 h FR (6 µmol/m2/s) (D+FR12 h) and subjected to anti-FHY1 ChIP-qPCR. AB, antibody.
(C) Morphology of FHY1 combined localization and phosphorylation mutants in FR. Two independent transgenic lines for each mutant as indicated (in fhy1-1 background) were grown in FR for 5 d. Measurements of hypocotyl lengths are shown as mean ± sd (n > 20). Immunoblots indicate expression of mutant GFP-FHY1 in corresponding transgenic lines, with RPT5 as a loading control. Bar = 1 mm.
(D) Nuclear P-mimic FHY1 fails to associate with the CHS promoter. GFP-FHY1Δnes/fhy1-1 (Δnes), GFP-FHY1AAΔnes/fhy1-1 (AAΔnes), and GFP-FHY1DDΔnes/fhy1-1 (DDΔnes) seedlings were examined by anti-FHY1 ChIP-qPCR as described in (A).
(E) Nuclear exclusion as well as phosphorylation of FHY1 abolishes CHS activation. Four-day etiolated seedlings of indicated genotypes were irradiated with FR (6 µmol/m2/s). Quantitative RT-PCR of CHS transcript was normalized against that of 18S.
All error bars represent ±sd of triplicate experiments.
Next, we generated transgenic plants carrying the NES/NLS mutations with or without the phosphorylation site mutations and introduced into the fhy1-1 background. Nuclear FHY1 (FHY1Δnes) could partially rescue the fhy1-1 phenotype, whereas cytoplasmic FHY1 (FHY1Δnls) could not (Figure 5C). Importantly, nuclear P-deficient FHY1 (FHY1AAΔnes) completely rescued fhy1-1, whereas the nuclear P-mimic FHY1 (FHY1DDΔnes) was essentially nonfunctional, with elongated hypocotyls comparable to fhy1-1 seedlings. These results demonstrate that FHY1 has a crucial function in addition to mediating phyA nuclear translocation and that both functions can be inactivated by phosphorylation on S39T61.
ChIP-qPCR assays showed that only FHY1Δnes and FHY1AAΔnes, but not FHY1DDΔnes, could associate with the CHS promoter on HY5 or PIF3 binding sites (Figure 5D). Similarly, the CHS gene was not expressed in FHY1DDΔnes transgenic seedlings, nor in fhy1-1 or FHY1Δnls seedlings, but was strongly induced by FR in FHY1Δnes and FHY1AAΔnes transgenic seedlings (Figure 5E). We conclude that phosphorylation of FHY1 at S39T61 can dislodge the protein from the promoter of light-responsive genes, such as CHS, which would explain the lack of functionality of the nuclear-localized P-mimic FHY1 mutant.
FHY1 Phosphorylation Abolishes the phyA-Mediated R Response
PhyA not only mediates the FR response, it also plays a dominant role in early R-responsive gene expression (Tepperman et al., 2004, 2006). We therefore tested the effect of FHY1 phosphorylation on the phyA-mediated R response. Anti-FHY1 ChIP-qPCR experiments showed that GFP-FHY1 and GFP-FHY1S39AT61A, but not GFP-FHY1S39DT61D, were on the CHS promoter upon R treatment (Figure 6A), though the association was weaker than in FR (Figures 4C and 4G). In the transient assay, GFP-FHY1 and GFP-FHY1S39AT61A, but not GFP-FHY1S39DT61D, coactivated CHS transcription with HY5 and PIF3 in R (Figure 6B). Similarly, R induced CHS expression (Figure 6C) in wild-type and phyA signaling-enhanced seedlings (GFP-FHY1 and GFP-FHY1S39AT61A), but not in phyA signaling-deficient seedlings (phyA, fhy1-1, and GPP-FHY1S39DT61D). These results suggest that FHY1, but not in its phosphorylated form, facilitates phyA function in mediating gene expression in response to R.
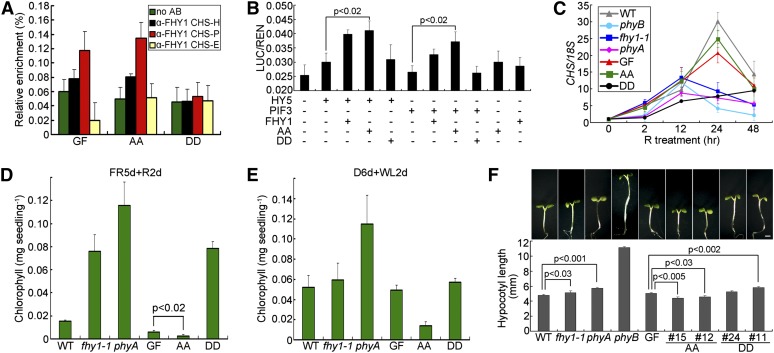
Figure 6.
Effect of FHY1 Phosphorylation on phyA Functions under R.
(A) Phosphorylation prevents R-induced FHY1 association with the CHS promoter. Four-day etiolated GFP-FHY1/fhy1-1 (GF), GFP-FHYS39AT61A/fhy1-1 (AA), and GFP-FHY1S39DT61D/fhy1-1 (DD) seedlings were irradiated with R for 12 h before anti-FHY1 ChIP-qPCR. AB, antibody.
(B) P-mimic FHY1 is deficient in transcription coactivation with HY5 and PIF3 in R. The LUC transcription reporter was coinfiltrated with constructs expressing the indicated proteins into tobacco leaves, followed by incubation for 1 d in R. AA, FHY1S39AT61A; DD, FHY1S39DT61D. Mean ± se (n = 6); P values are from Student’s t tests.
(C) FHY1 phosphorylation weakens R induction of CHS expression. Six-day-old etiolated seedlings of the indicated genotypes were exposed to R for the indicated time before RNA extraction. CHS transcript levels were normalized against that of 18S in RT-qPCR. WT, wild type.
(D) and (E) P-deficient FHY1 displays hypersensitivity to bleaching. Seedlings of the indicated genotypes were grown on Murashige and Skoog medium without Suc in FR for 5 d and switched to R (D) or in the dark for 6 d and switched to white light (E) for 2 d before chlorophyll measurement.
(F) P-deficient FHY1 inhibits hypocotyl elongation in R. Morphology of 5-d-old R-grown seedlings (Top). Measurements of corresponding hypocotyl lengths (Bottom). Two independent lines for GFPS39AT61A/fhy1 (AA) or GFPS39DT61D/fhy1 (DD) transgenic seedlings as described in Figure 1D were tested. Mean ± se (n = 20); P values are from Student’s t tests. Bar = 1 mm.
All error bars represent ±sd of triplicate experiments unless otherwise indicated.
R-Induced FHY1 Phosphorylation Is Necessary for Optimal Growth under R
Sustained phyA signaling can be detrimental to young seedlings, as shown in the extreme case of phyA-mediated blockage of greening by FR (Barnes et al., 1996). To determine whether weakening of phyA signaling by R-induced FHY1 phosphorylation may offer a physiological advantage, we examined the greening process of young seedlings. When grown in FR for 5 d then transferred to R for 2 d, mutants with defective phyA signaling, such as GFP-FHY1S39DT61D, phyA, and fhy1-1, survived with substantial chlorophyll accumulation, whereas wild-type, GFP-FHY1, and GFP-FHY1S39AT61A seedlings suffered heavy bleaching (Figure 6D). Notably, GFP-FHY1S39AT61A, which cannot undergo phosphorylation, was the most severely damaged. Similar results were obtained in a condition closer to the natural environment (Figure 6E) or with Suc-containing media (see Supplemental Figure 6 online), indicating that inability to phosphorylate FHY1 is disadvantageous during seedling deetiolation.
When grown in constant R, we noticed that the constitutively active mutant GFP-FHY1S39AT61A developed an R-hypersensitive response as indicated by a slight but significant shorter hypocotyl than GFP-FHY1 (Figure 6F). In short, while FHY1 P-mimic mutant exhibits insensitivity to FR (Figure 1D), FHY1 P-defective mutant displays a hypersensitivity to R (Figures 6D to 6F).
DISCUSSION
FHY1 plays an indispensable role in mediating FR responses by facilitating light-induced phyA nuclear translocation and by interacting with transcription factors (Yang et al., 2009; Rausenberger et al., 2011). Our data reinforce the close partnership of phyA and FHY1 throughout the entire phyA nuclear signaling pathway, from nuclear translocation to NB targeting, chromatin association, and transcription control of gene expression. Each of these steps can be blocked by phosphorylation of FHY1 at S39T61, highlighting the potency of FHY1 phosphorylation as an inactivation mechanism for phyA nuclear signaling. We further show that FHY1 phosphorylation, which is induced by the R signal, is part of the FR/R spectrum sensing mechanism that ensures its optimal response to specific light quality, which is particularly important during greening of etiolated seedlings.
FHY1 Is a Transcriptional Coactivator That Acts on Target Gene Promoters
Most of the studies on the molecular functions of FHY1 have been focused on its role in nuclear translocation of phyA (Hiltbrunner et al., 2005, 2006; Genoud et al., 2008; Rausenberger et al., 2011). This function of FHY1 was highlighted in a study showing that a constitutively nuclear-localized phyA could rescue the phyA phenotype in the absence of FHY1 (Genoud et al., 2008). However, this result did not take into account the presence of FHL, the FHY1 homolog that functions redundantly to FHY1. To address this issue, Yang et al. (2009) showed that an artificial FHY1 (NLS-YFP-SEP) transgene that allowed for phyA nuclear translocation was unable to rescue the fhy1 fhl double mutant, indicating FHY1/FHL have functions beyond nuclear translocation of phyA. Yang et al. (2009) further showed that FHY1 directly binds LAF1 and HFR1 and assembles various phyA-FHY1-transcription factor complexes. Here, we expand the role of FHY1 in the nucleus by showing that FHY1, along with phyA, is recruited to the chromatin loci via transcription factors HY5 and PIF3 and is capable of coactivating transcription with these factors (Figure 4). All of these functions of FHY1 on chromatin are subject to control by S39T61 phosphorylation. The physiological significance of this aspect of FHY1’s function is demonstrated by the lack of functionality of FHY1DDΔnes, the P-mimic mutant of FHY1 that localizes to the nucleus but is defective in chromatin association and fails to rescue the fhy1-1 mutant phenotype.
Currently at least four transcription factors (HY5, PIF3, HFR1, and LAF1) have been shown to function downstream of phyA-FHY1 signaling (this study; Shin et al., 2007; Yang et al., 2009), and each regulates a large number of genes (Lee et al., 2007; Zhang et al., 2011). It is conceivable that phyA-FHY1 regulates the expression of a broad range of genes via direct association with multiple transcription factors on target gene promoters. Consistent with this, multiple light-responsive genes are differentially regulated by FHY1 phosphorylation, including RBCS1A and PORA, as well as CHS.
FR and R Elicit Distinct Nucleocytoplasmic Trafficking Dynamics of FHY1
Our time-series snapshots on FHY1 and phyA distribution patterns following FR stimulation (Figures 2 and 3) agree with the dynamic FHY1 nuclear trafficking and recycling mathematical model (Rausenberger et al., 2011). Within a minute of FR exposure, there is robust phyA-FHY1 nuclear import and NB targeting. Afterwards, constant Pfr/Pr photoconversion and FHY1 recycling continue to drive the persistent nuclear translocation of phyA and the increasing nuclear localization of FHY1 that we have observed under FR.
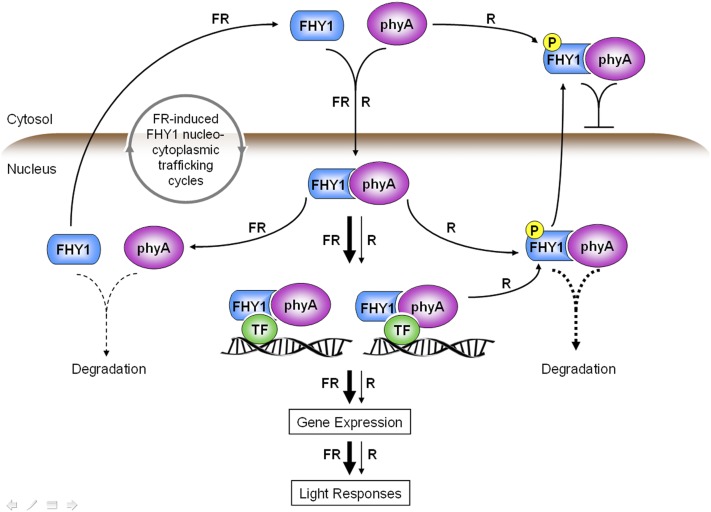
Our data shed light on the dynamic distribution of FHY1 in R, which we believe requires a modification of the FR model. Under R irradiation, the initial burst of nuclear import is followed by a FHY1 nucleocytoplasmic distribution that eventually reaches equilibrium (Figure 2). This subtly different dynamic distribution profile in R compared with FR is caused by R-induced phosphorylation of FHY1 at S39T61, which propels the balance of FHY1 nucleocytoplasmic trafficking toward cytoplasm. Mutations on phosphorylation site S39T61 render FHY1 totally blind to FR/R spectrum differences, with the P-defective mutant displaying a constitutive nuclear localization profile and the P-mimic mutant displaying a cytoplasmic localization profile regardless of FR or R irradiation. This finding indicates that FHY1 phosphorylation defines the distinct pattern of phyA-FHY1 signaling dynamics in response to FR/R spectrum signals. To describe precisely the behavior of phyA in ambient light, which contains both FR and R spectrums of light, phosphorylation of FHY1 must be taken into consideration (Figure 7).
Figure 7.
Schematic Model of phyA-FHY1 Signaling in FR and R.
R-induced FHY1 phosphorylation attenuates phyA signaling by promoting cytoplasmic localization of phyA/FHY1 and dislodging phyA/FHY1 from chromatin. Through these actions, FHY1 phosphorylation contributes to the differential dynamics of phyA signaling in FR and R. TF, transcription factor.
Implication of FHY1 Phosphorylation in phyA Signaling in R
Whereas both FR and R signals cause immediate nuclear translocation of phyA-FHY1, perception of R, but not FR, by phyA causes phosphorylation of FHY1 (Shen et al., 2009). In an extreme scenario in which the entire FHY1 pool becomes phosphorylated, theoretically the nuclear FHY1 would dislodge from chromatin and be depleted from the nucleus, and phyA signaling would be solely supported by FHL, the low-abundance FHY1 homolog. However, this does not happen because R irradiation can cause phosphorylation of only up to 40% of FHY1 pool at the peak point (30 min) (Shen et al., 2009). As a result, the exposure to R in young seedlings does not lead to systematic shutdown of phyA signaling. This would explain why phyA remains a dominant photoreceptor in mediating early R-responsive gene expression (Tepperman et al., 2006). In fact, the pool of nonphosphorylated FHY1 is required to facilitate phyA-mediated gene expression in the early R response (Figure 6). Presumably, equilibrium of phosphorylated and nonphosphorylated FHY1 pools would be established according to the ambient light conditions and modulate the strength of phyA signaling. Taken together, we propose that FHY1 phosphorylation is a key factor determining the level of phyA signaling activity in emerging seedlings.
The Pivotal Role of FHY1 Phosphorylation in Plant Adaption to an R-Enriched Environment
When a young plant seedling emerges into direct sunlight, which is a relatively R-enriched environment compared with under the soil or canopy, its rapid adaption to the higher ratio of R in the surrounding light environment is necessary for better growth and survival. We show in this study that the inability to downregulate FHY1 activity renders the seedlings hypersensitive to R irradiation and highly susceptible to photobleaching even during the dark-to-white-light transition (Figure 6). Thus, besides reduction of phyA levels (Clough and Vierstra, 1997), phosphorylation of FHY1 at S39T61 represents another mechanism by which the R signal weakens phyA nuclear function. Within the context where phyA nuclear activity is in overall decline during the transition from the phyA-dominated system of emerging seedlings to the phyB-E–mediated system of adult plants, R-induced phosphorylation of FHY1 provides a malleable mechanism for balanced and precise control of phyA signaling. This mechanism, which is regulated by the phyA photosensory system itself, helps to ensure a successful deetiolation process in preparation for photosynthetic autotrophic growth of plants.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis thaliana used in this study was of the Landsberg erecta ecotype, unless otherwise indicated. The fhy1-1 (Desnos et al., 2001), phyA-1 (Whitelam et al., 1993), hy5-ks50 (Oyama et al., 1997), and pif3-1 (Kim et al., 2003) mutants and the 35S:GFP-FHY1 (Shen et al., 2005), phyA-CFP/phyA-211 (Genoud et al., 2008), and phyA-GFP/phyA201 (Kim et al., 2000) transgenic plants were previously described. Seeds were surface sterilized and grown on Murashige and Skoog medium containing 1% Suc in darkness for 2 d at 4°C and then transferred to white light for 12 h at 22°C before treatment under the specific light conditions indicated in the text. The fluence rates were 62 µmol/m2/s for FR and 79 µmol/m2/s for R except indicated.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
Site-directed mutagenesis was introduced into the FHY1 open reading frame (ORF) (FHY1S39A, FHY1S39D, FHY1T61A, FHY1T61D, FHY1S39AT61A, FHY1S39DT61D, FHY1Δnls, FHY1Δnes, FHY1S39AT61AΔnes, and FHY1S39DT61DΔnes) by PCR (see Supplemental Table 1 online). To generate pUC18-35S-mGFP(S65T)-mutant FHY1 constructs, full-length mutant FHY1 cDNA was amplified with a forward primer containing a SalI restriction site (5′-TACACTAGTATGCCTGAAGTGGAAGT-3′) and a reverse primer containing a NotI restriction site (5′-TACGCGGCCGCTTACAGCATTAGCGTTGAG-3′) and then inserted between the SalI and NotI sites of pUC18-35S-mGFP (Chiu et al., 1996; Niwa et al., 1999). Primers with KpnI and SmaI restriction sites were used to generate pUC18-35S-wild type/mutant FHY1-mGFP constructs. A KpnI-EcoRI fragment was subcloned from the above-mentioned pUC18 constructs into a pJim19 vector (Zhang et al., 2008). pJim19-35S:GFP-mutant FHY1 and pJim19-35S:wild type/mutant FHY1-GFP constructs were introduced into fhy1-1 plants via Agrobacterium tumefaciens (GV3101 strain)–mediated transformation. Transgenic plants were selected with 200 mg/mL gentamicin (Sigma-Aldrich). To select single insertion lines, gentamicin-resistant/gentamicin-sensitive ratios in T2 were determined. Homozygous progenies of two representative single insertion lines for each construct were used for further studies.
Tobacco Transient Expression Assay
The following potential phosphorylation sites in FHY1 were mutated as indicated in Figure 1B: region 1 to 62: Ser-14, Ser-39, Ser-49, and Thr-61; region 63 to 88: Ser-69, Ser-76, Thr-80, Ser-83 and Ser-86; region 89 to 113: Thr-92, Ser-93, Ser-106, Ser-107, and Ser-109; region 114 to 164: Ser-117, Ser-120, Ser-135, and Ser-159; region 165 to 202: Ser-168, Ser-169, Thr-173, Thr-180, Thr-184, Thr-188, and Ser-198. Point-mutated FHY1 ORFs were amplified by primers containing KpnI and BamHI and inserted into a pCambia1300-221-Myc vector. Agrobacterium containing myc-tagged mutant FHY1 was infiltrated into Nicotiana benthamiana leaves with P19 as previously described (Liu et al., 2010). Tobacco leaves were homogenized in an extraction buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 4 M urea, and 1 mM PMSF. Proteins were separated in precast 10% Bis-Tris NuPAGE gels as directed by the manufacturer (Invitrogen). Anti-myc (Sigma-Aldrich) or anti-FHY1 (Shen et al., 2005) antibodies were used as primary antibodies to detect the proteins in immunoblot analysis. Immunoblots were quantified using ImageJ (http://rsb.info.nih.gov/ij/).
Immunoblot Analysis and Coimmunoprecipitation Assay
To detect phyA/FHY1 stability, 4-d-old dark-grown seedlings were exposed to R as indicated and homogenized in lysis buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1% Nonidet P-40, 1 mM PMSF, 40 µM MG132, and 1× complete protease inhibitor cocktail (Roche). Protein samples were analyzed by immunoblots as previously described (Shen et al., 2005) using anti-phyA (Xu et al., 1995), anti-FHY1, and anti-RPT5 (Kwok et al., 1999) antibodies. The in vitro pull-down assay was performed as previously described (Zhu et al., 2008). EcoRI-NotI fragments containing full-length FHY1, N-terminal FHY1 (1 to 113), or C-terminal FHY1 (114 to 202) were cloned into pGEX-4T1 (Amersham Biosciences). Full-length HY5 ORF was cloned into pET-28a (Novagen). GST-FHY1, GST-FHY1N, GST-FHY1C, and 6×His-HY5 fusion protein were expressed in Escherichia coli (DE3) and purified with glutathione beads (GE Healthcare) or nickel-nitrilotriacetic acid beads (Qiagen), respectively. Immunoprecipitated proteins were analyzed by immunoblots using anti-His (Qiagen) and anti-GST (GE Healthcare) antibodies. In vivo coimmunoprecipitation experiments were performed under dim green light. Four-day-old dark- or FR light–grown seedlings were homogenized in lysis buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% (w/v) glycerol, 10 mM NaF, 25 mM β-glycerophosphate, 2 mM Na3VO4, 0.1% Tween 20, 1 mM PMSF, and 1× complete protease inhibitor cocktail (Roche) and subjected to procedures described previously (Zhu et al., 2008). Anti-phyA, anti-FHY1, anti-RPT5, and anti-myc (Santa Cruz Biotechnology) antibodies were used in immunoblots analysis.
Confocal Imaging, Colocalization, and BiFC
Samples were scanned by a LSM 510 Meta confocal laser scanning microscope (Carl Zeiss). Arabidopsis cells were counted and pictures were taken within 10 min after R/FR irradiation. To image GFP-FHY1 subcellular localization in onion (Allium cepa) epidermal cells and in Arabidopsis hypocotyl cells, GFP and 4′,6-diamidino-2-phenylindole filter sets and a ×20 air objective were used. To capture NB images in Figures 3H, 4A, and 4B, a ×63 oil-immersion objective was used. For colocalization observations of phyA-CFP and GFP-FHY1, CFP fluorescence was excited with the 458-nm laser and detected with a 465- to 510-nm band-pass filter, while GFP fluorescence was excited with the 488-nm laser and detected between 520 and 555 nm. For BiFC, the full-length ORF sequences of FHY1, HY5, and PIF3 were amplified by PCR (see Supplemental Table 1 online) and subsequently inserted into the SalI-NotI sites of pSY728 or pSY738 vectors and into the SalI-BamHI sites of pSY736 or pSY735 vectors (Bracha-Drori et al., 2004). Pairwise combinations as indicated in Figures 5D and 5H were cobombarded into onion epidermal cells as described previously (Shen et al., 2009). Yellow fluorescent protein fluorescence was visualized under the confocal microscope.
ChIP-qPCR and Real-Time qPCR
For ChIP, materials were cross-linked with 1% formaldehyde in a vacuum for 30 min under dim green light. Following chromatin isolation, DNA was fragmented into an average size of ∼150 bp. Anti-FHY1, anti-HY5 (Osterlund et al., 2000), and anti-GFP (Clontech) antibodies were used for immunoprecipitation. An equal amount of sample without antibody was used as a mock control. ChIP DNA was analyzed by qPCR with power SYBR Green PCR Master Mix (Applied Biosystems) and specific primers as follows: CHS-H-F, 5′-CCCACCATTCAATCTTGGTAAG-3′; CHS-H-R, 5′-ACACCAACTTGGGTTTATTAGAG-3′; CHS-P-F, 5′-TATTAGATTAGTAGGAGCTAATGATGGAGT-3′; CHS-P-R, 5′-TTATTATGTCTTAAGATACGTATCGCTTG-3′; CHS-E-F, 5′-GTCTGCTCTGAGATCACAGCCG-3′; CHS-E-R, 5′-GAGATGAGGCCGGGAACATCCT-3′. Each ChIP value was normalized to its respective input DNA value (defined as 100%) (Guo et al., 2008). For real-time qPCR, total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) and reverse transcribed via the Taqman reverse transcription reagents kit (Applied Biosystems). qPCR was performed using predeveloped Taqman FAM primers for CHS (assay ID: At02199069_g1), RBCS1A (assay ID: At02334081_g1), and PORA (assay ID: At02321582_g1) genes. Expression levels were normalized to that of the 18S gene (Taqman VIC primers; Applied Biosystems). All ChIP-qPCR and quantitative RT-PCR experiments were independently performed in triplicate, and representative results are shown.
EMSA
EMSA was performed using the Lightshift Chemiluminescent EMSA kit (Pierce) according to the manual instructions. Oligonucleotides corresponding to the HY5 binding region on the CHS promoter (−151 to ∼−193) were biotin labeled as probes or unlabeled as a competitor. Proteins indicated (1 or 4 µg) were incubated together with probes or with competitor in 20-μL reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM DTT, and 50 ng/μL polydeoxyinosinate-polydeoxycytidylate for 20 min at room temperature and separated on 6% native polyacrylamide gels.
Transcription Dual-Luciferase Assay
Transcription dual-luciferase assay in N. benthamiana was performed as described previously (Hellens et al., 2005). A dual-luciferase reporter construct was generated, in which the CHS promoter (−1000 to ∼−1) was inserted before the LUC reporter gene into the KpnI-NcoI sites (see Supplemental Table 1 online) and the internal control REN reporter gene was driven by 35S promoter. The reporter construct was coinfiltrated with the indicated proteins. After infiltration, plants were left in darkness for 2 d and transferred to FR or R for 1 d. The LUC and REN activity of infiltrated leaf discs were measured via the Dual-Luciferase Reporter Assay System (Promega) on a GLOMAX 20/20 luminometer (Promega). Final transcriptional activity was expressed as LUC/REN. Six biological repeats were measured per sample.
Yeast Two-Hybrid Assay
Full length ORFs of FHY1 and HY5 were cloned into the EcoRI-XhoI sites of pB42AD vector (Clontech) or into the KpnI-XhoI sites of pGADT7 vector (Clontech) (see Supplemental Table 1 online). The CHS promoter (−50 to ∼−250) was cloned into the KpnI-XhoI sites of pLacZ2µ vector (Lin et al., 2007) to generate a reporter plasmid (CHSp:LacZ). Indicated combinations of AD fusion plasmids and empty vectors were cotransformed into the yeast strain (EGY48) containing CHSp:LacZ according to the Yeast Protocols Handbook (Clontech). Transformants were selected by SD/-Leu-Trp-Ura plates prior to X-Gal selection.
Anthocyanin Measurement
Approximately 250 mg of 5-d-old Arabidopsis seedlings were homogenized and incubated in 300 μL extraction buffer (methanol containing 1% HCl) overnight at 4°C in the dark. After centrifugation at 15,000g for 15 min, the supernatant was mixed with 200 μL water and 200 μL chloroform. The mixture was centrifuged at 15,000g for 30 min, and then the supernatant absorbance was measured at 530 and 657nm. Final anthocyanin content was calculated as (A530 − 0.25 × A657)/weight of fresh material (g). Three independent experiments were done for each sample.
Chlorophyll Measurement
Materials as indicated were grown on Murashige and Skoog medium without Suc. Approximately 50 seedlings were homogenized and incubated in 1 mL 80% acetone overnight at 4°C in the dark. After centrifugation at 15,000g for 10 min, the supernatant was measured at 660 and 647 nm. The final anthocyanin concentration was calculated as (7.15 × OD660 + 18.71 × OD647)/weight of fresh material (g). Three independent experiments were done for each sample.
Accession Numbers
Arabidopsis Genome Initiative numbers for genes discussed in this article are as follows: FHY1, AT2G37678; FHL, AT5G02200; PHYA, AT1G09570; PHYB, AT2G18790; HY5, AT5G11260; PIF3, AT1G09530; CHS, AT5G13930; PORA, AT5G54190; and RBCS1A, AT1G67090.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phosphorylation on Ser-39 and Thr-61 Inactivates FHY1.
Supplemental Figure 2. Transgenic GFP-FHY1S39DT61D Exhibits Dominant Traits over Wild-Type FHY1 in F1 Heterozygous Seedlings.
Supplemental Figure 3. P-Mimic FHY1 and phyA Colocalize in the Cytosol.
Supplemental Figure 4. FHY1 and phyA Associate with the CHS Promoter.
Supplemental Figure 5. Phosphorylation of FHY1 Affects the Expression of RBCS1A and PORA.
Supplemental Figure 6. FHY1 Phosphorylation Benefits the Greening Process of Etiolated Seedlings.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
This work is supported by a National Institutes of Health grant (GM47850) to X.W.D and in part by the Ministry of Science and Technology of China (2012CB910900) and the State Key Laboratory of Protein and Plant Gene Research at Peking University. We thank Christian Fankhauser and Andreas Hiltbrunner for providing phyA-CFP/phyA-1 seeds, Qi Xie for the pCambia1300-221-Myc vector and for assistance with the tobacco transient expression assay, Chentao Lin for vector pGreenII 0800-LUC and plasmid pSoup-P19, and Cynthia Nezames for critical reading of this article. We thank Joseph Wolenski for his assistance with the confocal microscope.
AUTHOR CONTRIBUTIONS
F.C. and X.W.D. designed the research. F.C., X.S., L.C., M.D., Z.Z, Y.S., J.L., and G.L. performed the research. F.C., N.W., and X.W.D. analyzed the data. F.C., N.W., X.W.D., X.S., and M.D. wrote the article.
Glossary
- R
red light
- FR
far-red light
- P
phosphorylation
- NB
nuclear body
- GFP
green fluorescent protein
- CFP
cyan fluorescent protein
- ChIP-qPCR
chromatin immunoprecipitation–quantitative PCR
- BiFC
bimolecular fluorescence complementation
- GST
glutathione S-transferase
References
- Barnes S.A., Nishizawa N.K., Quaggio R.B., Whitelam G.C., Chua N.H. (1996). Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Clough R.C., Vierstra R.D. (1997). Phytochrome degradation. Plant Cell Environ. 20: 713–721 [Google Scholar]
- Debrieux D., Fankhauser C. (2010). Light-induced degradation of phyA is promoted by transfer of the photoreceptor into the nucleus. Plant Mol. Biol. 73: 687–695 [DOI] [PubMed] [Google Scholar]
- Desnos T., Puente P., Whitelam G.C., Harberd N.P. (2001). FHY1: A phytochrome A-specific signal transducer. Genes Dev. 15: 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Casal J.J. (2004). Phenotypic characterization of a photomorphogenic mutant. Plant J. 39: 747–760 [DOI] [PubMed] [Google Scholar]
- Ferrer J.L., Austin M.B., Stewart C., Jr, Noel J.P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46: 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Zhou J., Elling A.A., Charron J.B., Deng X.W. (2008). Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 147: 2070–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczián A., Kunkel T., Kircher S., Schäfer E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L., Kircher S., Toth R., Adam E., Schäfer E., Nagy F. (2000). Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 22: 125–133 [DOI] [PubMed] [Google Scholar]
- Kircher S., Gil P., Kozma-Bognár L., Fejes E., Speth V., Husselstein-Muller T., Bauer D., Adám E., Schäfer E., Nagy F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S.F., Staub J.M., Deng X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285: 85–95 [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Mathews S. (2006). Phytochrome-mediated development in land plants: Red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 15: 3483–3503 [DOI] [PubMed] [Google Scholar]
- Nagatani A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7: 708–711 [DOI] [PubMed] [Google Scholar]
- Neff M.M., Van Volkenburgh E. (1994). Light-stimulated cotyledon expansion in Arabidopsis seedlings (the role of phytochrome B). Plant Physiol. 104: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y., Hirano T., Yoshimoto K., Shimizu M., Kobayashi H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18: 455–463 [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. (2010). Phytochromes. Curr. Biol. 20: R504–R507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825 [DOI] [PubMed] [Google Scholar]
- Sharrock R.A., Clack T. (2002). Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 130: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Feng S., Ma L., Lin R., Qu L.J., Chen Z., Wang H., Deng X.W. (2005). Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome. Plant Physiol. 139: 1234–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhou Z., Feng S., Li J., Tan-Wilson A., Qu L.J., Wang H., Deng X.W. (2009). Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21: 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Hudson M.E., Khanna R., Zhu T., Chang S.H., Wang X., Quail P.H. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38: 725–739 [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Hwang Y.S., Quail P.H. (2006). phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 48: 728–742 [DOI] [PubMed] [Google Scholar]
- Van Buskirk E.K., Decker P.V., Chen M. (2012). Photobodies in light signaling. Plant Physiol. 158: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G.C., Johnson E., Peng J., Carol P., Anderson M.L., Cowl J.S., Harberd N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Parks B.M., Short T.W., Quail P.H. (1995). Missense mutations define a restricted segment in the C-terminal domain of phytochrome A critical to its regulatory activity. Plant Cell 7: 1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.W., Jang I.C., Henriques R., Chua N.H. (2009). FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell 21: 1341–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M., Zhou Q., Sarda X., Yau C.P., Chua N.H. (2004). The nuclear localization signal and the C-terminal region of FHY1 are required for transmission of phytochrome A signals. Plant J. 40: 355–365 [DOI] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng S., Chen F., Chen H., Wang J., McCall C., Xiong Y., Deng X.W. (2008). Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20: 1437–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Hare P.D., Yang S.W., Zeidler M., Huang L.F., Chua N.H. (2005). FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 43: 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.H., Laubinger S., Saijo Y., Wang H., Qu L.J., Hoecker U., Deng X.W. (2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.