This work shows that the flv4-flv2 operon provides many β-cyanobacteria with a so far unknown photoprotection mechanism that evolved in parallel with oxygen-evolving photosystem II
Abstract
Synechocystis sp PCC 6803 has four genes encoding flavodiiron proteins (FDPs; Flv1 to Flv4). Here, we investigated the flv4-flv2 operon encoding the Flv4, Sll0218, and Flv2 proteins, which are strongly expressed under low inorganic carbon conditions (i.e., air level of CO2) but become repressed at elevated CO2 conditions. Different from FDP homodimers in anaerobic microbes, Synechocystis Flv2 and Flv4 form a heterodimer. It is located in cytoplasm but also has a high affinity to membrane in the presence of cations. Sll0218, on the contrary, resides in the thylakoid membrane in association with a high molecular mass protein complex. Sll0218 operates partially independently of Flv2/Flv4. It stabilizes the photosystem II (PSII) dimers, and according to biophysical measurements opens up a novel electron transfer pathway to the Flv2/Flv4 heterodimer from PSII. Constructed homology models suggest efficient electron transfer in heterodimeric Flv2/Flv4. It is suggested that Flv2/Flv4 binds to thylakoids in light, mediates electron transfer from PSII, and concomitantly regulates the association of phycobilisomes with PSII. The function of the flv4-flv2 operon provides many β-cyanobacteria with a so far unknown photoprotection mechanism that evolved in parallel with oxygen-evolving PSII.
INTRODUCTION
Flavodiiron proteins (FDPs), originally known as A-type flavoproteins (Flv) (Wasserfallen et al., 1998), function in detoxification of O2 and/or NO in many strict and facultative anaerobes (Vicente et al., 2008a). FDPs form a complex group of enzymes, and many of them have been thoroughly characterized. Purified FDPs have been shown to function as homodimers or homotetramers (Vicente et al., 2008b, 2009), and many structures have been solved by x-ray crystallography (Frazão et al., 2000; Silaghi-Dumitrescu et al., 2005; Seedorf et al., 2007; Di Matteo et al., 2008). All members of the FDP family share a common minimal core containing two structural domains. The N-terminal metallo-β-lactamase–like domain harbors a nonheme diiron center, and the C-terminal flavodoxin domain contains a flavin mononucleotide (FMN) moiety. The crystal structure has revealed a head-to-tail arrangement of the two FDP monomers, which is required for efficient electron transfer in the active site by bringing the diiron center from one monomer into close contact with the FMN moiety of the other monomer (Vicente et al., 2008a). Despite continuously accumulating knowledge, the definite answers on substrate specificity of many FDPs have remained unknown.
Besides the common sequence core, some of the FDPs have extra C-terminal extensions. Phylogenic analysis revealed that FDPs bearing the same C-terminal extensions cluster together (Saraiva et al., 2004). All cyanobacterial FDPs examined so far as well as some photosynthetic eukaryotic FDPs contain a C-terminal flavin reductase domain (Zhang et al., 2009). In Synechocystis sp PCC 6803 (hereafter Synechocystis), four genes encoding FDPs were identified. The flv1 (sll1521) and flv3 (sll0550) genes are dispersed in the genome, whereas flv2 (sll0219) and flv4 (sll0217) are organized in an operon (CyanoBase: http://genome.kazusa.or.jp/cyanobase/). Recombinant Synechocystis Flv3 homodimer has been shown to have NAD(P)H:oxygen oxidoreductase activity (Vicente et al., 2002). It was soon confirmed by in vivo studies that Synechocystis Flv1 and Flv3 function in the Mehler-like reaction, which acquires electrons after photosystem I (PSI) to reduce molecular oxygen to water like the real Mehler reaction but differs from that by no concomitant formation of reactive oxygen species (Helman et al., 2003). We recently showed that up to 60% of electrons from water-splitting photosystem II (PSII) can be directed to molecular oxygen via Flv1 and Flv3 upon inorganic carbon (Ci) starvation in Synechocystis 6803 (Allahverdiyeva et al., 2011).
Evolution of oxygenic photosynthesis exposed autotrophic organisms to a special demand of protection against reactive oxygen species to cope with highly oxidizing chemistry of PSII under constant changes in the light supply in natural environments. Several photoprotection mechanisms have evolved in oxygenic photosynthetic organisms, which rapidly modify the existing components to adjust the light-harvesting and energy dissipation functions of the photosynthetic apparatus (Niyogi, 1999; Kanervo et al., 2005; Bailey and Grossman, 2008; Tikkanen et al., 2011). State 1–state 2 transitions (state transitions) (reviewed in Allen, 1992; van Thor et al., 1998; Wollman, 2001; Mullineaux and Emlyn-Jones, 2005; Tikkanen et al., 2011) and nonphotochemical quenching (reviewed in Horton et al., 1996; Müller et al., 2001), both based on the light-harvesting antenna systems, are particularly important in short-term regulation of light harvesting in plants and green algae. In cyanobacteria, phycobilisomes (PBSs) function as a major light-harvesting antenna, and their quenching is mediated by a soluble orange carotenoid binding protein, OCP (El Bissati et al., 2000; Kirilovsky, 2007). The Flv2 and Flv4 proteins, unique for cyanobacteria, are also likely to function in photoprotection of PSII (Zhang et al., 2009). Light is an elusive substrate for all photoautotrophs, and photodamage to PSII occurs at all irradiance levels (Tyystjärvi and Aro, 1996). Cyanobacteria are particularly sensitive to light at low ambient CO2 levels (i.e., air level) when the electron acceptors are limiting photosynthesis. Noteworthy, these are the conditions that also strongly enhance the expression of the flv4-flv2 operon (Zhang et al., 2009).
The functional mechanisms of the flv4-flv2 operon (sll0217-19), encoding the Flv4, Sll0218, and Flv2 proteins, in apparent photoprotection of PSII have so far remained completely unknown. Here, we propose a unique mechanism for the Flv2, Flv4, and Sll0218 proteins in modulation of PSII function upon stress conditions.
RESULTS
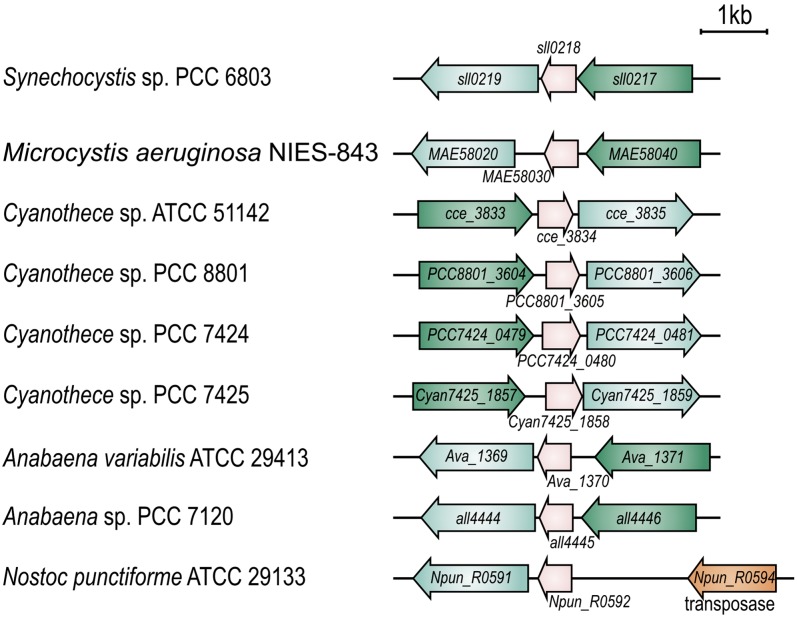
Occurrence of the Operon flv4-flv2 in Cyanobacteria
Cyanobacterial genomes harbor several copies of FDP genes that occur in pairs and cluster with their genetically close orthologs (Zhang et al., 2009). In Synechocystis, the flv4, sll0218, and flv2 genes form an operon that belongs to the low Ci stimulon (M. Eisenhut, J. Georg, S. Klähn, I. Sakurai, H. Silén, P. Zhang, W.R. Hess, and E.-M. Aro, unpublished data). Analysis of the sequenced cyanobacterial genomes revealed that the order of the genes in the operon is highly conserved (Figure 1). In this respect, only two exceptions were observed. One is Nostoc punctiforme ATCC 29,133 lacking the flv4 ortholog, which most probably results from the upstream transposase gene (Figure 1). The other exception is Thermosynechococcus elongatus BP-1, whose flv4 (tlr1088) and flv2 (tll1373) genes are scattered in the genome, and the sll0218 ortholog is absent.
Figure 1.
Organization of the flv4-flv2 Operon in Synechocystis sp PCC 6803 and Other Cyanobacterial Species.
Genes are shown in scale. The direction of transcription is indicated by block arrows. The gene numbers/names are given according to CyanoBase. Orthologs are shown by the same color: flv4 in green, sll0218 in pink, and flv2 in cyan.
Sll0218 is annotated as a hypothetical protein that belongs to the PsiE (for phosphate starvation inducible E) superfamily (http://www.ncbi.nlm.nih.gov/cdd) and whose function remains to be determined. Sll0218 orthologs, besides those located in the flv4-flv2 operons, can be found in many sequenced cyanobacteria, bacteria, and archaea. Phylogenic analysis (see Supplemental Figure 1 online) grouped the Sll0218 homologs into three clades. Those encoded by the flv4-flv2 operon are highly conserved and constitute one clade. Other bacteria and archaea sequences grouped into another clade. Interestingly, some filamentous cyanobacteria have two to three copies of Sll0218 homologs. Sll0218 homologs outside the flv4-flv2 operon in filamentous cyanobacteria form the third clade and are more related to those in bacteria and archaea. It is worth noting that the flv4-flv2 operon was not identified in Synechococcus species and Lyngbya, yet the high homology Sll0218 sequences were found, which grouped together with Sll0218 in the operon clade.
Cellular Location of the Gene Products of the flv4-flv2 Operon
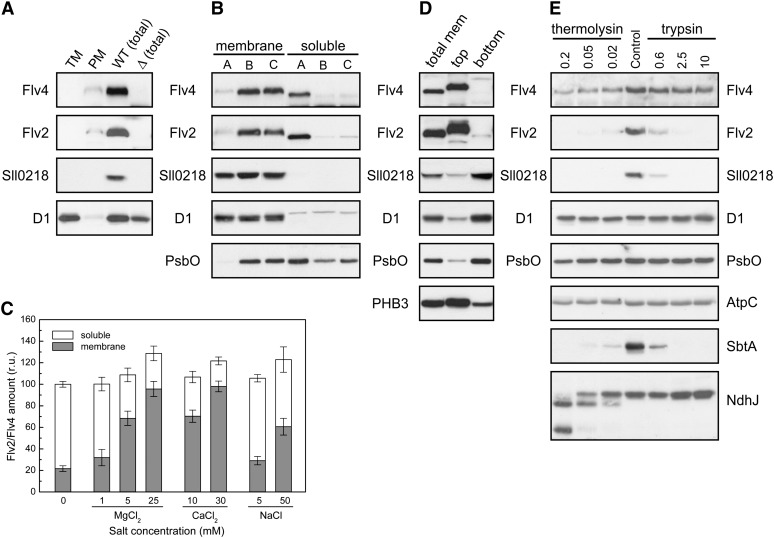
The transmembrane helix prediction program TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) provided evidence that Sll0218 is mainly composed of four transmembrane helices and, therefore, most probably resides in the membrane. Although no transmembrane helices were predicted in any of the FDP sequences, we also found a strong association of the Flv2 and Flv4 proteins with the membrane fraction (Zhang et al., 2009). To localize the Flv4, Sll0218, and Flv2 proteins precisely in the membrane fraction, the thylakoid and plasma membranes, purified by the two-phase partitioning method (Norling et al., 1998), were probed with protein specific antibodies. Unexpectedly, only a weak response was detected with Flv4 and Flv2 antibodies compared with that recorded in the total membrane fraction isolated by a standard method used in our laboratory (which includes high cation concentration; see Methods). However, no Sll0218 signal could be found in conventional two-phase fractions (Norling et al., 1998; Figure 2A).
Figure 2.
Cellular Locations of the Flv4 (Sll0217), Sll0218, and Flv2 (Sll0219) Proteins in Synechocystis.
(A) Thylakoid membrane (TM) and plasma membrane (PM) were purified from wild-type total membranes according to Norling et al. (1998), and total membranes from the wild type (WT [total]) and Δsll0217-18 mutant (Δ [total]) were isolated as described in Methods.
(B) Comparison of the distribution of the Flv2, Sll0218, and Flv4 proteins in the membrane and soluble fractions isolated by different buffer systems: buffer A (no Mg2+ or Ca2+), buffer B (30 mM CaCl2), and buffer C (25 mM MgCl2 was supplemented in buffer A).
(C) Relative distribution of the Flv2 and Flv4 proteins in the membrane and soluble fractions upon breaking the wild-type cells in the isolation buffer supplemented with different concentration of MgCl2, CaCl2, and NaCl. The amount of proteins was shown in relative units (r.u.), and the amount obtained by the buffer with no cation was set as 100. Flv2 and Flv4 proteins showed similar partitioning, which is highly dependent on the concentration of Mg2+ and Ca2+ ions.
(D) Distribution of the Flv4, Sll0218, and Flv2 proteins in the membrane fraction. Total membranes (mem) were isolated using buffer D (25 mM MgCl2) and further fractionated by a modified two-phase partitioning system as described in Methods.
(E) Stability of the Flv4, Sll0218, and Flv2 proteins in the membrane subjected to proteolysis by different concentration of trypsin (µg/mL) or thermolysin (mg/mL).
The cells were grown at air level of CO2. The Flv4, Sll0218, and Flv2 proteins were detected by protein immunoblot. D1 and PsbO of PSII were marker proteins of the thylakoid membrane and the lumen, respectively, and prohibitin 3 (PHB3) was used as a marker protein of the plasma membrane.
Nearly complete absence of the Flv4, Sll0218, and Flv2 proteins in the thylakoid and plasma membrane fractions purified by the conventional two-phase partitioning system (Norling et al., 1998) (Figure 2A) needed clarification. To this end, the Flv4, Sll0218, and Flv2 proteins were first analyzed from the total membrane (thylakoids plus plasma membrane) and soluble fractions of the wild-type cells. The fractions were isolated by two different buffer systems, buffer A and buffer B. Buffer A is a low ionic buffer used for the two-phase partitioning (Norling et al., 1998), whereas buffer B is a high ionic buffer generally used for PSII isolation (30 mM CaCl2). As shown in Figure 2B, the two buffer systems gave completely different results. The Flv4 and Flv2 proteins were largely released to the soluble fraction with buffer A but remained associated with the membrane with buffer B. When buffer A was supplemented with 25 mM MgCl2, the Flv4 and Flv2 proteins were mainly associated with the membrane fraction, similar to buffer B system. Indeed, the Flv4 and Flv2 proteins could be localized either in the membrane or the soluble fraction, the distribution being largely dependent on the concentration of cation, Ca2+ or Mg2+, in the isolation buffer (Figure 2C). Upon increasing the Mg2+ or Ca2+ concentration, the majority of the Flv2 and Flv4 proteins altered their location from the soluble fraction to the membrane (Figure 2C). On the contrary, the Sll0218 protein was detected only in the membrane fraction, independently of the buffer system.
To address the association of the Flv4, Flv2, and Sll0218 proteins with the thylakoid or the plasma membrane, membrane isolation was performed in HEPES at pH 7.5 by addition of 25 mM MgCl2, and the pH of the two-phase partitioning buffers was lowered from pH 7.8 to 7.4. As shown in Figure 2D, Flv4 and Flv2 were enriched in the top phase, whereas Sll0218 was enriched in the bottom phase. According to immunoblot analysis with several marker proteins, the plasma membrane was shown to be enriched in the top phase and the thylakoid membrane in the bottom phase, thus revealing the association of Flv4 and Flv2 at high cation concentration with the plasma membrane but being soluble at low cation concentration. Sll0218 was associated with the thylakoid membrane independently of the cation concentration (Figure 2B).
The question why Sll0218 could be detected neither in the thylakoid nor in the plasma membrane after fractionation in the original two-phase partitioning buffer system (Figure 2A) still remained unanswered. Recent proteomics study of Synechocystis thylakoid membrane fraction also failed to identify the Sll0218 protein (Pisareva et al., 2011). Such data suggest heterogeneity in the distribution of Sll0218 in the thylakoid membrane. To test this hypothesis, the total Synechocystis membrane fraction isolated in buffer B, which contains the thylakoid membrane mainly in right-side-out orientation (Figure 2B), was subjected to protease treatment using both trypsin and thermolysin. Intactness of the thylakoid during trypsin digestion was first evidenced by no loss of the PsbO protein, a lumenal extrinsic PSII subunit (Figure 2E). Digestion with thermolysin, on the contrary, decreased the amount of PsbO, suggesting leakage of the thylakoid membrane. Intactness upon trypsin digestion of the NdhJ subunit of NDH-1 on the cytoplasmic side of the thylakoid membrane indicated that this side of the thylakoid membrane was well sealed against trypsin. However, increasing concentration of thermolysin resulted in complete digestion of NdhJ, indicating that the enzyme had penetrated through the thylakoids and opened the membrane sealing completely. Interestingly, the Sll0218 protein was easily digested by both trypsin and thermolysin, opposite to the other integral membrane proteins like D1 of PSII and AtpC. This is in apparent contradiction with data in Figure 2B indicating that trypsin cannot access either the cytoplasmic or the lumenal side of the well-sealed thylakoid membrane during the treatment. Direct access of Sll0218 to proteolysis by both enzymes indicated that the distribution of the Sll0218 protein cannot be homogeneous throughout the thylakoid membrane in the cell. Rather, the Sll0218 protein is likely to be located in specific well-exposed regions of the thylakoid membrane. Such thylakoid regions may be largely lost during two-phase partitioning, which is dependent on membrane surface properties.
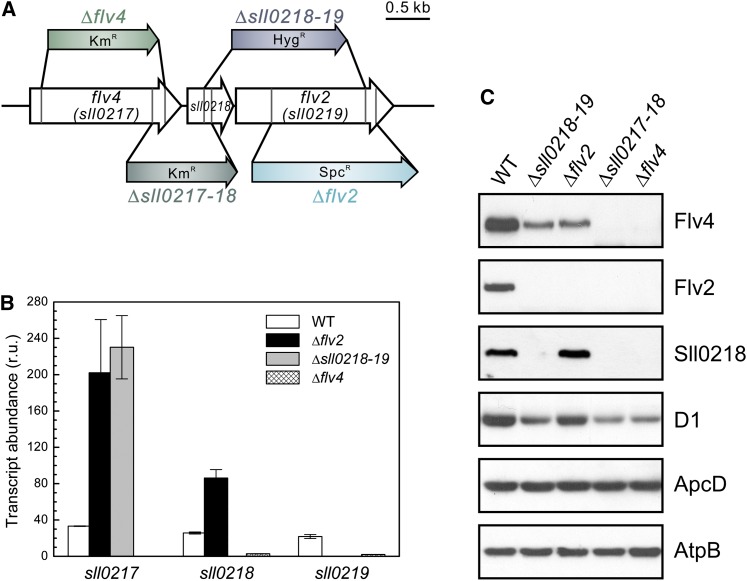
Expression of the flv4-flv2 Operon in Various flv Inactivation Mutants
It was next investigated whether the presence of the Flv4, Sll0218, and Flv2 proteins in Synechocystis cells is independent of the other partner proteins encoded by the flv4-flv2 operon or whether their stability at protein level relies on cotranscribed partners. To this end, the expression of the flv4-flv2 operon was investigated in the wild type and mutants with interruption of the operon at different positions (Figure 3). Inactivation of flv2, or both the flv2 and the sll0218 gene, repressed the expression of the Flv4 protein to <50% of that in the wild type (Figure 3C), despite the fact that the transcripts of the flv4 gene were upregulated fivefold compared with the wild type (Figure 3B). On the contrary, inactivation of the flv2 gene resulted in elevated expression of the sll0218 gene at both the transcript and the protein level. These data indicated a strong regulation of the Flv4 protein, at least an order of magnitude, by the presence or absence of the Flv2 protein. Such coregulation at protein level seems to be independent of the Sll0218 protein. Strong coregulation of the Flv4 and Flv2 proteins and their colocalization both in the presence or absence of cations (Figure 2) suggested a possibility for Flv2/Flv4 heterodimer formation in the cell.
Figure 3.
Description of flv Inactivation Mutants and the Expression of the Operon flv4 -flv2 at Air Level of CO2.
(A) Schematic description of the four flv inactivation mutants mostly used in this study and the location of the kanamycin- (Km), hygromycin- (Hyg), or spectinomycin (Spc)-resistant cassette at different positions.
(B) Transcript accumulation of sll0217-19 in the wild type (WT) and the three flv mutants analyzed by real-time quantitative RT-PCR. Transcription abundance is shown as relative units (r.u.). The transcript level of the rnpB gene is used as a reference. The results are the mean from three independent experiments ± sd.
(C) Protein immunoblot demonstrating the protein accumulation of Flv4, Sll0218, and Flv2 in the total cell extract from wild type and the four flv mutants.
[See online article for color version of this figure.]
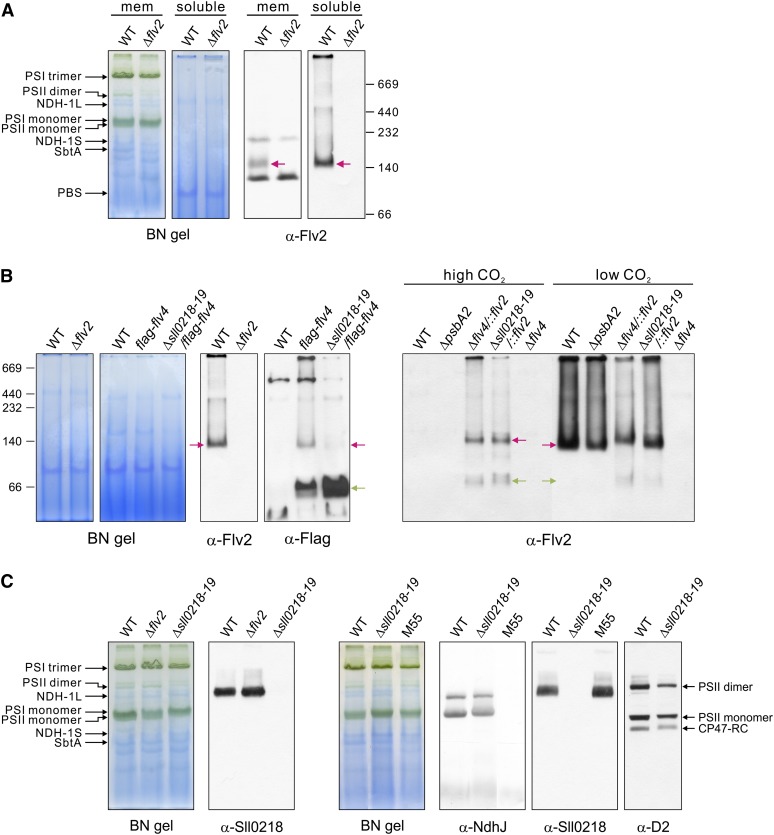
Protein Complexes Formed by the Flv4, Sll0218, and Flv2 Proteins
All studied FDPs in anaerobic microbes have a homodimer or homotetramer structure in vitro, and the dimer formation indeed is a necessity for enzymatic activity (Vicente et al., 2008a). It is conceivable that cyanobacterial Flv2 and Flv4 also form dimers. To verify this assumption, the thylakoids were isolated in buffer B (with 30 mM CaCl2), and protein complexes were solubilized by n-dodecyl-β-d-maltoside (DM), separated by blue native (BN)-PAGE, and subjected to protein immunoblotting as described (Zhang et al., 2004). However, the extraction of the Flv4 and Flv2 proteins from the membranes by DM or other nonionic detergents turned extremely inefficient (see Supplemental Figure 2 online). To help the identification of the weak signal obtained from the membrane fraction by the Flv2 antibody, we analyzed the soluble fraction as well. As shown in Figure 4A, the Flv2 protein (molecular mass = 65 kD) formed a complex of ∼140 kD, which run in BN gels slightly faster than the SbtA complex, a sodium bicarbonate antiporter involved in carbon concentration (Zhang et al., 2004). The immunosignal obtained with the Flv2 antibody in BN blots was verified by protein immunoblot of the two-dimensional (2D) BN/SDS gel (see Supplemental Figure 3 online).
Figure 4.
Protein Complexes of the Flv2, Flv4, and Sll0218 Proteins in the Membrane and Soluble Fractions.
Cells were grown at air level of CO2 (low CO2) if not otherwise specified. Membrane (mem) and soluble fractions were isolated from wild-type (WT) and the flv mutant cells and separated by BN-PAGE of different acrylamide concentration (5 to 12% or 6 to 13%). BN gels and respective protein immunoblots demonstrating the Flv2 complex in the wild type (A). The Δflv2 mutant is shown as a control. The Flv2/Flv4 heterodimer in the wild type, the flag-flv4 strains, and ΔpsbA2 as well as Flv2 homodimer and monomer in Δflv4/::flv2 and Δsll0218-19/::flv2 (B). Association of the Sll0218 protein in a large membrane protein complex and demonstration of this complex not being the NDH-1 complex (α-NdhJ) (C). The red arrows demonstrate the positions of the dimeric Flv2 and Flv4 complexes, and the green arrow demonstrates the position of monomeric FLAG-Flv4. See Figure 3A for detailed description of the flv inactivation mutants. NdhB subunit is deleted in the M55 mutant (Ogawa, 1991), which is unable to assemble NDH-1L and NDH-1M complexes (Zhang et al., 2004).
The size of the Flv2 protein complex in BN-PAGE (Figure 4A) corresponded to that of an Flv dimer. To dissect whether the band represented an Flv2 homodimer or an Flv2/Flv4 heterodimer, the Flv4 protein had to be identified. However, the Flv4 polyclonal antibody was not compatible with BN gels and no specific immunoresponse was detected. To solve this problem, the Flv4 protein was fused with a FLAG-tag, and the FLAG-Flv4 protein was expressed both in the wild type and the Δsll0218-19 mutant. Using the antibody against FLAG-tag revealed two specific bands in the BN gel blot of the flag-flv4 strain, corresponding to a dimer (red arrow) and a monomer (green arrow) (Figure 4B). The specificity of the FLAG-tag immunosignals to FLAG-Flv4 was verified by protein immunoblot analysis of the 2D BN/SDS gel (see Supplemental Figure 3 online). Different from the flag-flv4 strain, the dimeric FLAG-Flv4 form was nearly missing from the Δsll0218-19/flag-flv4 strain (Figure 4B), providing evidence that the FLAG-Flv4 protein did not form dimers in the absence of the Flv2 protein. Thus, the dimeric Flv4 complex in flag-flv4 was assigned as an Flv2/Flv4 heterodimer. Although a considerable amount of monomeric FLAG-Flv4 was detected in the flag-flv4 and Δsll0218-19/flag-flv4 strains, the Flv2 protein was detected only as a dimeric form in the wild type (Figures 4A and 4B). Two possible reasons can lead to this situation: Increase in Flv4 expression in the FLAG-tag strains causes accumulation of excess FLAG-Flv4 protein for stoichiometric formation of the Flv2/Flv4 heterodimer, or introduction of the FLAG-tag might hamper the dimer formation of Flv4. Enhancement of the accumulation of the Flv4 protein in the flag-flv4 and Δsll0218-19/flag-flv4 strains is shown in Supplemental Figure 4A online.
To investigate the possibility of Flv2 homodimer formation in vivo, the Flv2 protein was expressed under strong psbA2 promoter in the Δflv4 and Δsll0218-19 mutants. The expression pattern of the Flv2 and Flv4 proteins is shown in Supplemental Figure 4B online. Under high CO2, where the native flv4-flv2 promoter is suppressed and, thus, the Flv4 protein is absent, ∼60% of the Flv2 proteins were present as homodimers and 40% as monomers, similarly in both complemented strains. When the cells were grown under air level of CO2, the pattern of Flv2 in Δflv4/::flv2 was similar to that observed in cells grown under high CO2, whereas the amount of monomeric Flv2 largely decreased in the Δsll0218-19/::flv2 strain (i.e., in the presence of the Flv4 protein) (Figure 4B). It is worth noting that the dimeric Flv2 separated by BN-PAGE from the wild type, ΔpsbA2, and Δsll0218-19/::flv2 strains grown under air level of CO2 migrates slightly faster than the Flv2 dimer from high-CO2-grown Δflv4/::flv2 and Δsll0218-19/::flv2 cells, also suggesting that the Flv2 dimer complex in the wild type is the Flv2/Flv4 heterodimer (Figure 4B). Interestingly, the accumulation of the Flv2 protein in Δsll0218-19/::flv2 was higher when grown under air level of CO2 than under high CO2 (see Supplemental Figure 4B online), which contradicts with the higher expression level of PpsbA2 under high CO2. This provides further support for coregulation of Flv2 and Flv4 at protein level and, thus, for the formation of the heterodimer in vivo.
Contrary to the apparent heterodimer association of the Flv2 and Flv4 proteins, the Sll0218 protein was always found to be associated with a much larger complex, higher than 500 kD in size, which in BN-PAGE was found to be located between the PSII dimer and the NDH-1L complex (Zhang et al., 2004; Figure 4C). The expression of the NDH-1 complex (probed by the NdhJ antibody) in Δsll0218-19 was similar to that in the wild type, and the Sll0218 complex was normal also in the M55 strain (lacking both the NDH-1L and NDH-1M), suggesting that the Sll0218 protein is not associated with the NDH-1L complex. Nevertheless, a faint band, overlapping with the α-Sll0218 band, was observed with the D2 antibody particularly in the wild-type cells, suggesting a possible association of Sll0218 with an intermediate stage of PSII assembly.
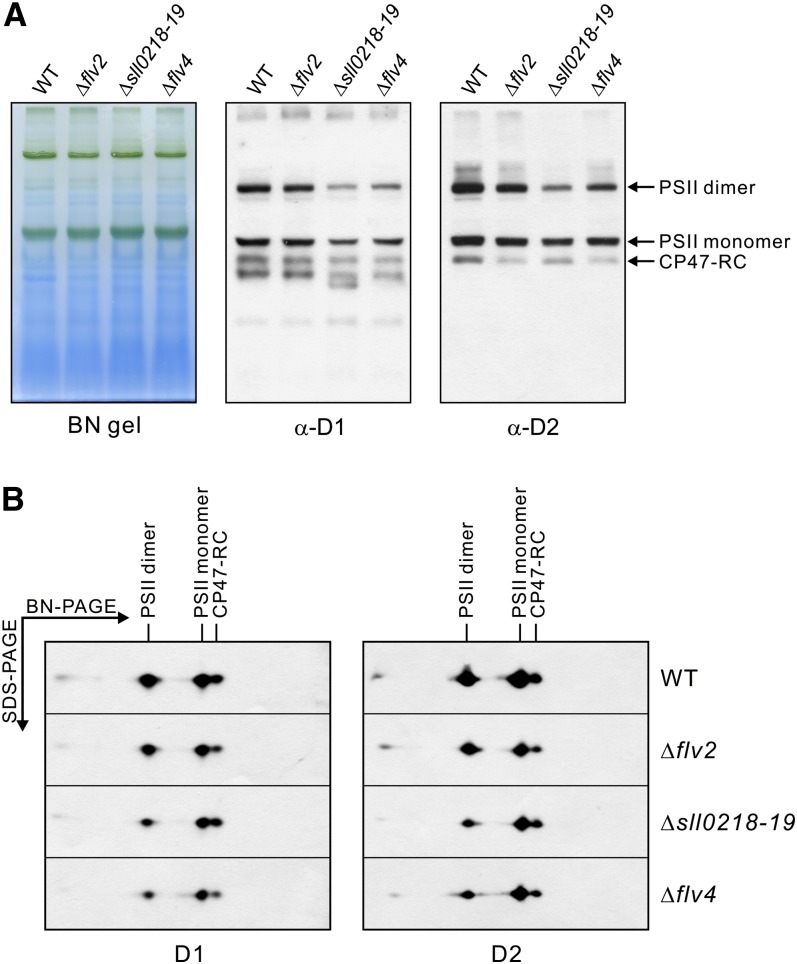
PSII Complexes in the flv Mutants
To clarify the effects at the level of the PSII complexes, the membranes of the wild type and the various flv mutants were subjected to BN/SDS-PAGE analysis (Figure 5). After BN-PAGE, three major different forms of the PSII complexes were detected: the dimer, the monomer, and the CP47-RC monomer. To quantify the amounts of the different forms of the PSII complexes, the PSII core proteins D1 and D2 were quantified by immunobloting the 2D BN/SDS gels. The wild type had almost equal amounts of the PSII dimer and monomer, which together consisted of >80% of the total PSII complexes. All the flv mutants had lower amounts of PSII than the wild type. The total PSII content in Δflv2 was ∼70% of that in the wild type, but the proportions of different PSII complexes were similar to that in the wild type. The PSII content in Δsll0218-19 and Δflv4 was only ∼50% of that in the wild type. Moreover, the proportion of the PSII dimer was significantly lower, and the proportion of the monomeric PSII complexes (both monomer and CP47-RC) was higher compared with the wild type and Δflv2. The quantification of the PSII complexes in the wild type and the flv mutants is shown in Table 1. The absence of the Sll0218 protein significantly reduced the relative content of PSII dimers.
Figure 5.
Different Forms of PSII Complexes in the Wild Type and the flv Mutants.
Membrane samples were isolated from the cells grown at air level of CO2 and were applied to BN/SDS-PAGE. After electrotransfer, the polyvinylidene fluoride membranes were probed with the D1 and D2 antibodies. WT, the wild type.
(A) BN gels and respective protein immunoblots demonstrating the different PSII complexes.
Table 1. Quantification of the Different Forms of PSII Complexes in the Wild Type and the flv Mutants.
| Strain | Total PSII (%) | Different Forms of PSII (%) |
||
|---|---|---|---|---|
| Dimer | Monomer | CP47-RC | ||
| The wild type | 100 | 42 ± 3 | 42 ± 4 | 16 ± 2 |
| Δflv2 | 73 ± 4 | 41 ± 3 | 43 ± 2 | 16 ± 4 |
| Δsll0218-19 | 49 ± 8 | 24 ± 2 | 50 ± 5 | 26 ± 3 |
| Δflv4 | 53 ± 8 | 29 ± 6 | 51 ± 6 | 20 ± 2 |
The quantification was performed from four independent BN/SDS-PAGE blots. The values are the mean of the D1 and D2 signals ± sd.
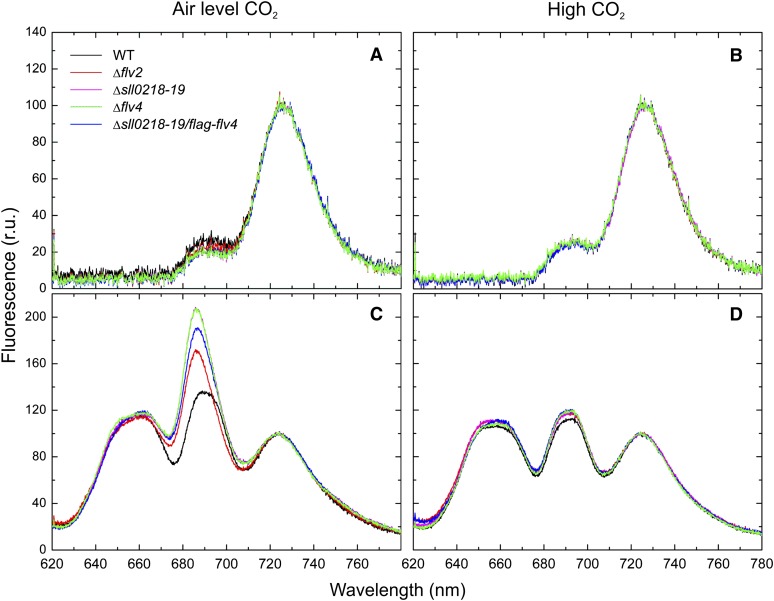
Energy Transfer from PBS to Photosystems in the Wild Type and the flv Mutants
To investigate whether the proteins encoded by the flv4-flv2 operon affect the efficiency of energy transfer from PBS to the photosystems, the 77K fluorescence emission spectra were recorded from the cells grown under high or air level of CO2 (Figure 6). When chlorophyll a was preferentially excited using 440-nm light, the fluorescence emission spectra showed two main peaks characteristic to PSII (685 and 695 nm) and PSI (723 nm), and differences between the wild type and all the flv mutants were only marginal in the cells grown both at high CO2 and in the air (Figures 6A and 6B). When 580-nm light was used to preferentially excite PBS, three fluorescence emission maxima were recorded (Figures 6C and 6D). They originate from PBS (650 and 665 nm), PSII (685 and 695 nm), and PSI (723 nm) emission, respectively. When the spectra were normalized to PSI fluorescence, the emission from PBS was similar in the wild type and all the flv mutants; however, the apparent PSII fluorescence emission (685 and 695 nm) was greatly enhanced from all the flv4-flv2 operon mutants when compared with that of the wild type grown at air level of CO2 (Figure 6C). Such a difference between the wild type and the flv mutants disappeared when using high-CO2-grown cells (Figure 6D). The flag-flv4 strain behaved similarly to the wild type (data not shown).
Figure 6.
77K Fluorescence Emission Spectra of the Wild Type and Various flv Mutant Strains.
(A) Air level of CO2 grown cells excited by 440-nm light.
(B) High-CO2-grown cells excited by 440-nm light.
(C) Air level of CO2 grown cells excited by 580-nm light.
(D) High-CO2-grown cells excited by 580-nm light.
The spectra were averaged from three to five independent measurements. Each spectrum was normalized to PSI fluorescence peak at 723 nm (set as 100), and the fluorescence was shown as relative units (r.u.). WT, the wild type.
The 77K fluorescence peak from PSII contains two subpeaks at 685 and 695 nm. The peak at 695 nm is assigned to emission from the internal antenna chlorophylls in CP47. The emission at 685 nm contains contributions from both the core antenna chlorophylls in CP43 and from the allophycocyanin emitters (Vernotte et al., 1992; Karapetyan, 2008). Due to the contribution of the terminal phycobilin emitters to the PSII fluorescence maximum at 685 nm, the spectra were deconvoluted, and the fluorescence yields were calculated (see Supplemental Figure 5 and Supplemental Table 1 online). As shown in Supplemental Table 1, the FPBS/FPSI ratio of the fluorescence yields was not significantly different between the wild type and any of the flv mutant strains. On the contrary, the 695-nm emission peak did not increase proportionally with the 685-nm peak in the flv mutants. Therefore, the increase in 685-nm fluorescence peak is likely to be largely due to terminal phycobilin emitters rather than to the core CP43 antenna chlorophylls (Vernotte et al., 1992). These results implied an apparent inefficiency in energy transfer from PBS to PSII in the flv mutants grown under air level CO2 conditions.
The 77K fluorescence spectra demonstrated that the flv4-flv2 operon mutants have distorted energy transfer from the terminal PBS emitters to PSII in cells grown in ambient air, whereas at high CO2 where the flv4-flv2 operon is suppressed, neither the wild type nor the flv mutants showed any abnormalities in 77K fluorescence emission at 685 nm. Moreover, the 77K fluorescence emission spectrum of the Δsll0218-19/flag-flv4 mutant cells was similar to that of the Δsll0218-19 mutant grown at air level of CO2 (see Supplemental Table 1 online), supporting the notion that the Flv2/Flv4 heterodimer is the functional form in vivo.
PSII Function in flv4-flv2 Operon Mutants
The photosynthetic electron transfer capacity was monitored by oxygen evolution measurements using different electron acceptors (Table 2). When the cells were grown under high CO2, there were no significant differences in the capacity of oxygen evolution between the wild type and the flv mutants, similar to our previous report (Zhang et al., 2009). To get further insights into the function of PSII, two different artificial electron acceptors were used; 2,6-dimethyl-p-benzoquinone (DMBQ), which accepts electrons from the plastoquinone (PQ) pool, and 2,6-dichloro-p-benzoquinone (DCBQ), which has a high affinity to the QB site and is suggested to accept electrons in the QB pocket (Graan and Ort, 1986). Generally, lower rates of oxygen evolution in the flv mutants than the wild type grown in the air level of CO2 and measured in the presence of DCBQ or DMBQ are in agreement with lower amounts of the PSII centers in the flv mutants (Figure 3C; Zhang et al., 2009).
Table 2. Oxygen Evolution Rates of Wild-Type and flv Mutant Cells Measured with Different Electron Acceptors.
| Oxygen Evolution (μmol O2 mg-1 Chlorophyll h−1) |
|||
|---|---|---|---|
| Strain and Growth Conditions | Water to CO2 | Water to DCBQ | Water to DMBQ |
| High CO2 | |||
| The wild type | 414 ± 27 | 1156 ± 35 | 849 ± 19 |
| Δflv2 | 443 ± 20 | 1048 ± 11 | 854 ± 10 |
| Δflv4 | 411 ± 30 | 1197 ± 16 | 870 ± 09 |
| Air level of CO2 | |||
| The wild type | 437 ± 15 | 547 ± 12 | 663 ± 12 |
| Δflv2 | 485 ± 18 | 504 ± 11 | 482 ± 17 |
| Δflv4 | 444 ± 10 | 508 ± 10 | 457 ± 21 |
Steady state oxygen evolution (μmol O2 mg-1 chlorophyll h−1) was measured by O2 electrode under saturating light. The cells grown under high (3%) or air level of CO2 were suspended in the growth medium. The measurements were performed in the presence of 10 mM NaHCO3. The results are presented as a mean ± sd from three independent cultures and four measurements from each culture.
The most intriguing result was a different behavior of the wild type and the flv mutants when DCBQ and DMBQ were used as artificial PSII electron acceptors (Table 2). The wild-type cells grown in the air level of CO2 showed higher oxygen evolution rate with DMBQ as an artificial electron acceptor than with DCBQ. On the contrary, the oxygen evolution rates of all the flv4-flv2 operon mutants, as well as the high-CO2-grown wild-type cells, were higher when measured with DCBQ as an electron acceptor compared with those measured with DMBQ. Indeed, after testing several wild-type Synechocystis sp PCC 6803 strains of different laboratories (data not shown), all the wild-type cells grown in the air (but not the cells grown under high CO2) showed similar behavior that strictly differs from that of all the flv mutants grown in the air level of CO2 (Table 2). These results provided evidence for a presence of an additional electron exit site in wild-type cells grown in the air level of CO2 located on, or in a close vicinity to, the QB site. Such electron transfer apparently becomes inhibited by binding of DCBQ to the QB site, but not by the naturally functioning PQ.
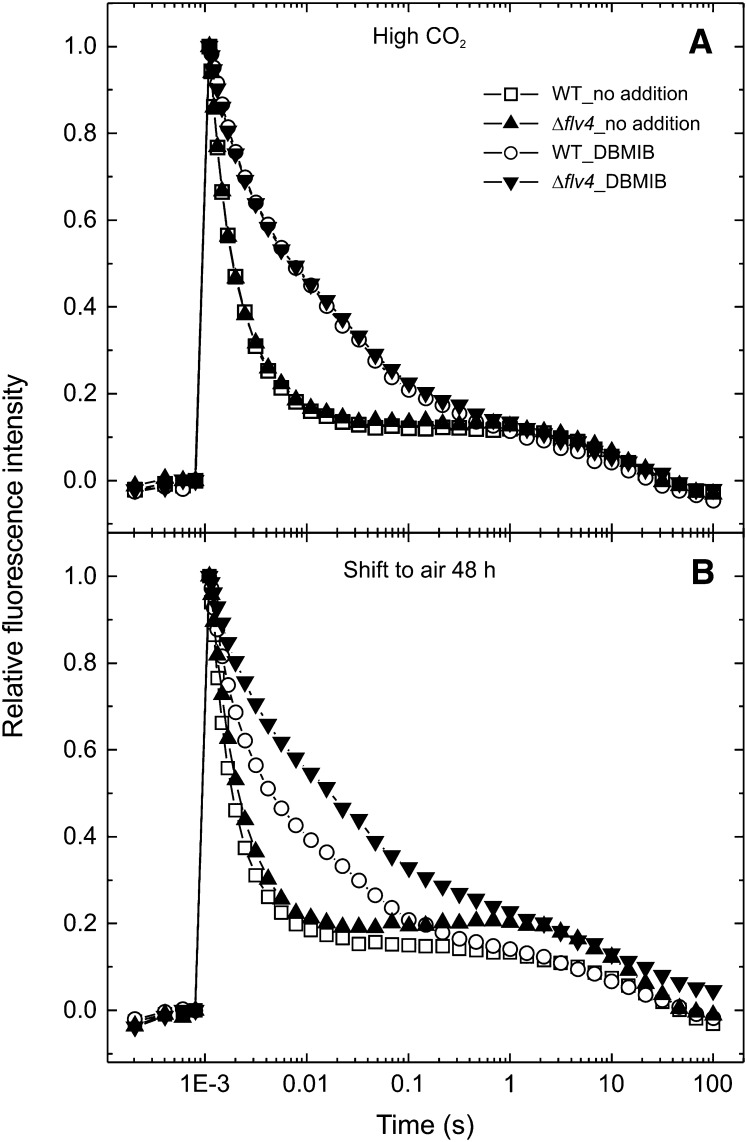
The possible alternative electron transport pathway may take electrons from the QB site of the PSII complex and transfer them to the Flv2/Flv4 proteins. To test this hypothesis, flash-induced chlorophyll fluorescence measurements were performed (Figure 7), which reflect electron transport processes at the acceptor side of PSII. Illumination of the samples by a short saturating light pulse transfers an electron from the water-oxidizing complex of PSII to the QA quinone electron acceptor, which induces an increase of the fluorescence yield. Subsequent decay of fluorescence in the dark reflects the reoxidation of QA− by various processes (Vass et al., 1999). Forward electron transfer to PQ, which is bound at the QB site at the moment of the flash, yields a 400- to 500-µs relaxation phase (fast phase). Whereas, binding of PQ to the QB site in centers, which did not contain QB at the moment of the flash, yields a 3- to 5-ms relaxation phase (middle phase). The electron that is stabilized on QB− can also recombine with the S2 state of the water-oxidizing complex via charge equilibrium between the QA−QB ↔ QAQB− states, leading to a 10- to 15-s relaxation phase (slow phase).
Figure 7.
Relaxation of Flash-Induced Chlorophyll Fluorescence. The cells were grown under high CO2 (A) and after a shift from high CO2 to air for 48 h (B). The wild type, open symbols; Δflv4, closed symbols. The fluorescence traces are shown after normalization to the same initial amplitude. WT, the wild type.
Comparison of the fluorescence decay kinetics between the wild type and the Δflv4 mutant showed that in high-CO2-grown cells, the relaxation is practically identical in the wild type and mutant stains (Figure 7A). After shifting the cells from high CO2 to air level of CO2, the overall fluorescence decay became slower in the Δflv4 mutant (Figure 7B), mainly due to the decrease of the fast phase amplitude and the higher time constant of the slow phase (Table 3). When the cells were grown in air level CO2, the slower fluorescence decay in the Δflv4 mutant relative to the wild type became even more clear (Table 3). These data showed that in the absence of the Flv2/Flv4 proteins, the reoxidation process of QA− is slowed down. This effect provided further support for the presence of an alternative electron transfer pathway to the FDPs. However, modification of electron transport at the acceptor side of PSII due to enhanced photoinhibition may possibly also contribute to this behavior. The existence of an alternative electron transfer route in PSII should influence the redox state of the PQ pool by providing a pathway to keep the PQ pool oxidized. To test this hypothesis, a 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) treatment was applied, which blocks the main oxidation route of plastoquinol (PQH2) in the PQ pool via inhibiting the Qo site of the cytb6f complex. As shown in Table 3, under these conditions the fluorescence relaxation was slowed down in the high-CO2-grown wild type as well as the Δflv4 mutant cells, but the curves of both strains still had the same relaxation kinetics (Figure 7A). Nevertheless, after transferring the cells from high to the air level CO2 conditions, the DBMIB effect became more pronounced in the Δflv4 mutant (Figure 7B). These data provide further evidence for the existence of an alternative electron transport pathway, which can channel out electrons from PSII and keep the PQ pool oxidized. It is worth noting that this effect was observed 48 h after the transfer of cells from high to air level of CO2, although the Flv2, Sll0218, and Flv4 proteins appear much earlier, possibly indicating that only those PSII centers synthesized in the presence of the Sll0218 proteins have a capability for alternative electron transfer to the Flv2/Flv4 heterodimer.
Table 3. Kinetic Data of Flash-Induced Fluorescence Relaxation Components in the Wild Type and Δflv4 Mutant Grown at High CO2, Air Level of CO2, and 48 h after a Shift from High to Air Level of CO2.
| Strains and Growth Conditions | Fast |
Middle |
Slow |
|||
|---|---|---|---|---|---|---|
| T1 (ms) | A1 (%) | T2 (ms) | A2 (%) | T3 (s) | A3 (%) | |
| High CO2 |
No addition |
|||||
| The wild type | 0.476 ± 0.017 | 58 ± 1.5 | 3.6 ± 0.29 | 26 ± 1.6 | 17.7 ± 1.2 | 16 ± 0.3 |
| Δflv4 | 0.469 ± 0.007 | 59 ± 1.7 | 3.7 ± 0.15 | 24 ± 1.9 | 14.7 ± 1.2 | 17 ± 0.5 |
| + DBMIB | ||||||
| The wild type | 1.360 ± 0.020 | 44 ± 1.5 | 33.6 ± 0.3 | 36 ± 1.7 | 4.3 ± 0.7 | 20 ± 0.2 |
| Δflv4 | 1.310 ± 0.040 | 45 ± 1.8 | 36.5 ± 0.3 | 35 ± 2.3 | 3.6 ± 0.7 | 20 ± 0.5 |
| Shift to air (48 h) | No addition | |||||
| The wild type | 0.503 ± 0.010 | 64 ± 1.6 | 4.3 ± 0.1 | 20 ± 0.7 | 11.6 ± 3.7 | 16 ± 0.9 |
| Δflv4 | 0.548 ± 0.045 | 49 ± 2.7 | 3.1 ± 0.5 | 30 ± 1.7 | 12.4 ± 2.8 | 21 ± 0.5 |
| + DBMIB | ||||||
| The wild type | 1.015 ± 0.030 | 52 ± 0.3 | 30.6 ± 2.7 | 30 ± 0.6 | 4.7 ± 0.9 | 18 ± 0.3 |
| Δflv4 | 1.430 ± 0.015 | 40 ± 1.3 | 39.8 ± 0.6 | 35 ± 2.1 | 4.0 ± 0.6 | 25 ± 0.9 |
| Air (growth) | No addition | |||||
| The wild type | 0.643 ± 0.025 | 65 ± 0.5 | 6.8 ± 0.4 | 19 ± 0.2 | 8.4 ± 1.1 | 16 ± 0.4 |
| Δflv4 | 0.822 ± 0.028 | 48 ± 2.1 | 7.7 ± 0.3 | 25 ± 1.6 | 3.5 ± 0.2 | 27 ± 1.3 |
| + DBMIB | ||||||
| The wild type | 1.030 ± 0.038 | 46 ± 1.5 | 32.1 ± 0.2 | 35 ± 1.8 | 2.2 ± 0.3 | 19 ± 0.4 |
| Δflv4 | 1.384 ± 0.097 | 30 ± 1.8 | 35.2 ± 2.6 | 36 ± 2.1 | 1.3 ± 0.2 | 34 ± 0.8 |
Measurements were made in the absence and presence of the electron transfer inhibitor DBMIB that binds to the Q0 site of the Cytb6f complex. The fitting parameters of fluorescence relaxation curves are presented as a mean ± se from three independent cultures and two to three measurements from each culture.
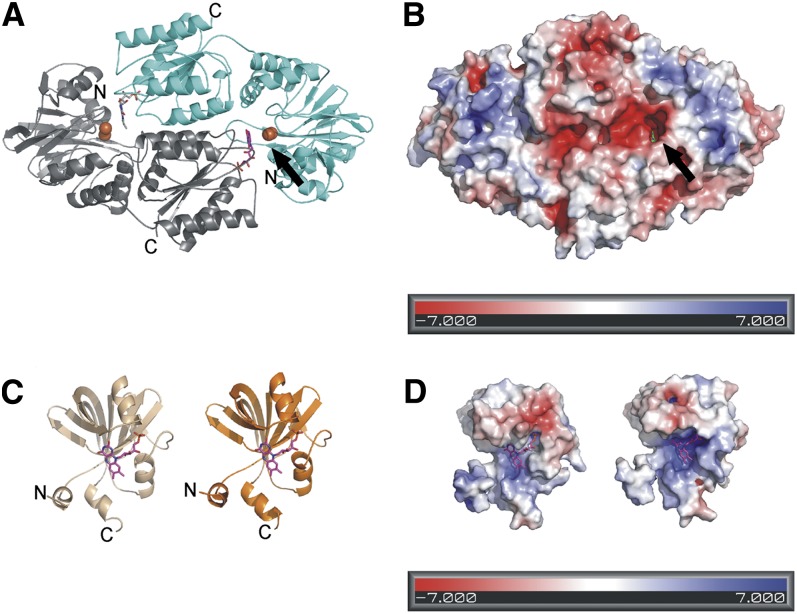
Homology Models of the Flv2/Flv4 Heterodimer
To analyze the Flv2/Flv4 heterodimer formation and to study how the protein(s) might dock to the membrane, the homology models of Flv2/Flv4 were constructed (Figure 8). Synechocystis Flv2 and Flv4 consist of three structural domains. The heterodimeric Flv2/Flv4 models were constructed for the β-lactamase–like and the flavodoxin domains. The β-lactamase–like domain (amino acids 28 to 270 in Flv2 and 19 to 250 in Flv4) contains the diiron site and the flavodoxin domain (amino acids 271 to 421 in Flv2 and 251 to 400 in Flv4) contains the FMN binding site. Conserved metal binding sites on the surface of the Flv2/Flv4 heterodimer model are summarized in Supplemental Table 2 online.
Figure 8.
Homology Model of the Flv2/Flv4 Heterodimer.
(A) The overall fold of the Flv2/Flv4 heterodimer. The functional reactive site is seen on the right with FMN (magenta) from the Flv2 monomer (gray) and diiron site from the Flv4 monomer (cyan). The irons are shown as orange spheres.
(B) The electrostatic surface of the Flv2/Flv4 heterodimer. The FMN (green) is visible in the reactive site cavity.
(C) The homology models of the flavin reductase domains with bound FMN; Flv2 is to the left (beige) and Flv4 to the right (orange).
(D) The electrostatic surface of the flavin reductase domains.
To investigate the structural changes of the enzyme during electron transfer, the Flv2/Flv4 dimer was modeled in two different oxidation states based on the structures of F420H2 oxidase from Methanothermobacter marburgensis (Seedorf et al., 2007) in the active reduced state with the switch loop in the closed conformation (Protein Data Bank [PDB] ID: 2OHI) and in the inactive oxidized state with the switch loop in the open conformation (PDB ID: 2OHJ). The sequence identity between M. marburgensis F420H2 oxidase and Synechocystis Flv2 and Flv4 is 29 and 26%, respectively. We also modeled the closed conformation of the Flv2/Flv4 heterodimer based on Moorella thermoacetica FprA (PDB ID: 1YCF), which has a sequence identity of 26% with Flv2 and 28% with Flv4. Since the sequence identities are relatively low, the structure-based sequence alignment of the crystallized FDPs, Synechocystis Flv2, and Flv4 together with cyanobacterial sequences (see Supplemental Figure 6 online) was used to increase the reliability of modeling. Furthermore, the structures in the FDP family are known to represent very similar three-dimensional folds, despite sharing a sequence identity of 30 to 40%.
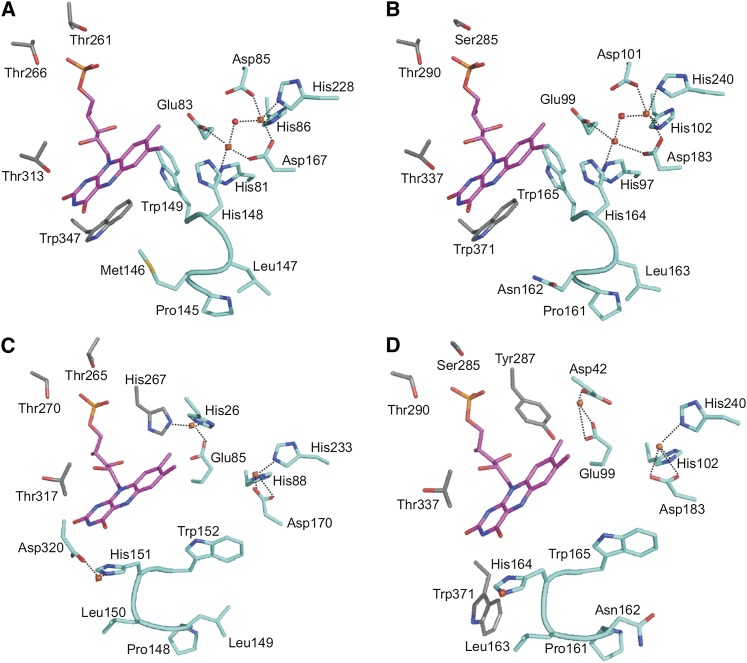
The Flv2/Flv4 heterodimer is formed by head-to-tail arrangement; therefore, the iron binding site in the Flv2 monomer faces the FMN binding site in the Flv4 monomer and vice versa (Figure 8A). Based on our analysis, the binding site that consists of the FMN binding site of the Flv2 monomer and the iron binding site of the Flv4 monomer (indicated with an arrow in Figures 8A and 8B) contains 10 conserved residues. In the FMN binding site of the Flv2 monomer, six FMN binding residues (Ser-285, Thr-290, and Thr-337 from Flv2 and Glu-99, His-164, and Trp-165 from Flv4) are conserved or substituted with similar residues in M. thermoacetica FprA (Thr-261, Thr-266, and Thr-313 from one monomer and Glu-83, His-148, and Trp-149 from the other monomer) (Figures 9A and 9B). In the closed form of the heterodimer, the iron binding residues of the Flv4 monomer (first iron, Asp-101, His-102, Asp-183, and His-240; second iron, His-97, Glu-99, and Asp-183) are totally conserved with those in M. thermoacetica FprA (first iron, Asp-85, His-86, Asp-167, and His-228; second iron, His-81, Glu-83, and Asp-167) (Figures 9A and 9B).
Figure 9.
The FMN and Diiron Binding Sites.
(A) M. thermoacetica FprA in the closed conformation.
(B) Synechocystis Flv2/Flv4 heterodimer in the closed conformation
(C) M. marburgensis F420H2 oxidase in the open conformation.
(D) Flv2/Flv4 heterodimer in the open conformation.
Only the conserved active site composed of the FMN binding site of Flv2 and the diiron site of Flv4 of Synechocystis Flv2/Flv4 heterodimer is shown ([B] and [D]). Trp-165 of the switch loop (161-PNLH-164 in Flv4; [B] and [D]) and Trp-371 (in Flv2) turn away from the FMN, forming a cavity next to the FMN. Trp-371 is replaced by a Gly in M. marburgensis F420H2 oxidase. FMNs are shown in magenta and irons in orange. Residues participating from the other monomer are shown in gray.
In almost all members of the FDP family, a conserved Trp stacks with the FMN isoalloxazine ring, but in methanogens and some cyanobacterial, it is replaced by Gly (Vicente et al., 2008a; pink in Supplemental Figure 6 online). This Trp is conserved in Flv2 (Trp-371) and M. thermoacetica FprA (Trp-347), but Flv4 and M. marburgensis F420H2 oxidase have a Gly. In the open conformation of the Flv2/Flv4 model (Figure 9D), which is based on the open conformation of M. marburgensis F420H2 oxidase (Figure 9C), Trp-371 is turned away, enlarging the binding site. Additionally, the second iron is moved and coordinated by Asp-42 and Glu-99 from Flv4 and Tyr-287 from Flv2 (Figure 9D). Furthermore, a third iron stabilizes the open conformation of the highly conserved switch loop (161-PNLH-164; Figure 9D), moving Trp-165 in Flv4 away and creating a cavity next to the FMN (Figures 9C and 9D). In the binding site formed by the FMN binding site of the Flv4 monomer and the iron binding site of the Flv2 monomer, only three residues are conserved (corresponding to His-97, Trp-165, and His-240 in Figure 9B) (see Supplemental Figure 7 online). In conclusion, the two reactive sites in the Flv2/Flv4 heterodimer differ from each other; the binding site formed by the FMN binding site of the Flv2 monomer and the iron binding site of the Flv4 monomer is more conserved than the other site and, thus, clearly functional. The FMN binding site of the Flv4 homodimer does not have all the conserved residues, whereas the ion binding sites are not conserved in the Flv2 homodimer (see Supplemental Figure 7 online). Thus, the functional binding site of the Flv2/Flv4 heterodimer structurally is even more conserved than either of the Flv homodimers.
The flavin reductase domain enables FDPs in cyanobacteria to transfer electrons directly from NAD(P)H to the catalytic iron binding site. The reaction, which in other organisms requires several steps, is condensed to a single protein (Vicente et al., 2002). Due to unavailability of structures with all three domains, the flavin reductase domains of Flv2 and Flv4 were modeled as separate monomers (Figures 8C and 8D) based on Archaeoglobus fulgidus FeR (PDB ID: 1I0S) (Chiu et al., 2001). It has a sequence identity of 19 and 22% to Flv2 and Flv4, respectively. The A. fulgidus FeR dimer binds one FMN and NADP+. The flavin reductase domains of Flv2 and Flv4 were modeled with bound FMN, which is bound mainly by main chain interactions, supporting the previous data that many of the residues involved in binding are not conserved among FeR homologs (Chiu et al., 2001).
DISCUSSION
Synechocystis Flv4 and Flv2 Form a Functional Heterodimer in Vivo
A particular feature of cyanobacterial-type FDPs is the presence of multiple (two to six) genes encoding different FDPs in one organism. Further analyses showed that these paralogs are present in pairs, belonging to either the FlvA or the FlvB cluster (Zhang et al., 2009). Since all FDPs studied so far function as a homodimer or a homotetramer in vitro (Vicente et al., 2008b, 2009), it is conceivable that the FDPs form dimers also in cyanobacteria. In Synechocystis, there are two pairs of FDP genes. The flv1 (sll1521) and flv3 (sll0550) genes are dispersed in the genome, yet both the Flv1 and Flv3 proteins are required for the function of Mehler-like reaction (Helman et al., 2003; Allahverdiyeva et al., 2011). Here, we focused on the flv2 (sll0219) and flv4 (sll0217) genes, which are organized in an operon (flv4-sll0218-flv2), strongly induced upon low Ci and high light conditions (Zhang et al., 2009). Moreover, the gene order of the flv4-sll0218-flv2 operon is highly conserved among cyanobacterial species (Figure 1). This also strongly suggests an interaction of the encoded proteins (Dandekar et al., 1998; Shi et al., 2005).
Interrelationship in the stability of the Flv2 and Flv4 proteins became evident from expression analysis of the flv4-flv2 operon at both the transcript and protein levels in the wild type and various flv mutants. Strong downregulation of the Flv4 protein occurs in the absence of the Flv2 proteins (Δflv2 and Δsll0218-19 mutants), even though the amount of flv4 transcripts is much higher in both mutants than in the wild type (Figure 3). Much less of the Flv2 protein was detected in the Δsll0218-19/::flv2 mutant grown under high CO2 than in the air level of CO2 (see Supplemental Figure 4B online), despite the fact that the psbA2 gene is expressed in an opposite manner. Such interdependency in the stability of Flv4 and Flv2 highly suggested the Flv2/Flv4 heterodimer formation. Finally, direct evidence for heterodimer formation came from a biochemical experiment with the flag-flv4 strain (Figure 4B), which revealed the existence of the Flv2/FLAG-Flv4 heterodimer. Moreover, the Flv4 and Flv2 proteins clearly favor the heterodimer formation over the homodimer formation. Only monomeric FLAG-Flv4 was detected in Δsll0218-19/flag-flv4, indicating that the FLAG-Flv4 can hardly dimerize in vivo. Flv2, on the contrary, was found to be capable of forming homodimers in the absence of Flv4. Nevertheless, the proportion of Flv2 monomers largely decreased when Flv4 was available for heterodimer formation. Thus, we conclude that both Flv2 and Flv4 favor the Flv2/Flv4 heterodimer formation.
Molecular modeling provided further support for the Flv2/Flv4 heterodimer formation. According to the constructed homology models of the Flv2/Flv4 heterodimer, only the active site composed of the FMN binding site of Flv2 and the diiron site of Flv4 is conserved and functional, while the less-conserved active site (composed of the FMN binding site of Flv4 and diiron site of Flv2) seems not to be functional. The conserved residues from the Flv2 and Flv4 monomers together form a conserved FMN binding site in the Flv2 monomer, which in the heterodimer configuration is even more conserved than in the homodimer models (Figure 9; see Supplemental Figure 7 online). Thus, based on the analysis of the binding sites in the homology models, the Flv2/Flv4 heterodimer is capable of rapid electron transfer between FMN and the diiron center, but only one of the active sites seems to be functional.
A similar conclusion of heterodimer formation in vivo can be drawn from a functional point of view. The Δsll0218-19 mutant behaved almost similar to the Δflv4 mutant (Figures 5 and 6; Zhang et al., 2009), suggesting that the Flv4 protein is malfunctioning in the absence of Flv2. Nonfunctional Flv2 homodimer, on the other hand, can be deduced from slow kinetics of flash-induced fluorescence decay in Δflv4/::flv2 cells grown under air level of CO2 compared with ΔpsbA2 and Δsll0218-19/::flv2 cells grown under similar conditions (see Supplemental Table 3 online).
Putative Metal Binding Sites on the Surface of the Flv2/Flv4 Heterodimer Mediate Membrane Association and Weak Protein–Protein Interactions
Strong association of the Flv2 and Flv4 proteins with the membrane fraction, despite the absence of predictable transmembrane helices (Zhang et al., 2009), caused severe problems for characterization of the Flv4 and Flv2 proteins. It was first speculated that such a membrane association occurs via the Sll0218 protein, which is an integral membrane protein with four predicted transmembrane helices. However, this turned out not to be the case as Flv4 largely remained in the membrane fraction even in the absence of the Sll0218 protein (the Δflv2, Δsll0218-19, and Δsll0218-19/flag-flv4 mutants) (data not shown). Moreover, the Sll0218 protein is associated with a big membrane complex independently of the Flv2/Flv4 heterodimer (Figure 4). Finally, by modifying the two-phase partitioning method for membrane subfractionation, the Sll0218 protein was localized to the thylakoid membrane, whereas Flv4 and Flv2 in that system tend to associate with the plasma membrane (Figure 2D). The latter association appeared to be strongly cation dependent (Figures 2B and 2C). Similar behavior was recently reported for another Synechocystis protein, although the mechanism for the membrane association is unclear (Carmel et al., 2011). Based on the Flv2/Flv4 heterodimer model and the sequence alignment, some putative metal binding sites involving His, Asp, and Glu residues were recognized on the protein surface. Surface-bound metals were found also in homologous structures. For example, the flavodoxin-like domain from Synechococcus sp (PDB ID: 3HLY) has a Ca2+, and M. thermoacetica FprA (PDB ID: 1YCF, 1YCG, and 1YCH) has several Zn2+ bound at the surface (Silaghi-Dumitrescu et al., 2005). The metal ions on the structures originate from the crystallization solution, but some of the metal binding residues are conserved or substituted with similar residues in our model (see Supplemental Table 2 online). Taken together, these residues might be responsible for the cation-dependent association of Flv2 and Flv4 to the membrane fraction.
Based on analysis of the electrostatic surface potential of the model, the Flv4 monomer has a more negatively charged surface (Figure 8B) and contains more putative metal binding sites on the surface than the Flv2 monomer (see Supplemental Table 2 online). Taking into consideration the fact that the cation concentration used in the isolation buffer is much higher than the real physiological conditions (Figures 2B to 2D), the binding of Flv2/Flv4 to the membrane in vivo might be transient and reversible. Specificity of Flv2/Flv4 to plasma membrane upon cell breakage (Figure 2D) may also relate to its surface charge since the inner surface of the plasma membrane is more negative than the right-side-out thylakoid membrane (Barber, 1982; Körner et al., 1985). However, the surface charge of the thylakoid membrane can be dramatically changed during photosynthesis, which may mediate transit binding of Flv2/Flv4 also with the thylakoid membrane.
Sll0218 Protein Resides in the Thylakoid Membrane and Stabilizes the PSII Dimer in Low Ci Condition
The Sll0218 protein was unambiguously localized to a high molecular mass complex in the thylakoid membrane. However, the strange behavior of the Sll0218 protein in the conventional two-phase partitioning system of the membrane subfractions suggested that the Sll0218 protein is not evenly distributed in the thylakoid membrane. It is associated with a large complex, which is just marginally smaller than the major PSII dimer. Decrease of the ratio of PSII dimer to PSII monomer complexes happens only in the mutants lacking the Sll0218 protein (Δsll0218-19 and Δflv4), but not in Δflv2 (Figure 5, Table 1). Therefore, we postulate that the Sll0218 may function as a chaperon in the stabilization of the PSII dimer assembled under conditions of air level of CO2.
Although the PSII complex has been isolated both in the monomeric and dimeric form (Rögner et al., 1987; Hankamer et al., 1997; Adachi et al., 2009), it is widely accepted that in vivo the PSII complex functions as a dimer in cyanobacteria as well as in higher plants (Hankamer et al., 1999; Kuhl et al., 2000), whereas the monomeric PSII may be an intermediate of the normal assembly and repair of PSII (Aro et al., 2005; Nixon et al., 2010). Although the in vivo dimerization of PSII in cyanobacteria has been questioned due to the use of detergents for solubilization and in vitro analysis of the thylakoid complexes (Takahashi et al., 2009; Watanabe et al., 2009), there is compelling evidence for dimer formation by mutant approaches. Indeed, small PSII subunits PsbM and PsbT, located in monomer-monomer interface, have been shown crucial for proper assembly and repair of PSII (Ohnishi and Takahashi, 2001; Iwai et al., 2004; Bentley et al., 2008). Since the Sll0218 protein is expressed only under low (i.e., air level) CO2 conditions, we postulate that the assembly of PSII dimers differs depending on the presence or absence of the Sll0218 proteins (see below).
Flv2/Flv4 Heterodimer and Sll0218 Protein Are Functionally Linked in Photoprotection of PSII
Optimization of photosynthesis requires strict regulation between absorption and conversion of solar energy into chemical energy and its utilization by downstream metabolic pathways. For aquatic photoautotrophs, such as cyanobacteria, CO2 is an essential but often deficient substrate. In natural environments, insufficient availability of CO2, especially when combined with high irradiation, results in high excitation pressure on photosystems (Huner, 1998) and limits the rate of photosynthesis. To cope with this, several CO2-concentrating mechanisms are heavily induced at low Ci conditions in cyanobacteria to enhance the CO2 concentration around ribulose-1,5-bis-phosphate carboxylase/oxygenase and at the same time induce cyclic electron flow to generate more ATP (reviewed in Kaplan and Reinhold, 1999; Giordano et al., 2005; Badger et al., 2006; Price et al., 2008). Microarray and proteomic studies have revealed a low-CO2 stimulon (Wang et al., 2004; Eisenhut et al., 2007; Battchikova et al., 2010), also including the operon sll0217-19 (flv4-flv2), which is regulated as strongly as the inducible CO2-concentrating mechanism genes. The flv2 and flv4 genes also have a high light response (Hihara et al., 2001; Zhang et al., 2009). The flv4 and flv2 inactivation mutants revealed a high susceptibility to photoinhibition of PSII under air level of CO2 conditions (Zhang et al., 2009). Moreover, our recent studies on the expression of the operon revealed a tight regulation of this operon flv4-flv2 by small regulatory RNAs (M. Eisenhut, J. Georg, S. Klähn, I. Sakurai, H. Silén, P. Zhang, W.R. Hess, and E.-M. Aro, unpublished data). Such strong induction and regulation by antisense RNAs of the flv4-flv2 operon together with observed protection of PSII under air level of CO2 and high light conditions suggest a pivotal role for all three proteins under carbon-limiting conditions.
According to the results presented here, the gene products of sll0217 and sll0219, the two FDPs Flv4 and Flv2, form a heterodimer and reside in the cytoplasm, although they may also occasionally be associated with the membrane in vivo. Low efficiency of energy transfer from PBS to PSII, most probably due to uncoupled PBS, is typical of all flv operon mutants (Figure 6B), suggesting that the Flv2/Flv4 heterodimer also is involved in dynamic interactions between PBS and PSII. It is important to note that this is occurring only under low ambient CO2 conditions where the protection of PSII against photodamage by the proteins encoded by the flv4-flv2 operon also occurs.
When energy transfer from PBS to reaction centers was studied by fluorescence spectra at 77K (Figure 6), we found a significant increase of F685 in all flv inactivation mutants grown at air level of CO2. Involvement of a chlorophyll binding protein, IsiA (Bibby et al., 2001; Boekema et al., 2001; Sandström et al., 2001), was first eliminated by showing normal fluorescence emission at 685 nm when 440-nm light was used for excitation. Similar 77K fluorescence peak at 685 nm in all the flv mutants and the wild type upon excitation of chlorophyll suggested that the unusual fluorescence pattern in the mutants is due to PBS. Preferential transfer of energy from PBS to PSI in the flv mutants was excluded since the FPBS/FPSI ratio is similar in the wild type and the flv mutants (see Supplemental Table 1 online). Involvement of nonphotochemical quenching was likewise eliminated, as it was not induced in the flv mutants under our standard growth conditions (data not shown). Thus, the increase of fluorescence emission at 685 nm in the flv mutants is concluded to result from partial uncoupling of the terminal emitters of PBS from PSII. Under normal growth conditions, the PBSs are generally associated with reaction centers and uncoupled PBSs are present in a very low amount (Mullineaux and Holzwarth, 1991). However, the flv mutants are exceptions and decoupled PBSs are present in low Ci growth conditions in much higher amounts than in the wild type.
Evidence has been provided indicating that PBSs are directly connected with the PSII dimer (Barber et al., 2003). However, a flexible interaction of PBS with both PSII and PSI is required for efficient energy transfer upon changes in the light regime (Mullineaux, 2008). It was suggested that PBS interacts with the thylakoid membrane via multiple weak charge–charge interactions, which allow flexible dissociation and reassociation with the reaction centers. The ApcE protein was shown to be of particular importance for the interaction of PBS with the membrane and the reaction centers (MacColl, 1998). However, the nature of the interaction between ApcE and the membrane remains obscure. We speculate, however, that the interaction between PBS and PSII, as influenced by Flv2/Flv4, is rather indirect and due to perturbation of the integrity of PSII dimers. Such perturbation also results from mutagenesis of specific subunits of PSII leading to a decrease in energy transfer efficiency from PBS to PSII (Funk et al., 1998; Veerman et al., 2005).
Although the gene products of flv4, sll0218, and flv2 reside in different cellular locations and are associated with their own interaction partners, we assume that a functional linkage tightly connects them together. This is compatible with the conserved gene order of the operon among a number of cyanobacterial species (Figure 1). A hypothetic model of the photoprotection mechanism by a coordinated function of the Flv2/Flv4 heterodimer and the Sll0218 protein is presented in Figure 10B. We postulate that the assembly and function of the PSII complexes require much more precise control under the deficiency of terminal electron acceptors in air level of CO2 compared with the process under high CO2. The Sll0218 protein is assumed to be involved in the assembly process leading to a subtle change in the conformation of the PSII dimer, which facilitates the interaction with the Flv2/Flv4 heterodimer and energy transfer between PBS and PSII. Moreover, due to a peculiar distribution of Sll0218 in the thylakoid membrane, our results provide further evidence for the presence of specific biogenesis sites of the PSII complexes (Rengstl et al., 2011).
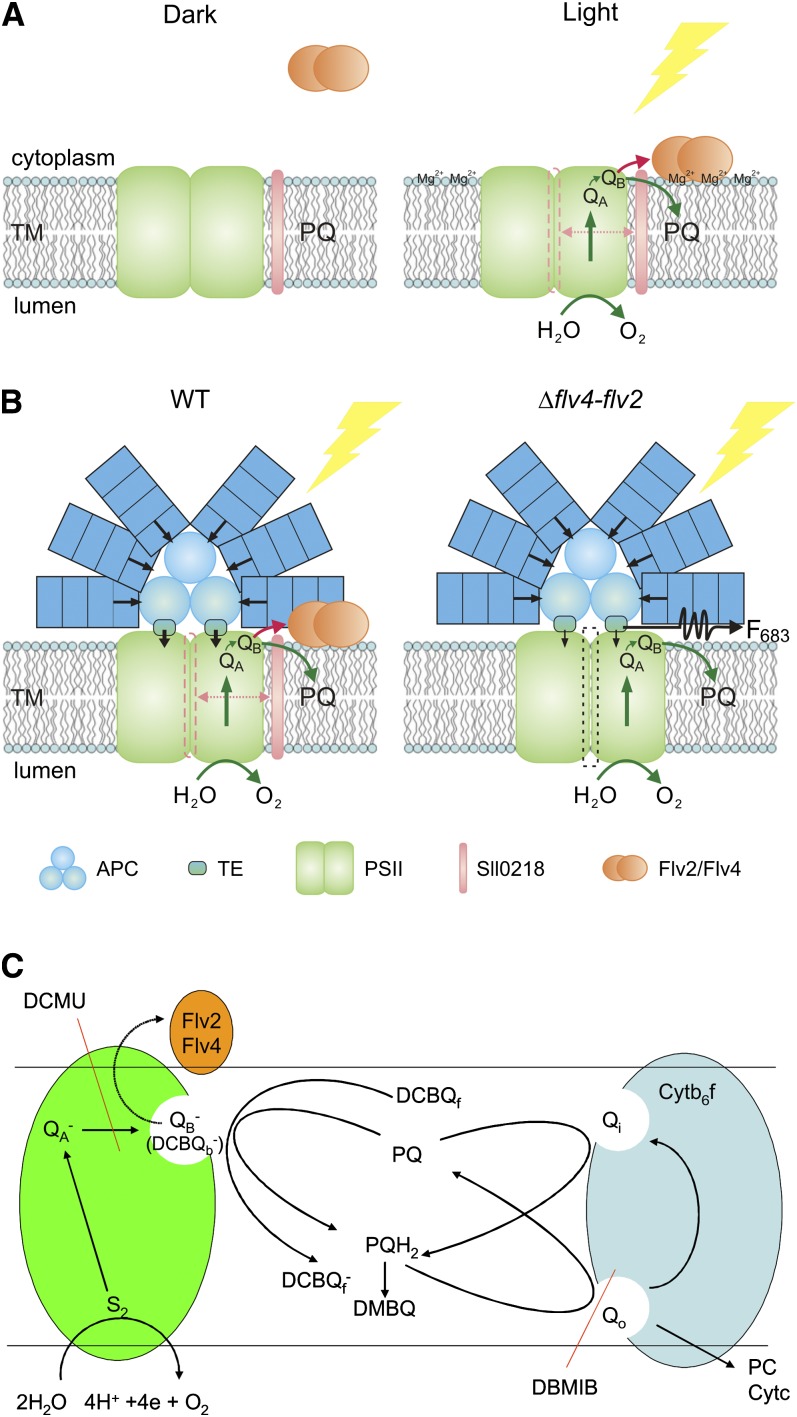
Figure 10.
Working Hypothesis on the Function of the Flv2/Flv4 Heterodimer and the Sll0218 Protein during Photosynthesis at Low Ci (Air Level of CO2) Conditions.
(A) In darkness, the Flv2/Flv4 heterodimer is mostly in soluble form in the cytoplasm. However, transient binding of the Flv2/Flv4 heterodimer to the thylakoid membrane (TM) is postulated to occur upon increase in Mg2+ concentration on the cytoplasmic surface of the thylakoid membrane when the lights are turned on. This creates an alternative electron transfer route from PSII to the Flv2/Flv4 heterodimer that is made possible by Sll0218-induced subtle changes in PSII.
(B) Hypothetical model for the involvement of the Flv2/Flv4 and Sll0218 proteins in energy transfer from PBS to PSII and electron transfer from PSII to the Flv2/Flv4 heterodimer at low Ci. Mg2+-induced attachment of the Flv2/Flv4 heterodimer to the thylakoid membrane coordinates both the energy transfer from PBS to PSII and the electron transfer from PSII to the Flv2/Flv4 heterodimer in PSII centers slightly modified by the Sll0218 protein upon the assembly process. WT, the wild type.
(C) Scheme of electron transport processes in the presence of the Flv2/Flv4 heterodimer and the functional mode of the artificial electron acceptors of PSII, DCBQ, and DMBQ.
The Flv2/Flv4 heterodimer, induced concomitantly with the Sll0218 protein, is likely to bind to the cytoplasmic side of PSII and take electrons from the QB site. Binding of the Flv4/Flv2 heterodimer to PSII likely occurs via charge interaction between the heterodimer and the PSII cytoplasmic side (Larom et al., 2010). The role of Flv2/Flv4 in interaction of PBS with PSII is likely to be linked to their electron transfer properties. Considering the electron transfer properties of FDPs, the fully reduced form has four-electron reduction capacity. However, our homology model of the Flv2/Flv4 heterodimer suggests that only one of the two active sites is functional. The Flv2/Flv4 heterodimer is thus likely to be capable of catalyzing a two electron transfer reaction.
One of the key observations regarding electron transport characteristics in the absence of the products of the flv4-flv2 operon is the slower kinetic of flash-induced fluorescence decay in the Δflv4 mutant grown in the air level of CO2 relative to the wild type, when the fluorescence curves are measured without any electron transport inhibitor (Table 3). This effect is mainly due to the increased amplitude of the middle phase of fluorescence decay, which reflects the amount of PSII centers without permanently bound PQ at the QB site. Occupancy of the QB site by PQ is determined by the amount of oxidized PQ in the pool, which is obviously decreased when higher proportion of the PQ molecules is in the fully reduced PQH2 state. Therefore, the change of the fluorescence decay kinetic indicates an increase of PQ pool reduction in the absence of Flv2 and Flv4 proteins. A more significant manifestation of this effect is observed when PQH2 reoxidation by the cytb6f complex is blocked by DBMIB (Figure 7B). These data point to the existence of an alternative electron transport pathway, which involves the Flv2/Flv4 proteins and plays a significant role in the regulation of the redox level of the PQ pool. However, the possibility that modification of the QB binding site due to enhanced photoinhibitory damage also contributes to the change of the fluorescence decay kinetics cannot be fully excluded at this stage.
The site where the alternative pathway branches out from the electron transport route resides most likely in the PSII complex. This idea is supported by the differential effect of the DCBQ and DMBQ artificial acceptors on the electron transport and oxygen evolution rates in the wild type and the flv mutant strains when cultured in the air level of CO2 (Table 2). DCBQ accepts electrons directly from the QB site and usually supports higher O2 evolution rate than DMBQ, which takes electrons mainly from the PQ pool. This is seen here in samples that lack the Flv2/Flv4 proteins (i.e., in the mutant strains) as well as in the wild type when grown at high CO2. However, in the wild type grown in the air, the DCBQ supported rate is smaller than the DMBQ supported rate, which indicates that occupancy of the QB site with DCBQ blocks the alternative electron transport pathway toward Flv2/Flv4. This idea is further supported by the kinetics of fluorescence decay (Table 3). In cells grown in the air, the PQ pool is more reduced than in the high-CO2-grown cells due to the efficient dark reduction of PQ via the NDH-1 complex. The higher reduction level of the PQ pool leads to decreased amount of bound PQ at the QB site, which is also reflected in the slower decay of the flash-induced fluorescence signal. However, the observation that the fast phase is accelerated in the wild type grown in the air level of CO2 relative to that of the Δflv4 mutant indicates that in the wild-type cells the presence of the Flv2/Flv4 proteins accelerates QA− reoxidation. This process may take place either via electron transfer from QA− to Flv2/Flv4 in parallel with the QA− to QB process or via acceleration of QB− reoxidation by Flv2/Flv4, which competes with the backward electron flow from QB− to QA and therefore accelerates the overall reoxidation of QA−.
Based on these results, we propose the existence of an electron transport pathway from PSII to the Flv2/Flv4 proteins, which serves to decrease the high reduction level of the PQ pool under air level of CO2 conditions. High redox level of the PQ pool results from substrate limitation and facilitates photodamage to PSII. We have a working hypothesis (Figure 10C) suggesting that the alternative electron transport pathway toward the Flv2/Flv4 proteins originates from the PSII complex, possibly from QB−. The inhibition of this process by DCBQ can arise from a structural modification of the QB site, which is induced by DCBQ binding. An alternative possibility is the difference in the binding affinities of QB−, which is very high and leaves sufficient time for the electron to go to the alternative route, and of DCBQ−, which probably has low affinity and limits the electron transfer toward Flv2/Flv4. It should be noted that analysis of the fluorescence relaxation curves is complicated by the increased level of PQ pool reduction in the Flv2/Flv4 mutants, which leads to similar changes in the fluorescence kinetics that are expected from the lack of the alternative electron transfer pathway. Therefore, the possibility that the electrons are transferred directly from PQH2 in the lipid phase of the membrane to Flv2/Flv4 cannot be completely excluded at this stage. More precise characterization of this very interesting process, including the determination of exact redox potentials and pathways as well as their rates will require further investigation.
In conclusion, we suggest an electron transfer pathway from PSII in cyanobacteria grown under air level of CO2. It involves a strong induction of the flv4-flv2 operon under low Ci and high light conditions, whose protein products modify both the PBS–PSII interaction and the exit of electrons from PSII, which in turn decreases the excitation pressure on PSII in β-cyanobacteria. This is made possible by a subtle modification of the PSII dimer by the Sll0218 protein, which then enables the electron transfer from PSII to the Flv2/Flv4 heterodimer. Compared with the electron donation to molecular oxygen via Flv1- and Flv3-mediated Mehler-like reaction, the electron acceptor of the Flv2/Flv4 heterodimer is not oxygen. Indeed, the electron transfer pathway from PSII to Flv2/Flv4 is a completely novel electron sink providing flexibility to PSII electron transfer, while the final electron acceptor from Flv2/Flv4 remains to be identified. Existence of such an electron transfer route may open up novel possibilities for using cyanobacteria for biotechnological purposes.
METHODS
Synechocystis Strains and Cell Culture Conditions
The Synechocystis sp PCC 6803 glucose-tolerant strain (Williams, 1988) was used as the wild type. The various sll0217-19 inactivation mutants were generated by disruption of the operon by insertion of an antibiotic-resistant cassette into different positions (Figure 3A). Δsll0217-18 has a kanamycin-resistant cassette replacing the positions between 1384 and 2055 (Helman et al., 2003). The Δsll0218-19 (earlier called Δflv2) has a hygromycin-resistant cassette replacing 188 to 1467 of the sll0219 gene (Helman et al., 2003). We verified by PCR that the deletion region also partially covers the sll0218 gene. Therefore, we renamed it as Δsll0218-19. The Δflv4 mutant (inactivation of the entire flv4-flv2 operon) was generated by insertion of a kanamycin-resistant cassette into 129 to 1529 of the sll0217 gene. A new Δflv2 mutant was generated by insertion of a spectinomycin-resistant cassette into 403 to 1585 of the sll0219 gene. Segregation of the flv inactivation mutants was verified by PCR. The flag-flv4 and Δsll0218-19/flag-flv4 strains were generated by transforming the wild type and the Δsll0218-19 cells with self-replicating plasmid pVZ-flag-flv4. The plasmid was constructed by fusing the FLAG encoding sequence at the C terminus of the flv4 gene, including the original flv4 promoter region. The complementary strains Δsll0218-19/::flv2 and Δflv4/::flv2 were constructed by transformation of the flv2 gene under psbA2 promoter into Δsll0218-19 and Δflv4 background, respectively, and the construct was integrated into the chromosome by replacing the psbA2 gene. ΔpsbA2 was served as a control strain for these complementary strains.
The wild-type and mutant strains were grown in BG-11 Na2CO3-free medium buffered with 20 mM HEPES-NaOH, pH 7.5, except for M55 (Ogawa, 1991), which was grown in the same medium buffered with 10 mM TES-KOH, pH 8.3. The cells were illuminated under continuous photon flux density of 50 μmol photons m−2 s−1, at 30°C. The cultures were aerated by shaking at 120 rpm at air level of CO2 (0.038% CO2 in the air, low CO2). In specific cases, air enriched with 3% CO2 (high CO2) was applied. The mutant strains were grown in the presence of the proper antibiotics.
RNA Isolation and Real-Time Quantitative RT-PCR Analysis
Total RNA was isolated by Trizol method according to McGinn et al. (2003). After removal of genomic DNA, the first-strand cDNA was synthesized as described before (Zhang et al., 2007). The primers for analyzing the transcripts of the flv4 and flv2 genes, as well as the reference gene rnpB were the same as in (Zhang et al., 2009). The primers 5′-CTCTCAACCAATGTGGATTCG-3′ and 5′-CCAGACTGACGAATTTGATGG-3′ were used to analyze the sll0218 transcripts. The primers were designed for generating a similar length (∼400 bp) of amplicons. The real-time quantitative RT-PCR was performed on a Bio-Rad IQ5 system. The annealing temperature was optimized, and the efficiency of each reaction was calculated as described earlier (Sicora et al., 2006). Gene expression of each sample was normalized to the expression level of rnpB. The melting curve analysis was performed to ensure the specificity of the products.
Isolation of the Membrane and Soluble Cell Fractions
The membrane and soluble fractions of Synechocystis cells, if not further specified, were routinely isolated as described earlier (Zhang et al., 2009) with slight modifications. In brief, the cells were pelleted from batch cultures and broken with glass beads by vortexing at 4°C in buffer B (50 mM HEPES-NaOH, pH 7.5, 30 mM CaCl2, 800 mM sorbitol, and 1 mM ε-amino-n-caproic acid [ACA]). The cell debris and glass beads were removed by 5 min centrifugation at 3000g. The membrane and soluble fractions were separated by centrifugation at 110,000g for 30 min. The membrane pellet was resuspended in 50 mM HEPES-NaOH, pH 7.5, 600 mM Suc, 30 mM CaCl2, and 1 M glycinebetaine. To study the effects of cations in the isolation buffer, the cells were broken in buffer A (20 mM potassium phosphate, pH 7.8) or in buffer C (20 mM potassium phosphate, pH 7.8, and 25 mM MgCl2). To optimize the Mg2+ concentration, the buffer for disrupting the cells (50 mM HEPES-NaOH, pH 7.5, 800 mM sorbitol, and 1 mM ACA) was supplemented with different amounts of MgCl2 (0, 1, 5, and 25 mM), CaCl2 (0, 10, and 30 mM), or NaCl (0, 5, and 50 mM). For compatibility of the solution, buffer D (50 mM HEPES-NaOH, pH 7.5, 25 mM MgCl2, 800 mM sorbitol, and 1 mM ACA) was applied for isolation of the membrane fraction when subjected to two-phase partitioning.
Aqueous Two-Phase Partitioning
Two-phase partitioning was performed according to Norling et al. (1998). Additionally, a rough purification of the thylakoid and plasma membranes was performed with some modifications of the original protocol. Briefly, the total membranes were isolated from low-Ci-grown wild-type cells in buffer D and resuspended in the two-phase buffer (5 mM potassium phosphate, pH 7.4, and 0.25 M Suc) in the presence of 1 mM pefabloc. A 10-g two-phase system (5.8% [w/w] Dextran T-500, 5.8% [w/w] polyethylene glycol 3350, 5 mM potassium phosphate, pH 7.4, 0.25 M Suc, and 1 mM pefabloc) was constituted by adding 3.75 g of total membranes (1.5 mg chlorophyll) to a 6.25-g polymer mixture as described (Norling et al., 1998). A 30-g repartitioning system with the same concentrations but without the sample was prepared. After gently inverting the tube 35 times at 3°C, the phases were settled by centrifugation at 1000g for 4 min. The upper phase and the lower phase of the sample tube were collected separately. A fresh lower phase solution (5.8% Dextran) was added to the upper phase sample, and a fresh upper phase solution (5.8% polyethylene glycol) was added to the lower phase sample. After three cycles of repartitioning, the final upper and lower phases were both diluted with two-phase buffer to ∼30 mL and centrifuged at 125,000g for 45 min. The pellets were washed with 20 mL of the two-phase buffer, and the centrifugation was repeated. The membranes were resuspended in a small volume of 20 mM Tricine-NaOH, pH 7.5, 10 mM NaCl, 10 mM MgCl2, and 500 mM Suc.
Electrophoresis and Immunoblotting
Protein complexes in the membrane and soluble fractions were analyzed by BN-PAGE, which was performed as described by Herranen et al. (2004) with minor modifications. The membrane samples were washed with 50 mM BisTris, pH 7.0, 330 mM sorbitol, and 1 mM pefabloc, pelleted by centrifugation at 18,000g for 10 min, and resuspended in 25 mM BisTris, pH 7.0, 20% glycerol (w/v), 1 mM pefabloc, and 10 mM MgCl2 to a protein concentration 15 μg/μL. The protein samples were solubilized by gently adding an equal volume of 2.5% DM in the same buffer, followed by 20 min incubation on ice and a subsequent 10-min incubation at room temperature. To remove membrane-bound DNA from the samples, 0.02 units/μL RNase-free DNase was added during solubilization. Insoluble materials were removed by centrifugation at 18,000g for 20 min. The supernatant was mixed with one-tenth volume of sample buffer containing 5% (w/v) Serva blue G, 100 mM BisTris, pH 7.0, 30% (w/v) Suc, and 500 mM ACA and loaded to the BN gel. Solubilization was not needed for the samples of soluble fraction; instead, they were diluted in 4× BN buffer containing 100 mM BisTris, pH 7.0, 80% (w/v) glycerol, and 4 mM pefabloc and were ready to apply to BN-PAGE. BN-PAGE was performed according to Zhang et al. (2004). Gradient polyacrylamide gels from 5 to 12% or 6 to 13% were used in this study. SDS-PAGE was applied to analyze denatured samples. The protein samples were solubilized in Laemmli sample buffer (Laemmli, 1970) with 5% β-mercaptoethanol and 6 M urea at room temperature for 2 h and separated in 9 or 12.5% polyacrylamide gel containing 6 M urea. After electrophoresis, the proteins were electrotransferred to a polyvinylidene fluoride membrane and detected by protein-specific antibodies. The Sll0218 polyclonal antibody was raised against amino acids 145 to 158 and 159 to 172 of Synechocystis Sll0218 protein. The antibodies against Flv4, Flv2, NdhJ, D1, SbtA, and PHB3 were described earlier (Zhang et al., 2004, 2007, 2009; Boehm et al., 2009). The monoclonal anti-FLAG M2 peroxidase (horseradish peroxidase) was a commercial product from Sigma-Aldrich.
Membrane Digestion by Proteases
Membranes of the wild type were washed with 50 mM HEPES-NaOH, pH 7.5, 5 mM MgCl2, 10 mM NaCl, and 600 mM Suc and resuspended in the same buffer to final protein concentration of 2.5 µg/µL. Membranes (in final volume of 40 µL) were digested on ice for 30 min in the presence of either 0.4 μL trypsin solution (in 1 mM HCl) or 2 μL thermolysin (in 50 mM HEPES-NaOH, pH 7.5, 100 mM sorbitol, and 10 mM CaCl2). The final protease concentrations were 0.06, 2.5, and 10 µg/mL for trypsin and 0.02, 0.05, and 0.2 mg/mL for thermolysin. The treatments were terminated by adding an equal volume of Laemmli sample buffer and immediately heated at 65°C for 5 min.
Fluorescence Spectrometry
Low-temperature fluorescence emission spectra at 77K of the whole cells were measured by a USB4000-FL-450 spectrofluorometer. The cells were harvested at OD750 = 0.6 to 0.8 and resuspended in fresh BG-11 (Na2CO3-free) medium at a chlorophyll concentration of 5 μg/mL. The resuspended cultures were acclimated under the same growth conditions for 1 h. The cells were then frozen in liquid nitrogen directly from the growth light. The cells were exited by 580 or 440 nm light obtained via 10-nm-wide filters. The fluorescence spectra were analyzed with Gaussian deconvolution, and the areas of all fluorescence sub-bands were calculated.
Flash-induced fluorescence increase and the subsequent decay of chlorophyll fluorescence yield were measured using a fluorometer FL 3500 (PSI Instruments) according to Vass et al. (1999). The cells were resuspended in fresh BG-11 medium with chlorophyll concentration 5 µg/mL and excited with a single, short saturation flash. A fitting function with three components [F(t) − F0 = A1 × exp(−t/T1) + A2 × exp(−t/T2) + A3/(1 + t/T3)] was used for deconvolution of the measured curves according to Cser et al. (2008). F(t) is the variable fluorescence yield at time t, F0 is the basic fluorescence level before the flash, A1 to A3 are the amplitudes, and T1 to T3 are the time constants. The Joliot model (Joliot and Joliot, 1964) was used to correct the nonlinear correlation between fluorescence yield and the redox state of QA with a value of 0.5 for the energy transfer parameter between the PSII units.
Oxygen Evolution Measurements
Steady state oxygen evolution was measured with a Clark-type oxygen electrode (DW1; Hansatech) at 30°C under saturating light. Before measurements, the cells were collected and resuspended in fresh growth medium at a chlorophyll concentration of 4 µg/mL. The measurements were performed by exposure of 1-mL cell suspension to 1500 μmol photons m−2 s−1 white light. The whole-chain photosynthesis electron transfer rates (water to CO2) were measured in the presence of 10 mM NaHCO3. The PSII electron transfer rates (water to quinone) were measured in the presence of an artificial electron acceptor, either 0.5 mM DCBQ or 2 mM DMBQ. These measurements were performed also in the presence of 10 mM NaHCO3.
Homology Modeling of Flv2/Flv4 Heterodimer
The amino acid sequences of Synechocystis Flv2 (Sll0219) and Flv4 (Sll0217) were retrieved from Cyanobase (http://genome.kazusa.or.jp/cyanobase). All alignments were constructed with MALIGN (Johnson and Overington, 1993) in the Bodil visualization and modeling package (Lehtonen et al., 2004). Visualization of the alignments was done by ESPript 2.2 implemented in ENDscript 1.1 (Gouet et al., 1999). The homology models were done with MODELER (Sali and Blundell, 1993). Ten models were created for each constructed homology model, and the one with the best objective function was chosen for further analysis. The models were evaluated by Procheck (Laskowski et al., 1993). Visualization of the models was done by Pymol (http://www.pymol.org; deLano, 2002). The crystal structures for the multiple structure-based alignment were retrieved by BLAST searches from the PDB (http://www.rcsb.org/) using Synechocystis Flv2 and Flv4 as query sequences. The cyanobacterial sequences were retrieved by BLAST from the UniProt Knowledgebase (http://www.ebi.ac.uk/uniprot/). The three-dimensional models for the open and closed conformation of the β-lactamase–like and flavodoxin domains of Synechocystis Flv2/Flv4 heterodimer and homodimers were constructed based on the alignment and the crystal structure of F420H2 oxidase from the methanogenic archaea Methanothermobacter marburgensis (closed conformation, PDB ID: 2OHI; open conformation PDB ID: 2OHJ) (Seedorf et al., 2007). A homology model of the Flv2/Flv4 heterodimer was also constructed based on Moorella thermoacetica FprA (PDB ID: 1YCF), which is in the closed conformation.
To model the C-terminal flavin reductase domains of Flv2 and Flv4 BLAST searches against PDB were performed, and a multiple structure-based alignment of the retrieved sequences was created. A three-dimensional model of the flavin reductase domains of Synechocystis Flv2/Flv4 was constructed based on the alignment and the crystal structure of Archaeoglobus fulgidus ferric reductase (PDB ID: 1I0S) (Chiu et al., 2001).
Phylogenetic Analysis
A rooted phylogenic tree was constructed from alignment of selected amino acid sequences of Sll0218 homologs using ClustalX2.1 (Larkin et al., 2007) and TreeView1.6.6.
Accession Numbers
Sequence data from this article can be found in The International Nucleotide Sequence Database Collaboration under the following accession numbers: Flv4, NP_440050.1; Sll0218, NP_440049.1; Flv2, NP_440048.1; and PsbA2, NP_439906.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenic Analysis of Sll0218 in Cyanobacteria, Bacteria, and Archaea.
Supplemental Figure 2. Solubility of the Flv4, Sll0218, and Flv2 Proteins by Nonionic and Zwitterionic Detergents.
Supplemental Figure 3. BN/SDS-PAGE Demonstrating the Heterodimer Formation by Flv2 and Flv4.
Supplemental Figure 4. Expression of the FLAG-Flv4, Sll0218, and Flv2 Proteins in the Wild Type, flv Mutants, and the flag-flv4 Strains.
Supplemental Figure 5. An Example of the Gaussian Sub-band Deconvolution of 77K Fluorescence Emission Spectra Excited at 580 nm in Synechocystis Wild-Type Cells.
Supplemental Figure 6. Multiple Structure-Based Alignment of Methanothermobacter marburgensis F420H2 Oxidase (PDB ID: 2OHI), Synechocystis Flv2, Synechocystis Flv4, Moorella thermoacetica FprA (PDB ID: 1YCF), Desulfovibrio gigas Rubredoxin Oxygen:Oxidoreductase (PDB ID: 1E5D), Giardia intestinalis Flavoprotein (PDB ID: 2Q9U), and Thermotoga maritima Flavoprotein (PDB ID: 1VME).
Supplemental Figure 7. The FMN and Diiron Binding Sites of the Flv2 and Flv4 Homodimers.
Supplemental Table 1. Ratios of Low Temperature (77K) Fluorescence Yields of PBS, PSI, and PSII (F685 + F695) of the Wild Type and Various flv Mutants Excited at 580-nm Light.
Supplemental Table 2. Conserved Metal Binding Sites on the Flv2/Flv4 Heterodimer Surface.
Supplemental Table 3. Kinetic Data of Flash-Induced Fluorescence Relaxation Components of ΔpsbA2 and Two flv2 Complemented Strains Grown at High and Air Level of CO2.
Supplementary Material
Acknowledgments
We thank Wolfgang R. Hess from the University of Freiburg for kindly providing the pVZ-flag-flv4 construct. The PsbO antibody was a kind gift from Roberto Barbato (Università del Piemonte Orientale). We also thank Mark Johnson for excellent computing facilities and for the availability of the Bio-center Finland infrastructure (bioinformatics and translational activities) at Åbo Akademi University. This work was supported by the Academy of Finland (Projects 118637 and 133299), by European Union FP7/Energy Network Projects SOLAR-H2 (contract 212508) and DirectFuel (contract 256808), by the Sigrid Juselius Foundation, Tor, Joe, and Pentti Borg Foundation, and by the National Doctoral Program in Informational and Structural Biology.
AUTHOR CONTRIBUTIONS
P.Z. performed most experiments, contributed greatly to the design of experiments, wrote the first version of the article, and made all revisions as advised by others. M.E. contributed to experiments of Sll0218 and flag-flv4, designed experiments, and revised the article. A.-M.B. made structural modeling of Flv2/Flv4 and wrote the corresponding part of the article. D.C. constructed Δflv2 and Δflv4 mutants and contributed to revising the article. H.M.S. constructed ΔpsbA2, Δflv4/::flv2, and Δsll0218-19/::flv2 strains and analyzed the accumulation of Flv2 and Flv4 proteins in these mutants. I.V. analyzed flash-induced fluorescence curves and constructed the model of electron transfer from PSII to Flv2/Flv4. Y.A. supervised the 77K fluorescence measurements and contributed to revising the article. T.A.S. supervised structural modeling and contributed to revising the article. E.-M.A. was mainly responsible for experimental design and for writing of the final article.
Glossary
- FDP
flavodiiron protein
- PSII
photosystem II
- PSI
photosystem I
- Ci
inorganic carbon
- PBS
phycobilisome
- DM
n-dodecyl-β-d-maltoside
- BN
blue native
- 2D
two-dimensional
- DMBQ
2,6-dimethyl-p-benzoquinone
- DCBQ
2,6-dichloro-p-benzoquinone
- DBMIB
2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone
- ACA
ε-amino-n-caproic acid
- PDB
Protein Data Bank
- PQ
plastoquinone
- PQH2
plastoquinol
- FMN
flavin mononucleotide
References
- Adachi H., Umena Y., Enami I., Henmi T., Kamiya N., Shen J.-R. (2009). Towards structural elucidation of eukaryotic photosystem II: Purification, crystallization and preliminary X-ray diffraction analysis of photosystem II from a red alga. Biochim. Biophys. Acta 1787: 121–128 [DOI] [PubMed] [Google Scholar]
- Allahverdiyeva Y., Ermakova M., Eisenhut M., Zhang P., Richaud P., Hagemann M., Cournac L., Aro E.M. (2011). Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J. Biol. Chem. 286: 24007–24014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098: 275–335 [DOI] [PubMed] [Google Scholar]
- Aro E.-M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., Rintamäki E. (2005). Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56: 347–356 [DOI] [PubMed] [Google Scholar]
- Badger M.R., Price G.D., Long B.M., Woodger F.J. (2006). The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J. Exp. Bot. 57: 249–265 [DOI] [PubMed] [Google Scholar]
- Bailey S., Grossman A. (2008). Photoprotection in cyanobacteria: Regulation of light harvesting. Photochem. Photobiol. 84: 1410–1420 [DOI] [PubMed] [Google Scholar]
- Barber J. (1982). Influence of surface charges on thylakoid structure and function. Annu. Rev. Plant Physiol. 33: 261–295 [Google Scholar]
- Barber J., Morris E.P., da Fonseca P.C.A. (2003). Interaction of the allophycocyanin core complex with photosystem II. Photochem. Photobiol. Sci. 2: 536–541 [DOI] [PubMed] [Google Scholar]
- Battchikova N., Vainonen J.P., Vorontsova N., Keränen M., Carmel D., Aro E.-M. (2010). Dynamic changes in the proteome of Synechocystis 6803 in response to CO(2) limitation revealed by quantitative proteomics. J. Proteome Res. 9: 5896–5912 [DOI] [PubMed] [Google Scholar]
- Bentley F.K., Luo H., Dilbeck P., Burnap R.L., Eaton-Rye J.J. (2008). Effects of inactivating psbM and psbT on photodamage and assembly of photosystem II in Synechocystis sp. PCC 6803. Biochemistry 47: 11637–11646 [DOI] [PubMed] [Google Scholar]
- Bibby T.S., Nield J., Barber J. (2001). Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412: 743–745 [DOI] [PubMed] [Google Scholar]
- Boehm M., Nield J., Zhang P., Aro E.-M., Komenda J., Nixon P.J. (2009). Structural and mutational analysis of band 7 proteins in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 191: 6425–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema E.J., Hifney A., Yakushevska A.E., Piotrowski M., Keegstra W., Berry S., Michel K.P., Pistorius E.K., Kruip J. (2001). A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412: 745–748 [DOI] [PubMed] [Google Scholar]
- Carmel D., Mulo P., Battchikova N., Aro E.-M. (2011). Membrane attachment of Slr0006 in Synechocystis sp. PCC 6803 is determined by divalent ions. Photosynth. Res. 108: 241–245 [DOI] [PubMed] [Google Scholar]
- Chiu H.J., Johnson E., Schröder I., Rees D.C. (2001). Crystal structures of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus and its complex with NADP+. Structure 9: 311–319 [DOI] [PubMed] [Google Scholar]
- Cser K., Deák Z., Telfer A., Barber J., Vass I. (2008). Energetics of photosystem II charge recombination in Acaryochloris marina studied by thermoluminescence and flash-induced chlorophyll fluorescence measurements. Photosynth. Res. 98: 131–140 [DOI] [PubMed] [Google Scholar]
- Dandekar T., Snel B., Huynen M., Bork P. (1998). Conservation of gene order: A fingerprint of proteins that physically interact. Trends Biochem. Sci. 23: 324–328 [DOI] [PubMed] [Google Scholar]
- deLano W.L. (2002). The PyMOL Molecular Graphics System. (San Carlos, CA: DeLano Scientific)
- Di Matteo A., Scandurra F.M., Testa F., Forte E., Sarti P., Brunori M., Giuffrè A. (2008). The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis. J. Biol. Chem. 283: 4061–4068 [DOI] [PubMed] [Google Scholar]
- Eisenhut M., von Wobeser E.A., Jonas L., Schubert H., Ibelings B.W., Bauwe H., Matthijs H.C.P., Hagemann M. (2007). Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 144: 1946–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bissati K., Delphin E., Murata N., Etienne A.-L., Kirilovsky D. (2000). Photosystem II fluorescence quenching in the cyanobacterium Synechocystis PCC 6803: Involvement of two different mechanisms. Biochim. Biophys. Acta 1457: 229–242 [DOI] [PubMed] [Google Scholar]
- Frazão C., et al. (2000). Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat. Struct. Biol. 7: 1041–1045 [DOI] [PubMed] [Google Scholar]
- Funk C., Schröder W.P., Salih G., Wiklund R., Jansson C. (1998). Engineering of N-terminal threonines in the D1 protein impairs photosystem II energy transfer in Synechocystis 6803. FEBS Lett. 436: 434–438 [DOI] [PubMed] [Google Scholar]
- Giordano M., Beardall J., Raven J.A. (2005). CO2 concentrating mechanisms in algae: Mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 56: 99–131 [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D.I., Métoz F. (1999). ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Graan T., Ort D.R. (1986). Detection of oxygen-evolving photosystem II centers inactive in plastoquinone reduction. Biochim. Biophys. Acta 852: 320–330 [Google Scholar]
- Hankamer B., Morris E.P., Barber J. (1999). Revealing the structure of the oxygen-evolving core dimer of photosystem II by cryoelectron crystallography. Nat. Struct. Biol. 6: 560–564 [DOI] [PubMed] [Google Scholar]
- Hankamer B., Nield J., Zheleva D., Boekema E., Jansson S., Barber J. (1997). Isolation and biochemical characterisation of monomeric and dimeric photosystem II complexes from spinach and their relevance to the organisation of photosystem II in vivo. Eur. J. Biochem. 243: 422–429 [DOI] [PubMed] [Google Scholar]
- Helman Y., Tchernov D., Reinhold L., Shibata M., Ogawa T., Schwarz R., Ohad I., Kaplan A. (2003). Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr. Biol. 13: 230–235 [DOI] [PubMed] [Google Scholar]
- Herranen M., Battchikova N., Zhang P., Graf A., Sirpiö S., Paakkarinen V., Aro E.-M. (2004). Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol. 134: 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y., Kamei A., Kanehisa M., Kaplan A., Ikeuchi M. (2001). DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Ruban A.V., Walters R.G. (1996). Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 655–684 [DOI] [PubMed] [Google Scholar]
- Huner N.P.A. (1998). Energy balance and acclimation to light and cold. Trends Plant Sci. 3: 224–230 [Google Scholar]
- Iwai M., Katoh H., Katayama M., Ikeuchi M. (2004). PSII-Tc protein plays an important role in dimerization of photosystem II. Plant Cell Physiol. 45: 1809–1816 [DOI] [PubMed] [Google Scholar]
- Johnson M.S., Overington J.P. (1993). A structural basis for sequence comparisons. An evaluation of scoring methodologies. J. Mol. Biol. 233: 716–738 [DOI] [PubMed] [Google Scholar]
- Joliot A., Joliot P. (1964). Etude cinétique de la réaction photochimique libérant l’oxygène au cours de la photosynthèse. C. R. Acad. Sci. 258: 4622–4625 [PubMed] [Google Scholar]
- Kanervo E., Suorsa M., Aro E.-M. (2005). Functional flexibility and acclimation of the thylakoid membrane. Photochem. Photobiol. Sci. 4: 1072–1080 [DOI] [PubMed] [Google Scholar]
- Kaplan A., Reinhold L. (1999). CO2-concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 539–570 [DOI] [PubMed] [Google Scholar]
- Karapetyan N.V. (2008). Protective dissipation of excess absorbed energy by photosynthetic apparatus of cyanobacteria: Role of antenna terminal emitters. Photosynth. Res. 97: 195–204 [DOI] [PubMed] [Google Scholar]
- Kirilovsky D. (2007). Photoprotection in cyanobacteria: The orange carotenoid protein (OCP)-related non-photochemical-quenching mechanism. Photosynth. Res. 93: 7–16 [DOI] [PubMed] [Google Scholar]
- Kuhl H., Kruip J., Seidler A., Krieger-Liszkay A., Bünker M., Bald D., Scheidig A.J., Rögner M. (2000). Towards structural determination of the water-splitting enzyme. Purification, crystallization, and preliminary crystallographic studies of photosystem II from a thermophilic cyanobacterium. J. Biol. Chem. 275: 20652–20659 [DOI] [PubMed] [Google Scholar]
- Körner L.E., Kjellbom P., Larsson C., Møller I.M. (1985). Surface properties of right side-out plasma membrane vesicles isolated from barley roots and leaves. Plant Physiol. 79: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Larom S., Salama F., Schuster G., Adir N. (2010). Engineering of an alternative electron transfer path in photosystem II. Proc. Natl. Acad. Sci. USA 107: 9650–9655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–291 [Google Scholar]
- Lehtonen J.V., et al. (2004). BODIL: A molecular modeling environment for structure-function analysis and drug design. J. Comput. Aided Mol. Des. 18: 401–419 [DOI] [PubMed] [Google Scholar]
- MacColl R. (1998). Cyanobacterial phycobilisomes. J. Struct. Biol. 124: 311–334 [DOI] [PubMed] [Google Scholar]
- McGinn P.J., Price G.D., Maleszka R., Badger M.R. (2003). Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp. strain PCC6803. Plant Physiol. 132: 218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux C.W. (2008). Phycobilisome-reaction centre interaction in cyanobacteria. Photosynth. Res. 95: 175–182 [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W., Emlyn-Jones D. (2005). State transitions: An example of acclimation to low-light stress. J. Exp. Bot. 56: 389–393 [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W., Holzwarth A.R. (1991). Kinetics of excitation energy transfer in the cyanobacterial phycobilisome-photosystem II complex. Biochim. Biophys. Acta 1098: 68–78 [Google Scholar]
- Müller P., Li X.-P., Niyogi K.K. (2001). Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. (Lond.) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi K.K. (1999). Photoprotection revisited: Genetics and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 333–359 [DOI] [PubMed] [Google Scholar]
- Norling B., Zak E., Andersson B., Pakrasi H. (1998). 2D-isolation of pure plasma and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 436: 189–192 [DOI] [PubMed] [Google Scholar]
- Ogawa T. (1991). A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 88: 4275–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi N., Takahashi Y. (2001). PsbT polypeptide is required for efficient repair of photodamaged photosystem II reaction center. J. Biol. Chem. 276: 33798–33804 [DOI] [PubMed] [Google Scholar]
- Pisareva T., Kwon J., Oh J., Kim S., Ge C., Wieslander Å., Choi J.S., Norling B. (2011). Model for membrane organization and protein sorting in the cyanobacterium Synechocystis sp. PCC 6803 inferred from proteomics and multivariate sequence analyses. J. Proteome Res. 10: 3617–3631 [DOI] [PubMed] [Google Scholar]
- Price G.D., Badger M.R., Woodger F.J., Long B.M. (2008). Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): Functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59: 1441–1461 [DOI] [PubMed] [Google Scholar]
- Rengstl B., Oster U., Stengel A., Nickelsen J. (2011). An intermediate membrane subfraction in cyanobacteria is involved in an assembly network for photosystem II biogenesis. J. Biol. Chem. 286: 21944–21951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rögner M., Dekker J.P., Boekema E.J., Witt H.T. (1987). Size, shape and mass of the oxygen-evolving photosystem II complex from the thermophilic cyanobacterium Synechococcus sp. FEBS Lett. 219: 207–211 [Google Scholar]
- Sali A., Blundell T.L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Sandström S., Park Y.I., Öquist G., Gustafsson P. (2001). CP43′, the isiA gene product, functions as an excitation energy dissipator in the cyanobacterium Synechococcus sp. PCC 7942. Photochem. Photobiol. 74: 431–437 [DOI] [PubMed] [Google Scholar]
- Saraiva L.M., Vicente J.B., Teixeira M. (2004). The role of the flavodiiron proteins in microbial nitric oxide detoxification. Adv. Microb. Physiol. 49: 77–129 [DOI] [PubMed] [Google Scholar]
- Seedorf H., Hagemeier C.H., Shima S., Thauer R.K., Warkentin E., Ermler U. (2007). Structure of coenzyme F420H2 oxidase (FprA), a di-iron flavoprotein from methanogenic Archaea catalyzing the reduction of O2 to H2O. FEBS J. 274: 1588–1599 [DOI] [PubMed] [Google Scholar]
- Shi T., Bibby T.S., Jiang L., Irwin A.J., Falkowski P.G. (2005). Protein interactions limit the rate of evolution of photosynthetic genes in cyanobacteria. Mol. Biol. Evol. 22: 2179–2189 [DOI] [PubMed] [Google Scholar]
- Sicora C.I., Appleton S.E., Brown C.M., Chung J., Chandler J., Cockshutt A.M., Vass I., Campbell D.A. (2006). Cyanobacterial psbA families in Anabaena and Synechocystis encode trace, constitutive and UVB-induced D1 isoforms. Biochim. Biophys. Acta 1757: 47–56 [DOI] [PubMed] [Google Scholar]
- Silaghi-Dumitrescu R., Kurtz D.M., Jr, Ljungdahl L.G., Lanzilotta W.N. (2005). X-ray crystal structures of Moorella thermoacetica FprA. Novel diiron site structure and mechanistic insights into a scavenging nitric oxide reductase. Biochemistry 44: 6492–6501 [DOI] [PubMed] [Google Scholar]
- Takahashi T., Inoue-Kashino N., Ozawa S., Takahashi Y., Kashino Y., Satoh K. (2009). Photosystem II complex in vivo is a monomer. J. Biol. Chem. 284: 15598–15606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M., Grieco M., Aro E.-M. (2011). Novel insights into plant light-harvesting complex II phosphorylation and ‘state transitions’. Trends Plant Sci. 16: 126–131 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi E., Aro E.-M. (1996). The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc. Natl. Acad. Sci. USA 93: 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Thor J.J., Mullineaux C.W., Matthijs H.C.P., Hellingwerf K.J. (1998). Light-harvesting and state transitions in cyanobacteria. Bot. Acta 111: 430–443 [Google Scholar]
- Vass I., Kirilovsky D., Etienne A.-L. (1999). UV-B radiation-induced donor- and acceptor-side modifications of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 38: 12786–12794 [DOI] [PubMed] [Google Scholar]
- Veerman J., Bentley F.K., Eaton-Rye J.J., Mullineaux C.W., Vasil’ev S., Bruce D. (2005). The PsbU subunit of photosystem II stabilizes energy transfer and primary photochemistry in the phycobilisome-photosystem II assembly of Synechocystis sp. PCC 6803. Biochemistry 44: 16939–16948 [DOI] [PubMed] [Google Scholar]
- Vernotte C., Picaud M., Kirilovsky D., Olive J., Ajlani G., Astier C. (1992). Changes in the photosynthetic apparatus in the cyanobacterium Synechocystis sp. PCC 6714 following light-to-dark and dark-to-light transitions. Photosynth. Res. 32: 45–57 [DOI] [PubMed] [Google Scholar]
- Vicente J.B., Carrondo M.A., Teixeira M., Frazão C. (2008a). Structural studies on flavodiiron proteins. Methods Enzymol. 437: 3–19 [DOI] [PubMed] [Google Scholar]
- Vicente J.B., Gomes C.M., Wasserfallen A., Teixeira M. (2002). Module fusion in an A-type flavoprotein from the cyanobacterium Synechocystis condenses a multiple-component pathway in a single polypeptide chain. Biochem. Biophys. Res. Commun. 294: 82–87 [DOI] [PubMed] [Google Scholar]
- Vicente J.B., Justino M.C., Gonçalves V.L., Saraiva L.M., Teixeira M. (2008b). Biochemical, spectroscopic, and thermodynamic properties of flavodiiron proteins. Methods Enzymol. 437: 21–45 [DOI] [PubMed] [Google Scholar]
- Vicente J.B., Testa F., Mastronicola D., Forte E., Sarti P., Teixeira M., Giuffrè A. (2009). Redox properties of the oxygen-detoxifying flavodiiron protein from the human parasite Giardia intestinalis. Arch. Biochem. Biophys. 488: 9–13 [DOI] [PubMed] [Google Scholar]
- Wang H.-L., Postier B.L., Burnap R.L. (2004). Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 279: 5739–5751 [DOI] [PubMed] [Google Scholar]
- Wasserfallen A., Ragettli S., Jouanneau Y., Leisinger T. (1998). A family of flavoproteins in the domains Archaea and Bacteria. Eur. J. Biochem. 254: 325–332 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Iwai M., Narikawa R., Ikeuchi M. (2009). Is the photosystem II complex a monomer or a dimer? Plant Cell Physiol. 50: 1674–1680 [DOI] [PubMed] [Google Scholar]
- Williams J.K.G. (1988). Construction of specific mutations in PSII photosynthetic reaction center by genetic engineering. Methods Enzymol. 167: 766–778 [Google Scholar]
- Wollman F.-A. (2001). State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J. 20: 3623–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Allahverdiyeva Y., Eisenhut M., Aro E.-M. (2009). Flavodiiron proteins in oxygenic photosynthetic organisms: photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. PLoS ONE 4: e5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Battchikova N., Jansen T., Appel J., Ogawa T., Aro E.-M. (2004). Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16: 3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Sicora C.I., Vorontsova N., Allahverdiyeva Y., Battchikova N., Nixon P.J., Aro E.-M. (2007). FtsH protease is required for induction of inorganic carbon acquisition complexes in Synechocystis sp. PCC 6803. Mol. Microbiol. 65: 728–740 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.