This article provides information about the interaction between the phycobilisomes and the Orange Carotenoid Protein needed for photoprotection. Its red light–activated form has an open structure that allows the interaction of its N-terminal domain, containing the Arg155, with the phycobilisome, permitting a closer interaction between the carotenoid and the phycobilisome chromophores.
Abstract
Most cyanobacteria, under high light conditions, decrease the amount of energy arriving at the reaction centers by increasing thermal energy dissipation at the level of the phycobilisome, the extramembranous antenna. This mechanism is induced by photoactivation of the Orange Carotenoid Protein (OCP). To identify how the activated OCP interacts with phycobilisomes (PBs), several OCP mutants were constructed, and the influence of mutations on photoactivity, stability, and binding to PBs was characterized. The disruption of the salt bridge between Arg155 and Glu244, which stabilizes the interaction between the N- and C-terminal domains, increased the rate of photoactivity and the stability of the photoactivated OCP, suggesting that the activated OCP has an open structure with decreased interdomain interaction. Changing Glu244 to leucine had no effect on OCP binding to PBs. By contrast, substitution of Arg155 with a neutral or a negatively charged amino acid largely decreased OCP binding to the PBs, whereas substitution with a lysine slightly perturbed the interaction. These results strongly suggest that the surface of the N-terminal domain, containing the Arg155, interacts with the PB and that the positive charge of Arg155 plays a key role in photoprotection.
INTRODUCTION
Excess light is harmful to photosynthetic organisms, because the surplus energy arriving at the reaction centers induces the formation of dangerous oxygen species that first damage the photosynthetic apparatus and then the membranes, DNA, and translational and transcriptional machineries, culminating in cell death. A safety valve must be opened at the level of the antenna under high irradiance to decrease the energy arriving at the reaction centers. In cyanobacteria, light is mainly harvested by an extramembrane complex, formed by phycobiliproteins covalently binding red and blue phycobilins, the phycobilisomes (PBs) (reviewed in Glazer, 1984; Grossman et al., 1993; MacColl, 1998; Adir, 2005). Intense illumination activates a carotenoid binding protein, the soluble Orange Carotenoid Protein (OCP) (Wilson et al., 2008), which was first described by Holt and Krogmann (1981). The activated protein interacts with PBs (Wilson et al., 2006; Wilson et al., 2008; Gwizdala et al., 2011) to increase the thermal dissipation of excess energy; this results in a decrease in energy arriving at both reaction centers (Scott et al., 2006; Wilson et al., 2006; Rakhimberdieva et al., 2010; Gorbunov et al., 2011). This process is accompanied by a decrease of the PB fluorescence and is called the OCP-dependent nonphotochemical quenching mechanism (reviewed in Kirilovsky, 2007; Kirilovsky and Kerfeld, 2012). Another protein, the Fluorescence Recovery Protein (FRP), is needed to recover the full antenna capacity when the light intensity decreases (Boulay et al., 2010). FRP interacts with the activated OCP and accelerates its deactivation and its detachment from the PB (Boulay et al., 2010; Gwizdala et al., 2011).
OCP consists of an all α-helical N-terminal domain, not known to be present outside of the phylum Cyanobacteria, and an α/β C-terminal domain, which is a member of the nuclear transport factor2 superfamily (Kerfeld et al., 2003; Wilson et al., 2010) (Figure 1). Both domains are joined by a linker (residues 163 to 193) that seems to be flexible, potentially allowing changes in the interaction between the domains. The ketocarotenoid 3′-hydroxyechinenone (hECN) spans the N- and C-terminal domains of the protein, with its carbonyl end embedded in and hydrogen bonded to the C-terminal domain. The carotenoid is almost entirely buried; only 4% of the hECN is solvent-exposed.
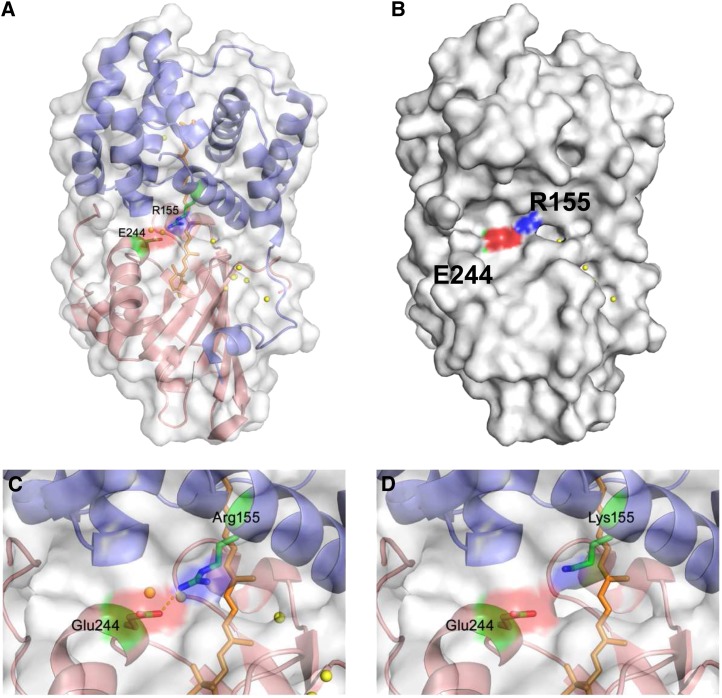
Figure 1.
The OCP Structure and Position of the Mutated Amino Acids Arg155 and Glu244.
(A) N-terminal domain is in blue, and the C-terminal domain is in red. The carotenoid is shown as orange sticks, and Arg155 (N-terminal domain) and Glu244 (C-terminal domain) are shown as sticks colored green for sidechains, red for oxygen atoms, and blue for nitrogen atoms.
(B) Surface view with similar orientation and coloring as in (A).
(C) Close-up of the central interdomain interface and the salt bridge between Arg155 and Glu244. Also shown are structurally conserved water molecules (yellow spheres).
(D) Close-up of the central interdomain interface with Lys substituted for Arg155. The Lys rotamer with the closest approach to Glu 244 was used in the model.
OCP is a photoactive protein (Wilson et al., 2008). Illumination of OCP by strong blue-green light induces changes in the carotenoid, converting the inactive orange dark form (OCPo) into an active red form (OCPr) (Wilson et al., 2008). In OCPo, hECN is in an all-trans configuration (Kerfeld et al., 2003; Polívka et al., 2005). In OCPr, it is also in an all-trans configuration, but the apparent conjugation length of the carotenoid increases, resulting in a less distorted, more planar structure (Wilson et al., 2008). Fourier transform infrared (FTIR) spectra showed that these changes in the carotenoid induce conformational changes in the protein, leading to a less rigid helical structure and a compaction of the β-sheet (Wilson et al., 2008). The changes in OCP are essential for the induction of the photoprotective mechanism. Only OCPr is able to bind to the PBs and induce fluorescence quenching and the photoprotective mechanism (Wilson et al., 2008; Punginelli et al., 2009; Gorbunov et al., 2011; Gwizdala et al., 2011). Because the photoactivation of OCP has a very low quantum yield (0.03 [Wilson et al., 2008]), the concentration of activated protein is zero or very low in darkness or under low light conditions (Wilson et al., 2008; Gorbunov et al., 2011). Thus, the photoprotective mechanism functions only under high light conditions. Although the rate of photoactivation of OCP depends on light intensity, the binding of OCPr to PBs is light-independent (Gwizdala et al., 2011). This was also reported to occur in vivo in Synechocystis sp strain PCC 6803 (Gorbunov et al., 2011; Rakhimberdieva et al., 2011). The amplitude of fluorescence quenching depends on the concentration of OCPr and on the strength of binding. In vitro (Gwizdala et al., 2011) and in vivo (Wilson et al., 2006), the same concentration of OCP induces a smaller fluorescence quenching when it interacts with the PB core than when it interacts with the whole PB. The presence of rods stabilizes the binding of OCPr in the whole PB (Gwizdala et al., 2011).
However, the conversion to OCPr is not sufficient to induce the photoprotective mechanism. The R155L mutant OCP is photoactive. Its light-absorption spectrum is similar to that of red wild-type OCP, indicating that similar changes are induced in the mutated and wild-type OCPs. However, strong blue light was unable to induce the photoprotective mechanism in mutant cells (Wilson et al., 2010). Arg155 is in the interface between the N- and C-terminal domains (Figure 1) and forms a salt bridge with Glu244 that stabilizes the interaction between the two domains. The structure of the R155L mutant was determined at 1.7 Å, but a glycerol molecule occupied the space vacated by the Arg sidechain and formed a hydrogen bond with Glu244 (Wilson et al., 2010). This resulted in a similar interaction between the two domains as observed in the structure in the wild-type OCP. Despite not altering the structure, replacing Arg155 with Leu has two important functional consequences: the salt bridge between Arg155 and Glu244 is disrupted, and the positive charge is lost. To determine which of these factors renders R155L-OCP unable to induce the photoprotective mechanism in the cells, we constructed three additional mutants: R155E, R155K, and E244L. The mutated OCPs were isolated, and the influence of the mutations on carotenoid binding, protein stability, photoactivity, red form stability, and PB binding was studied using the recently developed in vitro reconstitution system. Characterization of these mutants indicates that the N-terminal domain of OCP, specifically Arg155, interacts with the PBs to mediate photoprotection.
RESULTS
Effect of Mutations on Carotenoid Binding and the Light-Induced Carotenoid Conformational Changes
Synechocystis mutants overexpressing His-tagged R155K-OCP, R155L-OCP, R155E-OCP, and E244L-OCP were constructed using a ΔCrtR Synechocystis mutant as a recipient strain. In this mutant, the crtr gene coding for the β-carotene hydroxylase (Masamoto et al., 1998) was interrupted with an antibiotic cassette, resulting in a lack of zeaxanthin and hECN (Wilson et al., 2010). Thus, the mutant OCP isolated from these strains binds only echinenone. We have already demonstrated that the kinetics of photoactivation and dark recovery of echinenone-OCP is similar to that of hECN-OCP (Wilson et al., 2010). Mutant OCPs isolated from strains overexpressing OCP and containing zeaxanthin bind between 60 to 85% of zeaxanthin (Wilson et al., 2010). Because zeaxanthin-OCP is photoinactive (Punginelli et al., 2009), it is difficult to differentiate the effect of point mutations from that of zeaxanthin.
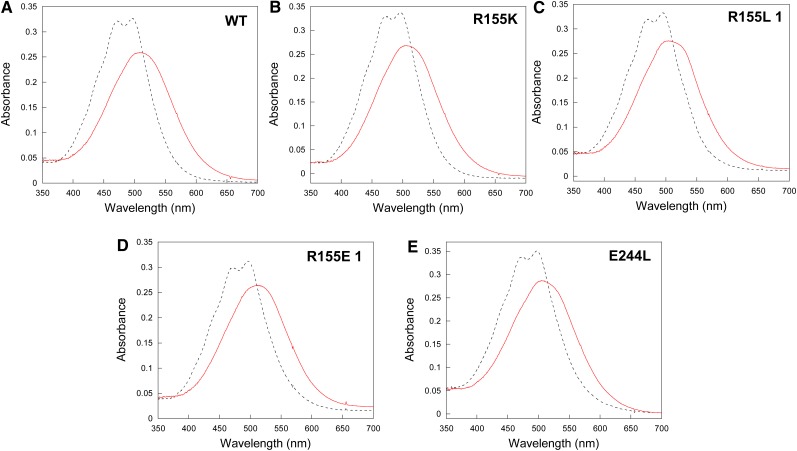
The mutant OCPs were isolated by nickel affinity and ion exchange chromatography (Wilson et al., 2008). The spectra of the OCPo and OCPr forms of R155K and E244L mutants are similar (but not identical) to those of wild-type OCP (Figure 2). Two different fractions of OCP were obtained from the mutants overexpressing R155L-OCP and R155E-OCP. The OCPs in the major fractions have absorption spectra similar to that of wild-type OCP (Figures 2C and 2D). By contrast, OCPo and OCPr spectra of the minor fractions are different (see Supplemental Figures 1A and 1B online). OCPo spectra contain a shoulder at 550 nm, representing an additional vibrational band, absent in wild-type OCPo spectrum. Wild-type OCPr loses the resolution of the vibrational bands and has an absorption maximum at 510 nm. Instead, the R155L-OCPr and R155E-OCPr minor forms have three vibrational bands at 467, 496, and 550 nm. All the in vitro experiments described in this article were conducted using the OCPs present in the major fractions.
Figure 2.
Absorbance Spectra of OCPo and OCPr Forms.
Wild-type (A), R155K (B), R155L (C), R155E (D), and E244L (E) mutants. To obtain the red form, OCP was illuminated for 5 min with 5000 μmol photons m−2 s−1 of white light. WT, wild type.
[See online article for color version of this figure.]
Effect of the Mutations on the Light-Induced Protein Conformational Changes of OCP
Light-induced FTIR difference spectroscopy was used to detect protein structural changes induced by strong blue light in the mutated OCPs. In the past, in the light-minus-dark FTIR spectra of wild-type OCP, large changes were observed in the spectral region extending between 1750 to 1600 cm−1, in which Amide I protein modes are expected to contribute (Wilson et al., 2008) (Figure 3). This region is the most informative for monitoring protein structure; specific frequencies are related to different secondary structures (loop, α-helix, and β-sheet) (Krimm and Bandekar, 1986). We have concluded that during photoactivation, wild-type OCP undergoes a loosening of α-helices and a strengthening of the β-sheet accompanied by some loop changes (Wilson et al., 2008). The light-minus-dark FTIR difference spectra recorded after illumination show similar spectral features in the so-called amide I region for wild-type and mutant OCPs, implying a very similar structural response of the proteins to light (Figure 3).
Figure 3.
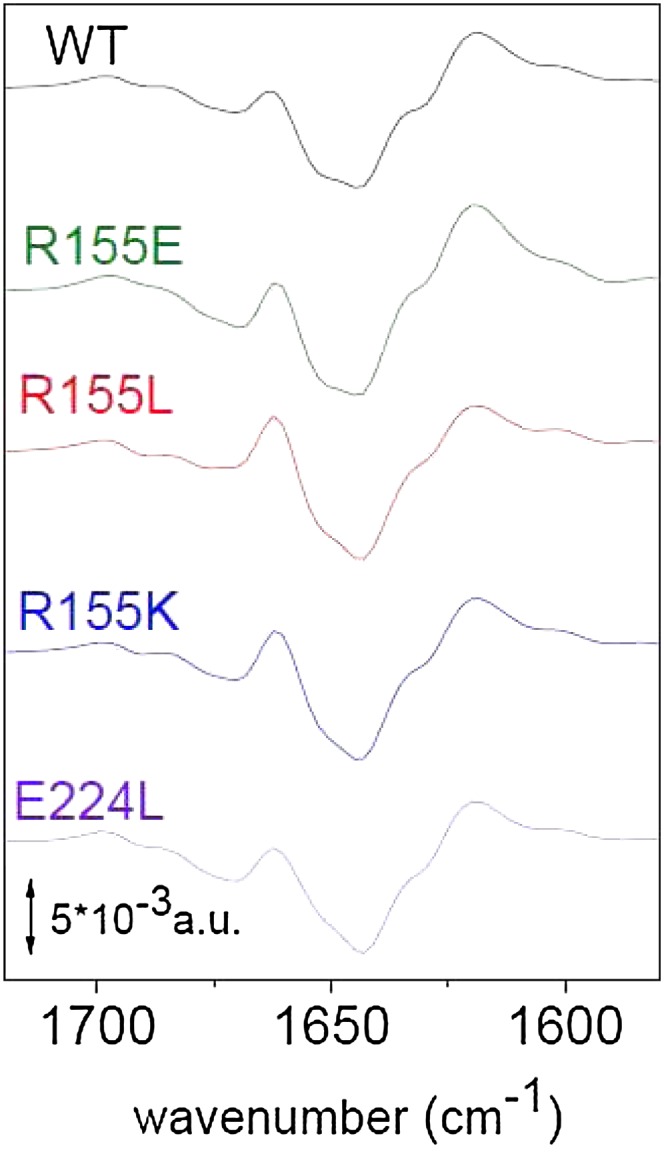
Light-Minus-Dark FTIR Spectra of Wild-Type and Mutated OCPs in Water after Illumination with a Blue Light–Emitting Diode in the Carbonyl Region between 1750 and 1550 cm−1 (Amide I Region).
The differential pattern is assigned to hydrogen bond loosening (upshift) of the loops and α-helices carbonyl modes together with a hydrogen bond strengthening (downshift of carbonyl modes) of the β-sheet as described in detail in Wilson et al. (2008). WT, wild type.
[See online article for color version of this figure.]
Effect of Mutations in Photoactivity and OCPr Stability
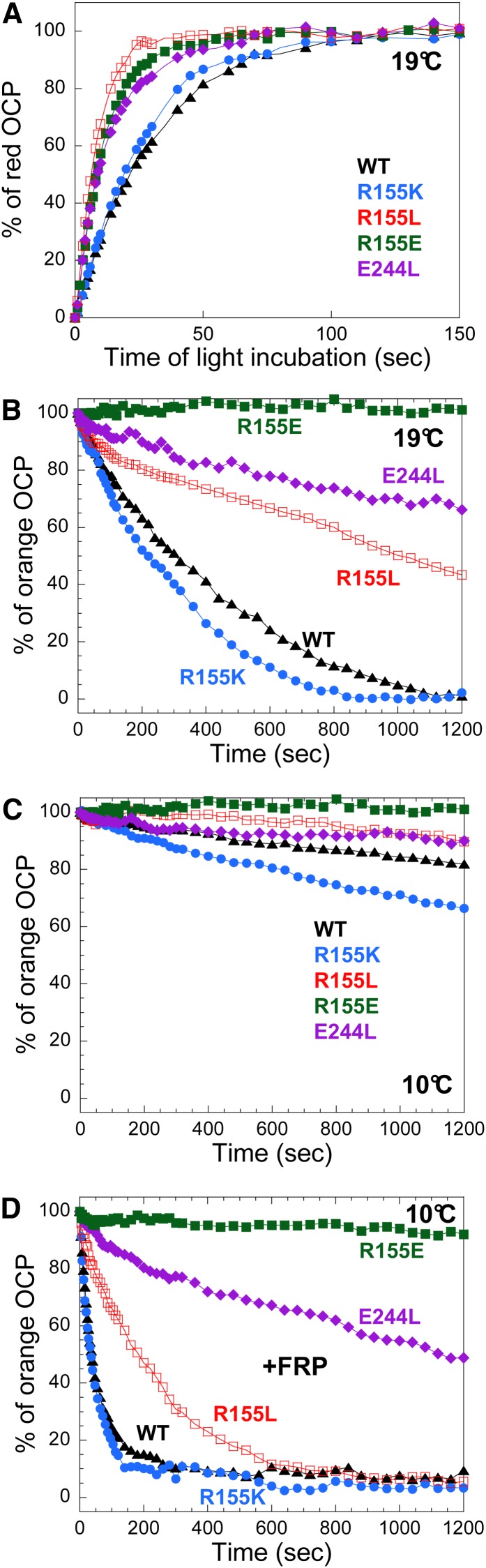
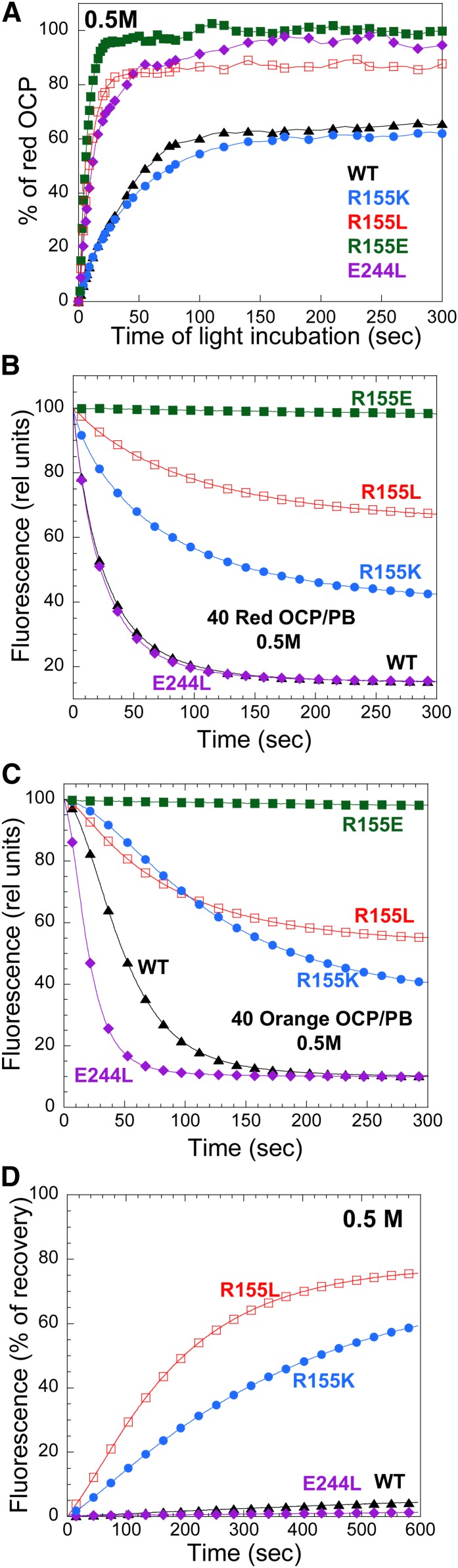
The effect of mutations on the kinetics of photoactivation is clearly visible in Figure 4A. Although R155K-OCP kinetics were similar to those of wild-type OCP, the OCPo-to-OCPr conversion was faster in all other mutants, with R155L-OCP being the fastest.
Figure 4.
Photoactivity and OCPr Stability of Wild-Type and Mutant OCPs.
(A) Light-induced conversion from OCPo to OCPr at 19°C in Tris-HCl pH 8.0 (40 mM) of wild-type OCPs (WT; triangle), R155K-OCPs (circle), R155E-OCPs (filled square), E244L-OCPs (rhomboid), and R155L-OCPs (empty square).
(B) to (D) Stability of wild-type and mutated OCPr at 19°C (B) and 10°C [(C) and (D)] in the absence [(B) and (C)] and in the presence of FRP (D). Kinetics of dark OCPr-to-OCPo conversion of wild-type OCPs (triangle), R155K-OCPs (circle), R155E-OCPs (filled square), E244L-OCPs (rhomboid), and R155L-OCPs (empty square). The ratio of FRP to OCP was 1. The same symbols are used in all figures.
[See online article for color version of this figure.]
The stability of OCPr was tested by measuring the rates of the dark recovery to OCPo at 10 and 19°C and in the absence and presence of FRP (Figures 4B to 4D). R155E-OCPr was the most stable. No OCPr-to-OCPo conversion was observed during 20 min of dark incubation at both temperatures (Figures 4B and 4C). In this mutant, the persistence of the OCPr form is likely caused by the repulsion between the two negative charges at the N- and C-terminal domain interface. E244L-OCPr and R155L-OCPr were more stable than wild-type OCPs and R155K-OCPs at 19°C (Figure 4B). At 10°C, all OCPrs were rather stable (Figure 4C). At this temperature, FRP greatly accelerates the conversion of wild-type OCPrs, R155K-OCPrs, and R155L-OCPrs; however, the effect was much less pronounced with E244L-OCP and was null with R155E-OCP (Figure 4D). Nevertheless, at 19°C, FRP largely accelerated the E244L OCPr-to-OCPo dark conversion and was able to induce the dark recovery of R155E-OCPr (see Supplemental Figure 2 online). It is notable that the E244L mutation has a greater effect on OCPr stability and possibly on FRP interaction than the R155L mutation, even though in both mutants, the potential for R155–E244 interdomain hydrogen bonds is lacking. These data suggested that in OCPr, the surface of the N-terminal domain that is buried in OCPo becomes exposed; likewise, the R155–E244 hydrogen bond stabilizes the OCPo form.
Effect of Mutations on PB Fluorescence Quenching and OCP Binding
Two types of Synechocystis PBs were utilized in this study: wild-type PBs and CK phycobilisomes (CK-PBs). Wild-type PBs consist of a core from which six rods radiate. The core contains 12 allophycocyanin (APC) trimers (eight emitting at 660 nm [APC660], and four emitting at 680 nm [APC680]) organized in three cylinders, and the rods contain phycocyanin hexamers. The CK-PBs are formed only by the core (Ajlani et al., 1995). High concentrations of phosphate are needed to isolate intact and energetically coupled PBs of both types. Concentrations of 0.8 or 0.5 M phosphate were used in most of the reconstitution experiments to maintain the integrity of Synechocystis PBs.
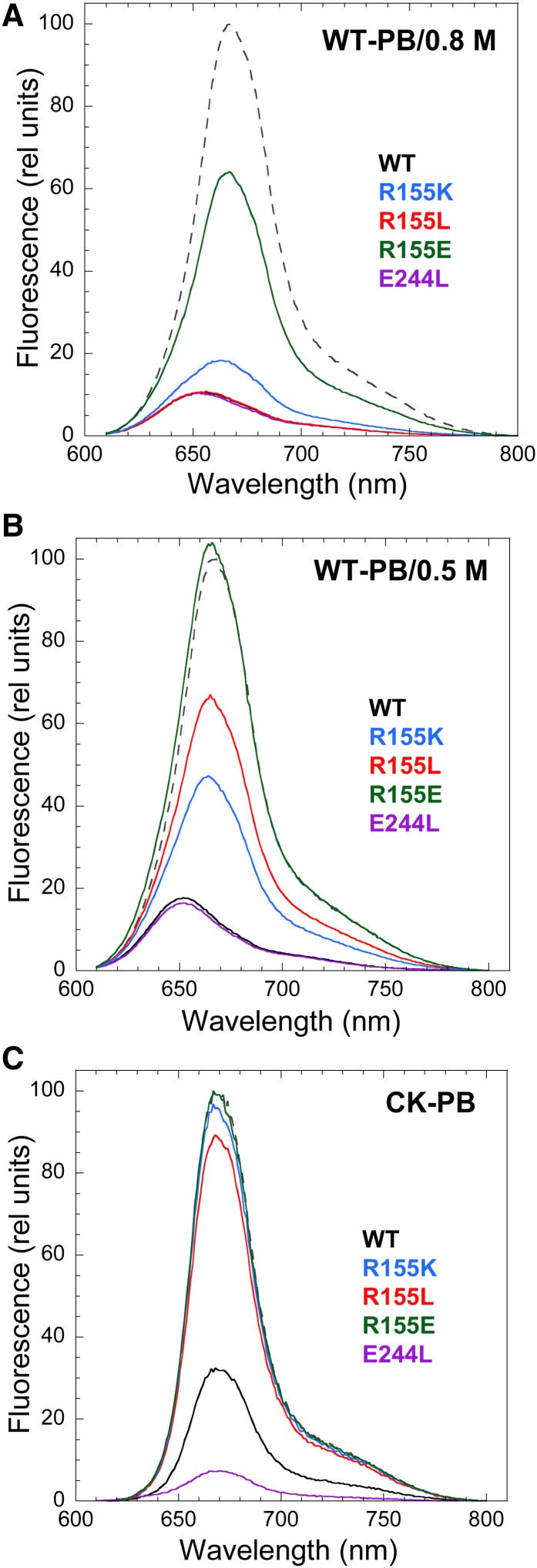
The room temperature fluorescence spectra of isolated wild-type PBs and CK-PBs present a large peak with maximum at 665 nm (related to APC660 trimers) and a shoulder at 680 nm (related to APC680 trimers) (Figure 5). When wild-type PBs were illuminated with strong white light in the presence of an excess of wild-type or mutated OCPs at 0.8 M phosphate, the PB fluorescence decreased in all cases (Figure 5A). However, although wild-type OCPs, E244L-OCPs, R155K-OCPs, and R155L-OCPs induced a decrease of ∼90% of the fluorescence, R155E-OCP induced only ∼35 to 40% of quenching. These results indicated that the mutated OCPs, when bound to the PBs, are able to induce fluorescence quenching. At 0.8 M phosphate, the binding of the OCP to the PBs is very strong, almost irreversible (Gwizdala et al., 2011). Thus, at this phosphate concentration, only very large differences in binding strength will affect the amplitude of the fluorescence quenching. Because OCP binding to PBs is weaker at 0.5 M phosphate than at 0.8 M phosphate (Gwizdala et al., 2011), this lower phosphate concentration was used to detect any differences in fluorescence quenching between the mutants. At 0.5 M phosphate, the integrity of PBs is maintained for the duration of the experiment (Gwizdala et al., 2011). Indeed, Figure 5B shows that only wild-type OCPs and E244L-OCPs induced 85% of fluorescence quenching at 0.5 M phosphate, whereas R155K-OCP induced 55%, and R155L induced only 35%. R155E-OCP was unable to induce any quenching at 0.5 M phosphate.
Figure 5.
Fluorescence Quenching Induced by Activated OCP.
PB fluorescence spectra of whole PBs [(A) and (B)] and PB cores (C) illuminated in the presence of wild-type (WT; black) or mutated OCPs: R155K (blue), R155L (red), R155E (green), and E244L (violet) at 0.8 M phosphate [(A) and (C)] or 0.5 M phosphate (B). The dashed spectra are those of wild-type PBs [(A) and (B)] or CK-PBs (C) in darkness. Illumination of PBs in the absence of OCP did not induce any fluorescence quenching.
The attachment of wild-type OCP to CK-PBs is weaker than to wild-type PBs, because the rods stabilize OCP binding (Gwizdala et al., 2011). At 0.5 M phosphate, even wild-type OCP is unable to bind to CK-PBs and to quench their fluorescence (Gwizdala et al., 2011). When CK-PBs were illuminated (at 0.8 M phosphate) in the presence of an excess of OCP, only wild-type OCPs and E244L-OCPs were able to induce a large fluorescence quenching (Figure 5C). Almost no quenching was induced by the other OCP mutants (Figure 5C). These results suggested that the replacement of Arg155 by other amino acids affected the strength of OCP binding to the PBs, whereas the replacement of Glu244 did not affect binding.
To further characterize the influence of mutations on OCP-induced fluorescence quenching, kinetics of fluorescence quenching and recovery were measured using a pulse amplitude modulation (PAM) fluorometer. The rate and amplitude of PB fluorescence quenching depend on the concentration of OCPr and on the strength of the OCPr binding to the PB (Gwizdala et al., 2011).
In a previous study, it was shown that high phosphate concentrations substantially affect the wild-type OCPo–to–wild-type OCPr conversion without affecting the dark conversion (Gwizdala et al., 2011). Increasing phosphate concentration decreases the initial rate of the light conversion and the steady state concentration of OCPr (Gwizdala et al., 2011). R155K-OCP and wild-type OCP are similarly affected by phosphate: At 0.8 M phosphate, only 20% of the OCPo was converted to OCPr (see Supplemental Figure 3A online), and at 0.5 M phosphate, only 60% of OCP was red after 200 s of illumination (Figure 6A). By contrast, the R155L- and E244L-OCP mutants were much less affected. Only a slight decrease of initial rates was observed, and 80 to 90% of OCPr was accumulated after illumination. R155E-OCP was not affected at all.
Figure 6.
Kinetics of PB Fluorescence Quenching Induced by Strong Blue-Green Light in the Presence of Wild-Type and Mutated OCPs.
PBs (0.013 μM) were illuminated in the presence of an excess of OCPr (B) or OCPo (C) (0.53 μM [40 OCP per PB]) with blue-green light (400 to 550 nm, 870 μmol photons m−2 s−1) at 0.5 M phosphate. WT, wild type.
(A) The effect of 0.5 M phosphate on OCPr accumulation is shown. The wild-type OCPs (triangle), R155K-OCPs (circle), R155E-OCPs (filled square), E244L-OCPs (rhomboid), and R155L-OCPs (empty square) were illuminated with white light (5000 μmol photons m−2 s−1) at 19°C. The same symbols were used in all figures.
(B) The OCP was first illuminated (5000 μmol photons m−2 s−1, white light, 10°C) in 0.08 M phosphate buffer and completely converted to OCPr, and then the PBs in 0.5 M were added, and the mixture was illuminated at 15°C.
(D) Kinetics of dark PB fluorescence recovery. Quenched whole wild-type PBs were incubated in darkness at 0.5 M phosphate. The decrease and increase of fluorescence was followed using a PAM fluorometer.
[See online article for color version of this figure.]
Thus, to look for differences in OCP binding, all OCP preparations were first converted to the red form by illuminating them with strong white light in Tris-HCl buffer and then were added to PBs and illuminated with strong blue light. The experiment was done at 15°C to minimize OCPr-to-OCPo conversion during the 5-min illumination. Under these conditions, the initial concentration of OCPr was identical for all of the OCPs. Wild-type OCPr and E244L-OCPr induced a fast and complete fluorescence quenching (Figure 6B; see Supplemental Figure 3B online) and the kinetics of fluorescence quenching were similar. R155K- and R155L-OCPr induced smaller quenching (60 and 30%, respectively, at 0.5 M phosphate), with slower kinetics at both phosphate concentrations (Figure 6B; see Supplemental Figure 3B online). R155E-OCPr was unable to induce any fluorescence quenching in 0.5 M phosphate (Figure 6B). However, at 0.8 M phosphate, it induced 35% of fluorescence quenching after 5 min of illumination with strong blue light (see Supplemental Figure 3B online). As expected, the kinetics were different when OCPo was added to PBs and then illuminated (Figure 6C). In this case, the kinetics of OCPr accumulation influenced the kinetics of fluorescence quenching. E244L-OCP induced fluorescence quenching faster than wild-type OCP, because E244L-OCPr accumulated faster than wild-type OCPr. During the first seconds of illumination, R155L-OCP induced a faster and stronger quenching compared with R155K-OCP, whereas at longer durations, R155K-OCP induced a larger fluorescence quenching. This result can be explained by the combination of a faster R155L-OCPr accumulation with a possible weaker binding to PBs compared with R155K-OCP.
At 0.5 M phosphate, PBs incubated with R155L-OCP or R155K-OCP rapidly recovered the lost fluorescence, whereas in PBs incubated with wild-type OCP or E244L-OCP, almost no fluorescence recovery was observed (Figure 6D). The fluorescence was recovered faster in PBs incubated with R155L-OCP than in those incubated with R155K. At 0.8 M phosphate, only PBs incubated with R155E-OCP partially recovered the lost fluorescence (see Supplemental Figure 3C online).
To compare the strength of the binding of the OCP mutants to the PBs, wild-type PBs were illuminated in the presence of an excess of the different OCPs in 0.5 and 0.8 M phosphate. The quenched OCP-PB complexes were reisolated by centrifugation in a Suc gradient containing 0.8 M phosphate. The PBs were recovered in the 0.75 M Suc fraction, whereas the unbound OCP remained in the upper phase (Gwizdala et al., 2011). When PBs were illuminated in 0.5 M phosphate, only the presence of wild-type OCPs and E244L-OCPs was observed in Coomassie blue–stained gels of the PBs. No traces of the other OCPs were detected (Figure 7). Moreover, the quenched R155-mutated OCP-PB complexes were no longer quenched after the reisolation on the Suc gradient. These results indicated that the mutated OCPs, with the exception of E244L-OCP, detached from the PBs during the reisolation, confirming their weaker binding to PBs. The presence of wild-type OCP and mutated OCPs was detected in Coomassie-blue stained gels in all the PBs when they were illuminated in 0.8 M phosphate (see Supplemental Figure 3D online). Nevertheless, the quantity of OCP bound to PBs depended on the OCP used. Wild-type OCP-PB and E244L-OCP-PB complexes contained the highest concentration of OCP (see Supplemental Figure 3D online). Then, decreasing OCP concentrations were observed in the complexes in the following order: R155K-PBs, R155L-PBs, and R155E-PBs.
Figure 7.
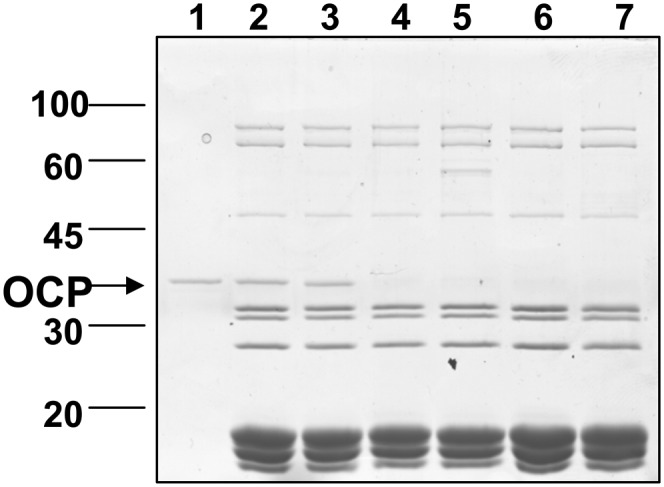
Detection of Wild-Type and Mutated OCPs Attached to PBs after Illumination at 0.5 M Phosphate.
PBs were incubated in the presence of excess OCP under strong illumination and then loaded on a Suc gradient for reisolation. SDS-PAGE of the polypeptide composition of the blue band obtained in the Suc gradient after illumination of wild-type PBs in the presence of wild-type OCP (2), E244L-OCP (3), R155L (4), R155K (5), or R155E (6). (1) OCP alone. (7) PBs without OCP. In each lane, 10 μL of a solution containing 0.5 μM PBs was loaded. The OCP band is marked by an arrow.
In Vivo Phenotype of the Synechocystis Mutants Overexpressing the Mutated OCPs
The replacement of the Arg155 or Glu244 substantially affected the concentration of OCP in the cells. Protein gel blot analysis showed that the strains overexpressing the mutated OCPs accumulate much less OCP than the strain overexpressing wild-type OCP (Figure 8): only 15 to 20% for R155E and E244L mutants, 30 to 35% for R155L, and 70 to 80% for R155K. Most likely, the lower concentrations of mutated OCPs are associated with a lower stability of carotenoid binding and, as a consequence, greater instability of the protein. This instability was also suggested by the small quantities of mutant OCPs that could be isolated from the overexpressing OCP strains. Using 15 L of Synechocystis-overexpressing OCP cells at OD800 = 0.8, 50 to 55 mL of wild-type OCP (100%), 30 to 35 mL of R155K-OCP (60 to 70%), 10 to 15 mL of R155L (20 to 30%), and only 2 mL of R155E or E244L (4%) at OD496 = 3 were obtained. The quantities of mutant OCPs obtained were lower than expected from the OCP content in the cells, suggesting protein degradation during the isolation procedure.
Figure 8.
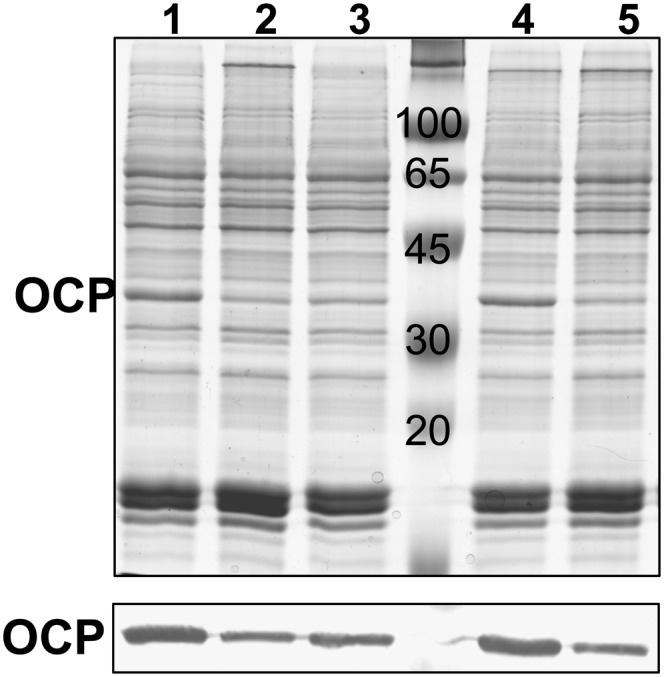
OCP Content in MP Complexes Isolated from Synechocystis Strains Overexpressing the Wild-Type (1) and Mutant OCPs: E244L (2), R155L (3), R155K (4), and R155E (5).
Coomassie blue–stained gel electrophoresis (Top) and immunodetection of OCP (Bottom). All cellular OCP is present in the MP complexes (Wilson et al., 2006). Each MP sample loaded contains 0.5 μg chlorophyll. Three independent experiments were done, and the density of the OCP bands in the immunoblots was measured. Considering the density of the wild-type OCP band as 100%, the density of the mutated OCP bands was 15 to 20% for R155E and E244L mutants, 30 to 35% for R155L, and 70 to 80% for R155K.
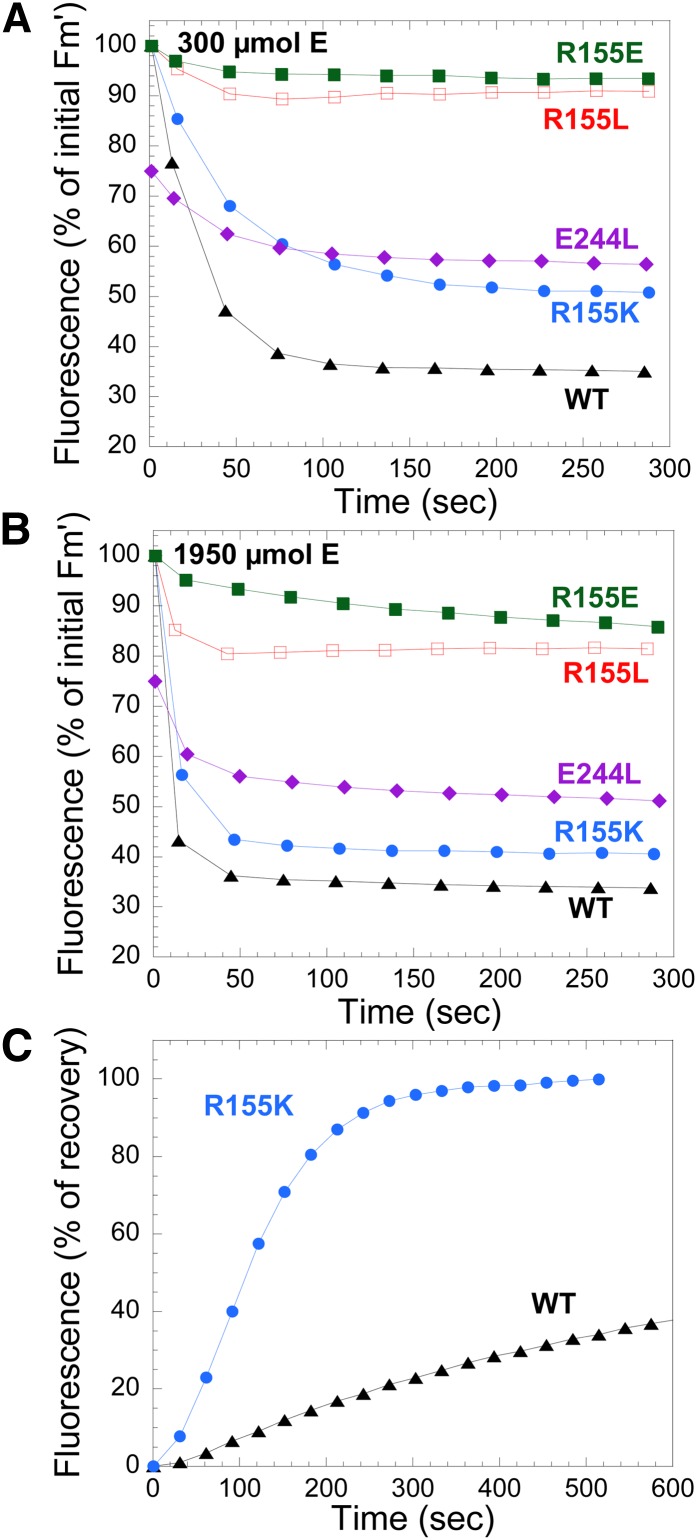
Figures 9A and 9B show the kinetics of fluorescence quenching induced by two intensities of blue-green light (300 and 1950 μmol photons m−2 s−1) followed with a PAM fluorometer. Dark-adapted cells were first illuminated with low intensities of blue-green light to induce State 1 transition and then by strong blue-green light. All Synechocystis strains expressing mutant OCPs, with the exception of the overexpressing E244L-OCP, showed similar light maximum fluorescence (Fm′) (∼2200 relative units) and dark minimum fluorescence (∼1100 relative units) values after illumination with low intensities of blue-green light. These values were similar to those observed in wild-type Synechocystis cells, indicating that growth light intensities did not induce a permanent or semipermanent fluorescence quenching despite the high concentration of OCP in the overexpressing cells. Exposure of cells to strong blue-green light induced a large fluorescence quenching only in strains overexpressing wild-type OCPs and R155K-OCPs. The rate and amplitude of the fluorescence quenching was slower and smaller in the strain overexpressing R155K-OCP. This can be partially explained by the lower concentration of OCP in this strain. Nevertheless, we propose that the phenotype is related to the weaker binding of R155K-OCP to the PB that leads to a faster fluorescence recovery. Indeed, Figure 9C shows a very fast dark fluorescence recovery in the strain containing R155K-OCP. Little to no fluorescence quenching was induced by strong blue-green light in the strains overexpressing R155E-OCPs and R155L-OCPs. The fact that in the first strain, no fluorescence quenching was observed can be explained by the very weak attachment of R155E-OCP to PBs, which was demonstrated by the in vitro experiments, and by the low concentration of OCP in this strain. R155L-OCP binds more strongly than R155E-OCP to PBs, and at high light intensities, a low fluorescence quenching was observed (Figure 9B). The weaker binding of R155L-OCP to PBs compared with that of R155K-OCP and lower concentration could explain why strong blue-green light induced fluorescence quenching in the R155K mutant strain and not in the R155L mutant strain.
Figure 9.
Blue-Green Light–Induced Fluorescence Quenching in Wild-Type and Mutant Synechocystis Cells and Fluorescence Recovery in Wild-Type and R155K Strains.
(A) and (B) Decrease of Fm′. Low light (80 μmol photons m−2 s−1, blue-green light) adapted wild-type (WT) and mutant Synechocystis cells (at 3 μg chlorophyll mL−1) were illuminated with high intensities [300 μmol photons m−2 s−1 (A) and 1950 μmol photons m−2 s−1 (B) of blue-green light (400 to 550 nm)]. Fluorescence yield changes were detected with a PAM fluorometer, and saturating pulses were applied to measure Fm′ levels. Overexpressing strains were wild-type ΔCrtR OCP (triangles), R155K-ΔCrtR (circles), R155L-ΔCrtR empty squares), R155E-ΔCrtR (filled squares), and E244L-ΔCrtR (rhomboids).
(C) Increase of Fm during exposure to low blue-green light after high irradiance of cells of wild-type (triangles) and R155K-ΔCrtR (circles) mutant.
[See online article for color version of this figure.]
In Synechocystis cells overexpressing E244L-OCP, the dark maximum fluorescence and dark minimum fluorescence values and the Fm′ and variable fluorescence values under low intensities of blue-green light were always lower (25 to 35%, approximately 1650 and 900 relative units, respectively) than those measured in the other strains, suggesting that growth light (60 μmol photons m−2 s−1) induced an almost permanent quenching. Three hours of dark incubation allowed the recovery of only 10% of the lost fluorescence. Exposure of these cells to strong blue-green light induced an additional fluorescence quenching (Figures 9A and 9B). The lower level of fluorescence quenching induced compared with wild-type OCP can be explained by the low OCP concentration in the cells.
DISCUSSION
When this research was begun, nothing was known about how OCPr interacts with PBs. The development of the in vitro reconstitution system (Gwizdala et al., 2011) and the construction of new OCP mutants now reveal that OCP binding to PBs involves the N-terminal domain of OCP, specifically the surface occluded by the interdomain salt bridge between Arg155 and Glu244. Arg155 and Glu244 are strongly conserved in the primary structure of the OCP. Of 110 full-length homologs of OCP found in sequenced genomes, only 10 have conservative substitutions for Arg155 (nine Gln, one His); the few substitutions observed at position Glu244 are Asp.
Role of the Hydrogen Bond between Arg155 and Glu244 in Photoactivity and Protein Stability
Arg155 and Glu244 are positioned in the central interface between the N- and C-terminal domains (Wilson et al., 2010). Both are located on helices in their respective domains (Figures 1A and 1C). In addition, they flank the surface depression in which the carotenoid is slightly solvent-exposed in OCPo. The loss of the salt bridge by mutation of the Arg155 or the Glu244 is expected to destabilize the interaction of the two domains, possibly increasing their relative movement with respect to each other; eight structurally conserved water molecules may also facilitate disruption of this interface. In this study, it is shown that, indeed, when substitutions that preclude formation of the salt bridge were made (R155L, R155E; E244L), OCPr accumulated faster, and its dark stability largely increased. By contrast, when Arg155 was replaced by a Lys, the photoactivity and the stability of OCPr were not affected, suggesting the importance of the electrostatic interaction between domains in stabilizing the dark, orange form. Figure 1D shows a close-up of the interdomain interface of a model of the R155K mutant OCP. Even with selection of the Lys rotamer with closest approach to Glu244, the separation distance is 4.5 Å, suggesting a salt bridge would be unlikely to form between Lys155 and Glu244. However, weaker, more transient interactions could still occur between Lys155 and Glu244 or between the helix dipoles. We cannot exclude the possibility that the domains could move closer together to allow formation of a salt bridge.
Our results are consistent with a model in which the bond between Arg155 and Glu244 is broken in OCPr, allowing separation between the two domains of the protein. This leads to an increase of the solvent accessibility of the carotenoid and, possibly, permits a closer interaction between the carotenoid and the chromophore of the APC. The larger separation between the two domains of the protein may also allow the binding of OCP to APC trimers through the resulting exposed surface of the N-terminal domain.
In all of the mutants, the concentration of OCP was lower than in the wild type, and only small quantities could be isolated. This instability could be related to a weaker binding of the carotenoid to the protein. It is not known how the carotenoid is inserted into OCP, but it is clear that carotenoid binding is stabilized in the orange form, because it is almost completely embedded in the protein. We hypothesize that the larger exposure of the carotenoid to solvent after loss of the R155-E244 salt bridge could increase the lability of carotenoid–protein interactions.
Role of E244 and R155 in OCP–FRP Interaction
Mutations of Arg155 and Glu244 also modified the effect of FRP on the kinetics of the OCPr-to-OCPo reversion. The R155E- and E244L-OCPs were the most affected mutants. Nothing is known about the site of interaction between OCP and FRP. The FRP structure has not been determined, although FRP crystals that diffract to 3 Å have been reported (Liu et al., 2011). Our results may be interpreted to suggest that the interface between the N- and C-terminal domains is involved in the OCP–FRP interaction. However, our results could also be explained by FRP binding OCPr elsewhere on the protein to lower the energy of activation of OCPr-to-OCPo reversion. This energy of activation is much higher for R155E- and E244L-OCPs than for wild-type OCP. This is substantiated by the fact that at 10°C, FRP had almost no effect on the kinetics, whereas at 19°C, FRP was able to accelerate the E244L and R155E OCPr-to-OCPo conversion (see Supplemental Figure 2 online). The resolution of the structure of FRP and the construction of FRP mutants will help identify the amino acids involved in the FRP–OCP interaction.
Role of the N-Terminal Domain and the Positive Charge at Arg155 in OCP Binding to PBs
The structures of the dark orange R155L-OCPs and wild-type OCPs are almost identical (Wilson et al., 2010). FTIR measurements showed that protein conformational changes induced by strong blue-green light were similar in wild-type and mutant OCPs (this work). Thus, we hypothesize that the structures of the red wild-type and mutated OCPs are also similar. Therefore, the differences in OCP binding to PBs cannot be attributed to large changes in the structure of the different OCPrs.
Replacing Glu244 with a Leu had almost no effect on the strength of binding of OCP to PBs, indicating that this amino acid is not directly involved in the interaction. By contrast, a neutral amino acid in the place of Arg, as in the R155L mutant, largely decreased the strength of binding, whereas a negative charge in place of the positive charge (R155E) almost completely abolished binding of OCPr to PBs. This is particularly striking, because Gln is one of the few conservative substitutions for Arg155; thus, the deleterious effect of this substitution is most likely caused by the negative charge. The importance of the positive charge of Arg155 for photoprotection is substantiated by the relatively small loss of fluorescence quenching by R155K-OCP.
Thus, our results demonstrated that the surface of the N-terminal domain of OCP that contains Arg155 is directly involved in the binding of OCPr to APC trimers. Moreover, the positive charge of Arg155 is an essential element of this process. In the past, we suggested that OCP interacts with the PB via its C-terminal domain (Wilson et al., 2008). This suggestion was based on the similarity of the structures of the OCP C-terminal domain and the Lc7.8 core linker protein (Wilson et al., 2008) and on the fact that once the His-tagged OCP attached to the PB, it was no longer bound to a nickel column (Gwizdala et al., 2011). The demonstration of the involvement of the N-terminal domain, and specifically the essential role of Arg155 in PB binding, suggests that this model may need to be revised. The data presented here suggest that the R155-E244 salt bridge between the N- and C-terminal domains precludes the binding of OCPo to PBs. Although Arg155 is slightly exposed to solvent in OCPo form, it is unable to bind to the PB until the salt bridge is broken. In OCPr, breakage of the salt bridge with concomitant domain motion exposes the surface of the N-terminal domain and may increase the exposure of Arg155 for the interaction between OCP and negative charges in the APC trimer close to one of the bilin chromophores (McGregor et al., 2008). The weaker binding of R155K-OCP to PBs compared with that of wild-type OCP could be explained by less precise orientation of the positive charge and decreased hydrogen bonding potential of the sidechains of Lys relative to Arg.
In conclusion, our results demonstrated that (1) the breakage of the salt bridge between the Arg155 and the Glu244 likely decreases the interdomain interaction and strongly suggests that in the red form, this interaction is weaker or nonexistent, (2) that this region of the N-terminal domain of the OCP is involved in interaction with PBs for fluorescence quenching, and (3) that the positive charge of Arg155 is directly involved in OCP binding to PBs. Finally, the in vitro study of the isolated OCP and of the OCP–PB interaction could explain the observed phenotype of the Synechocystis OCP mutant cells.
METHODS
Culture Conditions
The mesophylic freshwater cyanobacteria Synechocystis sp strain PCC 6803 wild-type and mutants were grown photoautotrophically in a modified BG11 medium (Herdman et al., 1973) but containing double the amount of sodium nitrate. Cells were kept in a rotary shaker (120 rpm) at 30°C, illuminated by fluorescent white lamps giving a total intensity of 60 μmol photons m−2 s−1 under a CO2-enriched atmosphere. The cells were maintained in the logarithmic phase of growth and were collected at OD800 = 0.6 to 0.8. For OCP and PB isolation, cyanobacteria cells were grown in 3-liter Erlenmeyer flasks in a rotary shaker under a light intensity of 90 to 100 μmol photons m−2 s−1. The cells were harvested at OD800 = 1.
Construction of Mutant Strains
The construction of the Synechocystis PCC 6803 ΔCrtR strain, which served as the receptive strain of the plasmids overexpressing the genes coding for wild-type and mutated OCPs, was described in Wilson et al. (2011). The construction of the plasmid containing the C-terminal His6-tagged OCP gene (slr1963) under the control of the psbAII promoter was described in Wilson et al. (2008). The point mutations R155 and E244 were introduced into the slr1963 gene in the overexpressing plasmid by site-directed mutagenesis using the Quickchange XL site-directed mutagenesis kit (Stratagene) and synthetic oligonucleotides (primers used in this work are listed in Supplemental Figure 4 online). The presence of the point mutation was confirmed by DNA sequencing. The plasmids were used to transform by double recombination the ΔCrtR mutant of Synechocystis PCC 6803.
In the strains overexpressing the mutated OCPs, the original wild-type OCP was suppressed by partial deletion of the slr1963 gene. A 2.2-kb XhoI-SpeI fragment containing the slr1963 (encoding for OCP) and slr1964 (encoding for FRP) was amplified with oligonucleotides car7 and car6 and cloned into SK(+) ampicillin-resistant vector (see Supplemental Figure 5 online). A deletion of a large part of the slr1963- and slr1964-encoding sequence was obtained by removing a 770–base pair fragment with restriction enzymes HincII and HindIII and replacing it with a 2-kb spectinomycin- and streptomycin-resistance cassette previously digested by HincII. The resulting plasmid was used to transform overexpressing OCP mutants of Synechocystis by double recombination (see Supplemental Figure 5 online). Segregation of all mutants was confirmed by PCR (primer sequences can be found in Supplemental Figure 4 online).
OCP and FRP Purification
OCP was isolated from the ΔCrtR strain overexpressing C-terminal His-tagged wild-type and mutated OCPs. Its purification was performed as previously described (Wilson et al., 2008). The construction of the Escherichia coli strain overexpressing the His-tagged short FRP-encoding gene of Synechocystis and the isolation of the FRP from this strain were described in Boulay et al. (2010).
Absorbance Measurements
Cell absorbance was monitored with an Uvikon XL spectrophotometer (SECOMAN). Chlorophyll content was determined in methanol using the extinction coefficient at 665 nm of 79.24 mg mL−1 cm−1. The orange-to-red OCP photoconversion was monitored in a Specord S600 (Analyticjena) spectrophotometer during illumination of the OCP with 5000 μmol photons m−2 s−1 of white light at 10, 19, and 23°C.
Fluorescence Measurements
Fluorescence quenching and recovery were monitored with a PAM fluorometer (101/102/103-PAM; Walz). All measurements were performed in a stirred cuvette of 1 cm diameter. In most of the experiments, the fluorescence quenching was induced by 870 μmol photons m−2 s−1 of blue-green light (400 to 550 nm) at 23°C. Recovery of PB fluorescence was recorded in the darkness.
Fluorescence emission spectra at room temperature were carried in a CARY Eclipse fluorescence spectrophotometer (Varian) fluorometer. The measurements were performed in a stirred cuvette of 1 cm diameter at 23°C. The fluorescence quenching was induced by 5 min of illumination with 5000 μmol photons m−2 s−1 of white light. All in vitro fluorescence measurements were performed at a PB concentration of 0.013 to 0.014 μM.
Isolation of Membrane-Bound PBs, PB, and OCP-PB Complexes
Purification of membrane-PB (MP) complexes from wild-type and OCP mutants and of PBs from wild-type and CK Synechocystis PCC 6803 were performed as previously described (Wilson et al., 2006; Gwizdala et al., 2011). The OCP-PB complexes were prepared by illumination of isolated PBs (diluted 10 times with 0.8 M K-phosphate buffer) with 2 × 1300 μmol photons m−2 s−1 (two light sources) of white light during 20 min at 23°C in the presence of OCP (OCP:PB ratio = 20).
OCP Immunodetection
Proteins from MP fractions were analyzed by SDS-PAGE on a 12% polyacrylamide/2 M urea in a TRIS/MES system (Kashino et al., 2001). The OCP protein was detected by a polyclonal antibody against OCP.
FTIR Experiments
FTIR difference spectra have been recorded on a Bruker IFS88 spectrometer equipped with a photoconductive MCT-A detector and Opus software. The photoreaction was induced by a blue light–emitting diode. The temperature was set to 293K using a cryostat (Oxford Instruments) and checked by a thermocouple. Before the measurements, the sample was allowed to stabilize for at least 4 h. Stability of the sample was checked by recording FTIR difference spectra in the dark.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers: OCP (slr1963), NP_441508; FRP (slr1964), NP_441509.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Absorbance Spectra of Minor Fractions of R155L-OCPs and R155E-OCPs.
Supplemental Figure 2. Effect of FRP on OCPr to OCPo Conversion at 19°C.
Supplemental Figure 3. OCP-Related Fluorescence Quenching Induced by Strong Blue-Green Light at 0.8 M Phosphate.
Supplemental Figure 4. Oligonucleotides Used in the Construction of OCP Mutants.
Supplemental Figure 5. Genome Region Including the slr1963 and slr1964 Genes Encoding the OCP and the FRP, Respectively, and Position of Mutations.
Supplementary Material
Acknowledgments
We thank Sandrine Cot for technical assistance, Ghada Ajlani for the CK mutant, and Seth Axen for help with figure preparation. This article was supported by grants from l’Agence Nationale de la Recherche (program CYANOPROTECT) and from Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique, and HARVEST European Union Seventh Framework Programme Marie Curie Research Training Network. The work of C.A.K. is performed under the auspices of the U.S. Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract number DE-AC02-05CH11231, and the Lawrence Livermore National Laboratory under contract number DE-AC52-07NA27344. C.A.K. also acknowledges support of the National Science Foundation (Molecular and Cellular Biosciences 0851070).
AUTHOR CONTRIBUTIONS
A.W., M.G., A.M., and M.A. performed research and contributed to the analysis of the data, C.A.K. contributed to the analysis of data and to writing of the article, and D.K. designed and performed the research, analyzed data, and wrote the article.
Glossary
- PAM
pulse amplitude modulation
- PB
phycobilisome
- FTIR
Fourier transform infrared
- APC
allophycocyanin
- hECN
3′-hydroxyechinenone
- OCPo
inactive orange dark form of OCP
- OCPr
active red form of OCP
- CK-PB
CK phycobilisome
- Fm′
maximum fluorescence
- Fm
to be defined
- pb
to be defined
- MP
membrane-phycobilisome
References
- Adir N. (2005). Elucidation of the molecular structures of components of the phycobilisome: Reconstructing a giant. Photosynth. Res. 85: 15–32 [DOI] [PubMed] [Google Scholar]
- Ajlani G., Vernotte C., DiMagno L., Haselkorn R. (1995). Phycobilisome core mutants of Synechocystis PCC 6803. Biochim. Biophys. Acta 1231: 189–196 [Google Scholar]
- Boulay C., Wilson A., D’Haene S., Kirilovsky D. (2010). Identification of a protein required for recovery of full antenna capacity in OCP-related photoprotective mechanism in cyanobacteria. Proc. Natl. Acad. Sci. USA 107: 11620–11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer A.N. (1984). Phycobilisome—a macromolecular complex optimized for light energy transfer. Biochim. Biophys. Acta 768: 29–51 [Google Scholar]
- Gorbunov M.Y., Kuzminov F.I., Fadeev V.V., Kim J.D., Falkowski P.G. (2011). A kinetic model of non-photochemical quenching in cyanobacteria. Biochim. Biophys. Acta 1807: 1591–1599 [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Schaefer M.R., Chiang G.G., Collier J.L. (1993). The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 57: 725–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdala M., Wilson A., Kirilovsky D. (2011). In vitro reconstitution of the cyanobacterial photoprotective mechanism mediated by the Orange Carotenoid Protein in Synechocystis PCC 6803. Plant Cell 23: 2631–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdman M., Delaney S.F., Carr N.G. (1973). A new medium for the isolation and growth of auxotrophic mutants of the blue-green alga Anacystis nidulans. J. Gen. Microbiol. 79: 233–237 [Google Scholar]
- Holt T.K., Krogmann D.W. (1981). A carotenoid-protein from cyanobacteria. Biochim. Biophys. Acta 637: 408–414 [Google Scholar]
- Kashino Y., Koike H., Satoh K. (2001). An improved sodium dodecyl sulfate-polyacrylamide gel electrophoresis system for the analysis of membrane protein complexes. Electrophoresis 22: 1004–1007 [DOI] [PubMed] [Google Scholar]
- Kerfeld C.A., Sawaya M.R., Brahmandam V., Cascio D., Ho K.K., Trevithick-Sutton C.C., Krogmann D.W., Yeates T.O. (2003). The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure 11: 55–65 [DOI] [PubMed] [Google Scholar]
- Kirilovsky D. (2007). Photoprotection in cyanobacteria: The orange carotenoid protein (OCP)-related non-photochemical-quenching mechanism. Photosynth. Res. 93: 7–16 [DOI] [PubMed] [Google Scholar]
- Kirilovsky D., Kerfeld C.A. (2012). The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Biochim. Biophys. Acta 1817: 158–166 [DOI] [PubMed] [Google Scholar]
- Krimm S., Bandekar J. (1986). Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 38: 181–364 [DOI] [PubMed] [Google Scholar]
- Liu T., Shuai Y., Zhou H. (2011). Purification, crystallization and preliminary X-ray diffraction of fluorescence recovery protein from Synechocystis PCC 6803. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67: 1627–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacColl R. (1998). Cyanobacterial phycobilisomes. J. Struct. Biol. 124: 311–334 [DOI] [PubMed] [Google Scholar]
- Masamoto K., Misawa N., Kaneko T., Kikuno R., Toh H. (1998). Beta-carotene hydroxylase gene from the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 39: 560–564 [DOI] [PubMed] [Google Scholar]
- McGregor A., Klartag M., David L., Adir N. (2008). Allophycocyanin trimer stability and functionality are primarily due to polar enhanced hydrophobicity of the phycocyanobilin binding pocket. J. Mol. Biol. 384: 406–421 [DOI] [PubMed] [Google Scholar]
- Polívka T., Kerfeld C.A., Pascher T., Sundström V. (2005). Spectroscopic properties of the carotenoid 3′-hydroxyechinenone in the orange carotenoid protein from the cyanobacterium Arthrospira maxima. Biochemistry 44: 3994–4003 [DOI] [PubMed] [Google Scholar]
- Punginelli C., Wilson A., Routaboul J.M., Kirilovsky D. (2009). Influence of zeaxanthin and echinenone binding on the activity of the orange carotenoid protein. Biochim. Biophys. Acta 1787: 280–288 [DOI] [PubMed] [Google Scholar]
- Rakhimberdieva M.G., Elanskaya I.V., Vermaas W.F.J., Karapetyan N.V. (2010). Carotenoid-triggered energy dissipation in phycobilisomes of Synechocystis sp. PCC 6803 diverts excitation away from reaction centers of both photosystems. Biochim. Biophys. Acta 1797: 241–249 [DOI] [PubMed] [Google Scholar]
- Rakhimberdieva M.G., Kuzminov F.I., Elanskaya I.V., Karapetyan N.V. (2011). Synechocystis sp. PCC 6803 mutant lacking both photosystems exhibits strong carotenoid-induced quenching of phycobilisome fluorescence. FEBS Lett. 585: 585–589 [DOI] [PubMed] [Google Scholar]
- Scott M., McCollum C., Vasil’ev S., Crozier C., Espie G.S., Krol M., Huner N.P., Bruce D. (2006). Mechanism of the down regulation of photosynthesis by blue light in the Cyanobacterium synechocystis sp. PCC 6803. Biochemistry 45: 8952–8958 [DOI] [PubMed] [Google Scholar]
- Wilson A., Ajlani G., Verbavatz J.M., Vass I., Kerfeld C.A., Kirilovsky D. (2006). A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell 18: 992–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Kinney J.N., Zwart P.H., Punginelli C., D’Haene S., Perreau F., Klein M.G., Kirilovsky D., Kerfeld C.A. (2010). Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria. J. Biol. Chem. 285: 18364–18375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Punginelli C., Couturier M., Perreau F., Kirilovsky D. (2011). Essential role of two tyrosines and two tryptophans on the photoprotection activity of the Orange Carotenoid Protein. Biochim. Biophys. Acta 1807: 293–301 [DOI] [PubMed] [Google Scholar]
- Wilson A., Punginelli C., Gall A., Bonetti C., Alexandre M., Routaboul J.M., Kerfeld C.A., van Grondelle R., Robert B., Kennis J.T., Kirilovsky D. (2008). A photoactive carotenoid protein acting as light intensity sensor. Proc. Natl. Acad. Sci. USA 105: 12075–12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.