Biochemical and genetic characterization of two petunia CoA ligases reveals that subcellular compartmentalization of enzymes determines their involvement in the benzenoid metabolic network. Additional evidence shows that formation of cinnamoyl-CoA in peroxisomes is the committed step in the synthesis of benzoyl-CoA via the β-oxidative pathway.
Abstract
Biosynthesis of benzoic acid from Phe requires shortening of the side chain by two carbons, which can occur via the β-oxidative or nonoxidative pathways. The first step in the β-oxidative pathway is cinnamoyl-CoA formation, likely catalyzed by a member of the 4-coumarate:CoA ligase (4CL) family that converts a range of trans-cinnamic acid derivatives into the corresponding CoA thioesters. Using a functional genomics approach, we identified two potential CoA-ligases from petunia (Petunia hybrida) petal-specific cDNA libraries. The cognate proteins share only 25% amino acid identity and are highly expressed in petunia corollas. Biochemical characterization of the recombinant proteins revealed that one of these proteins (Ph-4CL1) has broad substrate specificity and represents a bona fide 4CL, whereas the other is a cinnamate:CoA ligase (Ph-CNL). RNA interference suppression of Ph-4CL1 did not affect the petunia benzenoid scent profile, whereas downregulation of Ph-CNL resulted in a decrease in emission of benzylbenzoate, phenylethylbenzoate, and methylbenzoate. Green fluorescent protein localization studies revealed that the Ph-4CL1 protein is localized in the cytosol, whereas Ph-CNL is in peroxisomes. Our results indicate that subcellular compartmentalization of enzymes affects their involvement in the benzenoid network and provide evidence that cinnamoyl-CoA formation by Ph-CNL in the peroxisomes is the committed step in the β-oxidative pathway.
INTRODUCTION
Benzoic acid (BA) and its derivatives are important biosynthetic building blocks in a wide spectrum of plant compounds varying from primary metabolites, such as aromatic cytokinins and the ubiquitous plant hormone salicylic acid, to lineage-specific compounds with chemotherapeutic activities (e.g., the anticancer agent taxol) (Bjorklund and Leete, 1992; Walker and Croteau, 2000). Numerous specialized metabolites containing benzyl (derived from benzylalcohol) or benzoyl (derived from BA) moieties also play essential ecological roles in plant life cycles, acting as defense compounds (Yang et al., 1997; Kliebenstein et al., 2007; Qualley and Dudareva, 2008) or as attractants of pollinators and seed dispersers (Dobson, 2006). Volatile benzenoids contribute to aroma and scent properties of many plant species and represent one of the largest classes of plant volatile metabolites (Knudsen and Gershenzon, 2006). BA is believed to be a precursor of salicylic acid (2-hydroxy BA; León et al., 1995) produced under biotic (Ogawa et al., 2006) and abiotic stresses (Ogawa et al., 2005; Pan et al., 2006; Sawada et al., 2006), although an alternative route to salicylic acid formation directly from isochorismate (bypassing Phe) has recently been demonstrated in pathogen-infected Arabidopsis thaliana (Wildermuth et al., 2001) and Nicotiana benthamiana (Catinot et al., 2008).
Despite the importance and prevalence of plant benzenoids, the biochemical pathways to BA are still poorly understood. Formation of BA via the Phe pathway requires shortening the side chain by a C2 unit, which can occur via a β-oxidative pathway with formation of four CoA-ester intermediates (trans-cinnamic acid → cinnamoyl-CoA → 3-hydroxy-3-phenylpropanoyl-CoA → 3-oxo-3-phenylpropanoyl-CoA → benzoyl-CoA → BA), a nonoxidative side-chain cleavage of the hydrated form of trans-cinnamic acid (3-hydroxy-3-phenylpropanoic acid) with formation of benzaldehyde as an intermediate before oxidation to BA (Figure 1), or a variation of the latter pathway that uses cinnamoyl-CoA instead of cinnamic acid to produce benzaldehyde (Jarvis et al., 2000; Hertweck et al., 2001; Long et al., 2009; Van Moerkercke et al., 2009).
Figure 1.
Proposed Biosynthetic Pathways Leading to the Formation of Benzoyl-CoA and Benzenoid/Phenylpropanoid Volatile Compounds in Petunia Flowers.
Solid arrows indicate established biochemical reactions, and broken arrows depict possible steps not yet described. Stacked arrows show the involvement of multiple enzymatic steps. Gray arrow shows a potential step. Volatile benzenoid/phenylpropanoid compounds are highlighted with a blue background. The CoA-dependent β-oxidative pathway leading to benzoyl-CoA formation is localized in peroxisomes and shown with a pink background. BA-CoA, benzoyl-CoA; BAlc, benzylalcohol; BAld, benzaldehyde; BALDH, benzaldehyde dehydrogenase; BB, benzylbenzoate; BPBT, benzoyl-CoA:benzylalcohol/2-phenylethanol benzoyltransferase; BSMT, BA/salicylic acid carboxyl methyltransferase; CA, cinnamic acid; CA-CoA, cinnamoyl-CoA; CFA, caffeic acid; CFA-CoA, caffeoyl-CoA; C3H, p-coumarate-3-hydroxylase; C4H, cinnamate-4-hydroxylase; CCoAOMT, caffeoyl-CoA 3-O-methyltransferase; 4CL1, 4-coumaroyl-CoA ligase; Eug, eugenol; FA, ferulic acid; FA-CoA, feruloyl-CoA; IEug, isoeugenol; 3H3PP, 3-hydroxy-3-phenylpropionic acid; 3H3PP-CoA, 3-hydroxy-3-phenylpropionyl-CoA; KAT, 3-ketoacyl-CoA thiolase; MB, methylbenzoate; 3O3PP-CoA, 3-oxo-3-phenylpropionyl-CoA; OMT, O-methyltransferase; PAAS, phenylacetaldehyde synthase; pCA, p-coumaric acid; pCA-CoA, p-coumaroyl-CoA; PEB, phenylethylbenzoate; PhA, phenylacetaldehyde; PhEth, 2-phenylethanol.
All proposed pathways leading to BA formation begin from the conversion of l-Phe to trans-cinnamic acid by action of the well-known enzyme Phe ammonia lyase (PAL) and diverge thereafter. The first step in the β-oxidative pathway is the formation of cinnamoyl-CoA, a reaction that could conceivably be catalyzed by a member of the 4-coumarate:CoA ligase family (4CL; EC 6.2.1.12). The 4CL enzymes in general have broad substrate specificity and convert a range of hydroxy- or methoxy-derivatives of cinnamic acid into the corresponding CoA thioesters (Hamberger and Hahlbrock, 2004). However, trans-cinnamic acid itself is a poor substrate for most plant 4CL enzymes so far characterized with exception of a 4CL2 from raspberry (Rubus idaeus), which shows a strong preference for trans-cinnamic acid (153% of activity with coumarate) (Kumar and Ellis, 2003). No cinnamate-specific ligase (CNL) has been reported yet.
To establish the routes by which benzenoid compounds are made in planta, we used petunia (Petunia hybrida) flowers as a model system, which emit large amounts of methylbenzoate, the product of BA methylation, as well as other volatiles containing benzoyl or benzyl moieties (Kolosova et al., 2001; Negre et al., 2003; Verdonk et al., 2003; Boatright et al., 2004). We previously identified and biochemically characterized enzymes responsible for the formation of some of the emitted benzenoids, including methylbenzoate (Negre et al., 2003), benzylbenzoate, and phenylethylbenzoate (Boatright et al., 2004). Using computer-assisted metabolic flux analysis in combination with in vivo stable isotope labeling, we previously showed that both the β-oxidative and nonoxidative pathways contribute to the formation of these benzenoid compounds (Orlova et al., 2006). Here, we report the isolation and characterization of cinnamate:CoA ligase involved in the first step of the β-oxidative pathway and show that this step is localized in the peroxisomes.
RESULTS
Isolation of Potential CoA Ligases Responsible for Cinnamoyl-CoA Formation
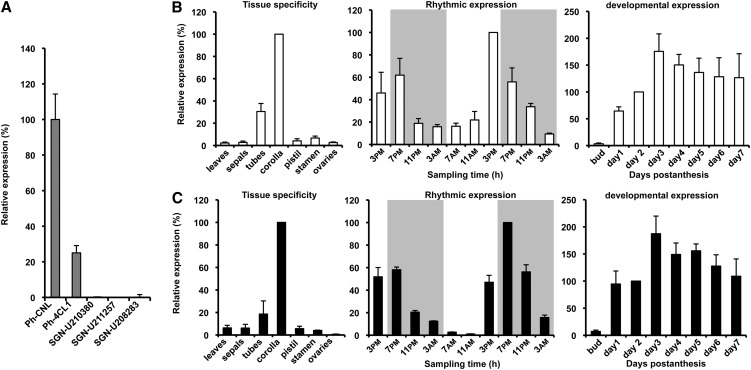
To isolate genes responsible for the formation of cinnamoyl-CoA, a tBLASTn search (threshold < e−10) of a petunia petal-specific EST database (Boatright et al., 2004) as well as the Sol genomics network (http://solgenomics.net) for sequences with homology to Arabidopsis 4CL1 (At1g51680) (Ehlting et al., 1999; Costa et al., 2005) was performed. The search of a petal-specific database revealed four ESTs representing transcripts of the same gene encoding a protein with high sequence similarity to known plant 4CLs, and four other ESTs representing a second gene that encodes a protein with a lower similarity to 4CLs but with higher similarity to other proteins in a superfamily of the acyl-activating enzymes (AAEs) that share a common reaction mechanism for forming an activated adenylate-substrate intermediate prior to esterification with CoA (Shockey and Browse, 2011). Based on the work presented below, we designated the first gene as Ph-4CL1 and the second as Ph-CNL (for P. hybrida cinnamate:CoA ligase). The search of Sol genomics network revealed three additional ESTs (SGN-U210380, SGN-U211257, and SGN-U208283) with high sequence similarity to plant 4CLs. Since expression of genes involved in formation of benzenoid compounds are very likely to be the highest in corollas, the scent-producing organ of petunia flowers (Boatright et al., 2004), we analyzed the expression of all five cDNA clones in this particular floral tissue of 2-d-old petunia flowers using quantitative RT-PCR (qRT-PCR) with gene-specific primers to select potential candidates. Out of the five potential candidates, three genes displayed low to no expression in corolla limbs (Figure 2A) with an unspecific expression pattern relative to floral scent formation (see Supplemental Figure 1 online) and were not further considered for a role in formation of benzenoid floral volatiles. On the other hand, 4CL1 and CNL were predominantly expressed in corollas and thus further analyzed (Figures 2B and 2C, left panels). Correlating with drastic nocturnal increases in phenylpropanoid/benzenoid internal and volatile emission pools in petunia corollas (Kolosova et al., 2001; Boatright et al., 2004), expression levels of both CNL and 4CL1 in corolla limbs changed rhythmically during a daily light/dark cycle, with a maximum at 3 to 7 pm (Figures 2B and 2C, middle panels). Over flower development, these genes also exhibited similar expression patterns, positively correlating with levels of produced benzenoid compounds (Figures 2B and 2C, right panels).
Figure 2.
Expression Profiles of Putative 4CL Candidates as well as CNL and 4CL1 in Petunia.
All transcript levels were determined by qRT-PCR and obtained relative to the reference gene (elongation factor 1-α). Data are means ± se (n = 3 biological replicates). Expression of SGN-U210380, SGN-U211257, SGN-U208283, CNL, and 4CL1 in corollas of flowers collected on day 2 postanthesis at 3 pm is shown relative to the highest expressed candidate, CNL (A). Tissue-specific expression of CNL (B) and 4CL1 (C) is shown relative to the corresponding transcript levels in corolla ([B] and [C], left panels). Rhythmic changes in CNL and 4CL1 mRNA expression in corollas of flowers 1 to 3 d postanthesis during a normal light/dark cycle are shown relative to the corresponding transcript levels on day 2 postanthesis at 3 pm for CNL and 7 pm for 4CL1 (middle). Gray and white areas correspond to dark and light periods, respectively. Developmental changes in CNL and 4CL1 transcript levels in corollas from mature buds to day 7 postanthesis are shown relative to the corresponding levels on day 2 postanthesis (far right).
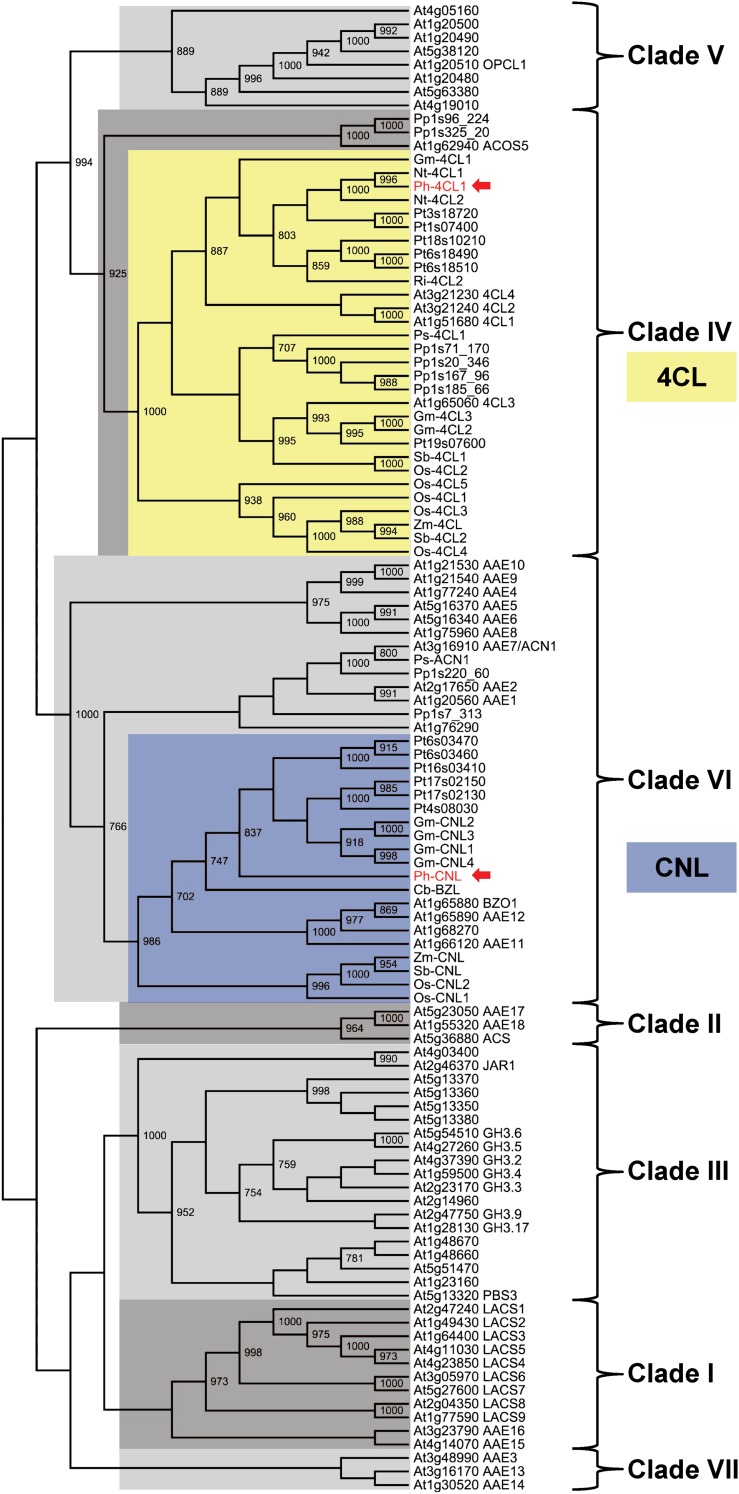
To predict and distinguish Ph-4CL1 and Ph-CNL functions further, we reconstructed their phylogeny within the AAE superfamily, initially using the defined Arabidopsis members (Shockey and Browse, 2011) to serve as the framework for the alignment. The alignment was then populated using the most closely related proteins to Ph-4CL1 and Ph-CNL based on homology searches and previous biochemical characterization of enzymes (see Supplemental Table 1 online). The resulting neighbor-joining tree revealed that Ph-4CL1 and Ph-CNL belong to strikingly different subfamilies (Figure 3). Ph-4CL1 is a member of clade IV, which primarily contains 4CL enzymes and exhibits ∼90% identity to tobacco (Nicotiana tabacum) 4CL1 and 4CL2 proteins (Lee and Douglas, 1996), as well as ∼70% identity to three characterized Arabidopsis 4CL enzymes (Ehlting et al., 1999). By contrast, Ph-CNL belongs to clade VI and is closely related to benzoyl-CoA ligase/benzoyloxyglucosinolate (BZL/BZO) proteins from Clarkia breweri (61% identical and 74% similar), whose flowers emit high levels of benzenoid compounds (Raguso and Pichersky, 1995; Dudareva et al., 1998), and Arabidopsis (At1g65880; 60% identical and 73% similar), recently shown to have CoA ligase activity with BA (Kliebenstein et al., 2007). Thus, the phylogenetic analysis suggested that Ph-CNL may have a distinct function from Ph-4CL1, and based on its relatedness to C. breweri BZL and Arabidopsis BZO1 (Figure 3), might be involved in the β-oxidative pathway of benzenoid biosynthesis.
Figure 3.
Ph-4CL1 and Ph-CNL Belong to Distantly Related Subfamilies within the AAE Superfamily Defined by Shockey and Browse (2011).
A nonexhaustive BLASTp search was performed to retrieve additional Ph-4CL1 and Ph-CNL orthologs. Protein sequences were aligned using ClustalO (version 1.0.3; Sievers et al., 2011; see Supplemental Data Set 1 online). The resulting alignment was used to generate a neighbor-joining cladogram with the PHYLIP package (version 3.69; Felsenstein, 2009). Bootstrap values (1000 replicates) are indicated at branch points with ≥70% support. See Supplemental Table 1 online for sequence accession numbers. Species abbreviations: At, Arabidopsis; Cb, C. breweri; Gm, G. max; Nt, N. tabacum; Os, Oryza sativa; Ph, P. hybrida; Pp, P. patens; Ps, P. sitchensis; Pt, P. trichocharpa; Ri, R. idaeus; Sb, Sorghum bicolor; Zm, Zea mays.
Functional Characterization of Ph-4CL1 and Ph-CNL Proteins
For functional characterization of the isolated potential CoA ligases, the coding region of each gene was expressed in Escherichia coli as an inducible fusion protein containing a hexahistidine (His)6-tag. Both active enzymes are monomers as was determined by comparisons of the molecular masses of recombinant proteins obtained by gel filtration chromatography (59 and 63 kD for 4CL1 and CNL, respectively) with the calculated protein masses. Affinity-purified recombinant proteins were tested with several substrates, including cinnamic acid and several of its hydroxy- or methoxy-derivatives, for their ability to form the corresponding CoA thioesters. Whereas both 4CL1 and CNL catalyzed the formation of CoA esters of cinnamic acid, 4-coumaric acid, and caffeic acid, only 4CL1 was also able to use ferulic and BA as substrates (Table 1). The maximum enzyme activities with cinnamic acid as a substrate were observed at pH 8.5 and 8.0 for 4CL1 and CNL, respectively.
Table 1.
Kinetic Parameters of Ph-4CL1 and Ph-CNL Proteins
| Enzyme | Substrate (Acid) | Km (µM) | Vmax (pkat mg−1) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|---|
| Ph-4CL1 | t-Cinnamic | 149.4 ± 13.7 | 14680 ± 409 | 0.847 ± 0.02 | 6.43 ± 0.86 |
| 4-Coumaric | 16.4 ± 3.0 | 25797 ± 1994 | 1.488 ± 0.12 | 84.32 ± 9.00 | |
| Caffeic | 47.8 ± 5.4 | 63200 ± 2020 | 3.645 ± 0.12 | 70.14 ± 5.95 | |
| Ferulic | 15.2 ± 2.7 | 41377 ± 879 | 2.39 ± 0.05 | 145.37 ± 27.21 | |
| Benzoic | 9008 ± 1795 | 95.9 ± 8.5 | 0.0007 ± 0.0001 | 0.0008 ± 0.0001 | |
| CoA | 9.9 ± 3.1 | 8451 ± 4017 | 0.487 ± 0.23 | 49.46 ± 6.51 | |
| Ph-CNL | t-Cinnamic | 285.7 ± 63.9 | 7401.7 ± 117.2 | 0.472 ± 0.01 | 1.82 ± 0.36 |
| 4-Coumaric | 550.4 ± 127.5 | 3890.7 ± 460.1 | 0.248 ± 0.03 | 0.512 ± 0.14 | |
| Caffeic | 1042.4 ± 95.9 | 3742.7 ± 3.6 | 0.238 ± 0 | 0.232 ± 0.02 | |
| Ferulic | n.d. | n.d. | n.d. | n.d. | |
| Benzoic | n.d. | n.d. | n.d. | n.d. | |
| CoA | 775.2 ± 34.3 | 7179.7 ± 126.8 | 0.458 ± 0.01 | 0.607 ± 0.06 |
All values represent mean ± se, n = 3; n.d., not determined, below detection limit.
Determination of the kinetic parameters of 4CL1 revealed that the apparent Km values for 4-coumaric and ferulic acids were very similar (16.4 and 15.2 μM, respectively) and threefold lower than that for caffeic acid (47.8 μM; Table 1). The 4CL1 apparent Km value for cinnamic acid was 149.4 μM, and the enzyme could also activate BA to its thioester, but with an apparent Km value of 9 mM (Table 1). The highest catalytic efficiency (kcat/Km ratio) of the 4CL1 enzyme was found toward ferulic acid, followed by 4-coumaric acid and caffeic acid. Its catalytic efficiency toward cinnamic acid was more than 20-fold lower than that with ferulic acid. The ability of Ph-4CL1 to convert 4-coumaric, caffeic, and ferulic acids to the corresponding CoA esters suggests that this enzyme is a bona fide 4CL.
Determination of the kinetic parameters of Ph-CNL showed that the enzyme has higher affinity toward cinnamic acid than 4-coumaric acid and caffeic acid, with the corresponding apparent Km values of 285, 550, and 1042 μM. Its apparent catalytic efficiency with cinnamic acid was 3.6 and 8 times higher than that with 4-coumaric acid and caffeic acid, respectively, indicating that trans-cinnamic acid is the preferred substrate. Thus, we designated this enzyme CNL, for cinnamate:CoA ligase. Time-of-flight liquid chromatography–mass spectrometry (LC/TOF-MS) analysis confirmed that the product formed from cinnamic acid by Ph-CNL, as well as by Ph-4CL1, had identical mass and retention characteristics as cinnamoyl-CoA standard synthesized using Nicotiana tabacum 4CL (Beuerle and Pichersky, 2002a) (see Supplemental Figure 2 online).
The Ph-CNL activity with cinnamic acid was strictly dependent on K+ ions with half the maximal activity observed at 10.5 mM KCl. The absence of K+ ions during both protein purification and assays resulted in complete loss of enzyme activity with cinnamic acid and could only be restored by the addition of K+, but not Na+, to the assay buffer (∼24% of the activity was restored). This K+ dependency was previously observed with 3-hydroxypropionate CoA synthase from Chloroflexus aurantiacus (Alber and Fuchs, 2002), acetyl-CoA synthase from Acetobacter aceti (O’Sullivan and Ettlinger, 1976), and butyryl-CoA synthase from Paecilomyces varioti (Takao et al., 1987), all of which also require K+ for their activities in addition to Mg2+ (Alber and Fuchs, 2002). The lack of K+ during protein purification and assays also had a severe effect on the Ph-CNL activity with 4-coumaric acid, and to a lesser extent with caffeic acid, reducing them by 73 and 14%, respectively.
Ph-CNL and Ph-4CL1 display drastic differences in their catalytic efficiency with 4-coumaric acid and caffeic acid, which were 165- and 305-fold higher for the 4CL1 protein, respectively (Table 1). Moreover, although cinnamic acid was the preferred substrate for Ph-CNL, the apparent catalytic efficiency of Ph-CNL with this substrate was 3.5-fold lower than that of Ph-4CL1. Furthermore, Ph-CNL had lower affinity toward CoA than Ph-4CL1 with apparent Km values of 775 and 9.9 μM, respectively.
Effect of RNA Interference–Mediated Suppression of 4CL1 and CNL Expression on Benzenoid Production
Since the overall 4CL1 and CNL expression profiles and enzyme substrate specificities did not provide us with sufficient information to differentiate which gene product(s) potentially contributes to the benzenoid volatile formation in petunia flowers, transcript levels of 4CL1 and CNL were downregulated in P. hybrida cv Mitchell using an RNA interference (RNAi) strategy to elucidate the specific role of each enzyme in planta. RNAi constructs were generated using 431- and 473-bp gene-specific fragments within the 5′ ends of the 4CL1 and CNL coding regions, respectively, and expressed under the control of the petal-specific LIS promoter (Cseke et al., 1998). Twenty-five and 16 independent transformants were obtained for 4CL1 and CNL, respectively. Multiple independent lines for each gene with the greatest reduction in target gene expression (>60% reduction) were selected for detailed metabolic profiling of emitted floral volatiles as well as internal pools of intermediates and end products (Figure 4; see Supplemental Figure 3 online). It is noteworthy that CNL silencing did not affect 4CL1 expression and vice versa (see Supplemental Figure 4 online).
Figure 4.
Effect of CNL-RNAi Suppression on CNL Expression, Emission, and Internal Pools of Benzenoid/Phenylpropanoid Compounds in Corollas of Petunia Flowers.
CNL mRNA levels were determined by qRT-PCR in corollas of control (C; white bars) and three independent CNL RNAi lines (1, 3, and 17, black bars) harvested at 3 pm, 2 d postanthesis (top left corner). Expression values for transgenic lines are shown as a percentage of CNL expression in control petals, which is set at 100%. Data are means ± se (n = 3 biological replicates). Emission rates of volatile compounds are shown in the graphs with white backgrounds, while internal pools for the corresponding volatiles are shown in the graphs with gray backgrounds. Scent collections were performed from detached flowers 2 d postanthesis for 12 h starting at 8 pm. Emission rates are expressed on a per hour basis assuming a constant emission rate over the 12-h period. Tissue for internal pools was collected at 8 pm. Data are means ± se (n ≥ 10 biological replicates). *P < 0.01 and **P < 0.05 by Student's t test of transgenics relative to control. FW, fresh weight.
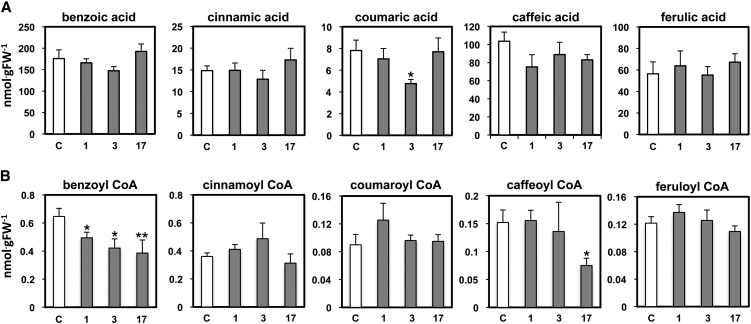
The decrease in CNL transcript levels led to a reduction in emission of benzylbenzoate and phenylethylbenzoate by ∼27 and ∼77% (on average), respectively (Figure 4), both of which require benzoyl-CoA for their biosynthesis (Boatright et al., 2004). These transgenic flowers emitted 72 and 27% more benzylalcohol and phenylethanol, the other two acyl acceptors for benzylbenzoate and phenylethylbenzoate formation, respectively. The Ph-CNL-RNAi flowers also showed a slight but significant decrease (12%) in methylbenzoate emission with an exception of unaltered level in transgenic line 3 relative to control flowers, while emissions of benzaldehyde, phenylacetaldehyde, eugenol, and isoeugenol remained unchanged (Figure 4). In general, the levels of emitted volatiles in transgenic and control plants positively correlated with their corresponding internal pools, with the exception of benzylalcohol and phenylethanol, whose internal pools were not increased in transgenic flowers (Figure 4). The activities of benzoyl-CoA:benzylalcohol/phenylethanol benzoyltransferase (Boatright et al., 2004) and BA/salicylic acid carboxyl methyltransferase (Negre et al., 2003; Underwood et al., 2005) responsible for benzylbenzoate/phenylethylbenzoate and methylbenzoate formation, respectively, as well as other scent-producing enzymes and 4CL, were unchanged in Ph-CNL-RNAi transgenic corollas when compared with control flowers (see Supplemental Table 2 online). A decrease in CNL expression had no effect on the internal pools of free hydroxycinnamic acids (Figure 5A) but led to a 38% (on average) decrease in the level of benzoyl-CoA, the final product of the β-oxidative pathway (Figure 5B). CoA esters of other hydroxycinnamic acids remained unchanged in the Ph-CNL-RNAi transgenic flowers (Figure 5B).
Figure 5.
Effect of CNL-RNAi Suppression on Internal Pools of Free Hydroxycinnamic Acids and Their Corresponding CoA Esters in Corollas of Petunia Flowers.
Free organic acids (A) and hydroxicinnamoyl-CoA esters (B) were extracted from corollas of control (white bars) and CNL-RNAi lines (gray bars) 2 d postanthesis harvested at 8 pm. Data are means ± se (n ≥ 6 biological replicates). *P < 0.01 and **P < 0.04 by Student’s t test of transgenics relative to control. C, control; FW, fresh weight.
RNAi suppression of 4CL1 gene expression by 60% reduced the activity of 4CL1 enzyme in crude extracts from transgenic petals by 61% (38.26 ± 7.35 and 15.05 ± 4.15 pkat/mg protein in control and transgenics, respectively). Ferulic acid was used as a substrate in these assays to distinguish between 4CL1 and CNL enzymes since the latter shows no activity with this substrate (Table 1). The decrease in 4CL1 transcript levels had no effect on emission of benzenoid compounds (see Supplemental Figure 3 online).
Subcellular Localization of Ph-4CL1 and Ph-CNL Proteins
A protein targeting prediction program, PSORT, predicted that Ph-CNL is a peroxisomal protein due to a putative peroxisomal targeting signal 1 (PTS1) AKL at the C terminus (Reumann, 2004), whereas Ph-4CL1 has no such targeting signal and is predicted to localize in the cytosol. To determine experimentally the subcellular localization of Ph-CNL, the CNL coding region, including the putative PTS1, was fused to the C-terminus of the coding sequence of the green fluorescent protein (GFP) reporter. This construct was transferred to Arabidopsis protoplasts, and the localization of the corresponding transient GFP expression was analyzed by confocal laser scanning microscopy (Figure 6). The GFP-Ph-CNL protein exhibited a punctate fluorescence pattern that was expected for proteins localized in the peroxisomes (Figures 6A and 6D). The overlay of chloroplast autofluorescence measured at 637 nm using a far-red filter (displayed in blue color in Figures 6C and 6F) demonstrated that GFP was not localized in the chloroplasts.
Figure 6.
Subcellular Localization of Ph-CNL Protein.
Schematic diagram of the GFP fusion constructs is shown on the left, and the corresponding transient expression in Arabidopsis protoplasts detected by confocal laser scanning microscopy is shown on the right. GFP fluorescence is shown in green and chlorophyll autofluorescence in blue. Mitochondria were stained with MitoTracker-Red to distinguish them from peroxisomes and are shown in red with the exception of (E). In (E), YFP fluorescence of OPCL1 [3-oxo-2-(2′-[Z]-pentenyl)cyclopentane-1-octanoic acid (OPC-8:0) CoA Ligase1] used as a peroxisomal marker is shown in red. Merged panel shows combined fluorescence from GFP, mitochondria, chloroplasts, and peroxisomes. Numbers in the fusion constructs correspond to amino acids positions. ARL is the peroxisomal targeting signal found in Ph-CNL. For each construct, results of one of the two independent transformations are presented. Bars = 50 μm.
To distinguish between peroxisomes and mitochondria, and to exclude the possibility that the observed fluorescence comes from mitochondrial targeting, mitochondria were first stained with MitoTracker-Red in the samples transformed with GFP-Ph-CNL (Figure 6B). There was no overlap between the GFP and the red color of MitoTracker, providing evidence that the Ph-CNL protein is not localized in the mitochondria (Figure 6C). Moreover, Ph-CNL protein was colocalized with Arabidopsis AAE OPCL1, which is known to localize in peroxisomes (Koo et al., 2006) (Figures 6E and 6F), thus confirming the peroxisomal localization of Ph-CNL protein. To verify whether the PTS1 ARL motif in the C terminus of Ph-CNL protein is responsible for this localization, truncated Ph-CNL with the tripeptide ARL deleted from the C terminus was fused to GFP. Elimination of the tripeptide ARL changed the subcellular localization of the protein from the peroxisomes to the cytosol (Figures 6G to 6I). In addition, fusion of the GFP to the C terminus of the Ph-CNL protein blocked the PTS1 and abolished peroxisomal targeting, leading to cytosolic localization of the resulting fusion protein (Figures 6J to 6L). By contrast, the Ph-4CL1 fusion protein was always localized in the cytosol independent of the position of GFP at the N or C terminus of the Ph-4CL1 coding region (Figures 7A to 7F), confirming its predicted cytosolic localization.
Figure 7.
Subcellular Localization of Petunia 4CL1.
Schematic diagram of the GFP fusion constructs is shown on the left, and the corresponding transient expression in protoplast is shown on the right. GFP fluorescence is shown in the “Green” panels, and “Red” panels show chloroplast autofluorescence. “Merged” panels show combined fluorescence from GFP and chloroplasts. Plasmid p326-sGFP was used as a control for cytosolic localization. Bars = 50 μm.
To confirm biochemically the peroxisomal localization of Ph-CNL (Figure 6), we measured CoA-ligase activity in peroxisomes prepared from petunia flowers (see Supplemental Table 3 online). Similar to recombinant Ph-CNL (Table 1), the peroxisomal extracts had no detectable activity toward ferulic acid (marker activity for 4CL). Additionally, activity toward cinnamic acid in peroxisomes was 2.4-fold higher relative to 4-coumaric acid (81.8 ± 7.3 pkat/mg protein versus 34.2 ± 5.5 pkat/mg protein, respectively), which was very close to the ratio of these activities for the recombinant Ph-CNL protein (Vmax with cinnamic acid/Vmax with coumaric acid = 1.9; Table 1). Moreover, CNL activity was reduced by 67% in peroxisomes isolated from petals of the CNL-RNAi transgenic plants (27 pkat/mg protein) when compared with that in control flowers (82 pkat/mg protein), consistent with the 60% reduction of CNL mRNA expression in the transgenic lines. Given the peroxisomal localization of Ph-CNL, we also analyzed whether it can be involved in the β-oxidation of fatty acids. Short-, medium-, and long-chain fatty acids were tested as substrates using recombinant Ph-CNL and none of them were converted to the corresponding CoA ester to any significant extent (see Supplemental Table 4 online).
DISCUSSION
Differences and Similarities between Petunia 4CL1 and CNL Enzymes
BA biosynthesis in plants remains an important unresolved question despite numerous attempts at its elucidation (Wildermuth, 2006). Several lines of evidence have been presented that peroxisomal β-oxidation contributes to BA biosynthesis in plants. For example, Arabidopsis plants carrying the chy1 mutation, which diminishes β-oxidation in peroxisomes, have reduced levels of benzoylated glucosinolates in the seeds (Ibdah and Pichersky, 2009), and petunia flowers with decreased expression of a gene encoding peroxisomal keto-acyl thiolase have reduced levels of volatiles formed from benzoyl-CoA (Van Moerkercke et al., 2009).
The first committed step in the proposed β-oxidative pathway of BA biosynthesis from Phe is the formation of the cinnamoyl-CoA intermediate, which requires a CoA ligase for its biosynthesis (Figure 1). Bona fide 4CL enzymes with differential activities toward a variety of hydroxy- and methoxy-substituted cinnamic acids have been functionally characterized from numerous plant species, including Arabidopsis (Ehlting et al., 1999; Hamberger and Hahlbrock, 2004; Costa et al., 2005), tobacco (Lee and Douglas, 1996), soybean (Glycine max; Lindermayr et al., 2002), parsley (Petroselinum crispum; Lozoya et al., 1988), aspen (Populus tremuloides; Hu et al., 1998), poplar (Populus trichocarpa × Populus deltoides; Allina et al., 1998), and raspberry (Kumar and Ellis, 2003). However, most of them showed little or no activity toward cinnamic acid itself. A search for cinnamoyl-CoA ligase in petunia flowers identified five potential CoA ligases, two of which (Ph-4CL1 and Ph-CNL) displayed expression profiles consistent with production of volatile benzenoid compounds (Figure 2). In contrast to bona fide plant 4CLs, and similar only to the raspberry 4CL2 (Kumar and Ellis, 2003), both enzymes can activate cinnamic acid to the corresponding CoA ester. However, the apparent catalytic efficiency is higher for Ph-4CL1 than for Ph-CNL (Table 1). Although Ph-4CL1 uses cinnamic acid as a substrate, its catalytic efficiency is lower than that with ferulic, 4-coumaric, and caffeic acids (Table 1). By contrast, Ph-CNL has strong preference for cinnamic acid and complete lack of activity with ferulic acid, the preferred substrate for Ph-4CL1, both of which make it distinct from bona fide 4CLs.
Despite overlapping substrate specificities, albeit with significant differences in their enzymatic properties (Table 1), Ph-4CL1 and Ph-CNL share only 25% amino acid identity. Moreover, they belong to two phylogenetically distinct subfamilies of the AAE superfamily (Figure 3; Shockey and Browse, 2011), indicating convergent evolution of function. Whereas Ph-4CL1 is a member of the well-characterized 4CL subfamily (clade IV, Figure 3; Shockey and Browse, 2011), which includes enzymes responsible for the formation of CoA thioesters used for biosynthesis of numerous phenylpropanoid compounds, Ph-CNL belongs to a plant-specific subfamily of CoA ligases with mostly unknown functions (clade VI, Figure 3; Shockey and Browse, 2011). The closest homologs of Ph-CNL are C. breweri BZL and Arabidopsis BZO1, the latter shown to convert BA to benzoyl-CoA (Kliebenstein et al., 2007). The phylogenetic proximity to Ph-CNL suggests that At-BZO1 should be reinvestigated for activity with cinnamic acid, especially in light of the requirement of K+ ion for catalysis (Table 1). The coexistence of 4CL and CNL types of proteins does not appear to be unique to petunia as both homologs are present throughout dicots and monocots (Figure 3). Interestingly, the closest Ph-CNL homologs in Physcomitrella patens (Pp1s220_60, Pp1s7_313; moss with sequenced genome available; Banks et al., 2011) and Picea sitchensis (Ps-ACN1; conifer with >6.464 full-length cDNA available; Ralph et al., 2008) are more closely related to the Arabidopsis acetyl-CoA ligase ACN1 (Figure 3), which uses acetate as a substrate (Turner et al., 2005). This may suggest that CNL does not exist outside of flowering plants. Therefore, if nonflowering plants use the β-oxidative pathway for BA biosynthesis, then they likely do not use CNL enzymes.
Peroxisomal CoA Ligase Is Responsible for the First Committed Step in BA Biosynthesis via the β-Oxidative Pathway
Another major difference between the isolated enzymes is their subcellular localization. Ph-4CL1 is cytosolic (Figure 7), while Ph-CNL is present in the peroxisomes (Figure 6). Ph-CNL has an apparent Km value for CoA 78-fold higher than that of Ph-4CL1 (Table 1). This may indicate that free CoA levels are high in peroxisomes, which is consistent with this organelle being the site of fatty acid β-oxidation and containing predicted high levels of acetyl-CoA (Perera et al., 2009). In addition, a low affinity of Ph-CNL for cinnamic acid relative to the affinity of bona fide 4CL for their substrates (Table 1) suggests that there is an uptake mechanism of cinnamic acid into peroxisomes allowing efficient Ph-CNL catalysis as no peroxisomal PAL isoform has been discovered.
Ph-4CL1 and Ph-CNL have similar expression profiles (Figure 2), and the corresponding enzymes are able to activate cinnamic acid, with the apparent catalytic efficiency of Ph-CNL being 3.5-fold lower than that of Ph-4CL1 (Table 1). However, downregulation of Ph-CNL expression, but not of Ph-4CL1 expression, reduces the emission of benzylbenzoate and phenylethylbenzoate (Figure 4), both of which require benzoyl-CoA, and in effect the upstream intermediate cinnamoyl-CoA, for their biosynthesis (Figure 1). This reduction in emission was consistent with a 38% decrease in the internal pool of benzoyl-CoA in the petals of Ph-CNL-RNAi transgenic plants (Figure 5). There were no changes in internal pools of cinnamic acid and cinnamoyl-CoA in Ph-CNL-RNAi petals (Figure 5), which is likely due to a redundancy in the cinnamoyl-CoA ligase activities. Ph-4CL1, similar to Ph-CNL, can convert cinnamic acid to cinnamoyl-CoA (Table 1; see Supplemental Figure 2 online); however, this reaction takes place in the cytosol (Figure 7). Since the measured pool represents a total cinnamoyl-CoA level in the cell and cannot distinguish between cytosolic and peroxisomal pools, the lack of change is not surprising.
Metabolic flux analysis of the benzenoid network in petunia flowers showed that benzoyl-CoA is synthesized primarily via the β-oxidative pathway with a small contribution of flux from BA formed through the non-β-oxidative pathway (Boatright et al., 2004). In addition, feeding of deuterium-labeled benzylalcohol confirmed existence of this small flux from BA to benzoyl-CoA (benzylalcohol → benzaldehyde → BA → benzoyl-CoA; Boatright et al., 2004). If RNAi downregulation of Ph-CNL redirects flux from trans-cinnamic acid to BA via the non-β-oxidative pathway, thereby increasing its contribution to benzoyl-CoA formation, it might explain the observed discrepancy between reduction in the benzoyl-CoA internal pool and Ph-CNL activity in Ph-CNL-RNAi petunia flowers (38 and 67%, respectively).
Metabolic control analysis of the benzenoid network recently conducted in petunia flowers revealed that only the enzymes involved in benzylbenzoate biosynthesis and degradation determine flux toward benzylbenzoate and can significantly alter its emission (Colón et al., 2010). Indeed, RNAi downregulation of 3-ketoacyl-CoA thiolase1, which likely converts 3-oxo-3-phenylpropionyl-CoA to benzoyl-CoA in the β-oxidative pathway of BA biosynthesis, resulted in a significant reduction of benzylbenzoate and phenylethylbenzoate in transgenic petunia flowers (Van Moerkercke et al., 2009). Since Ph-CNL is unable to convert BA to benzoyl-CoA (Table 1), our results suggest that this enzyme can only be involved in benzoyl-CoA formation via the β-oxidative pathway in peroxisomes by catalyzing formation of cinnamoyl-CoA in planta (Figure 1).
Previously, it was shown that Arabidopsis lines carrying mutations in the BZO1 gene are deficient in the formation of benzoyloxyglucosinolates in their seeds. Analysis of At-BZO1 activity revealed that it possesses BA:CoA ligase activity (Kliebenstein et al., 2007). Based on these results, it was suggested that At-BZO1 acts as a BA:CoA ligase in planta to produce the substrate that is used by a BAHD-type acyltransferase to synthesize benzoylated glucosinolates (Kliebenstein et al., 2007). However, At-BZO1 was not tested with cinnamic acid nor was BA reported to accumulate in bzo1 mutant seeds. Interestingly, Arabidopsis BZOI also contains a PTS1 peroxisomal targeting sequence (Kliebenstein et al., 2007), while biosynthesis of glucosinolates most likely occurs in the cytosol. Thus, the data from the analysis of the Arabidopsis bzo1 mutants, taken together with the data presented here, are consistent with Arabidopsis BZO1 potentially acting as cinnamic acid:CoA ligase. Therefore, Arabidopsis bzo1 mutants would not be able to produce cinnamoyl-CoA and thus be deficient in the synthesis of BA via the β-oxidation pathway, leading to a lack of BA needed for the benzoylation of glucosinolates. Direct biochemical characterization of At-BZO1 together with metabolic analysis of the compounds accumulating in bzo1 mutants will test this hypothesis.
Reduction of CNL activity in transgenic petunia flowers also affected emission of methylbenzoate, which is the result of the methylation of BA, but to a lesser extent when compared with benzylbenzoate and phenylethylbenzoate (Figure 4). The rather slight (∼12%) reduction in methylbenzoate emission could be the result of a combination of two factors: a decrease in the endogenous pool of benzylbenzoate that was previously shown to be an intermediate in BA formation from Phe in petunia flowers (Boatright et al., 2004; Orlova et al., 2006) and redirection of the flux from trans-cinnamic acid to BA via the non-β-oxidative pathway.
Whereas benzoyl-CoA formation occurs in the peroxisomes (Figure 6; Van Moerkercke et al., 2009), biosynthesis of benzylbenzoate and phenylethylbenzoate, for which benzoyl-CoA is the immediate precursor, takes place in the cytosol (Boatright et al., 2004). Current evidence in mammals and yeast indicates that acyl-CoA esters are imported across the peroxisomal membrane via a family of ATP binding-cassette transporters (Hettema et al., 1996; Verleur et al., 1997; Hunt and Alexson, 2008). However, to date there is no evidence for transport of acyl-CoA esters out of peroxisomes.
Taken together, our results show that although Ph-4CL1 and Ph-CNL have very similar apparent catalytic efficiency toward cinnamic acid and similar expression profiles, only Ph-CNL is located in the peroxisomes and therefore is the CoA ligase that directs cinnamic acid into the β-oxidative pathway producing benzoyl-CoA and its derivatives. It remains to be determined how the peroxisomally synthesized benzoyl-CoA is exported into the cytosol and eventually used in the synthesis of benzenoid volatiles in petunia.
METHODS
Plant Material
Petunia hybrida cv Mitchell (Ball Seed) was used as genetic background for the production of 4CL1 and CNL RNAi transgenic petunia plants. Control and transgenic plants were grown under normal greenhouse conditions (Dudareva et al., 2000) and in vitro to provide material of the same age and physiological conditions for experiments.
Phylogenetic Analysis
Amino acid sequences were aligned using the ClustalO program (http://www.ebi.ac.uk/Tools/msa/clustalo/; Sievers et al., 2011). An unrooted neighbor-joining cladogram with bootstrap support (1000 replicates) was constructed using the PHYLIP package (version 3.69; Felsenstein, 2009) and visualized with FigTree (version 1.3.1; http://tree.bio.ed.ac.uk/software/figtree/). Default parameters were used for all applications.
Expression and Purification of Recombinant Proteins
The coding region of petunia 4CL1 was amplified by PCR using the forward and reverse primers 5′-AACCCATATGCCGATGGAGACTGAAAC-3′ and 5′-GGCATATGTTAATTTGGAACACCAGCTGC-3′, respectively, and subcloned into the NdeI site of the expression vector pET-28a containing a cleavable N-terminal (His)6-tag (Novagen). The coding region of Ph-CNL was amplified using the forward and reverse primers 5′-ATCATATGGACGAGTTACCAAAATGTGGAGC-3′ and 5′-ATGCGGCCGCCAGACGAGCTGGCAAATCAAG-3′, respectively, and subcloned into the NdeI-NotI site of the pET-32a expression vector in frame and upstream of the (His)6-tag. Sequencing revealed no errors had been introduced during PCR amplifications.
For functional expression, Escherichia coli Rosetta cells were transformed with recombinant plasmids and corresponding pET vectors without insert as controls and grown in Luria-Bertani medium with 50 μg/mL kanamycin for pET28a or 100 μg/mL ampicilin for pET32a at 37°C. When the cultures’ density reached A600 = 0.5, the expressions of Ph4-CL1 and Ph-CNL were induced by addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.4 mM. After a 16-h incubation on a rotary shaker (200 rpm) at 18°C, the E. coli cells were harvested by centrifugation and resuspended in buffer containing 50 mM potassium phosphate, pH 8.0, 500 mM NaCl, 10% (v/v) glycerol, 1 mM EDTA, and 7 mM β-mercaptoethanol for Ph-4CL1 and 50 mM potassium phosphate, pH 8.0, 150 mM KCl, 20% (v/v) glycerol, and 1 mM phenylmethanesulfonyl fluoride for Ph-CNL. Cells were lysed by the addition of lysozyme (0.5 mg/mL) and DNase I (10 μg/mL in 4 mM MgCl2, final), incubated on ice for 30 min, and sonicated for 3 min. Extracts were centrifuged for 30 min (20,000g, 4°C), and the recombinant enzymes were purified by affinity chromatography on prepacked Ni Sepharose 6 Fast Flow columns (HisTrap FF crude 1 mL column; GE Healthcare) using FPLC (ÄKTA, GE Healthcare Bio-Sciences) at 0.5 mL/min flow rate. Columns were washed by a step gradient of 50 mL 20 mM imidazole followed by 50 mL 40 mM imidazole, both in a buffer containing 50 mM potassium phosphate, pH 8.0, and 500 mM NaCl. The (His)6-tagged enzymes were eluted by 500 mM imidazole in the same buffer. Eluted fractions (3 mL each) with the highest protein levels were desalted on Econo-Pac 10 DG columns (Bio-Rad) into the buffer containing 20 mM potassium phosphate, pH 8.0, and 20% (v/v) glycerol. Desalted enzymes were aliquoted, frozen in liquid N2, and stored at −80°C until use. Protein purity was determined by densitometry of the SDS-PAGE gels after Coomassie Brilliant Blue staining and ranged from 60% for Ph-4CL1 to 85% for Ph-CNL, which was taken into account for determination of kcat values. The protein concentration was determined by the Bradford method (Bradford, 1976).
Enzyme Assays
CoA ligase activity with trans-cinnamic acid was determined radiochemically and spectrophotometrically and gave comparable results. The standard radiochemical assay reaction (100 μL total volume) contained 2.7 μg Ph-CNL and 13.2 nCi [U-14C] cinnamic acid in 50 mM Tris/K phosphate/Na citrate, pH 8.0, 2.5 mM ATP, 2.5 mM MgCl2, 50 mM KCl, and 2 mM CoA. Enzymatic reactions were initiated by addition of CoA and incubated for 20 min at room temperature. The reactions were terminated by adding 5 μL 50% trichloroacetic acid, and unincorporated substrate was removed by extraction with 180 μL ethyl acetate three times. Radiochemical incorporation rates were determined by scintillation counting 50 μL of the aqueous phase. Controls included assays containing all reaction components in the presence of boiled proteins or without proteins added. For Ph-4CL1, the reaction mixture contained 6.3 μg of purified protein, 13.2 nCi [U-14C] cinnamic acid in 50 mM Tris/K phosphate/Na citrate buffer, pH 8.5, 2.5 mM ATP, 5 mM MgCl2, and 0.4 mM CoA. [U-14C] cinnamic acid was enzymatically synthesized from [U-14C] Phe (specific activity 370 mCi/mmol; American Radiolabeled Chemicals) with PAL (EC 4.3.1.5; Sigma-Aldrich) as previously described (Beuerle and Pichersky, 2002b).
The standard spectrophotometric assays (500 μL) for CoA ligase activity with trans-cinnamic acid and its derivatives were performed in the same conditions as radiochemical assays only with nonradioactive substrates and contained from 2.47 to 24.7 μg of Ph-4CL1 or 6.0 μg of Ph-CNL. Reactions were initiated by adding organic acids (trans-cinnamic acid, p-coumaric acid, caffeic acid, and ferulic acid), and formation of the corresponding CoA esters was determined by monitoring the increase in absorption maximum at wavelengths of 311, 333, 346, and 345 nm for cinnamoyl-CoA, p-coumaroyl-CoA, caffeoyl-CoA, and feruloyl-CoA, respectively (Lee et al., 1997). The extinction coefficients 22.000, 21.000, 18.000, and 19.000 mL mmol−1cm−1 were used for calculations of cinnamoyl-CoA, p-coumaroyl-CoA, caffeoyl-CoA, and feruloyl-CoA esters formed, respectively (Lee et al., 1997).
For CoA ligase activity with BA, radiochemical assays were performed in 100 μL of assay buffer containing 50 mM Na phosphate, pH 7.5, 6.3 μg of purified Ph-4CL1, 2.5 mM ATP, 2.5 mM MgCl2, 0.4 mM CoA, and 0.01 to 2 μmol (0.024 to 3.5 μCi) [U-14C]BA (specific activity 55 mCi/mmol; American Radiolabeled Chemicals) for 20 min at room temperature. The reactions were terminated by adding 5 μL 50% trichloroacetic acid, and unincorporated substrate was removed by extraction with 180 μL ethyl acetate three times. Radiochemical incorporation rates were determined by scintillation counting 50 μL of the aqueous phase.
CoA ligase activity with short-, medium-, and long-chain fatty acids was determined by coupled enzyme assays described previously (Ziegler et al., 1987; Schneider et al., 2005) with slight modifications. Activation of tested substrates to the corresponding CoA esters by Ph-CNL was determined by measuring AMP formation in a coupled spectrophotometric assay with myokinase, pyruvate kinase, and lactate dehydrogenase. The reaction mixture (500 µL) contained 50 mM Tris-HCl, pH 8.0, 2 mM EDTA, 80 mM KCl, 2.5 mM ATP, 20 mM MgCl2, 2 mM CoA, 0.48 mM NADH, 2 mM phosphoenolpyruvate, 12 nkat of myokinase, 7 nkat of pyruvate kinase, 9 nkat of lactate dehydrogenase, and 50 mM carboxylic acid substrate. Highly lipophilic substrates, such as long-chain fatty acids, were dissolved in DMSO with 2% Triton X-100. The reaction was initiated by addition of 6 µg of purified enzyme, and oxidation of 2 mol of NADH per mol of substrate activated was followed at 340 nm (extinction coefficient for NADH is 6.22 mM−1 cm−1). Controls included assays containing all reaction components without protein added.
PAL, benzaldehyde dehydrogenase, isoeugenol synthase, eugenol synthase, BA/salicylic acid carboxyl methyltransferase, benzoyl-CoA:benzylalcohol/phenylethanol benzoyltransferase, and phenylacetaldehyde synthase activities were measured as described previously in petal crude extracts prepared from control and transgenic flowers collected at 8 pm on day 2 postanthesis (Maeda et al., 2010).
Product Verification by LC-MS
The products formed during standard spectrophotometric assays were analyzed by LC-MS. Cinnamoyl-CoA formed by recombinant tobacco (Nicotiana tabacum) 4CL (Beuerle and Pichersky, 2002a) under the standard spectrophotometric assay conditions was used as a positive control. The chromatography was conducted on a Waters Alliance 2795 HPLC system equipped with a column oven and a Waters 2996 PDA detector. Separation was performed using an Agilent 4.6 mm × 150 mm × 3.5 µm ZORBAX SB-C18 column, maintained at 40°C with a flow rate of 1.0 mL/min. The mobile phase contained solvents A and B, where A was 0.1% ammonium acetate (w/v) in double distilled water and B was 0.1% ammonium acetate (w/v) in methanol. The column was eluted with a linear gradient of 15 to 85% B over 0 to 30 min, reduced from 85 to 15% from 30 to 32 min, then held in isocratic mode at 15% from 32 to 42 min. The HPLC system was coupled to a Micromass QTof-micro mass spectrometer equipped with an electrospray source operated in negative ion mode. Following separation, the column effluent flow was split 1:3 (retained: waste) prior to introduction into the mass spectrometer. The source temperature was set at 120°C with a cone gas flow of 50 L/h, a desolvation gas temperature of 325°C, and a nebulization gas flow of 700 L/h. The capillary voltage was set at 3.0 kV and the cone voltage to 40 V. A scan time of 920 ms with an interscan delay of 100 ms was used. Data were collected from mass-to-charge ratio 65 to 1500 in continuum mode using Waters MassLynx (version 4.0 SP4) software.
Kinetic Properties
For kinetic analysis, an appropriate enzyme concentration was chosen so that the reaction velocity was proportional to the enzyme concentration and linear during the incubation time period. Kinetic data were evaluated by hyperbolic regression analysis (HYPER.EXE, version 1.00, 1992). Triplicate assays were performed for all data points.
Determination of Native Molecular Mass
The molecular mass of the recombinant proteins was determined by gel filtration on a Superdex 200 HR column (GE Healthcare; 1.6 × 60 cm) calibrated with the standard native protein markers ranging from 1.35 kD (vitamin B12) to 670 kD (thyroglobulin) (Bio-Rad). Standard assay buffer was used for column equilibration and elution. Fractions of 1 mL were collected at a flow rate 0.5 mL/min and analyzed for CoA ligase activity. Denaturing SDS-PAGE was performed on 13% gels to determine the subunit molecular weight. The gels were calibrated with molecular mass standards ranging from 7.4 to 208 kD (Bio-Rad).
Enzyme Characterization
The pH optimum for Ph-4CL1 and Ph-CNL activities was determined using 50 mM Tri/K phosphate/Na citrate buffer with pH ranging from 5.5 to 9.0. Enzyme assays were performed with one of following divalent cations present in the assay buffer at a final concentrations of 5 and 2.5 mM for Ph-4CL1 and Ph-CNL, respectively: Ca2+, Cu2+, Fe2+, Mg2+, Mn2+, Ni2+, or Zn2+. The effect of K+ was tested only for Ph-CNL, and the concentration of the cation ranged from 1 to 100 mM. All results represent an average of three independent assays.
Generation of RNAi Silencing Constructs and Plant Transformation
Ph-CNL and Ph-4CL1 RNAi constructs driven by the Clarkia breweri LIS (linalool synthase) promoter (Orlova et al., 2006) were generated in the following way. First, the LIS promoter (1038-bp in size) was amplified using the forward 5′-AAGCTTATCTAATAATGTAT-3′ and reverse 5′-CCCGGGATGGTTGTCTTGT-3′ primers and integrated into the pART27 vector (Gleave, 1992). Next, the two polylinker sites in the RNAi silencing cassette of pRNA69 vector were replaced by Gateway cloning sites in opposite orientation, and the entire segment was transferred to the modified pART27 vector downstream of the LIS promoter. Ph-CNL and Ph-4CL1 cDNA fragments of 473 and 441 nucleotides in size, corresponding to nucleotides 96 to 568 and 214 to 654, respectively, were first spliced into the Gateway entry vector pENTR/D-TOPO (Invitrogen) and then transferred into both GATEWAY sites (i.e., in opposite orientation to each other) in the modified pART27 by an LR clonase reaction.
Transgenic petunias were obtained via Agrobacterium tumefaciens (strain GV3101 carrying plasmids pART27:LIS-Ph-CNLi and pART27:LIS-Ph-4CL1i) leaf disc transformation (Horsch et al., 1985). Rooted plants were screened for the presence of the kanamycin resistance ntpII gene by PCR using the following forward and reverse primers: 5′-GAGAGGCTATTCGGCTATTCGGCTATGACTG-3′ and 5′-ATCACGGGTAGCCAACGCTATGTC-3′. Positive transformants were transferred to the greenhouse. T0 and T1 lines were manually self-pollinated, and the resulting seeds were collected and analyzed for segregation by germinating on Murashige and Skoog medium supplemented with kanamycin (200 mg/L).
Analysis of Floral Volatiles
Floral volatiles were collected from control, CNL RNAi, and 4CL1 RNAi flowers using a closed-loop stripping method as described previously (Orlova et al., 2006). Since P. hybrida cv Mitchell is a nocturnally emitting plant, scent collections began at 8 pm and continued until 8 am the next day. To determine the internal pools of volatiles, 1 g of 2-d-old corolla tissue was collected from each transgenic and control plants at the same time of the day (8 pm) to minimize the effect of rhythmicity. Volatiles were extracted with 10 mL of dichloromethane, concentrated to 180 µL, and analyzed by gas chromatography–mass spectrometry (Orlova et al., 2006).
Analysis of Organic Acids and CoA Esters
To determine the internal pools of organic acids and CoA esters, corolla tissues from transgenic and control flowers were collected at 8 pm on day 2 postanthesis to minimize the effect of rhythmicity. Analyses were performed as described previously (Orlova et al., 2006; Maeda et al., 2010; Qualley et al., 2012).
Peroxisome Isolation from Petunia Petal Tissue
Peroxisomes were isolated from 10 g of petunia petals of 2-d-old flowers harvested at 8 pm using the method previously described (Reumann et al., 2007) with some modifications. All steps were performed at 4°C. Tissue was homogenized in 25 mL of grinding buffer (20 mM tetrasodium pyrophopsphate, pH 7.5, 1 mM EDTA, 0.3 M d-mannitol, and 25 mg BSA), using a glass/Teflon tissue grinder and an overhead stirrer (Wheaton Science Products) to minimize frothing. The homogenized tissue was strained through two layers of Miracloth and centrifuged at 5000g for 10 min. The resulting supernatant was centrifuged in a swinging bucket rotor at 13,000g for 35 min. The pellet was gently resuspended in ∼1 mL resuspension buffer (10 mM HEPES, pH 7.2, and 0.3 M d-mannitol) using a natural bristled paintbrush and added on top of a Percoll gradient (top to bottom: 6 mL 14% Percoll, 4 mL 28% Percoll, both prepared in 2× resuspension buffer, 2 mL 60% Suc cushion) and centrifuged in a swinging bucket rotor at 40,000g for 45 min (without brake). The peroxisomal fraction (visible as a white band just above the Suc cushion) was collected, washed to remove Percoll by diluting fivefold in resuspension buffer, and centrifuging at 14,000g for 2 min in a microcentrifuge. The resulting pellet was resuspended in 500 μL 20 mM potassium phosphate, pH 7.5, and 10% (v/v) glycerol. Protein concentration was determined by the Bradford (1976) method. Enrichment of peroxisomes was assessed by measuring activities of marker enzymes: peroxisomal catalase (Aebi, 1984), mitochondrial fumarase (Gibon et al., 2004), cytosolic alcohol dehydrogenase (Smith and ap Rees, 1979), and chlorophyll content (Hipkins and Baker, 1986; Weiss et al., 1988) for plastids since plastidial NADP-glyceraldehyde-3-phosphate dehydrogenase (Kang and Rawsthorne, 1994) was below detection limit in petal crude extracts as previously reported (Miernyk, 1989). Less than 1% of fumarase activity and no alcohol dehydrogenase activity were detected in the purified peroxisomes (see Supplemental Table 4 online).
RNA Isolation and Analysis
Total RNA was isolated from leaves and different floral organs of 2-d-old petunia flowers and from corollas at different stages of flower development and 10 time points during a daily light/dark cycle using an RNeasy plant mini kit (Qiagen). Unless specified, tissues were harvested at 3 pm. Total RNA was treated with DNaseI to eliminate genomic DNA using the TURBO DNA-free kit (Ambion), and 1 μg of treated RNA was reverse-transcribed to cDNA in a total volume of 100 μL using the High Capacity cDNA reverse transcription kit (Applied Biosystems). qRT-PCR was performed using gene-specific primers: Ph-CNL forward 5′-AGTGGCAGTCCGAATGGAACAAAC-3′ and reverse 5′-TTCACATCAACATCTTCAAGTGCTAGTACG-3′ (each at 900 nM final concentration); Ph-4CL1 forward 5′-TCCTTTTGGCTTTTAACGTGTT-3′ and reverse 5′-TCTCCATCGGCATCTTGGTT-3′ (each at 500 nM final concentration); SGN-U210380 forward 5′-TGGACCAGTGTTGGCAATGT-3′ and reverse 5′- TCTGGTCACCTCTGATGCAAAT-3′ (each at 900 nM final concentration); SGN-U211257 forward 5′-TGGTGGCAATGGCGAAGT-3′ and reverse 5′-CCGCAAGCCAGCAGTTTC-3′ (each at 900 nM final concentration); and SGN-U208283 forward 5′-CTGGTGAGGTTCCAATTGCA-3′ and reverse 5′- GCAACCTGATCTGCGATGAA-3′ (each at 500 nM final concentration). For relative quantification, Elongation factor 1-alpha (EF1α) was used as a reference gene (Mallona et al., 2010) and amplified with forward 5′-CCTGGTCAAATTGGAAACGG-3′ and reverse 5′-CAGATCGCCTGTCAATCTTGG-3′ primers at final concentrations of 300 nM each. All primers showed 90 to 100% efficiency. For absolute quantification of Ph-CNL and Ph-4CL1 transcript levels, their coding regions were amplified by PCR as described above using the pCR4-TOPO vectors carrying Ph-CNL and Ph-4CL1 as templates, and the resulting fragments were purified from agarose gels with a Qiaquick gel extraction kit (Qiagen). Purified DNA fragments were diluted to 1 and 4.8 pg/µL for Ph-CNL and Ph-4CL1, respectively, and used to obtain standard curves in qRT-PCR with gene-specific primers. Individual qRT-PCR reactions contained 5 μL of the SYBR Green PCR master mix (Applied Biosystems), 3 μL of 50-fold diluted cDNA, and 1 μL of 5 μM forward and reverse primers. Two-step qRT-PCR amplification (40 cycles of 95°C for 3 s followed by 60°C for 30 s) was performed using the StepOnePlus real-time PCR system (Applied Biosystems). Based on the standard curves, absolute quantities of Ph-CNL and Ph-4CL1 transcripts were calculated and expressed as a percentage of the total RNA or relative to a control sample. Each data point represents an average of three to four independent biological samples.
Subcellular Localization of CoA Ligases
Plasmid p326-sGFP containing the cauliflower mosaic virus 35S promoter was used for the generation of GFP fusion constructs. For N-terminal GFP fusions, GFP-Ph-4CL1 and GFP-Ph-CNL, the open reading frames of Ph-4CL1 and Ph-CNL were amplified by PCR using forward primers 5′-CTGTACAAGATGCCGATGGAGACTGAAACAAATC-3′ and 5′-CTGTACAAGATGGACGAGTTACCAAAATGTG-3′, respectively, each of which introduced a BsrGI site (underlined) upstream of ATG codon, in combination with corresponding reverse primers 5′-GGCGGCCGCTTAATTTGGAACACCAGCTGCAAG-3′ and 5′-GGCGGCCGCCTACAGACGAGCTGGCAAATCAAG-3′, introducing NotI sites (underlined) after stop codons. A truncated version of the GFP-Ph-CNL (GFP-trunc-Ph-CNL) lacking the C-terminal PTS1 (ARL tripeptide) was obtained using the same GFP-Ph-CNL forward primer in combination with the following reverse primer 5′-GGCGGCCGCCTATGGCAAATCAAGAATCTGGGAC-3′ with NotI site (underlined) after the introduced stop codon shown in italics. For construction of the C-terminal GFP fusions (Ph-4CL1-GFP and Ph-CNL-GFP), the open reading frames of Ph-4CL1 and Ph-CNL were PCR amplified using forward primers 5′-GTCTAGAATGCCGATGGAGACTGAAAC-3′ and 5′-GTCTAGAATGGACGAGTTACCAAAATGTG-3′, respectively, which introduced XbaI sites (underlined) at the initiating ATG codon, in combination with corresponding reverse primers 5′-CGGATCCATTTGGAACACCAGCTGCAAG-3′ and 5′-CGGATCCCAGACGAGCTGGCAAATCAAG-3′ containing BamHI sites (underlined). The PCR fragments were cloned into TOPO-TA vector (Invitogen) and checked by sequencing for absence of errors during PCR amplifications. The BsrGI-NotI and XbaI-BamHI fragments were subcloned into the p326-sGFP vector in frame with the GFP. Arabidopsis thaliana protoplasts were prepared and transformed as described previously (Nagegowda et al., 2008). Transient expression of GFP fusion proteins was observed 16 to 20 h after transformation, and images were acquired using a Radiance 2100 MP Rainbow (Bio-Rad) on a TE2000 inverted microscope (Nikon) using a ×60 oil 1.4–numerical aperture lens. MitoTracker-Red, GFP, and chlorophyll fluorescence images were collected sequentially to avoid any possible bleed through. The MitoTracker-Red was excited at 543 nm using the green HeNe laser, and the fluorescence emission between 560 and 600 nm was collected. GFP and yellow fluorescent protein (YFP) were excited with the 488- and 514-nm line of the four-line argon, respectively, and the emission was collected with a 500LP, 540SP filter combination. Chlorophyll was excited by the 637-nm red diode laser, and the emission greater than 660 nm in wavelength was collected.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Tissue-Specific Expression Profiles of Putative 4CL Candidates.
Supplemental Figure 2. LC-MS Analysis of Product Formed by Recombinant Ph-CNL and Ph-4CL1 from trans-Cinnamic Acid.
Supplemental Figure 3. Effect of 4CL1-RNAi Suppression on 4CL1 Expression and Emission of Benzenoid/Phenylpropanoid Compounds in Corollas of Petunia Flowers.
Supplemental Figure 4. Expression of 4CL1 and CNL in Ph-CNL-RNAi and Ph-4CL1-RNAi Lines, Respectively.
Supplemental Table 1. Accession Numbers of Protein Sequences Used for Phylogenetic Analysis in Figure 3.
Supplemental Table 2. Activities of Enzymes Involved in Benzenoid Biosynthesis in Petunia Petals of Control and CNL-RNAi Lines.
Supplemental Table 3. Subcellular Fractionation of Petunia Petal Crude Extracts and Activities of Marker Enzymes and Ph-4CL.
Supplemental Table 4. Substrate Specificity of Ph-CNL with Short-, Medium-, and Long-Chain Fatty Acids.
Supplemental Data Set 1. Alignment of Protein Sequences Used to Generate the Phylogeny Presented in Figure 3.
Supplementary Material
Acknowledgments
We thank Inhwan Hwang for the GFP vectors, Gregg A. Howe for the YFP-OPCL1 construct, and Jennifer Sturgis for the assistance with confocal laser scanning microscopy. Confocal microscopy data were acquired at the Purdue Cancer Center Analytical Cytometry Laboratories supported by the Cancer Center National Cancer Institute Core Grant NIH NCI-2P30CA23168. LC-MS analysis was supported by the National Science Foundation (Grant DBI-0421102). This project was supported by National Research Initiative Competitive Grant 2005-35318-16207 from the USDA Cooperative State Research, Education, and Extension Service to N.D. and D.R., by a grant from the National Science Foundation (MCB-0919987 to N.D.), and by U.S.–Israel Binational Agriculture Research and Development funds (Grant US-4322-10 to E.P.).
AUTHOR CONTRIBUTIONS
A.K., Y.K., D.R., E.P., and N.D. designed the research. A.K., Y.K., A.Q., D.A.N., J.R.W., I.O., A.K.S., G.T., C.M.K., B.R.C., and J.C.D. performed research. A.K., Y.K., A.Q., J.R.W., D.R., E.P., and N.D. analyzed data. A.K., J.R.W., E.P., and N.D. wrote the article.
Glossary
- BA
benzoic acid
- PAL
Phe ammonia lyase
- 4CL
4-coumarate:CoA ligase family
- CNL
cinnamate-specific ligase
- AAE
acyl-activating enzyme
- qRT-PCR
quantitative RT-PCR
- LC-MS
liquid chromatography–mass spectrometry
- RNAi
RNA interference
- GFP
green fluorescent protein
- YFP
yellow fluorescent protein
References
- Aebi H. (1984). Catalase in vitro. Methods Enzymol. 105: 121–126 [DOI] [PubMed] [Google Scholar]
- Alber B.E., Fuchs G. (2002). Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 277: 12137–12143 [DOI] [PubMed] [Google Scholar]
- Allina S.M., Pri-Hadash A., Theilmann D.A., Ellis B.E., Douglas C.J. (1998). 4-Coumarate:coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes. Plant Physiol. 116: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks J.A., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuerle T., Pichersky E. (2002a). Enzymatic synthesis and purification of aromatic coenzyme a esters. Anal. Biochem. 302: 305–312 [DOI] [PubMed] [Google Scholar]
- Beuerle T., Pichersky E. (2002b). Purification and characterization of benzoate:coenzyme A ligase from Clarkia breweri. Arch. Biochem. Biophys. 400: 258–264 [DOI] [PubMed] [Google Scholar]
- Bjorklund J.A., Leete E. (1992). Biosynthesis of the benzoyl moiety of cocaine from cinnamic acid via (R)-(+)-3-hydroxy-3-phenylpropanoic acid. Phytochemistry 31: 3883–3887 [Google Scholar]
- Boatright J., Negre F., Chen X., Kish C.M., Wood B., Peel G., Orlova I., Gang D., Rhodes D., Dudareva N. (2004). Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135: 1993–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Catinot J., Buchala A., Abou-Mansour E., Métraux J.P. (2008). Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett. 582: 473–478 [DOI] [PubMed] [Google Scholar]
- Colón A.M., Sengupta N., Rhodes D., Dudareva N., Morgan J. (2010). A kinetic model describes metabolic response to perturbations and distribution of flux control in the benzenoid network of Petunia hybrida. Plant J. 62: 64–76 [DOI] [PubMed] [Google Scholar]
- Costa M.A., et al. (2005). Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 66: 2072–2091 [DOI] [PubMed] [Google Scholar]
- Cseke L., Dudareva N., Pichersky E. (1998). Structure and evolution of linalool synthase. Mol. Biol. Evol. 15: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Dobson H.E.M. (2006). Relationship between floral fragrance composition and type of pollinator. In Biology of Floral Scent, N. Dudareva and E. Pichersky, eds (Boca Raton, FL: CRC Press, Taylor & Francis Group), pp. 147–198.
- Dudareva N., Murfitt L.M., Mann C.J., Gorenstein N., Kolosova N., Kish C.M., Bonham C., Wood K. (2000). Developmental regulation of methylbenzoate biosynthesis and emission in snapdragon flowers. Plant Cell 12: 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N., Raguso R.A., Wang J.H., Ross J.R., Pichersky E. (1998). Floral scent production in Clarkia breweri. III. Enzymatic synthesis and emission of benzenoid esters. Plant Physiol. 116: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J., Büttner D., Wang Q., Douglas C.J., Somssich I.E., Kombrink E. (1999). Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (2009). PHYLIP - Phylogeny Inference Package, Version 3.69. (Seattle, WA: University of Washington)
- Gibon Y., Blaesing O.E., Hannemann J., Carillo P., Höhne M., Hendriks J.H.M., Palacios N., Cross J., Selbig J., Stitt M. (2004). A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hamberger B., Hahlbrock K. (2004). The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 101: 2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck C., Jarvis A.P., Xiang L., Moore B.S., Oldham N.J. (2001). A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid beta-oxidation. ChemBioChem 2: 784–786 [DOI] [PubMed] [Google Scholar]
- Hettema E.H., van Roermund C.W., Distel B., van den Berg M., Vilela C., Rodrigues-Pousada C., Wanders R.J., Tabak H.F. (1996). The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 15: 3813–3822 [PMC free article] [PubMed] [Google Scholar]
- Hipkins M.F., Baker N.R. (1986). Photosynthesis energy transduction. In Spectroscopy, M.F. Hipkins and N.R. Baker, eds (Oxford, UK: IRL), pp. 51–105.
- Horsch R.B., Fry J.E., Hoffman N.L., Eichholts D., Rogers S.G., Fraley R.T. (1985). A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Hu W.J., Kawaoka A., Tsai C.J., Lung J., Osakabe K., Ebinuma H., Chiang V.L. (1998). Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc. Natl. Acad. Sci. USA 95: 5407–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M.C., Alexson S.E. (2008). Novel functions of acyl-CoA thioesterases and acyltransferases as auxiliary enzymes in peroxisomal lipid metabolism. Prog. Lipid Res. 47: 405–421 [DOI] [PubMed] [Google Scholar]
- Ibdah M., Pichersky E. (2009). Arabidopsis Chy1 null mutants are deficient in benzoic acid-containing glucosinolates in the seeds. Plant Biol. (Stuttg.) 11: 574–581 [DOI] [PubMed] [Google Scholar]
- Jarvis A.P., Schaaf O., Oldham N.J. (2000). 3-Hydroxy-3-phenylpropanoic acid is an intermediate in the biosynthesis of benzoic acid and salicylic acid but benzaldehyde is not. Planta 212: 119–126 [DOI] [PubMed] [Google Scholar]
- Kang F., Rawsthorne S. (1994). Starch and fatty acid synthesis in plastids from developing embryos of oilseed rape (Brassica napus L.). Plant J. 6: 795–805 [Google Scholar]
- Kliebenstein D.J., D’Auria J.C., Behere A.S., Kim J.H., Gunderson K.L., Breen J.N., Lee G., Gershenzon J., Last R.L., Jander G. (2007). Characterization of seed-specific benzoyloxyglucosinolate mutations in Arabidopsis thaliana. Plant J. 51: 1062–1076 [DOI] [PubMed] [Google Scholar]
- Knudsen J.T., Gershenzon J. (2006). The chemical diversity of floral scent. In Biology of Floral Scent, N. Dudareva and E. Pichersky, eds (Boca Raton, FL: CRC Press, Taylor & Francis Group), pp. 27–52.
- Kumar A., Ellis B.E. (2003). The 4-coumarate:CoA ligase gene family in Rubus idaeus: cDNA structures, expression during fruit development and substrate specificity of a divergent member. Plant Mol. Biol. 31: 327–340 [DOI] [PubMed] [Google Scholar]
- Kolosova N., Gorenstein N., Kish C.M., Dudareva N. (2001). Regulation of circadian methyl benzoate emission in diurnally and nocturnally emitting plants. Plant Cell 13: 2333–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A.J.K., Chung H.S., Kobayashi Y., Howe G.A. (2006). Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281: 33511–33520 [DOI] [PubMed] [Google Scholar]
- Lee D., Douglas C.J. (1996). Two divergent members of a tobacco 4-coumarate:coenzyme A ligase (4CL) gene family. cDNA structure, gene inheritance and expression, and properties of recombinant proteins. Plant Physiol. 112: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Meyer K., Chapple C., Douglas C.J. (1997). Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9: 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León J., Shulaev V., Yalpani N., Lawton M.A., Raskin I. (1995). Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc. Natl. Acad. Sci. USA 92: 10413–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Möllers B., Fliegmann J., Uhlmann A., Lottspeich F., Meimberg H., Ebel J. (2002). Divergent members of a soybean (Glycine max L.) 4-coumarate:coenzyme A ligase gene family. Eur. J. Biochem. 269: 1304–1315 [DOI] [PubMed] [Google Scholar]
- Long M.C., Nagegowda D.A., Kaminaga Y., Ho K.K., Kish C.M., Schnepp J., Sherman D., Weiner H., Rhodes D., Dudareva N. (2009). Involvement of snapdragon benzaldehyde dehydrogenase in benzoic acid biosynthesis. Plant J. 59: 256–265 [DOI] [PubMed] [Google Scholar]
- Lozoya E., Hoffmann H., Douglas C.J., Schulz W., Scheel D., Hahlbrock K. (1988). Primary structures and catalytic properties of isoenzymes encoded by the two 4-coumarate: CoA ligase genes in parsley. Eur. J. Biochem. 176: 661–667 [DOI] [PubMed] [Google Scholar]
- Maeda H., Shasany A.K., Schnepp J., Orlova I., Taguchi G., Cooper B.R., Rhodes D., Pichersky E., Dudareva N. (2010). RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell 22: 832–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallona I., Lischewski S., Weiss J., Hause B., Egea-Cortines M. (2010). Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk J. (1989). Leucoplast isolation. In Physiology, Biochemistry, and Genetics of Nongreen Plastids, C.D. Boyer, I.C. Shannon, and R.C. Hardison, eds (Rockville, MD: The American Society of Plant Physiologists), pp 15–23.
- Nagegowda D.A., Gutensohn M., Wilkerson C.G., Dudareva N. (2008). Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 55: 224–239 [DOI] [PubMed] [Google Scholar]
- Negre F., Kish C.M., Boatright J., Underwood B., Shibuya K., Wagner C., Clark D.G., Dudareva N. (2003). Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. Plant Cell 15: 2992–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D., Nakajima N., Sano T., Tamaoki M., Aono M., Kubo A., Kanna M., Ioki M., Kamada H., Saji H. (2005). Salicylic acid accumulation under O3 exposure is regulated by ethylene in tobacco plants. Plant Cell Physiol. 46: 1062–1072 [DOI] [PubMed] [Google Scholar]
- Ogawa D., Nakajima N., Seo S., Mitsuhara I., Kamada H., Ohashi Y. (2006). The phenylalanine pathway is the main route of salicylic acid biosynthesis in tobacco mosaic virus-infected tobacco leaves. Plant Biotechnol. 23: 395–398 [Google Scholar]
- Orlova I., Marshall-Colón A., Schnepp J., Wood B., Varbanova M., Fridman E., Blakeslee J.J., Peer W.A., Murphy A.S., Rhodes D., Pichersky E., Dudareva N. (2006). Reduction of benzenoid synthesis in petunia flowers reveals multiple pathways to benzoic acid and enhancement in auxin transport. Plant Cell 18: 3458–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan J., Ettlinger L. (1976). Characterization of the acetyl-CoA synthetase of Acetobacter aceti. Biochim. Biophys. Acta 450: 410–417 [DOI] [PubMed] [Google Scholar]
- Pan Q., Zhan J., Liu H., Zhang J., Chen J., Wen P., Huang W. (2006). Salicylic acid synthesized by benzoic acid 2-hydroxylase participates in the development of thermotolerance in pea plants. Plant Sci. 171: 226–233 [Google Scholar]
- Perera M.A., Choi S.Y., Wurtele E.S., Nikolau B.J. (2009). Quantitative analysis of short-chain acyl-coenzymeAs in plant tissues by LC-MS-MS electrospray ionization method. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 877: 482–488 [DOI] [PubMed] [Google Scholar]
- Qualley A.V., Cooper B.R., Dudareva N. (2012). Profiling hydroxycinnamoyl-coenzyme A thioesters: Unlocking the back door of phenylpropanoid metabolism. Anal. Biochem. 420: 182–184 [DOI] [PubMed] [Google Scholar]
- Qualley A.V., Dudareva N. (2008). Aromatic volatiles and their involvement in plant defense. In Induced Plant Resistance to Herbivory, A. Schaller, ed (Berlin: Springer), pp. 409–432.
- Raguso R.A., Pichersky E. (1995). Floral volatiles from Clarkia breweri and C. concinna (Onagraceae): Recent evolution of floral scent and moth pollination. Plant Syst. Evol. 194: 55–67 [Google Scholar]
- Ralph S.G., et al. (2008) A conifer genomics resource of 200,000 spruce (Picea spp.) ESTs and 6,464 high-quality, sequence-finished full-length cDNAs for Sitka spruce (Picea sitchensis). BMC Genomics 9: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S. (2004). Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 135: 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., Antonicelli G.E., Rasche N., Lüder F., Weckwerth W., Jahn O. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada H., Shim I.S., Usui K. (2006). Induction of benzoic acid 2-hydroxylase and salicylic acid biosynthesis – Modulation by salt stress in rice seedlings. Plant Sci. 171: 263–270 [Google Scholar]
- Schneider K., Kienow L., Schmelzer E., Colby T., Bartsch M., Miersch O., Wasternack C., Kombrink E., Stuible H.P. (2005). A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J. Biol. Chem. 280: 13962–13972 [DOI] [PubMed] [Google Scholar]
- Shockey J., Browse J. (2011). Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J. 66: 143–160 [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D.G., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., ap Rees T. (1979). Pathways of carbohydrate fermentation in the roots of marsh plants. Planta 146: 327–333 [DOI] [PubMed] [Google Scholar]
- Takao S., Ito T., Tanida M. (1987). Purification and kinetic properties of butyryl-CoA synthetase from Paecilomyces varioti. Agric. Biol. Chem. 51: 145–152 [Google Scholar]
- Turner J.E., Greville K., Murphy E.C., Hooks M.A. (2005). Characterization of Arabidopsis fluoroacetate-resistant mutants reveals the principal mechanism of acetate activation for entry into the glyoxylate cycle. J. Biol. Chem. 280: 2780–2787 [DOI] [PubMed] [Google Scholar]
- Underwood B.A., Tieman D.M., Shibuya K., Dexter R.J., Loucas H.M., Simkin A.J., Sims C.A., Schmelz E.A., Klee H.J., Clark D.G. (2005). Ethylene-regulated floral volatile synthesis in petunia corollas. Plant Physiol. 138: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Moerkercke A., Schauvinhold I., Pichersky E., Haring M.A., Schuurink R.C. (2009). A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. Plant J. 60: 292–302 [DOI] [PubMed] [Google Scholar]
- Verdonk J.C., Ric de Vos C.H., Verhoeven H.A., Haring M.A., van Tunen A.J., Schuurink R.C. (2003). Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry 62: 997–1008 [DOI] [PubMed] [Google Scholar]
- Verleur N., Hettema E.H., van Roermund C.W., Tabak H.F., Wanders R.J. (1997). Transport of activated fatty acids by the peroxisomal ATP-binding-cassette transporter Pxa2 in a semi-intact yeast cell system. Eur. J. Biochem. 249: 657–661 [DOI] [PubMed] [Google Scholar]
- Walker K., Croteau R. (2000). Taxol biosynthesis: molecular cloning of a benzoyl-CoA:taxane 2α-O-benzoyltransferase cDNA from taxus and functional expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 97: 13591–13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D., Schönfeld M., Halevy A.H. (1988). Photosynthetic activities in the petunia corolla. Plant Physiol. 87: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M.C. (2006). Variations on a theme: Synthesis and modification of plant benzoic acids. Curr. Opin. Plant Biol. 9: 288–296 [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu H., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defense. Nature 417: 562–565 [DOI] [PubMed] [Google Scholar]
- Yang Y., Shah J., Klessig D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11: 1621–1639 [DOI] [PubMed] [Google Scholar]
- Ziegler K., Braun K., Bockler A., Fuchs G. (1987). Studies on the anaerobic degradation of benzoic acid and 2-aminobenzoic acid by a denitrifying Pseudomonas strain. Arch. Microbiol. 149: 62–69 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.